Abstract
Childhood B-cell precursor (BCP) ALL is thought to be caused by a delayed immune response to an unidentified postnatal infection. An association between BCP ALL and HLA class II (DR, DQ, DP) alleles could provide further clues to the identity of the infection, since HLA molecules exhibit allotype-restricted binding of infection-derived antigenic peptides. We clustered >30 HLA-DPB1 alleles into six predicted peptide-binding supertypes (DP1, 2, 3, 4, 6, and 8), based on amino acid di-morphisms at positions 11 (G/L), 69 (E/K), and 84 (G/D) of the DPβ1 domain. We found that the DPβ11-69-84 supertype GEG (DP2), was 70% more frequent in BCP ALL (n=687; P<10−4), and 98% more frequent in cases diagnosed between 3 and 6 years (P<10−4), but not <3 or >6 years, than in controls. Only one of 21 possible DPB1 supergenotypes, GEG/GKG (DP2/DP4) was significantly more frequent in BCP ALL (P=0.00004) than controls. These results suggest that susceptibility to BCP ALL is associated with the DP2 supertype, which is predicted to bind peptides with positively charged, nonpolar aromatic residues at the P4 position, and hydrophobic residues at the P1 and P6 positions. Studies of peptide binding by DP2 alleles could help to identify infection(s) carrying these peptides.
Keywords: HLA-DPB1 , supertypes, BCP ALL, case–control comparison, allele frequency, peptide-binding pockets
Acute lymphoblastic leukaemia (ALL) is the most common childhood malignancy in developed countries, where it constitutes over 30% of childhood cancers (Stiller et al, 1998; Smith et al, 1999). The striking age-incidence peak between 2 and 5 years of age consists mainly of common, B-cell precursor (BCP) ALL (Greaves et al, 1993, 1985; McKinney et al, 1993; Buckley et al, 1994). Molecular data indicate that BCP ALL can arise in utero in association with acquired chromosomal rearrangements that result in covert preleukaemic clones (Wiemels et al, 1999; Greaves, 2006), but progression to clinical ALL requires additional clonal genetic abnormalities, accumulated in a variable postnatal latent period. These may arise under the influence of an immune response to delayed infection (McNally and Eden, 2004; Greaves, 2006), but lack of information incriminating a specific infectious agent (Greaves, 2006; MacKenzie et al, 2006) has hindered verification of this causal pathway.
Insights into the role of infection in the aetiology of BCP ALL could be provided by associations with HLA class II alleles (Dorak et al, 1995, 1999; Taylor et al, 1995, 1998, 2002). The highly polymorphic HLA DR, DQ, and DP loci are encoded by genes in the human major histocompatibility complex (MHC), and are responsible for the binding and presentation of infection-derived peptides to CD4+ T cells, leading to adaptive immune responses to infections (Cooke and Hill, 2001). The affinity of different HLA class II allotypes for infection-derived peptides is influenced by a series of discrete peptide-binding pockets (PBP) embedded in the antigen-binding groove of the HLA class II α/β heterodimer (Hammer et al, 1997).
Since T-cell responses to infection in the presymptomatic phase of BCP ALL are not readily accessible to functional analysis, HLA class II alleles provide a potential PBP ‘footprint’ of the infection that may be involved in this disease. However, tight linkage between the HLA-DR and DQ loci makes it difficult to distinguish the primary contribution of alleles at these loci. Contrasting patterns of DR-DQ allelic linkage disequilibrium (LD) in different ethnic groups (Oksenberg et al, 2004) could resolve this problem, but such studies have yet to be reported in childhood leukaemia. Since the HLA-DP locus is only weakly linked to DR-DQ (Begovich et al, 1992; Cullen et al, 2002), analysis of DP alleles in BCP ALL should identify associations independent of DR-DQ. We and others have previously reported associations between DP alleles and human leukaemia (Pawelec et al, 1988; Taylor et al, 1995, 2002). Furthermore, DP alleles are known to be associated with, or to act as restriction elements for a number of parasitic (Meyer et al, 1994; May et al, 1998), microbial and viral diseases, including hepatitis B and rabies (Celis and Karr, 1989; Celis et al, 1990), herpes simplex (Koelle et al, 2000), streptococcus (Dong et al, 1995), dengue virus (Kurane et al, 1993; Okamoto et al, 1998), Epstein–Barr virus (Voo et al, 2002), respiratory syncytial virus (RSV) (De Graaf et al, 2004; De Waal et al, 2004), and HIV (Cohen et al, 2006).
Peptide binding by HLA class II allotypes, including DP, is the outcome of interactions between the amino acid side chains of the peptide and four major peptide-binding pockets (1, 4, 6, and 9; Hammer et al, 1997). Since different alleles can have overlapping peptide-binding properties, depending on the number of PBP that they share (Southwood et al, 1998), this has permitted DR alleles with the same amino acid polymorphisms lining specific peptide-binding pockets to be clustered into supertypes (Sette and Sidney, 1998; Southwood et al, 1998; Doytchinova and Flower, 2005). Using a similar approach, Castelli et al (2002) defined three DP supertype clusters with shared amino acid residues in the P1 (β84) and P6 (β11) PBP. However, the P4 peptide-binding pocket, at position β69, also makes an important contribution to antibody and peptide-binding (Arroyo et al, 1995; Chicz et al, 1997), T-cell responses (Berretta et al, 2003; Diaz et al, 2003) and disease susceptibility (Potolicchio et al, 1999; Wang et al, 1999). For this reason we clustered >30 DPB1 alleles into six supertypes based on polymorphisms in three PBP, at positions 11, 69, and 84 of the β1 domain (i.e., pockets 6, 4, and 1). We compared their frequencies in childhood BCP ALL, non-BCP leukaemia and solid tumours recruited as part of the UK Childhood Cancer Study (2000) with newborn controls. We discuss the implications of our findings in relation to an infectious aetiology for BCP ALL.
MATERIALS AND METHODS
Cases and controls
Childhood leukaemia cases were recruited between 1992 and 98 as part of the UK Childhood Cancer Study (UKCCS, 2000). Leukaemias were classified as BCP ALL (CD10+, CD19+; n=687) or non-BCP acute leukaemia. The non-BCP leukaemias were the sum of Pro-B ALL (CD10−, CD19+), T-ALL (CD2/CD7+, CD19−, DR−), and AML (n=208). Diagnostic immunophenotyping was carried out according to the protocol for UK Medical Research Council leukaemia trials (UKCCS, 2000). Childhood solid tumour cases (n=409) were also recruited as part of the UKCCS (UKCCS, 2000). Umbilical cord blood samples from a cross-sectional series of normal white UK newborns (n=864) born in Manchester UK between 1991 and 1997 were used as controls (Taylor et al, 2002). Blood sample collection and HLA molecular typing were carried out with national and local ethical consent. UKCCS patient data (diagnoses, gender, ages, ethnic background) were validated by the UKCCS data centre at the Epidemiology and Genetics Unit, University of York.
