Abstract
Krp1, also called sarcosin, is a cardiac and skeletal muscle kelch-repeat protein hypothesized to promote the assembly of myofibrils, the contractile organelles of striated muscles, through interaction with N-RAP and actin. To elucidate its role, endogenous Krp1 was studied in primary embryonic mouse cardiomyocytes. While immunofluorescence showed punctate Krp1 distribution throughout the cell, detergent extraction revealed a significant pool of Krp1 associated with cytoskeletal elements. Reduction of Krp1 expression with siRNA resulted in specific inhibition of myofibril accumulation with no effect on cell spreading. Immunostaining analysis and electron microscopy revealed that cardiomyocytes lacking Krp1 contained sarcomeric proteins with longitudinal periodicities similar to mature myofibrils, but fibrils remained thin and separated. These thin myofibrils were degraded by a scission mechanism distinct from the myofibril disassembly pathway observed during cell division in the developing heart. The data are consistent with a model in which Krp1 promotes lateral fusion of adjacent thin fibrils into mature, wide myofibrils and contribute insight into mechanisms of myofibrillogenesis and disassembly.
Keywords: kelch, heart, myofibrillogenesis, α-actinin, actin, myosin
Introduction
Krp1, encoded by the murine Kbtbd10 gene, is a member of the BTB-kelch family of proteins, which are classified by the presence of an N-terminal BTB (Broad-Complex, Tramtrack, and Bric a brac) domain and a C-terminal kelch domain consisting of four to seven copies of the kelch motif. The BTB region mediates protein-protein interactions whereas the kelch repeats form a beta-propeller tertiary structure that is a putative binding site for actin and other proteins [1]. Most members of the BTB-kelch family contain an intervening region that includes a third type of protein-binding motif, the BACK domain [2].
Over 50 BTB-kelch proteins have been identified in humans thus far [3]. Although relatively few have been functionally characterized, it is apparent that BTB-kelch proteins are involved in diverse cellular processes and that alteration of their expression is associated with various disease states. A subset of BTB-kelch proteins, including Keap1 and Klhl12, are substrate adaptors for Cullin3 ubiquitin ligases [4-6]. Other BTB-kelch proteins are implicated in cytoskeletal organization. These include Mayven, a brain-specific protein that binds actin and contributes to oligodendrocyte process extensions [7]; Klhdc2, a ubiquitously expressed protein that may be involved with cytoskeletal remodeling in myoblasts [8]; and Nd1-L, which is found in all tissues and appears to stabilize actin filaments [9].
Krp1, also called sarcosin, is primarily expressed in skeletal and cardiac muscle [10, 11], but is also upregulated in transformed rat fibroblasts [10]. Krp1 overexpression in transformed fibroblasts results in co-localization of the protein with actin at the tips of pseudopodia and elongation of the processes [10], while reduction of Krp1 expression inhibits formation of pseudopodial extensions [12]. Krp1’s effect on cytoskeletal organization and pseudopod elongation is mediated by binding to LASP-1 (LIM and SH3 protein-1) [12], a nebulin repeat protein with an N-terminal LIM domain that is highly homologous to that found in N-RAP [13].
The function of Krp1 in cardiac and skeletal muscle has not been reported, but binding and localization studies lead us to hypothesize that Krp1 plays a role in the assembly of myofibrils, the contractile organelles of striated muscles [14]. The structure of the myofibril is well characterized, but the process of its assembly, termed myofibrillogenesis, is less well understood. Several models of myofibrillogenesis propose a stepwise process for the arrangement of actin, myosin, and titin-associated filaments into mature contractile units [15, 16]. These models emphasize actin-based stress fibers and nonmyofibrillar cytoskeletal elements as templates for sarcomeric organization [17], short premyofibril structures containing α-actinin, actin, and nonmuscle myosin that elongate and fuse laterally as muscle myosin is incorporated [18], and the role of the giant protein titin in integrating and organizing the different elements of the sarcomere during assembly [19-21].
More recent studies have emphasized the role of proteins that transiently associate with developing myofibrils to promote key steps in assembly. These include N-RAP, a LIM domain and nebulin repeat protein found in the premyofibril regions of cardiomyocytes and at the Z-lines and M-lines of newly formed myofibrils [14, 22, 23], and Krp1, which binds N-RAP and is found between laterally fusing myofibrils in spreading chick cardiomyocytes [14]. During myofibril assembly, N-RAP organizes actin and α-actinin into I-Z-I structures (symmetrical actin filaments with their barbed ends anchored at a central Z-body or Z-line containing α-actinin) [24, 25], while Krp1 has been hypothesized to promote lateral fusion of premyofibril structures [14].
Primary cultures of embryonic mouse cardiomyocytes are an excellent model for studying myofibril assembly and maintenance. These cells contain mature myofibrils as well as nascent structures that are easily identified by immunostaining with antibodies against sarcomeric α-actinin. The attached cardiomyocytes spread and accumulate additional myofibrils over several days, thus allowing for analysis of the dynamic progression of myofibril assembly [26]. Here we assess the localization of Krp1 in primary cultures of mouse embryonic cardiomyocytes and test the hypothesis that Krp1 is necessary for myofibril assembly by using RNA interference to specifically reduce endogenous Krp1 expression. The present study demonstrates that Krp1 plays a specific role late in assembly, promoting the alignment and fusion of thin myofibrils to form mature, wide structures typical of fully developed striated muscle. Furthermore, blocking the lateral alignment and fusion of premyofibril structures leads to accumulation of very thin myofibrils followed by disassembly through a scission mechanism that is distinct from the myofibril disassembly pathway observed during cell division in the developing heart.
Materials and Methods
Primary Culture and Transfection of Embryonic Mouse Cardiomyocytes
All procedures involving animals were conducted in accordance with an animal study protocol approved by the NIAMS Animal Care and Use Committee. Mouse embryonic cardiomyocytes were cultured as described previously with minor modifications [22, 26]. Briefly, embryonic day 17.5 hearts were isolated and cultured by successive digestion with 0.25% trypsin (Gibco) and 0.2% collagenase type II (Sigma). Fibroblast contamination was minimized by preplating cells onto tissue culture dishes (Falcon) for 90 minutes. The cardiomyocyte-enriched cultures were plated onto laminin-coated (Invitrogen) 6-well dishes (Falcon) and 2-chambered wells on glass coverslips (Nunc Nalgene) at a density of 2 × 104 cells per cm2. Growth medium consisted of 25 mM glucose DMEM lacking L-glutamine (Gibco) supplemented with 10% fetal bovine serum (Hyclone) and 1% gentamycin, 1% penicillin/streptomycin, and 1% antimycotic (all from Gibco).
Chemically synthesized double stranded siRNAs targeting Krp1 (Krp1 siRNA 1 target 5′-CCCACTGAAGTCAATGACATA-3′ or Krp1 siRNA 2 target 5′-CAGGCATGGAATGTTTGTTAA-3′) or a control siRNA without homology to any known mouse gene sequence (control target 5′-AATTCTCCGAACGTGTCACGT-3′) were purchased from Qiagen and transfected into cardiomyocytes at a final concentration of 20 nM using Hiperfect reagent (Qiagen) according to the manufacturer’s instructions. Transfection efficiency was ∼90% as determined from confocal images of cells treated with fluorescently labeled control siRNA [26]. Mock-transfected cells were treated identically except for the omission of siRNA.
RNA Isolation and Real-Time PCR
Total RNA was prepared from transfected cells using the RNEasy Mini kit (Qiagen). Genomic DNA contamination was reduced by on-column DNase digestion (Qiagen) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 500 ng total RNA using random hexamer primers and the Advantage RT-for-PCR kit (Clontech). cDNA corresponding to 10 ng starting RNA was then used for quantitative real-time PCR analysis (qPCR) using the Brilliant SYBR Green QPCR Master Mix and an MX3000p Real-Time PCR machine (Stratagene). Primers were designed to amplify mouse Krp1 (forward, 5′-CTACAACCCCAAGAAAGGAGACTG-3′ and reverse, 5′-CCGTCATCACTTCCCACTTATTG-3′ at 300 nM to amplify a 179 bp product) and mouse Keap1 (forward, 5′-CAAGCAGGAGGAGTTCTTCAACC-3′ and reverse, 5′-AAGTAACCGCCCGCTGTGTAGATG-3′ at 400 nM to amplify a 360 bp product). Primers to amplify N-RAP, sarcomeric α-actinin and 18S rRNA were previously described [26].
