Abstract
Medulloblastoma is an embryonal tumor thought to arise from the granule cell precursors (GCPs) of the cerebellum. PATCHED (PTCH), an inhibitor of Hedgehog signaling, is the best-characterized tumor suppressor in medulloblastoma. However, <20% of medulloblastomas have mutations in PTCH. In the search for other tumor suppressors, interest has focused on the deletion events at the 17p13.3 locus, the most common genetic defect in medulloblastoma. This chromosomal region contains HYPERMETHYLATED IN CANCER 1 (HIC1), a transcriptional repressor that is a frequent target of epigenetic gene silencing in medulloblastoma. Here we use a mouse model of Ptch1 heterozygosity to reveal a critical tumor suppressor function for Hic1 in medulloblastoma. When compared with Ptch1 heterozygous mutants, compound Ptch1/Hic1 heterozygotes display a fourfold increased incidence of medulloblastoma. We show that Hic1 is a direct transcriptional repressor of Atonal Homolog 1 (Atoh1), a proneural transcription factor essential for cerebellar development, and show that ATOH1 expression is required for human medulloblastoma cell growth in vitro. Given that Atoh1 is also a putative target of Hh signaling, we conclude that the Hic1 and Ptch1 tumor suppressors cooperate to silence Atoh1 expression during a critical phase in GCP differentiation in which malignant transformation may lead to medulloblastoma.
[Keywords: HIC1, PTCH, ATOH1, Math1, medulloblastoma]
Medulloblastoma is the most common malignant CNS tumor in children (Packer 1999). Many medulloblastomas are thought to arise from a well-characterized neuronal progenitor population in the developing cerebellum, a component of the CNS essential for motor coordination in vertebrates (Kho et al. 2004; Oliver et al. 2005). During cerebellar development, this progenitor population, known as granule cell precursors (GCPs), undergoes rapid expansion in the external granule cell layer (EGL). During the process of terminal differentiation, these cells descend through the molecular layer to form the mature granule cells that reside in the internal granule cell layer (IGL) (Wechsler-Reya and Scott 2001). Expansion of the GCP compartment is dependent on the Hedgehog (Hh) pathway, a highly conserved embryonic signaling system required for patterning and progenitor cell regulation in animal development (Dahmane and Ruiz-i-Altaba 1999). The Purkinje cells, a specialized neuronal population of the cerebellum, secrete the Hh pathway ligand Sonic Hedgehog (Shh), which tightly regulates the proliferation of the GCPs and is essential for normal cerebellar development (Kenney and Rowitch 2000; Kenney et al. 2003).
Because granule cell differentiation can be linked to migration from the EGL to the IGL, the cerebellum represents an ideal model of neuronal differentiation. Medulloblastoma, in turn, is an important model of how developmental pathways such as Hh signaling function in cancer (Wechsler-Reya and Scott 2001). The Hh pathway receptor PATCHED (PTCH), an essential inhibitor of Hh signaling, is perhaps the best understood tumor suppressor in medulloblastoma (Pietsch et al. 2004). Germline mutations in PTCH cause Gorlin’s syndrome, characterized by the development of medulloblastoma, basal cell carcinoma, and rhabdomyosarcoma (Pietsch et al. 2004). Mice bearing heterozygous mutations of Ptch1 also develop medulloblastoma (Goodrich et al. 1997). These tumors exhibit marked Hh pathway activation that is required for continued growth of the tumor in vivo and in vitro (Goodrich et al. 1997; Berman et al. 2002; Romer et al. 2004). Although Hh pathway activation seems to be important in many medulloblastomas, mutations in the Hh pathway, including PTCH, are seen in <25% of sporadic cases (Eberhart 2003).
Tumor suppressors known to be important in adult solid tumor biology, such as p53, are rarely mutated in medulloblastoma (Eberhart 2003). However, several investigators have demonstrated that loss of heterozygosity at the 17p13.3 locus distal to p53 is the most frequent genetic defect in sporadic medulloblastoma (Rood et al. 2002; Waha et al. 2003). This chromosomal region contains HIC1 (HYPERMETHYLATED IN CANCER 1), a POZ domain transcription factor that is a frequent target of promoter hypermethylation and epigenetic gene silencing in medulloblastoma (Rood et al. 2002; Waha et al. 2003; Lindsey et al. 2005). Heterozygous Hic1 knockout mice develop age-dependent malignancies associated with epigenetic gene silencing of the wild-type Hic1 allele (Chen et al. 2003). Since these animals do not develop medulloblastoma, we reasoned that the neuronal tumor suppressor function of Hic1 might be revealed in the Ptch1 knockout mouse model.
Here, we demonstrate a critical role of Hic1 as a neuronal tumor suppressor in the setting of heterozygous mutations in Ptch1. Hic1 heterozygosity dramatically increases the incidence of medulloblastoma on the Ptch1 heterozygous background. These tumors are characterized by silencing of the wild-type Ptch1 allele, as well as dense promoter hypermethylation of the wild-type Hic1 gene. Given that Hic1 is a well-characterized transcriptional repressor (Deltour et al. 1999, 2002; Pinte et al. 2004b; Chen et al. 2005), we hypothesized that it might regulate expression of an Hh-regulated gene that is important for GCP development. Using gene expression and chromatin immunoprecipitation (ChIP) analysis, we show that the proneural transcription factor Atoh1, commonly referred to as Math1, is a direct target of Hic1-mediated transcriptional repression. Atoh1 is a mammalian basic helix–loop–helix transcription factor required for the development of the EGL (Ben-Arie et al. 1997, 2000). Loss of Hic1 function in medulloblastoma cell lines derived from both our mouse model and human sporadic medulloblastoma is associated with marked overexpression of Atoh1, which we show is required for growth of these tumors in vitro. In cultured GCPs, we also demonstrate that Hic1 acts downstream from the Hh ligand Sonic Hedgehog (Shh) to repress Atoh1 expression. These data demonstrate that the efficiency of malignant GCP transformation in Ptch1 heterozygosity can be markedly increased through epigenetic silencing of Hic1, a gene that may play a critical role in terminating the GCP progenitor phenotype.
Results
Hic1/Ptch1 compound heterozygotes frequently develop medulloblastoma
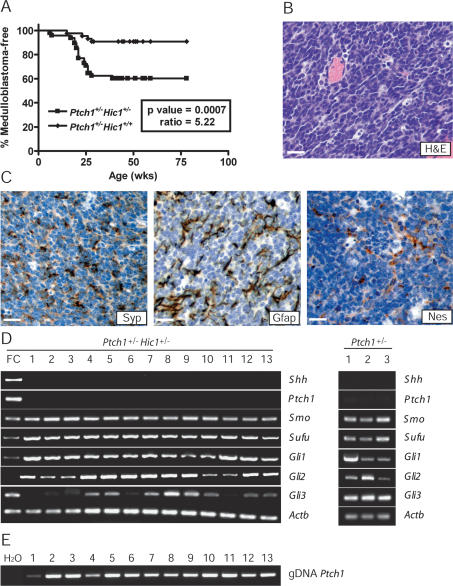
Mice heterozygous for a loss-of-function mutation in Ptch1 develop medulloblastoma at a frequency of 10%–15% (Goodrich et al. 1997). To investigate the importance of Hic1 as a tumor suppressor in the developing cerebellum, we created doubly heterozygous Ptch1+/− Hic1+/− animals and measured the frequency of medulloblastoma compared with Ptch1+/− littermates. Kaplan Meier analysis of 42 Ptch1+/− Hic1+/+ and 45 Ptch1+/− Hic1+/− heterozygotes demonstrated more than a fourfold increase in medulloblastoma incidence in the Ptch1+/− Hic1+/− animals, with a hazard ratio of 5.22 (P-value < 0.001, log-rank Kaplan Meier analysis) (Fig. 1A). Of the 19 tumors observed in the Ptch1+/− Hic1+/− mice, 16 of them developed in a similar time frame to their Ptch1+/− littermates, suggesting that there is a critical window of opportunity for GCP transformation that is not significantly accelerated by loss of Hic1. No other tumors were observed in these animals, although it should be noted that the animals were sacrificed prior to the age at which most Hic1+/− animals present with tumors (Chen et al. 2003).
Figure 1.
Hic1 interacts with Ptch1 in the pathogenesis of medulloblastoma. (A) Log-rank Kaplan Meier analysis of tumor incidence in Ptch1+/− Hic1+/− mice versus Ptch1+/− mice. (B) Hematoxylin/eosin staining of representative tumor from Ptch1+/− Hic1+/− animal. Bar, 20 μm. (C) Immunohistochemical stains for Synaptophysin (Syp), Glial fibrillary acid protein (Gfap), and Nestin (Nes) in a representative tumor, shown as DAB staining (brown), counterstained with hematoxylin (blue). Bars, 20 μm. (D) Semiquantitative Hh pathway expression profiling of 13 Ptch1+/− Hic1+/− and three Ptch1+/− tumors. Mouse fetal cerebellum (FC) (Clontech) used as positive control. (Shh) Sonic hedgehog; (Ptch1) Patched1; (Smo) Smoothened; (Actb) β-Actin. (E) Nonquantitative Ptch1 PCR on genomic DNA from 13 Ptch1+/− Hic1+/− tumors.
