Abstract
Cardiac chamber formation represents an essential evolutionary milestone that allows for the heart to receive (atrium) and pump (ventricle) blood throughout a closed circulatory system. Here, we reveal a novel transcriptional pathway between foxn4 and tbx genes that facilitates this evolutionary event. We show that the zebrafish gene slipjig, which encodes Foxn4, regulates the formation of the atrioventricular (AV) canal to divide the heart. sli/foxn4 is expressed in the AV canal, and its encoded product binds to a highly conserved tbx2 enhancer domain that contains Foxn4- and T-box-binding sites, both necessary to regulate tbx2b expression in the AV canal.
[Keywords: Atrioventricular canal, evolutionary development, Forkhead transcription factors, T-box transcription factors, calcium indicator, mutations]
The evolution of organs and adaptation of specialized structures utilizes the duplication of highly conserved transcription factors and their cis-regulatory elements. Because of its gradual progression from a single-chambered muscular tube with peristaltic contractility (invertebrates) to a complex multichambered structure with diverse specialized cardiomyocytes (vertebrates), the heart is ideal to study the genetic underpinnings of organogenesis as well as the evolutionary basis of morphological complexity. Division of the heart into chambers represents an essential evolutionary milestone that allows for the heart to receive and pump blood throughout a closed circulatory system (for review, see Olson 2006). Initially, the vertebrate heart develops as a linear tube without significant cellular or physiological differences; however, as the heart tube loops, distinct cardiac chambers begin to develop and acquire high gap junction density leading to faster conduction velocities. In contrast, the atrioventricular (AV) canal and inner curvature regions expand more slowly and maintain characteristics of embryonic tubular hearts including slow conduction velocity (for review, see Moorman and Christoffels 2003). Due to its primitive myocardial nature, the AV canal has evolved several specialized functions in heart formation and function including the formation of AV cushions as well as AV cardiac conduction delay. Here we reveal how these evolutionarily specialized AV structures are coupled developmentally through a novel transcriptional circuit to structurally (AV valves) and functionally (AV conduction delay) separate the atrium and ventricle.
Results and Discussion
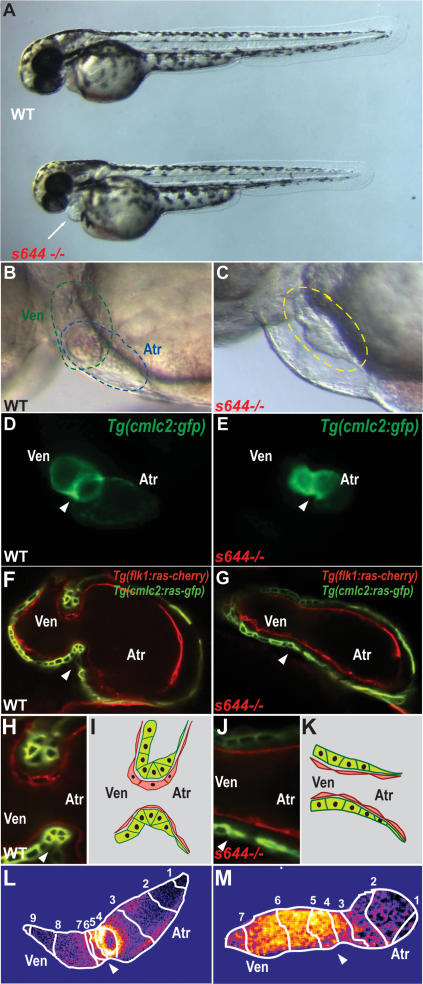
From a recent ENU mutagenesis screen (Beis et al. 2005), we identified a unique cardiovascular mutant, slipjig (sli) (Chen et al. 1996), which displays not only structural AV canal malformations but also AV conduction defects (Fig. 1). sli mutants appear indistinguishable from their wild-type siblings up to 30 h post-fertilization (hpf), displaying formation of a cardiac tube that pumps blood throughout the embryo (data not shown). However, by 36–48 hpf, sli mutants exhibit pericardial edema due to dysmorphic hearts that fail to loop and form an AV canal (Fig. 1A, and cf. B,D and C,E). Furthermore, these mutant hearts exhibit continued peristaltic pumping rather than coordinated and sequential beating of atrial and ventricular chambers as observed in wild-type hearts at similar stages (cf. Supplemental Movies S1 and S2). In addition to these cardiac defects, we observed that sli mutants become unresponsive to tactile stimuli at 60–72 hpf (data not shown).
