Abstract
Although internal water molecules are essential for the structure and function of many proteins, the structural and physical factors that govern internal hydration are poorly understood. We have examined the molecular determinants of internal hydration systematically, by solving the crystal structures of variants of staphylococcal nuclease with Gln-66, Asn-66, and Tyr-66 at cryo (100 K) and room (298 K) temperatures, and comparing them with existing cryo and room temperature structures of variants with Glu-66, Asp-66, Lys-66, Glu-92 or Lys-92 obtained under conditions of pH where the internal ionizable groups are in the neutral state. At cryogenic temperatures the polar moieties of all these internal side chains are hydrated except in the cases of Lys-66 and Lys-92. At room temperature the internal water molecules were observed only in variants with Glu-66 and Tyr-66; water molecules in the other variants are probably present but they are disordered and therefore undetectable crystallographically. Each internal water molecule establishes between 3 and 5 hydrogen bonds with the protein or with other internal water molecules. The strength of interactions between internal polar side chains and water molecules seems to decrease from carboxylic acids to amides to amines. Low temperature, low cavity volume, and the presence of oxygen atoms in the cavity increase the positional stability of internal water molecules. This set of structures and the physical insight they contribute into internal hydration will be useful for the development and benchmarking of computational methods for artificial hydration of pockets, cavities, and active sites in proteins.
INTRODUCTION
The ability of water to penetrate and dwell in pockets and cavities in the hydrophobic interior of proteins is well documented (1). Internal water molecules are usually found interacting among themselves and with buried polar groups; presumably they mitigate the energetic cost of transferring polar atoms from bulk water to the protein interior, especially when they are buried without a hydrogen bonding partner from the protein (2,3). Internal water molecules are essential in many biochemical processes, especially in catalysis (4,5), H+ transport (6–10) and in ion homeostasis (11,12). They can affect the pKa values of internal ionizable groups and stabilize transient internal charge during energy transduction processes (13). They can also modulate dynamic processes (14), and contribute toward the binding specificity and affinity of small molecules, drugs, peptides and other ligands by providing bridging interactions between ligand and protein (15). Despite the essential structural and functional roles of internal water molecules, their physical properties and the mechanisms of penetration are not well understood. In this study, we have used x-ray crystallography to elucidate some of the molecular determinants of internal hydration.
Many proteins have large ligand binding cavities that are occupied by water when the ligand is not bound (16). This study examines the hydration of small internal, polar cavities, such as those found in many active sites and in binding sites for ligands. These small cavities are especially useful to examine the roles of polarity, volume, flexibility, and environmental variables as determinants of internal hydration.
X-ray crystallography and NMR spectroscopy have been very useful for characterizing interactions between water and protein. Crystal structures can show the precise location of internal water molecules that are well ordered (17–19). However, the routine use of cryogenic methods in x-ray structure determination has raised a concern that the observed patterns of hydration are influenced by the low temperatures used to collect diffraction data (20). For this reason, we have pursued a detailed comparison of the patterns of internal hydration in structures obtained at low and room temperatures. Low temperatures are not a concern in NMR spectroscopy experiments, which are carried out at ambient temperatures. NMR experiments have been useful to confirm the presence of internal water molecules, to measure their lifetimes, and to describe kinetic barriers and other dynamical properties of proteins relevant to the process of water penetration (21–24). On the other hand, NMR experiments are not as useful as crystallography for identifying the precise locations where internal water molecules bind.
Despite progress in our understanding of molecular determinants of hydration of internal sites, many questions remain unanswered: Are empty cavities really empty or are they occupied by disordered water? How does temperature affect the patterns of hydration observed crystallographically? Are all polar groups equal in their capacity to bind and organize water molecules inside proteins? What factors can increase the organization of water molecules in cavities in proteins? Some of these questions are addressed by this study. Molecular dynamic (MD) simulations (15,17,25–27), statistical surveys (1–3), and equilibrium thermodynamic calculations (28) have already contributed some insight. For example, MD simulations suggest that water penetration in proteins is not unusual and that internal water molecules can be less constrained inside proteins than in bulk water. They have confirmed the important role of buried polar atoms in stabilizing internal water molecules and in modulating their kinetic and thermodynamic properties, and identified an important role for side chain flexibility (27). One of the goals of this study is to contribute structures and data useful to examine the validity of the insight contributed by computational methods.
The current analysis builds on our previous observations of internal water molecules associated with the carboxylic groups of Glu and Asp buried at positions 66 and 92 in staphylococcal nuclease (SNase) (29–31). We present cryo (100 K) and room temperature (298 K) crystal structures of SNase variants with Asn-66, Gln-66, and Tyr-66, and a comparison against the existing structures of variants with Asp-66, Glu-66, Glu-92, Lys-66, and Lys-92 that were obtained under conditions of pH where the internal ionizable moieties are in the neutral state. With this unique set of 16 related but independently determined structures it was possible to correlate small and systematic perturbations at a single internal site with differences in the patterns of internal hydration observed crystallographically. This study contributes physical insight into the factors that govern internal hydration, and a unique data set that will be useful for the design, testing and calibration of computational methods for the artificial hydration of proteins and active sites.
