Abstract
CCN5 is a secreted heparin- and estrogen-regulated matricellular protein that inhibits vertebrate smooth muscle cell proliferation and motility. CCN5 is expressed throughout murine embryonic development in most organs and tissues. However, after embryonic development is complete, we hypothesized that CCN5 distribution would be largely restricted to small set of tissues, including smooth muscle cells of the arteries, uterus, airway, and digestive tract. Because CCN5 inhibits proliferation of smooth muscle cells in vitro, it might function to prevent excessive growth in vivo. In contrast, another member of the CCN family, CCN2, promotes smooth muscle cell proliferation in vitro, and thus it was expected that its expression levels would be low in uninjured normal adult tissues. Frozen sections from adult tissues and organs were analyzed immunohistochemically using anti-CCN5 and anti-CCN2 antibodies. Both proteins were detected in arteries, the uterus, bronchioles, and the digestive tract as expected, and also in many other tissues including the pancreas, spleen, liver, skeletal muscle, ovary, testis, thymus, brain, olfactory epithelium, and kidney. CCN5 and CCN2 protein was found in smooth muscle, endothelial cells, epithelial cells, skeletal muscle, cells of the nervous system, and numerous other cell types. In many cells, both CCN5 and CCN2 was present in the nucleus. Rather than having opposite patterns of localization, CCN5 and CCN2 often had similar sites of expression. The wide distribution of both CCN5 and CCN2 suggests that both proteins have additional biological functions beyond those previously identified in specific cellular and pathological models.
Keywords: CCN family, CCN5, CCN2, CTGF, WISP-2, Nuclear localization, Matricellular, Rodents, Tissues, Immunohistochemistry
Introduction
The CCN family consists of six highly conserved, cysteine-rich cell surface and extracellular matrix (ECM)-associated proteins (Brigstock 2003; Perbal 2004). CCN proteins regulate cell proliferation and motility, as well as numerous other biological processes. They have been demonstrated to have key roles in a variety of pathophysiologic models (Brigstock et al. 2005). With the exception of CCN5, all CCN proteins have a distinctive sequence consisting of four polypeptide modules: (1) Insulin-Like Growth Factor-Binding Protein (IGF-BP), (2) von Willebrand Factor-C (VWC), (3) thrombospondin-1 (TSP1), and (4) a carboxy-terminal (CT) domain. CCN proteins are typically present on the surface of the cells that secreted them and they have been called matricellular proteins because of their strong association with the ECM. Some might serve as ECM-bound ligands for integrin-mediated signaling between the cells and the matrix (Brigstock et al. 2005).
The CCN5 gene, known previously as Cop-1 (Delmolino et al. 1997, 2001), rCop-1 (Zhang et al. 1998), Wisp-2 (Pennica et al. 1998), and CTGF-L (Kumar et al. 1999), encodes the only CCN protein that lacks the CT domain (Gray and Castellot 2004). Because of this structural difference, it was expected that many functions of CCN5 would be significantly different from those mediated by the four-domain CCN proteins such as CCN2. In some model systems, CCN5 seems to have opposite or complementary functions to those of the other CCN proteins, leading to the notion that CCN5 is an antagonist (either pharmacologically or biologically) of the four-domain CCN proteins (Gray and Castellot 2004).
Previous studies of animal tissues have revealed that CCN5 is normally expressed in the rat uterus in smooth muscle, glandular epithelium, and the endometrium (Mason et al. 2004), rat arterial smooth muscle and endothelium (Lake et al. 2003), and human fetal bone (Kumar et al. 1999). Studies of CCN5 mRNA expression by Northern blot, in situ hybridization (ISH), and quantitative polymerase chain reaction (Q-PCR) indicate that additional sites of CCN5 mRNA expression might include adult brain, heart, lung, colon, prostate gland, testis, ovary, skeletal muscle, and most cell types in bone (Delmolino et al. 2001; Kumar et al. 1999; Pennica et al. 1998).