Figure 1.
Failure of cardiac looping, malformation of AV canal, and loss of AV conduction delay in slipjig mutants. (A) Bright-field micrographs of 48-hpf wild-type (WT) and sli mutant (s644−/−) embryos. The white arrow points to pericardial edema. (B,C) Nomarski micrographs of wild-type and sli mutant hearts at 48 hpf. (B) Green circle delineates the wild-type ventricle, and blue circle indicates the wild-type atrium. (C) Yellow circle delineates the sli mutant heart with no discernible AV boundary. (D,E) Fluorescence micrographs of 48-hpf Tg(cmlc2:gfp) wild-type and sli mutant hearts. sli mutant hearts fail to loop and to form an AV canal. (F–H,J) Confocal micrographs of 40-hpf Tg(cmlc2:ras-GFP)s883; Tg(flk1:ras-cherry)s896 wild-type (F) and sli mutant (G) hearts and AV canal region (H,J). Myocardium and endocardium are in green and red, respectively. sli mutant AV myocardial and endocardial cells fail to undergo characteristic cell shape changes. (I,K) Schematic representation of wild-type and sli mutant AV region. (L,M) Forty-hour-post-fertilzation Tg(cmlc2:gCaMP)s878 wild-type and sli mutant heart optical maps of calcium-dependent fluorescence represented by isochronal lines every 60 msec. Numbers indicate temporal sequence of calcium activation in the heart. sli mutant hearts fail to develop an AV conduction delay by 40 hpf. The white arrowheads point to the AV canal. (Atr) Atrium; (Ven) ventricle.
To further investigate the cardiac defects, we analyzed sli mutants in several transgenic backgrounds. As visualized in the Tg(cmlc2:ras-GFP)s883; Tg(flk1:ras-cherry)s896 line, which outlines cardiomyocytes and endocardial/endothelial cells in green and red fluorescence, respectively, AV myocardial and endocardial cells display distinct cellular shapes, distinguishing them from chamber myocardial and endocardial cells at 40 hpf (Fig. 1F–K). Wild-type AV cardiomyocytes extend their basolateral surface and constrict their apical surface, resulting in a trapezoidal shape, whereas atrial and ventricular cardiomyocytes are squamous and cuboidal, respectively (Fig. 1F,H,I). Additionally, wild-type AV endocardial cells become cuboidal, while the chamber endocardial cells remain squamous (Beis et al. 2005). However, in 40-hpf sli mutants, all endocardial cells remain squamous, and AV myocardial cell shape changes also fail to occur (Fig. 1G,J,K).
Optical mapping of wild-type hearts reveals that AV conduction delay develops by 36–40 hpf where the AV canal begins to form (Fig. 1L; Supplemental Movie S3; Milan et al. 2006). However, optical mapping of 40-hpf Tg(cmlc2:gCaMP)s878 sli mutant hearts reveals the absence of an AV conduction delay, further supporting the observation that AV cardiomyocytes fail to differentiate in these mutants (Fig. 1M; Supplemental Movie S4). Thus, the lack of AV cellular changes in sli mutant hearts is accompanied by the failure of AV canal formation and cardiac looping, as well as electrophysiological abnormalities.
