Abstract
Background
Dietary restriction (DR) results in increased longevity, reduced fecundity and reduced growth in many organisms. Though many studies have examined the effects of DR on longevity and fecundity, few have investigated the effects on growth.
Results
Here we use Caenorhabditis elegans to determine the mechanisms that regulate growth under DR. We show that rather than a reduction in cell number, decreased growth in wild type C. elegans under DR is correlated with lower levels of hypodermal polyploidization. We also show that mutants lacking wild type sensory ciliated neurons are small, exhibit hypo-polyploidization and more importantly, when grown under DR, reduce their levels of endoreduplication to a lesser extent than wild type, suggesting that these neurons are required for the regulation of hypodermal polyploidization in response to DR. Similarly, we also show that the cGMP-dependent protein kinase EGL-4 and the SMA/MAB signalling pathway regulate polyploidization under DR.
Conclusion
We show C. elegans is capable of actively responding to food levels to regulate adult ploidy. We suggest this response is dependent on the SMA/MAB signalling pathway.
Background
Many animals change their life-history, size or shape in response to the environment; a phenomenon known as phenotypic plasticity [1,2]. One environmental factor that exerts great influence over the development and life history of an organism is that of nutrition, or 'dietary restriction' [3-8]. Studies in a variety of taxa have shown that restricting the nutrition of juveniles or adults reduces growth and fecundity, while increasing longevity [9-11].
Over the last decade the underlying cellular mechanisms that regulate the effect of DR on growth have been explored more extensively [12]. In metazoans, it appears that much of an organism's ability to respond to DR is determined by insulin-like signalling. For example, overexpression of Insulin-like Growth Factor Binding Protein-1 (IGFBP-1) is known to cause retardation of bone growth [13] and is found in DR rats at three times the normal level [14]. Drosophila and mice lacking components of the Insulin-like signalling pathway have greatly reduced body [15-19]. This reduction in size is due to a combination of reduced cell number and cell size [18,19]. In contrast, insulin-associated pathways in C. elegans are known to determine fat storage, diapause, and longevity, but their effect on body size is less evident [20-25]. However, genetic mechanisms of body size determination in C. elegans are known to involve DBL-1 signalling (TGF-β ligand homologous to Drosophila's Dpp and vertebrate's BMP). DBL-1 regulates normal growth in C. elegans through the SMA/MAB pathway [26], along with downstream components such as LON-1 [27,28]. It seems to us a reasonable hypothesis that the DBL-1 signalling may be involved in the DR response. Moreover, this relationship may extend to sensory-based regulation of growth. Mutant strains lacking properly formed and functional sensory ciliated neurons, such as the che mutants (cilia extension defects), together with downstream cGMP-dependent protein kinase EGL-4, exhibit alterations not only in longevity but also in body size [29-31].
In this study we investigate whether C. elegans undergoes a programmed regulation of growth in response to DR. First, we characterized life history responses, of wild type C. elegans, to DR, determining longevity, fecundity and body size. Second, we determined the role of the sensory system in growth regulation in response to DR. Thirdly, we examined the role of TGF-β signalling in DR mediated growth responses and determine how this relates to the sensory system.
Results
Dietary restriction in C. elegans reduces body size, hypodermal ploidy and fecundity but increases longevity
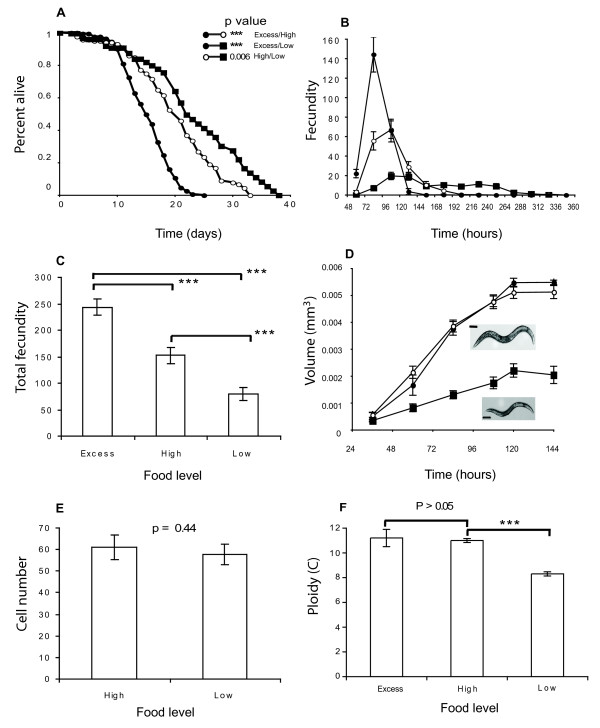
We first set up an experimental system for growing C. elegans under DR (also referred as "low food conditions"; see Materials and Methods). As we were not interested in the two adaptive responses of C. elegans larvae to DR, i.e. L1 arrest [32] and dauer formation [33], we exposed L3 animals grown in high food (see Materials and Methods) to DR. They produced adults with substantial differences with respect to their longevity (57% longer with DR; Figure 1A), fecundity (67% smaller with DR; Figures 1B and 1C) and body size (63% smaller with DR; Figure 1D).
