Abstract
Energy homeostasis in mammals is achieved through tight regulation of tissue-specific metabolic pathways that become dysregulated in metabolic diseases including diabetes and obesity. At the molecular level, main nutrient and hormonal signaling pathways impinge on expression of genes encoding for metabolic enzymes. Among the major components of this transcriptional circuitry are the PGC-1α transcriptional complexes. An important regulatory mechanism of this complex is through acetylation and SIRT1-mediated lysine de-acetylation under low nutrient conditions. Activation of SIRT1 can mimic several metabolic aspects of calorie restriction that target selective nutrient utilization and mitochondrial oxidative function to regulate energy balance. Thus, understanding the PGC-1α and SIRT1 pathways might have important implications for comprehending metabolic and age-associated diseases.
Keywords: PGC-1α, SIRT1, mitochondrial oxidation, glucose metabolism, lipid metabolism, aging
1. Molecular Mechanisms of PGC-1α and SIRT1 Function
The initial studies in lower organisms found that the gene SIR2 (Silent Information Regulator; ortholog of mammalian sirtuin SIRT1) potentially mediated the effects of calorie restriction on longevity [1], prompting different groups to investigate if SIRT1 had a role in metabolic function and longevity in mammals. Another active line of investigation was the search for genes that control mammalian energy and nutrient homeostasis. In this context, PGC-1α (Peroxisome Proliferator-Activated Receptor Gamma-Coactivator-1α) was identified as transcriptional coactivator that could define tissue-specific metabolic pathways in the adaptive response to environmental and nutritional stimuli [2]. Work by our group and Finkel’s group first identified the functional interaction and deacetylation of PGC-1α and SIRT1. Furthermore, this interaction and deacetylation could be regulated by energy fluctuations and nutrient levels, thus leading to direct transcriptional control of metabolic enzymes and pathways [3] [4]. We will discuss in this section how these two proteins function mechanistically in the context of controlling metabolic gene expression.
1.1. PGC-1α Functions as a Transcriptional Coactivator
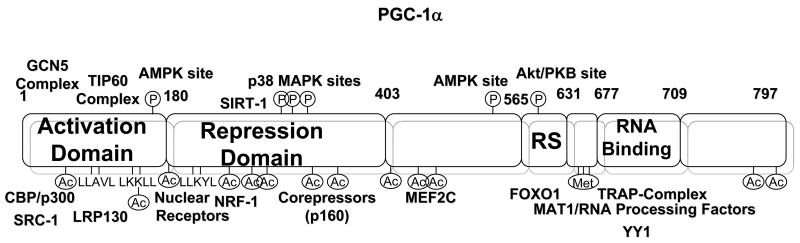
PGC-1α belongs to a small family of transcriptional coactivators, including PGC-1β and PRC which possess a common function in mitochondrial physiology, in addition to control over separate specific biological programs. PGC-1α was the first member identified as a cofactor for the nuclear hormone receptor PPARγ that is required for the adaptive thermogenic response to lower temperatures [2]. Until mid 90’s most of the regulation of gene expression and its biological implications were focused on transcription factors. During that time the discovery of the first transcriptional coactivators and corepressors for nuclear hormone receptors and other transcriptions factors initiated a novel concept centered around the control of specific genetic programs coordinated at the level of the transcriptional cofactor. PGC-1α as a transcriptional coactivator functions through direct physical interaction with transcription factors directly bound to DNA promoter regions. For example, LXXLL motifs in the PGC-1α N-terminus interact with different hormone nuclear receptors, including PPARs, HNF4α, GR and ERRα. In addition, other parts of the protein bind to other transcription factors such as NRF-1 in the 200–400 region, MEF2C in the 400–565 region or FoxO1 and YY1 in the RNA-processing C-terminal region (Fig. 1). Importantly, the ability of PGC-1α to interact with different transcription factors allows for the coordinated expression of gene sets in response to specific signals. However, another important and interesting implication is the possibility to activate gene expression in very specific contexts. As an example, PPARγ binding sites are present in UCP-1 and aP2 promoters, however PGC-1α only actives UCP-1, but not aP2 [2]. So far, the molecular basis of this specification is unknown, but it might involve DNA-binding affinities or facilitating interactions with other transcription factors or protein complexes to promote availability to access the promoter.
Fig. 1.
Architecture of the PGC-1α transcriptional Coactivator. PGC-1α contains several functional domains that correlate with the interactions with different proteins and complexes. Additionally, PGC-1α contains specific phosphorylated, acetylated and methylated amino acids which can modulate its activity. See text for further details.