MATERIALS AND METHODS
X-ray crystallography
The V66N, V66Q, and V66Y substitutions were engineered into a hyperstable form of SNase referred to as PHS after the three substitutions used to make it (P117G, H124L, and S128A). Proteins were overexpressed in Escherichia coli and purified as described (32). Proteins were crystallized using the vapor diffusion hanging drop method. All proteins crystallized in the P41 space group. For each experiment, 4 μL of protein solution were mixed with 4 μL of reservoir solution and suspended from a siliconized coverslip sealed over 1 mL of the reservoir solution. The reservoir solution consisted of 37–38% 2-methyl-2,4-pentanediol (MPD) and 25 mM potassium phosphate, pH 7.8. Initial protein concentrations were 8.3 mg/mL for V66N, 7.8 mg/mL for V66Q, and 9.5 mg/mL for V66Y. Single crystals grew in 1–2 weeks while incubated at 4°C. Independent structure determinations were made for each protein at two different temperatures. Crystals were selected from identical crystallization conditions for both room temperature and cryogenic temperature diffraction data collection. For room temperature data collection, each crystal was mounted in a 0.5 mm quartz capillary. For cryo temperature data collection, each crystal was suspended with mother liquor in a cryo loop and flash-cooled in a pool of liquid nitrogen before transfer to a chilled nitrogen stream maintained at 100 ± 0.1 K. Each data set consisted of diffraction data collected from a single crystal in 0.5° φ or ω oscillations using a Bruker APEX2 sealed-tube diffractometer system (Bruker/AXS, Madison, WI) with CuKα radiation. Reflections were integrated, scaled, and merged using APEX2 software (Bruker/AXS) to yield complete data sets to 2.1 Å resolution for all data sets. Each structure was solved by molecular replacement phasing methods using a search model modified from the cryo structure of the Glu-66 variant in PHS nuclease (29). In the search model, position 66 was mutated to Ala. Water molecules were removed and all B-factors were set to 20.0 Å2 before molecular replacement. Molecular replacement using crystalographic and NMR system (CNS) yielded a unique solution (33) with interpretable electron density maps to 2.1 Å resolution. Model building using the program O (34), and simulated annealing, positional, and B-factor refinement in CNS (33), were carried out iteratively for 3–5 cycles until each model converged. Water molecules were added to the model starting in the second round of model building to reflect electron density in both 2Fo–Fc and Fo–Fc maps (at 1.25 σ and 3.0 σ, respectively) that was within 3.5 Å of a hydrogen-bonding partner in the protein. The final model for each variant at each temperature contained residues 7–45 and 51–141 in addition to water molecules. Residues 46–50 were not included in the models because unambiguous electron density was not observed for these residues. Crystallographic statistics are summarized in Table 1.
TABLE 1.
Crystallographic statistics
| SNase variants
|
||||||
|---|---|---|---|---|---|---|
| V66N | V66N | V66Q | V66Q | V66Y | V66Y | |
| Crystallization | ||||||
| % MPD | 39 | 39 | 37 | 37 | 38 | 38 |
| pH | 7.8 | 7.8 | 7.8 | 7.8 | 7.8 | 7.8 |
| Data collection | ||||||
| Unit cell: a (Å) | 49.0 | 48.2 | 49.0 | 48.3 | 49.0 | 48.3 |
| Unit cell: b (Å) | 49.0 | 48.2 | 49.0 | 48.3 | 49.0 | 48.3 |
| Unit cell: c (Å) | 63.8 | 63.0 | 63.8 | 63.1 | 63.8 | 63.3 |
| Resolution range (Å) | 30.4–2.1 (2.2–2.1)* | 48.2–2.1 (2.2–2.1)* | 49.0–2.1 (2.2–2.1)* | 48.3–2.1 (2.2–2.1)* | 49.0–2.1 (2.2–2.1)* | 30.0–2.1 (2.2–2.1)* |
| Temperature (K) | 298 | 100 | 298 | 100 | 298 | 100 |
| Observations (n) | 75,581 | 107,494 | 72,857 | 106,979 | 70,056 | 74,910 |
| Unique reflections | 8893 (1140) | 8501 (1104) | 8881 (1141) | 8548 (1114) | 8913 (1146) | 8561 (1109) |
| Completeness (%) | 99.9 (99.9) | 99.9 (99.9) | 99.9 (99.9) | 99.9 (99.9) | 99.9 (99.9) | 99.9 (99.9) |
| I/σ | 32.8 (5.6) | 53.7 (13.4) | 32.9 (7.1) | 36.7 (7.0) | 24.3 (4.3) | 34.0 (5.2) |
| Redundancy | 8.5 (4.9) | 12.6 (9.2) | 8.2 (4.9) | 12.5 (8.9) | 7.9 (5.9) | 8.0 (6.0) |
| Rint | 0.037 (0.231) | 0.083 (0.136) | 0.037 (0.242) | 0.095 (0.226) | 0.053 (0.354) | 0.037 (0.247) |
| Refinement | ||||||
| Resolution range (Å) | 50.0–2.1 (2.18–2.10) | 50.0–2.1 (2.18–2.10) | 50.0–2.1 (2.18–2.10) | 50.0–2.1 (2.18–2.10) | 50.0–2.1 (2.18–2.10) | 50.0–2.1 (2.18–2.10) |
| Rfactor | 0.188 (0.227) | 0.210 (0.237) | 0.187 (0.218) | 0.207 (0.222) | 0.182 (0.214) | 0.222 (0.277) |
| Rfree | 0.225 (0.286) | 0.264 (0.332) | 0.230 (0.304) | 0.249 (0.261) | 0.215 (0.253) | 0.284 (0.360) |
| Water molecules (n) | 40 | 74 | 38 | 69 | 37 | 57 |
| Average B factors: protein (Å2) | 30.0 | 24.1 | 30.1 | 25.8 | 31.0 | 32.0 |
| Average B factors: water (Å2) | 34.7 | 28.8 | 36.5 | 30.6 | 34.9 | 35.6 |
| RMSDbond (Å) | 0.0079 | 0.0085 | 0.0087 | 0.010 | 0.012 | 0.010 |
| RMSDangle (°) | 1.5 | 1.6 | 1.5 | 1.6 | 1.7 | 1.6 |
| Luzzati error (Å) | 0.23 | 0.25 | 0.23 | 0.25 | 0.22 | 0.28 |
| Ramachandran | ||||||
| Most favored (%) | 88.5 | 88.5 | 91.2 | 87.6 | 88.5 | 88.5 |
| Additionally allowed (%) | 10.6 | 9.7 | 8.0 | 11.5 | 9.7 | 10.6 |
| Generously allowed (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Disallowed (%) | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 |
| PDB accession code | 2PZW | 2PZU | 2PYK | 2PZT | 2PW5 | 2PW7 |
Values in parentheses refer to the high resolution shell.
