Abstract
Survivin is a member of the inhibitor of apoptosis protein family. Survivin has splice variants with different biological functions associated with tumorigenesis. We investigated 134 non-small cell lung cancers (NSCLCs) to study the clinical significance of wild-type survivin, survivin-2B, and survivin-deltaEx3. Real-time PCR analyses were performed for their gene expressions. The subcellular localisation of survivin proteins was evaluated by immunohistochemistry. The Ki-67 proliferation index and the apoptotic index were also evaluated. The survivin-deltaEx3 gene expression was significantly higher in stage II–III than in stage I (P=0.0174), and significantly correlated with the nuclear pan-survivin expression (P<0.0001). The Ki-67 index was significantly higher in wild-type survivin-positive tumours (P<0.0001), survivin-deltaEx3-positive tumours (P<0.0001), and tumours with positive expression of the nuclear pan-survivin (P=0.0047). In contrast, the apoptotic index was significantly lower only in wild-type survivin-positive tumours (P<0.0001). Thus, the wild-type survivin gene expression was associated with apoptotic inhibition and tumour proliferation. Furthermore, the survivin-deltaEx3 gene expression was strongly associated with tumour proliferation, especially in advanced stage NSCLCs. In contrast, the survivin-2B gene expression did not correlate with tumour proliferation or tumour apoptosis.
Keywords: survivin, survivin-deltaEx3, nuclear survivin, proliferation, apoptosis, lung cancer
Non-small cell lung cancer (NSCLC) has become the leading cause of cancer death in Japan. Clarifying the mechanism of tumour biology is considered to be important for improving the clinical outcome of advanced NSCLC patients. Owing to recent developments in molecular biology, many molecular markers have proven to be associated with cell cycle regulation, apoptosis, and chemo-radio resistance. These molecular markers include survivin (Altieri, 2003), p53 (Vogelstein and Kinzler, 1992), and bcl-2 (Strasser et al, 1997).
Survivin was originally identified as a member of the inhibitor of apoptosis protein (IAP) family (Ambrosini et al, 1997). To date, survivin has at least five splice variants; including wild-type survivin (i.e. survivin itself), survivin-2α, survivin-2B, survivin-deltaEx3, and survivin-3B (Mahotka et al, 1999; Badran et al, 2004; Caldas et al, 2005a). Intriguingly, these splice variants are reported to have different biological functions (Mahotka et al, 2002a). Among them, recent studies found both wild-type survivin and survivin-deltaEx3 to demonstrate not only apoptotic inhibition but also an acceleration of cell proliferation (Honda et al, 2003; Song and Wu, 2005; Takashima et al, 2005). In contrast, survivin-2B is considered to attenuate the antiapoptotic function of wild-type survivin (Caldas et al, 2005b).
The ability to control tumour proliferation and apoptosis may enable us to develop new strategies for the treatments of NSCLC patients. Therefore, we performed a study on the gene expressions of wild-type survivin, survivin-2B, and survivin-deltaEx3 in relation to the subcellular localisation of survivin protein, the Ki-67 proliferation index (Gerdes et al, 1984), and the apoptotic index (Tanaka et al, 2001) to clarify the clinical significance of these expressions in NSCLCs.
MATERIALS AND METHODS
Clinical characteristics of patients
From June 1999 to December 2002, NSCLC patients, who underwent surgery at the Second Department of Surgery, Kagawa University or the Department of Thoracic Surgery, Japanese Red Cross Society Wakayama Medical Center, were studied. This study was approved by the institutional review board of Kagawa University (14-7, a clinical study of biological markers in NSCLCs). Signed, written informed consent was obtained from all patients before therapy was initiated. In total, 134 patients up to stage IIIB were investigated. The patients' clinical records and histopathological diagnoses were fully documented.