Tumors in both Ptch1+/− and Ptch1+/− Hic1+/− animals were similar in presentation, macroscopic appearance, and histopathology. Although the desmoplastic variant of medulloblastoma is typically associated with germline mutation in PTCH1 (Pietsch et al. 2004), the Ptch1+/− Hic1+/− tumors did not exhibit overt nodularity, instead resembling the more classic histology commonly associated with tumors arising in mouse models of the disease in which Ptch1 inactivation is a component of the genetic background (Fig. 1B; Goodrich et al. 1997; Berman et al. 2002) and often contained large exophytic areas growing on the surface of the brain along with more invasive regions (data not shown). Expression of the neural stem cell marker nestin, along with glial and neuronal lineage markers, was seen in superficial tumors lacking entrapped normal cells, establishing the diagnosis of a primitive neuroectodermal tumor (PNET) with the potential to differentiate along multiple lineages (Fig. 1C).
In order to clearly describe the resulting tumors from the Ptch1/Hic1 model of medulloblastoma, it is necessary to determine Hh pathway activity, especially given the ongoing controversy regarding the expression of wild-type Ptch1 in mouse models of medulloblastoma (Wetmore et al. 2000; Zurawel et al. 2000; Romer et al. 2004). Since the Ptch1 and Hic1 knockout alleles both contain a β-galactosidase expression cassette (Goodrich et al. 1997; Carter et al. 2000), we could not use β-galactosidase activity as a measure of Hh pathway expression as has been previously reported (Goodrich et al. 1997; Taipale et al. 2000; Berman et al. 2002). Instead, we assessed the status of Hh signaling by semiquantitative transcriptional profiling of known pathway targets, using β Actin as a reference gene to demonstrate that equivalent amounts of template were used for each tumor. In normal GCP development, Shh ligand binds to and inhibits the Ptch1 receptor, which in turn releases Smoothened (Smo), a seven transmembrane domain protein essential for Hh signaling, from Ptch1-mediated inhibition (Taipale et al. 2002). Active Smo leads to stabilization of the Gli transcription factors and expression of Hh target genes that include Gli1 and Ptch1 (Hooper and Scott 2005). The high-level expression of Ptch1, Smo, Gli1, and Gli2 in the absence of Hh ligand expression seen in this tumor model is characteristic of Ptch1 mutant tumors with unrestrained Smo activation (Fig. 1D; Goodrich et al. 1997; Berman et al. 2002; Kimura et al. 2005). Sufu is also highly expressed in all of the tumors isolated, consistent with prior observations in Ptch1+/− medulloblastoma demonstrating that its expression level is similar to that seen in the cerebellum of a 5-d-old mouse (Lee et al. 2007). Sufu is a known negative regulator of the Hh pathway (Ingham and McMahon 2001), yet its expression does not appear to impede pathway activity in these tumors. Targeted inactivation of Ptch1 in this model is achieved by replacing exon 2 with a β-galactosidase expression cassette (Goodrich et al. 1997), so we assessed the status of the wild-type Ptch1 allele using a PCR assay specific for exon 2 (Fig. 1D). This analysis reveals that the wild-type Ptch1 allele is intact but is not expressed (Fig. 1D,E), and these data support published studies demonstrating that the wild-type Ptch1 allele is not expressed in this medulloblastoma model (Oliver et al. 2005).
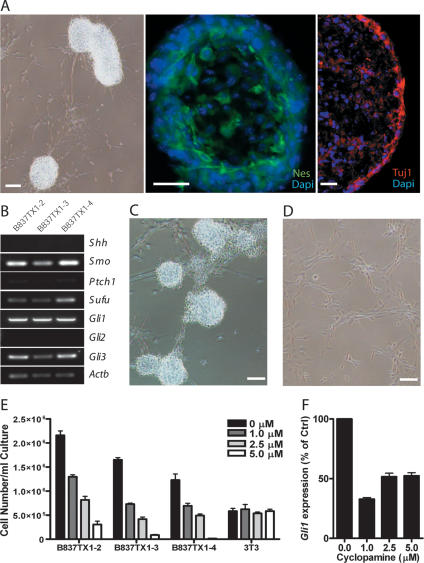
Tumors from Ptch1+/− Hic1+/− animals were propagated as subcutaneous allografts in NOD/SCID mice. Three separate cell lines were subsequently developed from one of the Ptch1+/− Hic1+/− allografts and were successfully cultured in low-serum conditions as tumor spheres. These spheres have an inner core expressing Nestin; Tuj1, a marker of neuronal differentiation, is found at the exterior (Fig. 2A); and they do not express Hic1 by real-time PCR (data not shown). The expression pattern of Hh pathway components in these cell lines replicated the expression pattern in the tumor from which they were derived (Fig. 2B). Treatment of these cell lines over 7 d with the Smo antagonist cyclopamine while challenging the proliferation potential of the cell lines by serially replating induced a switch from a spherical to an adherent morphology (Fig. 2C,D), a reduction in expression of Gli1, and a reduction in proliferation, whereas proliferation of control fibroblast cells (NIH-3T3) was unaffected (Fig. 2E,F). At higher concentrations of cyclopamine, we observed a slight increase in Gli1 mRNA expression (Fig. 2F), which may reflect a progressive loss of cells that are responsive to Hh pathway blockade. These data demonstrate that medulloblastoma cells from Ptch1+/− Hic1+/− animals are dependent on aberrant Hh pathway activation for growth and suggest that pathway inhibition induces terminal differentiation.
Figure 2.
Medulloblastomas from Ptch1+/− Hic1+/− mice are Hh pathway-dependent. (A) A tumor-sphere cell line derived from a Ptch1+/− Hic1+/− tumor: phase contrast image (bar, 50 μm), immunofluorescent staining for Neuronal class III β-Tubulin (Tuj1) and Nestin (Nes) expression (bars, 25 μm). Nuclei counterstained with DAPI (blue). (B) Hh pathway expression profiling in three cell lines derived from a Ptch1+/− Hic1+/− tumor. (C) Representative phase contrast image of Ptch1+/− Hic1+/− tumor-sphere cell line treated with vehicle control (4×). (D) Representative phase contrast image of Ptch1+/− Hic1+/− cell line treated with 5 μM cyclopamine (4×) (bars, 50 μm). (E) Cell concentrations of Ptch1+/− Hic1+/− medulloblastoma cell lines B837TX1-2–B837TX1-4, and mouse fibroblast cell line NIH-3T3 (3T3) after 7 d of culture with 0 μM, 1 μM, 2.5 μM, or 5 μM cyclopamine. (0 μM) Vehicle control. Error bars reflect SEM of five biological replicates. (F) Gli1 quantitative PCR performed on RNA isolated from B837TX1-3 following 7 d of culture in 0 μM, 1 μM, 2.5 μM, or 5 μM cyclopamine. Data are graphed in relation to untreated B837TX1-3. Error bars reflect SEM of five biological replicates.
