Abstract
Drosophila transient-receptor-potential (TRP) is a Ca2+ channel responsible for the light-dependent depolarization of photoreceptors. TRP is anchored to a macromolecular complex by tethering to inactivation-no-afterpotential D (INAD). We previously reported that INAD associated with the carboxyl tail of TRP via its third post-synaptic density protein 95, discs-large, zonular occludens-1 domain. In this paper, we further explored the molecular basis of the INAD interaction and demonstrated the requirement of the last 14 residues of TRP, with the critical contribution of Gly1262, Val1266, Trp1274, and Leu1275. We also revealed by pull-down assays that the last 14 residues of TRP comprised the minimal sequence that competes with the endogenous TRP from fly extracts, leading to the co-purification of a partial INAD complex containing INAD, no-receptor-potential A, and eye-protein kinase C (PKC). Eye-PKC is critical for the negative regulation of the visual signaling and was shown to phosphorylate TRP in vivo. To uncover the substrates of eye-PKC in the INAD complex, we designed a complex-dependent eye-PKC assay, which utilized endogenous INAD complexes isolated from flies. We demonstrate that activated eye-PKC phosphorylates INAD, TRP but not no-receptor-potential A. Moreover, phosphorylation of TRP is dependent on the presence of both eye-PKC and INAD. Together, these findings indicate that stable kinase-containing protein complexes may be isolated by pull-down assays, and used in this modified kinase assay to investigate phosphorylation of the proteins in the complex. We conclude that TRP associates with INAD via its last 14 residues to facilitate its regulation by eye-PKC that fine-tunes the visual signaling.
Keywords: eye-protein kinase C, inactivation-no-afterpotential D, macromolecular complex, phosphorylation, post-synaptic density protein 95, discs-large, zonular occludens-1 domain, transient-receptor-potential
Drosophila transient-receptor-potential (TRP) is the founding member of the TRP superfamily. Over 25 distinct TRP homologs have been identified, which have been implicated in diverse biological processes including sensory transduction, hot/cold sensation, and store-operated calcium entry (Ramsey et al. 2006; Venkatachalam and Montell 2007). Drosophila TRP is involved in visual transduction, a process that converts the signal of light into a change of membrane potential in photoreceptors of the eye (Pak and Leung 2003; Minke and Parnas 2006; Wang and Montell 2007). In the visual signaling cascade, light activates rhodopsins, which couple to the heterotrimeric Gq-protein to switch on a phospholipase Cβ4 (no-receptor-potential A; NORPA). Activated NORPA catalyzes the hydrolysis of phosphatidylinositol 4, 5-bisphosphate (PIP2) to generate inositol trisphosphate and diacylglycerol (DAG) (Hardie 2001; Pak and Leung 2003). DAG may be involved in opening the TRP channels (Chyb et al. 1999; Hardie 2004) leading to depolarization of photoreceptors. DAG is also a potent activator of the conventional protein kinase C’s (PKCs) (Newton 2001). In Drosophila, eye-PKC is a conventional PKC (Schaeffer et al. 1989) and mediates the negative regulation of the visual signaling (Smith et al. 1991; Hardie et al. 1993; Gu et al. 2005; Popescu et al. 2006). Eye-PKC is co-localized with TRP in a macromolecular complex organized by inactivation-no-afterpotential D (INAD) (Chevesich et al. 1997; Li and Montell 2000; Tsunoda et al. 2001). The INAD complex is important for the subcellular localization and the stability of these INAD-interacting proteins in photoreceptors possibly leading to fast kinetics of the visual signaling (Tsunoda et al. 1997; Scott and Zuker 1998).
The scaffolding protein INAD consists of five postsynaptic density protein 95, discs-large, zonular occludens-1 (PDZ) domains and interacts with the carboxyl terminus of TRP (Huber et al. 1996; Shieh and Zhu 1996; Chevesich et al. 1997; Tsunoda et al. 1997; Li and Montell 2000), eye-PKC (Huber et al. 1996; Tsunoda et al. 1997; Adamski et al. 1998; Kumar and Shieh 2001), and NORPA (Huber et al. 1996; Chevesich et al. 1997; Shieh et al. 1997; Tsunoda et al. 1997; van Huizen et al. 1998) via distinct PDZ domains. PDZ domains are modular protein interaction domains containing 80–100 amino acid residues (Hung and Sheng 2002), and usually associate with the last four residues of the binding proteins. Structural studies have revealed that each PDZ domain contains six β-strands and two α-helices giving rise to an overall structure of a six-strand β-sandwich (Doyle et al. 1996; Morais Cabral et al. 1996; Hillier et al. 1999). A binding pocket is formed between the second β-strand (βB) and the second α-helix (αB) for anchoring a carboxyl terminal peptide with the motif sequence, X (−3)-Ser/Thr/Φ/Tyr (−2)-X (−1)-Φ (0) (X, any amino acid and Φ, a hydrophobic residue, such as Ile, Leu, or Val). PDZ domains and the corresponding binding motifs can be divided into three classes, based on the side chain interactions between the −2 position of the binding motifs, and the first residue at the second α-helix (αB1) of the PDZ domain. In general, a class I PDZ domain contains a His at αB1 and recognizes a class I PDZ binding motif with Ser/Thr at the −2 position. The class II PDZ domains prefer a binding target containing a hydrophobic residue at the −2 position, because the class II PDZ domains have a hydrophobic residue at αB1. In contrast, the class III PDZ domains usually have a Tyr at αB1 and select a binding motif with an acidic residue, Glu/Asp, at the −2 position (Songyang et al. 1997; Hung and Sheng 2002). This classification of PDZ domains into three distinct groups has been recently challenged by a study from MacBeath’s group (Stiffler et al. 2007). By examining the interactions of 157 PDZ domains from the mouse genome, Stiffler et al. (2007) concluded that the selectivity of a PDZ domain was regulated by the interactions throughout the binding pocket, not simply by residues at the −2 position.
In Drosophila, the interaction between INAD and TRP has been well documented. Our laboratory first demonstrated the interaction by the use of protein overlay and co-immunoprecipitation, and uncovered that the third PDZ domain, PDZ3, of INAD was critical for the TRP interaction (Shieh and Zhu 1996). Moreover, we discovered that, because of a missense mutation in PDZ3, the mutant INAD expressed in InaDp215 was not able to bind TRP. We also showed that the carboxyl terminal sequence of TRP was required for the INAD interaction, and Asp substitutions of an internal Val in TRP, Val1266 drastically reduced the INAD interaction (Shieh and Zhu 1996). Recently, Li and Montell (2000) reported that Asp substitutions of Val1266 did not greatly affect the interaction, whereas removal of the last four residues in TRP led to a loss of the INAD association by pull-down assays. It remains to be determined if only the last four residues of TRP are required for the INAD interaction.
In this study, we report our findings supporting that the last 14 residues of TRP is involved in tethering to INAD by both protein overlay and pull-down assays. Importantly, we demonstrate that the carboxyl tail of TRP co-purifies NORPA and eye-PKC from fly extracts. The co-isolation allowed us to examine the eye-PKC dependent phosphorylation of TRP and NORPA in its native pre-assembled complex. We show that eye-PKC becomes active in the presence of phorbol ester and Ca2+ leading to phosphorylation of TRP and INAD, but not NORPA, and that the phosphorylation of TRP is dependent on co-purification of both INAD and eye-PKC. This complex-dependent protein kinase assay will be of great utilities to investigate phosphorylation of other protein kinases that target substrates in a protein complex.