Criteria used to identify water molecules
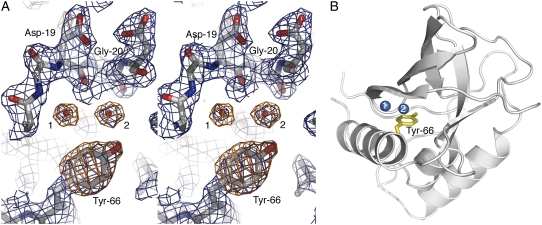
The criteria used to select water molecules for inclusion in the crystallographic model were an important consideration for this study. For the structures of variants with Tyr-66, Asn-66, and Gln-66 at room and cryo temperatures, and for the room temperature structure of the variant with Glu-66 (35), these criteria were imposed strictly: 1), spherical electron density in the 2Fo–Fc maps for each water at 1.25 σ contour; 2), spherical electron density in the Fo–Fc maps for each water at 3.0 σ contour; 3), at least one hydrogen-bonding partner from the protein within 3.5 Å; and 4), B-factor for the water molecules below 50.0 Å2 after successive refinement. Electron density from the relevant region in the variant with Tyr-66 is shown in Fig. 1 to illustrate these criteria graphically. Potential water molecules were predicted using the “water_pick” function in CNS (33) and subsequently screened manually in O (34) before inclusion in the model. The water molecules included in these models may be considered to be “strong” according to the definition by Zhang and Hermans (28). Use of less stringent criteria, such as a 2.0 σ 2Fo–Fc cutoff, would have led to inclusion of additional water molecules in the model, as noted by Zhang and Hermans for subtilisin Carlsberg (28,36). Higher B-factors and lower occupancies may be expected for the so-called weak waters that would be obtained if less restrictive criteria had been used. The internal water molecules considered in this study were all present with full occupancy.
FIGURE 1.
(A) Stereo view of electron density maps of the Tyr-66 protein at room temperature after molecular replacement, contoured over the final refined coordinates. The Tyr side chain and water molecules 1 and 2 are unambiguously shown in the 1.25 σ 2Fo–Fc (blue) and 3.0 σ Fo–Fc (orange) electron density. (B) Ribbon representation of the final structure of the variant with Tyr-66 at room temperature with water molecules 1 and 2 displayed.
RESULTS AND DISCUSSION
Patterns of internal hydration
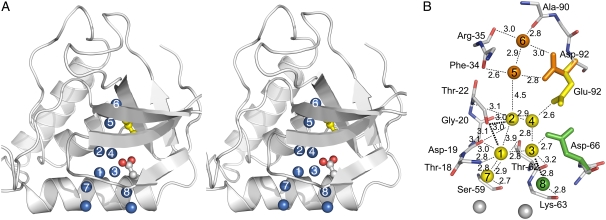
Fig. 2 is a composite image showing the various internal locations where water molecules have been observed in all available SNase structures. All possible hydrogen bonding partners of each water molecule are identified in Fig. 2 B, and average distances and the number of putative hydrogen bonds with protein atoms or with other internal water molecules are also listed in Table 2. The internal water molecules show extensive hydrogen bonding to each other, to the polar moiety of the group buried at position 66 or 92, and to the backbone, especially at the β-turn comprised of residues 18–22. Each of the internal water molecules has the potential to establish between 3 and 5 hydrogen bonds. Despite the moderate resolution of the structures, the location of the individual water molecules and the distance to hydrogen bonding partners remain remarkably constant from protein to protein, within coordinate error limit after Cα-based structural alignment. Polar atoms from the internal side chains in some of the variants occupy the positions occupied by the internal water molecules in other variants. For example, the carboxylic oxygen atoms of Glu-66 occupy the same positions as water molecules 3 and 4 in the structure with Glu-92.
FIGURE 2.
(A) Stereo view of composite representation of all the internal water molecules observed in variants of SNase with Glu, Gln, Asp, Asn, or Tyr at positions 66 or 92. Water molecules are represented as labeled blue spheres. Glu-66 is displayed, with carboxylic oxygens in red, and Ile-92 is shown in yellow. (B) Composite representation of the potential hydrogen bonding interactions of the internal water molecules as observed for waters in sites 1, 2, 3, 4, and 7 in the structure with I92E (yellow), for waters in sites 5 and 6 in the structure with I92D (orange), and for water in site 8 for the structure with V66D (green). All distances are expressed in Å.
TABLE 2.
Structural microenvironments of internal water molecules
| Sites* | SA† | Average BH2O (Å2)‡ | Average Bpartners (Å2) and range§ | Average distance to H bond partners (Å) and range | H bonds with protein (n) | H bonds with waters (n) |
|---|---|---|---|---|---|---|
| 1 | 0.01 | 21.7 | 24.2 (21.2–26.0) | 3.0 (2.8–3.2) | 3 | 2 |
| 2 | 0.01 | 25.5 | 23.8 (23.7–23.9) | 2.9 (2.6–3.2) | 3 | 1 |
| 3 | 0.01 | 17.9 | 13.5 (11.8–15.0) | 2.9 (2.8–3.2) | 1 | 2 |
| 4 | 0.02 | 11.8 | 13.1 (10.6–17.9) | 2.8 (2.6–2.9) | 1 | 2 |
| 5 | 0.01 | 10.6 | 5.4 (2.8–15.9) | 2.8 (2.6–2.9) | 2 | 1 |
| 6 | 0.01 | 5.7 | 8.2 (2.4–13.6) | 2.9 (2.8–3.0) | 3 | 1 |
| 7 | 0.03 | 19.2 | 24.4 (19.1–30.6) | 2.8 (2.7–2.9) | 1 | 2 |
| 8 | 0.01 | 20.8 | 22.6 (17.9–27.3) | 2.8 (2.7–2.9) | 1 | 2 |
Data for sites 1 and 2 is from the cryogenic structure of the V66E variant (29), for sites 3 and 4 it is from the cryogenic structure of the I92E variant (31), and for 7 and 8 it is from the cryogenic structure of the V66D variant (30).
Numbered with reference to Fig. 2.
Normalized solvent-accessible surface area.
Average temperature factors of water molecules.
Average temperature factors of protein hydrogen bonding partners.
It is noteworthy that with the exception of the water molecule bound at site 7, none of the other internal water molecules shown in Fig. 2 are present in any of the more than 100 structures of SNase and its variants. These internal water molecules are only observed when polar or ionizable moieties are introduced into the hydrophobic core of the protein by mutagenesis.