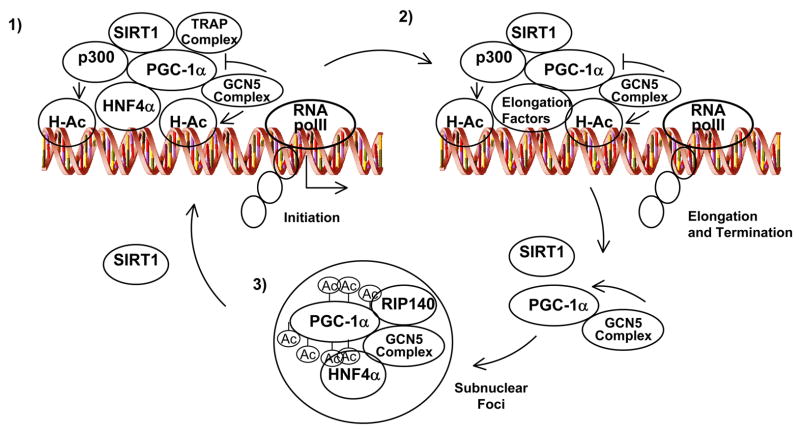
The mechanisms through which PGC-1α activates gene expression are poorly understood. Initial studies identified an extremely powerful autonomous transcriptional activity at the N-terminal region. This activation function correlates with the ability of PGC-1α to dock on this domain two other coactivators with acetyl transferase activity, SRC-1 and CBP/p300. In addition, the C-terminal region contains an SR motif and an RNA-binding domain. This region is required to induce expression of certain endogenous genes and interacts with proteins involved in RNA processing and the TRAP complex involved in transcriptional initiation [5]. Moreover, PGC-1α has been found in association with two additional different transcriptional complexes, the GCN5 and TIP60 acetyl transferases complexes and many of their associated protein components [6]. GCN5 directly acetylates PGC-1α at multiple lysine residues and negatively regulates its transcriptional activity, at least in part, through nuclear sublocalization. How spatially and temporally all these complexes are assembled to PGC-1α to control expression of genes is unknown. A current model is that PGC-1α binds to specific transcription factors at promoters, then, additional recruitment of p300 and TRAP complexes would open the chromatin through histone acetylation thereby allowing initiation of transcription through RNA polII. Although GCN5 may acetylate histone H3 in this complex, it would initially be refractory to acetylate PGC-1α. The fact that the PGC-1α complex contains several proteins involved in RNA elongation and processing also suggests that it might move with the elongating phospho-RNA polII and participate in fully maturation of the mRNAs [6]. To terminate gene expression, GCN5 would acetylate PGC-1α resulting in relocalization to repressive subnuclear foci where PGC-1α has been shown to co-localize with the transcriptional repressor RIP140 [6]. Conversely, SIRT1 activation will maintain PGC-1α in a deacetylated active form bound to the chromatin and increasing rates of transcription (Fig. 2) [4] [7]. However, this model is perhaps simplistic does not take into account the other PGC-1α modifications such as phosphorylation and methylation, as well as interaction with other proteins including corepressors. Three kinases have been demonstrated that directly phosphorylate PGC-1α. The stress-activated p38 MAP kinase phosphorylates PGC-1α at three residues (Thr262; Ser265; Thr298) in the 200–400 repression domain and correlates with a more active and stable protein [8]. In addition to enhanced stability, the activity is further increased due to the inability of the phospho-PGC-1α to bind the p160 corepressor [9]. AMP kinase directly phosphorylates PGC-1α at residues Thr177 and Ser538 resulting in a more active protein [10]. Moreover, PGC-1α is phosphorylated by Akt/PKB at Ser570 in the SR domain and leads to a more unstable protein with lower activity [11]. Therefore, it seems that a major mechanism by which phosphorylation functions is through control of PGC-1α protein degradation and/or interaction with corepressor proteins. It is not known how these specific phosphates control PGC-1α stability and the proteins involved in this regulation. Furthermore, PGC-1α is a target of the protein arginine methyltransferase 1 (PRMT1) that also coactivates nuclear hormone receptors. PRMT1 activates PGC-1α transcriptional activity through methylation at three arginine residues (Arg665, Arg667 and Arg669) at the C-terminus of the protein [12]. It is conceivable that PGC-1α functions in multiple protein complexes whose composition might depend on the specific target gene as well as the combination of different signals that are on and/or off. For example, LRP130 (Leucine Rich Protein 130), a gene mutated in Leigh Syndrome French Canadian, is part of the PGC-1α complex that regulates a specific set of PGC-1α target genes in liver including gluconeogenic genes and certain mitochondrial genes (Fig. 1) [13]. Further identification of the signals, specific protein components and chemical modifications controlling these proteins will certainly provide novel mechanistic information as to how the whole PGC-1α metabolic biochemical machinery operates.
Fig. 2.
Model for PGC-1α transcriptional gene expression activity. 1) PGC-1α is part of multiprotein complexes that contain histone acetyl transferase activity that open and remodels chromatin to allow the transcription factor to bind DNA. In the transcriptional initiation complex SIRT1 would maintain deacetylated PGC-1α. 2) PGC-1α and its associated proteins will move with RNA processing, elongation factors and RNA polII to transcribe the mRNA. 3) After the mRNA processing, GCN5 would acetylate PGC-1α localizing the whole complex to a RIP140 containing subnuclear repressive foci. To initiate another cycle, SIRT1 would deacetylate PGC-1α freeing it from the repressive foci, allowing it to become incorporated into protein complexes at promoter regions.
