Abstract
Appendage regeneration is defined by rapid changes in gene expression that achieve dramatic developmental effects, suggesting involvement of microRNAs (miRNAs). Here, we find dynamic regulation of many miRNAs during zebrafish fin regeneration. In particular, miR-133 levels are high in uninjured fins but low during regeneration. When regeneration was blocked by Fibroblast growth factor (Fgf) receptor inhibition, high miR-133 levels were quickly restored. Experimentally increasing amounts of miR-133 attenuated fin regeneration. Conversely, miR-133 antagonism during Fgf receptor inhibition accelerated regeneration through increased proliferation within the regeneration blastema. The Mps1 kinase, an established positive regulator of blastemal proliferation, is an in vivo target of miR-133. Our findings identify miRNA depletion as a new regulatory mechanism for complex tissue regeneration.
[Keywords: Zebrafish, fin, regeneration, miR-133, Fgf, Mps1]
Regeneration, the replacement of damaged or lost body parts, is a primary goal of stem cell research. Certain nonmammalian vertebrates like urodele amphibians and teleost fish restore complex tissues much more effectively than mammals, creating tantalizing examples of successful organ regeneration. For instance, the common laboratory model zebrafish regenerates heart muscle, retina, spinal cord, sensory hair cells, and appendages (Stoick-Cooper et al. 2007). Fin regeneration in zebrafish is a particularly efficient example of complex tissue regeneration, replacing bone, epidermis, blood vessels, nerves, connective tissue, and pigmentation within 2 wk of amputation of up to 95% of the organ. Following wound healing, spared mesenchymal cells disorganize, migrate distally, and accumulate to form the regeneration blastema, a proliferative mass of progenitor tissue. The blastema is maintained throughout the process of regenerative outgrowth, during which it creates new structures through a series of proliferation, patterning, and differentiation events. During fin regeneration and other examples of complex tissue regeneration, large-scale changes in gene expression programs occur to execute the rapid transformation of quiescent, differentiated tissue to proliferating, actively patterned tissue (Lien et al. 2006; Schebesta et al. 2006).
To implement these programmatic alterations in gene expression, it is almost certain that several modes of regulation are employed. These might include changes in chromatin accessibility and new production of transcriptional factors, as well as post-transcriptional mechanisms. Based on studies in other fields, microRNAs (miRNAs) present excellent candidates to mediate important post-transcriptional regulation during complex tissue regeneration. miRNAs are small, noncoding RNAs that base-pair with complementary sequences in the 3′ untranslated regions (UTRs) of target genes to promote mRNA degradation or inhibit protein translation (Kloosterman and Plasterk 2006). The first identified miRNAs, lin-4 and let-7, were discovered in genetic screens for mutations that disrupt developmental transitions in nematodes (Lee et al. 1993; Reinhart et al. 2000). miR-430 is the predominant miRNA species in early zebrafish embryos, where it refines the transition from maternal to zygotic mRNA utilization and helps balance levels of patterning factors (Giraldez et al. 2006; Choi et al. 2007). Investigators have also identified contributions by miRNAs to preventing or promoting the pathology of adult organs. In particular, miR-133 and miR-208 modulate pathologic cardiac muscle hypertrophy in mouse models of this common human cardiomyopathy, where they appear to have opposing functions (Care et al. 2007; van Rooij et al. 2007). Given the numerous roles for various miRNAs in different developmental processes like organogenesis, stem cell maintenance, and programmed cell death, as well as in adult pathogenesis, there is reason to suspect involvement in spectacular regenerative events of teleosts and urodele amphibians.
Here, we found that many miRNAs are differentially regulated during caudal fin regeneration in adult zebrafish. One of these miRNAs, miR-133, has relatively high levels in the uninjured fin, but these levels drop sharply during regeneration. By a combination of gain-of-function and loss-of-function experiments, our data indicate that miR-133 acts as a regenerative brake within a regulatory circuit for regeneration. Following amputation, Fibroblast growth factor (Fgf) signaling reduces miR-133 expression as part of the regeneration program, facilitating normal expression of targets like the mps1 kinase that direct blastemal proliferation and tissue renewal. Our findings identify miR-133 depletion as an important regulatory mechanism by which Fgfs promote appendage regeneration.