Replicate samples were amplified as follows: One cycle at 95 °C for 10 minutes; 40 cycles at 95 °C for 5 seconds, 60 °C for 1 minute, then a fluorescence reading followed by 72 °C for 30 seconds. Specificity of product amplification was determined by melting curve analysis and by verifying product sizes by gel electrophoresis. Amplification efficiency was determined for each primer pair and ranged from 94 to 108%. Data were analyzed using the 2-ΔΔCT method [27], with transcript levels first normalized to 18S rRNA levels in replicate wells (ΔCT) and then to mock-transfected cells from the same cultures (ΔΔCT).
Protein Isolation and Immunoblotting
Cell lysate preparation and immunoblotting were carried out as previously described [26]. A polyclonal antibody raised against the kelch repeats of Krp1 was a generous gift from Drs. Bradford Ozanne and Heather Spence (Beatson Institute for Cancer Research, Glasgow, UK) and was used at a 1:1000 dilution. PVDF strips were also probed with monoclonal antibodies against muscle myosin heavy chain (1:3000, clone MF20, Developmental Studies Hybridoma Bank at the University of Iowa), sarcomeric α-actin (1:1500, clone 5C5, Sigma), GAPDH (1:5000, clone 6C5, Abcam), or cleaved caspase-3 (1:500, clone 5A1, Cell Signaling Technology). Densitometric analysis was performed using Image J software (developed at the US National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). Protein levels were normalized to GAPDH in the same samples, and then expressed as a percentage of the level in mock-transfected controls prepared simultaneously.
For separation of cytosolic and insoluble fractions, cultured cardiomyocytes were scraped in PBS, then incubated on ice with 0.1% Nonidet P-40 in PBS for 10 minutes. An aliquot of total cell lysate was removed and briefly centrifuged to separate the extracted cytosolic supernatant from the unextracted insoluble pellet. Protein samples were analyzed by SDS-PAGE and immunoblotting, and relative protein amounts were quantitated by densitometric analysis.
Immunostaining and Confocal Microscopy
At the indicated time points, cells were fixed for 15 minutes with 4% formaldehyde (Ted Pella) and permeabilized for 2 minutes with 0.1% Nonidet-P-40 (Calbiochem). Following blocking with 5% horse serum for 30 minutes at 37 °C, samples were stained with primary antibodies against Krp1 (1:1250, as above), sarcomeric α-actinin (1:2000, clone EA-53, Sigma), cleaved caspase-3 (1:500, as above), muscle myosin heavy chain (1:40, clone F59, Developmental Studies Hybridoma Bank at the University of Iowa), myomesin (1:100, clone B4, Developmental Studies Hybridoma Bank), or caveolin-3 (1:1000, Clone 26, BD Transduction Laboratories). Secondary antibodies used were AlexaFluor 488 labeled goat anti-rabbit (1:1000, Invitrogen), AlexaFluor 555 labeled goat anti-rabbit (1:1000, Invitrogen), and TRITC conjugated rabbit anti-mouse IgG (1:1500, Sigma). Incubations were at 37 °C for 1 hour and samples were washed with PBS between incubations. For some samples, AlexaFluor 488-conjugated phalloidin was used to stain F-actin (1:800 for 20 min at room temperature, Invitrogen). Nuclei were stained with DAPI at a dilution of 1:8000 for 10 minutes at room temperature.
Cells were visualized with a Zeiss LSM 510 Meta confocal microscope using a Plan-Apochromat 63X / 1.4 Oil objective. Digital images were obtained using Zeiss LSM software. Morphometric analysis was performed using Image J software as previously described [24, 25].
Electron Microscopy
Cells were cultured in 60 mm Permanox dishes (Nunc Nalgene) and processed for transmission electron microscopy as previously described [28], with minor modifications as reported [29]. In brief, cultures were fixed at three or seven days post-transfection with glutaraldehyde and tannic acid (Mallinckrodt), post-fixed with osmium tetroxide, stained en bloc with uranyl acetate, ethanol dehydrated and Epon embedded. Chemicals were from Electron Microscopy Sciences, except as noted. Sections 70-90 nm thick were cut parallel to the adherent surface, stained with uranyl acetate and lead citrate, and viewed with a JEM 1200 EXII electron microscope (JEOL USA).
Results
Krp1 Localization in Primary Mouse Embryonic Cardiomyocytes
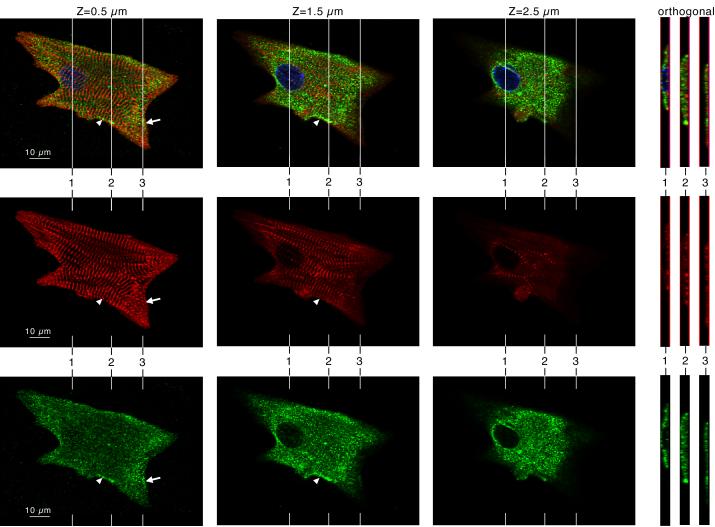
Krp1 localization in primary cultures of mouse embryonic cardiomyocytes was determined by immunostaining and confocal microscopy. Optical slices of a typical cardiomyocyte double stained with antibodies against Krp1 and sarcomeric α-actinin are shown in figure 1 (left 3 columns), and LSM software was used to generate orthogonal views of the Z stack (right panels). Sarcomeric α-actinin staining (middle row) shows that cultured cardiomyocytes contain organized striations corresponding to mature myofibrils, narrow myofibrils that appear to be fusing with mature structures (arrowhead), and closely spaced α-actinin dots (Z-bodies) that characterize premyofibrils (arrow). Krp1 is present as punctate staining throughout the cell, with most located above the plane of the myofibrils as shown in the software-generated orthogonal views (figure 1, right hand panels). Therefore, most of the Krp1 staining is not associated with α-actinin. However, punctate patches of Krp1 are also observed in the plane of the myofibrils, concentrated in premyofibril areas (arrow) and alongside narrow myofibrils fusing into more mature structures (arrowhead). In these areas, Krp1 staining is adjacent to developing myofibrillar structures, but is not colocalized with α-actinin.
Figure 1.
Krp1 (green, lower row) and α-actinin (red, middle row) localization in a single cultured mouse cardiomyocyte by confocal microscopy. DAPI labeling of the nucleus is also shown in the blue channel of the composite images (top row). Confocal imaging at planes 0.5, 1.5, and 2.5 μm above the adherent ventral surface of the cardiomyocyte are shown, as indicated. Orthogonal views through the cell are shown at the right, and their positions in the x-y plane are indicated in the main micrographs. Most myofibrils lie close to the ventral surface of the cell. Most of the Krp1 lies above the plane of the myofibrils, forming punctate dots in the cytoplasm. Punctate patches of Krp1 are also observed in the plane of the myofibrils, concentrated in premyofibril areas (arrow) as well as alongside narrow myofibrils fusing into more mature structures (arrowhead).
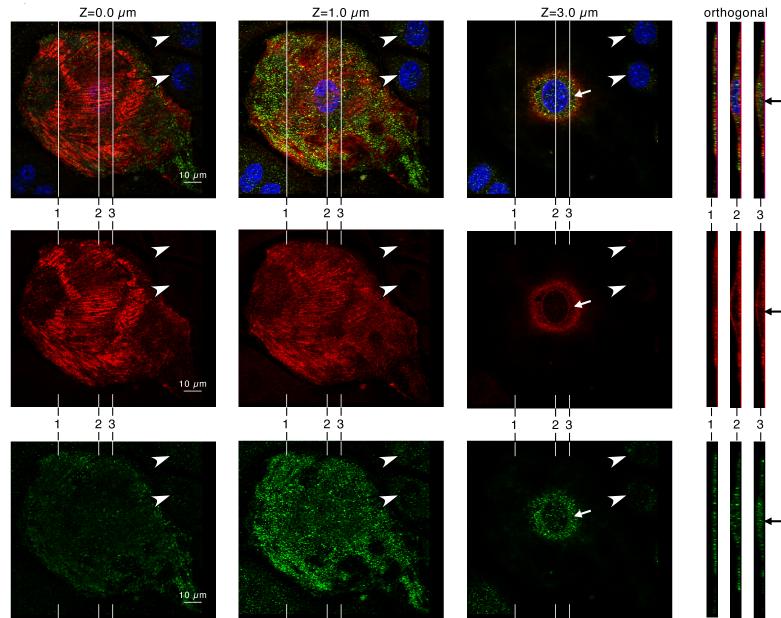
To better assess Krp1 localization with respect to the cell boundaries, cardiomyocytes were double stained with antibodies against Krp1 and caveolin-3 (figure 2). Caveolin-3 is an integral membrane protein linked with the dystrophin-associated membrane cytoskeleton [30], and its staining marks the ventral and dorsal surfaces of the cell, demonstrating that the spread cardiomyocytes are flattened, with a thicker area around the nucleus (figure 2). Punctate patches of Krp1 are clearly localized within the cytoplasm (arrows), between the cell boundaries defined by caveolin-3. Immunostaining is specific, as only low levels of background staining are seen in neighboring fibroblasts (figure 2, arrowheads).