1.2. SIRT1 Functions as a Protein Deacetylase
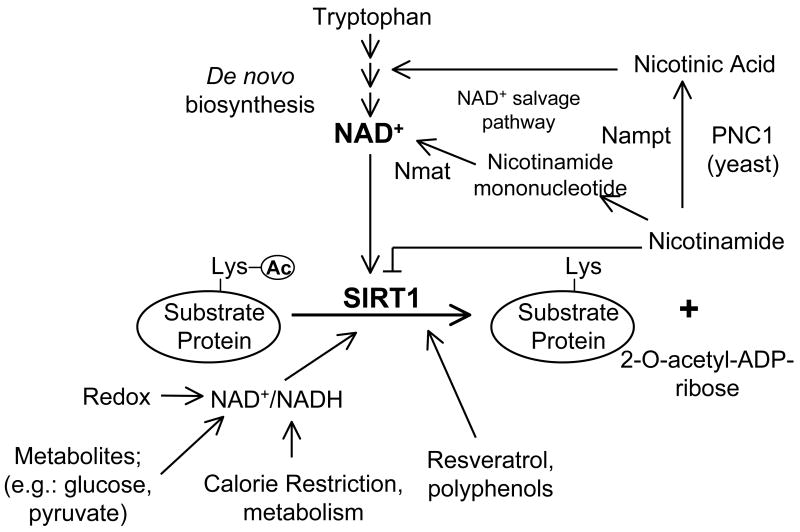
Sir2 was first identified as a factor which regulated silencing at the yeast mating type loci. Sir2 physically associates with chromatin in regions of the genome that were transcriptionally silent (mating type loci, telomere and centromeres) and also functions in preventing recombination of rDNA repeats [1]. Sir2, as well as SIRT1, are both NAD+-dependent deacetylase but neither used NAD+ as a redox acceptor. The NAD+ molecule is instead cleaved in the deacetylation reaction, transferring the acetyl group to the ribose sugar, thereby producing O-acetyl-ADP-ribose and nicotinamide (Fig. 3). The released nicotinamide ring can also function as an inhibitor of this reaction [14] [15]. As it relates to metabolism, the initial studies searching for yeast aging genes in the caloric restriction nutrient pathway identified Sir2. In worms and flies it appears that Sir2 is also involved in certain aspects of connecting calorie restriction to lifespan [16] [17].
Fig. 3.
Regulation of SIRT1 enzymatic activity. SIRT1 catalyzes a NAD+-dependent protein deacetylase activity. This biochemical reaction is regulated by different metabolic inputs as well as several polyphenols. Moreover, several metabolic pathways including NAD+ de novo biosynthesis from tryptophan and increasing the NAD+ salvage pathway influence SIRT1 enzymatic activity.
Most of the biological functions attributed to Sir2 or SIRT1 depend on the enzymatic activity. So far, the SIRT1 molecular mechanisms of function have been related to regulation of gene expression. In some cases, SIRT1 represses transcription and it is present in histone deacetylases and polycomb protein complexes [18]. SIRT1 specifically represses transcription factors such as PPARγ [19] and p53 [20]. The mechanism of repression is not completely understood but involves physical interaction with NCoR for PPARγ and direct deacetylation of p53 that decreases DNA-binding. In the case of FoxO proteins, SIRT1 can be either positive or negative depending on the type of target genes, but the mechanistic basis for this is not completely understood [21]. SIRT1 acts positively on activation of other genes through direct deacetylation of PGC-1α [4] [7] and HIV Tat [22]. SIRT1 activates Tat-mediated transcription of the HIV long terminal repeat. SIRT1 interacts with PGC-1α in the 200–400 region and deacetylates PGC-1α in at least 13 lysines in different domains of the proteins (Fig. 1). Mutation of these residues to arginine leads to a more active PGC-1α allele [23].
Interestingly, it is also possible that SIRT1 functions outside transcriptional control of gene expression. For example, AcetylCoA synthetases are specifically deacetylated by SIRT1 increasing its enzymatic activity [24], these enzymes seem to be very ancient substrates of Sir2 proteins that were originally identified bacteria [25].
Several mechanisms of regulation for SIRT1 have been described. First, nutrient deprivation (fasting or calorie restriction) upregulates SIRT1 protein levels and seems to be independent of its own mRNA induction [4]. In cultured hepatocytes, pyruvate, a metabolite increased in fasting conditions, induces SIRT1 protein levels affecting translation of the protein. In skeletal muscle cells, resveratrol also increases SIRT1 protein levels [26]. How SIRT1 protein levels are regulated and to what extent this is only at the translational level without degradation control is currently unknown. Second, fluctuations in NAD+ or ratios of NAD+/NADH as well as nicotinamide concentrations will directly affect SIRT1 enzymatic activity. Thus, redox state of the cell controlled by the activities of different metabolic pathways will change the ratio of NAD+/NADH. In addition, changes in activities of enzymes that control levels of these metabolites can affect SIRT1 activity. For example, NAD+ biosynthesis from nicotinamide is catalyzed in two reactions, one of the steps is controlled by the enzyme Nicotinamide phosphoribosyltransferase (Nampt) also known as PBEF or visfatin. Changes in the activity of this enzyme that occur in fasting conditions positively regulate SIRT1 activity. Finally, aside from these direct catalytic regulators, it is possible that endogenous or natural compounds might act similarly to resveratrol to increase SIRT1 activity (Fig. 3).