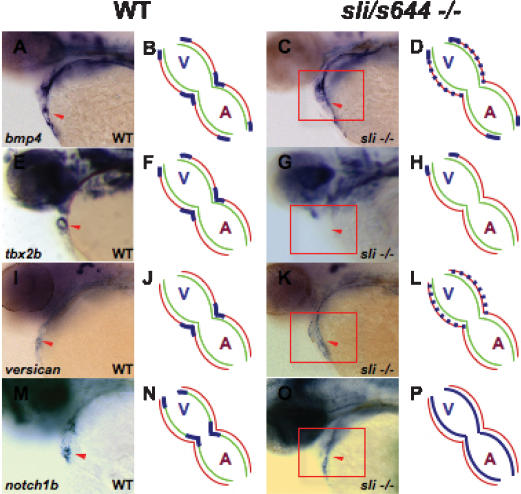
We examined the expression of several cardiac-specific genes to determine how sli affects AV canal formation. Expression of early genes for cardiogenesis (such as nkx2.5 and tbx20) and for myocardial differentiation (such as cmlc2, amhc, and vmhc) appears unaffected in sli mutants (Supplemental Fig. S1). On the other hand, sli mutants exhibit defects in the late expression of several AV canal genes including bmp4, tbx2b, versican, and notch1b. Initially, these AV boundary genes are expressed throughout the anteroposterior extent of the wild-type heart (Walsh and Stainier 2001; Hurlstone et al. 2003). By 48 hpf, the expression of these genes becomes restricted to the AV canal and outflow tract (Fig. 2A,E,I,M). However, in 48-hpf sli mutant hearts, bmp4 and versican expression is expanded throughout the ventricular myocardium, while tbx2b expression appears absent from AV cardiomyocytes (Fig. 2C,G,K). Additionally, endocardial expression of notch1b in sli mutants remains expanded throughout the atrium and ventricle (Fig. 2O). Thus, sli is required specifically for the precise patterning of the myocardium and endocardium within the AV canal.
Figure 2.
Molecular analyses of AV-specific cardiac genes reveal AV canal defects in sli mutant hearts. Schematized representations are shown to the right of whole-mount RNA in situ hybridization data. Red lines indicate myocardium, green lines indicate endocardium, and blue lines indicate gene expression. In 48-hpf wild-type hearts, bmp4 (A,B), tbx2b (E,F), and versican (I,J) are expressed in the AV myocardium and notch1b (M,N) is expressed in the AV endocardium. In 48-hpf sli mutant hearts, bmp4 (C,D) and versican (K,L) expression is expanded throughout the ventricular myocardium, tbx2b (G,H) expression is absent within the AV canal, and notch1b (O,P) expression is diffusely expanded throughout the endocardium. The red arrowhead points to the AV canal. The red box surrounds sli mutant hearts. (A) Atrium; (V) ventricle.
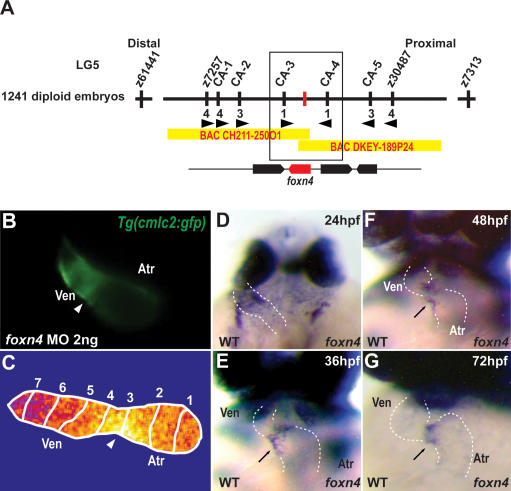
To better understand the molecular nature of the sli phenotype, we positionally cloned the gene disrupted by the sli mutation and found that it encodes the transcription factor Foxn4 (Fig. 3A; Supplemental Fig. S2). At 48 hpf, foxn4 is expressed within the AV myocardium, a pattern consistent with the observed mutant phenotypes (Fig. 3D,E). Injection of 2 ng of foxn4 morpholino (MO) into Tg(cmlc2:gfp) embryos recapitulated the sli mutant cardiac phenotype (Fig. 3B) (n = 138/152). Furthermore, optical mapping of foxn4 knocked-down Tg(cmlc2:gCaMP)s878 embryos showed a loss of AV conduction delay similar to that seen in sli mutants (Fig. 3C). Overall, the strong genetic linkage, identification of severe molecular lesions, and MO phenocopy indicate that foxn4 is the gene affected by the sli mutations.
Figure 3.
sli encodes Foxn4. (A) Genetic map of the sli region. Numbers below SSLP markers indicate recombination events. Two genes were identified within the critical region, which spans two BACs. (B) Knockdown of foxn4 by the injection of 2 ng of an ATG MO into Tg(cmlc2:GFP) embryos results in failure of the hearts to form an AV canal constriction (arrowhead) and absence of cardiac looping. (C) Calcium-dependent fluorescence optical map of 48-hpf Tg(cmlc2:gCaMP)s878 foxn4 MO knockdown heart reveals loss of AV conduction delay at 48 hpf. Isochronal lines depict the distance traveled every 60 msec. Numbers indicate temporal sequence of calcium activation in the heart. (D–G) foxn4 is expressed in the AV canal at 24, 36, 48, and 72 hpf. The black arrows point to the AV canal. The white dashed lines outline the heart. (D) Dorsal view. (E–G) Ventral view. (Atr) atrium; (Ven) ventricle.