Hic1 is epigenetically silenced in both the Ptch1+/− and Ptch1+/− Hic1+/− tumors
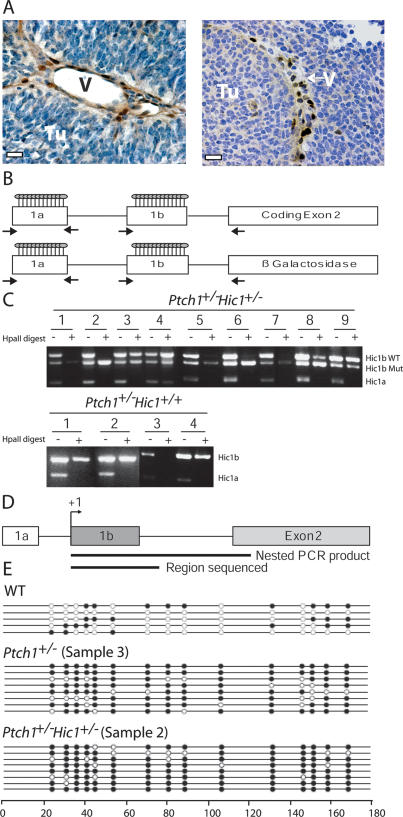
To determine whether Hic1 was expressed in the tumors, Hic1 mRNA expression was compared by RT–PCR in medulloblastoma from Ptch1+/− Hic1+/− and Ptch1+/− animals to expression in microscopically normal adjacent brain. In all cases, Hic1 mRNA was markedly reduced or absent in medulloblastoma but was readily detectable in normal brain (data not shown). To confirm these findings, we analyzed paraffin-embedded tissue for Hic1 protein expression by immunohistochemistry using a Hic1-specific antibody (Chen et al. 2003). Hic1 was absent in medulloblastoma cells but was evident in the nuclei of vessels and stromal fibroblasts (Fig. 3A; Supplemental Fig. 1). Hic1 has a complex gene structure in which transcription can initiate at three separate promoters, two of which, 1a and 1b, give rise to noncoding exons and 1c, which transcribes a 5′ untranslated region (Pinte et al. 2004a). Although the 1a transcript is most common in normal tissues, tumors that do not express HIC1 most often exhibit promoter 1b hypermethylation (Chen et al. 2003). A 1c transcript has only been found in RNA from mammary glands and ovaries, so we concentrated on the 1a and 1b promoters in our analysis of Hic1 in the brain (Pinte et al. 2004a). To analyze the methylation status of Hic1 in both the Ptch1+/− Hic1+/− and Ptch1+/− tumors, we used a previously described HpaII restriction enzyme assay in which genomic DNA from the tumors is first digested with the methylation-sensitive HpaII enzyme, and then a PCR reaction is used to amplify undigested (methylated) 1a and 1b promoter regions simultaneously, and the signal for the 1b region of the wild-type and targeted alleles could be further distinguished with allele-specific primers for Hic1 heterozygotes (Fig. 3B; Chen et al. 2003). The strength of the HpaII assay is that it is able to differentiate dense methylation from the mosaic methylation pattern found in normal cerebellum (Rood et al. 2002; Waha et al. 2003; Lindsey et al. 2005). Although Hic1 was not expressed in the 13 Ptch1+/− Hic1+/− and three Ptch1+/− Hic1+/+ tumors examined, none of the tumors exhibited Hic1 allelic deletion as shown by the mock-digested PCR; therefore, lack of expression is most likely due to promoter hypermethylation. However, only four of the Ptch1+/− Hic1+/− tumors tested strongly displayed dense wild-type Hic1b methylation, and only one had any discernible Hic1a dense methylation (Fig. 3C).
Figure 3.
Analysis of Hic1 expression and methylation status in tumors. (A) Hic1 immunohistochemistry in a representative tumor (bars, 20 μm), shown as DAB staining (brown), counterstained with hematoxylin (blue). (Tu) Tumor; (V) blood vessel. (B) Schematic for HpaII-based PCR dense methylation analysis of mouse Hic1 promoters showing alternative exons 1a and 1b, and downstream coding exon or mutation. Circles represent number and approximate location of CpGs analyzed in this assay, and arrowheads represent locations of PCR primers. The figure is not drawn to scale. (C) PCR results from HpaII assay for nine Ptch1+/− Hic1+/− and four Ptch1+/− Hic1+/+ tumors. Hic1b wild type (WT) represents the PCR product resulting from primers located in exons 1b and 2. Hic1b Mut represents the PCR product resulting from primers in exons 1b and β-galactosidase. Hic1a represents the PCR product resulting from primers in exon 1a. The minus sign (−) denotes a mock digestion, and the plus sign (+) means that the genomic DNA was digested with HpaII prior to PCR amplification. (D) Hic1 promoter 1b bisulfite sequencing schematic. The nested PCR reaction is allele- specific (Nested PCR product). The region used as a template for bisulfate sequencing is shown (Region sequenced). The figure is not drawn to scale. (E) Bisulfite sequencing data for Ptch1+/− Sample 3 and Ptch1+/− Hic1+/− Sample 2 from B; wild type (WT) is an age-matched C57Bl/6 cerebellum. Each row represents the sequence from a single allele with the 5′ end at the left. Empty circles represent unmethylated cytosine.
Given that only one unmethylated CpG residue will result in failure of PCR amplification in this assay, we performed bisulfite sequencing using a nested, allele-specific approach to exclusively amplify the wild-type Hic1 1b allele as a more sensitive approach to the detection of promoter hypermethylation. We performed this analysis on an age-matched wild-type cerebellum, and Sample 3 from the Ptch1+/− tumors and Sample 2 from the Ptch1+/− Hic1+/− tumors, both of which do not exhibit dense methylation according to the HpaII assay (Fig. 3D; Chen et al. 2003). The added benefit of bisulfite sequencing is that the results are specific for individual clones; hence the data take into consideration the cellular variability within a single tumor. For this reason, eight clones were chosen for each Sample 2 and Sample 3 to be confident in the Hic1-methylation status in tumor cells as the predominant cell type, as opposed to nontumor cells. The wild-type animal displayed the expected mosaic pattern of Hic1b methylation, and both Sample 3 and Sample 2 exhibited markedly increased methylation of the wild-type Hic1b promoter, suggesting that Hic1 is not expressed due to Hic1b promoter hypermethylation (Fig. 3E). These data demonstrate that epigenetic silencing of Hic1 is a potentially important mechanism in medulloblastoma derived from both the Ptch1+/− Hic1+/− and Ptch1+/− backgrounds, and models the methylation status of this gene in human tumors.
Hic1 is expressed during GCP differentiation
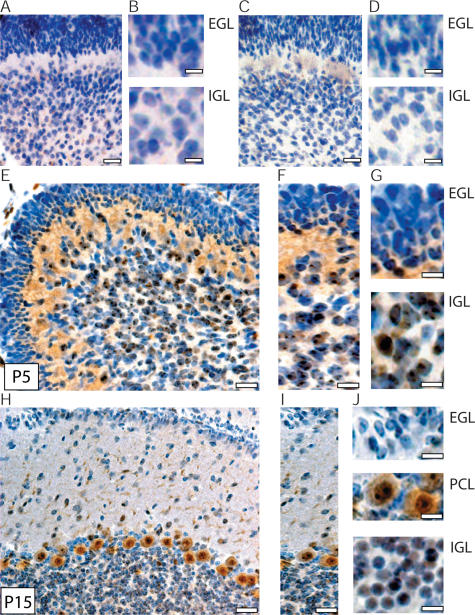
Heterozygous mutations in Hic1 markedly increases the incidence of medulloblastoma in Ptch1 heterozygotes; therefore, we hypothesized that Hic1 might play a role in normal cerebellar differentiation. Cerebellar development in mice proceeds rapidly from postnatal days 1–10 (P1–P10), the period of maximal Hh pathway-dependent proliferation and development of the EGL (Kho et al. 2004). When considering a potential role for Hic1 as a developmentally regulated transcriptional repressor, as well as a tumor suppressor, we examined the expression pattern of Hic1 by immunohistochemistry, specifically highlighting this critical period of cerebellar development. Since Hic1−/− animals die in utero prior to definitive cerebellar development, a genetic control is not possible for Hic1 antibody staining in the cerebellum. However, published studies demonstrate that Hic1−/− embryos have undetectable expression when stained with this Hic1 polyclonal antibody (Chen et al. 2003). For a cerebellar control, sections from a 5-d-old cerebellum were stained either with no primary antibody (Fig. 4A,B) or with a preblocked Hic1 antibody (Fig. 4C,D). Hic1 was absent in the outer EGL but is detectable in cells lining the inner EGL and was strongly positive in the cells of the molecular layer and IGL, as well as in Purkinje cells (Fig. 4E–J; Supplemental Fig. 2). It is uncertain at this time what role Hic1 may play in Purkinje cells, but its strong expression in this cell type implies another role for Hic1 in the cerebellum outside of neuronal development. These data are consistent with induction of Hic1 expression in developing cerebellar granule cells as they exit the cell cycle, lose expression of Nestin (Lendahl et al. 1990; Dahlstrand et al. 1995) and Atonal Homolog 1 (Atoh1) (Helms et al. 2000; Lumpkin et al. 2003), and descend through the molecular layer into the IGL.
Figure 4.
Hic1 expression in cerebellar development. Hic1 immunohistochemistry performed in C57Bl/6 animals aged to P5 and P15. All magnified sections are in the same orientation as original image. (A) Negative control P5 C57Bl/6 cerebellum, no primary antibody (bar, 20 μm). (B) Magnified EGL and IGL (bar, 6 μm). (C) Negative control P5 C57Bl/6 cerebellum stained with preblocked Hic1 antibody (bar, 20 μm). (D) Magnified EGL and IGL (bar, 6 μm). (E) P5 C57Bl/6 cerebellum (bar, 20 μm). (F) Magnified section of P5 cerebellum (bar, 10 μm). (G) Magnified EGL and IGL of P5 cerebellum (bar, 5 μm). (H) P15 C57Bl/6 cerebellum (bar, 30 μm). (I) Magnified section of P15 cerebellum (bar, 15 μm). (J) Magnified EGL, PCL, and IGL of P15 cerebellum (bar, 7 μm). Data are presented as DAB staining (brown), counterstained with hematoxylin (blue).