Real-time PCR for gene expressions
The total cellular RNA was extracted from frozen tissue specimens by the acid guanidinium thiocyanate procedure. First-strand cDNA synthesis was performed with 5 μg of total RNA using a cDNA synthesis kit (Pharmacia, Piscataway, NJ, USA). Because real-time PCR (RT–PCR) is not an appropriate method to discriminate between wild-type survivin and other splice variants, we carried out quantitative RT–PCR assays using densitometric analyses of gel electrophoresis to evaluate the gene expressions of wild-type survivin, survivin-2B, and survivin-deltaEx3. For the gene expressions of wild-type survivin, survivin-2B, and survivin-deltaEx3, oligonucleotides were synthesised as PCR primers based on the published reports (Yamada et al, 2003). The nucleotide sequence 5′-CCACCGCATCTCTACATTCA-3′ was used as the sense primer of wild-type survivin and survivin-deltaEx3, 5′-GAGGCTGGCTTCATCCACTG-3′ was used as the sense primer of survivin-2B, 5′-TATGTTCCTCTATGGGGTCG-3′ as the antisense primer of wild-type survivin, 5′-GTTCCTCTCTCGTGATCCG-3′ as the antisense primer of survivin-2B, and 5′-TTTCCTTTGCATGGGGTC-3′ as the antisense primer of survivin-deltaEx3. All subsequent assays were carried out using the parameters that yielded amplification of wild-type survivin, survivin-2B, survivin-deltaEx3, and β-actin DNA (the internal control) within a linear range. The reaction mixture of wild-type survivin or survivin-2B was subjected to 36 PCR amplification cycles of 60 s at 94°C, 60 s at 60°C, and 90 s at 72°C, and that of survivin-deltaEx3 was subjected to 36 PCR amplification cycles of 60 s at 94°C, 60 s at 55°C, and 90 s at 72°C, and that of β-actin was subjected to 31 PCR amplification cycles of 60 s at 94°C, 60 s at 60°C, and 90 s at 72°C. Preparations of a human adenocarcinoma cell line, A549, were used as positive controls for gene expressions of wild-type survivin, survivin-2B, and survivin-deltaEx3. The amplified DNA samples were run on a 1% agarose gel, and the bands were visualised with ethidium bromide. The densitometric value obtained for a wild-type survivin, survivin-2B, and survivin-deltaEx3 band in a given tumour sample was divided by the value of the β-actin, and the resultant ratio was referred to as the gene expression ratio. Thereafter, the expression ratio for a given tumour sample was divided by the expression ratio of A549 to obtain the standardised gene expression ratio.
Immunohistochemistry
We used a mouse monoclonal antibody against pan-survivin (sc17779; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) diluted at 1 : 50, and a mouse monoclonal antibody for the Ki-67 antigen (MIB-1; DAKO, Glostrup, Denmark) diluted at 1 : 40. Formalin-fixed paraffin-embedded tissue was cut into 4-μm sections and mounted on poly-L-lysine-coated slides. Sections were deparaffinised and rehydrated. The slides were then heated in a microwave for 10 min in a 10 μmol l−1 citrate buffer solution at pH 6.0. After quenching the endogenous peroxidase activity with 0.3% H2O2 (in absolute methanol) for 30 min, the sections were treated for 2 h with 5% bovine serum albumin to block nonspecific staining. Duplicated sections were incubated overnight with primary antibodies, respectively. Slides were then incubated for 1 h with biotinylated secondary antibodies (Vector Laboratories Inc., Burlingame, CA, USA). The sections were incubated with the avidin–biotin–peroxidase complex (Vector) for 1 h, and antibody binding was visualised with 3,3′-diaminobenzidine tetrahydrochloride. Finally, the sections were lightly counterstained with Mayer's haematoxylin.
All immunostained sections were independently evaluated by two authors (JN and MU), without knowledge of the patients' characteristics. At least 200 tumour cells were scored per × 40 field. Regarding pan-survivin expression, the nuclear staining and the cytoplasmic staining in each section were evaluated, respectively, in a semiquantitative manner that reflected both the intensity and percentage of cells staining at each intensity. The intensity was classified as 0 (no staining), +1 (weak staining), +2 (distinct staining), or +3 (very strong staining). A value designated as the ‘HSCORE’ thus was obtained for each slide by using the following algorithm: HSCORE=Σ (I × PC), where I and PC represent the intensity and the percentage of cells that stain at each intensity, respectively. The percentage of carcinoma cells with positive staining for Ki-67 antigen was scored as the Ki-67 proliferation index.
Detection of apoptosis
The presence of apoptotic cells was detected with the TUNEL method using the In Situ Apoptosis Detection Kit (Takara Biomedicals, Otsu, Japan). After sections were deparaffinised and rehydrated, the slides were treated for 15 min with 20 μg ml−1 Proteinase K. After quenching the endogenous peroxidase activity with 3% H2O2 for 5 min, the sections were then incubated for 90 min at 37°C with the TUNEL reaction mixture, including terminal deoxynucleotidyl transferase (TdT). Next, the sections were incubated for 30 min at 37°C with anti-FITC horseradish peroxidase conjugate. Staining was developed using 3,3′-diaminobenzidine tetrahydrochloride for 15 min. Finally, the sections were lightly counterstained with Mayer's haematoxylin. Sections incubated with the TUNEL reaction mixture without TdT were used as negative control slides. Apoptotic cells were determined based on observations of TUNEL-staining sections and serial HE-staining sections. Cells staining positive for TUNEL, if they represented the histologic features of necrosis in HE-staining sections, were not considered to be apoptotic cells. In each case, a total of 10 000 tumour cells were evaluated at high magnification by two authors (JN and MU) independently, without knowledge of the patients' characteristics. The apoptotic index was defined as the number of apoptotic cells per 1000 tumour cells.
