Abstract
Lucina pectinata is a bivalve mollusk that lives in the Southwestern coast of Puerto Rico and houses intracellular symbiotic bacteria. This peculiar organism contains three types of hemoglobin, each characterized by distinct physico-chemical properties. Hemoglobin I (HbI) is a sulfide-reactive protein that reacts with H2S to form ferric hemoglobin sulfide. In contrast, hemoglobin II and III are oxygen-reactive proteins that remain oxygenated in the presence of hydrogen sulfide. The partial coding region contained in the cDNA sequences we have cloned confirmed the Lucina pectinata HbIII amino sequence reported in the NCBI protein database) with a single amino acid difference (Asn72Asp; AsnE12Asp). The characterization of the full length mRNA coding for Lucina pectinata HbIII revealed an alternative polyadenylation site and an alternate transcription start site. The open reading frame (ORF) of the HbIII cDNA is composed of 459nts containing 153 codons. The initiation codon is preceded by 62 nts of untranslated region (5′UTR), whereas two 3′UTR regions of 640nt and 455nt long were identified, revealing the presence of alternative polyadenylation sites. Isoforms of the 3′UTR of HbIII only differed in the length of their sequences. It has been hypothesized that alternative polyadenylation acts through shortening of mRNA to regulate RNA localization, translation and stability. Interestingly, the HbIII mRNA is the only one of all the hemoglobin mRNAs from Lucina pectinata characterized so far with more than one 3′UTR. Primer extension products suggest two closely located start sites of HbIII mRNA transcription. We suggest that the Lucina pectinata hemoglobin genes may be under different cellular controls that direct them to exert their particular functions. These hypotheses need to be tested by functional studies and analysis of the regulatory elements of the cognate genes for Lucina pectinata hemoglobins.
Keywords: Alternative Polyadenylation, Invertebrate Hemoglobins, Lucina pectinata, mRNA isoform, HbIII
1. Introduction
The bivalve mollusk Lucina pectinata represents a peculiar organism belonging to the Lucinidae family. This bivalve mollusk lives in the Southwestern coast of Puerto Rico and houses intracellular symbiotic bacteria. The intracellular bacteria are considered a symbiotic sulfide-oxidizing chemoautotroph that needs to be supplied with both hydrogen sulfide and oxygen. Lucina pectinata contains three types of hemoglobin, each characterized by distinct physico-chemical properties. These hemoglobins transport oxygen and hydrogen sulfide from water to the bacterial endosymbiont, allowing the bacteria to sustain rapid hexose synthesis and supply the host animal with its entire carbohydrate nutrition (Arp, 1991; Kraus and Wittenberg, 1990). Hemoglobin I (HbI) is a sulfide-reactive protein that reacts with hydrogen sulfide (H2S) to form ferric hemoglobin sulfide (Kraus and Wittenberg, 1990). HbI is a monomeric hemoglobin of 142 amino acid residues. The HbI full-length cDNA sequence consists of 1322 nt (Antommattei et al., 1999). In contrast, Hb II and III are oxygen-reactive proteins that remain oxygenated in the presence of hydrogen sulfide (Kraus et al., 1990). HbII is a dimeric hemoglobin of 151 amino acid residues. The HbII full-length cDNA sequence consists of 2114 nt (Torres et al., 2003). Until now, none of L. pectinata hemoglobin genes have been characterized.
HbII and HbIII have very similar amino acid compositions (percent identity=64%) but differ significantly from HbI (percent identity=32%). The similarities of these hemoproteins were further demonstrated by the kinetics of their reactions with ligands. The association rates of O2 and CO with HbII and HbIII were found to be extremely slow in comparison with other monomeric hemoglobins. Likewise, dissociation rates of O2 from HbII and HbIII were found to be 0.11 and 0.07 s-1, respectively, which are very slow compared to the dissociation rate of this ligand from many other hemoglobins (Kraus and Wittenberg, 1990). The ligand binding kinetics of invertebrate hemoglobins are strongly influenced by the structure of the heme cavity, particularly the size and polarity of residues occupying the distal portion that exert steric and dielectric effects. In many invertebrate globins, the His E7 and Leu B10 residues are replaced by Gln and Tyr, resulting in a tight cage for O2 and much higher O2 binding affinities relative to vertebrate Myoglobin (Mb) (Peterson et al., 1997).
The L. pectinata HbII structure has been elucidated by X-ray crystallography (Gavira J.A. et al., 2006). Computer model predictions of HbIII structure based on its similarity with HbII, suggest that both have Gln E7 and Tyr B10 in the hemo distal pocket. Native HbII and native HbIII are similar in amino acid sequence and the kinetics of their reactions with oxygen. Nevertheless, these similarities may not extend to the mechanisms of oxygen binding. Mechanisms of oxygen binding for HbII have been proposed but the HbIII mechanisms remain to be determined.