Hic1 does not directly regulate the Hh pathway in vitro
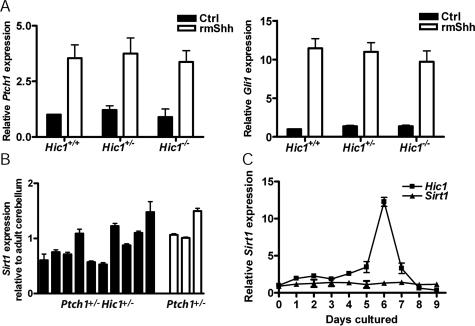
We considered three potential mechanisms by which Hic1 could play a role in the development of medulloblastoma: (1) Hic1 could directly influence the Hh pathway; (2) overexpression of Sirt1, a known Hic1 transcriptional target, could contribute to tumorigenesis (Chen et al. 2005); or (3) loss of Hic1 could cause the overexpression of a novel transcriptional target that could lead to tumorigenesis. To address the first possibility, we cultured mouse embryonic fibroblasts (MEFs) derived from Hic1+/+, Hic1+/−, and Hic1−/− embryos in the presence or absence of Hh ligand. In this experiment, we observed no significant difference in Shh-induced Ptch1 and Gli1 expression in cells lacking Hic1 (Fig. 5A). These data, in addition to our analysis of the Hh pathway in Ptch1+/− Hic1+/− medulloblastoma, suggest that Hic1 does not play a major role in Hh pathway regulation.
Figure 5.
Hh signaling and Sirt1 expression in medulloblastoma. (A) Ptch1 and Gli1 quantitative PCR in MEFs derived from Hic1+/+, Hic1+/−, and Hic1−/− E17.5 embryos. MEFs were either treated with recombinant mouse Shh or a PBS control. Ptch1 and Gli1 transcript levels are graphed relative to untreated Hic1+/+ MEFs. Error bars reflect SEM of three biological replicates. (B) Sirt1 quantitative PCR in 12 Ptch1+/− Hic1+/− and three Ptch1+/− tumors. Sirt1 RNA expression is graphed relative to Sirt1 expression in C57Bl/6 adult cerebellum. Error bars reflect SEM of three technical replicates. (C) Sirt1 quantitative PCR in GCPs cultured for 7 d. Error bars reflect SEM of biological replicates.
Sirt1 expression is a feature of normal and malignant cerebellar cells
SIRT1, a class III histone deacetylase that functions to protect cellular longevity in periods of oxidative stress and DNA damage, is under the direct transcriptional control of HIC1 (Chen et al. 2005). Loss of HIC1 leads to an accumulation of SIRT1, which then partially inactivates p53 by deacetylation (Luo et al. 2001; Vaziri et al. 2001; Chen et al. 2005). This interaction could potentially allow for a greater progression toward tumorigenesis, so we investigated whether this could be an explanation for the increased incidence of medulloblastoma in our mouse model. Sirt1 mRNA expression levels from Ptch1+/− Hic1+/− tumors, Ptch1+/− tumors, and normal age-matched cerebellum were similar (Fig. 5B). Because none of the tumors isolated from this study exhibited Hic1 expression, it was not possible to compare Sirt1 levels between medulloblastomas with and without inactivated Hic1. To quantitatively determine the relationship between Hic1 and Sirt1 in a similar, but nontumorigenic cell type, we used primary GCP cultures, which are well-characterized models of cerebellar progenitor differentiation (Wechsler-Reya and Scott 1999). Over 9 d of culture, Hic1 levels vary dramatically with peak expression at day 6, yet Sirt1 levels remain constant throughout the culture period. This suggests that Hic1 expression does not affect that of Sirt1. Sirt1 immunohistochemistry also showed no significant difference in protein levels in medulloblastoma cells as compared with normal IGL (Supplemental Fig. 3).
The lack of alterations in Sirt1 expression may reflect its importance in the CNS. Sirt1 has been implicated in the survival of neurons, and caloric restriction, which leads to an increase in Sirt1 activity, protects against neurodegenerative pathology in mouse models for Alzheimer’s and Parkinson’s diseases (Duan and Mattson 1999; Zhu et al. 1999; Patel et al. 2005). Sirt1 is expressed in cerebellar granule cells and has been linked to granule cell survival in periods of oxidative stress (Brunet et al. 2004). Given the connection between Sirt1 and organismal and cellular longevity, it is likely that Sirt1 is required in post-mitotic granule cells of the cerebellum to prevent apoptosis and therefore is not a mechanism for the development of medulloblastoma. These data also imply that Hic1 may not regulate Sirt1 in the CNS.
ATOH1 is a transcriptional target of HIC1
HIC1 functions as a transcriptional repressor (Deltour et al. 1999, 2002; Pinte et al. 2004b); consequently, we searched for transcriptional targets relevant to tumor suppression and neural differentiation using an array-based approach. We used a HIC1 adenovirus expression system in the D425 human medulloblastoma cell line to achieve high-level, transient overexpression (He et al. 1998). D425 was chosen for its Hh pathway expression (Fig. 6A) and HIC1 promoter hypermethylation (Lindsey et al. 2005). Cells were transduced with HIC1-expressing adenovirus or an adenovirus expressing a β-galactosidase control. Microarray analysis was performed on RNA harvested at 12 h and 24 h post-transduction (data not shown; Supplemental Table 1). A scatterplot was generated from the 24-h time point with data restricted to P-values ≤ 0.001, resulting in 932 highly significant gene changes among the 44,000 oligos represented on this array (data not shown). Consistent with the previously discussed Sirt1 data, SIRT1 expression was not significantly altered by exogenous expression of HIC1 in D425 according to the microarray analysis (Supplemental Table 1). We focused on one gene likely to explain, at least in part, the phenotype exhibited by the Ptch1+/− Hic1+/− animals. The human homolog of Atonal, a basic helix–loop–helix transcription factor is highly expressed in both GCPs and medulloblastoma, and its expression is significantly reduced by the exogenous expression of HIC1. The relationship between HIC1 and ATOH1 in human sporadic medulloblastoma is further demonstrated by expanding the cell lines used to include the medulloblastoma cell lines D283, D341, and Daoy, and the supratentorial PNET cell line, PFSK (Fig. 6A).
Figure 6.
Hic1 is a direct transcriptional repressor of Atoh1. (A) Semiquantitative PCR showing expression of Hedgehog pathway components; ATOH1 and HIC1 in human medulloblastoma cell lines D283, D341, D425 and Daoy; and human supratentorial PNET cell line PFSK. (B) HIC1 HpaII assay performed on human cell lines from A. HIC1b and HIC1a represent PCR products resulting from primers located in either exons 1b and 2, or exons 1a and 2, respectively. The minus sign (−) denotes a mock digestion, and the plus sign (+) means the genomic DNA was digested with HpaII prior to PCR amplification. (C) Semiquantitative PCR to validate ATOH1 microarray data in human medulloblastoma and supratentorial PNET cell lines. The minus sign (−) refers to transduction with a β-galactosidase-expressing adenovirus, and the plus sign (+) refers to transduction with a HIC1-expressing adenovirus. RNA was harvested 48 h post-transduction. (D) Schematic of ChIP experiment showing downstream Atoh1 enhancer region that contains sequence similar to the human HIC1-binding sequence. Solid arrows refer to PCR primer Pair 1, and dotted arrows refer to PCR primer Pair 2. The figure is not drawn to scale. (E) ChIP data from C57Bl/6 P7 and P14 cerebella immunoprecipitated with a Hic1 antibody and the denoted genes amplified by PCR. Representative PCR analyses done on the Hic1-immunoprecipitated (+), mock (−), and a 1:100 dilution of nonimmunoprecipitated (Input) DNA from both the P7 and P14 cerebella using β Actin (Actb) primers to show loading control and specificity of the ChIP.