Results and Discussion
Caudal fin regeneration is accompanied by alterations in miRNA expression
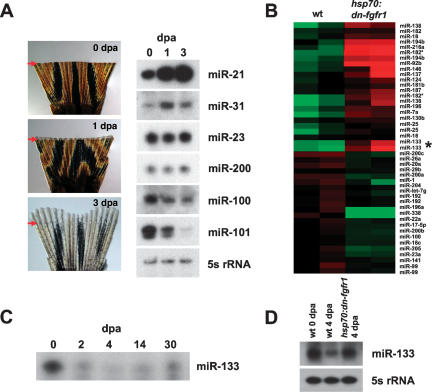
To identify miRNAs present during caudal fin regeneration, we performed miRNA microarray experiments using RNA collected from three phases of regeneration: (1) newly amputated fins (0 d post-amputation, dpa), (2) formation of the blastema (1 dpa, at 33°C), and (3) early regenerative outgrowth (3 dpa) (Fig. 1A). These analyses indicated dynamic miRNA regulation during regeneration. During blastema formation, 24 miRNAs showed significantly higher expression levels compared with uninjured fins, while 13 miRNAs exhibited lower expression levels (0 dpa compared with 1 dpa; 1.5-fold change). During regenerative outgrowth, 51 miRNAs showed significantly higher expression levels, and 14 miRNAs had lower levels than uninjured fins (3 dpa compared with 0 dpa; 1.5-fold change) (Supplemental Fig. 1A). Levels of some miRNAs differed between blastema formation and regenerative outgrowth, suggesting stage-specific functions, and many of these expression profiles were validated by Northern analysis (Fig. 1A). Thus, numerous miRNAs are differentially regulated during zebrafish fin regeneration.
Figure 1.
miRNAs are dynamically regulated during zebrafish caudal fin regeneration. (A, left) Tissue distal to a region one ray segment proximal to the amputation plane (red arrow) was collected from wild-type (wt) fins at 0, 1, or 3 dpa and used for RNA isolation and miRNA microarray analysis. (Right) Northern analyses of various miRNAs present during fin regeneration. (B) A heat map comparing relative miRNA expression in wild-type and hsp70:dn-fgfr1 4-dpa regenerates collected 5 h after a single heat shock at 38°C. (Green) Lower expression; (red) higher expression; (asterisk) miR-133. (C) Ribonuclease protection assays (RPAs) were performed using RNA collected from wild-type regenerates at the indicated stages. (D, top) RPA using RNA collected from wild-type or hsp70:dn-fgfr1 samples. miR-133 levels are reduced during regenerative outgrowth (compare wild type, 0 dpa vs. wild type, 4 dpa), but are restored during a brief period of Fgfr inhibition at 4 dpa. (Bottom) Northern analysis with a 5s rRNA probe was used as a control.
Fgfs control miRNA expression during fin regeneration
Multiple lines of experimental evidence have demonstrated that signaling by Fgfs is indispensable for fin regeneration (Poss et al. 2000; Lee et al. 2005; Whitehead et al. 2005; Thummel et al. 2006). These studies led to a regulatory model in which Fgf ligands like Fgf20a and Fgf24 induce blastema formation after amputation and then continue to maintain blastemal cell proliferation during regenerative outgrowth. To determine whether Fgf signaling controls expression of specific miRNAs during regenerative outgrowth, we used a transgenic zebrafish strain that carries a heat-inducible dominant-negative Fgf receptor (Fgfr; hsp70:dn-fgfr1). This construct is predicted to block signaling through all Fgfrs and effectively disrupts regeneration of fin and cardiac tissue in animals given daily heat shocks (Lee et al. 2005; Lepilina et al. 2006). In these experiments, wild-type and transgenic animals were allowed to regenerate normally for 4 d (at 26°C), followed by a single heat shock to induce dn-fgfr1 expression. Just 5 h after the heat shock was completed, RNA was collected from regenerates and hybridized onto miRNA microarrays. These experiments revealed 22 miRNAs that were expressed at lower levels during Fgfr inhibition than in wild-type regenerates (wild type, 4 dpa vs. hsp70:dn-fgfr1, 4 dpa; 1.5-fold), with 34 other miRNAs exhibiting higher expression levels (Fig. 1B; Supplemental Fig. 1B). To assist identification of Fgf-regulated miRNAs, we filtered these data with an additional data set of uninjured wild-type fins (0 dpa). Of those 22 miRNAs that were expressed at lower levels in hsp70:dn-fgfr1 4-dpa samples, six of these were also among the 27 miRNAs increased from 0 to 4 dpa in wild-type fins. Of the 34 miRNAs with higher levels during Fgfr inhibition, 16 were among the 54 miRNAs with levels that fall from 0 to 4 dpa in wild-type fins (Supplemental Fig. 1B). Northern analysis has confirmed several expression profiles from our filtered data set (data not shown). Thus, our microarray experiments point to a total of 22 miRNAs influenced by Fgf signaling during regeneration, six of which are induced or maintained by Fgf signaling, and 16 negatively regulated (Supplemental Fig. 1C).
