Abstract
During meiosis, DNA events of recombination occur in direct physical association with underlying chromosome axes. Meiotic cohesin Rec8 and cohesin-associated Spo76/Pds5 are prominent axis components. Two observations indicate that recombination complexes can direct the local destabilization of underlying chromosome axes. First, in the absence of Rec8, Spo76/Pds5 is lost locally at sites of late-persisting Msh4 foci, with a concomitant tendency for loosening of intersister and interhomolog connectedness at the affected sites. This loss is dependent on initiation of recombination. Second, in wild-type prophase, local separation of sister axes is seen at sites of synaptonemal complex-associated recombination nodules. Additional findings reveal that Rec8 localizes to both axis and bulk chromatin and is required for chromatin compactness. Further, Rec8 is essential for maintenance of sister cohesion, along arms and centromeres, during the pachytene-to-diplotene transition, revealing an intrinsic tendency for destabilization of sister cohesion during this period. This finding shows how the loss of sister connectedness, in arm and/or centric regions, could lead to the segregation defects that are seen in the human “maternal age effect” and how Rec8 could be a target of that effect. Finally, Rec8 plays related, but synergistic roles with Spo76/Pds5, indicating auxiliary roles for meiotic and mitotic cohesion-associated components.
[Keywords: Rec8, Spo76/Pds5, meiosis, recombination, chromosome axes, diffuse/dictyate stage]
One of the most interesting features of meiosis is occurrence of a prolonged prophase state devoted to interactions between homologous chromosomes. One prominent aspect of this process is recombination at the DNA level. Recombination initiates via programmed double-strand breaks (DSBs), and after a complex progression through several stages, a large number of initiated events finally yield a few crossovers (COs), which occur with highly particular spatial patterning (for review, see Jones and Franklin 2006). These COs, in combination with intersister connections along arms, form linkages between homologs (chiasmata) that are required to ensure regular segregation of the recombined homologous chromosomes at the first of the two meiotic divisions (MI).
Another prominent feature of meiotic prophase is organization of chromosomes into linear arrays of chromatin loops, the bases of which define chromosome axes. When this organization is fully developed, sister linear loop arrays are co-oriented with their axes tightly juxtaposed. These axes are elaborated by a complex array of axis-specific components, some meiosis-specific and some also present on mitotic chromosomes (for reviews, see Zickler and Kleckner 1999; Page and Hawley 2004; Revenkova and Jessberger 2005).
These two features are intimately interrelated. For example, recombination-initiating DSBs form in DNA sequences that are, organizationally, within chromatin loops. However, from an early stage onward, the corresponding recombination complexes (recombinosomes) are associated with their underlying chromosome axes (Blat et al. 2002; Oliver-Bonet et al. 2005; Moens et al. 2007 and references therein).
The functional significance of recombinosome/axis association is only slowly being elucidated. This physical relationship is likely to be crucial because it permits spatial, temporal, and functional coordination of events at the DNA and axis levels throughout meiotic prophase (for review, see Kleckner 2006). For example, at early prophase, axis-associated DSB recombinosomes mediate presynaptic coalignment of homologs (Tesse et al. 2003; Henderson and Keeney 2004; Calvente et al. 2005). Then, at the leptotene/zygotene transition, a subset of recombinational interactions is specifically designated for eventual maturation into COs. This event is accompanied by an important axis change: Synaptonemal complex (SC) is nucleated at the sites of COs, after which, by spreading outward, it brings homolog axes close together all along their lengths (for review, see Henderson and Keeney 2005). Also, in accord with the fact that each CO occurs between only one chromatid of each homolog, exchange of chromatid arms involves not only local separation and differentiation of sister chromatids at the chromatin level, but also a corresponding exchange at the axis level.
The filamentous fungus Sordaria provides a particularly attractive experimental system for examination of recombinosome/axis interplay. In many organisms, complete axes do not appear in a cytologically detectable form until mid-prophase, by which time many important events have already occurred. In Sordaria, in contrast, axis emerge concomitant with DNA replication and occur all along the lengths of the chromosomes at early prophase, just before appearance of RecA homolog Rad51 foci, which likely form very soon after DSB formation. Exactly these advantages have permitted elucidation of the mechanism of presynaptic coalignment (Tesse et al. 2003). An additional attractive feature of the Sordaria system is that the progression of nuclei through the various stages of meiosis can be monitored independent of chromosome status, by progressive increase in ascus size (meiocytes), thus permitting a clear establishment of event time lines in mutant situations in comparison with wild-type meiosis (e.g., Storlazzi et al. 2003).
In the current study, we used a Sordaria mutant lacking the meiotic cohesin Rec8 as a reagent for cytological investigation of prophase axis status and its relationship to recombinosomes. The rec8Δ mutant phenotype reveals that recombination complexes can promote local axis modifications and loss of (axis-mediated) linkages both between sisters and between homologs. Analogously, in wild-type meiosis, we observe local axis destabilization along SCs at sites of axis-associated recombination complexes (late nodules).
Mutant phenotypes also confirm and extend our understanding of the diverse processes that are directly or indirectly dependent on Rec8 and reveal a synergistic relationship between Rec8 and the general axis component Spo76/BIMD/Pds5, which is found in association with the mitotic counterpart of Rec8 (Mcd1/Scc1/Rad21) (for review, see Nasmyth and Haering 2005). This study triggered further consideration of the possibility that the dramatic age-dependent increase in missegregation that occurs during human female meiosis represents a degradation of intersister relationships during the period when meiosis is physiologically arrested.
Results
Rec8 localizes along chromosome axes and to bulk chromatin throughout prophase and remains on centromeres through metaphase II
In Sordaria, meiotic DNA replication occurs in haploid nuclei, which then fuse (karyogamy). Prophase axis organization begins to develop prior to karyogamy and is complete at early leptotene, as shown by electron microscopic (EM) ultrastructural analysis (Zickler et al. 1992), by immunostaining of tagged axis-protein Spo76/Pds5 and with the MPM-2 antibody that is thought to recognize epitopes of the axis component Topoisomerase II, or by direct visualization of a Spo76-GFP fusion protein (van Heemst et al. 1999).
In the present study we examined localization of the meiosis-specific cohesin Rec8 via a Rec8-GFP fusion protein. The REC8-GFP construct was present at an ectopic location in a strain carrying the wild-type REC8 gene. The fusion protein gives an accurate assessment of Rec8 localization because (1) progression of meiosis in this strain is normal by all cytological and genetic criteria (for further details, see the Supplemental Material), (2) the REC8-GFP gene complements all rec8Δ meiotic and sporulation defects with the same efficiency as the REC8 complementing subclone, and (3) the same pattern of Rec8-GFP localization is observed in both wild-type and rec8Δ backgrounds.
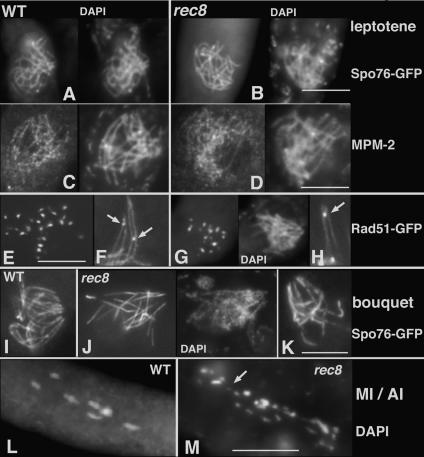
With respect to chromosome axes, the patterns of Rec8 localization observed during prophase (Fig. 1, left column and corresponding DAPI in right column) correspond exactly to those described previously for other axis markers: appearance prior to karyogamy as small lines/dots (thus, before/during S phase) (Fig. 1A), extension immediately after karyogamy to full length by early prophase (leptotene) (Fig. 1B,C), and persistence throughout pachytene (Fig. 1D). These images also illustrate the fact that the axes of homologs are concomitantly undergoing progressive juxtaposition, first to a distance of ∼400 nm (presynaptic alignment during leptotene) (Fig. 1C) and then to a closer distance (∼100 nm), which is full length at pachytene (Fig. 1D). At the EM level, synapsis involves installation of the SC, while at the light microscope level it involves merging of homolog axes into a single unit (Fig. 1D). Sordaria Rec8 localization corresponds to and further delineates previous observations from several organisms (e.g., budding and fission yeast in Klein et al. [1999]; Watanabe and Nurse [1999]; mammals in Eijpe et al. [2003]; Bannister et al. [2004]; Xu et al. [2005]; worm in Pasierbek et al. [2001]; Chan et al. [2003]; and Arabidopsis in Cai et al. [2003]; Chelysheva et al. [2005]).
Figure 1.
