Abstract
The role of the foraging (for) gene, which encodes a cyclic guanosine-3′,5′-monophosphate (cGMP)-dependent protein kinase (PKG), in food-search behavior in Drosophila has been intensively studied. However, its functions in other complex behaviors have not been well-characterized. Here, we show experimentally in Drosophila that the for gene is required in the operant visual learning paradigm. Visual pattern memory was normal in a natural variant rover (forR) but was impaired in another natural variant sitter (forS), which has a lower PKG level. Memory defects in forS flies could be rescued by either constitutive or adult-limited expression of for in the fan-shaped body. Interestingly, we showed that such rescue also occurred when for was expressed in the ellipsoid body. Additionally, expression of for in the fifth layer of the fan-shaped body restored sufficient memory for the pattern parameter “elevation” but not for “contour orientation,” whereas expression of for in the ellipsoid body restored sufficient memory for both parameters. Our study defines a Drosophila model for further understanding the role of cGMP-PKG signaling in associative learning/memory and the neural circuit underlying this for-dependent visual pattern memory.
Cyclic guanosine-3′,5′-monophosphate (cGMP)-dependent protein kinase (PKG or cGK) is a key responder that mediates signaling downstream of nitric oxide (NO) and cGMP, which are involved in many important functions—like smooth muscle relaxation, neurotransmitter release and uptake, axon guidance, and circadian rhythms—in a number of different species (Wang and Robinson 1997; Schafer 2002; Hofmann et al. 2006). It may exert its function by phosphorylating a series of substrates, including ion channels, cytoskeletal proteins, GTP binding proteins, phosphodiesterase (PDE), and nitric oxide synthase (NOS), with the latter two having significant effects on the intracellular levels of the cyclic nucleotides cAMP and cGMP (Wang and Robinson 1997).
In vertebrate brains, two forms of PKG, cGKI and cGKII, are complementarily distributed. cGKI is abundant in the hippocampus, whereas cGKII is present in regions with low cGKI levels, implying different roles of the two cGKs in regulating cGMP signaling pathways, at least in neurons. Indeed, a mixed pattern of results has implicated cGKI in hippocampal long-term potentiation (LTP) and cerebellar long-term depression (LTD) (Zhuo et al. 1994; Feil et al. 2003). Coincidently, hippocampal cGKI activity in rats was reported to be required for the early stages of memory formation in an inhibitory avoidance task (Bernabeu et al. 1997), and cGKI inactivation in mouse Purkinje cells caused deficits in motor learning (Feil et al. 2003), suggesting that cGKI might be integral to learning and memory in vertebrates. On the other hand, cGKII serves as a key component in a signal transduction pathway that is responsible for phase shifts of the circadian clock in the mammalian suprachiasmatic nucleus (Oster et al. 2003; Tischkau et al. 2003, 2004) and for anxiety-like behavior (Werner et al. 2004).
PKG is also involved in some behaviors in invertebrates. Studies in the honeybee suggested that PKG was required for phototaxis and division of labor (Ben-Shahar et al. 2002; Ben-Shahar 2005). In Caenorhabditis elegans, the cGKI homolog of egl-4 was reported to be required for olfactory adaptation and locomotor behavior (Fujiwara et al. 2002; L’Etoile et al. 2002; Schafer 2002). In Drosophila, there are two PKG-encoding genes, dg1 and dg2 (dg2 is also named foraging, or for) (Kalderon and Rubin 1989). DG1 has high homology with vertebrate cGKII and DG2 is similar to cGKI, based on primary sequence. dg1 expression has only been detected in the lamina of the optic lobes, and its neural function has not been well-characterized (Foster et al. 1996), while the prominent function of for appears to be related to larval foraging behavior (de Belle et al. 1989; Osborne et al. 1997). Biochemical analysis of for allelic variants suggested slight reductions in the amounts of for mRNA and protein, together with a ∼10% reduction in PKG activity, in the natural variant sitter (forS), which travels a shorter distance for food compared with another variant rover (forR). PKG has been shown to affect some forms of nonassociative learning in Drosophila (Scheiner et al. 2004). Recently, two papers reported that for was also involved in olfactory associative learning in larval and adult flies (Kaun et al. 2007; Mery et al. 2007). However, its effect on visual associative learning remains largely unknown.
In Drosophila, genetic and behavioral approaches have been applied to screen mutants with a dramatically reduced ability for learning and memory (Dubnau and Tully 1998; Waddell and Quinn 2001; McGuire et al. 2005). Analyses of these mutants have implicated the cAMP pathway, a second messenger system, in diverse learning paradigms. Type I adenylyl cyclase (rutabaga, rut), which catalyzes the conversion of ATP into cAMP, is thought to be a putative convergence site of unconditioned and conditioned stimuli in associative learning. Using the binary GAL4/UAS expression system, rut-dependent olfactory memory has been assigned to the mushroom bodies (MBs), and space memory to the neurons of the median bundle and the ventral ganglion (Zars et al. 2000a, b; McGuire et al. 2003; Mao et al. 2004). Recently, two distinct rut-dependent visual pattern memory traces were localized to small groups of neurons innervating two horizontal layers of the fan-shaped body, the largest part of the central complex (CX) (Liu et al. 2006). The CX is located centrally in the brain and consists of four characteristic substructures: the protocerebral bridge, the fan-shaped body (FB), the ellipsoid body (EB), and the noduli (Hanesch et al. 1989). The large-field neurons of the FB form six horizontal strata, among which the first layer (F1) and the fifth layer (F5) of stratified neurons correspondingly accommodate visual pattern memory for “contour orientation” and “elevation” (Liu et al. 2006). Likewise, the EB is composed of arborizations provided by four types of large-field ringlike neurons (R1–4) (Hanesch et al. 1989). Based on the radii of their concentric arborizations, R4-type neurons were further subdivided into distal (R4d) and medial (R4m) types (Renn et al. 1999). The roles of these distinguishable R-type neurons are not well understood.
In the current study, to find new genes that function in visual learning and memory, we screened P element insertion lines and obtained the PKG-encoding gene for as a candidate. We then tested a series of for allelic variants and transgenic flies in an operant conditioning paradigm to uncover the roles of PKG in visual associative learning and its functional localization. Our results indicated that short-term visual pattern memory is undermined in forS flies and that this defect can be rescued by either constitutive or temporary expression of for in the brain structures of the FB and, unexpectedly, the EB. These data suggested the involvement of Drosophila PKG in complex learning behavior and the implication of a new brain region involved in visual pattern memory.
Results
for allelic variants display distinct visual pattern memory
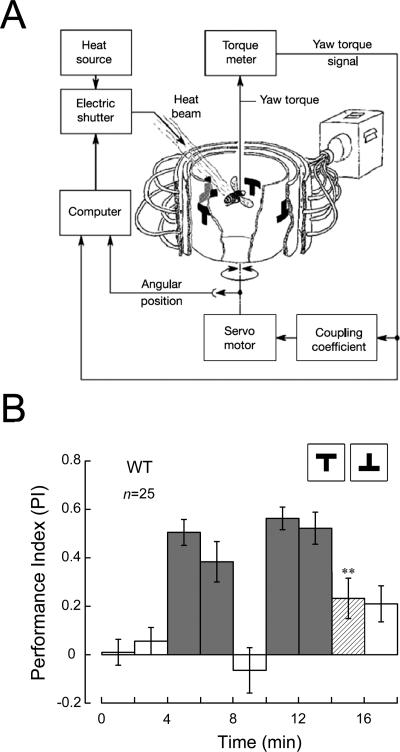
To identify new genes involved in visual pattern memory, we screened a collection of P[GawB] insertion lines in the flight simulator (Fig. 1A). In this behavioral test, individual flies were trained to avoid patterns associated with heat punishment and to fly toward those paired with the absence of heat punishment. This was followed by a 2-min test period without any heat punishment to check whether they remembered what they had learned during training (Fig. 1B).