miR-133 is depleted by Fgf signaling during fin regeneration
Because of its expression profile during regeneration, we focused our attention on miR-133, a highly conserved family of miRNAs that has been recently implicated in regulating cardiac and skeletal muscle development, as well as differentiation and function of dopaminergic neurons (Chen et al. 2006; Care et al. 2007; Kim et al. 2007). The miR-133 family has four members encoded by different genes that vary only by 2 nucleotides (nt) in the mature species, outside of the predicted seed sequence. miR-133b was the only member displaying strong expression of the precursor species in caudal fins by Northern analysis, suggesting that it is the predominant member in this tissue (data not shown). miR-133 was detectable at relatively high levels in the uninjured adult caudal fin, but its expression was 5.6-fold lower during regenerative outgrowth. A time course of miR-133 expression during wild-type fin regeneration showed that levels were reduced during blastema formation and remained low until 14 dpa (at 26°C) (Fig. 1C). By 30 dpa, a point at which fin regeneration has been completed, levels of miR-133 were not restored but appeared to be approaching those levels within uninjured fins.
Interestingly, when Fgfrs were briefly inhibited during regenerative outgrowth, a 4.5-fold increase in the level of miR-133 was observed (Fig. 1B,D; Supplemental Fig. 1A), restoring approximately the miR-133 level of uninjured fins. To determine if this response was specific to regeneration, we assessed the effects of Fgfr inhibition on miR-133 levels in uninjured adult fins. Blockade of Fgf signaling in uninjured fins did not elevate levels of miR-133, indicating regeneration-specific regulation (data not shown). Collectively, these expression data suggested a model in which Fgf signaling depletes miR-133 during fin regeneration.
miR-133 functions as a regenerative brake
miR-133 levels were higher in uninjured fins than during the regenerative process, suggesting that miR-133 attenuates growth mechanisms. To determine the effects of experimentally increasing levels of miR-133 during regeneration, we injected and electroporated miR-133 RNA duplexes into wild-type fin regenerates (Fig. 2A,B). We found that dorsal lobe regenerates electroporated with miR-133 duplexes had an ∼10% reduction in length compared with uninjected ventral lobe regenerates when measured 24 h after introduction, and an ∼13% reduction when measured 3 d after electroporation (Fig. 2B,D; Supplemental Fig. 3A). These measurements conservatively estimate the effects on regeneration, since the first 3 d of regeneration proceed normally prior to electroporation. Also, it is possible that the percentage of cells affected or relative miR-133 increases in these studies are low. Electroporation with a mutated miR-133 RNA duplex, containing 3-nt changes in the predicted seed region, had no significant effects on regeneration (Fig. 2C,D; Supplemental Fig. 2D). Intrafin comparisons of injected and uninjected fin lobes were also made to eliminate possible interfish variability in the lengths of regenerates. Regenerates injected with miR-133 RNA duplex showed significantly lower injected:uninjected length ratios (11%–14% at 1, 2, and 3 d post-injection) than regenerates injected with mutated RNA duplex (Fig. 2D). These results indicate that miR-133 slows regenerative outgrowth and support the idea that fin regeneration is assisted by regulated depletion of miR-133.
Figure 2.