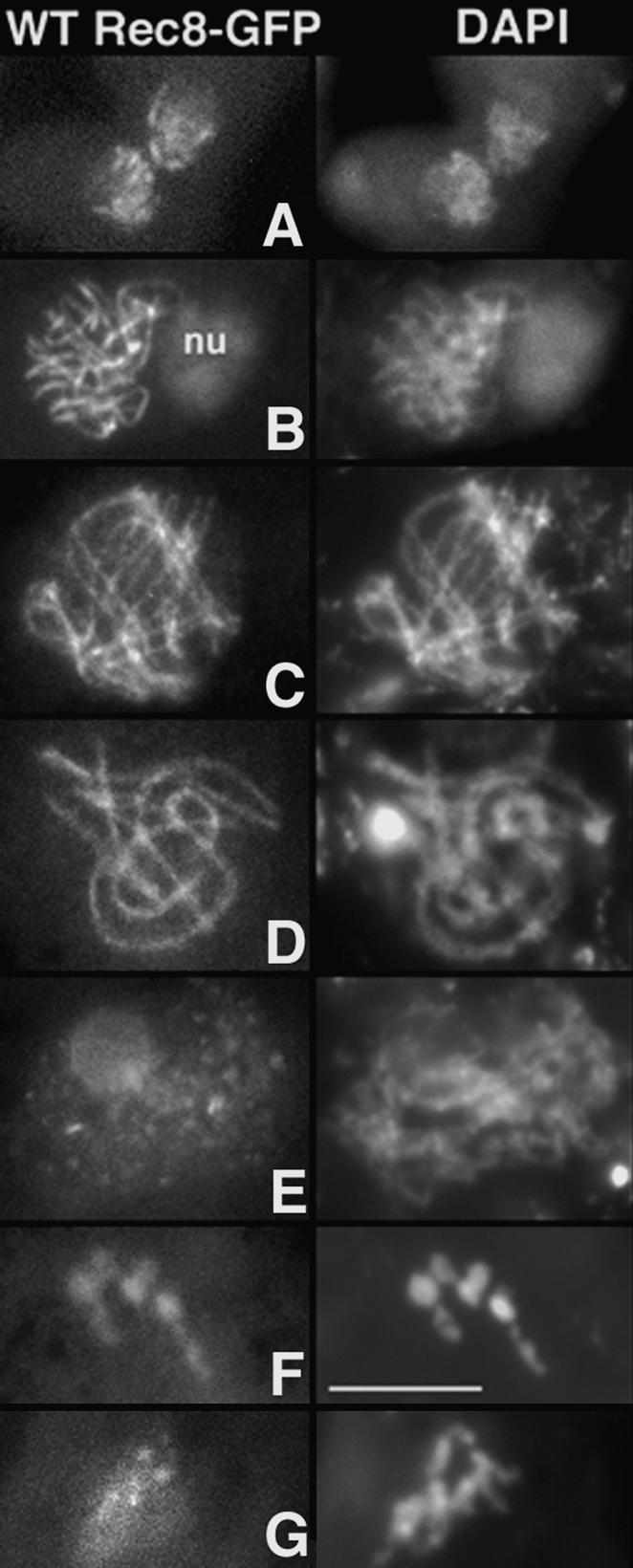
Rec8 localization in wild type. Rec8-GFP staining (left) and corresponding DAPI (right) from karyogamy (A; the two fusing haploid nuclei show short segments of Rec8) through early leptotene (B), late leptotene with aligned homologs (C), pachytene (D), diffuse stage (E), diplotene (F), and prophase of the second division (G). (nu) Nucleus. Bar, 5 μm.
As in most organisms, Sordaria pachytene is followed by a dramatic reorganization of the chromosomes. SC disappears and chromatin becomes completely decondensed at the so-called “diffuse stage.” Regular linear staining of Rec8 is lost during pachytene exit with the protein becoming broadly localized in a punctuate pattern on decompacted chromatin (Fig. 1E). Individualized chromosomes then re-emerge at diplotene and Rec8 bright staining is again observed on those compact chromosomes (Fig. 1F). Rec8 then disappears from chromosome arms at prometaphase I with staining becoming restricted to centromeric regions from that point through prophase II (Fig. 1G) and metaphase II, as is commonly observed (references above).
Two further features are revealed. First, while Rec8-GFP stains as bright continuous lines at leptotene (Fig. 1B), staining becomes progressively punctuate and less bright from zygotene on (Fig. 1, cf. B and C,D). Second, Rec8 localizes not only along axes but also on off-axis chromatin from S phase through pachytene (Fig. 1C,D; below).
Rec8 is not required for establishment of conjoined sister axes, initiation of recombination, or recombinosome-mediated presynaptic alignment
Analysis of mutant phenotypes can serve not only to probe the biochemical roles of individual molecules but, more generally, to probe functional relationships amongst relevant processes. To probe Sordaria meiosis from both perspectives, we carried out a complete cytogenetic analysis of a full rec8Δ-null mutant (this section and below). Throughout these studies, timing of progression through meiotic stages can be monitored independently of chromosomal morphology on the basis of nuclear volume and especially ascus (meiocyte) size, which increases in both wild type and rec8Δ from 20 μm at leptotene to 150 μm at diplotene.
Remarkably, despite the cytological prominence of Rec8 protein on meiotic chromosomes from early stages onward (above), axes in rec8Δ are virtually normal from onset through late leptotene. When compared with wild type, basic axial organization and robust sister axis cohesion are both normal, as shown by linearity staining of Spo76-GFP (Fig. 2A,B), MPM-2 (Fig. 2C,D), and corresponding DAPI. Thus, in Sordaria, Rec8 is not required for establishment of either of these aspects.
Figure 2.
Prophase phenotypes of wild type and rec8Δ. The two top rows show wild-type (A,C) and rec8Δ (B,D) leptotene nuclei stained by Spo76-GFP (A,B), and by MPM-2 and corresponding DAPI (C,D). (E–H) Rad51-GFP foci. They are numerous in early leptotene nuclei of wild type (E) and rec8Δ (G and corresponding DAPI). (F,H) In both strains, foci (arrows) are located between aligned homolog axes stained here by Spo76-GFP. (I–K) Bouquet formation in wild type (I) and rec8Δ (J,K). Note that wild-type chromosome ends are all clustered in a small area of the nuclear envelope (I), while mutant telomeres are only partially clustered (J,K). Wild-type metaphase I exhibits seven bivalents (L), while rec8Δ early anaphase I (M) shows scattered 28 chromatids and a smaller DAPI dot (arrow), corresponding to a broken chromatid. Bars, 5 μm.
Previous studies of DNA events in budding yeast, and of recombination-related foci in other organisms, have shown that Rec8 is not required for robust DSB formation (e.g., Klein et al. 1999; Xu et al. 2005). However, DNA analysis has revealed modest reductions in DSBs in fission yeast (with different intensities on a regional basis) (Ellermeier and Smith 2005) and budding yeast (A. Jordan, K. Kim, and N. Kleckner, unpubl.). In Sordaria, both wild type (Fig. 2E) and rec8Δ (Fig. 2G) form RecA homolog Rad51 foci, which mark the sites of DSBs (for review, see Hunter 2006). However, the mutant exhibits a ∼30% reduction in the number of leptotene Rad51 foci: 37 ± 6 compared with 57 ± 6 in wild type (n = 100 nuclei for each strain). Thus, DSB formation is likely reduced also in Sordaria, although another possibility, a defect in post-DSB loading of Rad51, is not excluded. A third possible explanation, faster turnover of Rad51 in the mutant, is probably not the case, because there is a similar decrease (∼46%) of Rad51 foci in both strains during zygotene (26 ± 5 in wild type; 17 ± 4 in rec8Δ; n = 100 for each strain) as compared with leptotene. Rec8 could potentially be involved directly in DSB formation. Alternatively, since replication and DSBs formation are normally coupled processes (e.g., Borde et al. 2000; Murakami et al. 2003), a decrease in DSBs could be a secondary consequence of earlier defects during that period (below).
Interestingly, also, the relationship between recombination complexes and their underlying chromosome axes is functionally normal at leptotene in rec8Δ. In wild-type meiosis, Rad51 foci appear at discrete positions in between coaligned axes (Fig. 2F) and mediate presynaptic coalignment (Tesse et al. 2003). In rec8Δ, presynaptic coalignment occurs with normal timing and with Rad51 foci observable between homolog axes (Fig. 2H).
Thereafter, a subset of Rad51 foci persist until pachytene in rec8Δ and, moreover, exhibit aberrant morphologies (line-like aggregates) rather than discrete foci as in leptotene, suggesting that some initiated recombinational interactions fail to progress; e.g., as in budding yeast (Klein et al. 1999).
Rec8 is required for normal karyogamy, proper chromatin organization, and bouquet dynamics
First, closer inspection of rec8Δ meiosis at early stages does reveal some subtle defects: (1) rec8Δ exhibits an excess of prekaryogamy nuclei (illustrated in Fig. 1A). In wild type, at a time when all of ∼150 asci per perithecium (fruiting body) are in meiosis, only approximately two are in prekaryogamy; in contrast, in rec8Δ, ∼20 are still at this stage (n = 100 perithecia for each strain). Further, the Spo76-GFP short-terminal segments formed during prekaryogamy (analogous to the Rec8-GFP segments seen in Fig. 1A) are longer in rec8Δ than in wild type (2–4 μm instead of 0.5–1 μm in wild type, n = 30). Their further elongation suggests that, like in budding yeast (Cha et al. 2000), S phase takes longer than normal in Sordaria rec8Δ. (2) From early leptotene on rec8Δ chromosomes exhibit slightly fuzzy DAPI staining, implying a defect in the degree of compactness, in functional correlation with chromatin-localized Rec8 (Fig. 2, top two rows, cf. DAPI of wild type and rec8Δ). Abrogation of DSB formation in rec8Δ spo11Δ (deleted for the DSB-catalyzing transesterase Spo11) has no effect on those prekaryogamy and leptotene defects (data not shown).