Figure 1.
The effects of dietary restriction on C. elegans life history traits. (A) Kaplan-Maier survival curves showing the longevity of C. elegans under excess (closed circles), high (open circles) and low food (closed squares) environments (see Material and Methods). Significance is shown for excess, high, and low food, from Log Rank tests n/censored individuals 175/22, 204/110 and 238/189 respectively. (B) Daily fecundity of C. elegans under excess (closed circles), high (open circles) and low food (closed squares) environments. (C) Total fecundity of C. elegans under excess, high and low food environments; n = 36, 44 and 35 respectively. (D) Growth curves of C. elegans under excess (closed circles), high (open circles) and low food (squares) environments, n = 49, 38, and 21 respectively. Images show representative adults from high food (upper panel) and low food (lower panel) treatments. Scale bar indicates 100 μm. (E) Hypodermal (hyp7) cell number of young adult C. elegans under high and low food environments; n = 10 and 9 respectively. (F) Hypodermal (hyp7) ploidy of C. elegans (120 h) under excess, high and low food environments; n = 19, 254, and 189 respectively. All error bars show 95% confidence intervals, and asterisk show level of significance, *** shows P < 0.0001, by ANOVA.
The reduced fecundity and extended longevity are consistent with previous studies on DR using C. elegans grown in liquid media [5]. They are also consistent with Drosophila's experiments where DR induces adults of smaller size [34,4]. However, unlike in Drosophila, where the reduction in size is due to a combination of reduced cell number and size, in C. elegans there is no alteration in cell number, at least in the hypodermis (Figure 1E), which secretes the cuticle, scales with body size and regulates it through TGF-β signalling [35]. Our data also show that the reduction in body size seen on DR is associated with reduced levels of hypodermal endoreduplication (Figure 1F), which we recently showed drives growth in adult worms [36].
Food consumption regulates body size but not hypodermal ploidy
How does food level control the endoreduplication and growth of worms? One possibility is that worms monitor the amount of food that they actually eat and adjust their ploidy and growth accordingly. To test this idea we first studied a mutant,eat-2(ad465), that has a defective pharynx and therefore cannot eat properly [37,38]. In effect, eat-2 mutants experience constitutive DR. We found that, when grown at high food levels, eat-2 (ad465) has a small body size but has wild type ploidy [Tables 1 and 2]. Under DR conditions, eat-2 (ad465) behaved like wild-type: its body size was even further reduced and its ploidy decreased by 24% (Table 2). This suggested to us that body size is at least partly controlled by the amount of food that a worm eats, but that hypodermal endoreduplication is not.
Table 1.
Effect of Dietary Restriction on Body Size. All genotypes show significant (p < 0.0001), wild type-like (genotype by environment interaction term; p > 0.05), reductions in volume under DR.
| Body size (mm3) | ||||
| Genotype | High Food | Low Food | n | % reduction |
| N2 | 0.0051 (± 1 × 10-4) | 0.0021 (± 1 × 10-4) | 147, 121 | 63 |
| che-2(e1033) | 0.0022 (± 1 × 10-4) | 0.0009 (± 2 × 10-4) | 106, 43 | 62 |
| che-3(e1124) | 0.0029 (± 2 × 10-4) | 0.0013 (± 8 × 10-5) | 19, 12 | 55 |
| che-11(e1810) | 0.0027 (± 1 × 10-4) | 0.0012 (± 1 × 10-4) | 41, 23 | 56 |
| che-13(e1805) | 0.0021 (± 3 × 10-4) | 0.0009 (± 5 × 10-5) | 11, 16 | 57 |
| osm-5(p813) | 0.0033 (± 2 × 10-4) | 0.0013 (± 2 × 10-4) | 26, 20 | 61 |
| egl-4(n478) | 0.0063 (± 2 × 10-4) | 0.0023 (± 1 × 10-4) | 127, 118 | 60 |
| dbl-1(nk3) | 0.0025 (± 1 × 10-4) | 0.001 (± 6 × 10-5) | 121, 80 | 62 |
| sma-2(e502) | 0.0020 (± 4 × 10-4) | 0.0010 (± 2 × 10-4) | 19, 15 | 50 |
| sma-3(wk20) | 0.0025 (± 2 × 10-4) | 0.0010 (± 1 × 10-4) | 32, 24 | 60 |
| sma-4(e729) | 0.0010 (± 1 × 10-4) | 0.0006 (± 1 × 10-4) | 33, 22 | 40 |
| sma-6(wk7) | 0.0019 (± 3 × 10-4) | 0.0009 (± 2 × 10-4) | 39, 22 | 53 |
| lon-1(e185) | 0.0050 (± 6 × 10-4) | 0.0017 (± 3 × 10-4) | 20, 9 | 66 |
| che-2(e1033);dbl-1(nk3) | 0.0019 (± 1 × 10-4) | 0.0007 (± 4 × 10-5) | 48, 35 | 58 |
| egl4(n478);dbl-1(nk3) | 0.0028 (± 3 × 10-4) | 9 | ||
| eat-2(ad465) | 0.0020 (± 1 × 10-4) | 0.0008 (± 4 × 10-5) | 71, 56 | 59 |
| eat-2(ad465);dbl-1(nk3) | 0.0011 (± 1 × 10-4) | 0.0005 (± 1 × 10-4) | 85, 45 | 52 |
Table 2.