HLA-DPB1 molecular typing
HLA-DPB1 molecular typing was carried out as previously described in detail (Taylor et al, 2002) by amplifying a 327 bp exon 2 product in each case and control genomic DNA sample using a single pair of generic DPB1 PCR primers, spotting aliquots of each PCR product onto 384 sample nylon filters, and hybridising replicate filters with a panel of 28 32P-labelled sequence specific oligonucleotide probes. Probe hybridisation was detected using real-time autoradiography, and alleles assigned from published DPB1 ideograms.
Data analysis
DPB1 alleles in cases and controls were grouped into the six supertype clusters defined in this study (see Table 2 and Results for further details). Supertype allele and genotype frequencies were compared in cases and controls using global and univariate statistical analysis. As discussed previously (Taylor et al, 2002) ethical constraints precluded the collection of samples from case-matched control children, so we used local white UK newborns as controls. DPB1 alleles with a cumulative frequency of <5% that did not fall within the supertype clustering system were excluded from the analysis. Only sequence variation in the three peptide-binding pockets (positions 11, 69, and 84; pockets 6, 4, and 1, respectively) used for supertype clustering was included in the analysis. Global case–control supertype frequencies were compared using the CLUMP programme of Sham and Curtis (1995), a Monte Carlo method that computes a Pearson χ2 statistic (T1) from a series of simulated case–control tables. In univariate analysis, cross-product odds ratios (ORs), and 95% confidence intervals were calculated from case–control supertype and genotype frequencies by the RERI program in the Linkage Utility Package, LINKUTIL, using the Sheehe method. The 2by2 programme in LINKUTIL was used to determine 2-sided P-values for case–control supertype and genotype differences using Fisher's Exact test. Six supertypes require an uncorrected P-value <0.008, and 21 supergenotypes an uncorrected value <0.002 to achieve significance (P=0.05). No correction for the total number of classical DP alleles was applied. POPGENE version 1.31 was used to test for two-locus linkage disequilibrium between DPB1 and DQA1, or DQB1 alleles.
RESULTS
Case and control characteristics
The UKCCS is an epidemiological case–control study designed to test the role of environmental factors in the aetiology of childhood cancer and leukaemia (UKCCS, 2000). As part of the UKCCS, we obtained HLA-DPB1 types for 982 cases of childhood leukaemia (Taylor et al, 2002). Ninety-one percent of the leukaemia cases were classified as white, based on parental information, the remainder being Asian (3.8%), Black (1%), mixed ethnicity (1.9%), other ethnic groups (0.5%) or unknown. Of 875 cases of ALL, 559 were identified as BCP ALL, and a further 228 ALL cases were unclassified (Taylor et al, 2002). Subsequent diagnostic information for the unclassified ALL cases enabled us to identify 128 additional BCP ALL, seven Pro-B ALL, and six T ALL cases. These were included in the present study, which therefore comprises 895 DP-typed cases of childhood leukaemia with a confirmed diagnosis, of which 687 were BCP ALL and 208 were non-BCP leukaemia cases (Table 1). A mixed diagnostic series of childhood solid tumour cases (n=409), not including childhood lymphoma (Taylor et al, 2002) is included for comparison. Of these, 405 cases had informative ethnic data, being classified as white in 91% of cases. Cord blood samples from a cross-sectional series of normal white UK term newborns (n=864) were used as controls. Male–female ratios were slightly higher in the leukaemia cases (1.22) than the solid tumours (1.14) and controls (1.01).
Table 1. Number of cases and controls in this study.
|
Number
|
Male/female | |||
|---|---|---|---|---|
| Study groupa | Total | Male | Female | ratio |
| Leukaemia | 895 | 492 | 403 | 1.22 |
| BCP ALL | 687 | 373 | 314 | 1.19 |
| Non-BCP leukaemia | 208 | 119 | 89 | 1.33 |
| Pro-B ALL | 26 | 7 | 19 | 0.37 |
| T ALL | 75 | 51 | 24 | 2.12 |
| AML | 107 | 61 | 46 | 1.33 |
| Solid tumour | 409 | 218 | 191 | 1.14 |
| Newborn controls | 864 | 436 | 428 | 1.01 |
Leukaemias are classified as B cell precursor ALL (BCP ALL) and non-BCP acute leukaemia including Pro-B ALL, T ALL and AML.
HLA-DPB1 supertypes
The majority (90%) of >30 DPB1 alleles in the cases and controls could be clustered into six supertypes (Table 2), consisting of three pairs, each pair differing at position 69 for a glutamic acid (E) or lysine (K) in pocket 4, but having the same residues at positions 11 (G or L; pocket 6) and 84 (G or D; pocket 1). We designated the six supertypes by their position 11-69-84 residues as GEG, GKG, LED, LKD, GED, and GKD, corresponding to dimorphisms in the P6-P4-P1 peptide-binding pockets. Using a modification of the hierarchical supertype clustering system for DP alleles developed by Doytchinova and Flower (2005), we have provisionally called these supertypes DP1 (GKD), DP2 (GEG), DP3 (LKD), DP4 (GKG), DP6 (LED), and DP8 (GED).
Table 2. DPB1 supertypes of DPB1 alleles.
| DPB1 supertype | Peptide-binding motifa | DPB1 alleles with this supertype |
|---|---|---|
| DP2 | GEG | 0201, 0202, 3301, 4801 |
| DP4 | GKG | 0401, 0402, 2301, 2401, 4901, 5101 |
| DP6 | LED | 0601, 0901, 1001, 1301, 1701, 2101, 3001 |
| DP3 | LKD | 0301, 1401, 2001, 2501, 2601, 3501 |
| DP8 | GED | 0801, 1601, 1901 |
| DP1 | GKD | 0101, 0501, 5001 |
DPB1 supertypes assigned from di-allelic amino acids at positions β111 (G,L), β169 (E,K) and β184 (G,D).
HLA-DPB1 supertype frequency in childhood leukaemia
In the total leukaemia case series (n=895) and the newborn controls (n=864), we identified 14 DPβ69E alleles, of which four are DP2 (GEG), seven are DP6 (LED), and three are DP8 (GED). Of 15 DPβ69K alleles, six are DP4 (GKG), six are DP3 (LKD), and three are DP1 (GKD). In global χ2 analysis, the supertype frequency in the total leukaemia series was significantly different (P<10−6) from the controls (Table 3), but there was only a marginal difference between the solid tumour cases and controls (P=0.04). In univariate analysis, DP2 (GEG) (OR, 95% confidence interval (CI): 1.6, 95%, CI, 1.2–2.0; 2 sided P=0.0002) and DP8 (GED) (OR, CI: 2.9, 1.4–6.3; P=0.006) were significantly more frequent in leukaemia cases than controls. DP6 (LED) (OR, CI: 1.3, 1.0–1.8; P=0.04) was only marginally significant without correction for six supertypes, while DP2 and DP8 were significant after correction.