All five cell lines express members of the GLI family of transcription factors, as well as PTCH1, consistent with Hh pathway activation. HIC1 is not expressed by any of the medulloblastoma cell lines, although it is expressed by the supratentorial PNET cell line. Interestingly, HIC1 expression correlates with a lack of ATOH1 expression. All of the medulloblastoma cell lines express ATOH1, although the levels are variable, and the supratentorial PNET does not. HIC1 expression in the cell lines also correlates with the level of HIC1b hypermethylation, with the only cell line expressing HIC1 (PFSK) also being the only one with no discernable dense HIC1 hypermethylation as determined by the HpaII assay (Fig. 6B). Similar to the Hic1+/− Ptch1+/− tumors, the intensity of the HIC1b band for Daoy is limited, but it still correlates with a complete lack of HIC1 expression as determined by quantitative PCR (data not shown). The D425 microarray data are verified by repeating the adenovirus treatments with all five cell lines. The exogenous expression of HIC1 in the human medulloblastoma cell lines causes a dramatic reduction in ATOH1 expression (Fig. 6C). If the mammalian Atonal homolog plays a critical role in the initiation and maintenance of medulloblastoma, this would constitute a mechanism to explain a potential interaction between Hh signaling and HIC1 function in pathogenesis of this tumor and could have implications for the role of HIC1 in normal cerebellar development. We hypothesized that Atoh1 was one candidate transcriptional target of Hic1 likely to explain the effect of Hic1 mutation seen in Ptch1 mutant mice. We based this contention on evidence demonstrating that (1) ATOH1 is highly expressed in human medulloblastoma (Lee et al. 2003); (2) Atoh1 is the earliest known marker of EGL and is required for the development of this progenitor population (Ben-Arie et al. 1997, 2000; Jensen et al. 2004); (3) Atoh1 is highly expressed in GCPs (Ben-Arie et al. 1997, 2000; Jensen et al. 2004), but its expression is extinguished following differentiation and descent into the IGL (Lumpkin et al. 2003); and (4) ATOH1 is a likely target of Hh signaling in cerebellar development and medulloblastoma (Kenney and Rowitch 2000; Berman et al. 2002; Kenney et al. 2003).
Transcription of ATOH1 is regulated by a 1.7-kb downstream enhancer region that faithfully reports ATOH1 expression in transgenic mice when used to drive expression of GFP (Helms et al. 2000). Analysis of this enhancer region reveals a potential Hic1-binding sequence very similar to the one described for human HIC1, including the critical spacing between the TGCC(A/C) flanking sequences allowing for concatamerization (Fig. 6D; Pinte et al. 2004b; Chen et al. 2005). We analyzed this enhancer region using ChIP on cerebellar cells purified from postnatal mice at different developmental stages. Cerebellum from a 14-d-old mouse shows that Hic1 binds the Atoh1 enhancer region in physiological conditions, whereas there was no detectable interaction at an earlier time point (Fig. 6E).
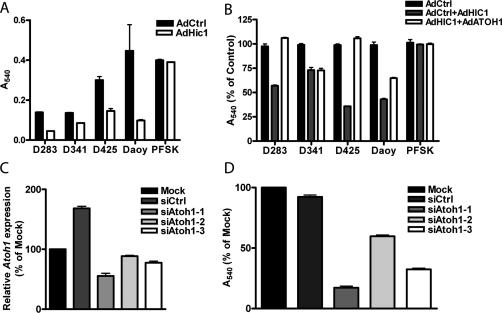
Treatment of the human medulloblastoma cell lines over 6 d with adenoviruses expressing either HIC1 or β-galactosidase and challenging the proliferation potential of the cell lines by serially replating resulted in a reduction in cell viability but not in the supratentorial PNET (Fig. 7A). To elucidate whether ATOH1 is the target of HIC1 that results in the reduced cell viability, an adenovirus was generated to exogenously express ATOH1 to be used as a potential rescue. To determine whether ATOH1 could rescue the HIC1-mediated effect, the cell lines were treated with adenoviruses expressing β-galactosidase, HIC1, and ATOH1 in combination as noted in Figure 7B and serially replated every 48 h for 6 d. Expression of ATOH1 in concert with HIC1 fully rescues the HIC1-mediated effect on growth in D283 and D425 cells (Fig. 7B); however, ATOH1 expression does not fully rescue D341 or Daoy cells, the two medulloblastoma cell lines tested with the least amount of endogenous ATOH1 qualitatively. Given the dramatic reduction in cell viability displayed by Daoy cells treated with HIC1 and the incomplete rescue by exogenous expression of ATOH1, it is very likely that there are additional targets for HIC1 in medulloblastoma and possibly for cerebellar development.
Figure 7.
ATOH1 is functionally important in medulloblastoma. (A) MTT analysis of human medulloblastoma cell lines D283, D341, D425, and Daoy, and human supratentorial PNET cell line PFSK either treated with adenovirus expressing β-galactosidase (AdCtrl) or HIC1 (AdHic1) for 6 d with replating every 48 h. Absorbance was measured at 540 nm (A540). Error bars reflect SEM of four biological replicates. (B) MTT analysis of human cell lines treated with adenovirus expressing β-galactosidase (AdCtrl), β-galactosidase and HIC1 (AdCtrl + AdHIC1), or HIC1 and ATOH1 (AdHIC1 + AdATOH1) for 6 d with replating every 48 h. Absorbance was measured at 540 nm (A540). Error bars reflect SEM of four biological replicates. (C) Representative quantitative PCR demonstrating success of siRNA knockdown of Atoh1 in B837TX1-2, a cell line derived from a Ptch1+/− Hic1+/− tumor. Data are graphed relative to the mock-transfected B837TX1-2 cell line. Cells were either transfected with a nontargeting siRNA (siCtrl) or Atoh1-targeting siRNAs (siAtoh1-1–siAtoh1-3). Error bars reflect SEM of three technical replicates. (D) MTT assay performed on B837TX1-2 cell line either mock-transfected (Mock), transfected with a nontargeting siRNA (siCtrl), or transfected with Atoh1-targeted siRNAs (siAtoh1-1–siAtoh1-3). Data are graphed relative to the mock-transfected cell line. Absorbance measured at 540 nm (A540). Error bars reflect SEM of three biological replicates.
To further determine the functional importance of ATOH1 in medulloblastoma, we used an siRNA-based approach to inhibit expression of Atoh1 in B837TX1-2, a cell line derived from a Ptch1+/− Hic1+/− tumor. Three unique siRNAs were used to specifically target Atoh1 with varying success. A twofold reduction in Atoh1 expression corresponded to a sixfold reduction in cell viability, whereas less dramatic decreases in cell viability were observed with the less efficacious siRNAs (Fig. 7C,D). These data suggest that the decrease in cell viability is specific to the success of the siRNA-mediated knockdown, as opposed to siRNA off-target effects. These data establish Atoh1 as necessary for growth of medulloblastoma cells arising in animals deficient for Ptch1 and Hic1.
Hic1 represses Atoh1 expression in GCPs
To further characterize the relationship between Hh signaling, Atoh1 and Hic1 in neural development, we used murine primary GCP cultures, which are well-characterized models of cerebellar progenitor differentiation (Wechsler-Reya and Scott 1999). GCPs were isolated from wild-type P7–P8 animals, a time point when proliferation and Hh pathway activity are maximal (Kho et al. 2004), and gene expression patterns were analyzed over 9 d in culture. Hic1 expression increases from Day 0, reaching a maximum at Day 6 of culture. This expression pattern coincided with loss of Atoh1 expression as well as the well-established time frame of GCP differentiation in this model (Fig. 8A,B; Gazit et al. 2004). It has been shown previously that when GCPs are cultured in the presence of Shh, there is a four- to fivefold increase in proliferative cells as determined by BrdU incorporation. The BrdU-positive cells colabel with Atoh1, and not with cells that are positive for markers of differentiated neurons (Zic1) or cells of glial lineage (Gfap and NG2), suggesting that the only cells capable of proliferating in vitro in response to Shh are Atoh1-positive immature GCPs. These data indicate that treatment of cultured GCPs with the Hh ligand Sonic hedgehog (Shh) prevents spontaneous differentiation and maintains expression of Atoh1 in addition to other GCP-specific genes (Kenney and Rowitch 2000; Kenney et al. 2003). We also demonstrate that activation of Hh signaling in GCPs maintains Atoh1 expression levels (Fig. 8C). However, adenoviral overexpression of Hic1 overcomes this effect and results in rapid inhibition of Atoh1 expression, which mimics the profile seen during spontaneous GCP differentiation (Fig. 8C). These data demonstrate that Hic1-mediated suppression of Atoh1 expression is dominant to Shh-mediated Atoh1 expression.
Figure 8.
Hic1 is epistatic to Atoh1. (A) Quantitative PCR tracking Hic1 expression in cultured GCPs for 9 d. Gene expression is relative to fresh GCPs. Error bars reflect SEM of three biological replicates. (B) Quantitative PCR tracking Atoh1 and Hic1 expression in cultured GCPs restricted to the first 4 d of culture. Gene expression is relative to fresh GCPs. Error bars reflect SEM of three biological replicates. (C) Quantitative PCR tracking Atoh1 expression in GCPs cultured for 4 d in the presence or absence (Untreated) of recombinant mouse Shh (R&D Systems). On Day 1, cells were either transduced with a Hic1-expressing adenovirus (Hh + AdHic1) or a β-galactosidase-expressing adenovirus (Hh + AdControl), or no adenovirus (rmShh). Gene expression is relative to fresh GCPs. Error bars reflect SEM of three biological replicates.