The CCN2 gene encodes a four-domain protein also known as connective tissue growth factor (CTGF) (Moussad and Brigstock 2000). CCN2 is mitogenic for fibroblasts, smooth muscle cells, vascular endothelial cells (Bradham et al. 1991), hepatic stellate cells (Gao et al. 2004), chondrocytes, and osteoblasts (Takigawa et al. 2003). In addition to proliferation, CCN2 also promotes cellular adhesion, migration (Babic et al. 1999), chemotaxis, and production of ECM molecules (Tong and Brigstock 2006). CCN2 has a role in tissue growth and regeneration, and has been studied extensively in skeletal and vascular systems. Over-expression of CCN2 has been implicated in pathological disorders, including angiogenesis in tumors (Rachfal et al. 2004; Ivkovic et al. 2003), atherosclerosis (Cicha et al. 2005), and fibrotic diseases (Leask 2004). CCN2 is normally expressed in vascular endothelial cells, osteoblasts, hypertrophic chondrocytes, brain, kidney, and numerous tissues in the embryo (Takigawa et al. 2003; Ivkovic et al. 2003; Friedrichsen et al. 2005).
To further describe the biological functions of the CCN5 protein, it is necessary to determine its normal protein distribution in vivo. The earlier studies of CCN5 expression patterns were based largely on Northern blot analyses of RNA samples prepared from isolated vertebrate tissues. Because CCN proteins are secreted, the pattern of mRNA expression might not match that of protein distribution. It is also possible that the degradation rate of the CCN5 protein is very slow, and CCN5 might persist in tissues long after the mRNA is destroyed. Thus, in order to fully describe CCN5 expression patterns, we examined CCN5 protein distribution in diverse rodent tissues using immunohistochemistry. We expected that (1): CCN5 protein would be found mostly in smooth muscle, glandular epithelium, and endothelial cells, and only slightly in other tissues. (2): CCN2 protein expression would be highest in endothelial cells, bone, cartilage, and connective tissue, and much lower in other tissues. (3): Because the biological effects of CCN5 expression have been observed to be opposite to that of CCN2, the two proteins would have opposite and non-overlapping patterns of expression. In contrast to our expectations, the immunohistochemistry results demonstrated that: (1) both CCN2 and CCN5 proteins are widely distributed in all tissues tested in both rat and mouse, (2) CCN2 and CCN5 have overlapping and very similar patterns of distribution, and (3) many cell types demonstrate intra-nuclear localization of both CCN5 and CCN2.
Materials and methods
Animals
Animal use protocols were reviewed and approved by the Institutional Animal Care and Use Committee at Tufts University. Albino Sprague–Dawley rats (200–225 g) and C57BL6/J mice were obtained from Charles River Laboratories (Wilmington, MA). Animals were maintained on a 14 h light and 10 h dark cycle with lights on at 0400 h and off at 1800 h. Food and water were available ad libitum. Prior to collection of tissues, animals were euthanized by carbon dioxide overdose followed by thoracotomy. Some mice were euthanized by thoracotomy after deep anesthesia with sodium pentobarbital as described below.
Tissue samples
Tissue samples and organs were dissected out and rinsed in PBS, and then fixed for 2–4 h in 4% paraformaldehyde in 0.05 M sodium phosphate buffer, pH 7.2. To obtain better fixation for some tissues (heart, kidney, lung, liver), some mice were deeply anesthetized with sodium pentobarbital and then perfused intracardially with PBS followed by 4% paraformaldehyde. Tissues and organs were then dissected out and post-fixed for 1 h in 4% paraformaldehyde. After fixation, tissues were rinsed in PBS, cryoprotected in 30% sucrose/PBS, and then frozen in OCT embedding medium (Tissue-Tek) on dry ice. Frozen tissue samples were sectioned (7 μM) using a Leica CM3050S cryostat and collected onto Fisher brand Plus slides and stored at −20°C. We analyzed frozen sections instead of sections embedded in paraffin to avoid any potential blocking of CCN protein antigens that could occur during dehydration and heating.
Immunohistochemistry
Immunohistochemical analysis was performed using the Vectastain Elite ABC kit (PK 6101; Vector Laboratories; Burlingame, CA). Slides were incubated with anti-CCN5 (1:500 in PBS; affinity purified polyclonal antibody to a rat peptide consisting of amino acids 103–117 of the CCN5 VWC domain) and anti-CCN2 (1:500 in PBS, affinity-purified polyclonal antibody to a mouse peptide that includes amino acids 223–348 of the CCN2 protein; ab6992; Abcam, Inc.; Cambridge, MA) overnight at 4°C. The anti-CCN5 antibody has been well-characterized in previous experiments (Lake et al. 2003; Mason et al. 2004). Its specificity has been verified by Western blot analysis, in which a prominent 28 kDa band is revealed in growth-arrested rat smooth muscle cells and in rat smooth muscle cells transfected with recombinant adenovirus that express the full-length rat CCN5 cDNA sequence (Lake et al. 2003). A second anti-CCN5 antibody against a rat peptide from the IGF-BP domain produced the same Western blot results as the anti-CCN5 antibody used in the present experiments (data not shown). The specificity of the CCN2 antibody used in the present experiments has been verified by Western blot analysis, in which a prominent 38 kDa band is detected in human fibroblasts and neurons (Ueberham et al. 2003).