2. Tissue-Specific Metabolic Functions
2.1. Liver
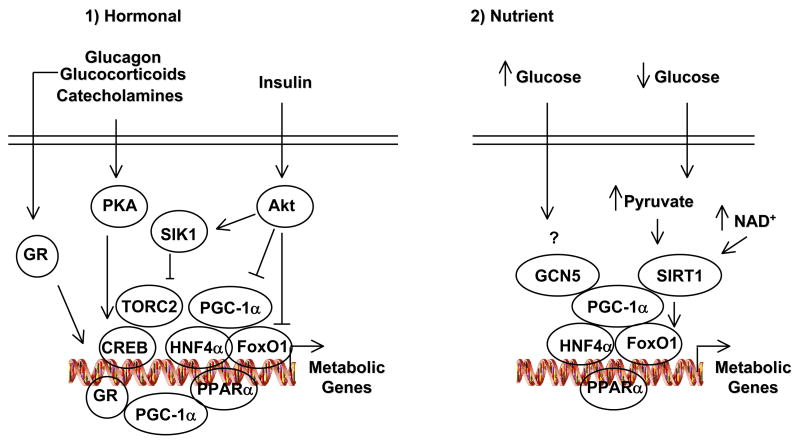
Liver is a major buffering tissue that ensures nutrient homeostasis in fed and fasting conditions. Metabolic adaptation to fasting requires an important control at the transcriptional level. PGC-1α is induced in the fasted liver to activate gluconeogenic and fatty acid oxidation genes. PGC-1α deficient mice lack this response and display hypoglycemia and hepatic steatosis [27] [28]. As part of the starvation response, signaling of the fasting hormones glucagon and glucocorticoids increases transcription of the PGC-1α gene. Glucagon signaling causes translocation and de-phosphorylation of the transcriptional coactivator TORC2 to the nucleus, where it then binds to and coactivates the CREB transcription factor which is present on the PGC-1α and gluconeogenic promoters [29]. Once PGC-1α is induced it will coactivate FoxO1 and HNF4α on the gluconeogenic genes such as PEPCK and G-6-Pase, increasing transcription of these genes (Fig. 4). The entirety of PGC-1α gluconeogenic activity requires these transcription factors, genetic ablation of HNF4α and FoxO1 completely abolishes PGC-1α’s ability to induce gluconeogenesis [30]. The counteregulatory effects of insulin impact this network through activation of Akt which phosphorylates at least three components of this signaling and transcriptional regulatory pathway. Akt directly phosphorylates FoxO1 at three residues and results in translocation to the cytoplasm forming a complex with 14-3-3 proteins [31]. Akt phosphorylates and activates the Ser/Thr kinase SIK2. Activated SIK2 induces TORC2 phosphorylation that is degraded by the 26S proteasome in association with COP1, a substrate receptor for an E3 ligase complex that promoted TORC2 ubiquitination and degradation [29]. PGC-1α is directly phosphorylated by Akt and increases PGC-1α protein degradation [11]. The effects of PGC-1α in hepatic lipid and mitochondrial metabolism are likely to function through the nuclear hormone receptors PPARα, ERRα and HNF4α. Moreover, lipin1a, a gene associated with lipodistrophy interacts with PGC-1α and positively controls fasted genes of fatty acid oxidation [32].
Fig. 4.
Hormonal and nutrient regulation of hepatic PGC-1α function. 1) Hormones of the fed (insulin) and fasted (glucagon, glucocorticoids and catecholamines) through different signaling cascades that target transcriptional complexes control expression of metabolic genes such as gluconeogenic enzymes. 2) Nutrient Regulation of PGC-1α through GCN5 Acetyl transferase and SIRT1 deacetylase. See the text for further detail.
Recently, our laboratory found that in parallel to this hormonal regulation there is a nutrient signaling that involves SIRT1 and PGC-1α. SIRT1 is induced in the fasted liver and interacts and deacetylates PGC-1α to activate gluconeogenic and fatty acid oxidation genes. This nutrient signaling involves increases in pyruvate and NAD+ levels resulting in elevated SIRT1 protein amounts as well as enzymatic activity. Expression of a PGC-1α acetylation mutant –in which 13 lysines have been mutated to arginines- maintains elevated expression of gluconeogenic genes in the fed state [4] [23]. These data imply that PGC-1α undergoes cycles of acetylation and deacetylation during the fed and fasted states that are dependent on SIRT1 activity. Furthermore, it also seems that the functions of SIRT1 might go beyond the fed/fasted response. This is evidenced by the observation that independently of the fed status of mice, hepatic SIRT1 controls systemic and hepatic cholesterol levels likely dependent on increases in LXRα and PGC-1β expression [23].