Figure 1.
Experimental apparatus and procedure for testing visual pattern memory in Drosophila. (A) Flight simulator. The fly is fixed to a torque meter, and its yaw torque determines the angular velocity of the arena, in which there are two groups of patterns. With a beam of infrared light as punishment, the fly can be conditioned to avoid a “hot” pattern and pursue a “cold” one (for more details, see Materials and Methods). (B) Time course of experiment. Bars show performance indices (PIs; for more details. see Materials and Methods) of nine consecutive 2-min periods including pretest (open bars, PI1 and PI2), training (dark gray bars, PI3, PI4, PI6 and PI7), and memory test (open bars, PI5 and PI9; hatched bars, PI8). Only the value of PI8 is shown in the following figures. If not specially mentioned, T-shaped patterns were used as visual stimuli. Error bars are SEMs throughout. **P < 0.01 (one-sample t-test; two-sided P-value against zero); (n) numbers of flies tested; (WT) wild-type Canton-S strain.
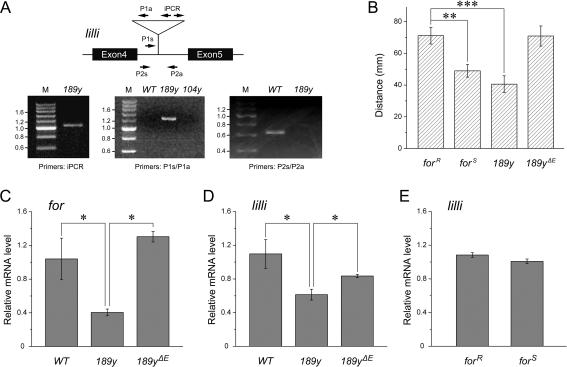
Using this experimental apparatus and procedure to screen the Gal4 library, we found that a Gal4 line, called 189y, showed defective performance in memory test. In homozygous 189y flies, the memory index PI8 (the eighth performance index; for details, see Materials and Methods) was not significantly different from zero (PI8 = 0.03 ± 0.07, one-sample t-test, two-sided P-value, t = 0.37, P = NS; Fig. 2A). This line was reported to carry a P element insertion in for, a Drosophila PKG-encoding gene, and to have a significantly reduced PKG level (Osborne et al. 1997). Inverse PCR and subsequent sequencing of the P element–flanking genomic DNA in our laboratory revealed that a single P element existed in the genome of 189y flies and that it was inserted not in the for coding region, but in the lilliputian (lilli) locus, which is 675 kb downstream of the for gene (Figs. 3A, 4A). This new locus was validated by PCR using primers specific for the P element–flanking DNA sequence and the P[GawB] sequence (Fig. 3A). Further quantitative PCR analysis indicated that both lilli and for mRNA expression were seriously disrupted by the P element in 189y flies (Fig. 3C,D). After precise excision of the P element, larval foraging behavior reverted from a sitter to a rover phenotype (Fig. 3B), and the mRNA levels of lilli and for also recovered (Fig. 3C,D). We did not observe any for mutations at the genomic level in the 189y flies used in this study, after careful examination (Z. Wang, L. Liu, and Z. Gong, unpubl.).
Figure 2.
Role of for in visual pattern memory. (A) for allelic variants showed different visual pattern memory. In the for allelic variant forR, the memory score is significantly higher than zero. In contrast, memory scores in the homozygous forS and 189y and the heterozygous forR/forS, forS/189y, forR/189y, and forR/Df flies are not significantly different from zero. Df is Df(2L)ed1 which is a deletion at for locus. (B) Dwelling time analysis of wild-type and for allelic variant flies. The mean dwelling time in “hot” quadrants during the first (hatched bars) and last (solid bars) training periods was calculated according to the original data used in A. No significant difference of the mean dwelling time can be observed between wild-type and for allelic variant flies. (C) Pattern discrimination of wild-type and for allelic variant flies. Fly’s pattern discrimination ability was evaluated by the discrimination value (D) during the two successive 2-min intervals of the pretest (PI1 and PI2). The discrimination values of wild-type and for allelic variant flies (hatched bars) are significantly different from the chance value (D = 1). As a control, in the experiment of wild-type flies with four identical patterns (solid bar) the discrimination value is not significantly different from the chance value. (n) Numbers of 2-min intervals in the pretest. (D) for allelic variants’ memory performances with arena oscillations as reinforcer instead of heat. The visual pattern memory score is significantly higher than zero in forR flies, whereas it is not in forS flies. The genotypes of tested flies are indicated. Error bars are SEMs. **P < 0.01; ***P < 0.001; (n) numbers of flies tested; (WT) wild-type Canton-S strain.
Figure 3.
Characterization of the P element in 189y flies. (A) The P element in 189y flies is inserted in lilli gene. In the schematic representation of part of lilli locus, exons are indicated as black boxes and P element insertion of 189y is indicated as triangle. Primers for PCR reactions are shown as arrows. The iPCR primers are used for inverse PCR. The primers P1s/P1a and the primers P2s/P2a are used for regular PCR. Inverse PCR of the self-ligated fragments from the 5′ end of P[GawB] in 189y showed a product of about 1.1 kb. This DNA fragment contains part of P[GawB] and flanking genomic DNA. After DNA sequencing and BLAST, the P element in 189y was localized to a site different from the previously described for locus (shown in Fig. 4A). PCR using primer P1s specific for genomic DNA upstream of the insertion site and primer P1a from the 3′ end of P[GawB] showed a 1.3-kb PCR product only in the 189y flies, as expected, but not in wild-type or 104y flies, an otherwise irrelevant Gal4 line. PCR with primers P2s/P2a specific for the genomic DNA flanking the 189y insertion site showed a 0.6-kb band in wild-type flies, as expected, but no detectable band in 189y flies. M, molecular weight markers measured in kb. (B) P element precise excision revert 189y larvae from sitter to rover. Larval foraging distance of forR, forS, 189y, and the precise excision strain 189yΔE on yeast were measured in millimeters (the same behavior paradigm described as in Osborne et al. 1997). There is no significant difference among their general locomotor ability (data not shown). Error bars are SEM. **P < 0.01; ***P < 0.001. n = 30 for all strains. (C–E) for and lilli mRNA expression levels in fly heads. for and lilli mRNA levels in 189y are significantly lower than in the precise excision strain 189yΔE and the wild-type control, while for and lilli mRNA levels in the excision strain are not significantly different from those in the wild-type control (C and D). lilli mRNA levels in forR and forS flies were not distinguishable (E). Error bars are SEMs. *P < 0.05; n = 6 for all; (WT) wild-type Canton-S.