Discussion
Several groups have shown that LOH of chromosome 17p13.3 is associated with poor prognosis in medulloblastoma patients and that HIC1, which maps to this site, is frequently hypermethylated in these malignant pediatric brain tumors (Batra et al. 1995; Steichen-Gersdorf et al. 1997; Rood et al. 2002; Lindsey et al. 2005). In this study, we provide evidence that epigenetic silencing of Hic1 is an important event in medulloblastoma pathogenesis using a mouse model in which the tumorigenic effect of Ptch1 mutation in the cerebellum can be markedly and specifically enhanced by loss of Hic1 function. Silencing of Hic1 in medulloblastomas from both the Ptch1+/− and Ptch1+/− Hic1+/− backgrounds has two important implications. First, it suggests that the increase in medulloblastoma incidence seen in compound heterozygotes results from more efficient Hic1 gene silencing when methylation of only one allele is needed to render the tumor Hic1-null. Secondly, this finding suggests that frequency of HIC1 gene silencing in human medulloblastoma, the majority of which are PTCH wild type, is reflective of a critical role for this gene as a tumor suppressor in the developing cerebellum.
The dynamic regulation of Hic1 expression during the descent of the GCPs into the IGL suggests that its importance is intimately linked to the process of differentiation. It has been suggested that Hic1 expression levels in normal neural stem cells and normal adult brain are similar to those seen in human medulloblastoma, thus questioning its importance as a tumor suppressor in medulloblastoma (Zwalik et al. 2006). Based on our data in cerebellar development and implications from recent studies in embryonic stem cells (Ohm et al. 2007), we postulate that lack of Hic1 expression is a characteristic feature of progenitor populations such as GCPs and neural stem cells until differentiation signals induce its expression. Our data suggest that the tumor suppressor function of Hic1 in the cerebellum is most important during the process of differentiation in cells such as GCPs. Several studies demonstrated that partial methylation of HIC1 is a frequent event in the adult brain (Rood et al. 2002; Waha et al. 2003; Lindsey et al. 2005), suggesting that the tumor suppressor function of HIC1 may not be needed once terminal differentiation is complete.
Given that our data link Hic1 expression to differentiation of GCPs, Hic1-mediated transcriptional repression of progenitor-specific genes is an attractive hypothesis to explain its function as a tumor suppressor. Although it is likely that additional important transcriptional targets of Hic1 are involved in GCP differentiation, Hic1-mediated repression of Atoh1 expression provides a potential explanation for the importance of Hic1 as a tumor suppressor and for its interaction with the Hh pathway and demonstrate that this is a critical mechanism in the tumorigenicity of medulloblastoma. These findings illustrate the potential importance of epigenetic events in medulloblastoma pathogenesis and demonstrate a clear functional role for Hic1 in neural tumor suppression and in normal cerebellar development. This conclusion may also explain a long-standing paradox in cerebellar development. Although GCPs require Purkinje cell-derived Shh for their growth, post-mitotic GCPs are exposed to the highest levels of Shh as they migrate past the Purkinje cell layer to take residence in the IGL (Dahmane and Ruiz-i-Altaba 1999). Our data demonstrate that Hic1-mediated repression of Atoh1 expression is dominant to the induction of Atoh1 by Shh. In other words, Hic1 can act downstream from Shh to repress Atoh1 expression; thus its expression may serve to render GCPs insensitive to Shh and therefore permit terminal differentiation despite exposure to high levels of Shh during their migration.
In colon cancer there is a clear multistep basis of tumorigenesis, in which multiple genetic deficits must be accumulated, often in a specific order, before leading to malignancy (Fearon and Vogelstein 1990). One interesting hallmark of Ptch1+/− mice is the accumulation of small clusters of EGL-like cells that persist on the surface of the cerebellum regardless of whether the animals develop medulloblastoma or not (Goodrich et al. 1997; Oliver et al. 2005). These EGL rests are most likely the precursor lesion for medulloblastoma, but additional genetic deficits must occur before these cells are transformed (Oliver et al. 2005). This could be the basis of one potential explanation for the increased incidence of medulloblastoma in Ptch1+/− Hic1+/− compound heterozygotes.
Interest in the connection between developmental processes and cancer has led to speculation that tumors can become “addicted” to lineage-specific transcription factors. These genes maintain a crucial role in normal development, and their persistent expression in tumors that arise from the associated lineage is required for tumor viability (Garraway and Sellers 2006). This so-called “lineage addiction” is exemplified by MITF, a transcription factor expressed in, and required for, the maintenance of melanocytes. Overexpression of MITF is a common event in melanomas, but its tumorigenicity is only revealed in the setting of aberrant MAPK pathway activation (Garraway et al. 2005). This is strikingly similar to what we observed in our Ptch1+/− Hic1+/− mouse model of medulloblastoma. It is possible that Atoh1 is not oncogenic in its own right, but instead as it directs cerebellar granule cell lineage survival during development and protects the proliferative potential and survival of medulloblastoma, and thus may function as a lineage-survival oncogene in medulloblastoma.
It is also important to note that it is very likely that Atoh1 expression is not only regulated by Hic1, but can also be influenced by both the Hedgehog pathway and also the Notch signaling pathway (Gazit et al. 2004). The potential influence by the Hedgehog pathway is based on (1) high-level expression of Atoh1 in GCPs of the EGL, where reception of Hh signaling is maximal; (2) induction of high-level Atoh1 expression in cultured GCPs in response to treatment with Shh ligand (Kenney and Rowitch 2000); (3) Atoh1 expression is down-regulated by Hh pathway blockade in mouse medulloblastoma cells (Berman et al. 2002); and (4) evidence of down-regulation of Atoh1 expression in medulloblastoma in vivo in mice treated with a novel, orally active Hh pathway antagonist (Romer et al. 2004).
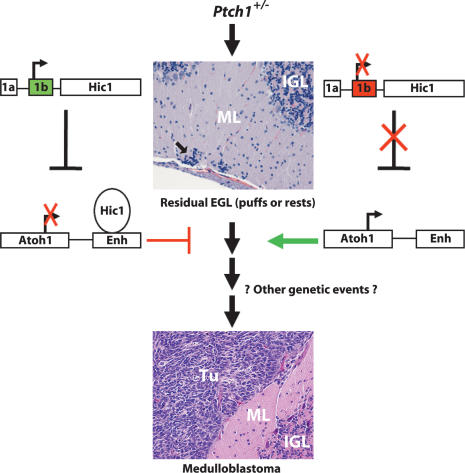
In our mouse model, loss of Ptch1 leads to the persistence of primitive GCP-like precursors, which are an ideal substrate for malignant transformation. In both the Ptch1+/− and Ptch1+/− Hic1+/− tumors, Hic1 is hypermethylated, indicating that epigenetic silencing of Hic1 may be one of the next deficiencies accumulated in a Ptch1+/−-induced precursor lesion and may contribute to the promotion or progression of malignancy. This model is depicted in Figure 9 and highlights how epigenetic events can interact with aberrant developmental signaling and other potential genetic events to promote tumorigenesis. Hedgehog pathway mutations can only be attributed to <25% of sporadic medulloblastomas in humans; however, loss of 17p is a frequent event. Another gene that localizes to this chromosomal arm at 17p13.2, REN, has also been implicated in medulloblastoma (Di Marcotullio et al. 2004). REN interacts with the Hh pathway by regulating the nuclear trafficking of Gli1; therefore, loss of REN could potentially lead to unrestricted Hh pathway expression, similar to loss of heterozygosity of Ptch1 in our mouse model. The potential consequences of deletion of 17p in humans are illustrated by the increased incidence of medulloblastoma in this Ptch1+/− Hic1+/− mouse model. Pharmaceutical approaches to reverse DNA promoter methylation are now used clinically in the treatment of myelodysplastic syndrome (Lubbert et al. 2001; Kuendgen et al. 2007); it is therefore possible that therapies targeted at epigenetic events may be of use in medulloblastoma.
Figure 9.
A model for tumorigenesis. (Arrow) Residual EGL occurs in the setting of Ptch1 heterozygosity. Unmethylated Hic1 (green promoter 1b) allows for Hic1 expression. Hic1 binds the Atoh1 enhancer leading to transcriptional repression and differentiation, which depletes the presumed precursor lesion. If Hic1 is hypermethylated (red promoter 1b), expression is silenced leading to uncontrolled Atoh1 expression and tumorigenesis is promoted as demonstrated by a representative image of a Ptch1+/− Hic1+/− tumor. (IGL) Internal granule cell layer; (ML) molecular layer; (Tu) medulloblastoma.