Figure 2.
Krp1 (green, lower row) and caveolin-3 (red, middle row) localization in a single cultured mouse cardiomyocyte by confocal microscopy. DAPI labeling of the nucleus is also shown in the blue channel of the composite images (top row). Confocal imaging at planes 0.0, 1.0, and 3.0 μm above the adherent ventral surface of the cardiomyocyte are shown, as indicated. Orthogonal views through the cell are shown at the right, and their positions in the x-y plane are indicated in the adjacent micrographs. The caveolin-3 staining marks the ventral and dorsal surfaces of the cell. Punctate patches of Krp1 are clearly localized within the cytoplasm (arrows), in between the cell boundaries marked by caveolin-3.
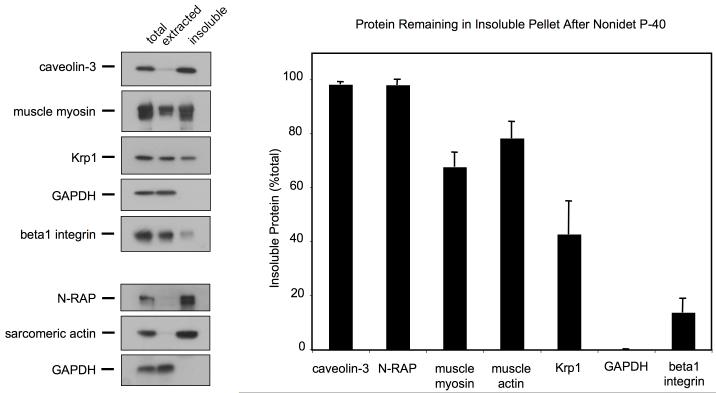
Krp1 association with cellular compartments was further explored by separation of extracted and insoluble fractions following treatment with Nonidet P-40. Cytosolic components were completely extracted, as indicated by the presence of all of the GAPDH in the extracted cytosolic fraction (figure 3). Some integral membrane proteins, such as ß1-integrin, were almost completely extracted, while others, such as caveolin-3, remained insoluble. The insoluble pellet fraction was enriched for cytoskeletal elements such as myosin, actin, and N-RAP. A large proportion of Krp1 (42%) remained in this insoluble pellet, suggesting that a sizable pool of Krp1 is strongly associated with the cytoskeleton.
Figure 3.
Immunoblot analysis of Nonidet P-40 extracted and insoluble fractions. Equal volumes of total cell lysate, extracted and insoluble fractions were analyzed by immunoblot (left) and densitometric analysis (right). Representative immunoblots are shown. The graph displays the means and SEM of three independent experiments. A large proportion of Krp1 remains in the insoluble pellet along with most of the cytoskeletal components (myosin, actin, N-RAP) and caveolin-3, although the majority of Krp1 is present in the extracted supernatant along with the cytosolic marker GAPDH and the integral membrane marker ß1-integrin.
We were unable to determine by immunofluorescence whether unextracted Krp1 is co-localized with actin since the cultured cells detached from the substrate during detergent extraction. A more gentle detergent treatment that preserves attachment failed to completely extract the cytosol as determined by GAPDH immunostaining (data not shown).
Transfection with Krp1 siRNA Specifically Reduces Krp1 Expression and Myofibril Accumulation
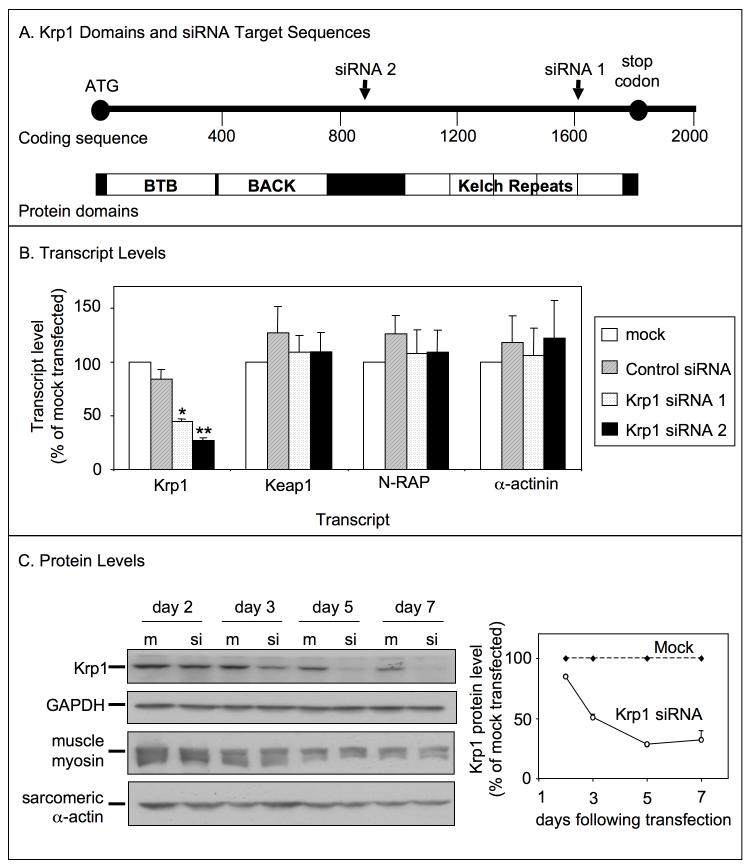
An RNA interference approach was used to gain insight into Krp1 function. Figure 4A shows a schematic diagram of the Krp1 mRNA, domain organization, and siRNA target areas. Cardiomyocytes were transfected with one of the Krp1 siRNAs 24 hours after plating, while replicate wells were mock-transfected or transfected with control siRNA. After 48 hours, total RNA was prepared and used for cDNA synthesis and quantitative RT-PCR analysis. Both of the Krp1 siRNAs significantly reduced Krp1 transcript levels (siRNA1: 55 ± 2% and siRNA2: 73 ± 2% decrease compared to mock transfected), while transfection with control siRNA had no effect (figure 4B, Krp1). The reduction of Krp1 expression is specific, as transcript levels of another kelch repeat protein, Keap1, and levels of muscle specific proteins N-RAP and α-actinin, were unchanged compared to mock-transfected controls (figure 4B). Unless otherwise noted, data reported in the following sections are from experiments using Krp1 siRNA 2, and the term “Krp1 siRNA” is used for simplicity; similar experimental results were obtained using Krp1 siRNA 1 (data not shown).
Figure 4.
Transfection with siRNA against Krp1 sequences specifically reduces Krp1 expression. (A) Krp1 protein is comprised of an N-terminal BTB-BACK domain and a C-terminal kelch repeat region. In the nucleotide sequence, Krp1 siRNA 1 targets an area within the kelch domain, while Krp1 siRNA 2 targets a sequence between the BTB-BACK and the kelch domains. (B) Transcript levels were analyzed by real time PCR 48 hours after transfection with control or Krp1 siRNA and normalized to levels in mock-transfected controls. Krp1 transcript levels were significantly decreased within 48 hours of transfection with either Krp1 siRNA 1 or 2, while control siRNA had no effect. Reduction of Krp1 expression was specific, as transcript levels of other genes were unchanged. p-values compared to control siRNA-transfected samples: *p<0.05; **p<0.01. Data are the means and SEM of three independent experiments. (C) Immunoblot analysis of specific proteins at the indicated times after mock (m) or Krp1 siRNA (si) transfection. Krp1 protein levels are significantly reduced by day three after transfection, whereas muscle myosin heavy chain and sarcomeric actin levels are unchanged compared to mock-transfected controls. Densitometric analysis of Krp1 protein levels relative to mock-transfected controls is shown on the right; data are the means and SEM from three independent experiments.