Statistical analysis
The ratio of the Ki-67 proliferation index divided by the apoptotic index was referred to as the growth index for each sample (Tsoli et al, 2002). The statistical significances of wild-type survivin, survivin-2B, and survivin-deltaEx3 gene expression in relation to several clinical and pathological parameters were assessed by t-test or an analysis of variance with Bonferroni/Dunn test. Because the wild-type survivin expression cutoff line of 1.0 demonstrated the most significance in relation to the Ki-67 proliferation index and the apoptotic index, the sample was classified as a wild-type survivin-positive tumour when the standardised wild-type survivin gene expression was >1.0. Because the survivin-deltaEx3 expression cutoff line of 0.5 demonstrated the most significance in relation to the Ki-67 proliferation index, the sample was classified as a survivin-deltaEx3-positive tumour when the standardised survivin-deltaEx3 gene expression was >0.5. The sample was classified as a survivin-2B-positive tumour when the standardised survivin-2B expression was >1.0 (a median value of survivin-2B gene expression). Samples were considered to have a positive expression of nuclear pan-survivin if the HSCORE of the nuclear staining of pan-survivin was more than 10, a positive expression of cytoplasmic pan-survivin if the HSCORE of the cytoplasmic staining of pan-survivin was more than 50. All P-values were based on two-tailed statistical analysis and a P-value of <0.05 was considered to indicate statistical significance.
RESULTS
Wild-type survivin gene expression in NSCLCs
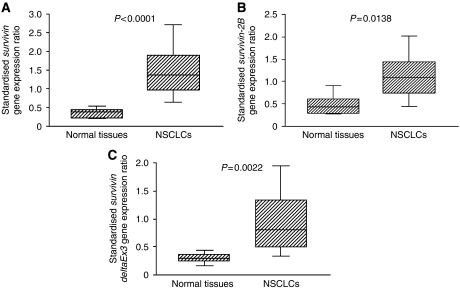
We studied the wild-type survivin gene expression in eight noncancerous samples to evaluate its expression in normal lung tissue. The standardised wild-type survivin gene expression ratio was low in normal lung tissue specimens (mean 0.363±0.135). In contrast, the standardised survivin gene expression ratio varied greatly among the 134 tumour tissue specimens (mean 1.531±0.795, Figure 1A). The standardised wild-type survivin gene expression ratio was significantly higher in tumour tissue specimens than in normal lung tissue specimens (P<0.0001, Figure 2A). Seventy-two carcinomas (53.7%) were wild-type survivin-positive tumours. Regarding tumour histology, the wild-type survivin gene expression was significantly higher in squamous cell carcinomas than in adenocarcinomas (1.690±0.892 vs 1.362±0.664, P=0.0181, Table 1). In addition, the wild-type survivin gene expression was significantly higher in moderately or poorly differentiated tumours than in well-differentiated tumours (1.614±0.807 vs 1.288±0.712, P=0.0383).
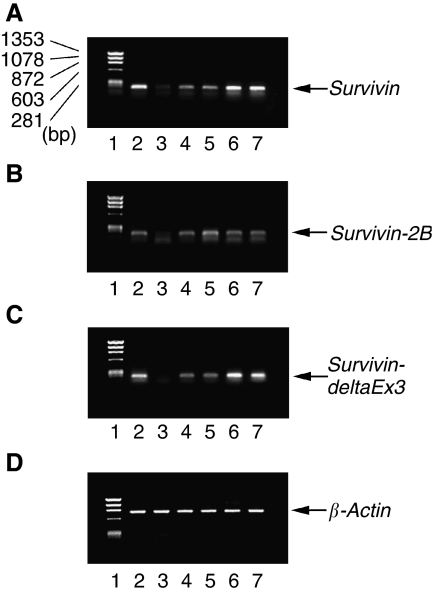
Figure 1.
Agarose gel electrophoresis of RT–PCR-amplified (A) wild-type survivin cDNA, (B) survivin-2B cDNA, (C) survivin-deltaEx3 cDNA, and (D) β-actin cDNA (internal PCR control). Lane 1, size marker; lane 2, A549 (positive control); lane 3, normal lung tissue; lanes 4 and 5, low expression of both wild-type survivin and survivin-deltaEx3; lanes 6 and 7, high expression of both wild-type survivin and survivin-deltaEx3.RT–PCR=real-time PCR.
Figure 2.
Standardised gene expression ratio in normal lung tissues and tumour tissues. (A) Wild-type survivin, (B) survivin-2B, and (C) survivin-deltaEx3.
Table 1. Gene expression of wild-type survivin, survivin-2B, and survivin-deltaEx3 in 134 NSCLCs according to clinical characteristics.