Experimental manipulation of miR-133 controls fin regeneration. (A) Design for RNA duplex (blue) and MO (red) electroporation studies. (B) Electroporation of miR-133 RNA duplex into the dorsal lobe (top, blue asterisk) slows regeneration in wild-type fins, as compared with a mutated miR-133 RNA duplex (bottom, black asterisk). (C) Electroporation of miR-133 MO into the dorsal lobe (top, red asterisk) enhances regeneration during Fgfr inhibition, as compared with a standard MO (bottom, black asterisk). (D,E) Quantification of average intrafin length ratios of injected versus uninjected fin lobes with miR-133 RNA duplex (D) or miR-133 MO (E). The injected:uninjected length ratio is significantly reduced by miR-133 duplex introduction, but significantly increased by miR-133 MO introduction (mean ± SEM; [*] P < 0.05, t-test; n = 10 per group).
miR-133 antagonism accelerates regeneration during Fgfr inhibition
To test the effects of antagonizing miR-133 function during regeneration, an antisense morpholino (MO) designed to disrupt miR-133 binding and activity was introduced into regenerating wild-type caudal fins. No significant effects on regenerate length were observed during the first 2 d following miR-133 MO electroporation; however, an ∼10% increase in length was evident by the third day (Supplemental Fig. 4). This minor effect may have been due to the fact that miR-133 levels are already quite low during regeneration. We hypothesized that we might achieve a greater effect during Fgfr inhibition, given that miR-133 levels increase under these conditions. Indeed, antagonizing miR-133 function in hsp70:dn-fgfr1 regenerates partially restored fin regeneration. miR-133 MO-treated dorsal lobe regenerates were ∼17% longer than uninjected ventral lobe regenerates by 1 d after electroporation (Fig. 2C). By 3 d after electroporation, miR-133 MO-treated regenerates were an average of ∼44% longer than uninjected regenerates (Supplemental Fig. 3). In contrast, electroporation of a standard MO that is not predicted to interact with an expressed RNA had no significant effects on regeneration. Intrafin data showed significantly higher injected:uninjected length ratios in miR-133 MO-treated fins versus standard MO-treated fins of ∼20%, ∼41%, and ∼50% at 1, 2, and 3 d post-injection (Fig. 2E). Thus, miR-133 is a significant regulatory target of Fgf signaling during fin regeneration. Antagonism of miR-133 activity is sufficient to partially rescue fin regeneration that has been inhibited by Fgfr blockade.
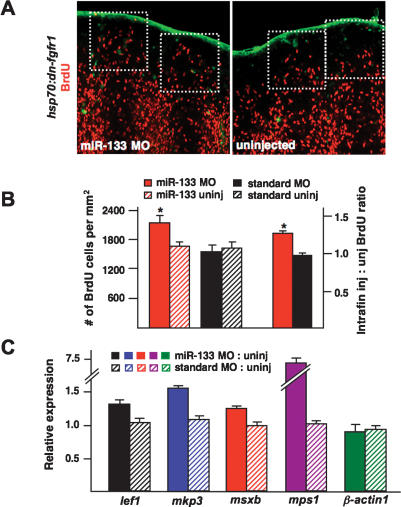
To determine the mechanism by which miR-133 impacts regeneration, we first quantified blastemal cell proliferation using BrdU incorporation assays. While a standard MO had no effect on the number of BrdU-labeled cells in hsp70:dn-fgfr1 regeneration blastemas, miR-133 MO-treated blastemas had ∼28% more BrdU-positive cells than uninjected blastemas (Fig. 3A,B). Intrafin comparisons showed an ∼29% increase in the injected:uninjected ratio of BrdU-positive cells in miR-133 MO-treated regenerates versus those treated with standard MO (Fig. 3A,B). To determine whether the rescue we observed included partial restoration of regeneration programs, we examined expression of markers that are normally activated during fin regeneration by quantitative PCR. We found that the Fgf target gene mkp3, the basal epidermal marker lef1, and the blastemal markers msxb and mps1 each displayed elevated expression in hsp70:dn-fgfr1 regenerates injected with the miR-133 MO, while β-actin1 levels showed no significant change (Fig. 3C). Because lef1, mkp3, and msxb lack a predicted binding site for miR-133, the effects of the miR-133 MO are most likely indirect. These results indicate that miR-133 antagonism partially rescues regeneration by increasing the expression of regeneration genes and enabling cellular proliferation.