Effects of temperature on patterns of internal hydration
One goal of this study was to examine how the patterns of internal hydration near the polar side chains at position 66 were affected by sample temperature during data collection. Consequently, care was taken to maintain all conditions constant during the crystallographic experiment to ensure that temperature was the only variable. The crystallographic statistics for the structures are listed in Table 1. All the proteins crystallized under identical conditions with isomorphous unit cells in space group P41. Data were collected on the same diffractometer and processed with the same software using analogous routines. The high-resolution limit for all data sets was truncated to 2.1 Å. A single crystallographer carried out molecular replacement, model building, and crystallographic refinement, using consistent software routines and assessment criteria.
Pairwise alignments of the coordinates were carried out using the lsq function in the O software. Each set of coordinates was superimposed onto the coordinates of the variant with Asn-66 obtained under cryogenic conditions, using Cα from residues 7 to 40 and 54 to 141. The break in the alignment sequence was required owing to missing residues 46–50 in some of the models. RMSD values were calculated using MMTSB (37). Pairwise comparison of Cα-aligned coordinates yielded an average RMSD of 0.2–0.4 Å, indicating strong structural conservation between the structures. Structural variability was observed in the backbone of residues 18–23, which consists of a β-turn near the protein's active site and which is near the side chain of residue 66 and hydrogen bonded to some of the water molecules of interest. However, the average RMSD of the 18–23 segment was only 0.3 Å, comparable to that of the full set of coordinates. Slight variations in rotamer positions were observed for residues 19, 21, and 22, even between structures of the same variant at different temperatures. Electron density for the different rotamers was clear in each case. Density for the side chain at position 66 and for numerous waters was apparent even in initial electron density maps. The quality of the electron density was excellent except for residues 1–6, 46–50, and 142–149, which were excluded from the model.
The total number of water molecules in the structures at 298 K and 100 K, respectively, were: 40 and 74 for the variant with Asn-66; 38 and 69 for the variant with Gln-66; and 37 and 57 for the variant with Tyr-66. The overall number of water molecules was similar for the different structures at both room and cryogenic temperatures. The water molecules of interest are the ones in the microcavity surrounding the side chain at position 66, labeled as 1 through 8 in Fig. 2. The distribution of internal water molecules in all the structures is also summarized in Table 3.
TABLE 3.
Summary of internal water molecules observed in crystal structures of SNase variants with internal ionizable groups
| Structure | Temp (K) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| Wild-type*‡ | 100 | − | − | − | − | − | − | + | − |
| V66D*§ | 298 | − | − | − | − | − | − | + | − |
| V66D*§ | 100 | + | − | + | − | − | − | + | + |
| V66N* | 298 | − | − | − | − | − | − | + | − |
| V66N* | 100 | + | − | + | − | − | − | + | + |
| V66E*¶ | 298 | + | − | − | − | − | − | + | − |
| V66E*¶ | 100 | + | + | − | − | − | − | + | − |
| V66Q* | 298 | − | − | − | − | − | − | + | − |
| V66Q* | 100 | + | − | − | − | − | − | + | − |
| V66Y* | 298 | + | + | − | − | − | − | + | − |
| V66Y* | 100 | + | + | − | − | − | − | + | − |
| V66K†‖ | 298 | − | − | − | − | − | − | − | − |
| V66K†‖ | 100 | − | − | − | − | − | − | − | − |
| I92K†** | 100 | − | − | − | − | − | − | − | − |
| I92D†** | 100 | − | − | − | − | + | + | + | − |
| I92E†** | 298 | − | − | − | − | − | − | + | − |
| I92E†** | 100 | + | + | + | + | − | − | + | − |
Water binding sites 7 and 8 are at the interface between the internal water molecules and bulk water, but they are still fully internal and solvent inaccessible (Table 2). The water molecule at site 7 is observed in all the structures under cryogenic or room temperature conditions, whereas the one at position 8 is observed only in cryo structures of variants with Asp-66 or Asn-66. Site 3 is also occupied only in the presence of Asp-66 or Asn-66 under cryogenic conditions. Not considering site 7, which is interfacial, site 1 is the one that is occupied most frequently; it is occupied under cryogenic conditions when position 66 is an Asp, Asn, Glu, Gln, or Tyr. At room temperature it is observed only when position 66 is a Glu or Tyr. Site 2, the innermost site, is occupied in the variants with Glu-66 or Glu-92 under cryogenic conditions, and in low and room temperature structures of the Tyr-66 variant. The water molecule in position 4 is observed only in the cryogenic structure of the variant with Glu-92, and water molecules in positions 5 and 6 are observed only in the cryogenic structure of the variant with Asp-66. In summary, the only internal water molecules observed at room temperature were at site 1 in the variant with Glu-66, and at sites 1 and 2 in the variant with Tyr-66.
Note that all the water molecules described in Table 2 and in Fig. 2 were present with full occupancy. If more relaxed standards for selection of water molecules had been applied, more internal water molecules would have been included. For example, if the requirements were loosened, one water molecule with less than full occupancy could be included at position 2 in the room temperature structure of the Glu-66 variant. Weak (nonspherical) Fo–Fc density was observed at 3.0 σ at this position and a potential hydrogen-bonding partner was identified, but no 2Fo–Fc density was observed, even at a baseline 0.8 σ contour. Consequently, no water was included in the room temperature model. Similarly, at site 1, weak Fo–Fc electron density was observed at 3 σ and 2.5 σ contours in maps of room temperature structures for variants with Asn-66 and Gln-66, respectively. However, no 2Fo–Fc density was seen at site 1 for either variant and waters were therefore not included at this position in the room temperature structures. These examples underscore the importance of consistency in crystallographic model building, and illustrate how these models should be interpreted. Waters with low B-factors, full occupancies, and multiple hydrogen-bonding partners are clearly present in the electron density maps despite the 2.1 Å resolution limit. These are the water molecules that are discussed in this study. Weakly bonded waters may also be present, but such water molecules, not included in the structures presented here, should be interpreted cautiously, especially given the resolution limit of these structures.