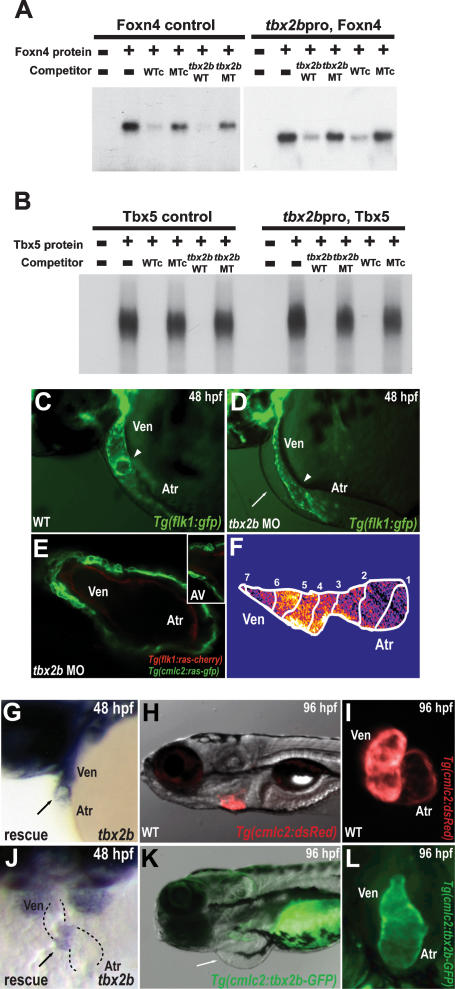
Genetic analyses in several species have revealed the crucial role of Tbx genes, including Tbx-1, Tbx-2, Tbx-3, Tbx-5, Tbx-18, and Tbx-20, in multiple aspects of cardiac development (for review, see Stennard and Harvey 2005; Olson 2006). In particular, Tbx2 has been shown to be critical for the patterning and formation of the AV canal as well as the outflow tract (Christoffels et al. 2004; Harrelson et al. 2004; Rutenberg et al. 2006; Kokubo et al. 2007). Interestingly, our analysis of the Tbx2 enhancer across vertebrates revealed perfectly conserved candidate binding sites for Foxn4 (Schlake et al. 1997) as well as Tbx5 (Mori et al. 2006), a transcription factor that regulates the appearance of AV conduction delay (Supplemental Fig. S3; Moskowitz et al. 2007). In electromobility shift assays (EMSA) to test DNA-binding affinity, Foxn4 and Tbx5 bound efficiently to their labeled putative binding sites, and binding was efficiently competed with excess unlabeled cognate competitors, but not with mutated oligonucleotides (Fig. 4A,B). Conversely, these unlabeled putative binding elements effectively competed with binding of Foxn4 and Tbx5 to the labeled canonical binding sites (Fig. 4A,B). Thus, these data suggest that Foxn4 and a T-box factor, possibly Tbx5, specifically interact with their consensus sites in the Tbx2 enhancer.
Figure 4.