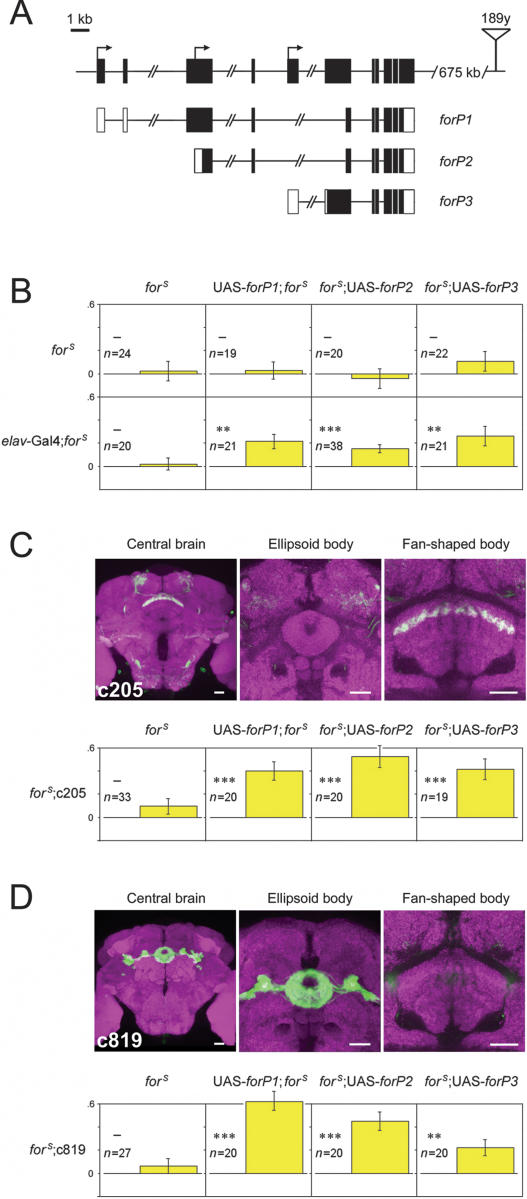
Figure 4.
Visual pattern memory rescue in forS flies expressing any of the three transcripts in the fan-shaped body or the ellipsoid body. (A) Schematic representation of for locus and transcripts. In the genome map at the top, exons and transcription start sites are shown as black boxes and arrows, respectively. The insertion site of 189y is represented by the triangle. Below, in the transcription map of the three major transcripts (forP1, forP2, and forP3), coding sequences are highlighted in black; 5′- and 3′-UTR are in white. Scale bar, 1 kb. (B–D) Flies with UAS-forP1, UAS-forP2, and UAS-forP3 inserted in the first, third, and third chromosomes, respectively, were genetically substituted into a forS background. The memory of forS flies was restored by the expression of each of the three wild-type for transcripts driven by elav-Gal4, whereas forS containing only elav-Gal4 or the UAS responders (elav-Gal4; forS, UAS-forP1; forS, forS;UAS-forP2/+, and forS;UAS-forP3/+) showed no visual pattern memory (B). When driven by c205 (C) or c819 (D), expression of for was able to rescue the memory defect in forS flies. The forS flies bearing only the Gal4 drivers (forS;c205/+ and forS;c819/+) used as a control, showed no memory (leftmost panels in C and D). The expression pattern of transgenes under the control of the drivers c205 (C) and c819 (D) in central brain are shown by GFP (green; overlay white). The synaptic neuropil is stained by anti-nc82 antibody (magenta). Scale bars are 20 μm for all. Y-axes represent PI8 of the memory test. In each panel of behavioral results, the maternal genotypes are shown at the top and the paternal genotypes are at the left. Error bars are SEM. —P ≥ 0.05; **P < 0.01; ***P < 0.001; (n) numbers of flies tested.
As the for gene might play a role in visual pattern memory in flies, we next focused on the role of for in this type of memory. Two for allelic variants, forR and forS, with the former having a higher PKG level than the latter (Osborne et al. 1997), were used to test this hypothesis. Intriguingly, as shown in Figure 2A, the visual pattern memory of forR flies appeared to be significantly higher than zero (PI8 = 0.27 ± 0.08, t-test, t = 3.40, P < 0.01) and similar to the memory level shown by wild-type flies, whereas the memory level of forS flies was not different from zero (PI8 = 0.02 ± 0.09, t-test, t = 0.27, P = NS [not significant]). Unlike larval foraging behavior, forS seemed not to be genetically recessive to forR in terms of visual pattern memory (forR/forS: PI8 = 0.01 ± 0.05, t-test, t = 0.27, P = NS). This was confirmed by the detection of memory loss in flies of two other genotypes: forR/189y (PI8 = −0.02 ± 0.03, t-test, t = 0.71, P = NS) and forR/Df(2L)ed1 (PI8 = 0.06 ± 0.05, t-test, t = 1.34, P = NS; Df(2L)ed1 is a deletion at for locus). forS flies did not exhibit normal visual pattern memory performance. This could be due to defects in pattern discrimination or in thermotolerance. To test the first possibility, we used Fourier analysis to evaluate flies’ abilities to discriminate between patterns (for details, see Materials and Methods). In principle, the discrimination value (D) with identical patterns is the chance value (D = 1). As expected, in experiments with four identical patterns, the discrimination of wild-type flies was not significantly different from the chance value (D = 1.28 ± 0.15, t-test, t = 1.82, P = NS). In experiments with different patterns, wild-type flies, as well as forR and forS flies retained their ability to recognize different patterns (WT: D = 2.46 ± 0.43, t-test, t = 3.43, P < 0.01; forR: D = 2.22 ± 0.31, t-test, t = 3.96, P < 0.001; forS: D = 2.19 ± 0.35, t-test, t = 3.42, P < 0.01), and their visual discrimination abilities appeared to be indistinguishable from each other (Fig. 2C). To exclude the latter possibility, we calculated the mean dwelling time in “hot” quadrants during training to analyze flies’ thermal-avoidance behavior (Dill et al. 1995). forR and forS flies spent similar amounts of time in the “hot” area to wild-type flies and did not show thermotolerance between two training periods (Fig. 2B). In addition, distinct memory performance was still observed in forR and forS flies when the reinforcement was substituted by arena oscillations (forR: PI8 = 0.28 ± 0.05, t-test, t = 4.70, P < 0.001; forS: PI8 = 0.02 ± 0.04, t-test, t = 0.50, P = NS; Fig. 2D). Finally, the memory defects in forS flies was neither complemented by 189y (forS/189y: PI8 = 0.07 ± 0.07, t-test, t = 1.05, P = NS; Fig. 2A) nor rescued by pan-neuronal expression of wild-type lilli cDNA (elav-Gal4;forS;UAS-lilli+: PI8 = 0.02 ± 0.07, t-test, t = 0.27, P = NS).
Taken together, these results indicated that short-term visual pattern memory in Drosophila requires for function and is impaired by reductions in the for-encoded PKG level.