In Lucina pectinata, HbIII tends to form dimers, and when it is in an equimolar mixture with HbII, tretramers are formed at high concentrations (>4mM) which suggests that the tetramer (HbII)2(HbIII)2 may exist in tissues (Kraus and Wittenberg, 1990). Several questions may be raised in the Lucina pectinata model system. What is the specific function of each Lucina pectinata hemoglobins in its symbiotic relationship with sulfide-oxidizing chemoautotrophic intracellular bacteria? Why are necessary two oxygen-reactive binding hemoglobins? What are the peculiarities of HbII and HbIII?
It is widely known that similarities at the protein level do not imply similarity at the genomic level. In order to clarify some of these questions, we decided to characterize the full-length mRNA coding for Lucina pectinata HbIII including determination of the length and the nucleotide sequences of the 5′ and 3′ untranslated regions (UTR) of the processed mRNAHbIII using RACE Methods. Surprisingly, some 3′ UTR segments can be more conserved than coding exons reflecting an unexpected selective pressure in this region (Hughes T. A., 2006). Phylogenetic studies of conserved poly (A) sites reported 500 genes with tandem conserved poly (A) sites (Moucadel V. et al., 2007). Moucadel et al. (2007) hypothesized that conservation of specific alternative 3′ends together with specific 3′UTR elements might reflect novel regulatory mechanisms. We hypothesize that the hemoglobin genes from Lucina pectinata may be under different cellular controls that direct them to exert their particular functions.
2. Materials and Methods
2.1 synthesis and cloning of cDNA
Total RNA from L. pectinata was extracted from ctenidias using Trizol reagent in a modification of the RNA isolation method developed by Chomczynski and Sacchi (Chomczynski et al. 1987). After designing degenerate oligonucleotides using the HbIII amino acid sequence reported in the NCBI protein database (GeneBank accession no. P41262), 1 μg of L. pectinata total RNA was reverse transcribed and amplified using the Thermostable rTth Reverse Transcriptase RNA PCR kit (Perkin Elmer). Total RNA was incubated with 1μL of 15 μM HbIIIrev2 primer (Fig. 2C) and 10.4 μL of RNase free water for 10 min at 70°C. Then, a PCR mix containing: 2 μL of 10x enzyme buffer, 2 μL of 10 mM MnCl2 solution, 0.4 μL of each 10 mM dNTP and 2 μL RTth DNA Polymerase (2.5 U/μL) was added and incubated at 55°C for 90 min. Polymerase Chain Reaction (PCR) was performed using the retrotranscriptase reaction products as template as recommended by manufacturer. The sequence of the primer HbIIIfow1 corresponds to amino acid residues NGTNFYM (positions 26-32). The sequence for the primer HbIIIrev2 corresponds to amino acid residues WEDFIAY (positions 136-139). The amplified product of 342 nt in size was cloned in the pCRII Topo vector (Invitrogen, Carlsbad, CA). The presence of the cloned insert was verified by colony PCRs. Plasmids were isolated using QIAprep Spin Miniprep Kits (Qiagen) and sequenced in both strands in an ABI 310 automated DNA sequencer using dye terminator chemistry (Big Dye V3 Dye Terminator Sequencing kit, Applied Biosystems). Analysis of the cDNA sequence was performed using the Basic Local Alignment Sequence Tool (BLAST http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al., 1997).
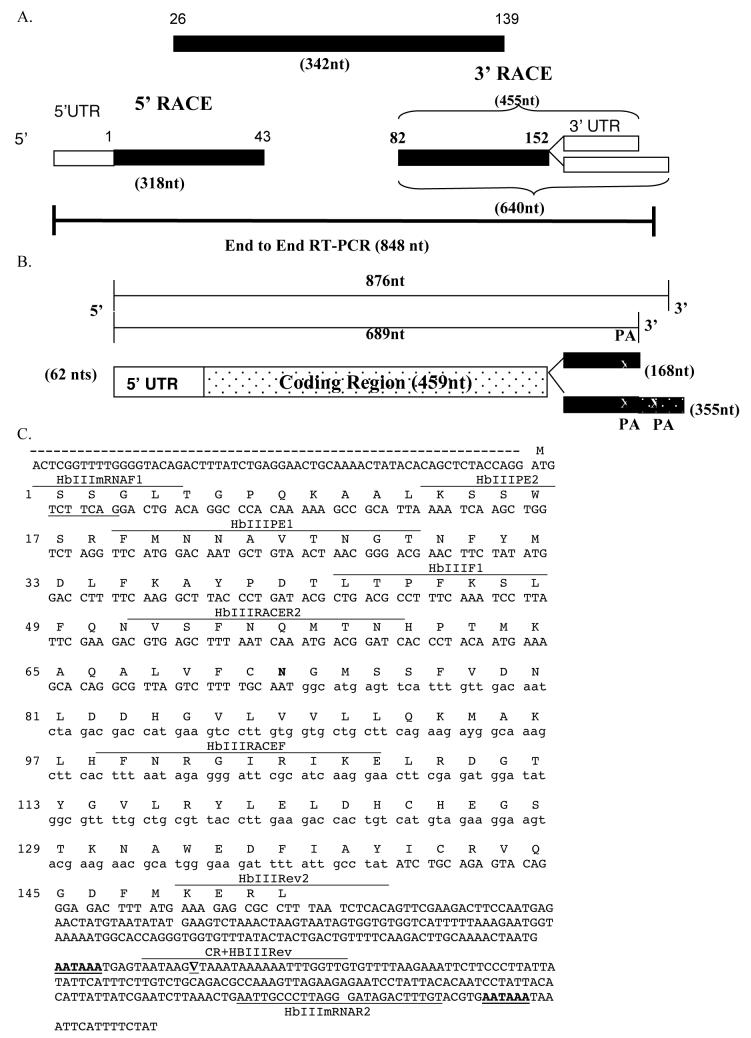
Figure 2. Complete nucleotide sequence for the HbIII cDNA.