Table 3. DPB1 supertype frequency in childhood leukaemia and solid tumours compared with controls.
|
Leukaemia
|
Solid tumour
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| DPB1 supertype | % | OR | 95% CI | P | % | OR | 95% CI | P | Newborn controls % |
| DP2 (GEG) | 10.3 | 1.6 | 1.2–2.0 | 0.0002* | 7.3 | 1.1 | 0.8–1.5 | 0.6 | 6.8 |
| DP4 (GKG) | 59.6 | 1.1 | 0.9–1.2 | 0.38 | 60.3 | 1.1 | 0.9–1.3 | 0.3 | 58.0 |
| DP6 (LED) | 6.8 | 1.3 | 1.0–1.8 | 0.04 | 6.5 | 1.3 | 0.9–1.8 | 0.2 | 5.2 |
| DP3 (LKD) | 12.0 | 0.9 | 0.7–1.1 | 0.29 | 12.8 | 0.9 | 0.8–1.2 | 0.8 | 13.2 |
| DP8 (GED) | 1.4 | 2.9 | 1.4–6.3 | 0.006* | 0.6 | 1.4 | 0.5–3.9 | 0.8 | 0.5 |
| DP1 (GKD) | 6.9 | 0.6 | 0.5–0.7 | <10−4* | 8.6 | 0.8 | 0.6–1.1 | 0.09 | 11.1 |
| Global χ2 | <10−6** | 0.04 | |||||||
| Number= | 895 | 409 | 864 | ||||||
*Significant (P<0.05) after correction for six supertypes.
**Significant (P<0.05) in global χ2 (CLUMP) analysis.
Stratification of the leukaemias into BCP ALL (n=687) and non-BCP acute leukaemia (n=208) revealed that DP supertypes in BCP ALL differed significantly from the newborn controls (P<10−6), but non-BCP leukaemia was only marginally significant (P=0.04) (Table 4). In univariate analysis, DP2 (GEG) (OR, CI: 1.7, 1.3–2.1; P<10−4) and DP8 (GED) (OR, CI: 3.2, 1.5–7.0; P=0.004) were significantly more frequent, after correction for six supertypes, than controls. DP6 (LED) was not significant in BCP ALL, but was significant in non-BCP leukaemia (OR, CI: 1.8, 1.2–2.7; P=0.007). DP1 (GKD) was significantly less frequent, after correction, than controls in BCP ALL (OR, CI: 0.5, 0.4–0.7; P<10−5), but not in non-BCP leukaemia.
Table 4. DPB1 supertype frequency in BCP ALL and non-BCP leukaemia compared with controls.
|
BCP ALL
|
Non-BCP leukaemia
|
|||||||
|---|---|---|---|---|---|---|---|---|
| DPB1 supertype | % | OR | 95% CI | P | % | OR | 95% CI | P |
| DP2 (GEG) | 10.8 | 1.7 | 1.3–2.1 | <10−4* | 8.4 | 1.3 | 0.9–1.9 | 0.3 |
| DP4 (GKG) | 60.0 | 1.1 | 0.9–1.2 | 0.27 | 57.9 | 0.9 | 0.8–1.2 | 1.0 |
| DP6 (LED) | 6.2 | 1.2 | 0.9–1.6 | 0.24 | 8.9 | 1.8 | 1.2–2.7 | 0.007* |
| DP3 (LKD) | 11.6 | 0.9 | 0.7–1.1 | 0.21 | 13.0 | 0.9 | 0.7–1.3 | 0.9 |
| DP6 (GED) | 1.5 | 3.2 | 1.5–7.0 | 0.004* | 1.0 | 2.2 | 0.7–6.6 | 0.4 |
| DP1 (GKD) | 6.4 | 0.5 | 0.4–0.7 | <10−5* | 8.4 | 0.7 | 0.5–1.1 | 0.1 |
| Global χ2 | <10−6** | 0.04** | ||||||
| Number= | 687 | 208 | ||||||
*Significant after correction for six supertypes.
**Significant (P<0.05) in global χ2 (CLUMP) analysis.
The association of BCP ALL with DP2 and DP8 raised the possibility of a chance finding. To test this, supertype frequencies in four BCP ALL case series were compared with controls: (1) cases included in our previous study (n=559; Taylor et al, 2002); (2) half of the cases in the present study (n=344); (3) half of the cases in the previous study combined with the ‘new’ cases (n=343); (4) the ‘new’ cases (n=128) alone. DP2 and DP8 were significant in all four case series, though only DP2 remained significant after correction (Table 5).
Table 5. DPB1 supertype-associated risk of BCP ALL in replicate series of cases compared with controls.
|
Series 1a
|
Series 2
|
Series 3
|
Series 4
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DPB1 supertype | OR | CI | P | OR | CI | P | OR | CI | P | OR | CI | P |
| DP2 | 1.6 | 1.2–2.1 | 0.001* | 1.8 | 1.3–2.4 | 0.003* | 1.6 | 1.1–2.1 | 0.007* | 1.9 | 1.3–2.9 | 0.006* |
| DP4 | 1.1 | 0.9–1.3 | 0.19 | 1.1 | 0.9–1.3 | 0.23 | 1.05 | 0.9–1.3 | 0.59 | 0.9 | 0.8–1.3 | 0.99 |
| DP6 | 1.3 | 0.9–1.7 | 0.17 | 1.2 | 0.8–1.7 | 0.48 | 1.3 | 0.9–1.8 | 0.26 | 1.02 | 0.6–1.8 | 0.99 |
| DP3 | 0.9 | 0.7–1.1 | 0.41 | 0.9 | 0.7–1.1 | 0.33 | 0.9 | 0.7–1.1 | 0.34 | 0.7 | 0.5–1.1 | 0.14 |
| DP8 | 3.0 | 1.3–6.8 | 0.01 | 3.1 | 1.3–7.6 | 0.02 | 3.4 | 1.4–8.2 | 0.01 | 4.4 | 1.6–12.5 | 0.03 |
| DP1 | 0.5 | 0.4–0.7 | <10−4* | 0.5 | 0.3–0.7 | <10−3* | 0.6 | 0.5–0.9 | 0.008* | 0.7 | 0.4–1.1 | 0.13 |
| Number= | 559 | 344 | 343 | 128 | ||||||||
Series 1: see Taylor et al (2002); series 2: 50% of series 1; series 3: 215 cases from series 1+128 cases from series 4; series 4: new cases in this study.