Materials and methods
Molecular biology: site-directed mutagenesis, subcloning, and DNA sequencing
Site-directed mutagenesis was performed by PCR-based overlap extension using mismatch oligonucleotide primers. The PCR products were gel-purified and subcloned into pCR2.1 vector using the TOPO cloning system (Invitrogen, Carlsbad, CA, USA). Desired point mutations were confirmed by automatic DNA sequencing. Clones containing the intended substitutions were subcloned into pGEMEX1/2 (Promega, Madison, WI, USA) or pGEX4T1.
Expression of T7 gene 10 fusion proteins in bacteria
Wild-type and modified TRP carboxyl terminal sequences were expressed as fusion proteins of T7 gene 10 as described previously (Shieh and Zhu 1996). Briefly, pGEMEX1/2 (Promega) derived plasmids were transformed into E. coli (HMS174) strain. Overnight cultures (1 mL) were prepared and used to inoculate a 5 mL Luria Broth broth containing ampicillin (50 µg/mL). Cultures were grown at 37°C for 2–3 h until the density of bacteria (OD600) reached 0.8–1.0. The expression of fusion proteins was induced by infection with CE6 phage (1 mL, 109 pfu) that expresses T7 RNA polymerase (Studier et al. 1990) after addition of MgSO4 to a final concentration of 10 mmol/L. Bacterial cultures were grown for additional 3 h and harvested by centrifugation at 5000 × g. Total bacterial lysates were prepared by solubilizing bacterial pellets with 2x loading dye and analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE).
Expression of GST fusion proteins in bacteria
DNA sequences encoding the carboxyl terminal sequences of trp or norpA were subcloned into pGEX4T1 or pGEX5X3 (GE Healthcare Life Sciences, Piscataway, NJ, USA) to generate DNA constructs for over-expression of glutathione S-transferase (GST) fusion proteins. The recombinant plasmids were introduced into E. coli (BL21) strain for expression. Overnight cultures (1 mL) were prepared and used to inoculate a 50 mL Luria Broth broth containing ampicillin. Cultures were grown at 37°C for 2–3 h until the density of bacteria (OD600) reached 0.6–0.8. The expression of fusion proteins was induced upon the addition of isopropyl-β-d-thiogalactopyranoside (0.1–1 mmol/L) and the bacteria cultures were harvested 3 h post-induction.
Preparation of fly head extracts
Fly heads were isolated in mass by the use of sieves following quick freeze in liquid nitrogen and vortexing to dislocate heads from bodies. One volume of fly heads was extracted with nine volumes of the extraction buffer (50 mmol/L Tris–HCl, pH 8.0, 150 mmol/L NaCl, and 1% Triton X-100 plus a cocktail of protease inhibitors containing 1 mmol/L phenylmethylsulfonyl fluoride, 1 µg/µL leupeptin and pepstatin A, 5 µg/µL aprotinin, 0.01 mmol/L benzamide, and 1 mmol/L benzamidine) for 1 h at 4°C after homogenization. Following extraction, the mixture was centrifuged at 5000 × g for 10 min. The supernatant was used as 1% Triton X-100 extracts in the pull-down assays. Protein concentrations of head extracts (about 3.5–5 µg/mL) were determined by BCA protein assay (Pierce, Rockford, IL, USA).
Pull-down assays using GST fusion proteins
Bacteria harvesting GST fusion proteins were lysed with a lysis buffer (50 mmol/L Tris, pH 7.5, 50 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L 1, 4-dithiothreitol, 1 µg/mL leupeptin, 1 µg/mL aprotinin, 1 µg/mL pepstain, 1 mmol/L benzamide, 1 mmol/L benzamidine, and 1 mmol/L phenylmethylsulfonyl fluoride). Bacterial lysates were incubated with 25 µL of glutathione Sepharose beads (GE Healthcare Life Sciences) to recover GST fusion proteins according to the protocol supplied by the manufacturer. Approximately 10 µg of affinity purified GST fusion proteins (or GST alone) were incubated with 500 µL of 1% Triton X-100 fly head extracts at 4°C for 1 h. Following incubation, the pull-down mixture containing GST fusion proteins and interacting proteins were briefly washed three times with the lysis buffer to remove nonspecific binding proteins and the mixture was analyzed by SDS/PAGE. Fusion proteins and co-purifying associating proteins were detected by Coomassie Blue staining or western blotting following transfer onto nitrocellulose filters. Alternatively, in vitro kinase assays were carried out following pull-down (see below).
In vitro kinase assay
The protein complexes were briefly rinsed with 100 µL of the kinase reaction buffer (50 mmol/L Tris–HCl, pH 8.0, 10 mmol/L MgCl2, 5 mmol/L 2-mercaptoethanol, 0.1 mmol/L dithiothreitol, 0.4 mmol/L EGTA, and 0.7 mmol/L CaCl2) followed by incubating with 50 µL reaction buffer containing phorbol-12-myristate-13-acetate (PMA) (1 µmol/L). The kinase reaction was initiated upon the addition of 3 µCi of carrier-free [γ-32P]-ATP (PerkinElmer, Waltham, MA, USA) in the presence of 100 µmol/L ATP at 30°C for 30 min. Kinase reactions were stopped by the addition of sample buffer and analyzed by SDS/PAGE (10%) followed by Coomassie Blue staining. Dried and stained gels were subject to autoradiography or PhosphorImager analysis (GE Healthcare Life Sciences).
Radiolabeling of proteins by coupled transcription and translation
Incorporation of 35S-methionine into INAD or PDZ3 (residues 347–450) was accomplished by in vitro transcription and translation using the transcription and translation system (Promega). In a standard reaction (50 µL), the following were included as recommended by the supplier: a plasmid (0.2–1 µg) containing the desired InaD cDNA in pBluescript SK vector (Stratagene, La Jolla, CA, USA), 35S-methionine (40 µCi, PerkinElmer), and rabbit reticulocyte lysates (25 µL). Synthesis of the radiolabeled proteins were monitored and quantitated by autoradiography following SDS/PAGE. 35S-labeled proteins were used directly for overlay assays without additional purification.
Protein overlay assay
Nitrocellulose filters containing various TRP T7 gene 10 fusion proteins were incubated with 35S-labeled INAD or PDZ3 in 1x phosphate-buffered saline (PBS), 5% milk powder, and 0.2% Triton X-100 at 4°C for 16 h with constant agitation (Shieh and Zhu 1996). In general, about 1–3 µL/mL of in vitro translated probes were used. Following incubation with probes, filters were rinsed with 1x PBS containing 0.2% Triton X-100 for 1 h (with 2–3 changes of the buffer) to remove nonspecific binding. Filters were then air-dried and radioactivity in the filter was analyzed by autoradiography and/or Phosphorimaging. At least three independent experiments were carried out for each amino acid substitution.
Western blotting
Following transfer, nitrocellulose filters were blotted with primary antibodies in 1x PBS, 5% milk powder, and 0.2% Triton X-100, followed by alkaline phosphatase-conjugated secondary antibodies as described previously (Shieh and Niemeyer 1995). Alkaline phosphatase activity was detected upon incubation with 5-bromo-4-chloroindoxyl phosphate and nitroblue tetrazolium. For quantitative western blotting, fluorophore-conjugated secondary antibodies (Alexa Fluor® 680 goat anti-rabbit IgG; Invitrogen) were used and the signals were visualized and quantified by the Odyssey Infrared Imaging system (LI-COR, Lincoln, NE, USA). Polyclonal antibodies against INAD (Shieh and Zhu 1996), eye-PKC (Adamski et al. 1998), and NORPA (Shieh et al. 1997) were obtained as described previously.