A current hypothesis poses that increased intracellular lipids and inefficiency to couple β-oxidation of fatty acids to mitochondrial respiration in tissues such as liver and skeletal muscle is a possible cause of insulin resistance and type 2 diabetes. In the liver, this metabolic defect leads to non-alcoholic fatty liver disease. Another main problem in diabetic patients is, although they display hepatic insulin resistance, the constant activation of lipogenesis also contributes to lipid accumulation and metabolic dysfunction. Mice lacking PGC-1α develop fasting hepatic steatosis probably due to lower rates of fatty acid oxidation that might play a causative role to develop insulin resistance [27] [28]. Intriguingly, PGC-1α KO mice are more insulin sensitive and resistant to high fat diet that could be related to hyperactivity. Alternatively, profound defects in fatty acid oxidation might be compensated with increases in glucose utilization to maintain energetic status. Moreover, lower rates of fatty acid oxidation and OXPHOS activity can also lead to decreases in rates of glucose synthesis that might improve glucose tolerance and insulin sensitivity [33] [34]. Notably, SIRT1 knockdown in the liver causes an increase of hepatic free fatty acids which might also compromise insulin signaling [23]. Consistent with that, it has been recently shown that SIRT1, similar to FoxO1, increases insulin signaling. In the case of SIRT1 part of these effects seems through inhibition of PTB1B [35].
One of the main contributors of hyperglycemia in diabetic patients is increased hepatic glucose production in both fed and fasted states. Knock-down of PGC-1α and SIRT1 in liver lowers blood glucose in normal and db/db mice [36] [23]. This reduction correlates with lower hepatic glucose production from pyruvate and increased glucose intolerance. Currently, due to this PGC-1α/SIRT1 dual effect in lipid and glucose metabolism is difficult to predict what it might be the final outcomes of the pharmacological activation of both proteins in the liver in diabetes. It is possible that facilitation of fatty acid oxidation and improvement in insulin sensitivity might be predominant over glucose production. Thus, in normal insulin sensitivity states, this hormone can effectively suppress PGC-1α activation of gluconeogenic genes. This would also be consistent with the increases in insulin sensitivity and normal blood glucose levels observed in mice treated with resveratrol [26].
2.2. Skeletal and Cardiac Muscle
Skeletal muscle represents a significant percentage of the total body mass and is energetically very active, especially in conditions of increased physical activity induction of mitochondrial oxidative function become essential to adapt and maintain the whole body energy balance. Several signaling pathways are important to activate mitochondrial function in skeletal muscle, for example Ca2+-regulated CAMKIV-calcineurin/NFAT and MEF2 axis, adrenergic and cholinergic signaling and AMPK activation. These signaling/transcription pathways can either induce and/or activate PGC-1α. Thus, skeletal muscle-tissue specific PGC-1α transgenic mice display higher mitochondrial content and a switch to oxidative type-1 fibers [37]. On the contrary, skeletal muscle-specific PGC-1α KO mice have deficient expression of mitochondrial proteins and develop myopathy that might be also related to its role to regulate neuromuscular gene expression [38]. In the heart, mitochondrial mass is also controlled by PGC-1α and is important to maintain cardiac function in response to a variety of stresses [39]. Similar to the liver, PGC-1α increases rates of oxidation of fatty acids that are important in fasting and neonatal conditions.
Little is known about the function of SIRT1 in skeletal muscle. In culture muscle cells SIRT1 seems to be involved in early steps of differentiation by interfering with myogenic factors [40]. Our group has found that in myotubes SIRT1 is required for the switch from glucose to fatty acid utilization that occurs in low nutrient conditions. Skeletal muscle cells trigger this switch in a cell autonomous manner depending on glucose concentrations. One of the mechanisms by which SIRT1 exerts these effects is through PGC-1α deacetylation. Consistent with SIRT1 activation, PGC-1α is deacetylated in fasting and in low glucose conditions to induce a whole battery of genes involved in mitochondrial fatty acid oxidation [7]. Intriguingly, resveratrol mimics the effects of low glucose by targeting SIRT1 and deacetylating PGC-1α. Mice treated with resveratrol have increased mitochondrial function in several tissues and display other metabolic changes that correlate with extension of life span [41]. In addition, resveratrol induces a genetic program in skeletal muscle that is consistent with higher energy expenditure that largely prevents increases in body weight under high fat diet [26]. It would be also of interest to investigate if the beneficial effects of PGC-1α in muscular degeneration are also improved with SIRT1 activation.