*Significant (P<0.05) after correction for six supertypes.
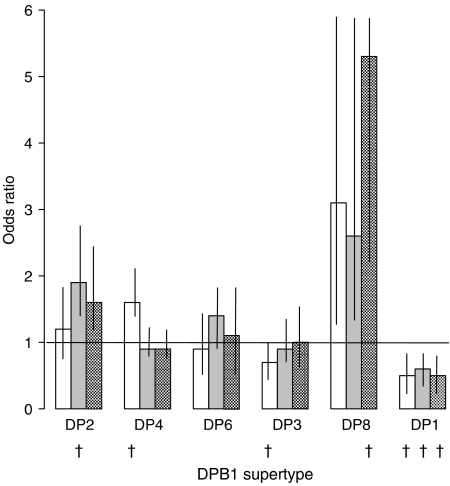
To determine the relationship between the age at diagnosis of BCP ALL and DP supertype, we compared the frequencies in cases diagnosed <3 years of age, >3–6 years, and >6 years, with controls. Figure 1 shows that the risk of BCP ALL was increased by 98% in DP2+ cases diagnosed at >3–6 years (OR, CI: 1.9, 1.4–2.6; P=10−4), but was not significant in BCP ALL diagnosed <3 or >6 years. DP4 was significantly increased in BCP ALL diagnosed <3 years, though not after correction. DP8 was not significant after correction, while DP1 protected from BCP ALL in all age groups.
Figure 1.
Odds ratios for DPB1 supertype frequencies compared with normal newborns in relation to the age at diagnosis of BCP-ALL. Ages at diagnosis: 0–<3 years (white bar), 3–6 years (grey bar), >6 years (checked bar). Vertical limits are 95% confidence intervals. †One-sided, corrected Fishers P-values: 0–<3 years: DP4=0.018, DP3=0.012; DP1=0.012. 3–6 years: DP2=0.0006, DP1=0.012. >6 years: DP8=0.018; DP1=0.03.
HLA-DPB1 supergenotype frequency
To determine which combination of supertype alleles was associated with BCP ALL, we compared the frequency of all 21 possible supergenotypes (six homozygous, 15 heterozygous) in BCP ALL, non-BCP leukaemia, and solid tumours with newborn controls (Table 6). Note that certain heterozygous DPB1 genotypes, such as DPB1*0201/0202 can have a homozygous supergenotype (DP2/DP2:GEG/GEG) (Table 2). Of the 21 supergenotypes, only one (DP2/DP4), was associated with a significantly increased risk (110%), after correction, of BCP ALL (OR, CI: 2.1, 1.5–2.9; P=0.00004). DP2/DP4 was associated with an increased risk (130%) of BCP ALL arising between 3 and 6 years of age (OR, 95% CI: 2.3, 1.4–3.8; P=0.04), but not <3 or >6 years of age. No DP2 supergenotypes were significant in non-BCP leukaemia or solid tumours. DP4/DP6 (GKG/LED) was significant in non-BCP leukaemia after correction (OR, CI: 2.7, 1.6–4.9; P=0.002), but not in BCP ALL or solid tumours. Homozygous DP1 (GKD/GKD) significantly protected, after correction, against BCP ALL (OR, CI: 0.2, 0.1–0.5; P=0.00004). No other DPB1 supergenotypes were significant at any of the diagnostic ages.
Table 6. Risk of BCP ALL, non-BCP leukaemia and paediatric solid tumours associated with DPB1 supergenotypes, compared with controls.
|
BCP ALL
|
Non BCP leukaemia
|
Solid tumour
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Supergenotype | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P |
| DP2/DP2 (GEG/GEG) | 0.9 | 0.4–1.9 | 0.99 | 0.9 | 0.3–2.8 | 0.99 | 1.02 | 0.4–2.4 | 0.99 |
| DP2/DP4 (GEG/GKG) | 2.1 | 1.5–2.9 | 0.00004* | 1.6 | 0.9–2.7 | 0.14 | 1.1 | 0.7–1.8 | 0.7 |
| DP2/DP6 (GEG/LED) | 2.5 | 1.0–5.8 | 0.06 | 2.5 | 0.8–7.7 | 0.3 | 1.5 | 0.5–4.5 | 0.6 |
| DP2/DP3 (GEG/LKD) | 1.5 | 0.7–3.1 | 0.42 | 0.8 | 0.3–3.6 | 0.99 | 1.1 | 0.4–2.8 | 0.99 |
| DP2/DP8 (GEG/GED) | 3.8 | 0.3–41.7 | 0.88 | 12.5 | 1.1–138.5 | 0.38 | — | — | — |
| DP2/DP1 (GEG/GKD) | 1.1 | 0.5–2.7 | 0.96 | 0.6 | 0.1–7.9 | 0.99 | 0.9 | 0.3–2.6 | 0.99 |
| DP4/DP4 (GKG/GKG) | 0.9 | 0.7–1.1 | 0.40 | 0.8 | 0.6–1.1 | 0.17 | 1.1 | 0.9–1.4 | 0.3 |
| DP4/DP6 (GKG/LED) | 1.9 | 1.2–3.0 | 0.006 | 2.7 | 1.6–4.9 | 0.002* | 1.7 | 1.0–2.9 | 0.06 |
| DP4/DP3 (GKG/LKD) | 1.1 | 0.8–1.4 | 0.80 | 1.2 | 1.2–14.6 | 0.1 | 1.1 | 0.7–1.5 | 0.8 |
| DP4/DP8 (GKG/GED) | 2.7 | 0.9–7.1 | 0.08 | 1.9 | 0.1–15.3 | 0.99 | 0.6 | 0.1–2.9 | 0.7 |
| DP4/DP1 (GKG/GKD) | 0.8 | 0.6–1.2 | 0.40 | 0.9 | 0.6–2.0 | 0.8 | 0.6 | 0.4–0.9 | 0.04 |
| DP6/DP6 (LED/LED) | 0.6 | 0.2–1.7 | 0.48 | 1.1 | 0.3–4.0 | 0.99 | 1.7 | 0.7–4.3 | 0.4 |
| DP6/DP3 (LED/LKD) | 0.5 | 0.2–1.1 | 0.09 | 1.1 | 0.04–3.1 | 0.67 | 0.2 | 0.07–0.81 | 0.02 |
| DP6/DP8 (LED/GED) | — | — | — | — | — | — | 6.3 | 0.6–70.2 | 0.6 |
| DP6/DP1 (LED/GKD) | 0.8 | 0.3–2.3 | 0.89 | 2.2 | 0.7–6.7 | 0.37 | 0.9 | 0.3–2.8 | 0.99 |
| DP3/DP3 (LKD/LKD) | 0.7 | 0.4–1.2 | 0.23 | 0.8 | 0.4–1.8 | 0.76 | 1.1 | 0.6–1.9 | 0.9 |
| DP3/DP8 (LKD/GED) | 3.3 | 0.8–12.8 | 0.16 | 2.5 | 0.4–15.0 | 0.95 | 2.1 | 0.4–10.5 | 0.7 |
| DP3/DP1 (LKD/GKD) | 0.5 | 0.2–1.2 | 0.15 | 1.6 | 0.5–3.9 | 0.85 | 1.2 | 0.5–2.7 | 0.8 |
| DP8/DP8 (GED/GED) | — | — | — | — | — | — | — | — | — |
| DP8/DP1 (GED/GKD) | 3.8 | 0.3–41.7 | 0.88 | — | — | — | — | — | — |
| DP1/DP1 (GKD/GKD) | 0.2 | 0.1–0.5 | 0.00004* | 0.4 | 0.1–1.06 | 0.06 | 0.8 | 0.4–1.5 | 0.6 |
| Number= | 687 | 208 | 409 | ||||||
*Significant (P<0.05) after correction for 21 supergenotypes.