To determine siRNA effects on protein levels, lysates were prepared at various times following transfection. Proteins were detected by SDS-PAGE and immunoblotting, and levels were quantitated by densitometric analysis. Three days following transfection, Krp1 protein levels were reduced 49 ± 3% compared to the mock-transfected control (figure 4C). Krp1 protein continued to decrease through day 5 and remained low over the period of study (reduced 68 ± 8% compared to mock at day 7). Again, the reduction of Krp1 expression is specific, as muscle myosin and sarcomeric actin levels were unchanged compared to mock transfected controls (figure 4C). At later time points, slight decreases in Krp1, myosin, and actin levels were observed in both mock and siRNA-transfected samples, likely due to cardiac fibroblast proliferation in the primary cultures.
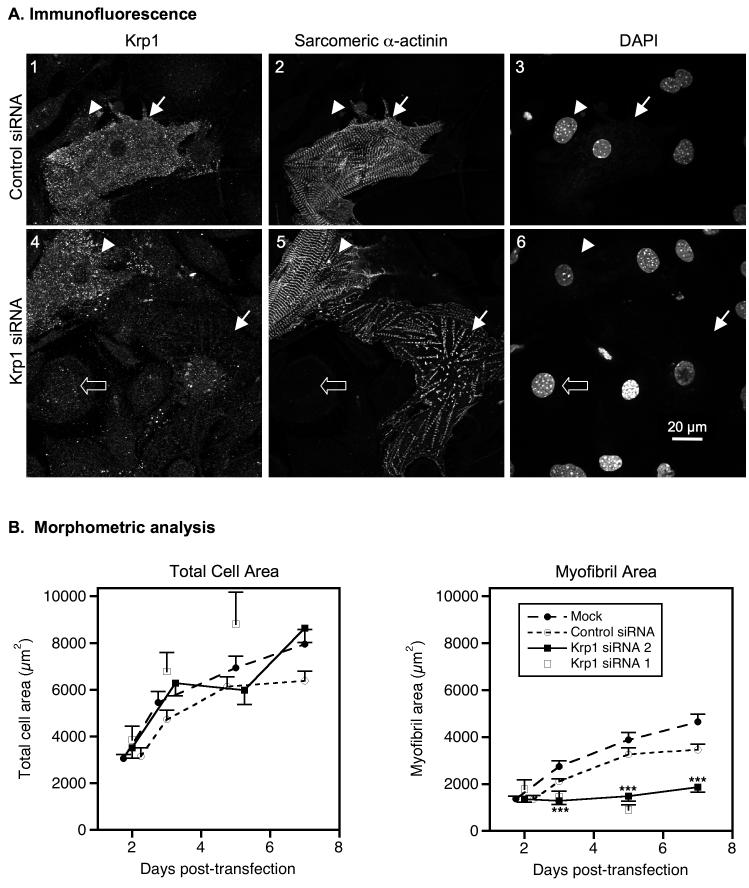
To assess the effects of Krp1 knockdown on myofibrillogenesis, cardiomyocytes were fixed five days after transfection and analyzed by confocal microscopy following immunostaining for Krp1 and sarcomeric α-actinin. Similar to untransfected cardiomyocytes (figure 1), control siRNA-transfected cardiomyocytes displayed bright punctate Krp1 staining throughout the cytoplasm (figure 5A, panel 1). These cells are frequently filled with mature myofibrils as indicated by α-actinin organization into wide Z-lines (figure 5A, panel 2). Fibroblasts in the culture exhibit background levels of Krp1 staining and do not contain sarcomeric α-actinin (figure 5A, panels 1-3, arrowheads). In Krp1 siRNA-transfected cardiomyocytes, Krp1 immunostaining is usually dim and similar in intensity to background staining of fibroblasts (figure 5A, panel 4, filled versus open arrows). Sarcomeric α-actinin staining is still present but rarely organized into wide Z-lines typical of mature myofibrils (figure 5A, panel 5, filled arrow). The correlation between Krp1 levels and mature myofibril content is typically observed within the population of siRNA treated cells. Cardiomyocytes with bright Krp1 staining frequently contain mature myofibrils, similar to control cells (figure 5A, panels 4 & 5, arrowheads) whereas cells with dim Krp1 staining often lack large areas of mature myofibrils (figure 5A, panels 4 & 5, filled arrows). These observations demonstrate a strong link between Krp1 levels and myofibril content.
Figure 5.
Krp1 knockdown decreases mature myofibril content. (A) Cells were fixed and stained for Krp1 and sarcomeric α-actinin 5 days after transfection with control siRNA (top row) or Krp1 siRNA (bottom row). Nuclei were counterstained with DAPI. Images are representative of cells evaluated from three independent cultures. Control cardiomyocytes stain positively for Krp1 and contain sarcomeric α-actinin organized into mature striations (panels 1-3, arrow). Decreased levels of Krp1 after siRNA treatment are associated with low levels of mature myofibrils (panels 4-6, arrow), while some cardiomyocytes retain normal Krp1 levels and exhibit normal striations (panels 4-6, arrowhead). Neighboring fibroblasts exhibit only background staining for Krp1 (panels 1-3, arrowhead; panels 4-6, open arrow). (B) Morphometric analysis of total cardiomyocyte areas (left) and mature myofibril areas (right) versus time. Each point represents the mean and SEM of 25-58 cardiomyocytes from at least 3 independent experiments, with the exception of the Krp1 siRNA 1 data where each point is the mean value of 10-23 cardiomyocytes from 1-3 independent experiments. Krp1 siRNA halts myofibril accumulation after 2 days, but cardiomyocyte spreading is unaffected. p-values compared to mock-transfected controls: ***p <0.001.
The effect of Krp1 knockdown on mature myofibril content was measured in cardiomyocytes fixed at various time points following transfection. Cells were stained with antibody against sarcomeric α-actinin and randomly chosen cardiomyocytes were imaged by confocal microscopy. Mature myofibril content was assessed by measuring areas containing α-actinin organized into wide Z-lines. At 2 days following transfection, the mean total area of mock-transfected cardiomyocytes was 3050 ± 183 μm2, increasing to 7953 ± 619 μm2 at 7 days post-transfection. Similar results were obtained from cardiomyocytes transfected with Krp1 siRNA (p-value not significant at any time point), indicating normal cell growth in cardiomyocytes with reduced Krp1 expression (figure 5B, left panel). In mock-transfected cells, mature myofibril content increased from 1373 ± 110 μm2 at 2 days after transfection to 4647 ± 330 μm2 at 7 days post-transfection. In contrast, the mature myofibril area in cells transfected with Krp1 siRNA was 1358 ± 132 μm2 at day 2, but remained low (1873 ± 225 μm2) at day 7 after transfection (figure 5B, right panel). Mature myofibril area was significantly reduced compared to both mock transfected and control siRNA treated cardiomyocytes at days 3, 5, and 7 after transfection (p-value < 0.001 for all). Thus, Krp1 is not required for normal cell spreading but is essential for accumulation of mature myofibrils in primary mouse embryonic cardiomyocytes. Furthermore, spontaneous beating was observed in both control and Krp1 siRNA treated cultures, suggesting that although accumulation of addiitonal myofibrils was halted by Krp1 knockdown, the existing myofibrils remained functional.
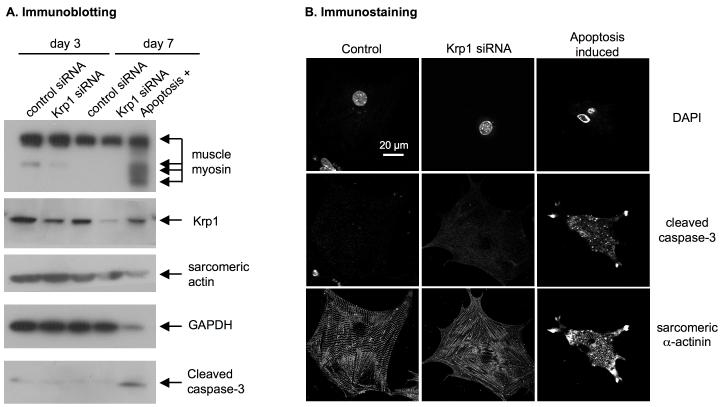
To determine if apoptosis is induced in cells lacking Krp1, cells were transfected with control or Krp1 siRNA and compared to those in replicate wells in which apoptosis was induced by incubation in serum-free, low glucose medium containing 20 mM 2-deoxy-glucose. Protein lysates were prepared and analyzed by SDS-PAGE and immunoblotting. Krp1 knockdown was verified, with reduced expression evident at three days and a further decrease observed seven days following transfection with Krp1 siRNA (figure 6A). Cleaved caspase-3, a positive marker for apoptosis [31], was easily detected in the glucose-deprived cells (figure 6A, apoptosis+ lane), but was not detected in cardiomyocytes transfected with control or Krp1 siRNA at either time point (figure 6A). Further analysis was performed on fixed cells stained for sarcomeric α-actinin and cleaved caspase-3. Nuclei were counterstained with DAPI. Apoptotic cardiomyocytes are smaller than normal, have fragmented nuclei with condensed chromatin, and cleaved caspase-3 immunostaining is bright and punctate (figure 6B, right panels). In contrast, nuclei of control and Krp1 siRNA-transfected cardiomyocytes are round and intact, and cleaved caspase-3 immunostaining is dim and diffuse (figure 6B, left and center panels).