Quantitation of the protein–protein interaction
The radioactivity retained following overlay assay was quantitated by ImageQuaNT™ following PhosphorImager analysis. The relative INAD binding of the TRP fusion proteins was calculated by taking into account of the fusion protein levels (moles), and represented as percentage using the binding of a wild-type TRP fusion protein containing the last 117 aa of TRP as 100%. The level of the TRP fusion proteins was estimated by the Coomassie Blue staining, using bovine serum albumin as a standard. Data from three independent experiments were analyzed, quantified, and represented as mean ± SEM in histograms.
Fly stocks
Drosophila stocks including wild-type (Oregon-R), InaDp215, and inaD1 (obtained from Dr Zuker, University of California San Diego, CA, USA) were maintained at 25°C in a 12-h dark/12-h light cycle.
Results
TRP associates with INAD via its last 14 residues and the terminal leucine is required for the interaction
Previously we showed that the third PDZ domain (PDZ3) of INAD interacted with the carboxyl terminal tail of TRP (Shieh and Zhu 1996). It also has been reported that deletion of the last four residues in TRP, aa1272–1275 (Li and Montell 2000), or an Asp substitution of an internal valine (Val1266) (Shieh and Zhu 1996), leads to a loss of the interaction. Based on these reports, it appears that additional residues beyond the terminal tetrapeptide motif are critical for the INAD association (Fig. 1a). To identify these critical residues for insights into the molecular basis of the INAD interaction, we first defined the minimal interacting domain in TRP by creating truncation mutants. These mutant TRP tails were expressed in bacteria as T7 gene 10 fusion proteins and analyzed by 35S-INAD or 35S-PDZ3 overlay. The binding results obtained were consistent and summarized in Fig. 1b. We demonstrate that bacterial fusion proteins containing the last 117, or 19 or 14 residues of TRP are sufficient for binding to INAD, whereas the last 13 residues display a drastically reduced association (Fig. 1c). Consistently, removal of the last two or six residues in TRP also disrupted the INAD interaction (Fig. 1c). These findings strongly support that the last 14 amino acids of TRP, TRP1262–1275, are critical for binding to PDZ3 of INAD.
Fig. 1.
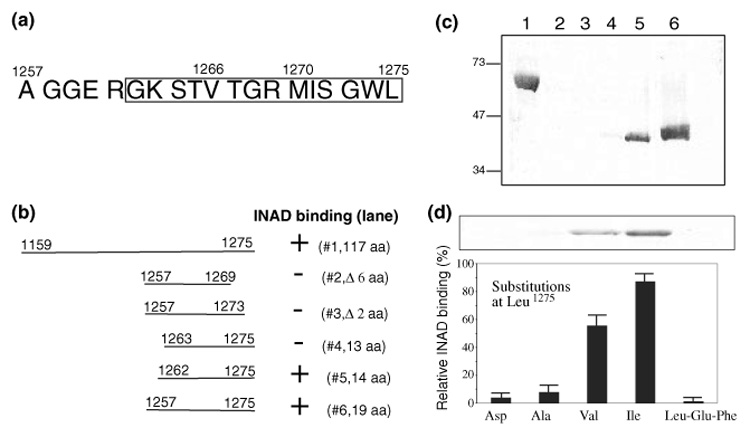
The last 14 residues of TRP are required for the INAD interaction by protein overlay assays (a) Shown are the last 19 amino acid residues of TRP. The last four residues represent a class II PDZ binding sequence. The last 14 residues encompassing the INAD interacting domain are boxed. (b) Mapping the INAD associating domain by protein overlay assays using T7 gene 10 fusion proteins containing portions of TRP. Shown is a summary of the deletion analysis. (c) The 35S-INAD overlay assay. Shown is a representative autoradiogram. Lane1, TRP1159–1275; lane 2, TRP1257–1269; lane 3, TRP1257–1273; lane 4, TRP1263–1275; lane 5, TRP1262–1275; lane 6, TRP1257–1275. T7 gene 10 alone (not shown) was used as a negative control. Molecular weight standards are indicated on the left (in kDa). (d) The mutagenesis study of the terminal Leu in TRP. Shown is a representative INAD overlay containing similar levels of TRP fusion proteins (Top). The relative INAD binding (mean ± SEM, n = 3) of various terminal substitutions is displayed in a histogram (Bottom).
The last 14 residues of TRP end with a hydrophobic residue (Fig. 1a) similar to a prototypical PDZ binding motif. To investigate whether the terminal Leu and its free carboxyl group are involved in the INAD association, we performed mutagenesis studies. We found that replacement of Leu1275 with either Asp or Ala led to a drastically reduced interaction (Fig. 1d). However, substitutions with either Val or Ile still supported the INAD binding (approximately 55 ± 6, and 87 ± 9%, respectively, mean ± SEM., n = 3). In contrast, extension of the carboxyl sequence by the addition of two residues, Glu-Phe, following the terminal Leu, also resulted in a loss of the INAD interaction (Fig. 1d). These findings indicate that a hydrophobic residue at the carboxyl terminus of TRP is essential for binding to INAD. Based on the results, we concluded that the INAD interaction involves an extended carboxyl terminus of TRP consisting of its last 14 residues and that the terminal hydrophobic residue is also critical for the INAD association.
Ser1264 in the putative internal motif of TRP, -Lys-Ser-Thr-Val, may be phosphorylated to modulate the INAD binding
As mentioned before, we showed that replacement of Val1266 with Asp resulted in a drastically reduced INAD interaction (Shieh and Zhu 1996), suggesting that the sequence, Lys-Ser-Thr-Val1266 may act as an internal tetrapeptide motif (Hillier et al. 1999; Hung and Sheng 2002) to regulate the interaction with PDZ3 of INAD. Indeed, Val1266 may be the equivalent of the terminal residue (at the 0 position) in a prototypical PDZ binding motif, which usually terminates with a hydrophobic amino acid (Hung and Sheng 2002). To explore the contribution of this internal motif, we performed site-directed mutagenesis for each residue and evaluated for the INAD interaction. By replacing Val1266 with Leu, we showed that this modified TRP retained a significant interaction (54 ± 7%) (Fig. 2a). This result suggests that a residue with hydrophobic side chain is preferred at Val1266. Thr1265 may correspond to the residue at the −1 position of the terminal binding motif, which usually has no direct contact with the backbone of a PDZ domain. However, we found that replacement of Thr1265 with Pro led to a reduced association (27 ± 3%), whereas substitutions with either Asp or Ala retained a significant INAD interaction (47 ± 4% and 64 ± 6%; Fig. 2a and b).
Fig. 2.