Second, Rec8 is also important for three-dimensional chromosome dynamics. In wild-type nuclei, chromosome ends cluster in one region of the nuclear envelope from late leptotene through early pachytene in the bouquet configuration (Fig. 2I). In rec8Δ, chromosome ends cluster also at late leptotene, but with mostly only one end of each chromosome participating (Fig. 2J,K). Specifically, in wild-type early pachytene nuclei, nine to 11 of the 13 chromosome ends (one end is in the nucleolus) are clustered together in a small area (Fig. 2I), while in rec8Δ (Fig. 2J,K), only five to eight ends are clustered (in 50 nuclei analyzed by Spo76-GFP and DAPI). Moreover, chromosomes ends remain clustered longer than in wild type as indicated by a higher fraction of bouquets among total prophase nuclei (two to three times more bouquets in rec8Δ: 112 of 522 and 123 of 514 mutant prophases in two independent experiments compared with 66 of 975 in wild type; P < 0.0001). Delayed bouquet exit is also observed in a budding-yeast strain (Trelles-Sticken et al. 2005). Nevertheless, in Sordaria rec8Δ, as in wild type, telomeres are redispersed by mid-pachytene.
Rec8 is required for sister cohesion after pachytene
In rec8Δ, axis staining of Spo76-GFP, and thus sister cohesion, remains robust through pachytene (albeit with appearance of unusual local gaps, as discussed below). However, Rec8 is absolutely required for maintenance of sister cohesion, along arms and in centric regions, during the diffuse stage. Chromosomes exit pachytene into the diffuse stage, where individualized chromosomes are no longer visible (Fig. 1E), and then reindividualize at diplotene into shorter, more compact chromosomes in which homologs are linked only by chiasmata (Fig. 1F). In wild type, axis staining of Spo76-GFP and Rec8-GFP is lost during pachytene exit with the proteins becoming progressively localized in a punctuate pattern on decompacted chromatin during the diffuse stage (see Fig. 1E for Rec8). Ensuing progressive loss of Spo76-GFP axis staining and the diffuse stage occur normally in rec8Δ, as does chromosome recondensation at diplotene. However, rather than the seven chiasma-linked homolog pairs seen in wild-type diplotene and metaphase I (Fig. 2L), 28 or more individual units are seen in rec8Δ (Fig. 2M). This phenotype implies that, in re-emergent chromosomes, sister chromatids are entirely separated from one another and, moreover, that a few of these are broken (Fig. 2M, arrow). Elimination of recombination in rec8Δ spo11Δ has no effect on diplotene precocious sister separation, but eliminates occasional broken chromosomes (data not shown), which are thus attributable to defective recombination, as also seen in worm and Arabidopsis (Pasierbek et al. 2001; Chelysheva et al. 2005). These findings show that while Rec8 is not essential for establishment of sister cohesion (above), it is essential for cohesion maintenance during this dramatic period of chromosome reorganization.
In the absence of Rec8, homologs synapse, but with concomitant appearance of local axial destabilization
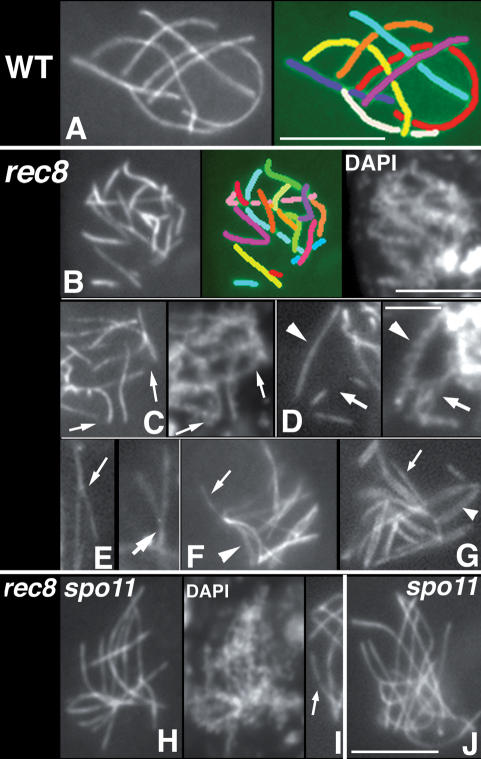
While rec8Δ chromosome axes and chromatin are only mildly defective through leptotene, they undergo important modifications concomitant with zygotene. In wild type, Spo76-GFP staining occurs as continuous lines along each chromosome from leptotene through pachytene, at which point it occurs as seven thick, continuous lines corresponding to seven synapsed bivalents (Fig. 3A). In contrast, in rec8Δ a larger number of shorter lines are observed from late zygotene on (14 ± 3; range, 10–21 in 115 nuclei from all relevant stages) (Fig. 3B). Most of these lines are of wild-type thickness, implying the presence of synapsed homolog axes (confirmed in Fig. 3B [right, by DAPI], see also G).
Figure 3.
Spo76-GFP staining of wild-type and rec8Δ axes. (A) Wild-type pachytene nucleus with seven bivalents plus corresponding drawing (shown at right). (B) rec8Δ pachytene nucleus with 16 Spo76-GFP segments, drawn with different colors (no correspondence with the wild-type colors) and corresponding DAPI (shown at right). In A and B, each line has its own color, but some are discontinuous to indicate that they lie in back of other chromosomes. (C–G) rec8Δ. (C) Arrows point to two gaps in Spo76-GFP lines on corresponding continuous axes by DAPI staining (shown at right). (D) Arrow points to a Spo76 gap that corresponds to three DAPI threads (arrow, right) indicating both local asynapsis and chromatid-cohesion defect; for comparison, arrowheads point to uninterrupted Spo76 line and DAPI. (E) Two examples of forks (arrows). At both zygotene (F) and early pachytene (G), asynapsed axes (arrows) and synapsed double Spo76 lines (arrowheads) are clearly visible. (H,I) rec8Δ spo11Δ: Spo76-GFP lines are continuous and chromatin (DAPI) is less diffuse than in rec8Δ (compare with DAPI in B, but in a few cases, Spo76 gaps remain (arrow in I). (J) spo11Δ single mutant at mid-prophase. Bars, 5 μm.
Comparison with corresponding DAPI images reveals that the rec8Δ phenotype results from gaps in Spo76-GFP staining along underlying continuous chromosomes (Fig. 3C,D, arrows). At ∼75% of gap sites, the underlying chromatin organization is morphologically normal and identical to that seen for chromosome segments that retain Spo76-GFP (Fig. 3C), suggesting that both basic axial structure along homologs and intimate cohesion of sister chromatids are still present at these gaps. At the remaining 25% of gaps, underlying chromosome structure is aberrant, with open DAPI threads suggesting local sister separation plus asynapsis of homologs (Fig. 3D, arrow, cf. normal morphology indicated by arrowhead).
In addition to these effects, a tendency for local asynapsis is also apparent: Approximately 15% of Spo76-GFP lines are Y-shaped, with the two arms being of half the normal width, implying absence of synapsis (Fig. 3E, arrows). An additional ∼35% of Spo76 lines are single-width along their entire length (Fig. 3F,G). These are likely to be regions between gaps, in which asynapsis has extended from the two flanking gap sites across the entire intervening segment (Fig. 3G, arrow).
The Spo76-GFP gaps seen in rec8Δ likely do not correspond to sites of centromeres because (1) the individual line lengths are highly variable from one nucleus to another; (2) in 60% of the 115 nuclei, the number of gaps is greater than seven, the number of centromeric regions at pachytene; and (3) the centromere of chromosome 2, which carries the nucleolar organizer region at one end, is located ∼2 μm from that region (Zickler et al. 1992). Among 25 rec8Δ nuclei with a clearly analyzable bivalent 2 (∼8 μm long), the distance between the nucleolar organizer region and the end of the associated Spo76 line varies from 1 to 6 μm.
The local nature of Spo76-GFP gaps is further emphasized by quantification of line lengths. Total bivalent-equivalent length of rec8Δ Spo76 lines (with single-width axes counted for half-lengths) in the late zygotene/pachytene nuclei is ∼80% of that seen in wild type (38 ± 5 μm compared with 47 ± 4 μm for wild type, P < 0.0001; 115 nuclei for each strain). Further, despite this structural difference, total Spo76 line lengths in both wild type and rec8Δ increase from early to late pachytene (32.1–59.3 μm and 22.6–52.3 μm, respectively, 150 and 95 nuclei), as also seen for wild-type in EM reconstructions (D. Zickler, unpubl.). We infer that Spo76-GFP gaps are local defects along otherwise normally behaving axes.
Spo76-GFP gaps are strongly, but not absolutely, dependent on recombination initiation
We examined Spo76-GFP staining in a rec8Δ strain where recombination initiation was abrogated. Among 100 spo11Δ rec8Δ nuclei at zygotene/pachytene, 48 nuclei exhibited 14 uninterrupted Spo76 lines (Fig. 3H) that are easily sorted into appropriate-length pairs, exactly the same phenotype as spo11Δ nuclei (Fig. 3J). An additional 49 exhibited a single gap (on one chromosome), while the remaining three showed two gaps (Fig. 3I, arrow).