In the context of type 2 diabetes, the role of PGC-1α and SIRT1 in the liver seems to be more complex due to the positive effects on both fatty acid oxidation and hepatic glucose production (note that these two processes are activated in nutrient deprivation conditions). However, the effects in skeletal muscle of pharmacological activation of PGC-1α and SIRT1 would be consistent with beneficial results of both proteins. Thus, increases in insulin sensitivity and energy expenditure are observed in mice treated with resveratrol. It is also important to highlight the role that these two proteins might play in exercise and physical activity. These energy-demanding conditions activate AMPK that directly phosphorylates PGC-1α at two residues (Thr177 and Ser53). Notably, increases of target genes of AMPK such as GLUT4 and cytochrome c totally depend on PGC-1α [10]. Full activation of PGC-1 α in skeletal muscle would allow efficient β-oxidation of fatty acids and coupling to mitochondrial oxidative phosphorylation. In addition, PGC-1α maintains higher number of active mitochondria and OXPHOS proteins that are decreased in type 2 diabetes. As it relates to SIRT1 in skeletal muscle, transgenic SIRT1 mice have increased physical activity and this also correlates with higher insulin sensitivity [42] and increase in physical activity during calorie restriction requires SIRT1 KO [43]. Furthermore, increases in mitochondrial function and fatigue resistance during exercise are also observed in mice treated with resveratrol [26].
The physiological role of SIRT1 in the heart is unclear. Transgenic mice with moderate expression of SIRT1 in heart are more resistant to oxidative stress under pressure overload. However, at high expression of SIRT1 mice develop cardiac hypertrophy. Similar phenotypes have also been observed with transgenic mice expressing PGC-1α in the heart [44]. Interestingly, mice treated with resveratrol under high fat diet have lower heart beat that could be related a decreases in locomotor spontaneous activities [26].
2.3. Brain
The brain constitutes perhaps the most vulnerable tissue to oxidative stress. One of the strong phenotypes observed in the PGC-1α KO are lesions in the striatal region of the brain that controls movement. These mice are hyperactive and display progressive loss of striatal neurons that is reminiscent of Huntington Disease [27]. Although is possible that some of this phenotype might be partially due to a defective mitochondrial gene expression and function, PGC-1α also activates genes that encode enzymes involved in ROS detoxyfication [45]. Thus, ROS induces PGC-1α expression and the lack of PGC-1α correlates with a loss of protection against oxidative stress damage. So far, the data of PGC-1α function in the brain is consistent with a major role in neuroprotection. Interestingly, SIRT1 plays a similar role in oxidative stress function in combination with FoxO transcription factors [21]. Moreover, studies with mice treated with resveratrol and ectopic expression of SIRT1 (that correlated with deacetylation of PGC-1α) in the hippocampus further support the neuroprotective role of SIRT1 in Alzheimer’s disease and amyotrophic lateral sclerosis [46]. Together, these studies suggest that PGC-1α and SIRT1 might be potential targets to treat neurodegenerative diseases.
Another important physiological aspect related to the brain is the role in energy balance through hypothalamic control of food intake, energy expenditure and physical activity. Since the hypothalamus controls food intake in fed and fasted conditions through leptin, insulin and nutrient signaling pathways, it is likely that PGC-1α and SIRT1 might play a role in modulating this response. However, PGC-1α KO mice, SIRT1 transgenic and resveratrol treated mice do not present any difference in food intake compared to control animals. Whether other compensatory mechanisms are involved in keeping this function is currently unknown.
A key component in energy balance controlled at the CNS is energy expenditure. The observation that PGC-1α is induced in conditions of elevated energy expenditure, such as cold via adrenergic stimulation, suggests that it might control part of this response. The conclusions about metabolic phenotypes in the PGC-1α and SIRT1 KOs should be taken with caution and would be need to validate with tissue-specific and inducible models. The total PGC-1α KO mice are cold-intolerant indicating defective energy expenditure at lower temperatures [27] [28]. However, it seems to become more complex when these mice are fed with high fat diet. The fact that these mice are hyperactive with high spontaneous locomotor activity seems to make them resistant to increases in body weight after high fat diet feeding. With regard to the role of SIRT1 in energy balance, the studies from the whole SIRT1 KO mice also provide little information; these mice are smaller and present several abnormalities. However, transgenic SIRT1 animals have higher physical activity [42] and mice treated with resveratrol display remarkably increase in energy expenditure [26]. In these two mouse models, whether the effects are due, at least in part, to the SIRT1 function in the brain is unknown. Although more data is needed from PGC-1α and SIRT1 tissue-specific KO or transgenic mice, it appears that functions of these two proteins correlate with increases in energy expenditure and physical activity.
2.4. Pancreas
Pancreatic β cells, together with a group of neurons in the hypothalamus, constitute very sensitive cellular sensors for systemic glucose levels. Small increases in glucose levels will stimulate β cells that will secrete insulin to the blood. Little is known about the role of PGC-1α in β cells, although initial studies indicated that overexpression of PGC-1α correlated with a decrease in insulin secretion [47]. However, further studies with PGC-1α KO mice should be performed to confirm this effect. This is important given the relevance of mitochondrial metabolism as well as ATP levels in insulin secretion. On the other hand, it appears that SIRT1 promotes insulin secretion through PPARγ-mediated repression of the uncoupling protein UCP2 and altering intracellular concentrations of ATP [48]. Similar to the skeletal muscle cells, β cells that lack SIRT1 do not fully sense fluctuations of glucose and release normal amounts of insulin. Given the protective effects of both SIRT1 and PGC-1α in other cell types it would be interesting to know whether these proteins might maintain β cell mass under oxidative stress conditions.