Materials and methods
Animals
Experiments were conducted in accordance with protocols approved by the Johns Hopkins Institutional Animal Care and Use Committee. Ptch1+/− mice (Goodrich et al. 1997) were crossed to Hic1+/− mice (Carter et al. 2000) on a C57Bl/6 background. A total of 180 mice were generated (47 Ptch1+/+ Hic1+/+, 42 Ptch1+/− Hic1+/+, 46 Ptch1+/+ Hic1+/−, and 45 Ptch1+/− Hic1+/−) and allowed to age to 18 mo. Medulloblastoma was detected in affected mice by daily observation for characteristic circling, hemineglect, and ataxia. In Ptch1+/− Hic1+/− weanlings, medulloblastoma presented as hydrocephalus, which was not seen in Ptch1+/− animals. Mice were sacrificed and the posterior fossa gently dissected in order to detect evidence of tumor. When tumors were not evident, entire cerebella were fixed and sectioned to look for histological evidence of medulloblastoma. Graphpad Prism 4 software was used to perform log-rank Kaplan Meier analysis on medulloblastoma incidence. All non-medulloblastoma-related deaths were entered as censored data.
RT–PCR
RNA was isolated either using Trizol (Invitrogen) or the RNeasy Mini Kit (Qiagen). Reverse transcription was preceded by DNase I treatment. Quantitative PCR was performed using the Bio-Rad SYBR green system and thermal cycler. All quantitative calculations were performed using the 2−ΔΔCt method using β Actin as a reference gene. β Actin transcription values did not vary significantly between samples for individual experiments. All primer sequences are available upon request.
Cell biology
B837TX1-(2–4) were created by manually mincing a medulloblastoma allograft followed by incubation in Dispase (BD Biosciences) for 15–20 min at 37°C. Cells were washed with PBS. Resuspended cells were allowed to settle for 5 min, and only the supernatant was used in culture. B837TX1-(2–4), D283, D341, D425, Daoy, and PFSK were maintained in Advanced RPMI (Invitrogen) supplemented with 1% FBS, 10 mM HEPES, and L-glutamine.
GCPs
GCPs were isolated and maintained as described (Bar et al. 2007). For the Hh–Hic1 epistasis experiment, 2 × 106 GCPs were plated per well in a poly-L-lysine-coated six-well plate with media supplemented with recombinant mouse Shh (R&D Systems, 464-SH-025). Twenty-four hours post-plating, adenoviruses expressing either HIC1 or β-galactosidase were added to the wells. RNA was harvested on each day, and quantitative PCR was performed.
MEFs
MEFs from embryonic day 17.5 (E17.5) mouse embryos were prepared using a standard protocol. We seeded early passage (typically p3 and p4) MEFs at a density of 2 × 106 per well in a six-well plate. The cells were maintained in DMEM (Invitrogen) supplemented with 10% BCS. When the cells were 100% confluent, either PBS or rmShh (R&D Systems, 464-SH-025) was added. Cells were cultured for an additional 24–48 h, and then RNA was harvested.
Ptch1+/− Hic1+/− cell line cyclopamine treatment
Cells (2 × 104) were plated per well in 24-well plates. To each well was added either 1 μM, 2.5 μM, or 5 μM cyclopamine, or an equivalent volume of vehicle control (EtOH). At 48 h, cells were washed with PBS and replated in 12-well plates in the same concentration of drug. At 96 h, cells were washed with PBS and replated in six-well plates with the same concentration of drug. At 168 h, cells were counted and RNA was extracted.
Adenovirus in human cells
ATOH1 adenovirus was generated using the Adeno-X ViraTrak DsRed-Express Expression System 2 (Clontech, 632516). The adenovirus was designed to express a Kozak sequence in addition to ATOH1 bases 1–1065 (NM_005172). Adenovirus was propagated in AD-293 cells (Stratagene, 240085) and purified using VivaPure AdenoPack100 (Vivascience, VS-AVPQ101). Expression in transduced cells was confirmed by semiquantitative PCR using the same ATOH1 primers as used to measure endogenous expression. Expression of the HIC1-expressing adenovirus was verified by immunoblotting.
For Figure 7A, cells were plated at 2 × 104 per well in a 24-well plate. Twenty-four hours post-plating, cells were either transduced with β-galactosidase-expressing adenovirus or HIC1-expressing adenovirus such that 75% of cells were expressing adenovirus at 48 h later, as determined by fluorescence affected by expressed adenoviral DNA. Forty-eight hours post-transduction, cells were expanded to 12-well plates. Ninety-six hours post-transduction, cells were expanded to six-well plates. At 144 h, an MTT assay was performed.
For Figure 7B, cells were plated similar to previous. Twenty-four hours post-plating, cells were transduced similar to previous, with the exception that half volumes of individual virus were used when cells were transduced by multiple adenoviruses to prevent adenoviral toxicity. Cultures were handled and assayed similar to previous.
Transfection
Cells were plated in a six-well plate. At 60% confluency, cells were transfected with 25 nM Atoh1 siRNA (Dharmacon, D-058607-01) or siCONTROL (Dharmacon, D-001210-01) using TransIT-TKO (Mirus) according to the manufacturer’s instructions. Twenty-four hours post-transfection, cells were transfected again. At 30 h, the cells were split 1:2. At 48 h, RNA was isolated and an MTT assay was performed according to standard protocols.
Immunohistochemistry/immunofluorescence
Hic1 immunohistochemistry was performed as described (Chen et al. 2003). For preblocked Hic1 antibody staining, Hic1 antibody was incubated overnight with a 1:10 concentration of Hic1 peptide matching the epitope recognition sequence. Vectastain ABC kits (Vector Laboratories) were used for all other immunohistochemistry. The Nestin antibody (Chemicon, MAB353) was processed using the DAKO ARK kit prior to proceeding with the Vectastain ABC reagent. The following antibodies were used for immunohistochemistry: GFAP (DAKO, Z0334), Synaptophysin (DAKO, A0010), Tuj1 (Chemicon, MAB5544), and Sir2 (Upstate Biotechnology, 07-131).
ChIP
ChIP was performed as described (McGarvey et al. 2006). Cells were isolated from C57Bl/6 P7 and P14 cerebella. HIC1 antibody was used at a concentration of 1:500. All primer sequences are available upon request.
Microarray
Total RNA was harvested from log-phase cells using Trizol (Invitrogen) followed by the RNA clean-up protocol from the RNeasy Mini kit (Qiagen), both according to the manufacturers’ instructions. RNA was analyzed using an Agilent DNA microarray system. More details are included in the Supplemental Material.
Acknowledgments
We thank Eli Bar (Pathology Department, Johns Hopkins University), Leslie Meszler and Lillian Dasko-Vincent (Cell Imaging Core Facility, The Sidney Kimmel Comprehensive Cancer Center), and Wayne Yu (The Sidney Kimmel Comprehensive Cancer Center Microarray Core Facility) for their assistance with this work. We also thank Infinity Pharmaceuticals for the gift of cyclopamine. This work was supported by The Children’s Brain Tumor Foundation House of Hope Award, The Sidney Kimmel Cancer Research Foundation, and the NIH/NINDS (R01N50540).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1640908.