Foxn4 and Tbx5 directly regulate expression of tbx2b, a gene required for AV canal formation. (A,B) EMSA reveals specific binding of Foxn4 and Tbx5 to their respective binding elements within the tbx2b enhancer. Lane 1 (−/−) contains reticulocyte lysate without recombinant Foxn4 or Tbx5 protein. Recombinant Foxn4 (A) and Tbx5 (B) proteins were combined with radiolabeled double-stranded oligonucleotides representing the canonical Foxn4-binding site (Foxn4 control) and Tbx5-binding site (Tbx5 control) as well as the tbx2b enhancer Foxn4-binding element (tbx2bpro, Foxn4) and Tbx5-binding element (tbx2bpro, Tbx5). Foxn4 and Tbx5 interactions to respective binding sites were further tested through competition assays using excess unlabeled canonical wild-type/mutant (WTc, MTc) and tbx2b enhancer wild-type/mutant (tbx2b WT, tbx2b MT), foxn4, and tbx5 oligonucleotides. (C–F) tbx2b MO knockdown hearts exhibit absence of AV canal and cardiac looping at 48 hpf. Fluorescence micrographs of 48-hpf control (C) and tbx2b MO-injected (D) Tg(flk1:gfp)843 embryos. The arrowheads point to the AV boundary. (E) Confocal micrograph of 48-hpf Tg(cmlc2:ras-GFP)s883; Tg(flk1:ras-cherry)s896 tbx2b MO knockdown heart. (Inset) AV boundary region. (F) Calcium-dependent fluorescence optical map of 48-hpf Tg(cmlc2:gCaMP)s878 tbx2b MO knockdown heart reveals loss of AV conduction delay at 48 hpf. Isochronal lines depict the distance traveled every 60 msec. Numbers indicate temporal sequence of calcium activation in the heart. (G,J) tbx2b expression in foxn4 mRNA rescued sli mutant heart. The black arrow points to the AV canal. (H,I,K,L) Cardiac-specific overexpression of tbx2b results in lack of cardiac looping and failure to form the AV canal. Fluorescence micrographs of 80-hpf Tg(cmlc2:dsRed)s879 wild-type (H,I) and Tg(cmlc2:Tbx2b-GFP)s900 (K,L) hearts. The white arrow points to pericardial edema.
To further test whether Foxn4 and Tbx5 directly regulate tbx2b AV canal expression in vivo, we generated zebrafish transgenics harboring a fluorescent reporter driven by the wild-type tbx2b conserved enhancer or the enhancer with mutations in the Foxn4 or Tbx5 site. The wild-type tbx2b conserved enhancer directed high expression of the dsRed fluorescent reporter in the posterior portion of the fin, nervous tissue, and heart/AV canal and outflow tract, nearly recapitulating the entire endogenous tbx2b expression pattern (Supplemental Fig. S4, cf. A,E and B,F). However, in the mutant Foxn4 site enhancer transgenic, cardiac expression appeared to be absent while fin, brain, and eye expression was maintained (Supplemental Fig. S4C,G). Additionally, the mutant Tbx5 site enhancer transgenic misexpressed the fluorescent reporter throughout the ventricle, while fin, brain, and eye expression was absent (Supplemental Fig. S4D,H). Thus, the putative Foxn4 and Tbx5 DNA-binding sites are required for the precise control of this highly conserved tbx2b enhancer during AV canal development.
To determine its role in AV canal formation and function, we performed MO knockdown of tbx2b. tbx2b MO-injected Tg(flk1:gfp)s843, Tg(cmlc2:ras-GFP)s883, and Tg(flk1:ras-cherry)896 embryos exhibited a lack of AV canal formation at 40 hpf, similar to the sli mutant phenotype (Fig. 4D,E) (n = 127/146). Furthermore, optical mapping of hearts of 40-hpf tbx2b MO-injected Tg(cmlc2:gCaMP)s878 embryos showed absence of AV conduction delay (Fig. 4F) (n = 51/63). Overall, these results indicate that tbx2b regulates AV canal formation by mediating foxn4 function in the heart.
Because of this epistatic relationship, we attempted to rescue the slis644 mutant cardiac phenotype with injection of foxn4 and tbx2b mRNA. After injecting foxn4 mRNA, ∼55% of slis644 mutant hearts exhibited complete rescue of the AV canal phenotype at 48 hpf (Supplemental Fig. S5A,B), including the pattern of tbx2b expression in the myocardium (Fig. 4G,J). On the other hand, tbx2b mRNA injections resulted in only 5% of slis644 mutant hearts forming the AV canal at 48 hpf. However, severe cardiac phenotypes were also observed in injected wild-type embryos, including absence of AV canal formation and cardiac looping (Supplemental Fig. S5A,B). Thus, failure to rescue a greater percentage of slis644 mutant hearts with tbx2b is likely due to cardiac defects caused by overexpression of this gene. To determine whether the myocardial phenotype from tbx2b mRNA overexpression was a cell-autonomous event, we misexpressed tbx2b throughout the linear heart tube using the Tg(cmlc2:tbx2b-gfp)s900 line. Similar to hearts in tbx2b mRNA-injected embryos, Tg(cmlc2:tbx2b-gfp)s900 hearts failed to form an AV boundary and undergo cardiac looping (Fig. 4K,L). Thus, as previously described in amniotes (Christoffels et al. 2004; Cai et al. 2005; Singh et al. 2005; Stennard et al. 2005; Takeuchi et al. 2005), tbx2b misexpression in the zebrafish myocardium appears to prevent the linear heart tube from forming chambers, resulting in loss of AV canal formation and cardiac looping.