The overlapping sequences of degenerate RT-PCR products with the 5′ RACE and 3′ RACE products were used to obtain the complete nucleotide sequence for the HbIII cDNA. Most of the HbIII cDNA sequence (97%) was confirmed by end to end RT-PCR. A. Schematic Organization of cDNA Fragments. An initial 342-nt cDNA clone encoding 114 amino acid residues of HbIII (amino acid 26 to 139) was amplified from total RNA using degenerate oligonucleotides. The 5′ RACE product contained a cDNA sequence encoding 43 amino acid residues of HbIII (amino acids 1 to 43) and 62 nt of the 5′ UTR. The 3′ RACE products contained a cDNA sequence encoding 71 amino acid residues of HbIII (amino acid 82 to 152) and two 3′UTR regions (168 and 355 nt long). The numbers at each end of the boxes identify the amino acid numbers of the HbIII aminoacid sequence included in the amplification product. Numbers in parentheses indicates the length in nucleotides of the cDNA regions indicated. Black boxes represent the coding regions and white boxes the UTRs. B. Schematic diagram of the HbIII cDNA. The untranslated region (5′UTR) is composed of 62 nt whereas two 3′UTR regions were isolated revealing the presence of alternative polyadenylation site. The shortest 3′UTR is composed of 168nt out of 689 nucleotides-long cDNA. The longest 3′UTR (355 nt long) is found in 876 nt-long cDNA. The PA indicates the poly-adenylation signal. Numbers in parentheses indicate the length in nucleotides of the cDNA regions indicated. C. Full length cDNA Sequence and Derived Amino Acid Sequence of Hemoglobin III from Lucina pectinata. A single amino acid difference (Asn72Asp) with the reported Lucina pectinata HbIII amino sequence is shown in bold. The two polyadenylation signals are underlined. The ▽ symbol represents the site where the poly (A)+ tail is added in the smaller cDNA. Primers used in RT-PCR, RACE and Primer Extension experiments are shown.
2.2 synthesis and cloning of cDNA ends
Gene specific primers (GSP) derived from the HbIII partial cDNA sequence were designed using the Primer3 software (http://fokker.wi.mit.edu/primer3/input.htm) to obtain the 5′ and 3′ end of the cDNA by the RACE method. The HbIIIRACER2 (GSP1) was used to obtain the 5′ end and HbIIIRACEF (GSP2) to obtain the 3′ end (Fig. 2C). Rapid Amplification of cDNA ends (RACE) reactions were carried out using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA) as recommended by the manufacturing. The cDNAs synthesized by RACE methods were cloned into the pST Blue-1 vector (Novagen) and sequenced as described in section 2.1.
2.3 primer extension analysis
Two oligonucleotide primers (HbIIIPE1 and HbIIIPE2) (Fig. 2C) complementary to a sequence within 101 and 72 nts downstream of the anticipated 5′ end of mRNA sequence were labeled at the 5′ end with γ 32P-ATP (GE Biosciences) and T4 polynucleotide kinase (New England Biolabs). Primer extension analysis was performed according to protocols published by Cadilla et al. (1992) with minor modifications: the thermostable rTth Reverse Transcriptase RNA PCR kit (Applied Biosystem) was used for RT reaction. (Cadilla et al., 1992).
2.4 end to end RT-PCR
End to end RT-PCR was performed for further confirmation of the coding sequence and untranslated regions as described in section 2.1. The HbIIImRNAF1 and HbIIImRNAR2 primers (Fig. 3C) derived from the overlapping RT-PCR products were used to amplify an 848nt product which was cloned and sequenced.
Figure 3. HbIII cDNA Primer Extension Analysis. (A). Schematic Diagram of Primers Design.