Linkage disequilibrium analysis
To test whether the DP supertype associations could be explained by LD with HLA-DQ alleles, we analysed the co-occurrence of DP and DQ alleles in 451 BCP ALL cases, using POPGENE. We detected only one DP allele, 1601, in LD with DQ (DQB1*0401; χ2=37.4; uncorrected P<10−4). Five BCP ALL cases (0.4%) typed for DPB1*1601, a frequency not significantly greater than in the controls, indicating that the DP-supertype results cannot be explained by LD between DP and DQ alleles.
DISCUSSION
Selective peptide binding by HLA allotypes is a prerequisite for the recognition of antigens by T cells leading to adaptive immunity (Madden, 1995). Such a mechanism may underpin the immune-mediated progression of pre-ALL to overt leukaemia following delayed postnatal infection (Greaves, 2006). In our previous study, we suggested that the presence in pocket 4 of a glutamic acid (E) residue at position 69 of the DPβ1 domain was associated with BCP ALL (Taylor et al, 2002). However, HLA class II allotype-associated peptide binding is not the property of a single PBP; rather, it is the sum of a series of key PBP forming a DP allotype-associated peptide-binding motif or ‘footprint’. Polymorphisms in PBP accommodating the P1, 4, 6, and 9 amino acid anchors appear primarily to influence the DP allotype footprint (Hammer et al, 1997; Diaz et al, 2003, 2005). Since pocket 9 is composed of polymorphisms in residues 9, 35, 36, 55, and 56 (Diaz et al, 2003), we excluded this level of complexity. Furthermore, grouping amino-acid polymorphisms at positions 36, 56, and 76 failed to define recognised supertypes, and were not associated with leukaemia (data not shown). Clustering of DP alleles into six supertypes based on amino acid dimorphisms at positions 84 (P1 pocket), 69 (P4 pocket), and 11 (P6 pocket) represents an expanded version of the scheme proposed by Castelli et al (2002) based on peptide binding, and a slightly modified version of the hierarchical clustering scheme proposed by Doytchinova and Flower (2005). We have provisionally denoted the six supertypes DP1 (GKD), DP2 (GEG), DP3 (LKD), DP4 (GKG), DP6 (LED), and DP8 (GED) since they broadly resemble those defined in the primed lymphocyte test (PLT) as DPw specificites (De Koster et al, 1991). Furthermore, HLA-DPw2 defined by PLT was previously reported to be associated with ALL (Pawelec et al, 1988).
The DPB1 locus is the second most polymorphic HLA class II locus after DRB1, with at least 120 alleles identified to date (http://anthonynolan.org.uk/HIG/lists/class2list.html). In a rare disease such as BCP ALL in which there are likely to be multiple aetiological factors, weak HLA associations potentially require hundreds of cases and controls to allow for correction for multiple testing. Supertype analysis, in which alleles are clustered according to common functional (i.e., peptide binding) properties, overcomes this problem. DPB1 alleles comprise combinatorial series of six variable regions (A-F) encoded by exon 2 (Bugawan et al, 1988), in which alleles with the same variable region polymorphisms have the same peptide-binding pockets. DP alleles with the same polymorphisms at position 11 in variable region A, position 69 in variable region D, and position 84 in variable region F (Bugawan et al, 1988) can be predicted to have similar immune functions, based on identical (P6, P4, and P1, respectively) PBP. Our supertype classification includes position 69 (P4 pocket) since this is known to influence antibody-binding (Arroyo et al, 1995), allorecognition and peptide binding (Diaz et al, 2005), and disease susceptibility (Potolicchio et al, 1999; Wang et al, 1999). Furthermore it allowed us to split β69E alleles into three supertypes (GEG (DP2), LED (DP6), GED (DP8)), and to compare these with three homologous β69K series (GKG (DP4), LKD (DP3), GKD (DP1)).
We observed a 70% increase in BCP ALL risk in children typing for DP2 (GEG), a 98% increase in DP2-associated risk between 3 and 6 years of age, and a 130% increased risk associated with a single supergenotype, DP2/DP4. This association was not present in BCP ALL diagnosed <3 or >6 years of age, and leads us to conclude that the peak of BCP ALL (Greaves et al, 1993, 1985) may be influenced by the immunological sequelae of age-related interactions between DP2/DP4 and a specific antigenic peptide derived from delayed infection.
Analysis of replicate case series, including the 128 BCP ALL cases new to this study, suggest strongly that the association with DP2 was unlikely to be due to chance. Furthermore, DP6, which also has E at position 69 was not associated with BCP ALL, but was associated with non-BCP leukaemia. Phylogenetic analysis suggests that the DPB1 peptide-binding motif may have undergone rapid recent diversification and β69E alleles, such as DPB1*0201 and DPB1*0601, are not all closely related (Gyllensten et al, 1996). Supertype analysis groups HLA alleles with convergent immunological properties (Hughes et al, 1996; Trachtenberg et al, 2003), based on common peptide-binding motifs, and may be more relevant to BCP ALL aetiology than individual alleles.
We measured the significance of case–control supertype frequency differences using Fisher's Exact tests, corrected for six supertypes or 21 supergenotypes. We did not correct for total DP alleles since our analysis was informed by the results of our previous study (Taylor et al, 2002) and would have been overly influenced by low frequency alleles. Nevertheless, our results require confirmation with independent case–control series.
Although associations between childhood ALL and DR, DQ and DP alleles have been reported in previous studies (Dorak et al, 1995, 1999; Taylor et al, 1995, 2002), there has been no test of the effect of LD between alleles at the different loci. We found no evidence that the association of BCP ALL with DP2 could be explained by LD with DQ alleles, suggesting that DP has a primary role in susceptibility to BCP ALL.