Figure 3.
miR-133 regulates blastemal cell proliferation. (A,B) hsp70:dn-fgfr1 animals were treated as in Figure 2A with miR-133 MO and injected with BrdU. (A, boxes) Dorsal regenerates treated with miR-133 MO show ∼28% greater blastemal BrdU incorporation than uninjected ventral regenerates. The standard MO had no significant effects on blastemal BrdU incorporation. (C) Quantitative PCR results indicating increased expression of the regeneration markers lef1, mkp3, msxb, and mps1 in hsp70:dn-fgfr1 regenerates treated with miR-133 MO. Expression levels of β-actin1 were unaffected by miR-133 MO treatment. Data are expressed as ratios of injected:uninjected lobes. (Solid bars) miR-133 MO; (hatched bars) standard MO. (Mean ± SEM; [*] P < 0.05; n = 14–16 fins per group for BrdU studies.)
mps1 is an in vivo target of miR-133
miRNAs typically have from several dozen to hundreds of different mRNA target genes (Lewis et al. 2005; Giraldez et al. 2006). We searched the miRanda and miRBase databases and identified many potential targets, as predicted by complementarity of miR-133 seed sequences with mRNA 3′ UTR sequences. These potential targets include factors that support cellular and molecular functions previously implicated in regeneration, including cell division and growth, metalloproteinase activity (Vinarsky et al. 2005), and ion flux (Supplemental Table 1; Adams et al. 2007). One gene containing a single predicted miR-133-binding site is mps1, which encodes a kinase that regulates multiple aspects of cell proliferation during morphogenesis, including centrosome duplication, spindle checkpoint activation, and normal mitotic progression (Fisk and Winey 2004). Most importantly, mps1 is one of only four genes to date that have been shown by forward genetic approaches to be essential for fin regeneration. mps1 expression is specifically induced in blastemal tissue during regeneration, and a temperature-sensitive mutation in mps1 arrests regeneration during outgrowth, the phase at which our data implicated miR-133 function (Poss et al. 2002).
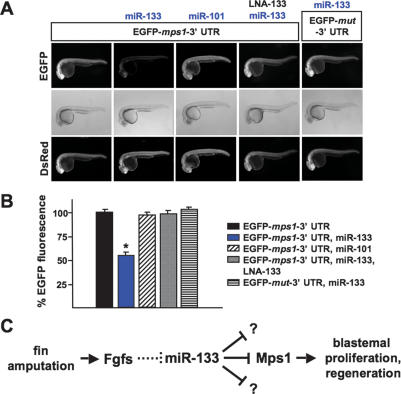
To determine if miR-133 regulates mps1 mRNA in vivo, we used zebrafish embryo sensor assays (Giraldez et al. 2005). We injected a sensor mRNA construct consisting of EGFP fused to the 3′ UTR of mps1 into one-cell zebrafish embryos, in the presence or absence of miR-133 RNA duplex (Fig. 4A). One day later, we quantified EGFP fluorescence as an indicator of Mps1 expression. Injections of the mps1 sensor mRNA alone resulted in high EGFP expression; however, this fluorescence was dampened almost 50% by coinjection of miR-133 RNA duplex (Fig. 4A). Three additional experiments provided evidence of specific interaction between miR-133 and the EGFP-mps1-3′ UTR sensor mRNA. First, no interaction was observed between miR-133 and a sensor mRNA containing a mutated miR-133-binding site. Second, coinjected miR-101 RNA duplex, not predicted to regulate the mps1 3′ UTR, did not reduce EGFP sensor fluorescence. Finally, coinjection of miR-133 MO or an LNA-133 antisense oligonucleotide effectively prevented miR-133-induced dampening of fluorescence (Fig. 4A,B). Northern blot analysis indicated little or no effect of miR-133 duplex on EGFP-mps1-3′ UTR sensor mRNA levels (data not shown). Therefore, we suspect that miR-133 regulates mps1 in these assays through translational repression. This finding suggests that the increases in mps1 mRNA levels we observed after antagonism of miR-133 during fin regeneration were likely contributed by other direct and indirect regulatory inputs (Fig. 3C). In summary, the known blastemal regulator mps1 is a target of miR-133.