In the electron density maps for variants with Asn-66 and Gln-66, there was no evidence that site 2 was occupied at either high or low temperatures. Lowering the contours of the Fo–Fc map to 2.5 σ and the 2Fo–Fc map to 0.8 σ showed a small volume of electron density in the vicinity of site 2 in the structure of the variant with Asn-66 at 100 K, but not at 298 K, and no electron density was observed at site 2 for the variant with Gln-66 at either temperature. Although these looser criteria are not the ones used to identify water molecules in this study, nor are they the ones considered standard for identification of crystallographic water molecules, they suggest that site 2 might be partially occupied at 100 K in the structure of the variant with Asn-66. At 100 K, the structures of variants with Asn-66 and Gln-66 both contained waters at positions 1 and 7; at 298 K only position 7 was occupied. In site 1 no 2Fo–Fc density was observed for room temperature structures, not even at 0.8 σ contour levels. Density was visible at 3.0 σ in the Fo–Fc electron density maps at this position for both mutants at room temperature; however, a water molecule was not included in the model because it failed to meet all selection criteria.
Low volume decreases disorder of internal water molecules
The microcavity where the side chain of Val-66 resides is quite small, but obviously sufficiently large to house a few water molecules. The cavity is visible on the molecular surface, but barely detectable on the water accessible surface calculated with a probe radius of 1.4 Å. Previous studies have suggested that the chemical activity of water (38) and the availability of polar groups for hydrogen bonding (17) are more critical determinants of internal hydration than the volume of the microcavities. In the microcavity of SNase there are many internal, backbone polar groups that internal water molecules could hydrogen bond to, especially in the neighborhood of the β-turn comprised of residues 18–22. The patterns of hydrogen bonding with the backbone, with the polar moieties of residues 66 and 92, and between the water molecules themselves is shown in Fig. 2 B. One reason that this cavity was suitable to examine how steric factors modulate the order of buried waters within a polar cavity was precisely the fact that the availability of polarity within the cavity was not a limiting factor.
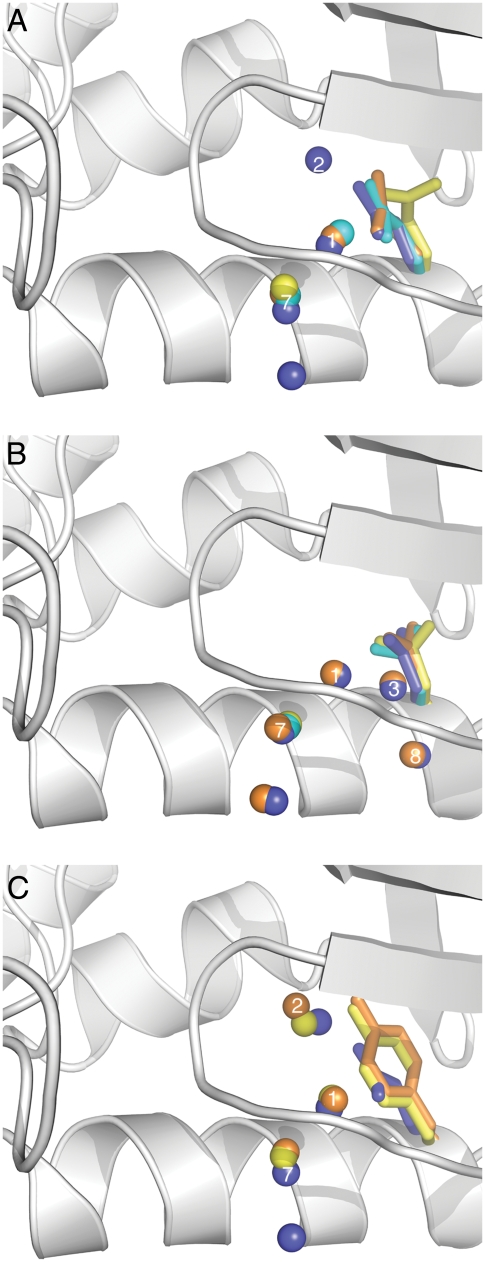
The comparison of the structures of variants with Glu-66 and Tyr-66 was particularly useful to assess the role played by volume cavity in stabilizing water molecules at fixed positions. Water sites 1 and 2 are occupied in the structure with Tyr-66 at both room and cryo temperatures (Fig. 3). The water at position 2 is stabilized by extensive hydrogen bonding with the backbone lining the cavity and also with the hydroxyl group of Tyr-66, which is at the appropriate distance (3.1 Å at 100 K and 2.7 Å at 296) and orientation to establish a hydrogen bond. In the cryogenic structure this water molecule makes contacts with the carbonyl oxygen atoms of Asp-19, Gly-20 and Thr-22 (Table 3 and Fig. 2 B). In the room temperature structure potential contacts are made with the carbonyl oxygen of Asp-19 and Gly-20, and with the water at position 1. The water at position 1 makes hydrogen bonds with the carbonyl oxygen from Asp-19 and Thr-22, and with waters at sites 2 and 7.
FIGURE 3.
(A) Superposition of side chains and water molecules near Glu-66 at 100 K (blue) and 298 K (cyan) and near Gln-66 at 100 K (orange) and 296 K (yellow). (B) Superposition of side chains and water molecules near Asp-66 at 100 K (blue) and 298 K (cyan) and near Asn-66 at 100 K (orange) and 296 K (yellow). (C) Superposition of side chains and water molecules near Glu-66 at 100 K (blue) and near Tyr-66 at 100 K (orange) and 296 K (yellow). Water molecules at sites 1, 2, 3, 7, and 8 are labeled.
The structure of Tyr-66 is the only one in which the pattern of internal hydration is not affected by temperature. Of the side chains inserted at position 66, Tyr has the largest volume, with 3 more heavy atoms than either Glu or Lys, and fewer degrees of freedom owing to the bulky aromatic ring. According to a statistical survey of distribution of water around amino acid residues, Tyr does not interact with water as frequently as Glu or Asp (1). In the presence of Tyr-66, waters are observed at positions 1 and 2 even at room temperature (Figs. 1 and 3). The superposition of the side chains of Tyr-66 and Glu-66 (Fig. 3 C) suggests that in the presence of Tyr-66 the volume of the void near water binding sites 1 and 2 is reduced. We speculate that this decrease in volume minimizes the number of possible states that the water molecules at site 2 can populate, thus minimizing the disorder that renders water molecules invisible at this site in the room temperature structure with Glu-66, and in both sites 1 and 2 in room temperature structures of the other variants.