Specific expression of for in the FB and the EB is sufficient to rescue visual pattern memory in forS flies
To further explore the spatiotemporal properties of for functions in visual pattern memory, three types of UAS-for+ transgenic flies were generated according to the coding sequences of three major for transcripts, to increase the PKG level in forS flies (Fig. 4A). These UAS lines were then crossed with the Gal4 lines used in the rescue experiments after breeding to a homozygous forS genetic background. As shown in Figure 4B, pan-neuronal expression of the three for transcripts driven by elav-Gal4 was able to effectively rescue the memory defect in forS flies (elav-Gal4/UAS-forP1; forS: PI8 = 0.22 ± 0.06, t-test, t = 3.35, P < 0.01; elav-Gal4;forS;UAS-forP2/+: PI8 = 0.15 ± 0.03, t-test, t = 4.40, P < 0.001; elav-Gal4;forS;UAS-forP3/+: PI8 = 0.26 ± 0.09, t-test, t = 3.05, P < 0.01), whereas neither elav-Gal4 nor UAS-for+ alone, on a homozygous forS background, was capable of this rescue (elav-Gal4;forS: PI8 = 0.02 ± 0.05, t-test, t = 0.35, P = NS; UAS-forP1;forS: PI8 = 0.03 ± 0.07, t-test, t = 0.38, P = NS; forS;UAS-forP2/+: PI8 = −0.04 ± 0.09, t-test, t = 0.50, P = NS; forS;UAS-forP3/+: PI8 = 0.11 ± 0.08, t-test, t = 0.13, P = NS). Previous results have suggested that the FB is necessary for visual pattern memory in Drosophila (Liu et al. 2006). Thus, we hypothesized that this region might also be the location of for function in visual pattern memory. To prove this hypothesis, the Gal4 line c205, in which F5 neurons are labeled, was assigned to drive the local expression of for. Indeed, effectors of the three for isoforms were sufficient to restore visual pattern memory in forS flies (forS;c205/+: PI8 = 0.10 ± 0.07, t-test, t = 1.43, P = NS; UAS-forP1;forS;c205/+: PI8 = 0.40 ± 0.08, t-test, t = 4.99, P < 0.001; forS;c205/UAS-forP2: PI8 = 0.53 ± 0.09, t-test, t = 5.60, P < 0.001; forS;c205/UAS-forP3: PI8 = 0.42 ± 0.09, t-test, t = 4.55, P < 0.001; Fig. 4C). These results are consistent with the observation that the visual pattern memory defect in the rut mutant is restored by expression of the wild-type rut cDNA in F5 neurons. Interestingly, a similar memory rescue was obtained in forS flies when for expression was enhanced in the EB, another component of the CX, using the Gal4 line c819, in which R2 and R4m ringlike layers in the EB are specifically marked (forS;c819/+: PI8 = 0.06 ± 0.07, t-test, t = 0.96, P = NS; UAS-forP1;forS;c819/+: PI8 = 0.62 ± 0.07, t-test, t = 8.74, P < 0.001; forS;c819/UAS-forP2: PI8 = 0.45 ± 0.08, t-test, t = 5.68, P < 0.001; forS;c819/UAS-forP3: PI8 = 0.22 ± 0.07, t-test, t = 3.17, P < 0.01; Fig. 4D). In both cases, rescued flies with each of the three for isoforms showed memory restoration, indicating that the FB and the EB, represented by the expression patterns in the c205 and c819 lines, are two important regions in which for-encoded PKG is involved in visual pattern memory. Overexpression of forP1 or DC0+ did not boost memory (UAS-forP1;;c819: PI8 = 0.31 ± 0.08, t-test, t = 3.75, P < 0.01; c819/UAS-DC0+: PI8 = 0.06 ± 0.07, t-test, t = 0.85, P = NS). It is worth noting that the ectopic expression of any of these isoforms led to memory recovery, probably because all three for-encoded PKG isoforms share a common catalytic domain, which is responsible for their kinase activity. Therefore, these results suggest that expression of for in certain neurons of the FB and the EB is sufficient for visual pattern memory.
Temporary expression of for in adulthood is sufficient to rescue visual pattern memory in forS flies
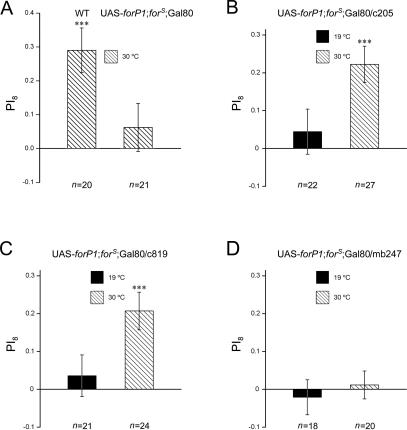
To exclude possible developmental effects in rescue experiments caused by constitutive Gal4 activity, we tried to use a temporal and regional gene expression targeting system (TARGET), in which Gal4 activity is suppressed by a temperature-sensitive Gal80ts at the permissive temperature (19°C) and temporarily induced by shifting to the restrictive temperature (30°C), to temporarily express a target gene (McGuire et al. 2003). The temperature change from 19°C to 30°C did not alter flies’ visual pattern memory (Fig. 5A; WT: PI8 = 0. 29 ± 0.07, t-test, t = 4.37, P < 0.001).
Figure 5.
Adult rescue of forS memory in the central complex. (A) Wild-type flies showed normal visual pattern memory and flies with genotype of UAS-forP1;forS; tub-Gal80ts showed no visual pattern memory, when raised at 19°C until adulthood and then shifted to 30°C for 36 h prior to memory test. (B–D) Three groups of flies (UAS-forP1;forS;c205 or c819 or mb247/tub-Gal80ts) were kept at 19°C and then transferred to 30°C before memory test. Flies in which for is temporarily expressed in the FB (c205) and the EB (c819) after heat-shock treatment show memory levels significantly higher than zero (B,C), but adult-limited expression of for in the mushroom bodies (MBs) (mb247) did not restore the memory defect in forS flies (D). The visual pattern memory of the corresponding control groups kept at 19°C before the memory test was not different from zero. The genotypes of flies are shown above the bars. Hatched bars indicate the memory levels of the flies kept at 19°C before shifting to 30°C for 36 h in adulthood, and solid bars indicate the memory levels of the flies kept at 19°C throughout. Error bars are SEMs. ***P < 0.001; (n) numbers of flies tested.
As the three major PKG isoforms encoded by for were functionally consistent in the rescue assay, one line carrying UAS-forP1 on the X chromosome was chosen as representative, to express for in adults. Therefore, we raised flies (UAS-forP1;forS; c205 or c819/tub-Gal80ts) at 19°C and then, in adulthood, transferred them to 30°C for 36 h prior to a memory test in the flight simulator. As a control group, flies of the same genotype were kept at 19°C until they were tested for visual pattern memory. As shown in Figure 5, B and C, the flies that had been kept at 30°C in adulthood showed almost complete rescue of the memory defect seen in forS flies (UAS-forP1;forS;c205/tub-Gal80ts: PI8 = 0.22 ± 0.05, t-test, t = 4.63, P < 0.001; UAS-forP1;forS;c819/tub-Gal80ts: PI8 = 0.21 ± 0.05, t-test, t = 4.16, P < 0.001), whereas the control groups for both Gal4 driver lines displayed significantly reduced memory performance indistinguishable from that of forS flies (UAS-forP1;forS;c205/tub-Gal80ts: PI8 = 0.04 ± 0.06, t-test, t = 0.75, P = NS; UAS-forP1;forS;c819/tub-Gal80ts: PI8 = 0.04 ± 0.06, t-test, t = 0.66, P = NS). To exclude the possibility that leaky expression of the UAS transgene was responsible for this rescue, we also raised flies (UAS-forP1;forS; tub-Gal80ts/+) at 19°C before transferring them to 30°C in adulthood. As shown in Figure 5A, no visual pattern memory was observed after inactivation of Gal80ts. These results indicated that temporary expression of for in either FB neurons (as in the c205 line) or EB neurons (as in the c819 line) was sufficient to rescue the memory defect seen in forS flies. Meanwhile, as a parallel control, an MB-specific Gal4 driver line mb247 was also used. Flies reared at 30°C in adulthood showed the same memory defect as controls lacking ectopic for gene expression at 19°C (UAS-forP1;forS;mb247/tub-Gal80ts 30°C: PI8 = 0.01 ± 0.04, t-test, t = 0.32, P = NS; 19°C: PI8 = −0.02 ± 0.05, t-test, t = 0.46, P = NS; Fig. 5D). This is consistent with a report that MBs are dispensable for visual pattern memory (Wolf et al. 1998). Collectively, these data support our previous results showing that for-dependent visual pattern memory is localized to the FB and the EB. Furthermore, temporally restricted expression of for in these regions is sufficient for memory rescue.