|
Wild-type survivin
|
Survivin-2B
|
Survivin-deltaEx3
|
|||||
|---|---|---|---|---|---|---|---|
| Variables | n | Standardised gene expression ratio | P-value | Standardised gene expression ratio | P-value | Standardised gene expression ratio | P-value |
| Tumour status | |||||||
| T1 | 46 | 1.407±0.786 | 0.3145 | 1.472±1.094 | 0.6335 | 0.922±0.614 | 0.3219 |
| T2 | 48 | 1.475±0.698 | 0.985±0.515 | 0.884±0.526 | |||
| T3 | 22 | 1.817±0.885 | 1.345±0.747 | 1.189±0.720 | |||
| T4 | 18 | 1.646±0.905 | 1.144±0.681 | 1.063±0.638 | |||
| Nodal status | |||||||
| N0 | 86 | 1.459±0.835 | 0.1613 | 1.265±0.748 | 0.5628 | 0.902±0.564 | 0.0781 |
| N1, N2, N3 | 48 | 1.660±0.707 | 1.175±0.914 | 1.096±0.673 | |||
| Pathological stage | |||||||
| Stage I | 60 | 1.385±0.819 | 0.0558 | 1.281±0.796 | 0.5626 | 0.833±0.512 | 0.0174 |
| Stage II, III | 74 | 1.649±0.759 | 1.194±0.860 | 1.084±0.661 | |||
| Differentiation | |||||||
| Well | 34 | 1.288±0.712 | 0.0383 | 1.318±0.832 | 0.4964 | 0.743±0.526 | 0.0110 |
| Moderately or poorly | 100 | 1.614±0.807 | 1.203±0.832 | 1.049±0.619 | |||
| Histology | |||||||
| Adenocarcinoma | 66 | 1.362±0.664 | 0.0181a | 1.192±0.736 | 0.5191a | 0.854±0.587 | 0.0377a |
| Squamous cell carcinoma | 65 | 1.690±0.892 | 1.285±0.923 | 1.075±0.616 | |||
| Large cell carcinoma | 3 | 1.780±0.570 | 0.850±0.507 | 1.321±0.620 | |||
| Total | 134 | 1.531±0.795 | 1.233±0.830 | 0.971±0.610 | |||
Abbreviation: NSCLCs=non-small cell lung cancers.
Squamous cell carcinoma vs adenocarcinoma.
Survivin-2B gene expression in NSCLCs
The standardised survivin-2B gene expression ratio was low in eight normal lung tissue specimens (mean 0.496±0.246). In contrast, the standardised survivin-2B gene expression ratio also varied greatly among the tumour tissue specimens (mean 1.233±0.830, Figure 1B). The standardised survivin-2B gene expression ratio was significantly higher in tumour tissues than in normal lung tissues (P=0.0138, Figure 2B). However, no difference was observed in the survivin-2B gene expression regarding tumour histology, tumour differentiation, or any other clinicopathological parameter (Table 1).
Survivin-deltaEx3 gene expression in NSCLCs
The standardised survivin-deltaEx3 gene expression ratio was low in eight normal lung tissue specimens (mean 0.296±0.098). In contrast, the standardised survivin-deltaEx3 gene expression ratio varied greatly among the tumour tissue specimens (mean 0.971±0.610, Figure 1C). The standardised survivin-deltaEx3 gene expression ratio was significantly higher in tumour tissues than in normal lung tissues (P=0.0022, Figure 2C). One hundred and one carcinomas (75.4%) were survivin-deltaEx3-positive tumours. Regarding tumour histology, the survivin-deltaEx3 gene expression was also significantly higher in squamous cell carcinomas than in adenocarcinomas (1.075±0.616 vs 0.854±0.587, P=0.0377, Table 1). Regarding tumour differentiation, the survivin-deltaEx3 gene expression was significantly higher in moderately or poorly differentiated tumours than in well-differentiated tumours (1.049±0.619 vs 0.743±0.526, P=0.0110). Regarding the clinicopathological characteristics, the survivin-deltaEx3 gene expression was significantly higher in stage II–III NSCLCs than in stage I NSCLCs (1.084±0.661 vs 0.833±0.512, P=0.0174).
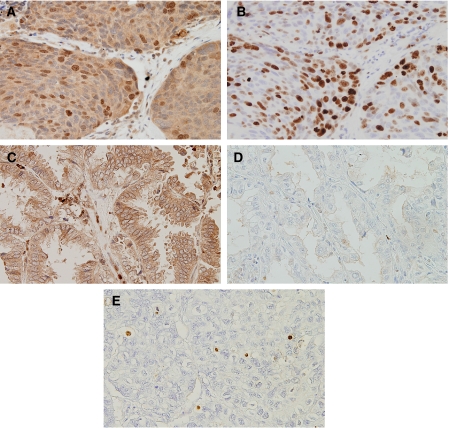
Subcellular localisation of the survivin protein expression in relation to wild-type survivin, survivin-2B, and survivin-deltaEx3 gene expressions. Immunostaining using the antibody against pan-survivin showed various patterns of cytoplasmic staining and nuclear staining (Figure 3A and C). Regarding the cytoplasmic staining, 98 carcinomas (73.1%) had a positive expression of cytoplasmic pan-survivin. However, the HSCORE of the cytoplasmic pan-survivin did not correlate with the gene expression of wild-type survivin, survivin-2B, or survivin-deltaEx3.
Figure 3.
Immunostaining of lung cancers. Squamous cell carcinoma with (A) strongly positive expression of both nuclear and cytoplasmic pan-survivin, (B) high Ki-67 index. Adenocarcinoma with only (C) positive expression of cytoplasmic pan-survivin, (D) low Ki-67 index. (E) Adenocarcinoma with high apoptotic index.