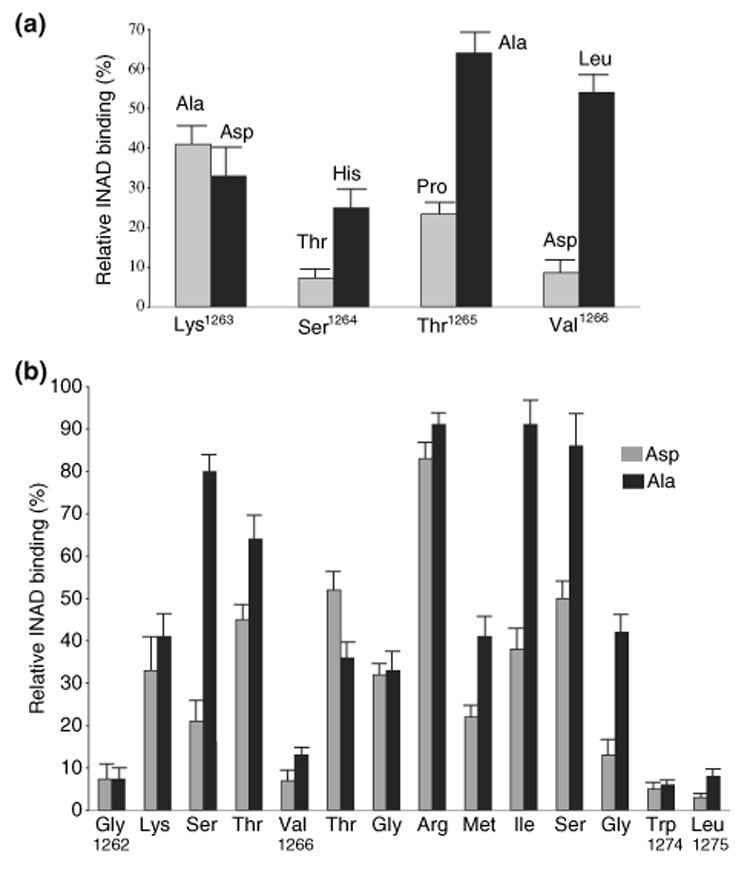
A mutational analysis in TRP1262–1275 (a) The mutagenesis study of the ‘internal peptide motif,’ Lys-Ser-Thr-Val. Mutant TRPs containing substitutions at Lys1263, Ser1264, Thr1265, or Val1266 in the TRP1159–1275 backbone were assayed by 35S-INAD protein overlay. The INAD binding was quantified by PhosphorImaging analysis. The relative INAD binding per mole of the fusion protein was represented as mean ± SEM (n = 3), and displayed in a histogram. (b) A summary of the mutagenesis analysis in TRP1262–1275. Each residue was replaced either with Ala (black) or Asp (gray) and the relative 35S-INAD binding of modified TRP was shown in a histogram (mean ± SEM, n = 3). In addition to Leu1275 (see Fig. 1d) and Val1266 (see Fig. 2a), mutations in Gly1262 and Trp1274 led to a drastic reduction (< 20%) of the relative INAD binding.
We also investigated the contribution of Ser1264, which may be equivalent to the residue at the −2 position of a class I PDZ binding motif (Hung and Sheng 2002). We demonstrate that Ala substitution at Ser1264 retained a significant interaction (80 ± 5%; Fig. 2b), however, replacement with Thr led to a drastic loss of the association (7 ± 3%; Fig. 2a). A reduced interaction was also observed when Ser1264 was substituted with His (Fig. 2a). These findings suggest that the hydroxyl side chain of Ser1264 is not crucial for binding to INAD. Similarly, we also examined the effect of substitutions at Lys1263 that may correspond to the residue at the −3 position in a typical class I motif. We revealed that both Ala and Asp mutations resulted in a minor reduction of the INAD association (Fig. 2a). Together, these results demonstrate that this tetrapeptide sequence does not behave like a typical internal PDZ recognition sequence.
Interestingly, Ser1264 is predicted to be a potential phosphorylation site (by PKC or Ca2+ and calmodulin-dependent protein kinase II) using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/). Therefore, we investigated how phosphorylation at Ser1264 in TRP might affect the INAD association. We show that substitution of Ser1264 with Asp mimicking a phosphorylated Ser resulted in a reduced INAD binding (21 ± 6%) (Fig. 2b), suggesting that phosphorylation at Ser1264 decreases the INAD interaction. Based on the finding, it is likely that phosphorylation at Ser1264 of TRP may reduce its interaction with INAD.
Substitution studies in TRP1262–1275 reveal two more critical residues, Trp1274 and Gly1262, required for the INAD interaction
To identify critical residues in the remaining residues of the putative INAD interacting domain, we carried out site-directed mutagenesis. We generated two mutants for each residue by replacing with either Ala or Asp and assayed for the INAD binding. We assumed that if the side chain of a residue is involved in chemical bonding with the backbone of the PDZ domain, substitutions that change polarity of the side chain may lead to a reduced protein–protein interaction. Results of the relative INAD binding in these modified TRP tails are summarized in Fig. 2b. We arbitrarily used 20% as the cut-off for considering as a significant interaction. Therefore, if both Asp and Ala mutations in a given residue result in below 20% INAD binding, we would conclude that this specific residue is essential for the INAD association.
Importantly, we demonstrate that substitutions of Gly1262 or Trp1274 with either Ala or Asp greatly reduced the INAD binding (< 20% relative INAD binding) (Fig. 2b). In contrast, Ala or Asp replacement in the remaining residues was tolerated leading to a significant interaction with INAD (> 20% relative INAD binding) (Fig. 2b). These findings strongly indicate that both Gly1262 and Trp1274 are critical for the INAD association.
Taken together, we conclude that there are four critical residues in the carboxyl tail of TRP, Gly1262, Val1266, Trp1274, and Leu1275, whose side chains may have direct contacts with INAD.
The last 14 residues of TRP are essential for binding to INAD by pull-down assays
To further confirm the involvement of the last 14 residues of TRP for the TRP-INAD interaction, we employed pull-down assays. Experimentally, immobilized GST fusion proteins containing carboxyl sequences of TRP were tested for interaction with INAD from fly extracts. The recovered INAD was detected by western blotting as shown in Fig. 3a. Consistently, we demonstrate that TRP fusion proteins containing either the last 117 or 14 residues (Fig. 3a, lanes 2 and 4) interacted with INAD, whereas the last 13 residues (lane 3), or TRP containing an Asp substitution at Leu1275 (Fig. 3, lane 5) displayed a drastically reduced INAD co-purification. These results are in agreement with the protein overlay results and further support the critical role of the last 14 residues of TRP to interact with INAD.
Fig. 3.