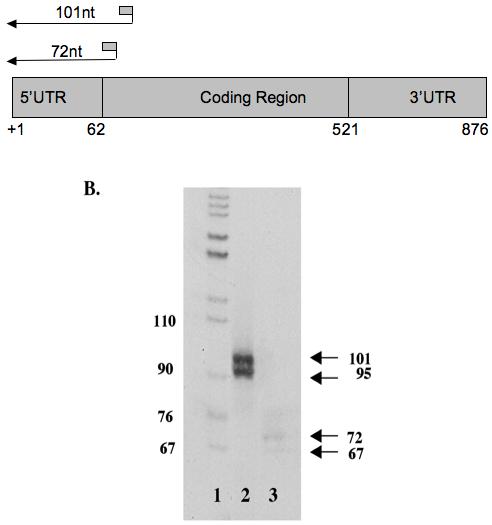
Two oligonucleotide primers complementary to a sequence within 72 and 101 nt downstream of the anticipated 5′ end of mRNA sequence were end labeled and used in primer extension reactions as described in section 2.6. (B) Primer Extension Analysis. 0.5-0.6×106cpm of radiolabeled primer was added to 15 μg of total RNA. Primer extension was done using the rTth Reverse Transcriptase. The radiolabeled cDNA was analyzed through an 8% denaturing polyacrylamide gel and co-electrophoresed with pBR322cut with MspI. Lane 1: 32P end-labeled pBR322 vector cut with MspI, Lane 2: Primer Extension using HbIIIPE1 and Lane 3: Primer Extension using HbIIIPE2. The sizes of fragments in marker and sample lanes are expressed in nucleotides (nt).
2.5 Lucina pectinata HbIII mRNA tissue distribution
RT-PCR was used to determine the tissue distribution of Lucina pectinata HbIII mRNA as described in section 2.1 using the HbIIImRNAF1/HbIIImRNAR2 and HbIIImRNAF1/CR+HbIIIrev primers (Figure 2C). Total RNA from L. pectinata was extracted from mantle, adductor muscle, foot and ctenidia, pericardial sac, digestive gland, gonad and intestines as described in section 2.1. The clam tissues that showed amplification product indicative of HbIII mRNA expression in RT-PCR were examined by the 3′ RACE method to determine the HbIII mRNA variants distribution, as described in section 2.2.
2.6 northern blot analysis
Northern blot analysis was performed by electrophoresis of increasing concentrations of total ctenidia RNA (5μg, 10μg, 20μg) in denaturing agarose gels (Sambrook et al., 1989), capillary transfer to nylon membranes, and hybridization with a 32P-labeled probe prepared by the random primer method. Pre-hybridization was performed in 50% formamide, 5X SSC, 1% SDS, 10X Denhardt solution, and 0.1 mg/ml of denatured salmon sperm DNA, at 42°C for 24 hours. The membrane was hybridized with 1μg of purified 32P-random-primer labeled HbIII cDNA RT-PCR fragment (DNA probe specific activity = 2×109cpm/μg) at 42°C overnight period. Blots were exposed to X-ray film at -80°C for 24 hours. The membrane was deprobed in 50% formamide, 2X SSPE for one hour at 65°C, and then rinsed briefly with 0.1X SSPE at room temperature. The same membrane was re-hybridized with an HbII random-labeled probe as a control for specific hybridization.
3. Results
3.1 full length cDNA sequence
An initial 342 nt cDNA clone encoding 114 amino acid residues of HbIII was amplified from total RNA by RT-PCR using degenerate oligonucleotides (Figure 1A). The partial coding region contained in the cDNA sequences we have cloned matched the Lucina pectinata HbIII amino sequence reported in the NCBI protein database (GenBank accession no. P41262) with only a single amino acid difference (Asn72Asp; AsnE12Asp). The 5′ and 3′ UTRs regions were isolated by RACE using gene specific primers (GSP) derived from the HbIII partial cDNA sequence (Figure 1B and 1C respectively). The complete nucleotide sequence for the HbIII cDNA was obtained by the overlapping of a 342-nts RT-PCR product obtained with degenerate oligonucleotides with the 5′ RACE and 3′ RACE products. The products of the 5′-RACE reaction were 318 nt in length, whereas 3′RACE products were 640 nt and 455nt long. The HbIII cDNA was confirmed by end to end RT-PCR. A total of 848 nt of the cDNA sequence from the 876 nt full length cDNA were amplified separately by RT-PCR using primers derived from the overlapping RT-PCR products for further confirmation of the coding sequence and untranslated regions. Schematic organization of cDNA fragments is shown in figure 2A.
Figure 1. 1% Agarose Gel Electrophoresis of Degenerative RT-PCR and PCR-RACE products.