Absence of DSBs in rec8Δ eliminates also the dramatic chromosome diffuseness observed after the leptotene–zygotene transition (Fig. 3, cf. DAPI in H and C,D). However, a residual defect in chromatin compactness still remains in rec8Δ spo11Δ. This finding implies both an additional effect of recombination on chromosome destabilization in rec8Δ and recombination-independent role for Rec8 in bulk chromatin status, in accord with its localization to off-axis chromatin (above).
These data permit two important conclusions. First, formation of gaps is strongly dependent on recombination, implying that events of recombination trigger local destabilization of the underlying axes. Second, there is still some tendency for gap formation and chromatin defects in rec8Δ, irrespective of recombination initiation.
In rec8Δ, late Msh4 foci are spatially correlated with Spo76-GFP gaps
MutS homolog Msh4 is known from studies in other organisms to form foci corresponding to recombination complexes during zygotene and pachytene (e.g., Higgins et al. 2004); i.e., during the periods when Spo76-GFP gaps are emerging and present in rec8Δ. To further investigate gaps/recombination relationship, we analyzed the position of Msh4 foci relative to wild-type and rec8Δ Spo76 lines. We utilized a Msh4-GFP fusion protein that, as for other Sordaria GFP-tagged proteins, was present ectopically in the genome. The fusion gene complements all msh4Δ meiotic and sporulation defects and has no discernible effect on the timing or efficiency of chromosome dynamics or any other aspect of meiosis in either wild type or rec8Δ (for details, see the Supplemental Material).
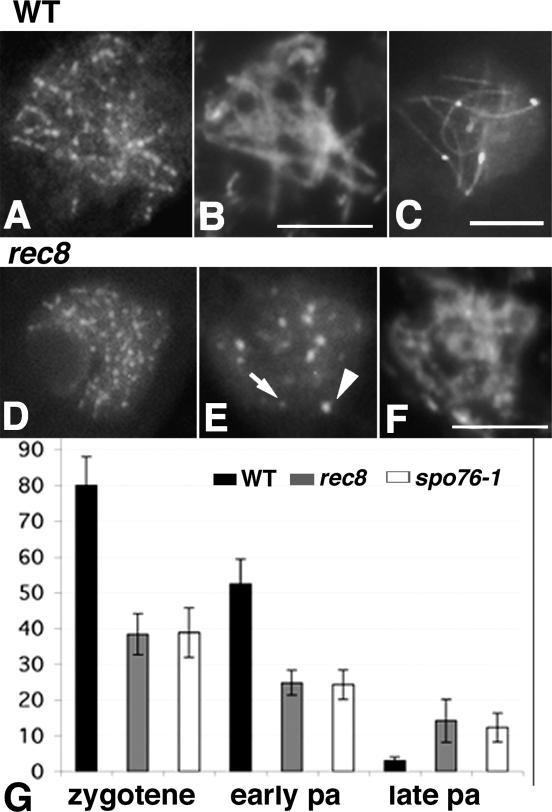
In wild-type meiosis, Msh4-GFP foci (Fig. 4A–C,G) appear in large numbers (80 ± 8; n = 100) during zygotene, diminish in number by early pachytene (52 ± 7; n = 100), and are essentially absent by late pachytene (Fig. 4C,G), analogously to other organisms (e.g., Higgins et al. 2004 for Arabidopsis and Moens et al. 2007 for mouse). In rec8Δ prophase, Msh4-GFP foci (Fig. 4D–G) appear also during zygotene, but as seen Figure 4G, in ∼47% the wild-type number (38 ± 6; n = 100), and diminish by early pachytene at the same rate as in wild type (24 ± 4; n = 100). However, unlike in wild type, Msh4 foci persist at ∼14 ± 6 per nucleus (n = 80) throughout pachytene (Fig. 4G). Moreover, while regular at early pachytene (Fig. 4D), Msh4 foci increase in size, irregularity, and brightness through pachytene (Fig. 4E). This failure to release Msh4 in a timely fashion again suggests, as also inferred from Rad51 foci above, that a subset of recombinational interactions has stalled. The deficit of Msh4 foci seen at earlier stages in rec8Δ is likely attributable to a reduction in the number of DSBs (above). However, as the reduction in the number of Msh4 foci is greater than the earlier reduction in the number of Rad51 foci, this deficit may be partly due to a later defect in the quality or quantity of Msh4-containing complexes and/or more rapid turnover of Msh4 complexes.
Figure 4.
Msh4 foci in wild type, rec8Δ, and spo76-1. (A–C) Wild type. Msh4-GFP foci are regular in shape from early pachytene (A,B, corresponding DAPI) through mid-late pachytene (C) (axes stained by Spo76-GFP). (D–F) rec8Δ. Foci are regular at early pachytene (D) but highly irregular in shape and brightness at mid-late pachytene (E, arrow points to small dot and arrowhead to large dot). (F) Corresponding DAPI. (G) Histogram shows the number (vertical axis) of Msh4 foci from zygotene to late pachytene (pa) in wild type (black bars), rec8Δ (gray bars), and spo76-1 (white bars). Error bars indicate standard deviation. Total nuclei scored: wild type, 350; rec8Δ, 300; spo76-1, 200. Bars, 5 μm.
We next analyzed the spatial relationship between Msh4-GFP foci and Spo76-GFP gaps. For this purpose, we analyzed wild-type and rec8Δ strains carrying both fusion constructs. Control experiments show that simultaneous presence of both tagged proteins caused no detectable change in any wild-type or rec8Δ phenotype as compared with the corresponding strains carrying either construct individually, or lacking any GFP-tagged construct (above; Supplemental Material). Spatial relationships of Msh4 foci relative to Spo76 lines and DAPI-stained chromosome segments were analyzed by a series of z-sections (∼25 per nucleus).
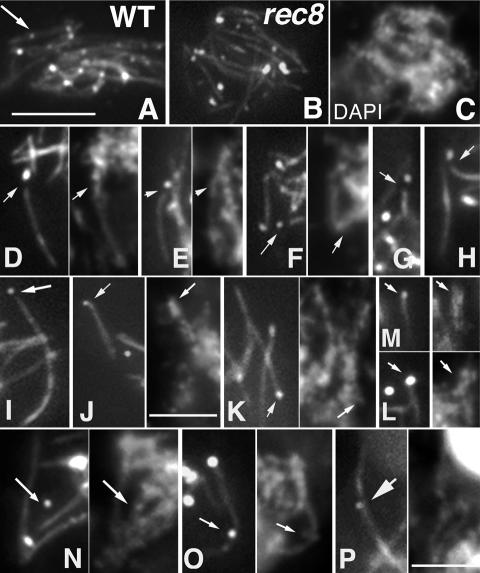
In a wild-type strain carrying both constructs, Spo76-GFP forms uninterrupted lines. All Msh4 foci are directly associated with those lines from late zygotene through early pachytene with none in the off-axis chromatin (Fig. 5A). Thus, in wild type, Msh4 foci are axis- and SC-associated at all stages until they disappear at mid-pachytene.
Figure 5.
Correspondence between Spo76-GFP gaps and Msh4 foci. (A) Wild-type pachytene nucleus. Msh4 foci (arrow) are located on continuous Spo76-GFP lines. (B,C) rec8Δ pachytene nucleus and corresponding DAPI. (D–P) Gallery of the four different Msh4 localizations. (D–H) Five examples of Msh4 foci (arrows) located in a gap of a Spo76 line as seen by corresponding DAPI in D–F. (I–M) Five examples in which the foci (arrows) located at the end of a Spo76 line correspond to a subterminal region of the chromosome as shown by corresponding DAPI (shown in J–M, right). (N) The Msh4 focus indicated by an arrow is not attached to a Spo76 line, but is clearly located on chromatin as seen by corresponding DAPI (arrow, shown at right). In contrast, the second focus (shown below) is in a Spo76 gap. (O,P) Msh4 foci located internally along a Spo76 line. (O) One focus is in the middle (arrow) and a second focus is at one end of a Spo76 line. Spo76 staining under the centrally located focus is faint, implying local destabilization within the line. Corresponding DAPI shows continuous chromatin, but with thread separation at the focus position (arrow). (P and corresponding DAPI) Nucleolar organizer chromosome (nucleolus on top) with one Msh4 focus: Spo76 staining shows a small gap above the focus (arrow). Bars, 5 μm.
For rec8Δ, we analyzed the positions of all Msh4 foci along the Spo76 lines in 35 late pachytene nuclei, where only late-persisting Msh4 foci are still present (Fig. 5B–P). The 512 Msh4 foci present in this sample are all associated with a segment of underlying DAPI-stained chromosomal DNA and can be assigned to four different configurations on the basis of their relationship to Spo76 lines. (1) In 52% of cases, the Msh4 focus occurs at the end of a Spo76 line (Fig. 5D–H) and that line can clearly be seen, by DAPI, bridged to another Spo76-GFP line (Fig. 5D–F). In these cases, therefore, the Msh4 focus occurred at one of a pair of bridged ends, and thus, at one edge of a gap. (2) Six percent of Msh4 foci localize at the end of a Spo76 segment that corresponds to the terminal region of a DAPI-defined chromosomal unit; i.e., a chromosome end (Fig. 5I–M). In wild-type meiosis, ∼10% of late recombination nodules do occur subterminally (Zickler et al. 1992). Thus, the 6% of Msh4 foci in this category are likely cases in which destabilization has occurred at a subterminal position with loss of Spo76-GFP from the site of destabilization out to the chromosome end. (3) Twenty-six percent of Msh4 foci were associated with a chromosome segment lacking any Spo76 line (Fig. 5N,J). In these cases, the underlying DAPI-staining segment is indistinguishable from chromosome segments that have an associated Spo76-GFP line (Fig. 5N, arrow). We infer that foci of this category are located within long gaps of Spo76-GFP. (4) Sixteen percent of foci were located at an internal position along a Spo76 line (Fig. 5O,P). As a weakening of Spo76-GFP staining at the site of the focus is often visible (Fig. 5O,P, arrows), some level of axis destabilization may also be present in these cases. Taken together, these data point to a strong correlation between the positions of Msh4 foci and the positions of gaps.