Figure 6.
Apoptosis is not induced after Krp1 knockdown. Cardiomyocytes were transfected with the indicated siRNA; as a positive control, apoptosis was induced in untreated cells by incubating for 1 day in serum-free, low glucose medium containing 20 mM 2-deoxy-glucose. (A) Immunoblot analysis at 3 and 7 days post-transfection. Krp1 protein levels are decreased after transfection with Krp1 siRNA, but muscle myosin and actin remain at normal levels and cleaved caspase-3 is not detected. (B) Immunofluorescence analysis demonstrates that apoptotic cardiomyocytes display an irregular shrunken morphology with fragmented nuclei and bright, punctate staining for cleaved caspase-3 (right panels). Krp1 siRNA transfected cardiomyocytes display a deficit of mature myofibrils, but their nuclei are intact and caspase-3 levels remain low. Data are representative of two independent experiments.
Characterization of Krp1 Knockdown Phenotypes
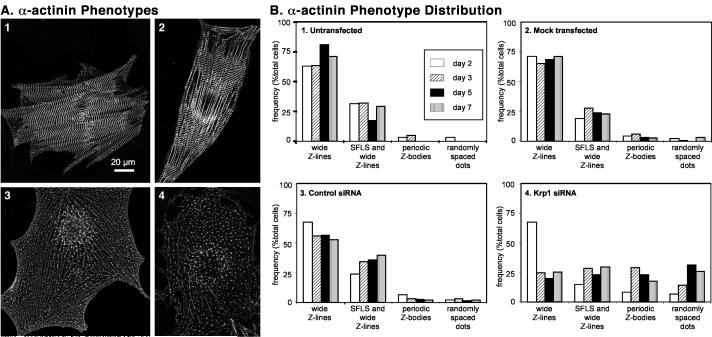
Although primary cardiomyocytes in culture are generally heterogeneous in size, shape, and myofibrillar organization, particular patterns of α-actinin organization were observed with increased frequency after Krp1 knockdown. The predominant patterns observed are illustrated in representative images of control and Krp1 siRNA-transfected cardiomyocytes that were fixed and stained with antibodies against sarcomeric α-actinin and Krp1. Patterns of sarcomeric α-actinin staining identified multiple structures in untransfected cardiomyocytes such as wide Z-lines of mature myofibrils, stress fiber-like structures (SFLS) with nearly continuous α-actinin staining, and periodically spaced Z-bodies characteristic of newly forming myofibrils (figure 7A1-A2 and figure 1). These patterns of α-actinin organization were also present in Krp1 siRNA-transfected cardiomyocytes. However, fewer cells were filled with mature myofibrils (figure 5), and a larger proportion of cells were filled with periodically spaced Z-bodies or narrow Z-lines (figure 7A3) and more randomly arranged α-actinin positive dots (figure 7A4). Lack of Krp1 immunostaining confirmed Krp1 knockdown in these cells (data not shown).
Figure 7.
Classification of cardiomyocytes according to α-actinin organization. Each cardiomyocyte was classified into one of four categories of α-actinin organization corresponding to the dominant phenotype observed by visual inspection. (A) Prototypical examples of α-actinin phenotypes in cultured cardiomyocytes. panel 1: wide Z-lines; panel 2: SFLS and wide Z-lines; panel 3: periodic Z-bodies; panel 4: randomly spaced dots. Panels 1 and 2 are untransfected cells, and cells in panels 3 and 4 were transfected with Krp1 siRNA. After Krp1 siRNA transfection, many cardiomyocytes are almost completely filled with long series of α-actinin dots resembling periodically-spaced Z-bodies or myofibrillogenesis intermediates (panel 3), or are filled with more randomly arranged dots of α-actinin (panel 4). (B) Prevalence of α-actinin phenotypes in cardiomyocytes. Cardiomyocytes were fixed at various times after transfection and analyzed by α-actinin immunostaining and confocal microscopy. Each bar represents the number of cells in the indicated category as a percentage of the total number of cardiomyocytes examined. Panel 1, Untransfected cardiomyocytes: N=120 cardiomyocytes scored from 5 independent experiments. Panel 2, Mock-transfected cardiomyocytes: N=480 cardiomyocytes scored from 10 independent experiments. Panel 3, Control siRNA-transfected cardiomyocytes: N= 355 cardiomyocytes scored from 9 independent experiments. Panel 4, Krp1 siRNA-transfected cardiomyocytes: N=548 cardiomyocytes scored from 11 independent experiments. Cardiomyocytes transfected with Krp1 siRNA were always evaluated in parallel with replicate samples of mock-transfected and/or control siRNA-transfected cells in each experiment.
To assess the prevalence of these phenotypes in control and Krp1 knockdown cells, we assigned cardiomyocytes to one of four categories depending on the dominant pattern of α-actinin organization assessed by visual inspection: Wide Z-lines filling the cell (figure 7B, wide Z-lines), patches of wide Z-lines interspersed with SFLS (figure 7B, SFLS and wide Z-lines), long series of periodic Z-bodies with very few wide Z-lines (figure 7B, periodic Z-bodies), and more randomly oriented or short series of α-actinin dots (figure 7B, randomly spaced dots). As expected, most untransfected cardiomyocytes were filled with wide Z-lines, and about a third were filled with patches of wide Z-lines interspersed with SFLS. Although periodic Z-bodies and randomly spaced α-actinin dots were frequently observed, they were typically present in small areas of the cell. Thus, few untransfected cardiomyocytes are scored as being predominantly filled with these structures (figure 7B1). Similar results were obtained for mock-transfected samples (Figure 7B2). In control siRNA-transfected cultures, the relative proportions in each category were similar to untransfected and mock-transfected cells at day 2 following transfection, while at later times the percentage of cells filled with wide Z-lines exhibited a modest decrease. This was accompanied by a small increase in the proportion of cells with SFLS and wide Z-lines (figure 7B3). The proportion of cells filled with Z-bodies and/or α-actinin dots remained low, accounting for less than 9% of cardiomyocytes at any time point (figure 7B3).
In Krp1 siRNA-transfected samples 2 days after transfection, the relative proportions of each phenotype were similar to control samples. However, the percentage of cells filled with wide Z-lines dropped sharply by day 3 after transfection (figure 7B4), in agreement with the morphometric data shown in figure 5B. Instead, a larger proportion of cells were filled with periodically spaced Z-bodies or narrow Z-lines, suggesting accumulation of myofibrillogenesis intermediates upon Krp1 knockdown. Interestingly, the proportion of cells with these structures peaked at day 3 and decreased at days 5 and 7, while the proportion of cells with more randomly spaced dots was greatest at days 5 and 7 following transfection (figure 7B4). These data suggest that the structures containing periodically spaced Z-bodies or narrow Z-lines disassembled into the structures represented by the more randomly spaced dots.
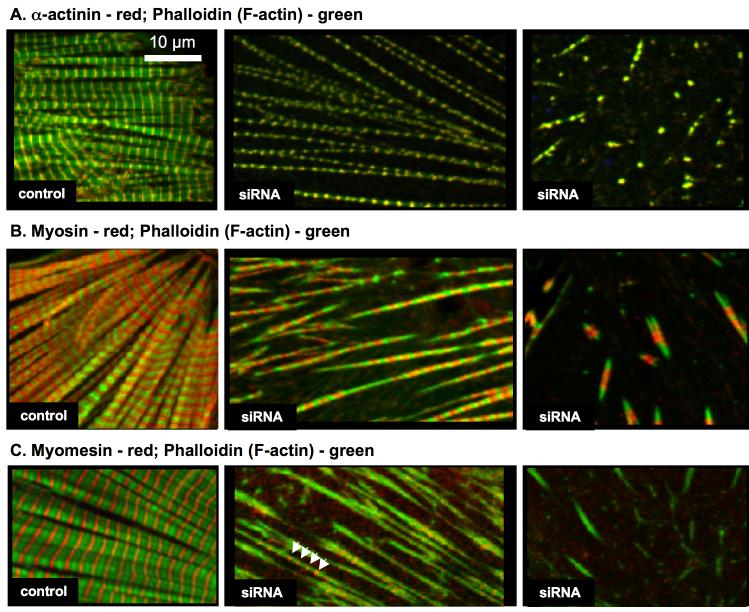
To characterize the structures that accumulate upon Krp1 knockdown, transfected cardiomyocytes were fixed and stained with antibodies against α-actinin, myomesin, or muscle myosin in combination with phalloidin for F-actin localization. In control cells, double staining revealed well-aligned myofibrils with the expected sarcomeric organizations of α-actinin at the Z-line, phalloidin marking F-actin in the I-band, myosin at the A-band, and myomesin at the central M-line (figure 8A-C, left panels). After Krp1 knockdown, double staining demonstrated that α-actinin organized in long series of periodic Z-bodies or thin Z-lines corresponded to thin, separated fibrils containing actin (figure 8A, center panel). The structures that appear as isolated or small groups of dots of α-actinin in random orientations are very short, sparse fibrils containing actin (figure 8A, right panel). Sarcomeric myosin is also associated with the long separated and the short sparse actin fibrils (figure 8B, center and right panels). Myomesin appears less organized than the other components, and is often absent from the shorter structures (figure 8C, center and right panels). The data demonstrate that the major sarcomeric components retain their normal longitudinal organization in very thin myofibrils that accumulate after Krp1 knockdown.