On the other hand, regarding the nuclear staining, 88 carcinomas (65.7%) had a positive expression of nuclear pan-survivin. Furthermore, the standardised survivin-deltaEx3 gene expression significantly correlated with the HSCORE for the nuclear staining of pan-survivin (r=0.642, P<0.0001). However, the HSCORE of the nuclear pan-survivin did not correlate with the gene expression of wild-type survivin or survivin-2B.
Ki-67 proliferation index in NSCLCs
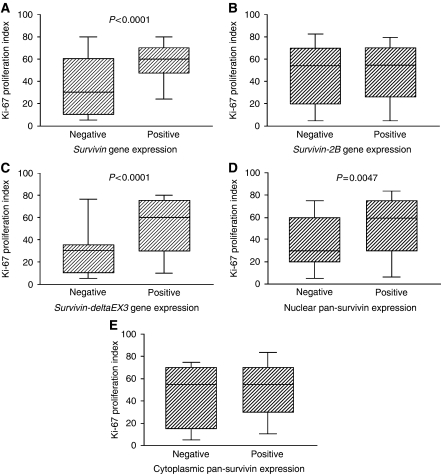
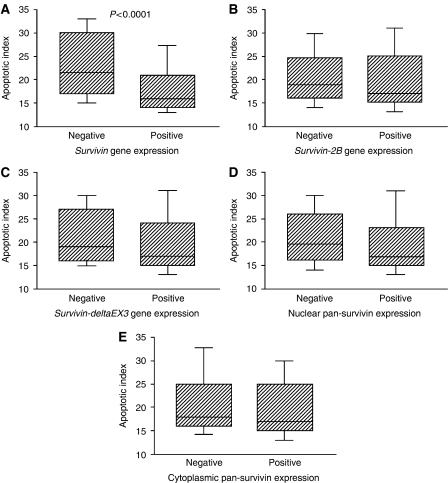
The Ki-67 proliferation index ranged from 2.0 to 95.0 (mean 47.6±27.3, Figure 3B and D). Regarding the gene expressions of splice variants of survivin, the Ki-67 proliferation index was significantly higher in the wild-type survivin-positive tumours than in the wild-type survivin-negative tumours (57.4±22.0 vs 36.2±28.5, P<0.0001, Figure 4A, Table 2). However, there was no difference in the Ki-67 proliferation index according to the survivin-2B gene expression (Figure 4B). In contrast, the Ki-67 proliferation index was also significantly higher in the survivin-deltaEx3-positive tumours than in the survivin-deltaEx3-negative tumours (53.4±25.5 vs 29.8±25.0, P<0.0001, Figure 4C). Regarding the subcellular localisation of pan-survivin protein, the Ki-67 proliferation index was significantly higher in tumours with a positive expression of nuclear pan-survivin than in tumours with a negative expression of nuclear pan-survivin (52.4±26.5 vs 38.5±26.6, P=0.0047, Figure 4D). In contrast, there was no difference in the Ki-67 proliferation index according to the cytoplasmic pan-survivin expression (Figure 4E).
Figure 4.
Ki-67 proliferation index in relation to wild-type survivin gene expression (A), survivin-2B gene expression (B), survivin-deltaEx3 gene expression (C), nuclear pan-survivin expression (D), and cytoplasmic pan-survivin expression (E).
Table 2. Biological behaviours in relation to gene expressions of survivin variants and survivin protein expressions.
| n | Ki-67 proliferation index | Apoptotic index | Growth index | ||||
|---|---|---|---|---|---|---|---|
| Wild-type survivin | |||||||
| Positive | 72 | 57.4±22.0 | P<0.0001 | 18.1±5.6 | P<0.0001 | 3.47±1.68 | P<0.0001 |
| Negative | 62 | 36.2±28.5 | 23.1±7.2 | 1.66±1.37 | |||
| Survivin-2B | |||||||
| Positive | 72 | 50.0±26.4 | P=0.1776 | 19.8±6.9 | P=0.3399 | 2.86±1.76 | P=0.0583 |
| Negative | 62 | 43.4±28.2 | 21.0±6.5 | 2.28±1.64 | |||
| Survivin-deltaEx3 | |||||||
| Positive | 101 | 53.4±25.5 | P<0.0001 | 20.0±6.7 | P=0.1872 | 3.01±1.76 | P<0.0001 |
| Negative | 33 | 29.8±25.0 | 21.8±7.1 | 1.48±1.33 | |||
| Nuclear survivin | |||||||
| Positive | 88 | 52.4±26.5 | P=0.0047 | 19.9±7.0 | P=0.2662 | 3.02±1.88 | P=0.0004 |
| Negative | 46 | 38.5±26.6 | 21.3±6.5 | 1.90±1.33 | |||
| Cytoplasmic survivin | |||||||
| Positive | 98 | 48.3±26.9 | P=0.6075 | 20.1±6.7 | P=0.4464 | 2.70±1.81 | P=0.4805 |
| Negative | 36 | 45.6±28.7 | 21.1±7.2 | 2.45±1.74 | |||
Apoptotic index in NSCLCs
The apoptotic index ranged from 10.0 to 40.0 (mean 20.7±7.4, Figure 3E). Regarding the gene expressions of splice variants of survivin, the apoptotic index was significantly lower in the wild-type survivin-positive tumours than in the wild-type survivin-negative tumours (18.1±5.6 vs 23.1±7.2, P<0.0001, Figure 5A). On the other hand, there was no difference in the apoptotic index according to the survivin-2B expression or survivin-deltaEx3 expression (Figure 5B and C). Regarding the pan-survivin protein expression, neither the nuclear pan-survivin expression nor the cytoplasmic pan-survivin expression was associated with the apoptotic index (Figure 5D and E).