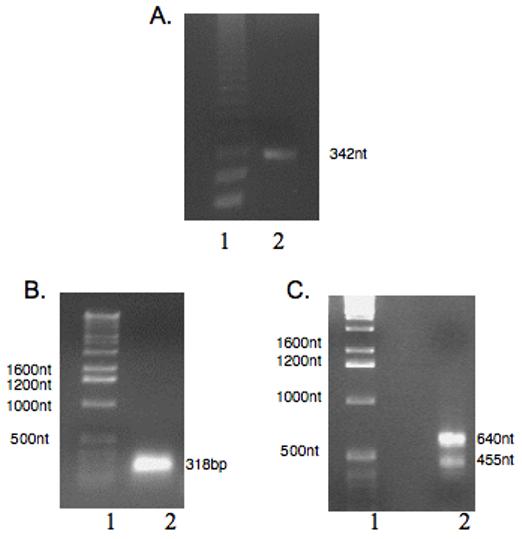
The reverse transcriptase-polymerase chain reaction (RT-PCR and Rapid amplification of cDNA ends (RACE) methods were employed to synthesize the cDNA fragments as described in section 2.2. A) Degenerative RT-PCR product. A 342nt fragment was obtained. Lane 1: Marker, Lane 2: RT-PCR product. B) 5′-RACE Product. A 318 nt fragment was obtained. Lane 1 Marker, Lane 2 5′-RACE Product (C) 3′-RACE products. Two DNA fragments of 640 nt and 455nt were obtained. Lane 1 Marker, Lane 2 3′-RACE Product
The open reading frame (ORF) of the HbIII cDNA is composed of 459 nts containing 153 codons. The HbIII molecular weight estimated from the cDNA-derived amino acid sequence after subtracting the N-terminal methionine is 17,410 Da.. Adding the mass of the heme prosthetic group gives a total molecular weight of 18,068 Da. This data agrees with the molecular weight of HbIII protein based on Edman Degradation (Kraus and Wittenberg, 1990). When this estimated molecular weight is compared with that obtained from MALDI-MS [17,494 Da (±5to10); Sanoguet Z., 1999], a difference of 84 Da is observed, which suggests that the first two N-terminal serine residues may be acetylated. In general, invertebrate globins are amino-acetylated (Dewilde et al., 1998) and it has been proposed that HbI and HbII of L. pectinata are acetylated (Antommattei et. al, 1999; Kraus et al., 1990, Torres et al., 2003).
The initiation codon was preceded by 62 nucleotides of untranslated region (5′UTR) whereas two 3′UTR regions were isolated revealing the presence of alternative polyadenylation site. Isoforms of the 3′ UTR of HbIII only differ in the length of their sequences. The shortest 3′UTR is composed of 168 nt out of 689 nucleotides-long cDNA. This isoform was identified in 27% of colonies screened (3/11). The other longer isoform includes the 168 nt and an additional 187 nt. The longest 3′UTR is found in 876 nt-long cDNA and was identified in 73% of colonies screened (8/11). These 3′ UTR’s are the shortest ones of all the hemoglobins from Lucina pectinata characterized so far (825nt for HbI and 1549nt for HbII). The polyadenylation site is usually located 600-800 nt downstream of the termination codon in mammalian cDNAs. The 3′UTR of the mollusk Yoldia eightsi globin cDNA contains 744 nt in the 3′UTR, excluding the polyadenylation tail (Dewilde et al., 2003) whereas the mollusk Biomphalaria glabrata has an exceptionally long 3′UTR (970 nt) (Dewilde et al., 1998). Nevertheless, the 3′ RACE data showed that polyadenylation signals in the Lucina pectinata HbIII cDNA are located only 156 and 334 nucleotides downstream of the termination codon. The presence of multiple mRNA isoforms is a common phenomenon in eukaryotes but the HbIII mRNA is the only one of all the Lucina pectinata hemoglobins with more than one 3′UTR characterized so far. A schematic diagram of the the Lucina pectinata HbIII cDNA is shown in figure 2B and the full-length cDNA sequence and derived amino acid sequence are shown in figure 2C. The full-length cDNA sequence of the Lucina pectinata HbIII has been submitted to Genbank with the accession number EU040120.
3.2 start site of RNA transcription
The start site of RNA transcription was confirmed by the primer extension method (Figure 3). Two oligonucleotide primers complementary to a sequence within 101 and 72 nt, downstream of the anticipated 5′ end of the HbIII mRNA sequence (HbIII PE1 and HbIII PE2 respectively) were used in these reactions. We obtained two extended products with each primer. When performing the primer extension reactions with the HbIIIPE1, we obtained the expected product of 101 nt and an additional one of 95 nt. With HbIIIPE2, we obtained the expected product of 72 nt and an additional one of 67 nt. These products suggest two start sites of RNA transcription for HbIII, confirming the data obtained by 5′ RACE where the initiation codon was preceded by 62 nucleotides of untranslated region (5′UTR) in 65% of colonies screened (15/23), 22% of colonies screened (5/23) showed a smaller 5′UTR of 51nt and 13% of the colonies (3/5) had very small inserts (5, 18, 38nt). The smaller 5′UTR of 51nt could represent an incomplete RACE product, but the remaining 13% of the colonies could not be confirmed as authentic 5′ ends of the HbIII gene transcripts. These small fragments could be produced during the initial reverse transcriptase step, where the reverse transcriptase is likely to have paused or terminated with low frequency at specific nucleotides before reaching the 5′ends of the transcripts.”.
3.3 Lucina pectinata HbIII mRNA tissue distribution
The tissue distribution of the Lucina pectinata HbIII mRNA was determined using end to end RT-PCR and 3′ RACE methods. Semi-quantitative RT-PCR for 28S rRNA of Lucina which served as an internal loading control, detected similar levels of RNA in each lanes confirming that cDNA was successfully prepared from each of these tissues, and that a similar amount of cDNA was included in each PCR reaction (Figure 4A). RT-PCR detected 673 nt-long amplification product indicative of HbIII mRNA expression in mantle, adductor muscle, foot, pericardial sac and ctenidia, but HbIII mRNA was not detected in the digestive glands, gonads and intestine (Figure 4B). The clam tissues that showed the highest levels of amplification products, indicative of HbIII mRNA expression, were examined by the 3′ RACE method to determine the HbIII mRNA variants distribution. Analysis of the 3′ RACE reaction products detected the two mRNA variants (640 and 455nt-long RACE products) in mantle, adductor muscle, foot and ctenidia (Figure 4C).