Figure 4.
miR-133 targets the blastemal regulator mps1. (A,B) EGFP embryo sensor assays demonstrating specific regulation of the mps1 3′ UTR in vivo by miR-133. For each experimental group in B, EGFP-mps1-3′ UTR fluorescence is expressed as a percentage of fluorescence observed from the sensor injected alone. (A, bottom) DsRed mRNA is injected as a control. (Mean ± SEM; [*] P < 0.05; n = 10 embryos per group.) (C) Model for miR-133 function during fin regeneration. Fin amputation triggers Fgf signaling, which attenuates miR-133 levels through unknown mechanisms. The reduction in miR-133 levels boosts expression of target genes important for blastemal proliferation like Mps1, optimizing regeneration.
Conclusions
In conclusion, our experiments implicate post-transcriptional regulation by miRNAs in the process of complex tissue regeneration. We identified many miRNAs in zebrafish appendages that showed sharp increases or decreases in expression during the transition from uninjured to regenerating tissue. Normal expression of several miRNAs during regeneration is dependent on intact Fgf signaling. Through both gain-of-function and loss-of-function experiments, our data reveal that miR-133 is a regenerative brake whose regulated depletion ensures optimal fin regeneration.
Our findings support a model in which miR-133 levels are diminished by Fgf signaling, a critical pathway for regeneration that is activated within hours of fin amputation, maintained throughout the process, and progressively subdued as final structures are replaced (Fig. 4C; Lee et al. 2005; Whitehead et al. 2005). How miR-133 depletion is triggered by Fgfs mechanistically remains to be determined and may be relevant to other Fgf-dependent developmental events. Lowering miR-133 levels augments blastemal proliferation through regulation of genes like mps1, optimizing regeneration. Thus, miRNA function is integrated into a regulatory circuit that commences via the Fgf signaling pathway and is consummated through a known regulator of blastemal proliferation (Fig. 4C). Given the cellular functions of other predicted miR-133 targets, as well as data from previous miRNA studies, we suspect that there are targets of miR-133 in addition to mps1 that are important for its effects on regeneration (Kloosterman and Plasterk 2006).
miR-133 depletion represents an intriguing form of regulation during complex tissue regeneration. We speculate that animals with enhanced regenerative capacity like zebrafish and axolotls carry an arsenal of tuning mechanisms that maintain the differentiated state and/or preclude unwelcome activation of growth mechanisms. In turn, these tuning mechanisms are repressed upon injury to facilitate regeneration. Thus, it is possible that regulated restriction of steady-state levels of specific miRNAs like miR-133 by early or persistent regeneration signals like Fgfs is a recurring theme in regenerative morphogenesis. Our observation that levels of multiple different miRNAs are lowered during fin regeneration supports this idea. Conversely, miRNAs we identified with increased levels during regenerative outgrowth might help restrict cellular differentiation programs and/or balance the formation and proliferation of blastemal cells. Indeed, we predict that there is an abundance of fascinating mechanistic information about regenerative events to be gained from functional exploration of the many miRNAs that are differentially expressed during fin regeneration. With respect to regenerative medicine, our findings suggest that tactical modulation of key miRNAs and their target populations may be sufficient to revise the regenerative capacity of vertebrate organs.
Materials and methods
Zebrafish caudal fin amputations
Zebrafish of the Ekkwill (EK) strain or EK/AB mixed strain 4–6 mo old were used for all experiments. Amputations were performed with a razor blade and removed one-half of the caudal fin. In Figure 1A experiments, samples were collected from wild-type animals and allowed to regenerate at 33°C, a temperature that accelerates the process. For all other experiments, regeneration occurred at 26°C. Heat shocks were given at 38°C to wild-type and hsp70:dn-fgfr1 animals as described (Lee et al. 2005).
Gene expression analysis
Total RNA was isolated (Tri-Reagent; Sigma) from tissue distal to a region one segment proximal to the amputation plane and used for miRNA microarray hybridizations, Northern analysis, quantitative PCR, and ribonuclease protection assays (see Supplemental Material).