FB and EB show different pattern specificities in visual memory
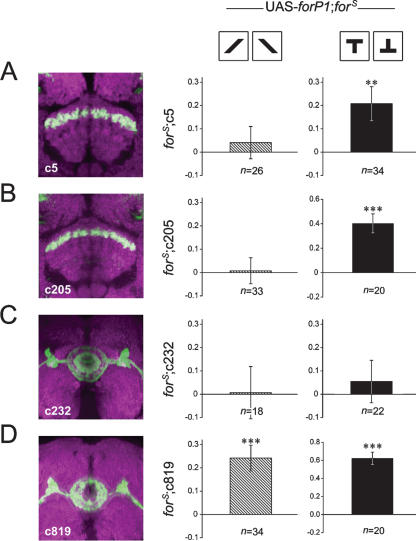
The FB is a structure of functional differentiation with a matrix of layers. Among these layers, rut-dependent short-term memory for the pattern parameters “elevation” and “contour orientation” is divergently localized (Liu et al. 2006). The patterns used in our experiments described so far exclusively addressed the parameter “elevation,” represented by upright and inverted T-shaped patterns. Thus, we wondered whether the same situation existed in for-dependent visual pattern memory. To address this question, wild-type for was again locally expressed to rescue the memory defect in forS flies. In rescue experiments with the c205 line (UAS-forP1;forS;c205/+), only memory for “elevation” was restored (Fig. 4C) but not that for “contour orientation” (PI8 = 0.01 ± 0.06, t-test, t = 0.13, P = NS). Similarly, another driver line, c5, in which F5 neurons in the FB are also marked, but probably with minor pattern differences from c205, rescued only the defect in memory for “elevation” (PI8 = 0.21 ± 0.07, t-test, t = 2.84, P < 0.01) but not the defect in memory for “contour orientation” (PI8 = 0.04 ± 0.07, t-test, t = 0.58, P = NS) (Fig. 6A,B). Next, we extended our study of this pattern-specific effect to the EB. Surprisingly, expression of for by the Gal4 line c819 sufficiently restored visual pattern memory for “contour orientation” (PI8 = 0.24 ± 0.06, t-test, t = 4.35, P < 0.001) in addition to that for “elevation” (Fig. 4D). However, another driver line, c232, in which R3 and R4d ringlike layers in the EB are labeled, failed to rescue the memory defect for either pattern parameter (elevation, PI8 = 0.05 ± 0.09, t-test, t = 0.60, P = NS; contour orientation, PI8 = 0.01 ± 0.11, t-test, t = 0.06, P = NS) (Fig. 6C,D). These results suggest that, in terms of for-dependent visual pattern memory, the F5 neurons in the FB underlie memory for “elevation” but not memory for “contour orientation,” whereas the EB neurons labeled in c819 flies are sufficient for both parameters.
Figure 6.
Memory rescue of two pattern parameters in a spatially specific manner. The Gal4 lines c5 (A), c205 (B), c232 (C), and c819 (D), driving forP1 transcript in a forS background, were tested for memory rescue using the single pattern parameters “contour orientation” or “elevation.” In the presence of the pattern parameter “contour orientation” (differently orientated bars), the visual pattern memory scores of three groups of flies (UAS-forP1;forS;c5 or c205 or c232/+) are not significantly higher than zero (A–C), whereas that of flies of the genotype UAS-forP1;forS;c819/+ is (D). In the presence of the pattern parameter “elevation” (the upright T and inverted T), flies of the genotype UAS-forP1;forS;c5/+, as well as UAS-forP1;forS;c205/+ and UAS-forP1;forS;c819/+ (duplicated from Fig. 4 for clearer comparison) show memory scores significantly higher than zero (A,B,D) whereas flies of the genotype UAS-forP1;forS;c232/+ do not (C). Hatched bars indicate the memory levels for the parameter “contour orientation,” and solid bars indicate the memory levels for the parameter “elevation.” The expression pattern of each Gal4 driver line in the central complex is shown by GFP (green; overlay white). The synaptic neuropil is stained by anti-nc82 antibody (magenta). Y-axes represent PI8 of the memory test. The maternal genotypes are shown at the top, and the paternal genotypes are at the left of the panel of behavioral results. Error bars are SEMs. **P < 0.01; ***P < 0.001; (n) numbers of flies tested.
Discussion
The PKG-encoding Drosophila gene foraging, which was named after its initially discovered function in food-search behavior, has been reported to be required for nonassociative learning and olfactory associative learning in flies (Engel et al. 2000; Scheiner et al. 2004; Kaun et al. 2007; Mery et al. 2007). In this paper, we presented evidence to show that for plays an important role in a complex form of behavior, visual operant conditioning. Reduced for-encoded PKG levels led to defects in visual pattern memory. Specifically, up-regulation of spatiotemporally restricted for expression could facilitate memory, indicating that the effect of for takes place during adulthood in both the FB and the EB. Moreover, just like the rutabaga adenylyl cyclase (Liu et al. 2006), for also affects visual memory in a pattern-specific manner.
The relationship between for and visual pattern memory was established through functional studies. First from an analysis of different for allelic variants and then from targeted up-regulation of for, visual memory was positively correlated with the for expression level. It is worth noting that all three major isoforms of for-encoded PKG were able to successfully rescue visual pattern memory defects, implying that they are functionally interchangeable. Nevertheless, it is possible that only one or a few forms of PKG are endogenously responsible for visual pattern memory because of the likely limited regional localization of each isoform.
In addition to the functional localization of for, the study of timing showed that for is temporarily sufficient for visual pattern memory. Using temperature-sensitive Gal80ts to regulate Gal4 activity, we were able to induce for expression during adulthood by raising the temperature, thereby rescuing the defective memory in forS flies. This acute effect means that for-encoded PKG and the corresponding biochemical pathways play fundamental roles in visual pattern memory and may even represent the underlying molecular mechanism. On the other hand, for may also affect developmental processes that facilitate visual pattern memory to a certain extent. This is not surprising, though, as PKG has been shown to be involved in the development of the nervous system in Drosophila and other species (Renger et al. 1999; Yamamoto and Suzuki 2005; Zhang et al. 2005).
One important but unanswered question in our work is how for-encoded PKG affects visual pattern memory. As a key factor in the NO and cGMP signaling pathways, PKG has plenty of potential substrates (Wang and Robinson 1997). It may phosphorylate protein substrates that modulate neurotransmission, neuronal excitability, and even gene expression (Wang and Robinson 1997). In our study, we focused only on short-term memory, which most likely involves PKG substrates like ion channels, cytoskeleton proteins, or calcium signaling-regulated proteins that subsequently lead to instant electrophysiological modifications in neurons. Nevertheless, it is reasonable to postulate that long-term memory also involves PKG, as many studies have established a role for PKG in LTP and LTD, which are generally thought to underlie long-term memory in mammals (Zhuo et al. 1994; Feil et al. 2003).
There are a number of possible explanations for the finding that the c819-driven expression of for in EB neurons successfully restored defects in the memory for the visual pattern parameters “elevation” and “contour orientation,” which have previously been shown to be mediated by FB neurons. First, it is possible that c819 is also expressed in FB neurons. This possibility can be largely excluded as we were unable to detect any signal in the FB when membrane-tethered GFP was expressed under the control of c819. Furthermore, if there is minimal c819 expression in the FB that was not detected, such a low level of expression might not be enough to restore for expression and subsequently rescue the memory defect. The second possible explanation for our finding is that the FB and the EB may work together to facilitate visual memory formation. If we take into account the possibility that flies may have different levels of memory (in our scoring system, only those flies with memory above a certain threshold were scored as having “normal memory,” and those under this threshold were scored as having “no memory”), the memory of flies may rely on the overall activity of FB and EB neurons. Indeed, it is possible that forS flies with residual PKG activity possess memory ability just slightly below the threshold level, so that elevation of PKG activity in the EB could restore EB neuron activity and subsequently enhance FB neuron activity followed by restoration of visual pattern memory. With regard to our previous finding that tetanus toxin and Gαs* block memory formation in the FB (Liu et al. 2006), both tetanus toxin and Gαs* might induce a significantly severe neuronal malfunction in FB neurons; because the overall function of FB and EB neurons would then be abnormal, memory would also be deficient. Further evidence at the circuit level and functional level is required to elucidate the details of this mechanism before reaching a final conclusion. The application of more Gal4 lines may help to dissect these neural circuits and behaviors in more detail.