Effect of Dietary Restriction on Hypodermal Ploidy. All genotypes, unless stated (NS, p > 0.05), show highly significant (p < 0.0001) alterations from wild type ploidy responses to DR.
| Hypodermal ploidy (xC) | ||||
| Genotype | High Food | Low Food | n | % reduction |
| N2 | 10.9 (± 0.3) | 8.4 (± 0.2) | 113, 94 | 23 |
| che-2(e1033) | 8.6 (± 0.3) | 7.5 (± 0.4) | 56, 25 | 13 |
| che-3(e1124) | 8.4 (± 0.5) | 7.5 (± 0.7) | 17, 12 | 11 |
| che-11(e1810) | 9.2 (± 0.3) | 8.5 (± 0.5) | 32, 21 | 8 |
| che-13(e1805) | 8.4 (± 0.5) | 7.5 (± 0.5) | 13, 13 | 11 |
| osm-5(p813) | 8.8 (± 0.4) | 7.6 (± 0.5) | 24, 17 | 14 |
| egl-4(n478) | 12.3 (± 0.3) | 11.6 (± 0.3) | 101, 88 | 5 |
| dbl-1(nk3) | 7.5 (± 0.5) | 6.9 (± 0.4) | 51, 32 | 8 |
| sma-2(e502) | 7.6 (± 0.7) | 7.0 (± 0.5) | 14, 11 | 8 |
| sma-3(wk20) | 8.2 (± 0.4) | 7.0 (± 0.4) | 39, 23 | 15 |
| sma-4(e729) | 7.4 (± 0.3) | 6.4 (± 0.5) | 35, 22 | 14 |
| sma-6(wk7) | 8.3 (± 0.3) | 7.1 (± 0.3) | 31, 18 | 14 |
| lon-1(e185) | 12.2 (± 0.9) | 8.5 (± 1.0) | 9, 8 | 30 NS |
| che-2(e1033);dbl-1(nk3) | 9.1 (± 0.5) | 7.8 (± 0.5) | 29, 18 | 14 |
| egl4(n478);dbl-1(nk3) | 8.9 (± 0.8)' | 6 | ||
| eat-2(ad465) | 10.1 (± 0.7) | 7.7 (± 0.4) | 24, 17 | 24 NS |
| eat-2(ad465);dbl-1(nk3) | 8.5 (± 0.4) | 7.0 (± 0.4) | 43, 23 | 18 |
Endoreduplication requires the sensation of food by ciliated neurons
If the amount of food that a worm actually eats does not control endoreduplication, why do DR worms have low hypodermal ploidies? One possibility is that worms regulate endoreduplication in response to the amount of food that they sense in their environments. Worms sense their environment by means of their amphids, two small sensory organs that are exposed to the environment through pores located near the worm's mouth. Each amphid has 12 neurons from which eight project into the channel that leads to the pore [39-42]. These eight neurons are ciliated and have specialised endings containing receptor proteins that interpret and distinguish between external stimuli [43].
To test whether sensory signals from the amphids are involved in the DR response, we measured body size in various mutants possessing malformed, non-functional, sensory cilia. Consistent with Fujiwara et al. [30], we found that che-2(e1033) is smaller than wild type worms under high food conditions (Table 1). This phenotype is shared with all the other sensory cilia mutants examined (Table 1): che-13(e1805), osm-5(p813), che-3(e1124) and che-11(e1810). We also investigated whether the sensory mutants become smaller under DR and found that they had wild type responses (Table 1).
Then, to determine if the small body size of the sensory cilia mutants was associated with reduced ploidy we examined the hypodermis of all the sensory mutants. All of these mutants showed a reduction in ploidy (p < 0.001) (Table 2). More importantly, when subjected to DR, their ploidy declined only by approximately 11%, compared to a 23% reduction of the wild type (Table 2). We found no significant differences between the hypodermal nuclei number of che-2(e1033) and wild type worms (data not shown). These results suggest that signals from the amphids partly control endoreduplication in response to DR.