It is unlikely that the association of BCP ALL with DP2 is due to a defect in the immune response to an oncogenic virus (immune evasion). There is no evidence that childhood BCP ALL is caused by an oncogenic virus (MacKenzie et al, 2006), and the positive association with DP2 suggests that binding of specific peptide(s) and T-cell activation are involved in causation, which is inconsistent with immune evasion by an oncogenic virus. The negative association of DP1 with BCP ALL may be due to the binding and recognition of TEL-AML1 peptide(s) in children with pre-ALL with this supertype, as discussed elsewhere (Taylor et al, 2008), since a TEL-AML1 junctional peptide has been shown to elicit a DPB1*0501-restricted (DP1) CD4+ T cell response (Yun et al, 1999).
The delayed response to infection hypothesis for BCP ALL (Greaves, 2006) proposes that a child carrying an in utero-initiated preleukaemic clone is vulnerable to the development of leukaemia if it is insulated from infection during the early postnatal period, but exposed at a later age. We previously reported that the risk of BCP ALL was greater in DPB1*0201 heterozygotes than homozygotes (Taylor et al, 2002), suggesting that BCP ALL might be the rare ‘down-side’ of the advantage that MHC-heterozygosity confers on immune responses to infection. Although evolution of HLA allelic diversity is thought to favour heterozygotes (Takahata and Nei, 1990), a recent study suggests that this advantage may be allele-specific (Lipsitch et al, 2003). Our finding that only one (DP2/DP4) of 15 heterozygous supergenotypes (GEG/GKG) is associated with BCP ALL fits this model.
Using DPB1*0201 peptide-binding data and molecular modelling (Diaz et al, 2005), it is possible to make predictions about the amino acid anchors at P1, P4, and P6 of peptides binding to DP2. Pocket 4 of DP2 is deeper, more negatively charged than DP4 (Diaz et al, 2003), giving it a greater affinity for positively charged nonpolar aromatic residues, such as glutamine (Q), arginine (R), and lysine (K). Furthermore, glycine (G) makes pocket 1 (β84) and pocket 6 (β11) deep and hydrophilic, preferentially-binding hydrophobic and aromatic amino acids, notably phenylalanine (F), and tyrosine (Y) (Berretta et al, 2003; Diaz et al, 2003, 2005). This predicts that infectious peptides with an 1FXXKXFXXA/V9 motif (where X is unknown, and P9 can be A or V) are likely to bind to DP2.
In this context, Van Steensel-Moll et al (1986) reported a negative (protective) association between childhood ALL and infections in the first year of life, and Rosenbaum et al (2005) documented a weak negative association between childhood ALL and bronchiolitis and pneumonia. Roman et al (2007) found a slight deficit in lower respiratory tract infection in the first year of life of UKCCS ALL cases diagnosed at 2–5 years. Together these findings suggest that the immune response to RSV infection may be a factor in BCP ALL. RSV is a highly contagious, weakly pathogenic, but strongly immunogenic virus that is widely distributed in the childhood population (Handforth et al, 2000; McNamara and Smyth, 2002). The G protein of RSV elicits CD4+ T-cell responses (De Graaf et al, 2004; De Waal et al, 2004), the peptide 162D-N179 containing two overlapping T-cell epitopes, 163FHFEVFNFV171 and 165FEVFNFVPC173 that are restricted by DPB1*0401 (DP4), and DPB1*0201 (DP2) (De Graaf et al, 2004). Both peptides have F at P1 and P6 suggestive of binding to GEG (DP2) and GKG (DP4), consistent with the association of BCP ALL with DP2/DP4. While this conclusion is speculative it points to a need for detailed sero-epidemiological studies of RSV in BCP ALL.
Acknowledgments
This study was funded by grants from the Kay Kendall Leukaemia Fund (to GMT and MFG), by support from Cancer Research UK (JMB, TE), and by the Leukaemia Research Fund (MFG). We are indebted to the children and families who took part in the UK Childhood Cancer Study. We thank J Simpson and Professor E Roman at the Epidemiology and Genetics Unit, University of York for providing diagnostic and other patient information, Mrs R Carter for blood sample documentation and the midwives at St Mary's Hospital, Manchester for cord blood samples. We are grateful to MD Robinson, Dr C Watson, Dr DA Gokhale, S P Dearden for sample processing, and DPB1 typing.
A complete list of UKCCS investigators and sponsors is given in: UK Childhood Cancer Study Investigators (2000).
References
- Arroyo J, Álvarez AM, Nombela C, Sánchez-Pérez M (1995) The role of HLA-DPβ residue 69 in the definition of antibody-binding epitopes. Human Immunology 43: 219–226 [DOI] [PubMed] [Google Scholar]
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148: 249–258 [PubMed] [Google Scholar]
- Berretta F, Butler RH, Diaz G, Sanarico N, Arroyo J, Fraziano M, Aichinger G, Wucherpfennig KW, Colizzi V, Saltini C, Amicosante M (2003) Detailed analysis of the effects of Glu/Lys human leukocyte antigen-DP polymorphism on peptide-binding specificity. Tissue Antigens 62: 459–471 [DOI] [PubMed] [Google Scholar]
- Buckley JD, Buckley CM, Ruccione K, Sather HN, Waskerwitz MJ, Woods WG, Robison LL, For The Children's Cancer Group (1994) Epidemiological characteristics of childhood acute lymphocytic leukemia. Analysis by immunophenotype. Leukemia 8: 856–864 [PubMed] [Google Scholar]
- Bugawan TL, Horn GT, Long CM, Mickelson E, Hansen JA, Ferrara GB, Angelini G, Erlich HA (1988) Analysis of HLA-DP allelic sequence polymorphism using the in vitro enzymatic DNA amplification of DP-α and DP-β loci. J Immunol 141: 4024–4030 [PubMed] [Google Scholar]
- Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Śégard H, Guillet J-G, Menez A, Georges B, Maillere B (2002) HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol 169: 6928–6934 [DOI] [PubMed] [Google Scholar]
- Celis E, Karr RW (1989) Presentation of an immunodominant T-cell epitope of hepatitis B surface antigen by the HLA-DPw4 molecule. J Virol 63: 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis E, Larson J, Otvos L, Wunner WH (1990) Identification of a rabies virus T cell epitope on the basis of its similarity with a hepatitis B surface antigen peptide presented to T cells by the same MHC molecule (HLA-DPw4). J Immunol 145: 305–310 [PubMed] [Google Scholar]