Figure 8.
Organization of sarcomeric components after Krp1 knockdown. Cardiomyocytes were fixed five days post-transfection and double stained with phalloidin to visualize actin filaments (green) and antibodies for either sarcomeric α-actinin (A), muscle myosin (B), or myomesin (C) (red). Examples of control cardiomyocytes (left panels) and cells transfected with Krp1 siRNA 1 (center and right panels) are shown. In control cardiomyocytes, the sarcomeric proteins are organized into mature, well-aligned myofibrils. In contrast, after Krp1 knockdown cells contain sparse, separated fibrils or very short fibrils, which still contain α-actinin, actin and myosin organized in the same banding pattern observed in mature myofibrils. Myomesin is present in the longer fibrils (arrowheads), but is often absent from the shorter fibrils (C, center and right panels, respectively).
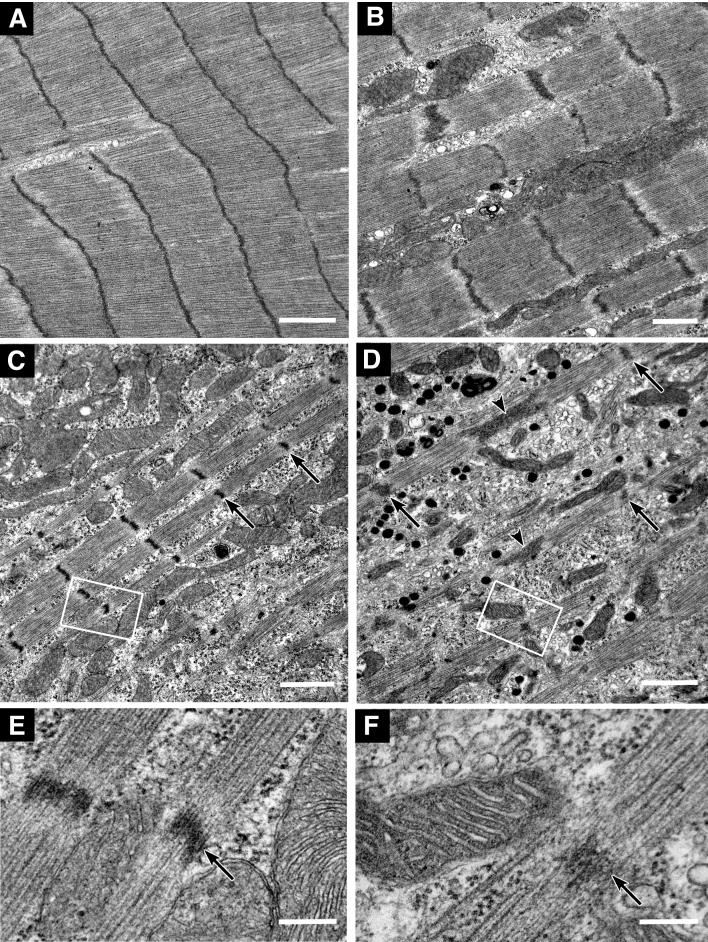
Ultrastructural characterization by transmission electron microscopy supports this conclusion. Laterally aligned, wide myofibrils are abundant in untransfected and control siRNA transfected cardiomyocytes (figure 9A-B). In cells transfected with Krp1 siRNA, thin fibrils containing Z-lines and thick myosin and thin actin filaments are commonly observed (figure 9C-D), corresponding to the double-stained images obtained by confocal microscopy. These thin myofibrils are sparse and the cytoplasm between them contains organelles, such as mitochondria and endoplasmic reticulum, which are indistinguishable from those in control cells. In addition, some cardiomyocytes contain many 100-200 nm diameter electron dense granules (figure 9D). These membrane bound organelles are numerous in approximately 10% of the cardiomyocytes examined, and their frequency was not significantly changed by Krp1 knockdown (data not shown). They resemble atrial natriuretic peptide secretory granules commonly found in the atrial cells of adult murine hearts and in both atrial and ventricular cardiomyocytes during embryonic development [32, 33].
Figure 9.
Electron micrographs showing examples of myofibrillar structure and organization in control siRNA transfected (A), untransfected (B), and Krp1 siRNA transfected (C-F) cardiomyocytes. Cells were fixed 3 days (A, C, E) or 7 days (B, D, F) after transfection. High magnification micrographs of the boxed areas in (C) and (D) are shown in (E) and (F), respectively. In Krp1 siRNA transfected myocytes, myofibrils are thinner and sparser than in controls. Wide contiguous Z-lines such as in (A) are seldom observed. However, the Z-lines (arrows), thick filaments and thin filaments are organized into normal sarcomeres, and the space between myofibrils remains filled with organelles such as mitochondria, polyribosomes and endoplasmic reticulum that appear normal in structure. Arrowheads in (D) indicate abnormally long Z-lines that further suggest immature myofibrillar structure. The myocyte shown in (D) also has numerous dense membrane-bound granules 100-200 nm in diameter, resembling atrial natriuretic peptide secretory granules. Scale bars = 1 μm (A-D) and 0.2 μm (E, F).
Discussion
Krp1 Promotes Lateral Fusion of Myofibril Assembly Intermediates
The identification of Krp1 as a striated muscle-specific protein [10, 11], its binding to the I-Z-I scaffolding protein N-RAP [14], and its concentration at sites of maturing myofibrils ([14] and this report) lead us to investigate its role in myofibril assembly. Our previous studies on cardiomyocytes isolated from chick or mouse hearts at embryonic day 17 demonstrated that preexisting myofibrils are largely disassembled when the cells are dissociated and plated, and that new myofibrils assemble as the cardiomyocytes spread in culture [22, 25, 26]. However, on subsequent days the cultured cardiomyocytes are heterogeneous with respect to cell size, shape, and myofibril organization. Therefore, conclusions regarding the effects of Krp1 knockdown on myofibril assembly can only be made based on quantitative analysis from many cells. The morphometric analysis of striated α-actinin provides a reproducible measure of Z-line assembly (figure 5B) [24-26], while assignment of cardiomyocytes according to patterns of α-actinin organization yields additional information concerning the organization of this protein at different times (figure 7). Using these quantitative assays, we examined the effects of Krp1 knockdown on accumulation of mature Z-lines and tracked the accumulation of normally low abundance assembly and disassembly intermediates in the cardiomyocyte population. The morphometry clearly demonstrated net myofibril accumulation between 2 and 7 days post transfection in control cells, and halting of net myofibril assembly when Krp1 protein levels were specifically reduced by RNA interference (compare figures 5B and 4C). At the same time as net Z-line assembly stopped, periodic Z-bodies accumulated (figure 7). Double staining experiments and electron microscopy demonstrated that these latter structures are long, thin fibrils containing actin, myosin, α-actinin, and myomesin, the major components of sarcomeric thin filaments, thick filaments, Z-lines, and M-lines, respectively, organized with longitudinal periodicities and banding patterns similar to those observed in mature myofibrils (figures 8 and 9). Despite the significant defect in myofibril accumulation, cardiomyocytes lacking Krp1 were otherwise healthy. Muscle myosin and sarcomeric actin levels were unaltered (figure 4C), normal cell spreading was observed (figure 5B), and apoptosis was not induced (figure 6). Furthermore, ultrastructural examination revealed no defects in non-myofibrillar structures; nuclei, mitochondria, and membrane systems appeared normal (figure 9). In addition, neonatal murine cardiomyocytes proliferate slowly in culture [34]. Although we did not carefully examine cardiomyocyte proliferation in this study, Krp1 knockdown was not accompanied by the appearance of abnormally large or multinucleated cardiomyocytes, ruling out an effect on cytokinesis. Together, these data demonstrate that Krp1 knockdown specifically affects the assembly, maintenance, or degradation of myofibrils.