Figure 5.
Apoptotic index in relation to wild-type survivin gene expression (A), survivin-2B gene expression (B), survivin-deltaEx3 gene expression (C), nuclear pan-survivin expression (D), and cytoplasmic pan-survivin expression (E).
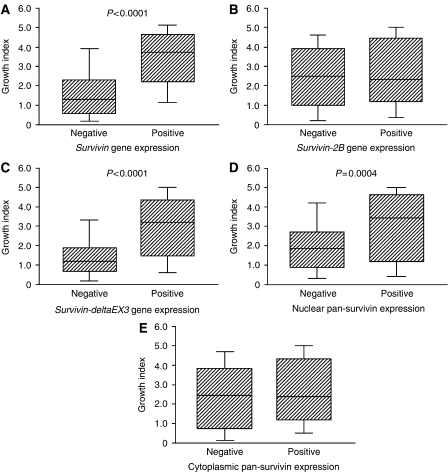
Growth index in NSCLCs
The growth index ranged from 0.05 to 9.00 (mean 2.62±1.79). The growth index was significantly higher in the wild-type survivin-positive tumours than in the wild-type survivin-negative tumours (3.47±1.68 vs 1.66±1.37, P<0.0001, Figure 6A). However, there was no difference in the growth index according to the survivin-2B gene expression (Figure 6B). In contrast, the growth index was also significantly higher in the survivin-deltaEx3-positive tumours than in the survivin-deltaEx3-negative tumours (3.01±1.76 vs 1.48±1.33, P<0.0001, Figure 6C). Regarding the subcellular localisation of pan-survivin protein, the growth index was significantly higher in tumours with a positive expression of nuclear pan-survivin than in tumours with a negative expression of nuclear pan-survivin (3.02±1.88 vs 1.90±1.33, P=0.0004, Figure 6D). In contrast, there was no difference in the growth index according to the cytoplasmic pan-survivin expression (Figure 6E).
Figure 6.
Growth index in relation to wild-type survivin gene expression (A), survivin-2B gene expression (B), survivin-deltaEx3 gene expression (C), nuclear pan-survivin expression (D), and cytoplasmic pan-survivin expression (E).
DISCUSSION
Survivin is reported to be expressed in most human malignancies (Kawasaki et al, 1998; Monzo et al, 1999; Sui et al, 2002), while its expression is absent in normal differentiated tissues (Salvesen and Duckett, 2002). Although survivin was initially identified as a member of the IAP family (Ambrosini et al, 1997), recent studies have demonstrated that survivin not only inhibits the caspase-dependent apoptotic pathway, but also accelerates cell proliferation (Honda et al, 2003).
Survivin is known to have at least five spliced variants. These splice variants have different biological functions (Mahotka et al, 2002a). Among them, recent experimental studies demonstrated that survivin-deltaEx3 also inhibits apoptosis and induces cell proliferation, similar to wild-type survivin (Song and Wu, 2005; Takashima et al, 2005). In contrast, survivin-2α and survivin-2B are considered to attenuate the antiapoptotic function of wild-type survivin (Caldas et al, 2005b).
As a result, these splice variants are variably involved in the biological functions associated with carcinogenesis, including cell proliferation, apoptosis, and chemo-radio resistance. Previous clinical studies have reported the survivin expression to be associated with tumour cell proliferation, apoptotic inhibition, and a poor prognosis in various human cancers, including NSCLCs (Martinez et al, 2004; Shinohara et al, 2005). However, only a few clinical reports studied these splice variants of survivin in human cancers (Mahotka et al, 2002a; Meng et al, 2004; Ling et al, 2005; Taubert et al, 2005). Therefore, to clarify the clinical significance of these splice variants of survivin in NSCLCs, we performed the present study on the gene expressions of wild-type survivin, survivin-2B, and survivin-deltaEx3 in relation to the subcellular localisation of survivin proteins, tumour cell proliferation, and apoptosis.