- Chicz RM, Graziano DF, Trucco M, Strominger JL, Gorga JC (1997) HLA-DP2. Self peptide sequences and binding properties. J Immunol 159: 4935–4942 [PubMed] [Google Scholar]
- Cohen WM, Pouvell-Moratille S, Wang X-F, Farci S, Munier G, Charron D, Menez A, Busson M, Maillère B (2006) Scanning the HIV genome for CD4+ T cell epitopes restricted to HLA-DP4, the most prevalent HLA class II molecule. J Immunol 176: 5401–5408 [DOI] [PubMed] [Google Scholar]
- Cooke GS, Hill AVS (2001) Genetics of susceptibility to human infectious disease. Nature Reviews Genetics 2: 967–977 [DOI] [PubMed] [Google Scholar]
- Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M (2002) High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet 71: 759–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graaf PMA, Heidema J, Poelen MC, Van Dijk MEA, Lukens MV, Van Gestel SPJ, Reinders J, Rozemuller E, Tilanus M, Hoogerhout P, Van Els CACM, Van der Most RG, Kimpen JLL, Van Bleek GM (2004) HLA-DP4 presents an immunodominant peptide from the RSV G protein to CD4 T cells. Virology 326: 220–230 [DOI] [PubMed] [Google Scholar]
- De Koster HS, Kenter MJH, D'Amaro J, Luiten RM, Schroeijers WEM, Giphart MJ, Termijtelen A (1991) Positive correlation between oligonucleotide typing and T-cell recognition of HLA-DP molecules. Immunogenetics 34: 12–22 [DOI] [PubMed] [Google Scholar]
- De Waal L, Yüksel S, Brandenburg AH, Langedijk JPM, Sintnicolaas K, Verjans GMGM, Osterhaus ADME, De Swart RL (2004) Identification of a common HLA-DP4-restricted T-cell epitope in the conserved region of the respiratory syncytial virus G protein. J Virol 78: 1775–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz G, Amicosante M, Jaraquemada D, Butler RH, Guillén MV, Sanchez M, Nombela C, Arroyo J (2003) Functional analysis of HLA-DP polymorphism: a crucial role for DPβ residues 9, 11, 35, 55, 56, 69, and 84–87 in T cell allorecognition and peptide binding. Int Immunol 15: 565–576 [DOI] [PubMed] [Google Scholar]
- Diaz G, Caňas B, Vazquez J, Nombela C, Arroyo J (2005) Characterization of natural peptide ligands from HLA-DP2: new insights into HLA-DP peptide-binding motifs. Immunogenetics 56: 754–759 [DOI] [PubMed] [Google Scholar]
- Dong R-P, Kamikawaji N, Toida N, Fujita Y, Kimura A, Sasazuki T (1995) Characterization of T cell epitopes restricted by HLA-DP9 in Streptococcal M12 protein. J Immunol 154: 4536–4545 [PubMed] [Google Scholar]
- Dorak MT, Lawson T, Machulla HK, Darke C, Mills KI, Burnett AK (1999) Unravelling an HLA-DR association in childhood acute leukaemia. Blood 94: 694–700 [PubMed] [Google Scholar]
- Dorak MT, Owen G, Galbraith I, Henderson N, Webb D, Mills KI, Darke C, Burnett AK (1995) Nature of HLA-associated predisposition to childhood acute lymphoblastic leukaemia. Leukemia 9: 875–878 [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR (2005) In silico identification of supertypes for class II MHCs. J Immunol 174: 7085–7095 [DOI] [PubMed] [Google Scholar]
- Greaves MF (2006) Infection, immune responses, and the aetiology of childhood leukaemia. Nat Rev Cancer 6: 193–203 [DOI] [PubMed] [Google Scholar]
- Greaves MF, Colman SM, Beard MEJ, Bradstock K, Cabrera ME, Chen P-M, Jacobs P, Lam-Po-Tang PRL, MacDougal LG, Williams CKO, Alexander FE (1993) Geographical distribution of acute lymphoblastic leukaemia subtypes: Second Report of the Collaborative Group Study. Leukemia 7: 27–34 [PubMed] [Google Scholar]
- Greaves MF, Pegram SM, Chan LC (1985) Collaborative group study of the epidemiology of acute lymphoblastic leukaemia subtypes: background and first report. Leuk Res 9: 715–733 [DOI] [PubMed] [Google Scholar]
- Gyllensten U, Bergstrom T, Josefson A, Sundval M, Erlich HA (1996) Rapid allelic diversification and intensified selection at antigen recognition sities of the Mhc class II DPB1 locus during hominoid evolution. Tissue Antigens 47: 212–221 [DOI] [PubMed] [Google Scholar]
- Hammer J, Sturniolo T, Sinigaglia F (1997) HLA class II peptide binding specificity and autoimmunity. Advances Immunol 66: 67–100 [DOI] [PubMed] [Google Scholar]
- Handforth J, Friedland JS, Sharland M (2000) Basic epidemiology and immunopathology of RSV in children. Paed Resp Reviews 1: 210–214 [DOI] [PubMed] [Google Scholar]
- http://anthonynolan.org.uk/HIG/lists/class2list.html
- Hughes AL, Yeager M, Carrington M (1996) Peptide-binding function and the paradox of HLA disease associations. Immunol Cell Biol 74: 444–448 [DOI] [PubMed] [Google Scholar]
- Koelle DM, Reymand SN, Chen H, Kwok WW, McClurkan C, Gyaltsong T, Petersdorf EW, Rotkis W, Talley AR, Harrison DA (2000) Tegument-specific, virus-reactive CD4 T cells localize to the cornea in Herpes simplex virus interstitial keratitis in humans. J Virol 74: 10930–10938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurane I, Dai L-C, Livingston PG, Reed E, Ennis FA (1993) Definition of an HLA-DPw2-restricted epitope on NS3, recognized by a Dengue virus serotype-cross-reactive human CD4+ CD8− cytotoxic T cell clone. J Virol 67: 6285–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINKUTIL, http://linkage.rockefeller.edu/soft/linkutil/
- Lipsitch M, Bergstrom CT, Antia R (2003) Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med Genet 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie J, Greaves MF, Eden TOB, Clayton RA, Perry J, Wilson KS, Jarrett RF (2006) The putative role of transforming viruses in childhood acute lymphoblastic leukemia. Haematologica 91: 240–243 [PubMed] [Google Scholar]
- Madden DR (1995) The three-dimensional structure of peptide-MHC complexes. Ann Rev Immunol 13: 587–622 [DOI] [PubMed] [Google Scholar]
- May J, Kremsner PG, Milovanovic D, Schnittger L, Loliger C-C, Bienzle U, Meyer CG (1998) HLA-DP control of Schistosoma haematobium infection. Am J Trop Med Hyg 59: 302–306 [DOI] [PubMed] [Google Scholar]
- McKinney PA, Alexander FE, Cartwright RA, Scott CS, Staines A (1993) Acute lymphoblastic leukaemia incidence in the UK by immunophenotype. Leukemia 7: 1630–1634 [PubMed] [Google Scholar]