After incubation with the secondary antibody (biotinylated goat anti-rabbit IgG) and the ABC reagent (30 min each at room temperature), and a 5-min staining with the DAB peroxidase substrate reagent (SK 4100; Vector Laboratories; Burlingame, CA), the slides were counterstained with Harris modified hematoxylin with acetic acid (Fisher) for 1 min. All slides were dehydrated and embedded in permanent mounting medium (#13510; DPX Mountant; Electron Microscopy Sciences; Hatfield, PA) and photographed using a microscope (Zeiss Axioscope) and a digital camera system (SPOT; Diagnostic Instruments). The magnification used for all examples was 200×, except Figs. 10 and 13 (630×).
Fig. 10.
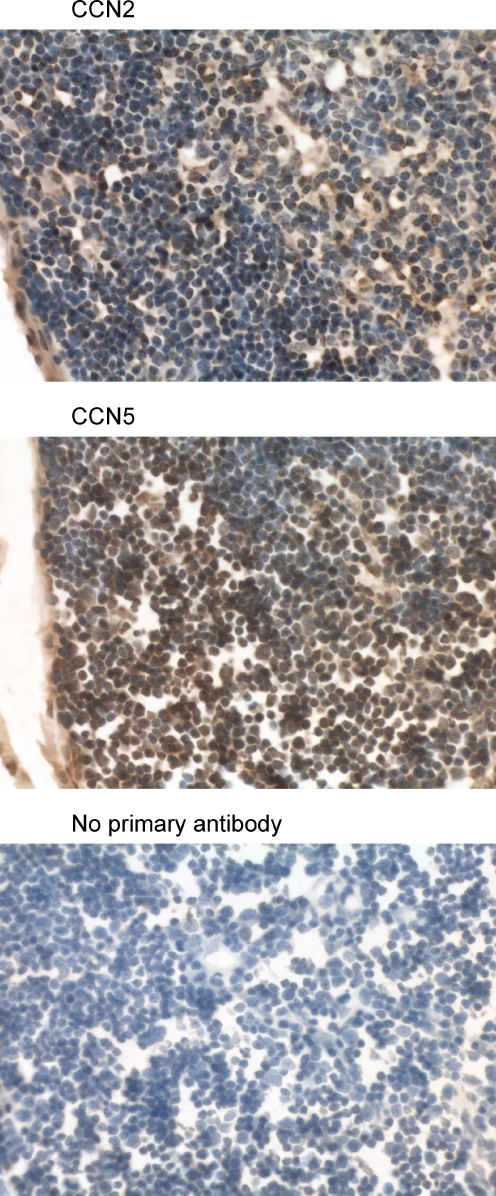
Immunohistochemical analysis of the mouse thymus using anti-CCN2 and anti-CCN5. The tissue sections (7 μm transverse) reveal numerous lymphocytes in the cortex and medulla. The nuclei are very large and almost completely fill each cell. Many lymphocytes in the mouse thymus demonstrate nuclear localization of both CCN2 and CCN5. (Magnification = 630×)
Fig. 13.
Nuclear localization of CCN2 and CCN5 in mouse and rat cells. The indicated tissue sections (7 μm parasagittal) were incubated with anti-CCN2 or anti-CCN5 and photographed at a magnification of 630×
Results
Immunohistochemistry analysis revealed CCN5 and CCN2 staining in numerous cell types of most tissues and organs analyzed, in both mouse and rat. The staining patterns of mouse tissue samples stained with anti-CCN5 were very similar to those obtained with anti-CCN2 (Figs. 1, 2, 4, 5, 6, 7, 10, 11, 13; Table 1). Rat tissues had very similar patterns of CCN5 and CCN2 staining (Figs. 3, 8, 9, 12, 13; Table 2).