Recently, Kaun et al. (2007) published an article on the effects of for on larval memory. Their report is consistent with our conclusion that for is involved in Drosophila memory, although their results suggested that for affected larval olfactory conditioning but not visual conditioning. The ostensible discrepancy between the function of for in larval visual memory that they reported, and its role in adult visual memory that we described here, can be explained by the huge difference between larval and adult fly visual information processing systems and behavioral paradigms. For example, the EB and the FB, which are important for adult fly visual memory, have not been reported to form in larval fly. Rather, the consistency between the two reports confirmed a common function of for in Drosophila memory.
While this paper was in preparation, another report was published suggesting that forR and forS larvae have different thermotolerances, as measured by mouth-hook movement and evoked excitatory junction potentials in neuromuscular junctions (Dawson-Scully et al. 2007); this report overtly confounds our conclusion, since heat was used in our behavioral paradigm as reinforcement. However, if analyzed carefully, the effect of thermotolerance does not reduce the relevance of our behavioral test. Even if forR and forS adult flies have different thermotolerance ability, this difference in thermotolerance does not affect visual learning in our paradigm, since forR and forS flies have similar thermosensation and thermal-avoidance abilities (Fig. 2B), and these are the real heat-related factors that could affect the learning behavior. Moreover, the possible nonspecific thermal effect of heating on visual learning and memory ability is negligible, since the effects of laser-induced heat are limited to the body surface and, in our experiment, to the abdomen; thus, the brain is almost certainly not affected. Furthermore, according to the report by Dawson-Scully et al. (2007), forS flies should be more resistant to the negative effect of heat, and thus perform better, than forR flies; however, our study clearly showed that forR flies had better memory than forS flies. Thus, the difference in performance between forR and forS flies resulted from different learning/memory abilities but not from a hypothetical difference in thermotolerance.
An interesting phenomenon is that the difference in behavioral performance between forR and forS flies was drastic, while the corresponding difference in PKG activity between them was previously reported to be only 10% (Osborne et al. 1997), which is generally considered to be tiny. We suggest that a 10% difference in overall PKG activity does not necessarily mean that the same 10% difference applies evenly to all PKG isoforms and all PKG-expressing brain regions. It is possible that the activity of certain PKG isoforms, or the PKG level in certain local brain regions, is reduced by more than 10% in forS flies compared with forR flies, and that such a difference leads to distinct behavioral performances in visual learning and memory. Additionally, it is also possible that some behavior, such as visual learning and memory, may be sensitive to PKG activity at a certain limited activity range, for example, the range between forR and forS flies, but not outside of this range. Further fine experimental investigation is needed to elaborate the details.
In our experiments, as 189y, which turned out a lilli allele, was used as a clue to derive the for story, it could be asked whether the forS allelic phenotypes in this study actually resulted from the contamination of lilli mutations, or from 189y with the contamination of forS allele. From our observations, overexpression of lilli was unable to restore the memory defects in forS flies in our visual learning paradigm. This result, together with the result that overexpression of for could restore the memory defect, indicated that the memory defect in forS flies is not caused by lilli abnormality, but by a variation in the for gene itself. In addition, forS-like larval foraging phenotype in 189y flies, as well as the expression of lilli and for, was completely reversed after precise P element excision, suggesting that 189y itself is a lilli allele, but not on a forS genetic background, and that for is somehow subsequently affected. We postulated that there could be some interaction between lilli and for, but the details of such an interaction are not yet clear.
Future studies on for function should identify the PKG substrates and putative crosstalk between for and lilli that contribute to learning and memory; it will be particularly interesting to clarify the relationship between the FB and the EB in terms of visual pattern memory. Unraveling this circuitry will be a big step toward a full understanding of the relationship between brain structures and complex behaviors in fruit flies.
Materials and Methods
Fly stocks
All flies were maintained at 25°C on standard corn meal/molasses medium (Guo et al. 1996) in a 12-h light/12-h dark cycle at 60% humidity. Two- to 6-day-old flies were used in the experiments. Two for allelic variant lines, forR and forS (kindly provided by M.B. Sokolowski, University of Toronto, Canada), were outcrossed for eight generations to get a wild-type CS genetic background and bred to homozygosity before use. The following fly strains were also used: wild-type Canton-S (CS); Df(2L)ed1; UAS-lilli+ (kindly provided by A. Muller, University of Dundee, Scotland); tub-Gal80ts; the Gal4 lines 189y, elav-Gal4, c205, c5, c819, c232, and mb247. 189yΔE is a P element precise excision line generated in our laboratory from 189y flies according to standard procedures.
Inverse PCR
Standard methods for inverse PCR were used to identify the P element insertion site of the Gal4 line 189y. Genomic DNA was prepared from ∼50 adult flies using a QIAamp DNA Mini Kit (QIAGEN) and digested with HpaII. DNA fragments were then self-ligated overnight at 16°C using T4 DNA ligase (Invitrogen). Ligated fragments containing the 5′ end of P[GawB] and flanking genomic DNA were amplified by PCR with the primer pair iPCR (5′-CTCCACAATTCCGTTGGATT-3′ and 5′-CTATCGACGGGAC CACCTTA-3′) and sequenced. The P[GawB] elements were superimposed on the genomic map of known or predicted genes by a BLAST search of the flanking DNA sequence against the Drosophila genome sequence, available from the Berkeley Drosophila Genome Project (BDGP).
To confirm the newly localized P element insertion sites in 189y flies, the following pairs of primers were used for regular PCR: P1s: 5′-CTAACGCCCCCGCTGACC-3′; P1a: 5′-GCACCG CCTACATACCTCGC-3′; P2s: 5′-CTGTGTTTGTCTCCGTCT-3′; P2a: 5′-TGCTTCACTTATTGGTTGTC-3′.
Generation of transgenic flies
Approximately 100 wild-type CS flies were homogenized in TRIzol reagent (Invitrogen) to isolate total RNA, from which mRNA was extracted using an Oligotex mRNA Mini Kit (QIAGEN). First-strand cDNA was synthesized from mRNA using a SuperScript II first-strand synthesis system (Invitrogen). It was then amplified by PCR reactions using pfu polymerase (Promega) and primers designed based on sequences from the FlyBase Consortium (http://www.flybase.org). After the relevant restriction digestion, DNA fragments spanning the transcripts of forP1 (with the help of the inner XhoI cut site) and forP3 coding regions were directly cloned into pUAST vectors (Brand and Perrimon 1993). A DNA fragment corresponding to the transcript of the forP2 coding sequence was first cloned into the pGEM-T vector (Promega) and then subcloned into the pUAST vector. All cloning steps were confirmed by restriction digestion and DNA sequencing. The primers used in PCR reactions were as follows (lower-case letters represent restriction sites): forP1 part 1 upper: 5′-CGGgaattcATG CGTTTCTGCTTTGAT-3′; forP1 part 1 lower: 5′-AATATGGC AGCCTTGATAAGTTCAC-3′; forP1 part 2 upper: 5′-CCACA CGCAAGTCGGGTCAG-3′; forP1 part 2 lower; 5′-AATAA TcggccgTCAGAAGTCCTTGTC-3′; forP2 upper: 5′-AAAgc ggccgcATGCAGAGTCTGCGGATCTCG-3′; forP2 lower: 5′-CGCCCGgctagcTCAGAAGTCCTTGTCCCATCC-3′; forP3 upper: 5′-AATATAcggccgATGAAAATCAAACATTATCCGGGC-3′; forP3 lower: 5′-GGTGGTctcgagTCAGAAGTCCTTGTCCCATC-3′.