- McNally RJ, Eden TO (2004) An infectious aetiology for childhood acute leukaemia: a review of the evidence. Br J Haematol 127: 243–263 [DOI] [PubMed] [Google Scholar]
- McNamara PS, Smyth RL (2002) The pathogenesis of respiratory syncytial virus disease in childhood. Br Med Bull 61: 13–28 [DOI] [PubMed] [Google Scholar]
- Meyer CG, Gallin M, Erttmann KD, Brattig N, Schnittger L, Gelhaus A, Tannich E, Begovich AB, Erlich HA, Horstmann RD (1994) HLA-D alleles associated with generalized disease, localized disease, and putative immunity in Onchocerca volvulus infection. Proc Natl Acad Sci 91: 7515–7519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto Y, Kurane I, Leporati AM, Ennis FA (1998) Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4 T cell clones. J Gen Virol 79: 697–704 [DOI] [PubMed] [Google Scholar]
- Oksenberg JR, Barcellos LF, Cree BA, Barazini SE, Bugawan TL, Khan O, Lincoln RR, Swerdlin A, Mignot E, Lin L, Goodin D, Erlich HA, Schmidt S, Thompson G, Reich DE, Pericak-Vance MA, Haines JL, Hauser SL (2004) Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet 74: 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G, Ehninger G, Muller C, Blaurock M, Schneider EM, Wernet P (1988) Human leukocyte antigen-DP in leukaemia. Cancer 61: 475–477 [DOI] [PubMed] [Google Scholar]
- POPGENE version 1.31: Microsoft Window-based freeware for population genetic analysis, http://www.ualberta.ca/~fyeh
- Potolicchio I, Festucci A, Hausler P, Sorrentini R (1999) HLA-DP molecules bind cobalt: a possible explanation for the genetic association with hard metal disease. Eur J Immunol 29: 2140–2147 [DOI] [PubMed] [Google Scholar]
- Roman E, Simpson J, Ansell P, Kinsey S, Mitchell CD, McKinney PA, Birch JM, Greaves M, Eden T, on behalf of United Kingdom Childhood Cancer Study Investigators (2007) Childhood acute lymphoblastic leukemia and infections in the first year of life: a report from the United Kingdom childhood cancer study. Am J Epidemiol 165: 496–504 [DOI] [PubMed] [Google Scholar]
- Rosenbaum PF, Buck GM, Brecher ML (2005) Allergy and infectious disease histories and the risk of childhood acute lymphoblastic leukaemia. Paediatr Perinat Epidemiol 19: 152–164 [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J (1998) HLA supertypes and supermotfis: a functional perspective on HLA polymorphism. Curr Opin Immunol 10: 478–482 [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59: 97–105 [DOI] [PubMed] [Google Scholar]
- Smith MA, Ries LAG, Gurney JG, Ross JA (1999) Leukemia. In Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program 1975–1995, Ries LAG, Smith MA, Gurney JG, Linet M, Tamra T, Young JL, Bunin GR (eds) Bethesda, MD: National Cancer Institute, SEER program, NIH pub. No. 99-4649, pp 17–34 [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio M-F, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A (1998) Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 160: 3363–3373 [PubMed] [Google Scholar]
- Stiller C, Allen M, Brownbill P, Draper G, Eatock E, Loach M, Vincent TJ (1998) National registry for childhood tumours, England and Wales, 1981–1990, pp 365–367. In International Incidence of Childhood Cancer, Parkin DM, Kramarova E, Draper GJ, et al (eds) Vol II, Lyon, France: IARC Scientific Publications, no. 144, International Agency for Research on Cancer [Google Scholar]
- Takahata N, Nei M (1990) Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GM, Dearden S, Payne N, Ayres M, Gokhale DA, Birch JM, Blair V, Stevens RF, Will AM, Eden OB (1998) Evidence that an HLA-DQA1-DQB1 haplotype influences susceptibility to childhood common acute lymphoblastic leukaemia in boys provides further support for an infection-related aetiology. Br J Cancer 78: 561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GM, Dearden S, Ravetto P, Ayres M, Watson P, Hussain A, Greaves M, Alexander F, Eden OB, and UKCCS Investigators (2002) Genetic susceptibility to childhood common acute paediatric lymphoblastic leukaemia is associated with polymorphic peptide-binding pocket profiles in HLA-DPB1*0201. Hum Mol Genet 11: 1585–1597 [DOI] [PubMed] [Google Scholar]
- Taylor GM, Robinson MD, Binchy A, Birch JM, Stevens RF, Jones PM, Carr T, Dearden S, Gokhale DA (1995) Preliminary evidence of an association between HLA-DPB1*0201 and childhood common acute lymphoblastic leukaemia supports an infectious aetiology. Leukemia 9: 440–443 [PubMed] [Google Scholar]
- Taylor M, Harrison C, Eden T, Birch J, Greaves M, Lightfoot T, Hussain A (2008) HLA-DPB1 supertype-associated protection from childhood leukaemia: relationship to leukaemia karyotype and implications for prevention. Cancer Immunol Immunother 57: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, Funkhauser R, Fugate M, Theiler J, Hsu YS, Kunstman K, Wu S, Phair J, Erlich H, Wolinsky S (2003) Advantage of rare HLA supertype in HIV disease progression. Nat Med 9: 928–935 [DOI] [PubMed] [Google Scholar]
- UK Childhood Cancer Study Investigators (2000) The United Kingdom childhood cancer study: objectives, materials and methods. Br J Cancer 82: 1073–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel-Moll HA, Valkenburg HA, Van Zanen GE (1986) Childhood leukaemia and infectious diseases in the first year of life: a register-based case–control study. Am J Epidemiol 124: 590–594 [DOI] [PubMed] [Google Scholar]
- Voo KS, Fu T, Heslop HE, Brenner MK, Rooney CM, Wang R-F (2002) Identification of HLA-DP3-restricted peptides from EBNA1 recognized by CD4+ T cells. Cancer Res 62: 7195–7199 [PubMed] [Google Scholar]
- Wang Z, White PS, Petrovic M, Tatum OL, Newman LS, Maier LA, Marrone BL (1999) Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and –DPA1 alleles. J Immunol 163: 1647–1653 [PubMed] [Google Scholar]
- Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF (1999) Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 354: 1499–1503 [DOI] [PubMed] [Google Scholar]
- Yun C, Senju S, Fujita H, Tsuji Y, Irie A, Matsuchita S, Nishimura Y (1999) Augmentation of immune responses by altered peptide ligands of the antigenic peptide in a human CD4+ T-cell clone reacting to TEL-AML1 fusion protein. Tissue Antigens 54: 153–161 [DOI] [PubMed] [Google Scholar]