EGL-4 mediates the response from sensory cilia
Previous studies have shown that EGL-4, a cGMP-dependent protein kinase, functions downstream of sensory ciliated neurons in wild type worms [30]. Furthermore, mutations in egl-4 result in increased body length, altered sensory perception and egg laying behaviour, without affecting cilia structure [44]. To determine whether EGL-4 is required for the regulation of body size and endoreduplication in response to DR, we first characterized the growth of a strong loss-of-function mutant, egl-4 (n478), under normal levels of food. We found these worms to be 21% larger than wild type (Table 1) and possess a 13% higher level of hypodermal endoreduplication (Table 2), while maintaining wild type cell numbers (data not shown).
Surprisingly, under DR, egl-4 exhibits a wild type reduction in volume, but importantly, it fails to show a wild type reduction in endoreduplication. Hypodermal polyploidization, in egl-4 worms, declines only 5% under DR compared to a 25% decline in N2 (Table 2). Therefore, egl-4 defective worms maintain a hyper-endoreduplicated state at their hypodermis even under DR. Their hyper-endoreduplicated state, their failure to show wild type declines in endoreduplication, and the placement of EGL-4 downstream of CHE-2 [30] (also see Table 1 &2), all together suggest that EGL-4 acts as a negative regulator of food dependent endoreduplication.
DBL-1 signalling regulates the DR endoreduplication response
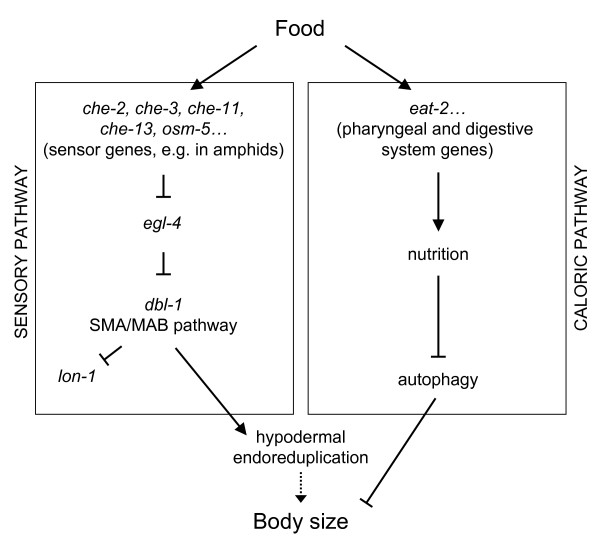
DBL-1 is known to be a dose-dependent regulator of body size and endoreduplication in C. elegans. This protein activates the SMA-6/DAF-4, Ser/Thr kinase receptor, which in turn is thought to activate the cytoplasmic effectors SMA-2, SMA-3 and SMA-4 [26]. Here we confirm that loss-of-function dbl-1(nk3) worms, as well as worms defective for downstream signalling components such as sma-6, sma-2, sma-3 and sma-4, all show a 60% reduction in body size when grown in normal food levels (Table 1), similarly to previously reported [45-48]. They also show a ~25% reduction in hypodermal polyploidization (Table 2). To determine the role of DBL-1 in the DR response of wild type worms, all these mutants were subjected to DR and their body size and hypodermal endoreduplication characterized. All mutants showed a marked decrease in size (40% – 60%), responding to DR in a wild type manner (63%; Table 1). More interestingly, when the effects of DR on endoreduplication were examined, mutants deficient for dbl-1, sma-6, sma-2, sma-3 and sma-4 all show a distinct non-wild type response: endoreduplication declines by approximately 12%, compared to the 23% seen in N2 (Table 2). These results suggest that DBL-1 signalling, as described previously for sensory cilia mutants and egl-4, is partially responsible for the regulation of endoreduplication as a response to DR. We note, however, that loss-of-function lon-1, placed downstream of the dbl-1 pathway [28], behaves as wild type under DR (Table 2). Therefore, we suggest that lon-1, despite its role in determining body size and hypodermal endoreduplication, is not part of the polyploidization response to nutrients availability (Figure 2).
Figure 2.
Model of body size regulation by nutrients availability in C. elegans (from L3 onwards). Our results suggest that food availability may regulate body size in at least two ways. First, by the "caloric pathway", that is, simply considering that food intake and its absorption by the digestive tract facilitates nutrition, which in turn may inhibit autophagy. Second, by the "sensory pathway", which refers to the sensing food through organs such as the amphids, with their ciliated neurons expressing genes like che-2, would inhibit EGL-4. Downstream, this cGMP-dependent protein kinase downregulates DBL-1 signalling, which in turn promotes hypodermal endoreduplication, upregulator of body size [36]. LON-1 inhibition by DBL-1 [28] would not influence ploidy upon nutrient activation. This model explains why the nutrient-dependent regulation that the sensory cilia proteins, EGL-4 and DBL-1 are all playing on hypodermal polyploidization has not been observed for body size; their role on body size, but not upon endoreduplication, may be obscured by the dominant influence of caloric restriction.