Separation of the heart into chambers evolutionarily necessitated the development of specialized structures, such as the cardiac valve leaflets and a cardiac conduction system, to coordinate the beating of these compartments and achieve antegrade blood flow (for review, see Olson 2006). The family of T-box transcription factors has been integral in orchestrating the development of these additional structures (for review, see Stennard and Harvey 2005). Similar to these T-box transcription factors, a cadre of Forkhead transcription factors has recently been shown to be critical for cardiovascular development (Yamagishi et al. 2003; von Both et al. 2004; Wang et al. 2004; Creemers et al. 2006; Ramakrishna et al. 2007). Interestingly, Foxa2, Foxc1, and Foxc2 mediate heart development through direct regulation of Tbx1 (Yamagishi et al. 2003). In our studies, we show that Foxn4, in concert with Tbx5, may exert its transcriptional effect on the regulatory apparatus of Tbx2, directing AV canal formation. Previous reports have indicated that in amniotes the AV myocardium fails to differentiate (de Jong et al. 1992). Our data show that the Foxn4–Tbx2b pathway leads to AV myocardial and endocardial cellular transformations that initiate the development of AV conduction delay and valve leaflets. Whether AV canal defects in Tbx2-null mice (Harrelson et al. 2004) are due to lack of AV cellular changes as observed in foxn4- and tbx2b-defective zebrafish embryos is worth exploring. Interestingly, Foxn4-null mutant mice die perinatally, but their cause of death is unclear (Li et al. 2004). Thus, analyzing how these AV cardiomyocyte changes in zebrafish translate to the mammalian AV specialized myocardium will be of great importance in understanding the evolution of the AV canal as well as human AV conduction block and congenital heart disease.
AV canal formation depends on an elaborate cross-talk between specialized myocardial and endocardial cells involving multiple signaling pathways including Wnt/β-catenin, bone morphogenetic protein (BMP), and Notch signaling (Yamada et al. 2000; Hurlstone et al. 2003; Milan et al. 2006; Rutenberg et al. 2006; Kokubo et al. 2007; Xin et al. 2007). BMP2/4 signaling participates in AV canal formation by inducing Tbx2 expression at the AV canal (Yamada et al. 2000), while Notch signaling may suppress this signal in the chambers to restrict Tbx2 expression (Rutenberg et al. 2006; Kokubo et al. 2007; Xin et al. 2007). Our data indicate a possible model in which foxn4 expressed specifically in the AV myocardium directs competent cardiomyocytes at the AV boundary to undergo cellular and molecular changes required for AV myocardial specialization. Wnt/β-catenin or BMP signaling may be the factors that establish this competence at the AV boundary. Alternatively, these signals may directly regulate foxn4 expression or function at the AV canal. Thus, these data provide a mechanistic framework to begin explaining how the combinatorial interactions of the Foxn4 and Tbx5 transcription factors culminate in transcription of Tbx2 to establish the AV boundary. Additionally, because of the complexity of T-box factors and their binding sites, other potential T-box proteins may also regulate tbx2b through the highly conserved Tbx5-binding element in its enhancer. Finally, given that human mutations in TBX5 (Holt-Oram syndrome) lead to cardiac septal and conduction defects (Basson et al. 1997), screening for FOXN4 or TBX2 mutations in humans with heart disease may provide new avenues for understanding the development of congenital heart diseases, leading to future therapeutic or preventive interventions.
Materials and methods
Zebrafish strains
Embryos and adult fish were raised and maintained under standard laboratory conditions. We used the following lines: slis644 (Chi et al. 2008), slitm117c (Chen et al. 1996), Tg(cmlc2:gfp) (Huang et al. 2003), Tg(flk1:gfp)s843 (Jin et al. 2005), Tg(cmlc2:gCaMP)s878 (Arnaout et al. 2007), Tg(cmlc2:dsRed)s879, Tg(cmlc2:ras-GFP)s883 (B. Jungblut, C. Munson, J. Huisken, L.A. Trinh, and D.Y. Stainier, in prep.), Tg(flk1:ras-cherry)s896, Tg(tbx2bpro:dsRed)s899, Tg(cmlc2:tbx2b-P2A-GFP)s900, bicistronic expression of tbx2b and GFP, Tg(tbx2bpro/foxn4MT:GFP)s901, and Tg(tbx2bpro/tbx5MT:cherry)s902.