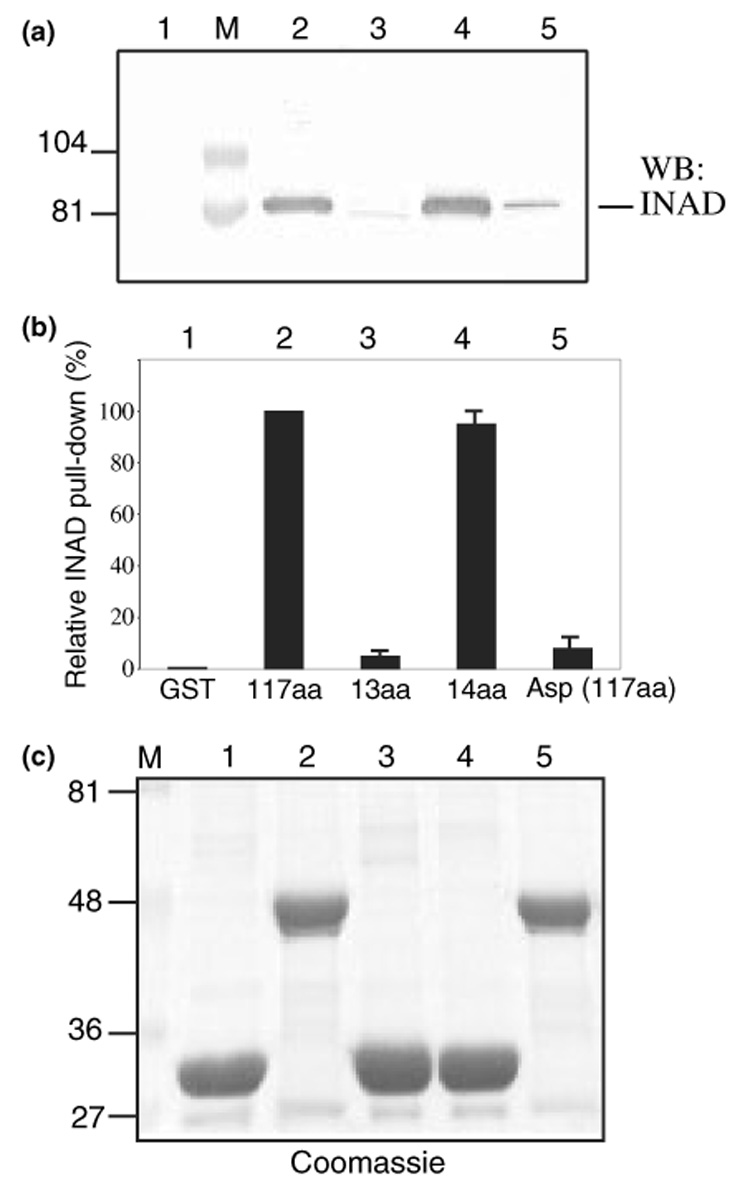
The last 14 residues of TRP pull-down INAD from wild-type fly extracts. (a) Western blot analysis of the TRP pull-down assays. Affinity purified GST fusion proteins containing TRP1159–1275 (117 aa, lane 2), TRP1263–1275 (13 aa, lane 3), TRP1262–1275 (14 aa, lane 4), and TRP (L1275D) (Asp/117 aa, lane 5) were used to isolate INAD from wild-type fly head extracts. GST alone was used as a negative control (lane 1). (b) The relative level of INAD pulled down by various TRP fusion proteins. Shown is a histogram (mean ± SEM, n = 3) depicting the level of the recovered INAD as % of pull-down by TRP1159–1275. (c) The Coomassie staining of the TRP fusion proteins following pull-down assays. Molecular weight standards (lane M) are indicated on the left (in kDa).
The carboxyl tail of TRP co-purifies eye-PKC and NORPA from fly extracts
It has been proposed that the interaction between TRP and INAD forms the ‘core complex’ that further directs INAD and its associated proteins to the rhabdomere, a specialized subcellular compartment in photoreceptors where visual transduction takes place (Li and Montell 2000; Tsunoda et al. 2001). We investigated if TRP fusion proteins that interact with INAD could also pull-down other proteins in the INAD complex including NORPA and eye-PKC. By analyzing the co-purifying proteins by SDS/PAGE followed by the Coomassie staining, we discovered that several proteins with apparent molecular weights ranging from 130 to 80 kDa were specifically co-purified by TRP fusion proteins that interact with INAD (Fig. 4a, lanes 2 and 4). In contrast, GST alone or TRP fusion containing its last 13 residues (Fig. 4a, lanes 1 and 3) co-isolated a protein of about 80 kDa, which represents a nonspecific binding protein of bacterial origin as it appeared prior to the incubation with fly extracts (see also Fig. 5b, lanes 5, 7, and 8).
Fig. 4.
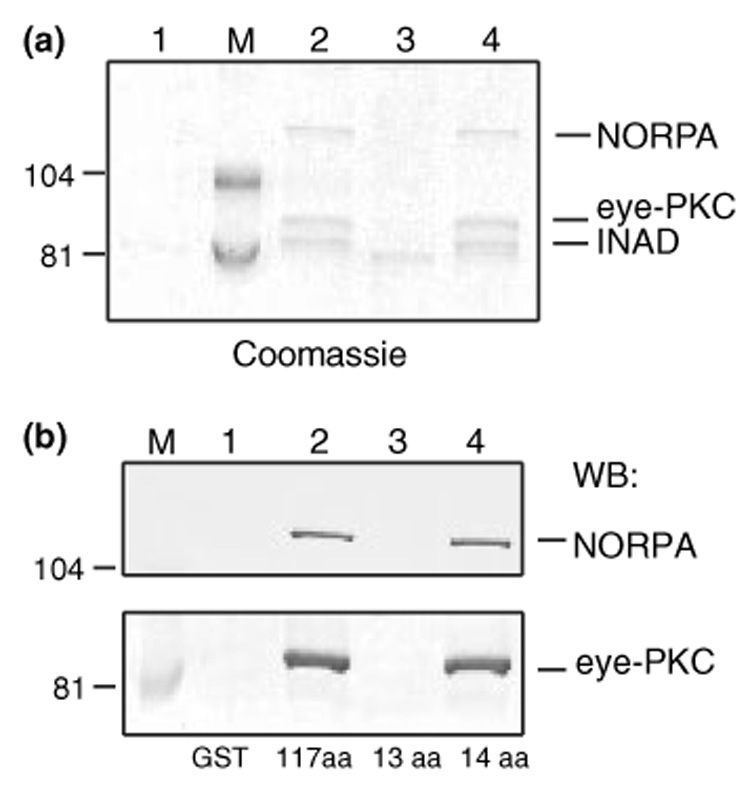
Co-isolation of eye-PKC and NORPA by TRP fusion proteins. (a) The high molecular weight proteins co-purified by TRP tails. Lane 1, GST alone; lane 2, TRP1159–1275 (117 aa); lane 3, TRP1263–1275 (13 aa); lane 4, TRP1262–1275 (14 aa). (b) Both NORPA and eye-PKC were pulled down by the TRP tails containing the terminal 117 or 14 residues. Shown is a western blot analysis using either anti-NORPA (Top) or anti-eye-PKC antibodies (Bottom). Molecular weight standards (lane M) are indicated on the left (in kDa).
Fig. 5.
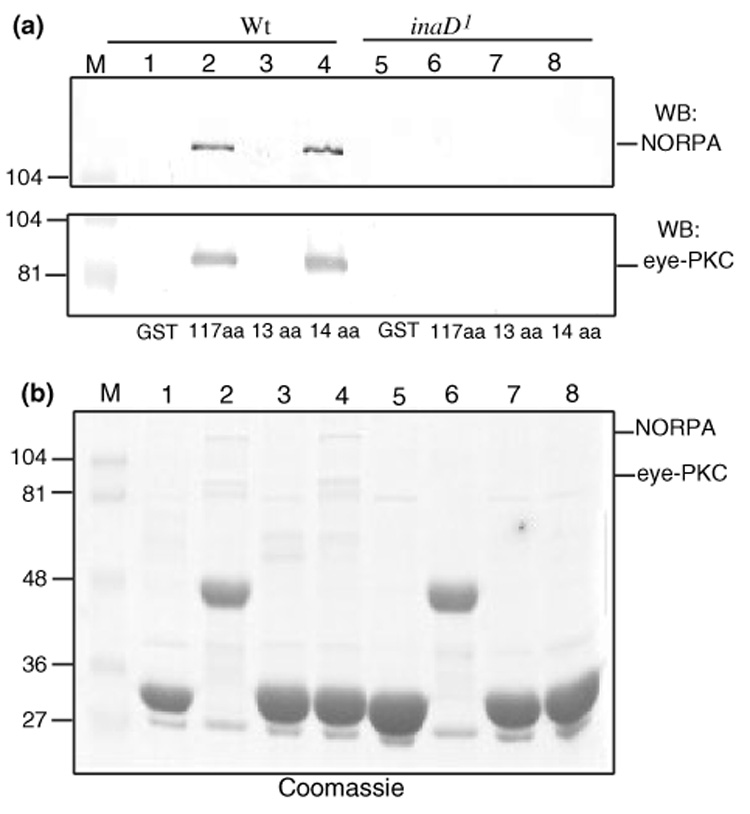
Co-isolation of eye-PKC and NORPA by the TRP tails is dependent on the presence of INAD. (a) Both eye-PKC and NORPA were co-purified by TRP using wild-type, but not inaD1 extracts. Shown is the western blotting of the pull-down assay using either wild-type (lanes 1–4) or inaD1 extracts (lanes 5–8). Top, anti-NORPA; bottom, anti-eye-PKC. TRP fusion proteins used included TRP1159–1275 (117 aa, lanes 2 and 6), TRP1263–1275 (13 aa, lanes 3 and 7), and TRP1262–1275 (14 aa, lanes 4 and 8). GST alone (lanes 1 and 5) was used as a negative control. (b) The Coomassie staining following the pull-down assay revealed TRP fusion proteins, as well as a lack of co-purification of both eye-PKC and NORPA when inaD1 extracts were used. Molecular weight standards (lane M) are indicated on the left (in kDa).