The simplest explanation of these data is that Krp1 knockdown inhibits a particular step in myofibril assembly, leading to accumulation of the assembly intermediates preceding this step. This interpretation leads to the conclusion that assembly of very thin, periodic myofibrillar structures occurs upstream from Krp1 action, which promotes lateral fusion of these structures. However, the molecular mechanism by which Krp1 promotes lateral fusion of assembly intermediates remains to be elucidated. Possible mechanisms include accelerating lateral association of premyofibril structures, stabilization of weakly associated structures long enough for extensive multimolecular complexes to form, or slowing of disassembly pathways. Regardless of the mechanism, the results show that Krp1 is specifically necessary to promote the lateral fusion of thin myofibrillar structures, but is not required for appropriate longitudinal organization of sarcomeric components.
Myofibril Assembly Pathways: Lateral and Longitudinal Organization of Myofibrillar Components
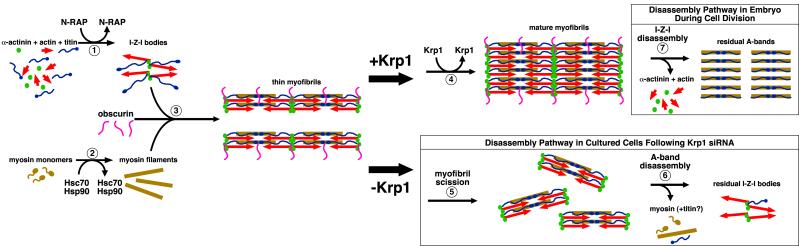
Figure 10 incorporates our findings regarding Krp1 into a putative pathway for myofibril assembly that emphasizes the role of proteins transiently associated with developing myofibrillar structures. Historically, myofibril assembly has been studied as the sequence of organization of the major proteins that form the sarcomeres [16]. However, the advent of gene targeting and silencing technologies permits dissection of the contribution of low abundance proteins to myofibril assembly, even when these proteins are only transiently associated with the assembling structures and do not form part of the mature myofibril. Our previous application of RNA interference to reduce N-RAP expression resulted in decreased accumulation of mature myofibrils without the apparent accumulation of intermediate myofibrillar structures; the long thin periodic fibrils observed after Krp1 knockdown (figures 7 and 8) were not observed after N-RAP knockdown [26]. The results were consistent with our previous work showing that N-RAP participates in assembly of α-actinin and actin, which occurs in the first steps of myofibrillogenesis (figure 10, step 1). Titin is also present during this step; its N-terminus assembles early in myofibrillogenesis with α-actinin and actin while its C-terminus assembles with myosin filaments that integrate later in assembly [20, 21]. It has long been known that muscle myosin filaments assemble separately from nascent Z-bodies and actin filaments [35]. Recent studies have shown that nascent myosin filaments are associated with the chaperone proteins Hsc70 and Hsp90 before incorporation into sarcomeres [36] (figure 10, step 2). The accumulation of abnormally thin striated myofibrils after Krp1 knockdown shows that Krp1 functions downstream of N-RAP, Hsc70 and Hsp90 in the assembly pathway to promote lateral fusion of thin myofibrils (figure 10, step 4). Krp1, N-RAP, Hsc70 and Hsp90 are all transiently associated with assembling myofibrillar structures, consistent with scaffolding molecules that promote specific steps in the assembly pathway (figure 10).
Figure 10.
A putative pathway for myofibril assembly (steps 1-4) highlighting the role of transiently associated proteins in organizing the major structural components. Disassembly pathways after Krp1 knockdown (steps 5-6) and during cell division in embryonic development (step 7) are also illustrated. (1) N-RAP promotes assembly of the I-Z-I structures containing actin, α-actinin, and N-terminal titin. (2) Myosin filaments form separately, with appropriate folding and assembly promoted by the Hsc70 and Hsp90 chaperone proteins. (3) Obscurin plays a role in promoting integration of the thick filaments with the I-Z-I structures, with titin associating with the myosin filaments along their length. This gives rise to thin myofibrils. (4) Finally, Krp1 promotes their lateral fusion to form mature myofibrils. (5-6) Krp1 knockdown results in scission of the thin myofibrils, followed by disassembly of the A-bands. The last organized structures observed contain α-actinin and actin, but not myosin. (7) In contrast, disassembly during cell division occurs by removal of α-actinin and actin, leaving organized A-bands. Subsequent A-band disassembly is not shown. See text for details.
Another protein recently linked to myofibril assembly is obscurin, a component of mature myofibrils thought to link with the sarcoplasmic reticulum through interaction with ankyrin 1 [37-40]. A role for obscurin in integrating myosin filaments into sarcomeres is supported by specific disruption of this process in cultured neonatal rat myotubes after overexpression of a C-terminal obscurin fragment [41] as well as by obscurin knockdown studies in adult rat cardiomyocytes [42]. In contrast, longitudinal organization of myosin was retained in Krp1 knockdown cardiomyocytes (figures 8 and 9). Taken together, these data suggest that obscurin-mediated incorporation of myosin filaments occurs downstream of N-RAP mediated assembly of α-actinin and actin but upstream of Krp1 mediated lateral fusion of fully formed thin myofibrils (figure 10, step 3). While the sequence of events depicted in figure 10 (steps 1-4) is consistent with much cell biological data, in vitro reconstitution experiments will be necessary to critically test the proposed pathway of assembly.
Myofibril Disassembly Pathways
In addition to long, thin myofibrils, cardiomyocytes also accumulate randomly arranged dots of α-actinin after Krp1 knockdown (figure 7). Double staining shows that these are a mixture of isolated α-actinin Z-bodies associated with actin or short runs of 1 to 3 thin sarcomeres (figure 8). Interestingly, the percentage of cells that have accumulated periodic Z-bodies or thin Z-lines characteristic of the long, thin fibrils peaked 3 days following transfection with Krp1 siRNA and then decreased over the next several days. In contrast, the percentage of cells dominated by randomly spaced dots of α-actinin characteristic of short runs of 1-3 thin sarcomeres peaked at day 5 post-transfection (figure 7B). These data suggest a biphasic effect of Krp1 knockdown in which myofibrillogenesis first advances to a stage where long thin fibrils form. These structures accumulate, but eventually begin to disassemble. The mechanism of disassembly after Krp1 knockdown appears to involve shortening or scission of the longer fibrils to shorter fibrils, and finally to single Z-bodies with associated actin (figure 10, steps 5 and 6).
During embryonic development, cardiomyocytes undergo cycles of myofibril assembly and disassembly that accompany each round of cell division [43]. During mitosis, Z-disks and actin filaments appear to disassemble before myosin filaments and myomesin (figure 10, step 7). In contrast, organized muscle myosin in the absence of sarcomeric actin was not observed after Krp1 knockdown (figure 8B), suggesting that the mechanism of disassembly in this study differs from the pathway that accompanies cell division during normal cardiac development. Identifying the signals mediating the switch from accumulation of thin myofibrils to disassembly may shed light on the mechanisms by which myocytes regulate disassembly under different conditions. Interestingly, Krp1 and N-RAP both bind to ubiquitin ligases. Krp1 has been shown to assemble with cullin-3 ubiquitin ligases in vitro [44], while N-RAP binds the muscle ring finger ubiquitin ligases MURF-1 and MURF-2 in yeast two-hybrid binding assays [45]. This presents the intriguing possibility that the myofibril assembly scaffold proteins N-RAP and Krp1 also regulate degradation of specific proteins through the proteasome pathway.