CHE-2 and DBL-1 act in the same pathway to regulate body size and hypodermal ploidy
In order to test the hypothesis that sensory signals and DBL-1 signalling act in the same pathway, we generated double che-2;dbl-1 mutants and analysed their size and ploidy levels under standard and DR conditions. When grown in high food conditions, che-2;dbl-1 was similar in size and ploidy (p > 0.05 for all comparisons), to dbl-1, che-2, or related genes (e.g. sma-6, che-13; Tables 1 and 2). The corresponding reduction for both characters under DR was also similar (Tables 1 and 2). This result suggests that dbl-1 and the amphid mutants act in the same pathway when controlling body size and hypodermal endoreduplication.
We also asked whether the regulation of body size by food intake per se was affected by DBL-1 signalling. To test this we examined eat-2;dbl-1 double mutants. We found that at high food levels, these worms are smaller than either eat-2 or dbl-1 (Table 1). This additive effect suggests that these genes regulate body size through different pathways, and is consistent with the finding that eat-2 worms have normal ploidy.
EGL-4 negatively regulates DBL-1
To confirm the effect of EGL-4 on the signalling of DBL-1 seen previously [30,31], epistasis analysis was carried out between null mutants dbl-1 and egl-4. The nature of these mutants allowed a relatively simple analysis because egl-4 worms are larger than wild type, whereas dbl-1 worms are smaller [45,46] (Table 1). The same thing can be said about hypodermal ploidy (Table 2). Examination of egl-4;dbl-1 worms revealed that, though slightly smaller, the double mutant did not significantly differ from dbl-1 worms in either adult volume or hypodermal ploidy (P > 0.05, for body size and ploidy; Tables 1 and 2, respectively), but it did with respect to egl-4 worms (P < 0.0001, for body size and ploidy).
Discussion
Growth is a fundamental part of biology, yet its regulation is still poorly understood [12]. The notion that growth responds passively to nutrient availability has been replaced with the idea that growth is actively regulated in response to constant monitoring of nutrient availability in the external environment. We observed that when C. elegans is exposed to a low food environment there is a reduction in adult body size, similar to the reductions seen in other organisms e.g. Drosophila and Daphnia [4,34,49,50]. However, in contrast to these organisms, the stunting in C. elegans is not due to a lack of cell proliferation, which implies that it is due to a reduction in cell size.
In order to investigate how DR controls adult body size in C. elegans, we studied the growth of wild type and mutant worms subjected to high and low food regimes. We found that all of our mutants became smaller by about the same amount (60%) at low food levels. This absence of interaction between food and genotype on growth might mean that none of the genes examined are involved in the dietary-dependent regulation of growth, but it could also simply mean that severe DR has additional effects.
For this reason we needed a more subtle way of examining the effects of DR on worm development. We have previously shown that hypodermal endoreduplication is required for growth in adult C. elegans [36]. Strikingly, we also found that DR inhibits hypodermal endoreduplication and so adult ploidy. This result gave us a sensitive assay for the effects of DR on the worm's development. We found that mutations in several genes mimic the DR response: even at high food levels, mutations that disrupt sensory or DBL-1 signalling show reduced ploidy and body size. That suggested to us that these genes might be involved in the DR endoreuplication response. This inference was confirmed when we examined these mutants under DR: in each case, the reduction in ploidy normally found at low food levels was largely abrogated. An even more striking lack of response to DR was also found in a large mutant that disrupts egl-4, a cGMP-dependent protein kinase previously associated with food sensing and food dependent behaviour [30].
These results, and our epistasis experiments, suggest a model in which the amphids monitor nutrient availability and activate a downstream signalling pathway involving the growth repressor EGL-4 (Figure 2). This kinase in turn regulates the DBL-1/SMA/MAB pathway, which positively regulates hypodermal endoreduplication. As the observed body size reduction under DR for both the sensory and DBL-1 signalling pathway mutants was similar to that of wild type (Table 1), we suggest that the main effects of DR on body size do not arise from the lack of endoreduplication, but rather from some other unknown pathway. A likely candidate could be what we call the "caloric pathway" in Figure 2. That is, the severe food restriction under DR could be masking the "sensory pathway" on body size when this one is impaired (e.g. in dbl-1(nk3); Figure 2). Reduction in food may prevent DBL-1 like mutants, whose endoreduplication levels do not drop as much as wild type under DR, from growing larger. Nevertheless, the reduced ploidy programmed by the sensing of lower levels of food (Table 2) must contribute to the stunting, since previous work shows a cause-and-effect relationship between endoreduplication and adult growth [36]. Consistent with our model, we showed that eat-2 mutants, one of the genes active in the feeding mechanism, has small size but normal ploidy, and that it reduces both characters in a wild type manner under DR (Tables 1 and 2). Recent work suggests that eat mutants have small body sizes due to increased autophagy [38], which is also included in our model (Figure 2).