Effects of polarity on patterns of internal hydration
Previous studies have suggested that internal water molecules are found in cavities that are large enough to accommodate them, but only when polar side chains are available to satisfy their requirements for hydrogen bonding (17). Our set of structures allowed the systematic comparison of the state of hydration of a microcavity in the presence of Asp, Asn, Glu, Gln, Tyr, and Lys side chains at the same position, enabling us to identify differences in the ability of different types of internal polar groups to interact with water molecules. A superposition of water molecules and side chains under cryo and room temperature conditions is shown in Fig. 3. Note in this figure that the side chain rotamers of residue 66 are different in the structures where the internal water molecules were or were not observed.
Overall, the data suggest that internal carboxylic groups are hydrated more strongly than the amides, and that amides are hydrated better than amines. The most interesting comparison is between Glu-66, Gln-66, and Lys-66; Glu-66 stabilizes more water than Gln-66 under both cryogenic and room temperatures. In turn, Gln stabilizes more internal water molecules than Lys. Similarly, under cryogenic conditions Glu-92 stabilizes water molecules at positions 1, 2, 3, 4, and 7, whereas no internal water molecules are present with Lys-92. We have never observed any internal water molecules in interaction with an internal Lys in SNase, not even under cryogenic conditions. The inability of the Lys side chain to impart order on the internal water molecules could be related to its greater inherent flexibility, which probably leads to enhanced disorder (27). However, in the case of Lys-66, for example, the B-factor of the NZ atom in the structure obtained under cryogenic conditions is 28.9 Å2, comparable to the average value of 26.4 Å2 for the rest of the side chain, and 24.1 Å2 for its backbone (29,39). Therefore, we deem it more likely that the inability of the Lys side chain to impart order on internal water molecules actually reflects the weak propensity of amines for hydration, especially relative to the carboxylic group, which are extremely well hydrated (40,41). Our observations are also consistent with a survey of the distribution of water molecules around ionizable groups in proteins, which identified the polar side chains of Glu and Asp as the ones that are hydrated most frequently (1,42).
CONCLUSIONS
The systematic comparison of a set of nearly identical structures that differ mainly in the nature of the ionizable or polar group at the internal positions 66 or 92 and in the number of internal water molecules that these groups stabilize, suggests that the following factors can be important determinants of internal hydration: i), Without exception in the set of structures obtained under cryogenic conditions, buried oxygen atoms are always found in complex with internal water molecules. ii), under cryogenic conditions the capacity of polar side chains to organize internal water molecules can be ranked as follows, from strongest to weakest: carboxylic in neutral state > amide > amine in neutral state. (iii) Fewer internal water molecules were observed in the room temperature structures than in the ones obtained under cryogenic conditions. However, the presence of internal water molecules in the structures with Glu-66 and Tyr-66 at room temperature, and the presence of residual electron density in the room temperature data in the same sites that are occupied by water molecules under cryogenic conditions, suggest that internal water molecules might be present in the microcavity at room temperature in all variants of SNase albeit in a disorganized state that renders them invisible in the x-ray diffraction experiment, at least with the criteria that were applied during refinement of these structures. (iv) The observation of internal water molecules in room temperature structures of two different variants suggests that the internal water molecules observed under cryogenic conditions are not an artifact of freezing, and that the main effect of cryogenic conditions is to minimize disorder among the internal water molecules, trapping them kinetically in fixed positions that presumably represent minima on the binding surface, allowing the waters to be identified in electron density maps. (v) In microcavities the volume available per water molecule is limited. Under these circumstances, sterics matter. Bulky side chains that reduce the volume accessible to the internal water molecules likely minimize the number of locations that the water molecules can occupy, thereby reducing disorder and rendering these water molecules detectable in the crystal structures. (vi) Internal Lys residues are not hydrated readily. Although this could be related to the flexibility of this long side chain, we deem it more likely that this reflects the low hydration propensity of amines in general. It remains to be established if any of these observations with SNase are relevant to describe the hydration of microcavities in other proteins, or in systems such as bacteriorhodopsin (10,43), cytochrome c oxidase (6,9,10,43), and other proteins where the internal water molecules actually play essential roles in H+ transfer.
Further elucidation of the positions and dynamic properties of ordered waters in internal cavities in proteins may be provided by neutron diffraction studies. Such studies have been extremely successful in describing statically disordered water molecules (44) and detailing complex patterns of hydration (45,46). Neutron diffraction studies carried out in concert with x-ray diffraction structures (47) and molecular dynamics simulations (44) have yielded insights into complex water orientations. However, growth of crystals large enough and of sufficiently high quality for neutral diffraction remains a considerable challenge, particularly for a survey of protein variants such as in this study.
Our original interest in internal hydration stems from the observation that many internal ionizable groups titrate with perturbed pKa values that report apparent dielectric constants of 10 or higher when analyzed with continuum electrostatics methods (29,48,30). These are high dielectric constants, considerably higher than the ones predicted for proteins by theory (49), and than the ones measured experimentally with dry protein powders (50). Dielectric constants of 10 or higher are consistent with the response of highly polarizable materials (51). It is likely that internal water molecules make important contributions to high apparent dielectric permittivity inside proteins, especially when they are free to reorganize in the presence of an electric field. For this reason they are likely to be important determinants of the pKa values of internal ionizable groups.
Recently our group and others have used MD simulations to examine the physical properties of internal water molecules and mechanisms of water penetration in proteins (15,25–27). The simulations suggest that water can penetrate readily into microcavities such as the one near position 66 in SNase, and that the internal water molecules can have residence times in the picosecond to nanosecond range. The simulations also show that water molecules are not stable in hydrophobic pockets – internal polar groups are necessary to stabilize internal water molecules at fixed positions (27,52). Consistent with our crystallographic observations, the MD simulations suggest that internal side chains can exhibit significant disorder, and that the probability of visualizing ordered water molecules is complex because of the interplay between order and disorder of both side chains and internal water molecules (53,27). Using the structures described in this article we have already identified a protocol for MD simulations that reproduces the patterns of hydration observed for these proteins under cryogenic conditions (27). It involves carrying out many short simulations starting from different states of internal hydration, instead of carrying out a single very long simulation, in which, apparently, water penetration and exit from the protein is not sampled sufficiently in the time scales that are currently accessible with MD methodology. We anticipate that the set of structures described in this article will be extremely useful for the development of other fast and accurate protocols for artificial hydration of cavities and pockets (54,28).