The identity of the co-purified proteins was confirmed by western blotting, demonstrating that both eye-PKC and NORPA were isolated by the TRP pull-down assays (Fig. 4b, lanes 2 and 4). Because the last 13 residues of TRP failed to pull-down eye-PKC and NORPA (Fig. 4b, lane 3), it indicates that co-isolation of NORPA and eye-PKC by the TRP tail is dependent on its interaction with INAD.
To further support the role of INAD in promoting the co-purification of eye-PKC and NORPA from fly extracts, we performed pull-down assay using extracts prepared from inaD1. inaD1 is a loss of function allele and completely lacks INAD (Tsunoda et al. 1997). We revealed that, when inaD1 extracts were used, both eye-PKC and NORPA were not co-purified (Fig. 5a, lanes 6 and 8), whereas co-purification was observed when wild-type extracts were used (Fig. 5a, lanes 2 and 4). These results strongly support that the recovery of both eye-PKC and NORPA by TRP pull-down is dependent on the presence of INAD and hence the INAD complex in the fly extracts. Our findings indicate that pull-down assays can also be employed to isolate stable protein complexes from cell extracts.
Complex-dependent phosphorylation of TRP by eye-PKC
The recovery of a partial INAD complex containing eye-PKC allowed us to examine the regulation of TRP phosphorylation by eye-PKC. To accomplish this, we combined pull-down with an in vitro kinase assay to develop into a novel complex-dependent kinase assay. We first validated the feasibility of the complex-dependent eye-PKC activity by exploring whether eye-PKC could phosphorylate TRP and INAD, both of which were shown to become phosphorylated by eye-PKC via an immunocomplex assay (Liu et al. 2000). Experimentally, we pull-downed a partial INAD complex consisting of INAD, TRP, and eye-PKC by using fusion proteins containing the carboxyl tail of NORPA (NORPA985–1095), which interacts with INAD (Shieh et al. 1997). The partial INAD complex was then subjected to an in vitro kinase reaction. First we found that the activity of the co-purified eye-PKC was greatly enhanced in the presence of Ca2+ and PMA (not shown), like other conventional PKCs. Moreover, active eye-PKC led to phosphorylation of INAD and TRP (Fig. 6a, lane 1).
Fig. 6.
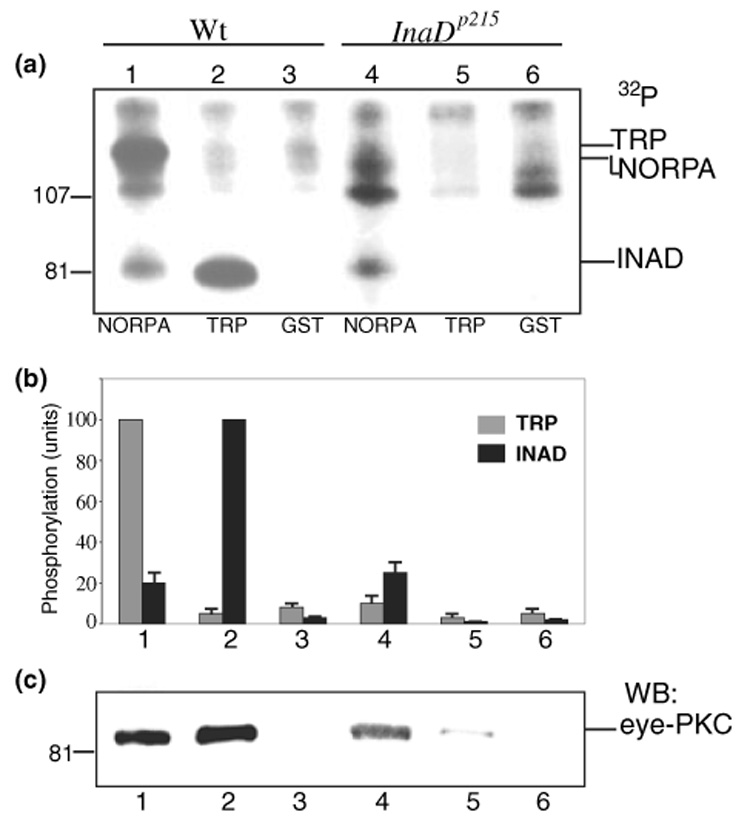
Phosphorylation of TRP and INAD by the complex-dependent eye-PKC assay. (a) Phosphorylation of TRP and INAD by eye-PKC is regulated by co-purification with eye-PKC. Shown is an autoradiogram depicting phosphorylation by the complex-dependent eye-PKC assay using wild-type (lanes 1–3) or InaDp215 extracts (lanes 4–6). The INAD complex was pulled down by GST fusion proteins containing the carboxyl tail of NORPA (lanes 1 and 4), or TRP1262–1275 (14 aa, lanes 2 and 5). GST alone was used as a negative control (lanes 3 and 6). (b) The relative level of phosphorylation in TRP and INAD by the complex-dependent eye-PKC assay. Shown is a histogram depicting the phosphorylation (mean ± SEM, n = 3) using either wild-type or InaDp215 extracts. The relative TRP phosphorylation was calculated by comparing to the phosphorylation of TRP by wild-type extracts in lane 1, and the relative INAD phosphorylation, to the phosphorylation of INAD in lane 2. (c) The co-isolation of eye-PKC is critical for the phosphorylation of TRP and INAD. Eye-PKC was detected by western blotting.
Once we established that eye-PKC was able to phosphorylate TRP and INAD, we investigated if eye-PKC could also phosphorylate NORPA. NORPA is a phospholipase Cβ (Bloomquist et al. 1988) that breakdowns phosphatidylinositol 4, 5-bisphosphate to generate DAG, which activates eye-PKC. Therefore, NORPA is a candidate for the feedback regulation by eye-PKC. To investigate whether NORPA can be phosphorylated by eye-PKC via the complex-dependent kinase assay, we pulled down a partial INAD complex consisting of NORPA, eye-PKC and INAD using fusion proteins containing the last 14 residues of TRP. We showed that, in the presence of Ca2+ and PMA, INAD became intensely phosphorylated, but NORPA did not (Fig. 6a, lane 2), indicating that NORPA may not be a substrate of eye-PKC. It is also possible that endogenous NORPA may be already phosphorylated to a high degree, and therefore it is not an ideal substrate for the kinase assay. Alternatively, NORPA may not be directly regulated by eye-PKC phosphorylation. Together, our results demonstrate that in the complex-dependent kinase assay endogenous eye-PKC can be activated to phosphorylate both INAD and TRP.