References
- Bar E.E., Chaudhry A., Farah M.H., Eberhart C.G. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am. J. Pathol. 2007;170:347–355. doi: 10.2353/ajpath.2007.060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra S.K., McLendon R.E., Koo J.S., Castelino-Prabhu S., Fuchs H.E., Kirscher J.P., Friedman H.S., Bigner D.D., Bigner S.H. Prognostic implications of chromosome 17p deletions in human medulloblastomas. J. Neurooncol. 1995;24:39–45. doi: 10.1007/BF01052657. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N., Bellen H.J., Armstrong D.L., McCall A.E., Gordadze P.R., Guo Q., Matzuk M.M., Zoghbi H.Y. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N., Hassan B.A., Bermingham N.A., Malicki D.M., Armstrong D., Matzuk M., Bellen H.J., Zoghbi H.Y. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Berman D.M., Karhadkar S.S., Hallahan A.R., Pritchard J.I., Eberhart C.G., Watkins D.N., Chen J.K., Cooper M.K., Taipale J., Olson J.M., et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deactylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Carter M.G., Johns M.A., Zeng X., Zhou L., Zink M.C., Mankowski J.L., Donovan D.M., Baylin S.B. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum. Mol. Genet. 2000;9:413–419. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- Chen W.Y., Zeng X., Carter M.G., Morrell C.N., Chiu Yen R.W., Esteller M., Watkins D.N., Herman J.G., Mankowski J.L., Baylin S.B. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat. Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- Chen W.Y., Wang D.H., Yen R.C., Luo J., Gu W., Baylin S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Dahlstrand J., Lardelli M., Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res. 1995;84:109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- Dahmane N., Ruiz-i-Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Deltour S., Guerardel C., Leprince D. Recruitment of SMRT/N-CoR–mSin3A–HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: The case of HIC-1 and γFBP-B. Proc. Natl. Acad. Sci. 1999;96:14831–14836. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltour S., Pinte S., Guerardel C., Wasylyk B., Leprince D. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 2002;22:4890–4901. doi: 10.1128/MCB.22.13.4890-4901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., De Smaele E., Argenti B., Mincione C., Zazzeroni F., Gallo R., Masuelli L., Napolitano M., Maroder M., et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc. Natl. Acad. Sci. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., Mattson M.P. Dietary restriction and 2-deoxyglucose adinistration improve behavioural outcome and reduce degeneration of dopaminerigc neurons in models of Parkinson’s disease. J. Neurosci. Res. 1999;57:195–206. doi: 10.1002/(SICI)1097-4547(19990715)57:2<195::AID-JNR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Eberhart C.G. Medulloblastoma in mice lacking p53 and PARP: All roads lead to Gli. Am. J. Pathol. 2003;162:7–10. doi: 10.1016/S0002-9440(10)63792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E.R., Vogelstein B. A genetic model of colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Sellers W.R. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Weir B.A., Zhao X., Widlund H., Beroukhim R., Berger A., Rimm D., Rubin M.A., Fisher D.E., Meyerson M.L., et al. ‘Lineage addiction’ in human cancer: Lessons from integrated genomics. Cold Spring Harb. Symp. Quant. Biol. 2005;70:25–34. doi: 10.1101/sqb.2005.70.016. [DOI] [PubMed] [Google Scholar]
- Gazit R., Krizhanovsky V., Ben-Arie N. Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development. 2004;131:903–913. doi: 10.1242/dev.00982. [DOI] [PubMed] [Google Scholar]
- Goodrich L.V., Milenkovic L., Higgins K.M., Scott M.P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- He T.-C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms A.W., Abney A.L., Ben-Arie N., Zoghbi H.Y., Johnson J.E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hooper J.E., Scott M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Ingham P.W., McMahon A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes & Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jensen P., Smeyne R., Goldwitz D. Analysis of cerebellar development in math1 null embryos and chimeras. J. Neurosci. 2004;24:2202–2211. doi: 10.1523/JNEUROSCI.3427-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.M., Rowitch D.H. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.M., Cole M.D., Rowitch D.H. Nmyc upregulation by Sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kho A.T., Zhao Q., Cai Z., Butte A.J., Kim J.Y., Pomeroy S.L., Rowitch D.H., Kohane I.S. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes & Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Stephen D., Joyner A., Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- Kuendgen A., Graf T., Zohren F., Hildebrandt B., Hunerliturkoglu A., Gattermann N., Haas R., Kobbe G. Induction of complete remission in a patient with acute myeloid leukemia refractory to high dose chemotherapy through treatment with 5-azacytidine. Leuk. Res. 2007;31:407–409. doi: 10.1016/j.leukres.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Lee Y., Miller H.L., Jensen P., Hernan R., Connelly M., Wetmore C., Zindy F., Roussel M.F., Curran T., Gilbertson R.J., et al. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- Lee Y., Kawagoe R., Sasai K., Li Y., Russell H.R., Curran T., McKinnon P.J. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- Lendahl U., Zimmerman L.B., McKay R.D. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Lindsey J.C., Anderton J.A., Lusher M.E., Clifford S.C. Epigenetic events in medulloblastoma development. Neurosurg. Focus. 2005;19:E10. doi: 10.3171/foc.2005.19.5.11. [DOI] [PubMed] [Google Scholar]
- Lubbert M., Wijermans P., Kunzmann R., Verhoef G., Bosly A., Ravoet C., Andre M., Ferrant A. Cytogenetic responses in high risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5-aza-2′-deoxyazacytidine. Br. J. Haematol. 2001;114:349–357. doi: 10.1046/j.1365-2141.2001.02933.x. [DOI] [PubMed] [Google Scholar]
- Lumpkin E.A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., Doetzlhofer A., White P., Groves A., Segil N., et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Brain Res. Gene Expr. Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- McGarvey K.M., Fahrner J.A., Greene E., Martens J., Jenuwien T., Baylin S.B. Silenced tumor suppressor genes reactivated by DNA demethylation do not fully return to a euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Ohm J.E., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., et al. A stem cell-like chromatic pattern may predispose tumor suppressor genes to DNA methylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T.G., Read T.A., Kessler J.D., Mehmeti A., Wells J.F., Huynh T.T., Lin S.M., Wechsler-Reya R.J. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- Packer R.J. Primary central nervous system tumors in children. Curr. Treat. Options Neurol. 1999;1:395–408. doi: 10.1007/s11940-996-0003-0. [DOI] [PubMed] [Google Scholar]
- Patel N.V., Gordon M.N., Connor K.E., Good R.A., Engelman R.W., Mason J., Morgan T.E., Finch C.E. Caloric restriction attenuates Aβ-deposition in Alzheimer transgenic models. Neurobiol. Aging. 2005;26:995–1000. doi: 10.1016/j.neurobiolaging.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Pietsch T., Taylor M.D., Rutka J.T. Molecular pathogenesis of childhood brain tumors. J. Neurooncol. 2004;70:203–215. doi: 10.1007/s11060-004-2750-7. [DOI] [PubMed] [Google Scholar]
- Pinte S., Guerardel C., Deltour-Balerdi S., Godwin A.K., Leprince D. Identification of a second G-C-rich promoter conserved in the human, murine and rat tumor suppressor genes HIC1. Oncogene. 2004a;23:4023–4031. doi: 10.1038/sj.onc.1207504. [DOI] [PubMed] [Google Scholar]
- Pinte S., Stankovic-Valentin N., Deltour S., Rood B.R., Guerardel C., Leprince D. The tumor suppressor gene HIC1 (Hypermethylated in Cancer 1) is a sequence-specific transcriptional repressor: Definition of its consensus binding sequence and analysis of its DNA binding and repressive properties. J. Biol. Chem. 2004b;279:38313–38324. doi: 10.1074/jbc.M401610200. [DOI] [PubMed] [Google Scholar]
- Romer J.T., Kimura H., Magdaleno S., Sasai K., Fuller C., Baines H., Connelly M., Stewart C.F., Gould S., Rubin L.L., et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/−p53−/− mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Rood B.R., Zhang H., Weitman D.M., Cogen P.H. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- Steichen-Gersdorf E., Baumgartner M., Kreczy A., Maier H., Fink F.M. Deletion mapping on chromosome 17p in medulloblastoma. Br. J. Cancer. 1997;76:1284–1287. doi: 10.1038/bjc.1997.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Chen J.K., Cooper M.K., Wang B., Mann R.K., Milenkovic L., Scott M.P., Beachy P.A. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Taipale J., Cooper M.K., Maiti T., Beachy P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Waha A., Koch A., Meyer-Puttlitz B., Weggen S., Sorensen N., Tonn J.C., Albrecht S., Goodyer C.G., Berthold F., Wiestler O.D., et al. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J. Neuropathol. Exp. Neurol. 2003;62:1192–1201. doi: 10.1093/jnen/62.11.1192. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R.J., Scott M.P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R., Scott M.P. The developmental biology of brain tumors. Annu. Rev. Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- Wetmore C., Eberhart D.E., Curran T. The normal patched allele is expressed in medulloblastomas from mice with heterozygous germ-line mutation of patched. Cancer Res. 2000;60:2239–2246. [PubMed] [Google Scholar]
- Zhu H., Guo Q., Mattson M.P. Dietary restrictions protects hippocampal neurons against the death-promoting action of a presenilin-1 muation. Brain Res. 1999;842:224–229. doi: 10.1016/s0006-8993(99)01827-2. [DOI] [PubMed] [Google Scholar]
- Zurawel R.H., Allen C., Wechsler-Reya R., Scott M.P., Raffel C. Evidence that haploinsufficiency of Ptch leads to medulloblastoma in mice. Genes Chromosomes Cancer. 2000;28:77–81. [PubMed] [Google Scholar]
- Zwalik I., Zakrzeweska M., Witusik M., Golanska E., Kulczycka-Wojdala D., Szybka M., Piaskowski S., Wozniak K., Zarkewski K., Papierz W., et al. KCTD11 expression in medulloblastoma is lower than in adult cerebellum and higher than in neural stem cells. Cancer Genet. Cytogenet. 2006;170:24–28. doi: 10.1016/j.cancergencyto.2006.04.014. [DOI] [PubMed] [Google Scholar]