2.5. Adipose Tissues
Among the first studies in mammalian cells that provided a metabolic functional role for PGC-1α as well as SIRT1 came from the two types of adipose tissues; PGC-1α in brown adipose tissue [2] and SIRT1 in white adipose tissue [19]. Cold is a potent activator of brown adipose tissue thermogenic function that maintains body’s temperature. PGC-1α is strongly induced in this situation and lack of PGC-1α correlates with impaired cold tolerance. In addition, cultured brown adipocytes lacking PGC-1α possess a deficient thermogenic program but are still able to differentiate into brown adipocytes [49]. This later function seems to be dependent on a new transcriptional cofactor named PRDM16 [50].
The thermogenic, similar to the gluconeogenic, response largely depends on the cAMP pathway and PGC-1α controls a large set of the genes regulated by this pathway [49]. In addition, the lack of the three β-adrenoreceptors also produces cold intolerance, supporting the necessity of this pathway to maintain corporal temperature. Thermogenic capacity in brown adipose tissue depends on a high number of mitochondria with uncoupling respiration that leads to heat production. This process depends on a very abundant mitochondrial protein termed UCP-1 which is exclusively expressed in brown adipocytes. Expression of this protein entirely depends on β-adrenergic stimulation and mice deficient in UCP-1, similar to PGC-1α and β-adrenoreceptors, are also cold intolerant.
In white adipose tissue PGC-1α is expressed at much lower levels, however in situations of remodeling or activation by β-agonists PGC-1α as well as its mitochondrial target genes are increased. Ectopic expression of PGC-1α in white adipocytes induces a thermogenic program strongly resembling that of brown adipocytes. Another metabolic target of PGC-1α in white adipocytes is glycerol kinase that might promote a futile cycle of triglyceride hydrolysis and fatty acid reesterification [51], especially in combination with lipolytic activators such as catecholamines.
SIRT1 is activated by low nutrient conditions in white adipose tissue and induces systemic fatty acid mobilization. Similar to pancreatic β cells it appears that the mechanism by which SIRT1 induces lipolysis and prevent triglyceride accumulation in white adipocytes is through repression of PPARγ. The mechanism involves SIRT1-dependent recruitment of the transcriptional corepressor NCoR on PPARγ targets such as the abundant aP2 fat-selective gene. Furthermore, similar to skeletal muscle cells, SIRT1 also interferes with the ability of white adipocytes to fully differentiate, most likely by suppressing PPARγ function [19]. However, how SIRT1 controls the adipocyte lipolytic action and whether PPARγ is involved is unknown. Recent transgenic mice in which SIRT1 is overexpressed in different tissues including white adipose tissue display some of the metabolic features of caloric restricted mice such as lower body weight, reduction in glucose and insulin and increased glucose tolerance [42]. The role of SIRT1 in brown adipose tissue and thermogenic function has not been explored. Emerging genomic studies suggest a common cell precursor between brown adipocytes and myotubes that coincides with SIRT1 ability to block myogenesis and promote brown adipocyte differentiation with increased mitochondrial biogenesis [52]. In fact, other genomic studies have correlated expression of SIRT1 with increases in mitochondrial genes that is fully consistent with our own finding [7]. Along the same lines, mice treated with resveratrol show higher PGC-1α deacetylation in brown fat and expression of thermogenic genes including UCP-1 [26]. However, future studies will need to address SIRT1 function in this tissue and the possibility that SIRT1 activators might be used to treat metabolic diseases by increasing uncoupled respiration in brown adipose tissue. In light of some new findings, the possibility of therapeutically targeting brown fat thermogenic activities in humans might take a new twist. It has been thought for a long time that brown adipose tissue depots were only important in humans at birth but not in adults, however, recent observations with PET (positron emission tomography) analysis seems to provide evidence that adult humans have active brown fat depots localized in different areas of the upper body [53].
3. Concluding Remarks and Implications for Metabolic Diseases
The abundance of human metabolic diseases gives prime illustration of what occurs when energetic homeostatic pathways become dysregulated. An interesting approach to the problem of these diseases seems to be the study of the beneficial health effects of prolonged calorie restriction and a more detailed analysis of tissue-specific metabolic pathways including how they function and adapt in normal physiology. In this context, we have concentrated on how the PGC-1α and SIRT1 pathways seem to largely function together to promote tissue-specific metabolic adaptation to nutrient fluctuations. It may be of no coincidence that dysregulation of many PGC-1α/SIRT1 targets occurs in, or is even causative to metabolic diseases.