In the present study, regarding the relationship of the expression of these splice variants of survivin, the survivin-deltaEx3 gene expression significantly correlated with the wild-type survivin gene expression (r=0.646, P<0.001). However, the correlation between the survivin-2B gene expression and the wild-type survivin gene expression was low (r=0.254). Regarding the biological significance of these splice variants of survivin, the present study also demonstrated the wild-type survivin gene expression to be associated with both tumour cell proliferation and apoptotic inhibition. In contrast, the survivin-deltaEx3 gene expression correlated with the nuclear pan-survivin expression. Furthermore, both the survivin-deltaEx3 gene expression and the nuclear pan-survivin expression were found to be associated with tumour cell proliferation, but not with apoptotic inhibition. However, the survivin-2B gene expression did not correlate with tumour cell proliferation or apoptotic inhibition. In total, the growth index was associated with both the wild-type survivin expression and the survivin-deltaEx3 expression. As a result, the present study revealed wild-type survivin to be associated with both tumour cell proliferation and apoptotic inhibition, while survivin-deltaEx3 was strongly associated with tumour cell proliferation.
In fact, previous clinical studies also revealed the survivin-deltaEx3 expression to be associated with a high proliferation rate (Takashima et al, 2005). Recent studies have revealed that the survivin-deltaEx3 overexpression is a significant poor prognostic factor in cancer patients, including NSCLCs (Ling et al, 2005; Taubert et al, 2005). Furthermore, the present study found the survivin-deltaEx3 gene expression to be significantly higher in advanced-stage NSCLCs. Therefore, the survivin-deltaEx3 gene overexpression could produce more aggressive tumours with a high proliferation rate in advanced-stage NSCLCs. Because the biomarkers associated with tumour proliferation are significant factors in advanced-stage NSCLCs (Huang et al, 2005), the level of the survivin-deltaEx3 gene expression could thus affect the clinical impact of NSCLCs, especially in the advanced stages. A recent study on soft tissue sarcoma also reported the overexpression of survivin-deltaEx3 gene to correlate with an advanced clinical stage and a poor prognosis (Taubert et al, 2005).
On the other hand, previous immunohistochemical studies revealed the nuclear expression of survivin proteins also to be associated with tumour cell proliferation and a poor prognosis in cancer patients, as was also shown in the present study, while the cytoplasmic expression of survivin proteins was not (Martinez et al, 2004; Huang et al, 2005; Ling et al, 2005; Shinohara et al, 2005; Taubert et al, 2005). These studies were performed using antibodies against pan-survivin, which cannot distinguish wild-type survivin from other spliced variants. In general, wild-type survivin is considered to be located both in the cytoplasm for apoptotic inhibition, and in the nuclei for cell proliferation. In contrast, survivin-deltaEx3 is probably located in the nucleus because of its nuclear localisation signal (Mahotka et al, 2002b; Song and Wu, 2005). The present study also demonstrated the survivin-deltaEx3 gene expression to correlate with the nuclear pan-survivin expression. As a result of the above findings, the nuclear expression of pan-survivin could be the representation of both wild-type survivin and survivin-deltaEx3, both of which are associated with tumour proliferation. In contrast, the cytoplasmic expression of pan-survivin might indicate the presence of wild-type survivin and survivin-2B (Mahotka et al, 2002b). In fact, the present study revealed that the cytoplasmic pan-survivin expression was not associated with the apoptotic index. This result might be partly because cytoplasmic survivin-2B can inhibit the antiapoptotic function of wild-type survivin. We performed additional immunohistochemistry to clearly identify the subcellular localisation of survivin-deltaEx3 using a commercially available specific antibody against survivin-deltaEx3; however, we were not successful (data not shown).
As a result, the present study demonstrated the wild-type survivin gene expression to be associated with not only apoptotic inhibition but also tumour proliferation. In contrast, the survivin deltaEx3 gene expression significantly correlated with the nuclear expression of pan-survivin and the Ki-67 proliferation index. However, the survivin 2B gene expression did not correlate with tumour proliferation or tumour apoptosis. Considering these results, the combined evaluations of wild-type survivin and survivin-deltaEx3 gene expression may thus be a useful strategy for evaluating NSCLCs. The RNA inhibition of survivin can induce apoptosis in human tumour cell lines (Ambrosini et al, 1998; Olie et al, 2000). Therefore, the RNA inhibition designed against common regions of both the wild-type survivin and the survivin-deltaEx3 could be a more effective strategy for NSCLCs not only to improve the efficacy of chemo-radio therapy but also to suppress tumour cell proliferation (Li et al, 1998; Lu et al, 2004).
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Japanese Society for Promotion of Science to CH (18390379).