Conclusion
How do our results relate to other animal models? Endoreduplication in Drosophila depends on a mitogen from the fat body that is regulated in a nutrition-dependent manner [51], which may suggest at least an underlying common plan beyond their differences (see Introduction)[52]. However, one of the proteins studied here, EGL-4, is a key regulator of nutrient responses not only in worms but, with the generic name of cGMP-dependent protein kinase, in organisms such as honeybees and fruit-flies controlling their foraging behaviour [53]. It is somewhat surprising that loss-of-function egl-4 has a change in hypodermal endoreduplication in high vs. low food which is half of what it is observed for the sensory ciliated or for the dbl-1-related mutants (Table 2). We think that this difference can be explained because egl-4's role in nutrient-dependent growth may be central, not shared with other proteins in parallel positions, whereas the various sensor genes investigated may be acting in parallel, either among themselves, or in relation to other genes or pathways (similarly for DBL-1 and the SMA/MAB pathway). In agreement with this, egl-4 is considered a highly pleiotropic gene, a main regulatory hub, not only mediating body size but longevity, locomotion feeding, and other processes [54].
Methods
Strains
Apart from the wild type strain N2, the following mutant strains were used, which were obtained from the Caenorhabditis elegans Genetics Center. Mutations are listed by linkage group: LGI: che-3(e1124), che-13(e1805); LGII: sma-6(wk7), eat-2(ad465); LGIII: lon-1(e185), sma-2(e502), sma-3(wk20), sma-4(e729); LGIV: egl-4(n478); LGV: che-11(e1810), dbl-1(nk3); LGX: che-2(e1033), osm-5(p813). We also used double mutants that we produced through crosses of the previous strains. Double mutants eat-2(ad465);dbl-1(nk3) were confirmed by PCR and sequencing using the following primers: 5'-eat-2: 5' TGATCACCCTAGTTGTCTGG; 3'-eat-2: 5' AGTGTAGAGGTACTGTATGG; 5'-dbl-1: 5' CATGGACAAACATCGGGGA; and 3'-dbl-1: 5' CGTGTACACAAATCTGTTCG. che-2(e1033);dbl-1(nk3) was generated by crossing heterozygous dbl-1(nk3) males with che-2(e1033) hermaphrodites. Then, their F1 progeny was PCR-screened for the nk3 allele, and double mutants in F2 were confirmed by PCR for both nk3 and e1033 alleles, and by DNA sequencing with oligonucleotides 5'- dbl-1, 3'- dbl-1, 5'-che-2: 5' AGATGGATGTTTACTGCC, and 3'-che-2: 5' GAGAATGACACAATGTGG.
All strains and experiments were maintained at 20°C.
Dietary restriction
We developed a novel method of dietary restriction (DR) on solid media. Three different food treatments are described within this study: excess, high, and low food treatment plates. Excess food plates: 100 μl of 5.19 × 108/ml E. coli (OP50)-Luria broth was spread around the centre of 5.5 cm NGM plates and left at room temperature for 24 hours before being killed by exposure to UV light for 1 hour. High food plates were prepared as excess food plates, but were exposed to UV light for 1 hour immediately after preparation. Low food plates were prepared as high food plates, but using a suspension of 3.95 × 107/ml E. coli (OP50)-Luria broth. For each experiment, the same E. coli culture was used for each food treatment. Treatment plates were replaced every 24 h during worm growth experiments to prevent depletion food source.
Body size analysis
Growth curves were determined for each strain, from worms grown individually on 5 cm Petri dishes. At 24 h intervals from 36 h to 120 h post hatching, images were captured using a video camera (JVC KY-F50) attached to a dissecting microscope (×50), and analyzed with OBJECT-IMAGE 1.62. Length and area were measured from pictures of individual worms and calibrated from a 1 mm graticule. Volume was calculated assuming cylindrical body shape using the formula (pi*length*(area/length)2/4) [36,46]. All comparisons of body size use Log-transformed data.
Hypodermal ploidy analysis
Upon completing growth (120 h), worms were fixed in Carnoy's solution for 24 h, stained in a 0.007 mg/ml solution of 4',6-diamidino-2-phenylindole dihydrochloride (DAPI) [36,48,55,56] and viewed under a Leitz epifluorescence microscope. Images of hypodermal and ventral cord nuclei were collected using a CV-M300 video camera, and analyzed using OBJECT-IMAGE 1.62. C values of hypodermal nuclei were estimated by dividing their DAPI-based densitometric quantifications by an average of those values from ventral cord nuclei (divided by two) in the same microscopic preparations [36].
Cell number analysis
Young adult worms were anesthetized with 0.1 M sodium azide [57], and viewed at ×1000 under differential interference contrast optics with a Nikon Eclipse E600 microscope. All nuclei, excluding neuronal and seam cells, between the posterior pharyngeal bulb and anus were counted. Images were captured with a CV-M300 camera and reconstructed by using Adobe PHOTOSHOP 4.0.