Nutrient signals impinge on the PGC-1α and SIRT1 to control expression of genes encoding for proteins that execute tissue-specific metabolic functions in response to this signal. In most cases, these proteins are metabolic enzymes or additional regulators that will change metabolite fluxes within the cell or tissue as well as impact functions of other tissues. Although the specific nutrient signals which the PGC-1α/SIRT1 pathways respond to remain elusive, it seems very likely that a nutrient signal that fluctuates in normal physiology would serve to synchronize the metabolic networks of the entire body. It is therefore very plausible that the metabolic dysregulation that occurs in disease might be due to the inability of the body to couple normal nutrient/hormonal fluctuation to the appropriate metabolic response. It is important also to mention that by investigating and identifying other genes and signals that are upstream or downstream of the PGC-1α and SIRT1 complex might provide novel insights into the pathology of these diseases.
Although we have concentrated in this review at the connection of the PGC-1α and SIRT1 pathways, other functions of these proteins could be independent of each other. In this regard, future studies of how PGC-1α acetylation functions and what sites are specific for different biological activities will also help to understand the implications of this protein complex in metabolic regulation. Furthermore, we also need to take into consideration other members of the PGC-1 family, for instance PGC-1β also targets mitochondrial genes and induces mitochondrial biogenesis. In fact, knock-down of both PGC-1α and PGC-1β results in a collapse in mitochondrial gene expression [49]. Another issue not covered in this review is the possible effects of PGC-1α and SIRT1 in other metabolic tissues or cells. For example, how it might these proteins contribute to energy balance its function in the thyroid or in macrophages that also have considerable metabolic importance.
A major defect in type 2 diabetes and obesity can be defined as a lack of metabolic stability in which the body, tissue and cells are not responsive to normal nutrient or hormonal fluctuations. A clear example is insulin resistance in type 2 diabetes. However, many pathways are also likely to be defective in these complex diseases. For example, old animals do not respond similarly to exercise as young, the blunted response of of AMPK and PGC-1α in old mice could exacerbate any metabolic complications. In addition, dysregulation of glucose and lipid metabolic pathways will completely change intracellular and systemic metabolite levels which will also further compromise the cells to respond to hormonal and nutrient signals appropriately. Related to this problem, it seems that at the “heart” of type 2 diabetes and obesity are inflammatory responses and chronic oxidative stress that leads to ROS production that might additionally complicate the pathology of these diseases. In this context, PGC-1α, SIRT1 and SIRT1 targets, such as FoxO proteins [21] and p53, control ROS formation and are important to maintain survival [20]. Notably, increases in free radical production have been causatively associated with insulin resistance. As it relates to the human genetics, several SNPs have been found in PGC-1α that in certain populations are associated with the different aspects of the metabolic syndrome including diabetes, obesity and hypertension. Recently SNPs for SIRT1 have also been identified in humans that were associated with energy expenditure [26]. Finally, further genetic and metabolic studies in different human populations with different susceptibilities to metabolic diseases will provide more information of how these pathways might be therapeutically exploited to treat these diseases.
Acknowledgments
We thank other members of the Puigserver laboratory for insightful discussions and comments on this manuscript. Work in this laboratory is supported in part by an Ellison Medical Foundation New Scholar Award, the American Diabetes Association, the U.S. Department of Defense, and National Institutes of Health Grant R01 DK069966 (to P.P.). Due to space limitations, we apologize for not including additional information or references that are related to studies discussed in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–80. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 3.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 5.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–49. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 6.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–38. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Gerhart-Hines Z, et al. Metabolic Control of Mitochondrial Function and Fatty Acid Oxidation Through PGC-1alpha/SIRT1. EMBO J. 2007;26:1902–1912. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puigserver P, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–82. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 9.Fan M, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–89. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–6. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 12.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–73. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper MP, Qu L, Rohas LM, Lin J, Yang W, Erdjument-Bromage H, Tempst P, Spiegelman BM. Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev. 2006;20:2996–3009. doi: 10.1101/gad.1483906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 15.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–45. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 16.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 17.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 21.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 22.Pagans S, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 26.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–9. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6:208–16. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 32.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Krebs HA, Hems R. Fatty acid metabolism in the perfused rat liver. Biochem J. 1970;119:525–33. doi: 10.1042/bj1190525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess SC, Leone TC, Wende AR, Croce MA, Chen Z, Sherry AD, Malloy CR, Finck BN. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J Biol Chem. 2006;281:19000–8. doi: 10.1074/jbc.M600050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 Improves Insulin Sensitivity under Insulin-Resistant Conditions by Repressing PTP1B. Cell Metab. 2007;6:307–19. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Koo SH, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10:530–4. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 38.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal Muscle Fiber-type Switching, Exercise Intolerance, and Myopathy in PGC-1{alpha} Muscle-specific Knock-out Animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 39.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 41.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007 doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 44.Alcendor RR, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–21. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 45.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 46.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. Embo J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon JC, et al. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 48.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–41. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzucotelli A, et al. The transcriptional coactivator peroxisome proliferator activated receptor (PPAR)gamma coactivator-1 alpha and the nuclear receptor PPAR alpha control the expression of glycerol kinase and metabolism genes independently of PPAR gamma activation in human white adipocytes. Diabetes. 2007;56:2467–75. doi: 10.2337/db06-1465. [DOI] [PubMed] [Google Scholar]
- 52.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–6. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–52. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]