Figure 4. L pectinata HbIII mRNA tissue distribution.
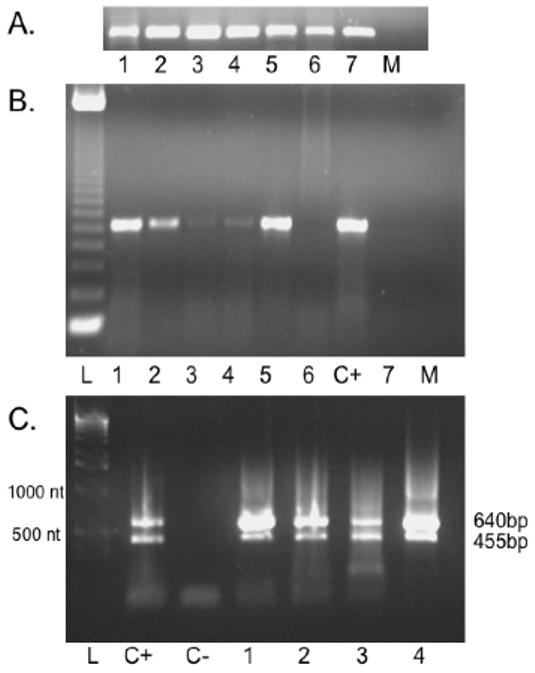
A. RT-PCR for 28S rRNA. Internal loading control detected similar levels of starting RNA in each lanes B. RT-PCR products. The tissue distribution of the Lucina pectinata HbIII mRNA was determined using RT-PCR using the HbIIImRNAF1 and HbIIImRNAR2 primers to amplify a 673 bp cDNA fragment. Lane L: 123 bp DNA ladder, Lane 1: mantle, Lane 2: adductor muscle, Lane 3: foot, Lane 4: pericardial sac, Lane 5: ctenidia, Lane 6: gonad and intestine, Lane C+: Positive Control (ctenidia RNA), Lane 7: digestive gland, Lane M: Mock (human mRNA) C. 1% Agarose Gel Electrophoresis of 3′ PCR-RACE products. RACE reactions were carried out as described in section 2.2. Lane L:1 kb bp DNA Ladder, Lane C+: Positive control, Lane C-: Negative control (no RNA), Lane 1: mantle, Lane 2: adductor muscle, Lane 3: foot, Lane 4: ctenidia
3.4 northern blot hybridizations
The Northern Blot hybridization revealed a single band of approximately 1.1 Kb for the HbIII mRNA (Figure 5). The smaller HbIII cDNA detected by RT-PCR and RACE was 689 nt long. This cDNA is too small to correspond to the band observed in Northern blot, even if we assume it has a ca. 200 bp long Poly A+ tail. The longest cDNA was 876 nt long, without taking into account the Poly A+ tail. The 876 nt long HbIII mRNA detected by RT-PCR and RACE correlates with the 1.1 Kb band revealed by Northern blot if we infer that the HbIIImRNA had a poly A+ tail of approximately 228 nt. The smaller cDNA variant was undetectable in Northern blot. This suggests that the longest mRNA variant is the predominant isoform in ctenidia, consistent with the results obtained in the 3′RACE reactions for ctenida and mantle, which showed a more intense signal for the longer isoform (Figure 4C) (Tian B., 2005 and Legendre M., 2003).
Figure 5. Northern Blot Analysis.
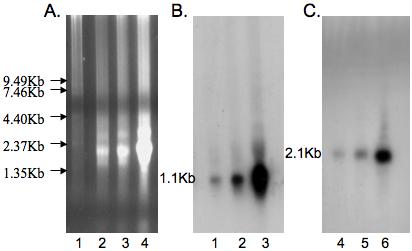
(A) Denaturing gel electrophoresis of total L. pectinata RNA. Lane 1: RNA ladder, Lane 2: 5 μg total RNA, Lane 3: 10 μg total RNA and Lane 4: 20 μg total RNA. B. HbIII Northern Blot analysis. The membrane was hybridized with a 32P- labeled 342 nt HbIII cDNA RT-PCR fragment. The estimated size of the HbIII mRNA was 1.1Kb. Increasing concentration of total RNA were used; Lane 1: 5 μg, Lane 2: 10 μg and Lane 3: 20 μg. C. HbII Northern Blot analysis. The same membrane was re-hybridized with a 32P labeled HbII cDNA RT-PCR fragment as a control for specific hybridization. The expected 2.1 Kb band was shown for HbII. Increasing concentration of total RNA were used; Lane 1: 5 μg, Lane 2: 10 μg and Lane 3: 20 μg.