Longevity analysis
We analysed Kaplan-Meier survival distributions, which are based on a discrete stepped survival curve, adding time specific data as each death occurs. Individuals that died from internal hatching of eggs (bagging), or crawled off the plate were censored. Censoring allows the inclusion of individuals that were lost to the study, and thus contribute towards knowledge of survivorship, but nothing to the knowledge of age at death. Log-rank tests were performed to determine if survival curves were significantly different from each other.
Egg-laying assays
Individual worms were placed OP50-seeded 5.5 cm NGM plates before adult moult occurred and transferred to a fresh plate every 24 h. Total fecundity was measured with only fertilized eggs and larvae being included in the count.
Statistical analyses
Data analysis was undertaken using JMP 3.2 (SAS Institute, Cary NC, USA). Body size and ploidy data were compared across food level and genotype using a standard two-way ANOVA, including a genotype by environment interaction term, to determine responses of each genotype to DR. A food level by genotype interaction term allowed the comparison of each mutant genotype's response to DR to that of wild type. Ratios, between high and low food groups, were not used in this analysis.
Authors' contributions
LST designed the study, created the dietary restriction methodology, performed body size, ploidy, cell number, longevity, egg laying and statistical analysis. EL performed body size and ploidy analysis. AGS did ploidy analysis. AML conceived the study and participated in its design. The paper was written by LST and AGS, with the help of AML and EL.
Acknowledgments
Acknowledgements
We thank the Caenorhabditis Genetics Center for providing strains, and Almudena de Vivero for helping us to measure some of the mutants and their levels of endoreduplication. Our support came primarily from NERC (UK) and BBSRC. LST was also supported by Parkinson's Diease Society (PDS). EL and AGS were also supported by programs I3P and Ramón y Cajal, respectively, and by Plan Nacional I+D (CGL2005-00307; all from the Ministry of Education and Science, Spain).
Contributor Information
Luke S Tain, Email: luke.tain@sheffield.ac.uk.
Encarnación Lozano, Email: elozano@mncn.csic.es.
Alberto G Sáez, Email: asaez@mncn.csic.es.
Armand M Leroi, Email: a.leroi@imperial.ac.uk.
References
- Pigliucci M. Phenotypic plasticity Beyond nature and nurture. Baltimore, The John Hopkins University Press; 2001. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. New York, Oxford University; 2003. [Google Scholar]
- McCay CM, Crowel MF, Maynard LA. The effect of growth upon the length of the life span and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Robertson FW. Studies in quantitative inheritance. XII. Cell size and number in relation to genetic and environmental variation of body size in Drosophila. Genetics. 1959;44:869–896. doi: 10.1093/genetics/44.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Caloric restriction. Aging (Milano) 1998;10:173–174. [PubMed] [Google Scholar]
- Tu MP, Tatar M. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell. 2003;2:327–333. doi: 10.1046/j.1474-9728.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford MD. The retardation of aging and disease by dietary restriction. Springfield, IL, Charles C Thomas Publisher; 1988. [Google Scholar]
- Chippindale AK, Leroi AM, Kim SB, Rose MR. Phenotypic Plasticity and Selection in Drosophila Life-History Evolution.1. Nutrition and the Cost of Reproduction. J Evol Biol. 1993;6:171–193. doi: 10.1046/j.1420-9101.1993.6020171.x. [DOI] [Google Scholar]
- Townshend TJ, Wootton RJ. Effects of food supply on the reproduction of the convict cichlid, Cichlasoma nigrofasciatum. J Fish Biol. 1985;24:91–104. doi: 10.1111/j.1095-8649.1984.tb04779.x. [DOI] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003:947–54. doi: 10.1016/S0531-5565(03)00161-X. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/S0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Ben Lagha N, Menuelle P, Seurin D, Binoux M, Lebouc Y, Berdal A. Bone formation in the context of growth retardation induced by hIGFBP-1 overexpression in transgenic mice. Connect Tissue Res. 2002;43:515–9. doi: 10.1080/713713462. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Nogami H, Ikeda T. Growth hormone and nutrition interact to regulate expressions of kidney IGF-I and IGFBP mRNAs. Kidney Int. 1995;48:65–71. doi: 10.1038/ki.1995.268. [DOI] [PubMed] [Google Scholar]