Distributions of Msh4 foci and Spo76 gaps are consistent with their occurrence at sites of CO interactions
While a large number of recombinational interactions are initiated via DSBs at leptotene, only a select subset of those interactions are designated for maturation into CO products, with the remainder being fated to mature as noncrossover (NCO) products. In Sordaria, the number of CO events is ∼20 (21 ± 3 per nucleus from late recombination nodules and chiasmata) (Zickler et al. 1992) and the total number of recombination interactions is approximately three to four times higher, as judged from the numbers of early recombination nodules (Zickler et al. 1992) and the numbers of both Rad51 and Msh4 foci (see above). Since the local recombination-provoked destabilizations seen in rec8Δ chromosome axes arise during the transition from leptotene through zygotene, and thus during or after the point at which a subset of recombinational interactions are designated for eventual maturation into COs (for review, see Kleckner 2006), the question arises whether Msh4 foci and Spo76-GFP gaps might correspond specifically to sites of CO-fated interactions. Two findings are consistent with this possibility.
First, in rec8Δ, the number of late-persisting gap/end-associated Msh4 foci is similar to the predicted numbers of COs. The average number of late-persisting Msh4 foci in the 35 late pachytene nuclei of rec8Δ is 15 ± 5 per nucleus, of which 84% (or ∼13) are associated with a discontinuity in Spo76 lines (above; Fig. 5D–N). The number of COs in rec8Δ should be significantly less than the 21 ± 3 observed in wild type (above). Based on either an ∼35% reduction of Rad51 foci or an ∼47% reduction of early Msh4 foci (Fig. 4G), either ∼14 or ∼10 COs would be predicted, very similar to the ∼13 gap/end-associated late-persisting Msh4 foci.
Second, there is also a good correlation between the number of Spo76-GFP lines and the number predicted if gaps occur at sites of COs. The reduction in Rad51 foci in rec8Δ predicts ∼14 COs, while the reduction in Msh4 foci predicts ∼10 COs per nucleus. Assuming that a Spo76 gap occurs at the site of every CO, these numbers predict, in turn, ∼21 and ∼17 Spo76 lines per nucleus, respectively. The number of Spo76 lines observed in 35 middle-late pachytene nuclei was 17 ± 3 (above), and thus well within the appropriate range. Further, the number of lines may be slightly underestimated in these nuclei for two potentially related reasons: (1) Seventy-eight percent of the 115 early pachytene nuclei exhibited Spo76 lines smaller than 1 μm (examples in Fig. 3D,G) and those short lines may no longer be visible at a later stage, and (2) the gaps corresponding to the late recombination nodules located close to the chromosome ends by EM (Zickler et al. 1992) may be no longer visible in the mutant.
The possibility that Spo76 gaps and associated Msh4 foci seen in rec8Δ correspond specifically to COs carries with it the implication that CO designation occurs normally in rec8Δ. This assumption is supported by DNA studies in budding yeast, which show that rec8Δ differentially blocks formation of COs but not NCOs, implying that CO/NCO differentiation has occurred (K. Kim, A. Jordan, and N. Kleckner, unpubl.).
Axis destabilization at sites of late recombination (CO) nodules is seen in wild type
CO-specific recombination-associated axis destabilization is also seen in wild-type meiosis. In the 10 pachytene nuclei reconstructed from serial-sectioned nuclei observed in EM, lateral elements (LEs) of SC segments that underlie “late” nodules, (which correspond to CO recombination complexes) exhibit morphological duality, implying a tendency for local separation of sister axes specifically around the position of the CO site (Fig. 6A–C, G–I). In contrast, LEs lacking nodules exhibit tight conjunction of sister LEs, which appear as a single morphological unit (Fig. 6C–E,J). Among the 208 nodules present in this sample, ∼75% exhibited local sister separation on both homologs (e.g., Fig. 6G,H), the remaining 25% showed clear separation on only one homolog (Fig. 6A–E). The maximum degree of sister axis separation in these regions of duality is 20–30 nm, which explains why they cannot be seen by light microscopy along wild-type axes. These observations support the existence of a tendency for separation of sister LEs, locally at sites of CO recombinational interactions in wild-type meiosis. Interestingly, the region of duality seen at these sites extends over ∼400 nm (Fig. 6A, G–I), which corresponds to the lengths of the majority of the Msh4-associated Spo76-GFP gaps (400–700 nm) seen in rec8Δ, pointing to a direct relationship between the phenomena observed in the wild-type and mutant cases.
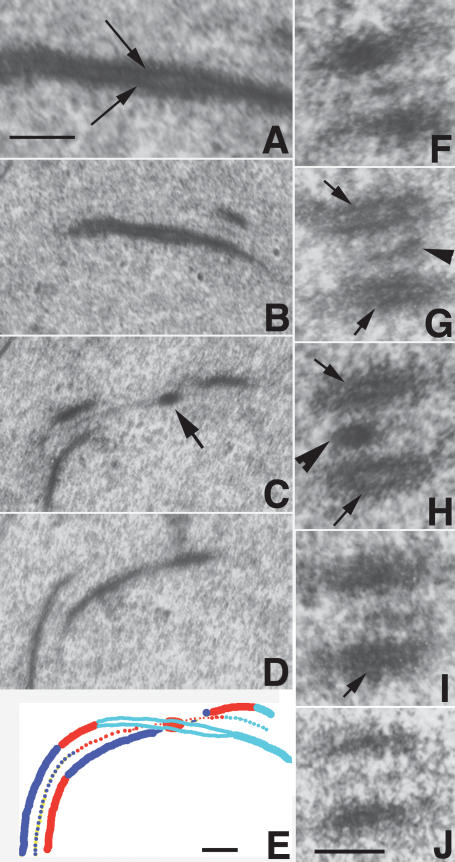
Figure 6.
Electron micrographs of wild-type SCs. (A) LE in longitudinal section. Arrows point to the local separation of the two chromatids. (B–D) Three consecutive serial sections of a SC segment. The recombination nodule (arrow in C) is located at the dual LE site (B) enlarged in A. (C,D) Note that LEs adjoining the nodule are single units. (E) Reconstruction drawing of B–D (B in cyan, C in red, D in blue) showing that the SC twists at the nodule site. (F–J) Five consecutive oblique/cross-sections through a SC segment exhibiting local separation of sister chromatids (arrows) on both homologs (G–I) at the nodule site (arrowhead in G,H); in contrast, in the two adjacent sections (F,J), LEs are single units. Bars, 100 nm.
Rec8 and Spo76 have partially distinct roles for chromosome status
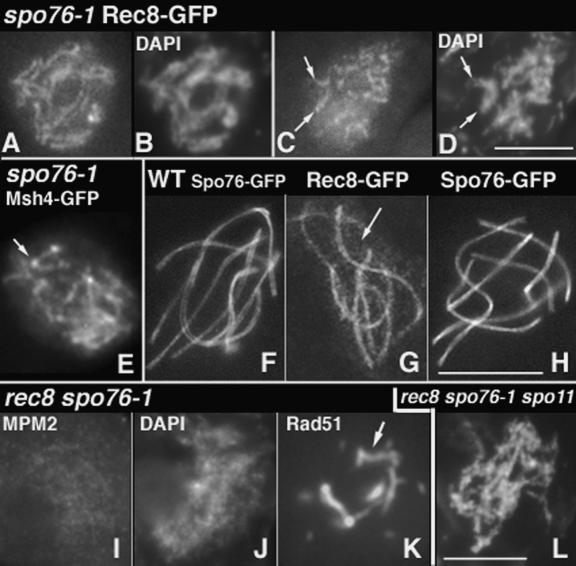
The axis destabilization observed above for rec8Δ is reminiscent of the previously described destabilization conferred by a nonnull allele of SPO76, spo76-1 (SPO76 is an essential gene in Sordaria). Both mutations confer chromosome axis destabilization during zygotene, with accompanying tendencies for coordinate splitting of sisters and homologs on a region-by-region basis (above; van Heemst et al. 1999). Immunofluorescence analysis confirms and extends the previous results seen by EM for spo76-1: Leptotene axes are continuous when stained by Rec8-GFP (Fig. 7A,B); pachytene chromosomes exhibit gaps of Rec8 staining (arrows in Fig. 7C,D); and these gaps are recombination-dependent because spo76-1 spo11Δ chromosomes show continuous Rec8-GFP staining (data not shown). Additionally, spo76-1, like rec8Δ, shows late-persisting Msh4-GFP foci (14 ± 4 per nucleus; n = 70) (Fig. 4G). In light of these similarities, we further investigated the functional relationships between Rec8 and Spo76.
Figure 7.