Role of INAD and eye-PKC in the complex-dependent phosphorylation
Once we established the complex-dependent kinase assay, we investigated how would INAD promote the complex-dependent phosphorylation of TRP. It has been shown that the interaction between INAD and TRP is greatly reduced in InaDp215, which expresses a modified INAD (INADM442K) with a point mutation in PDZ3 (Shieh and Zhu 1996). Therefore, we carried out the complex-dependent phosphorylation using InaDp215 extracts. Unlike wild-type extracts, by which both endogenous INAD and TRP were phosphorylated by eye-PKC (Fig. 6a, lane 1) as these proteins were recovered by the carboxyl tail of NORPA, only INAD (i.e. INADM442K) was recovered and significantly phosphorylated (Fig. 6a, lane 4) when InaDp215 extracts were used. The lack of a phosphorylated TRP band (Fig. 6a, lane 4) is due to the absence of the co-purified endogenous TRP as it failed to interact with INADM442K. This result indicates that phosphorylation of TRP by eye-PKC via the complex-dependent phosphorylation is dependent on co-purification of eye-PKC and TRP.
We further investigated the complex-dependent phosphorylation by the use of the terminal 14 residues of TRP. We revealed that the TRP tail failed to recover INADM442K and hence INADM442K was not phosphorylated when InaDp215 extracts were used (Fig. 6a, lane 5). The loss of interaction between INADM442K and TRP also resulted in a reduction of the eye-PKC co-purification (Fig.6c, lane 5), supporting that the eye-PKC phosphorylation of INAD is regulated by its co-purification with eye-PKC.
Together, we have demonstrated that that eye-PKC is required for the complex-dependent phosphorylation of INAD and TRP and that the detection of phosphorylated TRP in the in vitro assay used here is dependent on its interaction with INAD, which tethers eye-PKC. This complex-dependent kinase assay can be modified to investigate other protein kinases that regulate substrates via anchoring to an adaptor or scaffolding protein.
Discussion
Scaffolding proteins containing multiple protein-protein interaction modules such as PDZ domains are capable of tethering several proteins to organize a multi-molecular protein complex. Formation of the protein complex, which supports co-localization of the interacting proteins, may be involved in delineating the specificity of signaling mechanisms, as well as promoting efficient protein-protein interactions leading to fast kinetics of the signaling process. The critical role of a macromolecular complex in signaling transduction has been illustrated in studies on the INAD complex in Drosophila photoreceptors. It has been shown that the scaffolding protein INAD tethers eye-PKC, NORPA and TRP in the rhabdomere. Interactions with INAD are important for subcellular localization, as well as for promoting the stability of these three interacting proteins (Tsunoda et al. 1997). Moreover, the INAD signaling complex appears to facilitate fast kinetics possibly by coordinating both positive and negative regulation of the visual response (Tsunoda et al. 1997; Scott and Zuker 1998).
Previously we showed that the carboxyl sequence of TRP interacted with PDZ3 of INAD. To gain further insights into the basis of the association, here we demonstrate that the INAD interacting domain encompasses the last 14 residues of TRP. Moreover, we illustrate the critical contribution of Gly1262, Val1266, Trp1274, and Leu1275, as mutations in these residues lead to a drastic reduced INAD interaction. Importantly we show that the terminal residue, Leu1275, is essential for the INAD association, suggesting that the interaction is similar to that involves a terminal motif (Hung and Sheng 2002). However, unlike the terminal motif, the residue at the −2 position of the TRP tail is not critically involved, whereas the residue at the −1 position (Trp1274) is. It remains possible that the terminal residues of the TRP tail anchor to PDZ3 similar to a prototypical terminal motif, with the upstream residues (Gly1262 and Val1266) engaging in additional interactions.
Most of PDZ domains interact with the last four terminal residues; however, some PDZ domains display selectivity in residues beyond the tetrapeptide sequence, when tested in peptide libraries. Similarly, it has been shown that upstream residues beyond the tetrapeptide motif may be necessary for optimal binding or enhanced affinity (Hung and Sheng 2002). For example, Schultz et al. reported that the interaction between syntrophin and a terminal heptapeptide of sodium channel involved the residue at the −4 position of the peptide (Schultz et al. 1998). Similarly, the interaction between the third PDZ domain of post-synaptic density protein 95 and cysteine-rich PDZ-binding protein calls for more than six terminal residues (Niethammer et al. 1998). In another example, the six terminal amino acids of Fas have been shown to associate with the second PDZ domain of protein tyrosine phosphatase 1E (Kozlov et al. 2000). To sum up, the involvement of upstream residues for interactions with PDZ domain has been documented, however, this is the first report that supports the contribution of an extended 14 terminal residues for binding to PDZ3 of INAD.
It has been shown that the interaction between INAD and TRP targets the INAD complex to the rhabdomere, and this interaction is also critical for the stability of TRP in photoreceptors (Tsunoda et al. 1997, 2001; Li and Montell 2000). TRP is a Ca2+ channel that is open to initiate the light-dependent depolarization (Minke and Parnas 2006). We postulated that the TRP-INAD interaction also regulates the TRP channel activity. Indeed, we demonstrated that a loss of the INAD association led to slow deactivation of the visual response in InaDp215, which expresses a mutant INAD with a missense mutation in its PDZ3 (Shieh and Zhu 1996). Interestingly, the slow deactivation of InaDp215 photoreceptors is dependent on the extracellular Ca2+, because the deactivation phenotype becomes less severe when the extracellular Ca2+ is reduced (Shieh and Niemeyer 1995). This Ca2+-dependent deactivation of the visual signaling may be contributed in part via the modulation by eye-PKC, a conventional PKC that plays an essential role in the negative regulation of the visual signaling. A loss of eye-PKC leads to slow deactivation and response inactivation (Smith et al. 1991; Hardie et al. 1993; Peretz et al. 1994; Gu et al. 2005). Eye-PKC has been shown to phosphorylate TRP in vitro and in vivo. Recently, we reported that transgenic expression of a modified TRP missing an eye-PKC phosphorylation site resulted in an electroretinogram phenotype similar to that of InaDp215. Together, these findings strongly support that deactivation of TRP is regulated by the INAD association, which in part promotes phosphorylation of TRP by eye-PKC (Popescu et al. 2006). Interestingly Li and Montell (2000) reported that the TRP-INAD interaction was required only for retention of the INAD complex in the rhabdomere, but not for the regulation of the photo-response. These authors showed that transgenic expression of a truncated TRP missing the last four residues did not lead to a slow deactivation phenotype. It remains to be investigated if the slow deactivation phenotype is indeed caused by the loss of the INAD interaction or additional deficits associated with the expression of the modified INAD in InaDp215.
To follow up the regulation of TRP by eye-PKC phosphorylation, we designed a complex-dependent kinase assay. We show that a partial pre-assembled INAD complex containing INAD, TRP, and eye-PKC can be obtained, and eye-PKC can be activated to phosphorylate INAD and TRP. These results are in good agreement with the results obtained by the immunocomplex kinase assay (Liu et al. 2000). We also demonstrate the critical role of INAD and eye-PKC in promoting the complex-dependent phosphorylation of TRP. This complex-dependent eye-PKC assay can be used to define specific in vitro phosphorylation sites in TRP (Popescu et al. 2006) and in INAD. Importantly, this kinase assay can be applied for the investigation of other protein kinases that target to substrates in a protein complex.