4. Discussion
The full-length cDNA sequence we have cloned confirmed the Lucina pectinata HbIII amino sequence reported in the NCBI protein database (P41262) with a single amino acid difference (Asn72Asp; AsnE12Asp). The characterization of the full length mRNA coding for Lucina pectinata HbIII revealed an alternative polyadenylation site and alternate transcription start sites. Although the presence of multiple transcription mRNA isoforms is a common phenomenon in eukaryotes, the HbIII mRNA is the only one of all the hemoglobins from Lucina pectinata with alternate transcription start and polyadenylation sites reported so far. Isoforms of the 3′ UTR of HbIII only differ in the length of their sequences. The mRNA isoforms that differ only at their 5′ and 3′ ends and do not vary in their non-coding regions may cause differences in stabilities, RNA localization and translation rates (Moucadel et al., 2007). RT-PCR and 3′ RACE methods were used to detect expression of the HbIII transcripts in several tissues. The existence of two HbIII gene transcripts with different 3′UTR lengths were confirmed by the 3′ RACE method using total RNA from mantle, adductor muscle, foot and ctenidia. RT-PCR detected 673 nt-long amplification product indicative of HbIII mRNA expression in those tissues that are in contact with the hemolymph (mantle, adductor muscle, foot, pericardial sac and ctenidia) but not in internal tissues (digestive glands, gonads and intestine). It has been hypothesized that alternative polyadenylation acts through shortening of mRNA to regulate RNA localization, translation and stability (Tian B. et al., 2005). Among the Lucina pectinata hemoglobin mRNA, HbIII has the shortest 3′UTRs (168 or 355nt) whereas the HbII 3′UTR is markedly longer (1549nt.). The 3′UTR of HbI is 825nt long. As we mentioned before, Moucadel and collaborators studied phylogenetically conserved poly (A) sites and reported 500 genes with tandem conserved poly (A) sites (Moucadel et. al 2007). Some 3′ UTR segments can be more conserved than coding exons, reflecting an unexpected selective pressure in this region (Hughes, 2006). Moucadel hypothesized that conservation of specific alternative 3′ends together with specific 3′UTR elements might reflect novel regulatory mechanisms. Interestingly, the mRNA sizes of HbII and HbIII only differ in the length of their 3′UTRs since the 5′UTR’s and coding regions of the HbII and HbIII mRNA are identical in sizes (62 and 459 nt respectively) although not in sequence. The coding regions of HbII and HbIII show a high sequence identity (75%), but not the 3′ UTRs. The 5′ UTR and coding region of HbI are shorter (42 and 429nt respectively). We hypothesize that the Lucina pectinata hemoglobin genes may be under different cellular controls that direct them to exert their particular functions. This hypotheses need to be tested by functional studies and analysis of the regulatory elements of the cognate genes for L. pectinata hemoglobins.
We used the UTResource (http://www.ba.itb.cnr.it/UTR/) to identify potential regulatory regions present in the two variants of HbIII 3′ UTR. One internal ribosome entry sites (IRES) was detected in the smaller variant but not in the longer variant other. IRES are sequences usually in the 5′UTR of some mRNA that enable end-independent initiation to occur. Although the IRES is located more frequently in the 5′UTR, the literature reported a translation-enhancing activity of the 3′UTR of the mRNA that codes for the ß-subunit of mitochondrial H+-ATP synthase, this functionally resembling an IRES (Izquierdo J. M. and Cuezva J. M., 2000). Izquierdo and Cuezva suggested that the 3′UTR of ß-mRNA could behave very similarly to an IRES, despite is opposite positioning within the sequence of the mRNA. Analysis of the HbIII mRNA sequences using the RNA analyzer at http://wb2x01.biozentrum.uni-wuerzburg.de/ (Bengert and Dandekar, 2003) detected two snRNP binding motifs at bases 5-12 and 811-818 and a stem GG pair from bases 10-39 in the longest mRNA. The shorter mRNA had the same sequence motifs with the exception of the second snRNP binding motif.
Lucina pectinata is an unusual species because it is lives deeply burrowed in black reducing mud of mangrove swamps and it is the most resistant to stagnant conditions that imply anaerobic conditions (Read, 1965). It has been reported that initiation mediated by IRES allows translation under physiological circumstances when translation of most mRNAs is repressed, including hypoxia (Hellen C. and Sarnow P., 2001). Additional studies are necessary to reveal the relevance of the IRES in the 3′UTR of HbIII from L. pectinata.
Primer Extension reaction products suggest two start sites of HbIII mRNA transcription. The absence of a TATA Box in the HbIII gene promoter could cause that more than one start site of transcription be used. These hypotheses need to be tested by functional studies and analysis of the regulatory elements of the cognate genes for Lucina pectinata hemoglobins.