Phenotypes of spo76-1, rec8Δ spo76-1, and interdependent localization of Rec8 and Spo76. (A–E) spo76-1. Rec8-GFP forms continuous lines through leptotene (A; corresponding DAPI in B), but is patchy at zygotene through pachytene (C). (D) Arrows point to a bivalent with discontinuous Rec8-GFP but continuous DAPI. (E) Msh4-GFP foci are located on Spo76-1-GFP lines (arrow). (F–H) Comparison between Spo76 and Rec8 staining in wild type. Early pachytene nuclei was stained with Spo76-GFP (F) and Rec8-GFP (G). Note that, contrary to Spo76, Rec8 is also seen on chromatin (arrow) and stains axes in a punctuate pattern. (H) Spo76 staining at late pachytene. (I–K) rec8Δ spo76-1 double mutant. (I) MPM-2 staining is absent through prophase (compare with rec8Δ single in Fig. 2D). Chromatin is completely fuzzy (J), and Rad51-GFP (K) exhibits aberrant lines (arrow) in rec8Δ spo76-1-arrested prophases. (L) Absence of DSBs restores visible chromosomes in the rec8Δ spo76-1 spo11Δ triple mutant. Bars, 5 μm.
Despite these similarities, comparison of the rec8Δ single mutant phenotype described above with that described previously for the spo76-1 mutant reveals significant differences. (1) Contrary to rec8Δ, spo76-1 exhibits no defects in prekaryogamy, karyogamy, and leptotene chromatin morphology (van Heemst et al. 1999). (2) In spo76-1, axis destabilization at zygotene/pachytene occurs between the sites of CO interactions: The two to 11 SC “pieces” observed per nucleus mainly show an associated late nodule (van Heemst et al. 1999). In contrast, the rec8Δ data suggest that axis destabilization occurs at, rather than between, the sites of COs. (3) In contrast to rec8Δ where most Msh4 foci are associated with regions that have completely lost Spo76-GFP (above) in spo76-1, Msh4 foci are most often localized within a spo76-1 line, as shown by analysis with Spo76-1-GFP (arrow in Fig. 7E). Quantitative analysis of Msh4 foci versus axial lines and gaps is impeded in spo76-1, because Rec8-GFP lines are too irregular and “dotty” (Fig. 7C).
Rec8 and Spo76 also exhibit spatial and temporal differences in axis loading patterns. In wild type, Spo76-GFP staining is always bright and continuous along each chromosome when Rec8-GFP exhibits a punctuate staining from late leptotene on (Fig. 7, cf. F and G). Moreover, Rec8 axis staining becomes progressively less bright beginning at late leptotene and through pachytene (Fig. 1), while Spo76 remains uniformly bright until the end of pachytene (Fig. 6H).
We further find that, strikingly, the rec8Δ spo76-1 double mutant exhibits a much stronger meiotic defect than either single mutant. (1) The total number of asci per perithecium is severely reduced—from ∼150 in wild type and each single mutant to only ∼20 in rec8Δ spo76-1— suggesting that some nuclei fail to enter meiosis altogether. (2) Fifty percent of those asci show prekaryogamy nuclei versus ∼20% in rec8Δ and ∼2% in spo76-1 and wild type (100 perithecia analyzed for each strain). (3) From early through late leptotene, chromatin diffuseness is severe and no MPM-2 axis staining is detectable (Fig. 7I), versus no, or modest, defects for these features in spo76-1 and rec8Δ (Fig. 2D; van Heemst et al. 1999). (4) Rad51 foci appear on time during leptotene, but their number is further reduced as compared with the two single mutants at this and later stages: 22 ± 6 in rec8Δ spo76-1, 37 ± 6 in rec8Δ, 36 ± 5 in spo76-1, and 57 ± 6 in wild type (n = 100 for each strain). (5) Even more dramatic differences between single and double mutant phenotypes are seen at and after leptotene/zygotene. While rec8Δ and spo76-1 single mutants exhibit local or regional axis defects (above), the double mutant exhibits total destruction of all aspects of chromosome structure: Chromatin is diffuse, and individual chromosomes are no longer discernible by DAPI staining (Fig. 7J). Additionally, Rad51 (by GFP and anti-Rad51 antibody) forms aberrant lines from zygotene on (estimated by ascus size) (Fig. 7K). Msh4 foci, which are abundant at zygotene in both single mutants (38 ± 6 in rec8Δ and 39 ± 7 in spo76-1 in 100 and 80 nuclei, respectively) (Fig. 4G), are now absent (in 75 of 100 nuclei) or rare (one to three in the remaining 25 nuclei) at both zygotene and pachytene. In correspondence to these severe mid-prophase defects, the double mutant exhibits cell-wide arrest at an ascus size corresponding to early pachytene in wild type, rather than progressing through meiosis I, as do the two single mutants.
Remarkably, however, both structural and progression defects are largely ameliorated by elimination of DSBs. In spo11Δ rec8Δ spo76-1, univalents are formed at leptotene and persist through pachytene (Fig. 7L) like in spo11Δ single mutant, despite the absence of Rec8 and abnormal Spo76/Pds5, two important components of the chromosome axes. In contrast, bulk chromatin remains more diffuse than in spo11Δ. Also, lack of recombination does not suppress loss of chromatid cohesion through the diffuse stage, as indicated by the emergence of 28 condensed chromatids at diplotene in the triple mutant (data not shown).
Discussion
The primary general finding revealed by this study is that local destabilization of axes is found spatially associated with recombination complexes. This is observed in rec8Δ meiosis by patterns of Msh4 foci relative to interruptions in Spo76-GFP localization, and in wild-type meiosis, by EM analysis of LE structure in relation to the positions of recombination nodules at pachytene. Local destabilization of chromosomes at recombination sites, while predicted (for review, see Kleckner 2006), has not been observed previously.
Three additional conclusions of interest emerged from this study. First, Rec8 localizes to, and is important for the state of bulk (off-axis) chromatin. Second, Rec8 and Spo76/Pds5 share common functions, but also have distinct individual functions, as shown by analysis of single and double mutant phenotypes. Third, destabilization of sister cohesion is an intrinsic feature of the period that encompasses the transition from pachytene to diplotene, with possible implications for the human maternal age effect as discussed below.
Recombination and local destabilization of underlying chromosome axes
In Sordaria rec8Δ, Spo76-GFP is lost from chromosome axes, in gaps or longer segments, from zygotene on. While many affected regions still exhibit apparently normally organized chromosomes, loss of Spo76 is sometimes accompanied by a local loss of intersister cohesion and/or absence of synapsis. These effects occur in specific relation with axis-associated recombinosomes as revealed by (1) the dependence of Spo76-GFP gaps upon initiation of recombination, and (2) a tight spatial correlation between Msh4 foci and Spo76 gaps. Circumstantial evidence is consistent with the possibility that these gaps occur specifically at sites of CO-fated recombination sites. Axis destabilization is also detected at pachytene of wild-type meiosis as a tendency for separation of sister LEs, specifically at the sites of late/CO recombination nodules. Moreover, in 75% of the cases, both homologs show LE separation, which parallels the absence of Spo76 on both homologs seen in rec8Δ. These two sets of observations provide two independent lines of evidence that recombination complexes can direct the local destabilization of underlying axes, and suggest (rec8Δ) or show (wild type) that this is a specific effect of CO-fated recombinosomes. Further, SC analysis shows that such an effect is an intrinsic feature of CO recombination during wild-type meiosis.
Local destabilization of Spo76-GFP in rec8Δ and local sister LE duality at nodule sites in wild type could be unrelated phenomena. The effect observed in rec8Δ could be a peculiarity of the mutant condition. On the other hand, both effects (1) occur on pachytene chromosomes, (2) involve intersister destabilization and/or a function implicated in sister cohesion, and (3) affect regions of similar lengths (∼400 nm). These commonalities therefore point instead to the more economical possibility that the two sets of phenomena represent two aspects of the same basic effect. By this model, CO-designated recombination complexes would direct the local destabilization of underlying chromosome axes, and more specifically, intersister axis cohesion as a normal aspect of CO-specific recombination-related events. In wild type, this tendency would be substantially constrained by Rec8, with the subtle effects observed by EM analysis, but when Rec8 is absent, destabilization would occur in an exaggerated form that includes pronounced loss of sister and homolog connectedness.
We cannot exclude the alternative possibility that, in wild-type meiosis, recombination directs subtle modulation of local axis status irrespective of the presence or absence of Rec8, whereas the more dramatic destabilization observed in rec8Δ is specifically triggered by the mutant situation. In a rec8Δ background, for example, defective progression of recombination might trigger a “checkpoint” response, which includes local axis destabilization. The possibility of a surveillance-mediated effect might be supported by the fact that, in rec8Δ, appearance of axis gaps at zygotene is accompanied by increase of chromatin diffuseness, with elimination of recombination substantially reducing both defects. Also, in budding yeast, when cells arrested at mid-prophase by a recombination defect are returned to mitotic growth conditions, one of the first responses is elimination of SC components (e.g., Zenvirth et al. 1997) in accord with axis destabilization as a potential target for a surveillance response. On the other hand, it is notable that there is an intrinsic tendency for chromatin diffuseness and axis destabilization even in the absence of recombination, as seen by defect in chromatin compactness and occasional occurrence of Spo76 gaps in spo11Δ rec8Δ nuclei. If destabilization were exclusively a checkpoint response to stalled recombinational progression, this phenotype would not be expected. In contrast, such an effect is easy to understand if local axis destabilization were a normal intrinsic feature of wild-type meiosis with recombinosome/axis association enhancing the degree of axis destabilization and targeting that destabilization to appropriate specific positions.