- Van Wyk JJ, Smith P. Insulin-like growth factors and skeletal growth: possibilities for therapeutic interventions. J Clin Endocrinol Metab. 1999;84:4349–4354. doi: 10.1210/jc.84.12.4349. [DOI] [PubMed] [Google Scholar]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N. The translational initiator 4E-BP is an effector if PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Genetics of mouse growth. Int J Dev Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- Oldham S, Bohni R, Stocker H, Brogiolo W, Hafen E. Genetic control of size in Drosophila. Philos Trans R Soc Lond B Biol Sci. 2000;355:945–952. doi: 10.1098/rstb.2000.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Hafen E. Genetic control of cell size. Curr Opin Genet Dev. 2000;10:529–535. doi: 10.1016/S0959-437X(00)00123-4. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Chomiki N, Kachinskas D, Warden CH. Cloning and developmental regulation of a novel member of the insulin-like gene family in Caenorhabditis elegans. Biochem Biophys Res Commun. 1998;249:385–390. doi: 10.1006/bbrc.1998.9164. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Kawano T, Ito Y, Ishiguro M, Takuwa K, Nakajima T, Kimura Y. Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;273:431–436. doi: 10.1006/bbrc.2000.2971. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Ogg SS, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;369:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- McCulloch D, Gems D. Body size, insulin/IGF signaling and aging in the nematode Caenorhabditis elegans. Exp Gerontol. 2003;38:129–136. doi: 10.1016/S0531-5565(02)00147-X. [DOI] [PubMed] [Google Scholar]
- Patterson GI, Padgett RW. TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/S0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- Maduzia LL, Gumienny TL, Zimmerman CM, Wang H, Shetgiri P, Krishna S, Roberts AF, Padgett RW. lon-1 regulates Caenorhabditis elegans body size downstream of the dbl-1 TGF beta signaling pathway. Dev Biol. 2002;246:418–428. doi: 10.1006/dbio.2002.0662. [DOI] [PubMed] [Google Scholar]
- Morita K, Flemming AJ, Sugihara Y, Mochii M, Suzuki Y, Yoshida S, Wood WB, Kohara Y, Leroi AM, Ueno N. A Caenorhabditis elegans TGF-beta, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. Embo J. 2002;21:1063–1073. doi: 10.1093/emboj/21.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/S0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Hirose T, Nakano Y, Nagamatsu Y, Misumi T, Ohta H, Ohshima Y. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C. elegans. Development. 2003;130:1089–1099. doi: 10.1242/dev.00330. [DOI] [PubMed] [Google Scholar]
- Kao G, Nordenson C, Still M, Ronnlund A, Tuck S, Naredi P. ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell. 2007;128:577–587. doi: 10.1016/j.cell.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. Cold Spring Harbor Laboratory; 1997. [PubMed] [Google Scholar]
- Alpatov WW. Phenotypical variation in body and cell size of Drosophila melanogaster. Biol Bull. 1930;58:85–103. doi: 10.2307/1537121. [DOI] [Google Scholar]
- Wang J, Tokarz R, Savage-Dunn C. The expression of TGF-β signal transducers in the hypodermis regulates body size in C. elegans. Development. 2002;129:4989–4998. doi: 10.1242/dev.129.21.4989. [DOI] [PubMed] [Google Scholar]
- Lozano E, Sáez AG, Flemming A, Cunha A, Leroi AM. Regulation of growth by ploidy in C. elegans. Curr Biol. 2006;16:493–498. doi: 10.1016/j.cub.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. No reduction of metabolic rate in food restricted Caenorhabditis elegans. Exp Gerontol. 2002;37:1359–1369. doi: 10.1016/S0531-5565(02)00172-9. [DOI] [PubMed] [Google Scholar]
- Mörck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Ware RW, Crossland K, Russell RL, Clark DV. The nerve ring of the nematode C elegans: Sensory input and motor output. J Comp Neurol. 1975;162:71–110. doi: 10.1002/cne.901620106. [DOI] [Google Scholar]
- Lewis JA, Hodgkin JA. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J Comp Neurol. 1977;172:489–510. doi: 10.1002/cne.901720306. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans". Dev Biol. 1986:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/S0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Daniels SA, Ailion M, Thomas JH, Sengupta P. egl-4 acts through a transforming growth factor-beta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- Morita K, Chow KL, Ueno N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development. 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun CY, Baird SE, Padgett RW. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming AJ, Shen ZZ, Cunha A, Emmons SW, Leroi AM. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc Natl Acad Sci USA. 2000;97:5285–5290. doi: 10.1073/pnas.97.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FW. The ecological genetics of growth in Drosophila. 6. The genetic correlation between the duration of the larval period and body size in relation to larval diet. Genet Res. 1963;4:74–92. [Google Scholar]
- Beaton MJ, Hebert PDN. Shifts in postembryonic somatic ploidy levels in Daphnia pulex. Hydrobiologia. 1999;394:29–39. doi: 10.1023/A:1003567012950. [DOI] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Wagner GP. The developmental genetics of homology. Nat Rev Gen. 2007;8:473–479. doi: 10.1038/nrg2099. [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Dawson-Scully K, Sokolowski MB. The neurogenetics and evolution of food-related behaviour. Trends Neurosci. 2005;28:644–52. doi: 10.1016/j.tins.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Raizen DM, Cullison KM, Pack AI, Sundaram MV. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics. 2006;173:177–187. doi: 10.1534/genetics.106.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. The Nematode Caenorhabditis elegans. New York: Cold Spring Harbor Laboratory Press; 1988. Methods; pp. 587–606. [Google Scholar]
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horwitz HR. Post-embryonic lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]