Mapping
We mapped the mutation to linkage group 5 using a set of SSLP markers. For fine-mapping, 1241 mutant embryos were tested with SSLP markers in the critical interval (Fig. 3A). sli/foxn4 complementary DNA was isolated, sequenced, and analyzed from the two mutant alleles.
Histochemical methods
Whole-mount in situ hybridization was performed as described previously (Walsh and Stainier 2001; Hurlstone et al. 2003), using the following probes: amhc, vmhc, cmlc2, tbx20, nkx2.5, bmp4, versican, notch1b, foxn4, and tbx2b. For generating sli/foxn4 and tbx2b in situ probes, 750 and 2200 base pairs at the 3′ end of these genes were PCR-amplified. Embryos were embedded in low-melting-point agarose and imaged using either confocal microscopy or conventional fluorescence microscopy.
Videorecording/microscopy
Bright-field pictures and videos were taken using the Stemi SV11 dissecting microscope (Zeiss). Videos were captured using a standard CCD camera at 20 frames per second (fps).
Optical mapping by wide-field epifluorescence
Individual zebrafish embryos between 36 and 48 hpf were placed on a coverglass. Images were acquired and data analysis was performed as described previously (Arnaout et al. 2007). Isochronal lines at 60-msec intervals were obtained by identifying the maximal spatial gradient for a given time point.
MO antisense oligonucleotide injection and mRNA overexpression
We used a MO targeted against an ATG upstream of the translational start site of foxn4 and tbx2b with the following sequence: 5′-CGTG CAGTTTGCTCTGGACGGTCAT-3′ and 5′-GAGCGTGGAAAGG GTGGTAAGCCAT-3′, respectively. Embryos were injected at the one-cell stage with 2 ng of foxn4 or tbx2b MO and assayed between 40 and 48 hpf. For mRNA overexpression, one-cell-stage embryos were injected with 50–150 pg of sli/foxn4 or tbx2b mRNA.
EMSAs
EMSAs were performed as described previously (De Val et al. 2004). The truncated version of FoxN4, containing the DNA-binding domain, and the full-length Tbx5 were generated from the plasmids pCITE-FoxN4(tr) and pcDNA3-Tbx5, respectively, using the TNT Quick-coupled Transcription/Translation System as described (Promega). The Tbx5 and Foxn4 control and mutant binding sites were adapted as described previously (Schlake et al. 1997; Mori et al. 2006). The sense strand sequences of the tbx2b oligonucleotides used for EMSA were FoxN4tbx2bWT, 5′-GGTT GATTGCTGATTTGACGCTTTTTGGACCAA-3′; FoxN4tbx2bMT, 5′- GGTTGATTGCTGATTTTacaCTTTCTGGACCAA-3′; Tbx5tbx2bWT, 5′GGGGCGTCCGAGAAGGTGTCGGAAGCCTCAGG-3′; Tbx5tbx2bMT, 5′-GGGGCGTCCGAGAAGGatcCGGAAGCCTCAGG-3′.
Acknowledgments
We thank all the members of the Baier and Stainier laboratory screen teams for their support during the screens; Ana Ayala, Diana Kuo, Justin Bosch, and Koro Brand for expert help with the fish; and Takashi Mikawa and Markus Bussen for critical comments on the manuscript. N.C.C. and R.M.S. are supported by K08 grants (NHLBI), GSK Research and Education Foundation Cardiovascular Awards, and Fellow to Faculty AHA post-doctoral awards. S.D.V. is supported by a fellowship from the AHA. This work was supported in part by grants from the Packard Foundation and the NIH (NHLBI) to D.Y.R.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1629408.