Cytoplasmic and Cytoskeletal Pools of Krp1
In cultured mouse cardiomyocytes, Krp1 exhibits punctate localization throughout the cytoplasm as well as concentrations in areas with developing myofibrils (figures 1 & 2). This is consistent with the localization of Krp1 in primary chick cardiomyocyte cultures, in which Krp1 immunofluorescence revealed a general cytoplasmic distribution with pools of Krp1 localized to areas with laterally fusing myofibrils [14]. Subcellular fractionation demonstrated that a significant proportion of Krp1 remains in the cell, presumably bound to cytoskeletal structures, after extraction of membrane and cytosolic components with detergent (figure 3). The localization pattern of Krp1 in cardiomyocytes is reminiscent of its localization in transformed rat fibroblasts, in which a large pool of Krp1 is cytosolic and detergent-extractable, and the remainder is co-localized with actin at the tips of pseudopodia [10]. These data support the interpretation that the pool of Krp1 in cardiomyocytes that is resistant to extraction with detergent is associated with cytoskeletal elements. The significance of cytoskeletal and cytoplasmic pools of Krp1 and trafficking of Krp1 between subcellular compartments remains to be explored.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases and National Heart, Lung and Blood Institute. We thank Shajia Lu (Laboratory of Muscle Biology, NIAMS) for expert technical assistance. We also thank Drs. Evelyn Ralston and Kristien Zaal (Light Imaging Section, NIAMS) for instruction and guidance with the confocal microscopy. We are grateful to Drs. Bradford Ozanne and Heather Spence (Beatson Institute for Cancer Research, Glasgow, UK) for the gift of antibody against Krp1. We thank Dr. Kuan Wang (Laboratory of Muscle Biology, NIAMS) for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- [2].Stogios PJ, Prive GG. The BACK domain in BTB-kelch proteins. Trends Biochem Sci. 2004;29:634–637. doi: 10.1016/j.tibs.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [3].Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- [7].Jiang S, Avraham HK, Park SY, Kim TA, Bu X, Seng S, Avraham S. Process elongation of oligodendrocytes is promoted by the Kelch-related actin-binding protein Mayven. J Neurochem. 2005;92:1191–1203. doi: 10.1111/j.1471-4159.2004.02946.x. [DOI] [PubMed] [Google Scholar]
- [8].Neuhaus P, Jaschinsky B, Schneider S, Neuhaus H, Wolter A, Ebelt H, Braun T. Overexpression of Kelch domain containing-2 (mKlhdc2) inhibits differentiation and directed migration of C2C12 myoblasts. Exp Cell Res. 2006;312:3049–3059. doi: 10.1016/j.yexcr.2006.06.006. [DOI] [PubMed] [Google Scholar]
- [9].Sasagawa K, Matsudo Y, Kang M, Fujimura L, Iitsuka Y, Okada S, Ochiai T, Tokuhisa T, Hatano M. Identification of Nd1, a novel murine kelch family protein, involved in stabilization of actin filaments. J Biol Chem. 2002;277:44140–44146. doi: 10.1074/jbc.M202596200. [DOI] [PubMed] [Google Scholar]
- [10].Spence HJ, Johnston I, Ewart K, Buchanan SJ, Fitzgerald U, Ozanne BW. Krp1, a novel kelch related protein that is involved in pseudopod elongation in transformed cells. Oncogene. 2000;19:1266–1276. doi: 10.1038/sj.onc.1203433. [DOI] [PubMed] [Google Scholar]
- [11].Taylor A, Obholz K, Linden G, Sadiev S, Klaus S, Carlson KD. DNA sequence and muscle-specific expression of human sarcosin transcripts. Mol Cell Biochem. 1998;183:105–112. doi: 10.1023/a:1006824331819. [DOI] [PubMed] [Google Scholar]
- [12].Spence HJ, McGarry L, Chew CS, Carragher NO, Scott-Carragher LA, Yuan Z, Croft DR, Olson MF, Frame M, Ozanne BW. AP-1 differentially expressed proteins Krp1 and fibronectin cooperatively enhance Rho-ROCK-independent mesenchymal invasion by altering the function, localization, and activity of nondifferentially expressed proteins. Mol Cell Biol. 2006;26:1480–1495. doi: 10.1128/MCB.26.4.1480-1495.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Luo G, Zhang JQ, Nguyen TP, Herrera AH, Paterson B, Horowits R. Complete cDNA sequence and tissue localization of N-RAP, a novel nebulin-related protein of striated muscle. Cell Motil Cytoskeleton. 1997;38:75–90. doi: 10.1002/(SICI)1097-0169(1997)38:1<75::AID-CM7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [14].Lu S, Carroll SL, Herrera AH, Ozanne B, Horowits R. New N-RAP-binding partners α-actinin, filamin and Krp1 detected by yeast two-hybrid screening: implications for myofibril assembly. J Cell Sci. 2003;116:2169–2178. doi: 10.1242/jcs.00425. [DOI] [PubMed] [Google Scholar]
- [15].Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- [16].Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- [17].Dlugosz AA, Antin PB, Nachmias VT, Holtzer H. The relationship between stress fiber-like structures and nascent myofibrils in cultured cardiac myocytes. J Cell Biol. 1984;99:2268–2278. doi: 10.1083/jcb.99.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci U S A. 1997;94:9493–9498. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gregorio CC, Granzier H, Sorimachi H, Labeit S. Muscle assembly: a titanic achievement? Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- [20].Ehler E, Rothen BM, Hämmerle SP, Komiyama M, Perriard J-C. Myofibrillogenesis in the developing chicken heart: assembly of the z-disk, m-line and thick filaments. Journal of Cell Science. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- [21].Rudy DE, Yatskievych TA, Antin PB, Gregorio CC. Assembly of thick, thin, and titin filaments in chick precardiac explants. Dev Dyn. 2001;221:61–71. doi: 10.1002/dvdy.1125. [DOI] [PubMed] [Google Scholar]
- [22].Carroll SL, Horowits R. Myofibrillogenesis and formation of cell contacts mediate the localization of N-RAP in cultured chick cardiomyocytes. Cell Motil Cytoskeleton. 2000;47:63–76. doi: 10.1002/1097-0169(200009)47:1<63::AID-CM6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- [23].Lu S, Borst DE, Horowits R. N-RAP expression during mouse heart development. Dev Dyn. 2005;233:201–212. doi: 10.1002/dvdy.20314. [DOI] [PubMed] [Google Scholar]
- [24].Carroll S, Lu S, Herrera AH, Horowits R. N-RAP scaffolds I-Z-I assembly during myofibrillogenesis in cultured chick cardiomyocytes. J Cell Sci. 2004;117:105–114. doi: 10.1242/jcs.00847. [DOI] [PubMed] [Google Scholar]
- [25].Carroll SL, Herrera AH, Horowits R. Targeting and functional role of N-RAP, a nebulin-related LIM protein, during myofibril assembly in cultured chick cardiomyocytes. J Cell Sci. 2001;114:4229–4238. doi: 10.1242/jcs.114.23.4229. [DOI] [PubMed] [Google Scholar]
- [26].Dhume A, Lu S, Horowits R. Targeted disruption of N-RAP gene function by RNA interference: A role for N-RAP in myofibril organization. Cell Motil Cytoskeleton. 2006;63:493–511. doi: 10.1002/cm.20141. [DOI] [PubMed] [Google Scholar]
- [27].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [28].Olek AJ, Ling A, Daniels MP. Development of ultrastructural specializations during the formation of acetylcholine receptor aggregates on cultured myotubes. J Neurosci. 1986;6:487–497. doi: 10.1523/JNEUROSCI.06-02-00487.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Flucher BE, Andrews SB, Daniels MP. Molecular organization of transverse tubule/sarcoplasmic reticulum junctions during development of excitation-contraction coupling in skeletal muscle. Mol Biol Cell. 1994;5:1105–1118. doi: 10.1091/mbc.5.10.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Song KS, Scherer PE, Tang Z, Okamoto T, Li S, Chafel M, Chu C, Kohtz DS, Lisanti MP. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- [31].Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circ Res. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- [32].McKenzie JC, Kelley KB, Merisko-Liversidge EM, Kennedy J, Klein RM. Developmental pattern of ventricular atrial natriuretic peptide (ANP) expression in chronically hypoxic rats as an indicator of the hypertrophic process. J Mol Cell Cardiol. 1994;26:753–767. doi: 10.1006/jmcc.1994.1090. [DOI] [PubMed] [Google Scholar]
- [33].Navaratnam V, Woodward JM, Skepper JN. Specific heart granules and natriuretic peptide in the developing myocardium of fetal and neonatal rats and hamsters. J Anat. 1989;163:261–273. [PMC free article] [PubMed] [Google Scholar]
- [34].Kajstura J, Cheng W, Reiss K, Anversa P. The IGF-1-IGF-1 receptor system modulates myocyte proliferation but not myocyte cellular hypertrophy in vitro. Exp Cell Res. 1994;215:273–283. doi: 10.1006/excr.1994.1343. [DOI] [PubMed] [Google Scholar]
- [35].Holtzer H, Hijikata T, Lin ZX, Zhang ZQ, Holtzer S, Protasi F, Franzini-Armstrong C, Sweeney HL. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- [36].Srikakulam R, Winkelmann DA. Chaperone-mediated folding and assembly of myosin in striated muscle. J Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- [37].Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Borisov AB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: obscurin as an integrator of myofibrillar structure. J Histochem Cytochem. 2004;52:1117–1127. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- [40].Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am J Physiol Cell Physiol. 2004;287:C209–217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- [42].Borisov AB, Sutter SB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2006;125:227–238. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- [43].Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci. 2004;117:3295–3306. doi: 10.1242/jcs.01159. [DOI] [PubMed] [Google Scholar]
- [44].Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- [45].Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]