Proposed biological significance of CO-directed local axis destabilization
During meiosis, occurrence of exchange at the DNA/chromatin level must be accompanied by an analogous exchange at the structural level between the underlying non-sister chromatid axes at the corresponding positions. Exchange is apparent from images of chiasmata in diplotene chromosomes, which further show that sister chromatid axes are locally separated at these sites (see picture in Jones and Franklin 2006). Indeed, certain axial structural components, including cohesins, are already known to be specifically absent from chiasmata sites; e.g., Rec8 in mouse (Eijpe et al. 2003) and MCD1/SCC1/RAD21 plus SMC3 in grasshoppers (Valdeolmillos et al. 2007). Occurrence of axis changes being specific to sites of COs, it would be sensible for these local axis changes to begin at the time of CO designation, thus much earlier than when observed at chiasmata sites during diplotene, and then to proceed in concert with the DNA events of recombination (e.g., Blat et al. 2002). We therefore propose that the Spo76-GFP axis gaps observed in rec8Δ and the local separation of sister LEs seen in wild type are both manifestations of the onset of axis exchange at the sites of developing CO recombinational interactions. The idea of axis destabilization concomitant with CO designation at leptotene/zygotene as a general feature of meiosis is further supported by the fact that in Sordaria humana, which has tubular AEs, appearance of CO-correlated recombination nodules at zygotene/pachytene is accompanied by a spatially correlated change in the morphology of axes (bulges) around the site of the nodule, implying a local CO-directed change in axis status (Zickler and Sage 1981).
The proposed model is also attractive from another perspective. A notable feature of recombinosome-mediated axis destabilization is that it would occur at exactly the same time as nucleation of SC formation, which (also) occurs specifically at, and as a consequence of CO designation (for review, see Henderson and Keeney 2005). Thus, CO designation might trigger two apparently opposing local effects—i.e., both stabilization and destabilization—affecting chromosomes along and between sister and homolog axes. This possibility in turn suggests a new idea for a potential role of the SC: The SC, which is the stabilizing feature, might serve as a protective scaffolding, ensuring retention of basic chromosome organization structure across CO sites during crucial local changes involved in axis exchange, which are the destabilizing feature.
Molecular events producing the destabilization process remain to be determined. It is worth noting that the budding yeast Polo/Cdc5 kinase has been implicated in successful progression of CO formation and in the cohesin removal process through Rec8 phosphorylation (Clyne et al. 2003; Brar et al. 2006). These are the types of events that could be at the origin of the observed effects.
Rec8 and Spo76/Pds5 play both distinct and synergistic roles for basic chromosomal events
The current study emphasizes the pleiotropic and temporally modulated nature of REC8 and SPO76/PDS5 mutant defects, and thus further emphasizes how difficult it is to assign specific molecular roles to individual components of multifaceted complexes. Nonetheless, this analysis provides new information about the relationship between these two functions.
Spo76 and Rec8 clearly have distinguishable roles: They exhibit discernible differences in spatial and temporal axis staining patterns, and spo76-1 and rec8Δ single mutations confer similar, but not identical defects in many aspects of meiotic chromosome organization and function. Importantly, single mutant defects do not reflect interdependence of loading onto the chromosomes because, up until the leptotene/zygotene transition, Rec8 loading is normal in spo76-1 and Spo76 loading is normal in rec8Δ. The different patterns of destabilization observed in rec8Δ and spo76-1 mutants following recombination initiation raise the possibility that Rec8 may be devoted primarily to maintenance/modulation of axis status locally at sites of COs, while Spo76/Pds5 may be devoted primarily to maintenance of axis status between these sites. In both cases, destabilization is recombination-dependent, as seen in rec8Δ spo76-1 spo11Δ triple mutant (above) (Storlazzi et al. 2003). Therefore, the two proteins have distinct and specific roles, even as they remain involved in common processes.
The current study further reveals that the rec8Δ and spo76-1 mutations have dramatically synergistic effects when both are present. This phenotypic synergy could suggest that the two proteins have partially overlapping roles in the same basic process; e.g., sister chromatid cohesion. However, an equally alternative possibility is that the two proteins have distinct roles in processes that themselves are interacting; e.g., cohesion and DNA metabolism in general for Spo76, and CO-designated interactions specifically in meiosis for Rec8.
In this regard, it is further notable that (1) contrary to Rec8, Spo76/Pds5/BimD plays also important roles for mitotic chromosomes (e.g., van Heemst et al. 1999, 2001; Wang et al. 2003; Zhang et al. 2005, and references therein; Ding et al. 2006); (2) Pds5 is known to interact physically with Mcd1/Scc1/Rad21 (Losada et al. 2005 and references therein); and (3) Rec8’s mitotic counterpart Mcd1/Rad21 plays roles for meiosis in mouse and budding yeast (e.g., Parra et al. 2004; Xu et al. 2004; Kateneva et al. 2005). Given these considerations, as well as the fact that Rec8 is not the primary mediator of cohesion (above), the synergistic effects of rec8Δ and spo76-1 mutations could reflect a fundamental functional collaboration between meiosis-specific and general cohesion components. Rec8 and Mcd1/Scc1, associated or not with Spo76/Pds5, may have both overlapping and distinct functions during prophase through the diffuse stage, just as they are known to have at the meiosis I division (e.g., Yokobayashi et al. 2003 for fission yeast). By this view, the synergistic effects of rec8Δ and spo76-1 mutations for prophase events could be explained by synergistic functionality of the “meiotic” and “mitotic” kleisin homologs.
The nature of the pachytene-to-diplotene transition and implications for chromosome structure and the maternal age effect in human female
In human female meiosis, oocytes decades-long arrest occurs at the “dictyate” stage. At this stage, as during the diffuse stage of the standard meiotic program, the chromosomes are no longer visible by any of the known staining methods (above; see Garcia et al. 1987 for human meiosis). Further, the nearly universal diffuse stage is more transient but, like the dictyate stage, occurs between two stages in which chromosomes are clearly visible: pachytene (diffuse stage) or prediplotene (dictyate stage) and diplotene where homologs are condensed. Thus, the same overall change in chromosome organization must occur in both cases: It involves not only a diminution of interhomolog relationships (loss of the SC) but also loss of intersister relationships, which is accompanied at the molecular level by a concomitant decrease of cohesins (e.g., Prieto et al. 2004; Hodges et al. 2005 for mammalian oocytes). Thus, the dictyate stage is a stage at which there is a transition from robust to less-robust sister chromatid cohesion and therefore a period when sister connections are specifically “at risk.”
The current and previous studies (van Heemst et al. 1999) show that two cohesion molecules, Rec8 and Spo76, are both strongly delocalized during the diffuse stage. Moreover, this stage is also particularly sensitive to lack or defect in these molecules. Unlike previous stages, where mutations of REC8 or SPO76 confer only partial destabilization of sister connections, both proteins are absolutely required through the diffuse stage as shown by complete loss of both arm and centric cohesion in each of the corresponding single mutants. Taken together, these considerations suggest that there is an intrinsic tendency for destabilization of sister cohesion during passage through the diffuse stage, and that constraint of this intrinsic tendency requires both Spo76/Pds5 and Rec8.
In human female, the proportion of aneuploid meiotic products increases dramatically with age (e.g., Hassold and Hunt 2001; Pellestor et al. 2003) and the age-dependent aspect of meiosis is decades-long arrest at the dictyate stage, implying that aneuploidy results from progressive accumulation of some defect during this period. Age-related aneuploidies do exhibit aberrant CO patterns. However, formation of COs per se cannot be the sensitive process, because COs form at pachytene (Guillon and de Massy 2002), thus before the dictyate stage. Loss of sister cohesion, on the other hand, has been proposed to be a key determinant of age-related aneuploidies, with altered CO patterns among aneuploid products reflecting functional interplay between cohesion and CO patterns (see Hodges et al. 2005 and references therein). This possibility is attractive from two perspectives not previously considered. First, given that sister cohesion is intrinsically destabilized during the dictyate stage, it would make sense that even minor defects in cohesins and/or associated proteins would amplify the normally occurring process. Second, Rec8 is identified here as being destabilized during the pachytene-to-diplotene transition and, moreover, is shown to be a key meiosis-specific component for ensuring that destabilization of sister cohesion is properly constrained and channeled along arms and in centric regions. Minor defects in modulation of Rec8 activity, along with that of another meiotic-specific cohesin SMC1β (Hodges et al. 2005) would, therefore, be reasonable candidates to be the molecular basis of the maternal age effect. Other attractive molecular targets would be Sgo proteins and their regulators (e.g., PP2A), which appear to be specifically crucial for reductional versus equational functioning of centromeres and the response of kinetochores to microtubule-mediated tension (for review, see Lee et al. 2005).
Material and methods
Cloning and sequencing
REC8 (GenBank accession no. EF194064) was identified by PCR from a Sordaria macrospora indexed genomic library (Pöggeler et al. 1997). Primers design was based on Neurospora crassa genome (http://www.genome.wi.mit.edu). REC8 was sequenced on both strands using gene-specific primers (Sigma-Genosys or Primm).