Acknowledgements
We want to thank Mrs. Lilliam Villanueva of the UPR MSC RCMI Molecular Biology Facility for DNA Sequencing Services. This research was funded in part by NCRR RCMI grant G12RR03051 and MBRS RISE grant R25GM061838.
abbreviations
- AMV
Avian Myeloblastosis Virus
- ATP
adenosine 5′-triphosphate
- bp
base pair
- cDNA
complementary deoxyribonucleic acid
- CO
Carbon dioxide
- cpm
counts per min
- Da
dalton
- EDTA
ethylenediamine tetraacetic acid
- FTIR
Fourier transform infrared spectroscopy
- GSP
gene specific primers
- HbI
Hemoglobin I
- HbII
Hemoglobin II
- HbIII
Hemoglobin III
- MALDI-MS
Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry
- Mb
myoglobin
- mM
millimolar
- mRNA
Messenger Ribonucleic Acid
- NCBI
National Center for Biotechnology Information
- nt
nucleotides
- ORF
open reading frame
- PCR
Polymerase Chain Reaction
- RACE
Rapid Amplification of cDNA ends
- RPM
Revolutions per minute
- RT-PCR
reverse transcriptase- Polymerase Chain Reaction
- SDS
Sodium dodecyl sulfate
- 3′ UTR
3′-untranslated mRNA region
- 5′-UTR
5′-untranslated mRNA region
- UV
Ultraviolet-visible
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antommattei F, Rosado T, Cadilla CL, López-Garriga J. The cDNA Derived Amino Acid Sequence of Hemoglobin I from Lucina pectinata. J. Prot. Chem. 1999;18(8):831–836. doi: 10.1023/a:1020623011363. [DOI] [PubMed] [Google Scholar]
- Arp AJ. The role of heme compounds in sulfide tolerance in the echiurian worm Urechis caupo. In: Vinagradov SN, Kapp OH, editors. Structure and Function of Invertebrate Oxygen Carries. Springer-Verlag; New York: 1991. pp. 337–346. [Google Scholar]
- Bengert P, Dandekar T. A software tool-box for analysis of regulatory RNA elements. Nucleic Acids Res. 2003;31(13):3441–3445. doi: 10.1093/nar/gkg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadilla C, Isham KR, Lee KL, Ch’ang LY, Johnson AC, Kenney FT. Insulin increases transcription of rat gene 3 through cis-acting elements in 5′-flanking DNA. Gene. 1992;118(2):223–229. doi: 10.1016/0378-1119(92)90192-r. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dewilde S, Winnepenninckx B, Arndt MHL, Nascimento DG, Santoro MM, Kinght M, Miller a. N., Kerlavage AR, Geoghagen N, Van Marck E, Liu LX, Weber RE, Moens L. Characterization of the Myoglobin and Its Coding Gene of the Mollusc Biomphalaria glabrata. J. Biol. Chem. 1998;273(22):13583–13592. doi: 10.1074/jbc.273.22.13583. [DOI] [PubMed] [Google Scholar]
- Dewilde S, Anelini E, Kiger L, Marden MC, Beltramini M, Salvato B, Moens L. Structure and function of the globin and globin gene from the Antarctic mollusk Yoldia eightsi. Biochem. J. 2003;370:245–253. doi: 10.1042/BJ20020727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira JA, de Jesus W, Camara-Artigas A, López-Garriga J, García-Ruiz JM. Capillary crystallization and molecular-replacement solution of haemoglobin II from the clam Lucina pectinata. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62(Pt 3):196–9. doi: 10.1107/S1744309106002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Cuezva JM. Internal-ribosome-entry-site functional activity of the 3′ untranslated region of the mRNA for the ß subunit of mitochondrial H+-ATP synthase. Biochemistry Journal. 2000;348:849–855. [PMC free article] [PubMed] [Google Scholar]
- Kraus D, Wittenberg J. Hemoglobins of the Lucina pectinata/Bacteria Symbiosis I. Molecular properties, kinetics and equilibria of reactions with ligands. J. Bio. Chem. 1990;265(27):16043–16053. [PubMed] [Google Scholar]
- Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moucadel V, Lopez F, Ara T, Benecha P, Gautheret D. Beyond the 3′end: experimental validation of extended transcript isoforms. Nucl. Ac. Res. 2007:1–11. doi: 10.1093/nar/gkm062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ES, Huang S, Wang J, Miller LM, Vidugiris G, Kloek AP, Goldberg DE, Chance MR, Wittenberg JB, Friedman JM. A comparison of functional and Structural Consequences of the Tyrosine B10 and Glutamine E7 Motifs in Two Invertebrate hemoglobins (Ascaris suum and Lucina pectinata) Biochemistry. 1997;36:13110–13121. doi: 10.1021/bi971156n. [DOI] [PubMed] [Google Scholar]
- Read KR. The characterization of the hemoglobins of the bivalve mollusc Phacoides pectinatus (Gmelin) Comp Biochem Physiol. 1965;15(2):137–157. doi: 10.1016/0010-406x(65)90342-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab.; Cold Spring Harbor, NY: 1989. pp. 2.108–2.125. [Google Scholar]
- Sanoguet Z. MS Thesis. University of Puerto Rico; Mayaguez Campus: 1999. Isolation and Purification of Lucina pectinata Hemoglobins using Preparative Isoelectric Focusing. [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E, Renta JY, Rodríguez Y, López-Garriga J, Cadilla CL. The cDNA Derived Amino Acid Sequence of Hemoglobin II from Lucina pectinata. J. Prot. Chem. 2003;22(78):683–690. doi: 10.1023/b:jopc.0000008734.44356.b7. [DOI] [PubMed] [Google Scholar]