References
- Arnaout R., Ferrer T., Huisken J., Spitzer K., Stainier D.Y., Tristani-Firouzi M., Chi N.C. Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J., et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Beis D., Bartman T., Jin S.W., Scott I.C., D’Amico L.A., Ober E.A., Verkade H., Frantsve J., Field H.A., Wehman A., et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Cai C.L., Zhou W., Yang L., Bu L., Qyang Y., Zhang X., Li X., Rosenfeld M.G., Chen J., Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.N., Haffter P., Odenthal J., Vogelsang E., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., Hammerschmidt M., Heisenberg C.P., et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Chi N.C., Shaw R.M., Jungblut B., Huisken J., Arnaout R., Ferrer T., Scott I., Beis D., Xiao T., Baier H., et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biology. 2008 doi: 10.1371/journal.pbio.0060109. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V.M., Hoogaars W.M., Tessari A., Clout D.E., Moorman A.F., Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Creemers E.E., Sutherland L.B., McAnally J., Richardson J.A., Olson E.N. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- de Jong F., Opthof T., Wilde A.A., Janse M.J., Charles R., Lamers W.H., Moorman A.F. Persisting zones of slow impulse conduction in developing chicken hearts. Circ. Res. 1992;71:240–250. doi: 10.1161/01.res.71.2.240. [DOI] [PubMed] [Google Scholar]
- De Val S., Anderson J.P., Heidt A.B., Khiem D., Xu S.M., Black B.L. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev. Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Harrelson Z., Kelly R.G., Goldin S.N., Gibson-Brown J.J., Bollag R.J., Silver L.M., Papaioannou V.E. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- Huang C.J., Tu C.T., Hsiao C.D., Hsieh F.J., Tsai H.J. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Hurlstone A.F., Haramis A.P., Wienholds E., Begthel H., Korving J., Van Eeden F., Cuppen E., Zivkovic D., Plasterk R.H., Clevers H. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- Jin S.W., Beis D., Mitchell T., Chen J.N., Stainier D.Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Kokubo H., Tomita-Miyagawa S., Hamada Y., Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- Li S., Mo Z., Yang X., Price S.M., Shen M.M., Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Milan D.J., Giokas A.C., Serluca F.C., Peterson R.T., MacRae C.A. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- Moorman A.F., Christoffels V.M. Cardiac chamber formation: Development, genes, and evolution. Physiol. Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Mori A.D., Zhu Y., Vahora I., Nieman B., Koshiba-Takeuchi K., Davidson L., Pizard A., Seidman J.G., Seidman C.E., Chen X.J., et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Moskowitz I.P., Kim J.B., Moore M.L., Wolf C.M., Peterson M.A., Shendure J., Nobrega M.A., Yokota Y., Berul C., Izumo S., et al. A molecular pathway including id2, tbx5, and nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Olson E.N. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Kim I.M., Petrovic V., Malin D., Wang I.C., Kalin T.V., Meliton L., Zhao Y.Y., Ackerson T., Qin Y., et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev. Dyn. 2007;236:1000–1013. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- Rutenberg J.B., Fischer A., Jia H., Gessler M., Zhong T.P., Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake T., Schorpp M., Nehls M., Boehm T. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc. Natl. Acad. Sci. 1997;94:3842–3847. doi: 10.1073/pnas.94.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.K., Christoffels V.M., Dias J.M., Trowe M.O., Petry M., Schuster-Gossler K., Burger A., Ericson J., Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Stennard F.A., Harvey R.P. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Stennard F.A., Costa M.W., Lai D., Biben C., Furtado M.B., Solloway M.J., McCulley D.J., Leimena C., Preis J.I., Dunwoodie S.L., et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Takeuchi J.K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A.B., Mori A.D., Arruda E.P., Gertsenstein M., Georges R., Davidson L., Mo R., et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- von Both I., Silvestri C., Erdemir T., Lickert H., Walls J.R., Henkelman R.M., Rossant J., Harvey R.P., Attisano L., Wrana J.L. Foxh1 is essential for development of the anterior heart field. Dev. Cell. 2004;7:331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Walsh E.C., Stainier D.Y. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- Wang B., Weidenfeld J., Lu M.M., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- Xin M., Small E.M., van Rooij E., Qi X., Richardson J.A., Srivastava D., Nakagawa O., Olson E.N. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc. Natl. Acad. Sci. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Revelli J.P., Eichele G., Barron M., Schwartz R.J. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: Evidence for BMP2 induction of Tbx2. Dev. Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Maeda J., Hu T., McAnally J., Conway S.J., Kume T., Meyers E.N., Yamagishi C., Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes & Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]