MO and RNA duplex injections
Wild-type and hsp70:dn-fgfr1 caudal fins were amputated and allowed to regenerate for 3 d at 26°C. MOs (Gene-Tools) or RNA duplexes (IDT Technology; design based on miR-133b sequence) (see Supplemental Table 2; Supplemental Fig. 2) were injected into each ray of the dorsal lobe, followed by electroporation of the entire caudal fin using a 5-mm electrode (Thummel et al. 2006; Visvanathan et al. 2007). Fluorescein-tagged MOs were used to monitor electroporation efficiency. When hsp70:dn-fgfr1 regenerates were treated with MOs, electroporation occurred 5 h after a single heat shock at 38°C. Regenerates were imaged daily for 3 d following electroporation and measured from the amputation plane to the distal tip of regenerating rays 2 and 3 (with respect to the most lateral ray) for dorsal and ventral lobes, using Openlab software. The lengths of these rays were averaged to give one length value each for the dorsal and ventral regenerates per animal. Intrafin ratios represent injected dorsal regenerate length:uninjected ventral regenerate length, calculated for each animal at each time point. Student’s t-tests (two-tailed, unequal variance) were performed to determine P-values.
BrdU analysis
A 2.5 mg/mL BrdU solution was injected intraperitoneally into hsp70:dn-fgfr1 animals 5 h after MO injection. After 30 min, caudal fins were collected and processed for immunofluorescence as described (Lee et al. 2005). We quantified the number of BrdU-positive cells by counting within a 0.036-mm2 box, based on the average size of 4-dpa, BrdU-dense blastemas of wild-type animals that had been heat-shocked and electroporated with a standard control MO. Each square was aligned at the distal edge of the regenerate in hsp70:dn-fgfr1 MO-treated fin rays. For each fin, BrdU-positive cells in dorsal rays 2 and 3 were counted for MO effects, while ventral rays 2 and 3 were counted for the uninjected control.
EGFP sensor assays
One-cell zebrafish embryos were injected with 1–2 nL of a solution consisting of 150 ng/μL sensor mRNA and 100 ng/μL Dsred mRNA. When applicable, 10 μM miRNA duplex was added with or without 0.5 ng/μL miR-133 MO or 0.8 ng/μL LNA-133. EGFP fluorescence was quantified at 24–28 h post-fertilization as described (Giraldez et al. 2006).
Acknowledgments
We thank J. Bumgardner, O. Ighile, A. Lepilina, and C. Wheeler for excellent zebrafish care; Y. Lu for protocols; and K. Kikuchi, Y. Lee, A. Nechiporuk, and A. Wills for helpful comments on the manuscript. V.P.Y. was supported by NIH training grant 2 T-32 HL007101-29 and is a post-doctoral fellow of the American Heart Association. This work was supported by grants to K.D.P. from the National Institute of General Medical Sciences, Whitehead Foundation, and Pew Charitable Trusts.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1641808.
References
- Adams D.S., Masi A., Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134:1323–1335. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.Y., Giraldez A.J., Schier A.F. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Fisk H.A., Winey M. Spindle regulation: Mps1 flies into new areas. Curr. Biol. 2004;14:R1058–R1060. doi: 10.1016/j.cub.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M., Poss K.D. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A.N., Kikuchi K., Holdway J.E., Roberts R.W., Burns C.G., Poss K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lien C.L., Schebesta M., Makino S., Weber G.J., Keating M.T. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Shen J., Nechiporuk A., McMahon G., Thisse B., Thisse C., Keating M.T. Roles for Fgf signaling during zebrafish fin regeneration. Dev. Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Poss K.D., Nechiporuk A., Hillam A.M., Johnson S.L., Keating M.T. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Schebesta M., Lien C.L., Engel F.B., Keating M.T. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal. 2006;6 (Suppl. 1):38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper C.L., Moon R.T., Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes & Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- Thummel R., Bai S., Sarras M.P., Song P., McDermott J., Brewer J., Perry M., Zhang X., Hyde D.R., Godwin A.R. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev. Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vinarsky V., Atkinson D.L., Stevenson T.J., Keating M.T., Odelberg S.J. Normal newt limb regeneration requires matrix metalloproteinase function. Dev. Biol. 2005;279:86–98. doi: 10.1016/j.ydbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J.W., Lee S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes & Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead G.G., Makino S., Lien C.L., Keating M.T. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]