Strains, plasmids, and transformation of Sordaria
The homothallic S. macrospora wild-type strain is “St Ismier” strain FGSC 4818. In rec8Δ, a hygromycin resistance cassette replaces the REC8 coding region (exactly from ATG to the stop codon). Transformants carrying a deleted allele were selected for hygromycin resistance and confirmed by Southern blotting and PCR.
In all analyzed GFP fusions (REC8, RAD51, and MSH4) the GFP coding sequence (p-EGFP-1, Clontech) was fused just after the last C-terminal amino acid predicted from the ORF. For the spo76-1 allele, GFP is inserted exactly as in the wild-type SPO76-GFP fusion (van Heemst et al. 1999). GFP alleles were ectopically integrated in wild type by selection for hygromycin resistance. For details concerning complementation tests of Rec8-GFP and Msh4-GFP, see the Supplemental Material. In the absence of a rad51 mutant, we made parallel observations with the anti-Rad51 antibodies from Oncogene. The number of foci was identical with both methods (see the Supplemental Material). GFP fluorescence as shown in Figures 1, 2, 3, 4, 5, 6 was examined directly either on living cells or after fixation when double-staining with DAPI was necessary. The specificity of all patterns seen by GFP fluorescence in the EGFP-specific filter was confirmed with anti-GFP antibodies (Roche) on fixed material.
Cytology
Asci were processed for immunofluorescence as described (Thompson-Coffe and Zickler 1993). Primary antibodies were MPM-2 at 1:300, anti-Rad51 (Oncogene) at 1:400 and anti-GFP (Roche) at 1:500. Secondary antibodies were FITC anti-mouse and anti-rabbit or CyTM3 anti-rabbit (Jackson) at 1:100 and 1:4000. Chromatin was visualized with DAPI (0.5 μg/mL). Strains containing Rec8-GFP, Msh4-GFP, and Rad51-GFP were fixed in 4% paraformaldehyde and stained with DAPI (0.5 μg/mL). Imunofluorescence and GFP fluorescence were observed on a Zeiss Axioplan microscope with a CCD Princeton camera.
For EM, asci were fixed in 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 3 h, post-fixed in phosphate-buffered 2% osmium tetroxide for 1 h, and dehydrated through an alcohol series. Asci were embedded in Epon 812 for 24 h at 60°C. Serial sections were mounted on Formvar-coated single-hole grids and stained in aqueous uranyl acetate for 30 min, followed by lead citrate for 10 min.
Acknowledgments
This work, A.S., S.T., and G.R.-R. were supported by grants from the Centre National de la Recherche Scientifique (UMR 8621) to D.Z. and from the National Institutes of Health (GM25326) to N.K. S.G. was supported by Regione campania, Legge 28/03/2002 n.5.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.459308.
References
- Bannister L.A., Reinholdt L.G., Munroe R.J., Schimenti J.C. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Blat U., Protacio R.U., Hunter N., Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:1–12. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Borde V., Goldman A.S., Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Brar G.A., Kiburz B.M., Zhang Y., Kim J.E., White F., Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Cai X., Dong F., Edelmann R.E., Makaroff C.A. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 2003;116:2999–3007. doi: 10.1242/jcs.00601. [DOI] [PubMed] [Google Scholar]
- Calvente A., Viera A., Page J., Parra M.T., Gomez R., Suja J.A., Rufas J.S., Santos J.L. DNA double-strand breaks and homology search: Inferences from a species with incomplete pairing and synapsis. J. Cell Sci. 2005;118:2957–2963. doi: 10.1242/jcs.02391. [DOI] [PubMed] [Google Scholar]
- Cha R.S., Weiner B.M., Keeney S., Dekker J., Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes & Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chan R.C., Chan A., Jeon M., Wu T.F., Pasqualone D., Rougvie A.E., Meyer B.J. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423:1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., Belcram K., Rocques N., Marquez-Lema A., Bhatt A.M., Horlow C., et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Sakurai N., Katou Y., Itoh T., Shirahige K., Haraguchi T., Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C., Smith G.R. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Dietrich A.J.J., Freixa L., Vink A.C.G., Ponsa M., Egozcue J. Development of the first meiotic prophase stages in human fetal oocytes observed by light and electron microscopy. Hum. Genet. 1987;77:223–232. doi: 10.1007/BF00284474. [DOI] [PubMed] [Google Scholar]
- Guillon H., de Massy B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet. 2002;32:296–299. doi: 10.1038/ng990. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Henderson K.A., Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K.A., Keeney S. Synaptonemal complex formation: Where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C., Jones G.H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes & Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C.A., Revenkova E., Jessberger R., Hassold T.J., Hunt P.A. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat. Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination. In: Aguilera A., Rothstein R., editors. Homologous recombination, topics in current genetics. Springer-Verlag; Heidelberg, Germany: 2006. pp. 1–62. [Google Scholar]
- Jones G.H., Franklin C.H. Meiotic crossing-over: Obligation and interference. Cell. 2006;126:246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kateneva A.V., Konovchenko A.A., Guacci V., Dresser M.E. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J. Cell Biol. 2005;171:241–253. doi: 10.1083/jcb.200505020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Hayashi-Hagihara A., Orr-Weaver T.L. Roles and regulation of the Drosophila centromere cohesion protein MEI-S332 family. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:543–552. doi: 10.1098/rstb.2005.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Yokochi T., Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Moens P.B., Marcon E., Shore J.S., Kochakpour N., Spyropoulos B. Initiation and resolution of interhomolog connections: Crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- Murakami H., Borde V., Shibata T., Lichten M., Ohta K. Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res. 2003;31:4085–4090. doi: 10.1093/nar/gkg441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Oliver-Bonet M., Turek P.J., Sun F., Ko E., Martin R.H. Temporal progression of recombination in human males. Mol. Hum. Reprod. 2005;11:517–522. doi: 10.1093/molehr/gah193. [DOI] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Parra M.T., Viera A., Gomez R., Page J., Benavente R., Santos J.L., Rufas J.S., Suja J.A. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 2004;117:1221–1234. doi: 10.1242/jcs.00947. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F., Andreo B., Arnal F., Humeau C., Demaille J. Maternal aging and chromosomal abnormalities: New data drawn from in vitro unfertilized human oocytes. Hum. Genet. 2003;112:195–203. doi: 10.1007/s00439-002-0852-x. [DOI] [PubMed] [Google Scholar]
- Pöggeler S., Nowrousian M., Jacobsen S., Kuck U. An efficient procedure to isolate fungal genes from an indexed cosmid library. J. Microbiol. Methods. 1997;29:49–61. [Google Scholar]
- Prieto I., Tease C., Pezzi N., Buesa J.M., Ortega S., Kremer L., Martinez A., Martinez-A C., Hulten M.A., Barbero J.L. Cohesin component dynamics during meiotic prophase I in mammalian oocytes. Chromosome Res. 2004;12:197–213. doi: 10.1023/b:chro.0000021945.83198.0e. [DOI] [PubMed] [Google Scholar]
- Revenkova E., Jessberger R. Keeping sister chromatids together: Cohesins in meiosis. Reproduction. 2005;130:783–790. doi: 10.1530/rep.1.00864. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Tesse S., Gargano S., James F., Kleckner N., Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes & Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S., Storlazzi A., Kleckner N., Gargano S., Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Coffe C., Zickler D. Cytoskeletal interactions in the ascus development and sporulation of Sordaria macrospora. J. Cell Sci. 1993;104:883–898. [Google Scholar]
- Trelles-Sticken E., Adelfalk C., Loidl J., Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdeolmillos A.M., Viera A., Page J., Prieto I., Santos J.L., Parra M.T., Heck M.M., Martinez-A C., Barbero J.L., Suja J.A., et al. Sequential loading of cohesin subunits during the first meiotic prophase of grasshoppers. PLoS Genet. 2007;3:e28. doi: 10.1371/journal.pgen.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D., James F., Pöggeler S., Berteaux-Lecellier V., Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- van Heemst D., Käfer E., John T., Heyting C., van Aalderen M., Zickler D. BimD/Spo76 is at the interface between cell cycle progression, chromosome morphogenesis and recombination. Proc. Natl. Acad. Sci. 2001;98:6267–6272. doi: 10.1073/pnas.081088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yoder J., Antoshechkin I., Han M. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol. Cell. Biol. 2003;23:7698–7707. doi: 10.1128/MCB.23.21.7698-7707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Xu H., Beasley M., Verschoor S., Inselman A., Handel M.A., McKay M.J. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–384. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M.D., Warren W.D., van der Horst G.T., McKay M.J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S., Yamamoto M., Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D., Loidl J., Klein S., Arbel A., Shemesh R., Simchen G. Switching yeast from meiosis to mitosis: Double-strand break repair, recombination and synaptonemal complex. Genes Cells. 1997;2:487–498. doi: 10.1046/j.1365-2443.1997.1370335.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ren Q., Yang H., Conrad M.N., Guacci V., Kateneva A., Dresser M.E. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol. Microbiol. 2005;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- Zickler D., Sage J. Synaptonemal complexes with modified lateral elements in Sordaria humana: Development of and relationship to the ‘recombination nodules.’. Chromosoma. 1981;84:305–318. [Google Scholar]
- Zickler D., Moreau P.J., Huynh A.D., Slezec A.M. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]