UAS-forP1, UAS-forP2, and UAS-forP3 plasmids were then purified and subjected to germline transformation into w1118 embryos according to standard protocols, as previously described (Rubin and Spradling 1982). UAS-for+ transgenic lines with P elements on the first and third chromosomes were established and changed into a homozygous forS genetic background.
Quantitative PCR
Quantitative PCR (qPCR) was used in a standard way. In brief, 0.5 μL of cDNA mixtures prepared from the heads of 3- to 5-d-old flies, as described above, were used as templates and tested on a Chromo 4 system (MJ Research/Bio-Rad). For each fly strain, cDNA was obtained from six independent RNA preparations for repeating and averaging. The relative differences in for or lilli mRNA expression levels were quantified by comparing their levels with standard curves, which were constructed using the corresponding recombinant plasmids, and normalized to the level of actin. One-way analysis of variance (ANOVA) was used for the statistical analysis of relative mRNA levels. The primers used for qPCR were as follows: qPCR-actin upper, 5′-CAGGCGGT GCTTTCTCTCTA-3′; qPCR-actin lower, 5′-AGCTGTAACCGC GCTCAGTA-3′; qPCR-for upper, 5′-AGGCGGAGTACAGC GATTTC-3′; qPCR-for lower, 5′-CGCACTTTTCCCTTGGATATG 3′; qPCR-lilli upper, 5′-ACACCAACTGCCGGATAGTC-3′; qPCR-lilli lower, 5′-CCACTTCCAACCTGTGACCT-3′.
Immunohistochemistry
Dissection of adult brains was performed in cold PBS (phosphate buffer saline) to remove the cuticle and connective tissues. After a brief wash in PBS, samples were fixed in freshly prepared 4% paraformaldehyde in PBS for 2 h at room temperature, rinsed for 3 × 15 min in 0.5% PBT (PBS with 0.5% Triton X-100) followed by blocking with 10% PNT (10% normal goat serum in PBT) for 2 h at room temperature. Subsequently, samples were incubated with primary antibodies overnight at 4°C, washed 3 × 15 min with PBT, and then incubated in PNT containing secondary antibody. After 3 × 15 min washes in PBT, brains were mounted in Vectashield (Vector Laboratories) and viewed with an Olympus FV500 confocal microscope. Raw confocal images were imported into Leica Deblur software (AutoQuant Imaging, Inc.) to reconstruct 3D images. Modifications were made using Adobe Photoshop (Adobe Systems) to obtain optimal visual effects. Care was taken to ensure that brightness and contrast alterations were performed on the entire panel without losing any detail. Monoclonal mouse anti-nc82 (kindly provided by E. Buchner, University of Wuerzburg, Germany) primary antibody was used at a dilution of 1:10. TRITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch) was used at a 1:100 dilution.
Visual pattern memory assay
In the flight simulator, a single tethered fly was suspended at a torque meter to measure its yaw torque in the center of an arena. The arena is rotated by an electric motor such that its angular velocity is proportional to, but directed against, the fly’s yaw torque. The fly’s yaw torque determines the angular velocity of the arena instead of rotating the fly’s own body. This arrangement allows the tethered fly to stabilize and choose its flight orientation with respect to the arena by adjusting its yaw torque. A computer continuously registers yaw torque and the angular position of the arena. To test visual learning, a beam of infrared light was directed at the fly as an instantaneous source of heat. The beam can be intercepted by a computer-driven electric shutter. The arena was virtually divided into four quadrants with patterns at their respective centers. During training, the computer opened the shutter whenever the fly was heading into a quadrant with, for example, pattern A, and closed it when the fly was oriented toward one of the two adjacent quadrants with pattern B. Hence, half of all possible orientations in the arena were paired with heat, and the others were paired with ambient temperature. During tests, heat was permanently switched off. Angular position was recorded every 50 msec, and orientation preferences were calculated in nine consecutive 2-min periods (performance index, PI1–9). Pattern A was paired with ambient temperature during training, and pattern B with heat. Each of the two patterns was pattern A in half of the experiments. This procedure eliminates any effects of spontaneous or nonassociatively induced pattern preferences. If tA is the time the fly spends heading toward the quadrants with pattern A, and tB is the time heading toward pattern B-containing quadrants, then the performance index (PI) is calculated as
Mean PIs were obtained for nine consecutive 2-min periods (PI1–9), and PI8 was used as an index of short-term memory. Before each 2-min period, the arena was rotated to an arbitrary position. Upright and inverted T-shaped patterns were used in the test of pattern parameter “elevation,” while bars of two different orientations were used in the test of pattern parameter “contour orientation.” Unless specifically mentioned, only the T-shaped patterns were applied. For the experiments in which heat was not used as punishment, the arena was oscillated (peak-to-peak amplitude, App = ±7.5°; frequency, f = 5 Hz) as a negative reinforcement. All behavioral experiments were performed at 25°C. A one-sample t-test (two-sided P-value) was used for the statistical analysis of memory scores (PI8). NS: P ≥ 0.05.
Flies carrying tub-Gal80ts were cultured at 19°C. The experimental groups were shifted to 30°C for 36 h prior to behavioral tests, while the control groups without heat shock treatment were raised at 19°C throughout development and then left at 25°C for adaptation just before behavioral tests.
Flies’ pattern discrimination abilities were measured as previously described (Liu et al. 2006). In brief, the orientation distribution of an individual fly during the two successive 2-min intervals of the pretest (PI1 and PI2) was analyzed by Fourier transformation. The discrimination value (D), which was used to evaluate a fly’s pattern discrimination ability, is calculated as the rate of the amplitude of the 180° component to the mean amplitude of the (near-by) 120° and 72° components (D = 2A180/(A120 + A72); subscripts refer to Fourier components; A = amplitude). The evaluation of a 4-min pretest with four identical patterns (bars) in wild-type flies was also obtained in the same way. A one-sample t-test (two-sided P-value) was used for the statistical analysis of discrimination values against the chance value (D = 1).
Larval foraging assay
Third-instar larval foraging was assessed and quantified as described in Osborne et al. (1997). Strain differences in behavior on yeast were statistically analyzed using ANOVA.
Acknowledgments
We thank M.B. Sokolowski, A. Muller, J.A. Kiger, M. Saitoe, and E. Buchner for providing fly stocks and antibody. We also thank M. Heisenberg and R. Wolf for valuable comments on the manuscript, and H.Y. Gong for excellent technical assistance. This work was supported by the ‘973 Program’ (2005CB522804, L.L.), the National Natural Sciences Foundation of China (30621004, L.L., and 30770682, G.Z.-F.), and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-R-28, L.L.).
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.873008.