Acknowledgments
We thank Dr. Carolyn Fitch for carrying out solvent accessibility calculations.
This study was supported by National Institutes of Health (GM-061597) to B.G-M.E. J.L.S. was supported by a John F. Crowley fellowship from the United States Naval Academy. C.A. gratefully acknowledges the Office of Naval Research (N0001406WR20137) for partial support of this research. D.A.K. was supported by a predoctoral fellowship from the National Science Foundation, and M.A.D. was supported by a postdoctoral fellowship from the Burroughs-Wellcome Fund.
Michael A. Dolan's present address is Tripos Associates, St. Louis, Missouri.
Apostolos Gittis' present address is Dept. of Natural Sciences, Bowie State University, Bowie, Maryland.
Editor: Janos K. Lanyi.
References
- 1.Williams, M. A., J. M. Goodfellow, and J. M. Thornton. 1994. Buried waters and internal cavities in monomeric proteins. Protein Sci. 3:1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park, S., and J. G. Saven. 2005. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins Struct. Funct. Genet. 60:450–463. [DOI] [PubMed] [Google Scholar]
- 3.Rose, G. D., W. B. Young, and L. M. Gierasch. 1983. Interior turns in globular proteins. Nature. 304:655–657. [DOI] [PubMed] [Google Scholar]
- 4.Warshel, A. 1978. Energetics of enzyme catalysis. Proc. Natl. Acad. Sci. USA. 75:5250–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warshel, A. 1998. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J. Biol. Chem. 273:27035–27038. [DOI] [PubMed] [Google Scholar]
- 6.Gennis, R. B. 1998. How does cytochrome oxidase pump protons? Proc. Natl. Acad. Sci. USA. 95:12747–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu, J., and G. A. Voth. 2005. Computer simulation of explicit proton translocation in cytochrome c oxidase: The D-pathway. Proc. Natl. Acad. Sci. USA. 102:6795–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luecke, H., B. Schobert, H. T. Richter, J. P. Cartailler, and J. K. Lanyi. 1999. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291:899–911. [DOI] [PubMed] [Google Scholar]
- 9.Gennis, R. B., and T. G. Ebrey. 1999. Proton pump caught in the act. Science. 286:252–253. [DOI] [PubMed] [Google Scholar]
- 10.Lanyi, J. K. 2004. Bacteriorhodopsin. Annu. Rev. Physiol. 66:665–688. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 12.Kolbe, M., H. Besir, L. O. Essen, and D. Oesterhelt. 2000. Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science. 288:1390–1396. [DOI] [PubMed] [Google Scholar]
- 13.Gibas, C. J., and S. Subramaniam. 1996. Explicit solvent models in protein pKa calculations. Biophys. J. 71:138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, S., and C. S. Verma. 1999. Binding of buried structural water increases the flexibility of proteins. Proc. Natl. Acad. Sci. USA. 96:9613–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrone, P. M., and A. E. Garcia. 2004. MHC-peptide binding is assisted by bound water molecules. J. Mol. Biol. 338:419–435. [DOI] [PubMed] [Google Scholar]
- 16.Scapin, G., J. I. Gordon, and J. C. Sacchettini. 1992. Refinement of the structure of recombinant rat intestinal fatty acid-binding apoprotein at 1.2 Å resolution. J. Biol. Chem. 267:4253–4269. [DOI] [PubMed] [Google Scholar]
- 17.Xu, J., W. A. Baase, M. L. Quillin, E. P. Baldwin, and B. W. Matthews. 2001. Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme. Protein Sci. 10:1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, X. J., and B. W. Matthews. 1994. Conservation of solvent-binding sites in 10 crystal forms of T4 lysozyme. Protein Sci. 3:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattos, C., C. R. Bellamacina, E. Peisach, A. Pereira, D. Vitkup, G. A. Petsko, and D. Ringe. 2006. Multiple solvent crystal structures: probing binding sites, plasticity and hydration. J. Mol. Biol. 357:1471–1482. [DOI] [PubMed] [Google Scholar]
- 20.Halle, B. 2004. Biomolecular cryocrystallography: structural changes during flash-cooling. Proc. Natl. Acad. Sci. USA. 101:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denisov, V. P., and B. Halle. 1995. Hydrogen exchange and protein hydration: the deuteron spin relaxation dispersions of bovine pancreatic trypsin inhibitor and ubiquitin. J. Mol. Biol. 245:698–709. [DOI] [PubMed] [Google Scholar]
- 22.Otting, G., E. Liepinsh, and K. Wutrich. 1991. Proton exchange with internal water molecules in the protein BPTI in aqueous solution. J. Am. Chem. Soc. 113:4363–4364. [Google Scholar]
- 23.Denisov, V. P., J. Peters, H. D. Hörlen, and B. Halle. 1996. Using buried water molecules to explore the energy landscape of proteins. Nat. Struct. Biol. 3:505–509. [DOI] [PubMed] [Google Scholar]
- 24.Wiesner, S., E. Kurian, F. Prendergast, and B. Halle. 1999. Water molecules in the binding cavity of intestinal fatty acid binding protein: dynamic characterization by water 17O and 2H magnetic relaxation dispersion. J. Mol. Biol. 286:233–246. [DOI] [PubMed] [Google Scholar]
- 25.Damjanović, A., B. García-Moreno, E. E. Lattman, and A. E. Garcia. 2005. Molecular dynamics study of water penetration in staphylococcal nuclease. Proteins. 60:433–449. [DOI] [PubMed] [Google Scholar]
- 26.García, A. E., and G. Hummer. 2000. Water penetration and escape in proteins. Proteins. 38:261–272. [PubMed] [Google Scholar]
- 27.Damjanovic, A., J. L. Schlessman, C. A. Fitch, A. E. García, and B. García-Moreno. 2007. Role of flexibility and polarity as determinants of the hydration of internal cavities and pockets in proteins. Biophys. J. 93:2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, L., and J. Hermans. 1996. Hydrophilicity of cavities in proteins. Proteins. 24:433–438. [DOI] [PubMed] [Google Scholar]