In this study, we demonstrate the utility of a novel complex dependent kinase assay that employs endogenous protein complexes to address the regulation of the TRP phosphorylation by eye-PKC. We show that a partial INAD complex can be isolated from fly extracts and phosphorylation of TRP by activated eye-PKC can be observed. Moreover, phosphorylation of TRP is dependent on the co-purification of eye-PKC, which is regulated via the interaction with INAD. We also reveal that the INAD interacting domain encompasses the last 14 residues at the carboxyl terminus of TRP. This extended terminal binding motif of TRP may anchor to PDZ3 of INAD similar to a prototypical PDZ binding motif.
Acknowledgements
We thank Joel Schwartz for DNA constructs and Qin-Xia Chen for technical help. This work was supported by a grant from NIH (EY09743).
Abbreviations used
- DAG
diacylglycerol
- GST
glutathione S-transferase
- INAD
inactivation-no-afterpotential D
- NORPA
no-receptorpotential A
- PBS
phosphate-buffered saline
- PDZ, PSD95
discs-large, zonular occludens-1
- PKC
protein kinase C
- PMA
phorbol-12-myristate-13-acetate
- SDS/PAGE
sodium dodecyl sulfate/polyacrylamide gel electrophoresis
- TRP
transient-receptor-potential
References
- Adamski FM, Zhu MY, Bahiraei F, Shieh BH. Interaction of eye protein kinase C and INAD in Drosophila. Localization of binding domains and electrophysiological characterization of a loss of association in transgenic flies. J. Biol. Chem. 1998;273:17713–17719. doi: 10.1074/jbc.273.28.17713. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Chevesich J, Kreuz AJ, Montell C. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- Chyb S, Raghu P, Hardie RC. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature. 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Gu Y, Oberwinkler J, Postma M, Hardie RC. Mechanisms of light adaptation in Drosophila photoreceptors. Curr. Biol. 2005;15:1228–1234. doi: 10.1016/j.cub.2005.05.058. [DOI] [PubMed] [Google Scholar]
- Hardie RC. Phototransduction in Drosophila melanogaster. J. Exp. Biol. 2001;204:3403–3409. doi: 10.1242/jeb.204.20.3403. [DOI] [PubMed] [Google Scholar]
- Hardie RC. Regulation of Drosophila TRP channels by lipid messengers. Novartis Found. Symp. 2004;258:160–167. Discussion 167–171, 263–266. [PubMed] [Google Scholar]
- Hardie RC, Peretz A, Suss-Toby E, Rom-Glas A, Bishop SA, Selinger Z, Minke B. Protein kinase C is required for light adaptation in Drosophila photoreceptors. Nature. 1993;363:634–637. doi: 10.1038/363634a0. [DOI] [PubMed] [Google Scholar]
- Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- Huber A, Sander P, Gobert A, Bahner M, Hermann R, Paulsen R. The transient receptor potential protein (Trp), a putative store-operated Ca2+ channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD. EMBO J. 1996;15:7036–7045. [PMC free article] [PubMed] [Google Scholar]
- Huber A, Sander P, Bahner M, Paulsen R. The TRP Ca2+ channel assembled in a signaling complex by the PDZ domain protein INAD is phosphorylated through the interaction with protein kinase C (ePKC) FEBS Lett. 1998;425:317–322. doi: 10.1016/s0014-5793(98)00248-8. [DOI] [PubMed] [Google Scholar]
- van Huizen R, Miller K, Chen DM, Li Y, Lai ZC, Raab RW, Stark WS, Shortridge RD, Li M. Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling. EMBO J. 1998;17:2285–2297. doi: 10.1093/emboj/17.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from human phosphatase hPTP1E and its interactions with C-terminal peptides from the Fas receptor. Biochemistry. 2000;39:2572–2580. doi: 10.1021/bi991913c. [DOI] [PubMed] [Google Scholar]
- Kumar R, Shieh BH. The second PDZ domain of INAD is a type I domain involved in binding to eye protein kinase C. Mutational analysis and naturally occurring variants. J. Biol. Chem. 2001;276:24971–24977. doi: 10.1074/jbc.M103570200. [DOI] [PubMed] [Google Scholar]
- Li HS, Montell C. TRP and the PDZ protein, INAD, form the core complex required for retention of the signalplex in Drosophila photoreceptor cells. J. Cell Biol. 2000;150:1411–1422. doi: 10.1083/jcb.150.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Parker LL, Wadzinski BE, Shieh BH. Reversible phosphorylation of the signal transduction complex in Drosophila photoreceptors. J. Biol. Chem. 2000;275:12194–12199. doi: 10.1074/jbc.275.16.12194. [DOI] [PubMed] [Google Scholar]
- Minke B, Parnas M. Insights on TRP channels from in vivo studies in Drosophila. Annu. Rev. Physiol. 2006;68:649–684. doi: 10.1146/annurev.physiol.68.040204.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral JH, Petosa C, Sutcliffe MJ, Raza S, Byron O, Poy F, Marfatia SM, Chishti AH, Liddington RC. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg TM, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- Pak WL, Leung HT. Genetic approaches to visual transduction in Drosophila melanogaster. Recept. Channels. 2003;9:149–167. [PubMed] [Google Scholar]
- Peretz A, Sandler C, Kirschfeld K, Hardie RC, Minke B. Genetic dissection of light-induced Ca2+ influx into Drosophila photoreceptors. J. Gen. Physiol. 1994;104:1057–1077. doi: 10.1085/jgp.104.6.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu DC, Ham AJ, Shieh BH. Scaffolding protein INAD regulates deactivation of vision by promoting phosphorylation of transient receptor potential by eye protein kinase C in Drosophila. J. Neurosci. 2006;26:8570–8577. doi: 10.1523/JNEUROSCI.1478-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu. Rev. Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- Schaeffer E, Smith D, Mardon G, Quinn W, Zuker C. Isolation and characterization of two new Drosophila protein kinase C genes, including one specifically expressed in photoreceptor cells. Cell. 1989;57:403–412. doi: 10.1016/0092-8674(89)90915-x. [DOI] [PubMed] [Google Scholar]
- Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias MJ, Schmieder P, Schneider-Mergener J, Oschkinat H. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat. Struct. Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- Scott K, Zuker CS. Assembly of the Drosophila phototransduction cascade into a signalling complex shapes elementary responses. Nature. 1998;395:805–808. doi: 10.1038/27448. [DOI] [PubMed] [Google Scholar]
- Shieh BH, Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- Shieh BH, Zhu MY. Regulation of the TRP Ca2+ channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Shieh BH, Zhu MY, Lee JK, Kelly IM, Bahiraei F. Association of INAD with NORPA is essential for controlled activation and deactivation of Drosophila phototransduction in vivo. Proc. Natl Acad. Sci. USA. 1997;94:12682–12687. doi: 10.1073/pnas.94.23.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DP, Ranganathan R, Hardy RW, Marx J, Tsuchida T, Zuker CS. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science. 1991;254:1478–1484. doi: 10.1126/science.1962207. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimal across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sun Y, Suzuki E, Zuker C. Independent anchoring and assembly mechanisms of INAD signaling complexes in Drosophila photoreceptors. J. Neurosci. 2001;21:150–158. doi: 10.1523/JNEUROSCI.21-01-00150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]