References
- Altieri DC (2003) Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 22: 8581–8589 [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 3: 917–921 [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Sirugo G, Altieri DC (1998) Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem 273: 11177–11182 [DOI] [PubMed] [Google Scholar]
- Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T, Inuzuka M (2004) Identification of a novel splice valiant of the human anti-apoptosis gene survivin. Biochem Biophys Res Commun 314: 902–907 [DOI] [PubMed] [Google Scholar]
- Caldas H, Honsey LE, Altura RA (2005a) Survivin 2α: a novel Survivin splice valiant expressed in human malignancies. Mol Cancer 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM, Altura RA (2005b) Survivin splice variants regulate the balance between proliferation and cell death. Oncogene 24: 1994–2007 [DOI] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H (1984) Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 133: 1710–1715 [PubMed] [Google Scholar]
- Honda R, Korner R, Nigg EA (2003) Exploring the functional interactions between aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 14: 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu D, Masuya D, Nakashima T, Kameyama K, Ishikawa S, Ueno M, Haba R, Yokomise H (2005) Clinical application of biological markers for treatments of resectable non-small-cell lung cancer. Br J Cancer 92: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N (1998) Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res 58: 5071–5074 [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC (1998) Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396: 580–584 [DOI] [PubMed] [Google Scholar]
- Ling X, Yang J, Tan D, Ramnath N, Younis T, Bundy BN, Slocum HK, Yang L, Zhou M, Li F (2005) Differential expression of survivin-2B and survivin-deltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC). Lung Cancer 49: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Mu Y, Cao C, Zeng F, Schneider S, Tan J, Price J, Chen J, Freeman M, Hallahan DE (2004) Survivin as a therapeutic target for radiation sensitization in lung cancer. Cancer Res 64: 2840–2845 [DOI] [PubMed] [Google Scholar]
- Mahotka C, Krieg T, Krieg A, Wenzel M, Suschek CV, Heydthausen M, Gabbert HE, Gerharz CD (2002a) Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer 100: 30–36 [DOI] [PubMed] [Google Scholar]
- Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, Gerharz CD (2002b) Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ 9: 1334–1342 [DOI] [PubMed] [Google Scholar]
- Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD (1999) Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res 59: 6097–6102 [PubMed] [Google Scholar]
- Martinez A, Bellosillo B, Bosch F, Ferrer A, Marce S, Villamor N, Ott G, Montserrat E, Campo E, Colomer D (2004) Nuclear survivin expression in mantle cell lymphoma is associated with cell proliferation and survival. Am J Pathol 164: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Lu CD, Sun YL, Dai DJ, Lee SW, Tanigawa N (2004) Expression level of wild-type survivin in gastric cancer is an independent predictor of survival. Wold J Gastroenterol 10: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo M, Rosell R, Felip E, Astudillo J, Sanchez JJ, Maestre J, Martin C, Font A, Barnadas A, Abad A (1999) A novel anti-apoptosis gene: Re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol 17: 2100–2104 [DOI] [PubMed] [Google Scholar]
- Olie RA, Simoes-Wust AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U (2000) A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res 60: 2805–2809 [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Shinohara ET, Gonzalez A, Massion PP, Chen H, Li M, Freyer AS, Olson SJ, Andersen JJ, Shyr Y, Carbone DP, Johnson DH, Hallahan DE, Lu B (2005) Nuclear survivin predicts recurrence and poor survival in patients with resected nonsmall cell lung carcinoma. Cancer 103: 1685–1692 [DOI] [PubMed] [Google Scholar]
- Song Z, Wu M (2005) Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene 24: 2723–2734 [DOI] [PubMed] [Google Scholar]
- Strasser A, Huang DC, Vaux DL (1997) The role of the bcl-2/ced-9 gene family in cancer and general implications of defects in cell death control for tumourigenesis and resistance to chemotherapy. Biochim Biophys Acta 1333: F151–F178 [DOI] [PubMed] [Google Scholar]
- Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tokuda M (2002) Survivin expression and its correlation with cell proliferation and prognosis in epithelial ovarian tumors. Int J Oncol 21: 315–320 [PubMed] [Google Scholar]
- Takashima H, Nakajima T, Moriguchi M, Sekoguchi S, Nishikawa T, Watanabe T, Katagishi T, Kimura H, Minami M, Itoh Y, Kagawa K, Okanoue T (2005) In vivo expression patterns of survivin and its splicing variants in chronic liver disease and hepatocellular carcinoma. Liver Int 25: 77–84 [DOI] [PubMed] [Google Scholar]
- Tanaka F, Otake Y, Yanagihara K, Yamada T, Miyahara R, Kawano Y, Li M, Inui K, Wada H (2001) Apoptosis and p53 status predict the efficacy of postoperative administration of UFT in non-small cell lung cancer. Br J Cancer 84: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert H, Kappler M, Bache M, Bartel F, Kohler T, Lautenschlager C, Blumke K, Wurl P, Schmidt H, Meye A, Hauptmann S (2005) Elevated expression of survivin-splice variants predicts a poor outcome for soft-tissue sarcomas patients. Oncogene 24: 5258–5261 [DOI] [PubMed] [Google Scholar]
- Tsoli E, Zacharatos P, Dasiou-Plakida D, Peros J, Evangelou K, Zavras AI, Yannoukakos D, Konstantopoulou I, Asimacopoulos PJ, Kittas C, Gorgoulis VG (2002) Growth index is independent of microvessel density in non-small-cell lung carcinomas. Hum Pathol 33: 812–818 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW (1992) p53 function and dysfunction. Cell 70: 523–526 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kuroiwa T, Nakagawa T, Kajimoto Y, Dohi T, Azuma H, Tsuji M, Kami K, Miyatake S (2003) Transcriptional expression of survivin and its splice variants in brain tumors in humans. J Neurosurg 99: 738–745 [DOI] [PubMed] [Google Scholar]