- 29.Dwyer, J., A. Gittis, D. Karp, E. Lattman, D. Spencer, W. Stites, and B. García-Moreno. 2000. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 79:1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp, D. A., A. G. Gittis, M. R. Stahley, C. A. Fitch, W. E. Stites, and B. García-Moreno. 2007. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys. J. 92:2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, D. M., R. L. Reynald, A. G. Gittis, and E. E. Lattman. 2004. X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior. J. Mol. Biol. 341:565–574. [DOI] [PubMed] [Google Scholar]
- 32.Shortle, D., and A. Meeker. 1986. Mutant forms of staphylococcal nuclease with altered patterns of guanidine hydrochloride and urea denaturation. Proteins. 1:81–89. [DOI] [PubMed] [Google Scholar]
- 33.Brünger, A., P. Adams, G. Clore, W. DeLano, P. Gros, R. Grosse-Kunstleve, J. Jiang, J. Kuszewski, M. Nilges, N. Pannu, R. Read, L. Rice, T. Simonson, and G. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 54:905–921. [DOI] [PubMed] [Google Scholar]
- 34.Jones, T. A., J.-Y. Zhou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models into electron density maps and the location of errors in these models. Acta Crystallogr. A. 47:110–119. [DOI] [PubMed] [Google Scholar]
- 35.Denisov, V. P., J. L. Schlessman, B. García-Moreno, and B. Halle. 2004. Stabilization of internal charges in a protein: water penetration or conformational change? Biophys. J. 87:3982–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bode, W., E. Papamokos, and D. Musil. 1987. The high resolution crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur. J. Biochem. 166:673–692. [DOI] [PubMed] [Google Scholar]
- 37.Feig, M., J. Karanicolas, and C. L. Brooks III. 2004. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for structural biology. J. Mol. Graph. Model. 22:377–395. [DOI] [PubMed] [Google Scholar]
- 38.Collins, M. D., G. Hummer, M. L. Quillin, B. W. Matthews, and S. M. Gruner. 2005. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. USA. 102:16668–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stites, W. E., A. G. Gittis, E. E. Lattman, and D. Shortle. 1991. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J. Mol. Biol. 221:7–14. [DOI] [PubMed] [Google Scholar]
- 40.Kiriukhin, M. Y., and K. D. Collins. 2002. Dynamic hydration numbers for biologically important ions. Biophys. Chem. 99:155–168. [DOI] [PubMed] [Google Scholar]
- 41.Collins, K. D. 1995. Sticky ions in biological systems. Proc. Natl. Acad. Sci. USA. 92:5553–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, S. M., and M. M. Teeter. 1993. Patterns for prediction of hydration around polar residues in proteins. J. Mol. Biol. 229:419–427. [DOI] [PubMed] [Google Scholar]
- 43.Luecke, H., and J. K. Lanyi. 2003. Structural clues to the mechanism of ion pumping in bacteriorhodopsin. Adv. Protein Chem. 63:115–130. [DOI] [PubMed] [Google Scholar]
- 44.McDowell, R. S., and A. A. Kossiakoff. 1995. A comparison of neutron diffraction and molecular dynamics structures: hydroxyl group and water molecule orientations in trypsin. J. Mol. Biol. 250:553–570. [DOI] [PubMed] [Google Scholar]
- 45.Kossiakoff, A. A., M. D. Sintchak, J. Shpungin, and L. G. Presta. 1992. Analysis of solvent structure in proteins using neutron D2O–H2O solvent maps: pattern of primary and secondary hydration of trypsin. Proteins. 12:223–236. [DOI] [PubMed] [Google Scholar]
- 46.Shu, F., V. Ramakrishnan, and B. P. Schoenborn. 2000. Enhanced visibility of hydrogen atoms by neutron crystallography on fully deuterated myoglobin. Proc. Natl. Acad. Sci. USA. 97:3872–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finer-Moore, J. S., A. A. Kossiakoff, J. H. Hurley, T. Earnest, and R. M. Stroud. 1992. Solvent structure in crystals of trypsin determined by X-ray and neutron diffraction. Proteins. 12:203–222. [DOI] [PubMed] [Google Scholar]
- 48.Fitch, C. A., D. A. Karp, K. K. Lee, W. E. Stites, E. E. Lattman, and B. García-Moreno. 2002. Experimental pKa values of buried residues: analysis with continuum methods and role of water penetration. Biophys. J. 82:3289–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilson, M. K., and B. H. Honig. 1986. The dielectric constant of a folded protein. Biopolymers. 25:2097–2119. [DOI] [PubMed] [Google Scholar]
- 50.Harvey, S. C., and P. Hoekstra. 1972. Dielectric relaxation spectra of water adsorbed on lysozyme. J. Phys. Chem. 76:1987–1994. [DOI] [PubMed] [Google Scholar]
- 51.Schutz, C. N., and A. Warshel. 2001. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins. 44:400–417. [DOI] [PubMed] [Google Scholar]
- 52.Vaitheeswaran, S., H. Yin, J. C. Rasaiah, and G. Hummer. 2004. Water clusters in nonpolar cavities. Proc. Natl. Acad. Sci. USA. 101:17002–17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taraphder, S., and G. Hummer. 2002. Dynamic proton transfer pathways in proteins: role of sidechain conformation fluctuations. Physica A. 318:293–301. [Google Scholar]
- 54.Imai, T., R. Hiraoka, A. Kovalenko, and F. Hirata. 2007. Locating missing water molecules in protein cavities by the three-dimensional reference interaction site model theory of molecular solvation. Proteins. 66:804–813. [DOI] [PubMed] [Google Scholar]
- 55.Chen, J., Z. Lu, J. Sakon, and W. E. Stites. 2000. Increasing the thermostability of staphylococcal nuclease: implications for the origin of protein thermostability. J. Mol. Biol. 303:125–130. [DOI] [PubMed] [Google Scholar]
- 56.García-Moreno B., J. Dwyer, A. Gittis, E. Lattman, D. Spencer, and W. Stites. 1997. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys. Chem. 64:211–224. [DOI] [PubMed] [Google Scholar]