References
- Ben-Shahar Y. The foraging gene, behavioral plasticity, and honeybee division of labor. J. Comp. Physiol. A. 2005;191:987–994. doi: 10.1007/s00359-005-0025-1. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y., Robichon A., Sokolowski M.B., Robinson G.E. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- Bernabeu R., Schroder N., Quevedo J., Cammarota M., Izquierdo I., Medina J.H. Further evidence for the involvement of a hippocampal cGMP/cGMP-dependent protein kinase cascade in memory consolidation. Neuroreport. 1997;8:2221–2224. doi: 10.1097/00001756-199707070-00026. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- de Belle J.S., Hilliker A.J., Sokolowski M.B. Genetic localization of foraging (for): A major gene for larval behavior in Drosophila melanogaster. Genetics. 1989;123:157–163. doi: 10.1093/genetics/123.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson-Scully K., Armstrong G.A., Kent C., Robertson R.M., Sokolowski M.B. Natural variation in the thermotolerance of neural function and behavior due to a cGMP-dependent protein kinase. PLoS ONE. 2007;2:e773. doi: 10.1371/journal.pone.0000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill M., Wolf R., Heisenberg M. Behavioral analysis of Drosophila landmark learning in the flight simulator. Learn. Mem. 1995;2:152–160. doi: 10.1101/lm.2.3-4.152. [DOI] [PubMed] [Google Scholar]
- Dubnau J., Tully T. Gene discovery in Drosophila: New insights for learning and memory. Annu. Rev. Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- Engel J.E., Xie X.J., Sokolowski M.B., Wu C.F. A cGMP-dependent protein kinase gene, foraging, modifies habituation-like response decrement of the giant fiber escape circuit in Drosophila. Learn. Mem. 2000;7:341–352. doi: 10.1101/lm.31600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Hartmann J., Luo C., Wolfsgruber W., Schilling K., Feil S., Barski J.J., Meyer M., Konnerth A., De Zeeuw C.I., et al. Impairment of LTD and cerebellar learning by Purkinje cell-specific ablation of cGMP-dependent protein kinase I. J. Cell Biol. 2003;163:295–302. doi: 10.1083/jcb.200306148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J.L., Higgins G.C., Jackson F.R. Biochemical properties and cellular localization of the Drosophila DG1 cGMP-dependent protein kinase. J. Biol. Chem. 1996;271:23322–23328. doi: 10.1074/jbc.271.38.23322. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Sengupta P., McIntire S.L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Guo A., Li L., Xia S.Z., Feng C.H., Wolf R., Heisenberg M. Conditioned visual flight orientation in Drosophila: Dependence on age, practice, and diet. Learn. Mem. 1996;3:49–59. doi: 10.1101/lm.3.1.49. [DOI] [PubMed] [Google Scholar]
- Hanesch U., Fischbach K.F., Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 1989;257:343–366. [Google Scholar]
- Hofmann F., Feil R., Kleppisch T., Schlossmann J. Function of cGMP-dependent protein kinases as revealed by gene deletion. Physiol. Rev. 2006;86:1–23. doi: 10.1152/physrev.00015.2005. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Rubin G.M. cGMP-dependent protein kinase genes in Drosophila. J. Biol. Chem. 1989;264:10738–10748. [PubMed] [Google Scholar]
- Kaun K.R., Hendel T., Gerber B., Sokolowski M.B. Natural variation in Drosophila larval reward learning and memory due to a cGMP-dependent protein kinase. Learn. Mem. 2007;14:342–349. doi: 10.1101/lm.505807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Etoile N.D., Coburn C.M., Eastham J., Kistler A., Gallegos G., Bargmann C.I. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- Liu G., Seiler H., Wen A., Zars T., Ito K., Wolf R., Heisenberg M., Liu L. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- Mao Z., Roman G., Zong L., Davis R.L. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc. Natl. Acad. Sci. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McGuire S.E., Deshazer M., Davis R.L. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog. Neurobiol. 2005;76:328–347. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Mery F., Belay A.T., So A.K., Sokolowski M.B., Kawecki T.J. Natural polymorphism affecting learning and memory in Drosophila. Proc. Natl. Acad. Sci. 2007;104:13051–13055. doi: 10.1073/pnas.0702923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne K.A., Robichon A., Burgess E., Butland S., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Oster H., Werner C., Magnone M.C., Mayser H., Feil R., Seeliger M.W., Hofmann F., Albrecht U. cGMP-dependent protein kinase II modulates mPer1 and mPer2 gene induction and influences phase shifts of the circadian clock. Curr. Biol. 2003;13:725–733. doi: 10.1016/s0960-9822(03)00252-5. [DOI] [PubMed] [Google Scholar]
- Renger J.J., Yao W.D., Sokolowski M.B., Wu C.F. Neuronal polymorphism among natural alleles of a cGMP-dependent kinase gene, foraging, in Drosophila. J. Neurosci. 1999;19:RC28. doi: 10.1523/JNEUROSCI.19-19-j0002.1999. http://www.jneurosci.org/cgi/content/full/19/19/RC28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S.C., Armstrong J.D., Yang M., Wang Z., An X., Kaiser K., Taghert P.H. Genetic analysis of the Drosophila ellipsoid body neuropil: Organization and development of the central complex. J. Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- Rubin G.M., Spradling A.C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schafer W.R. PKG and the neural basis for behavioral phenotypes. Neuron. 2002;36:991–993. doi: 10.1016/s0896-6273(02)01135-2. [DOI] [PubMed] [Google Scholar]
- Scheiner R., Sokolowski M.B., Erber J. Activity of cGMP-dependent protein kinase (PKG) affects sucrose responsiveness and habituation in Drosophila melanogaster. Learn. Mem. 2004;11:303–311. doi: 10.1101/lm.71604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkau S.A., Weber E.T., Abbott S.M., Mitchell J.W., Gillette M.U. Circadian clock-controlled regulation of cGMP-protein kinase G in the nocturnal domain. J. Neurosci. 2003;23:7543–7550. doi: 10.1523/JNEUROSCI.23-20-07543.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkau S.A., Mitchell J.W., Pace L.A., Barnes J.W., Barnes J.A., Gillette M.U. Protein kinase G type II is required for night-to-day progression of the mammalian circadian clock. Neuron. 2004;43:539–549. doi: 10.1016/j.neuron.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Waddell S., Quinn W.G. Flies, genes, and learning. Annu. Rev. Neurosci. 2001;24:1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- Wang X., Robinson P.J. Cyclic GMP-dependent protein kinase and cellular signaling in the nervous system. J. Neurochem. 1997;68:443–456. doi: 10.1046/j.1471-4159.1997.68020443.x. [DOI] [PubMed] [Google Scholar]
- Werner C., Raivich G., Cowen M., Strekalova T., Sillaber I., Buters J.T., Spanagel R., Hofmann F. Importance of NO/cGMP signalling via cGMP-dependent protein kinase II for controlling emotionality and neurobehavioural effects of alcohol. Eur. J. Neurosci. 2004;20:3498–3506. doi: 10.1111/j.1460-9568.2004.03793.x. [DOI] [PubMed] [Google Scholar]
- Wolf R., Wittig T., Liu L., Wiestmann G., Eyding D., Heisenberg M. Drosophila mushroom bodies are dispensable for visual, tactile, and motor learning. Learn. Mem. 1998;5:166–178. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Suzuki N. Expression and function of cGMP-dependent protein kinase type I during medaka fish embryogenesis. J. Biol. Chem. 2005;280:16979–16986. doi: 10.1074/jbc.M412433200. [DOI] [PubMed] [Google Scholar]
- Zars T., Fischer M., Schulz R., Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000a;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- Zars T., Wolf R., Davis R., Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: In search of the engram. Learn. Mem. 2000b;7:18–31. doi: 10.1101/lm.7.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Beuve A., Townes-Anderson E. The nitric oxide-cGMP signaling pathway differentially regulates presynaptic structural plasticity in cone and rod cells. J. Neurosci. 2005;25:2761–2770. doi: 10.1523/JNEUROSCI.3195-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo M., Hu Y., Schultz C., Kandel E.R., Hawkins R.D. Role of guanylyl cyclase and cGMP-dependent protein kinase in long-term potentiation. Nature. 1994;368:635–639. doi: 10.1038/368635a0. [DOI] [PubMed] [Google Scholar]