Fig. 1.
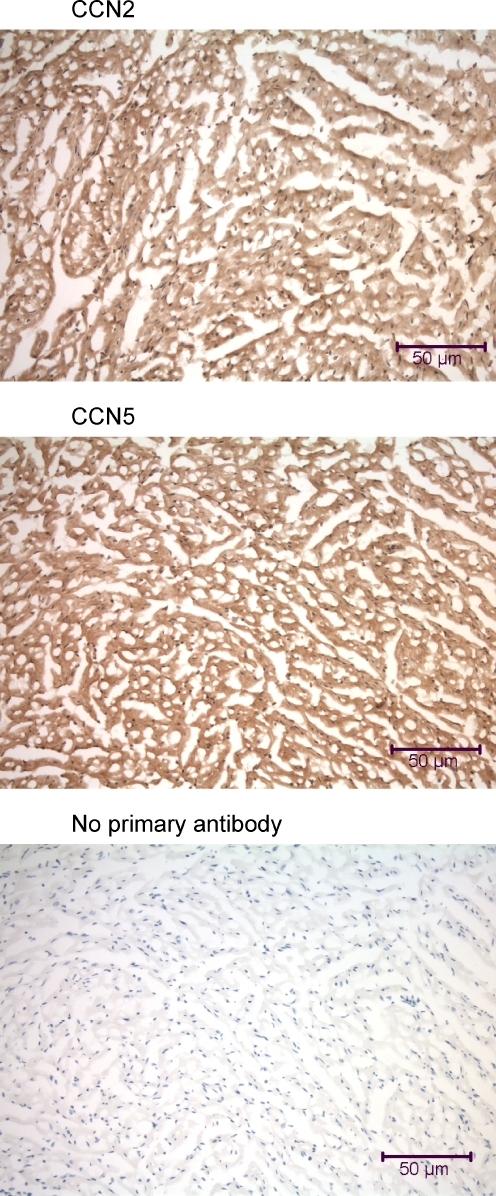
Immunohistochemical analysis of the mouse heart using anti-CCN2 and anti-CCN5. Left ventricular myocardium samples from a mouse were fixed by intracardial perfusion immediately post-mortem. The 7 μm frozen tissue sections are transversely oriented before incubation with anti-CCN2 and anti-CCN5. Some myocardial cells demonstrate nuclear localization of both CCN2 and CCN5
Fig. 2.
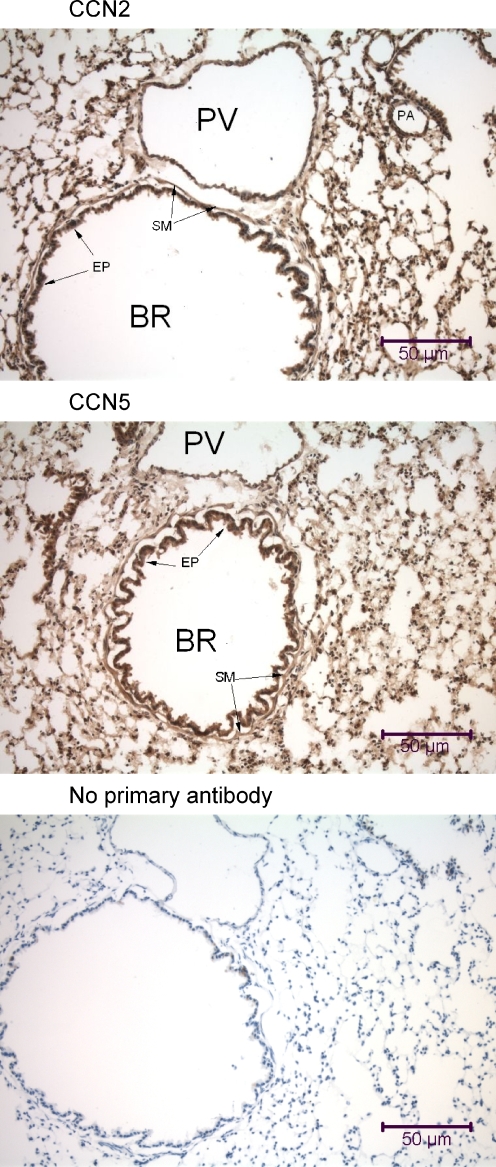
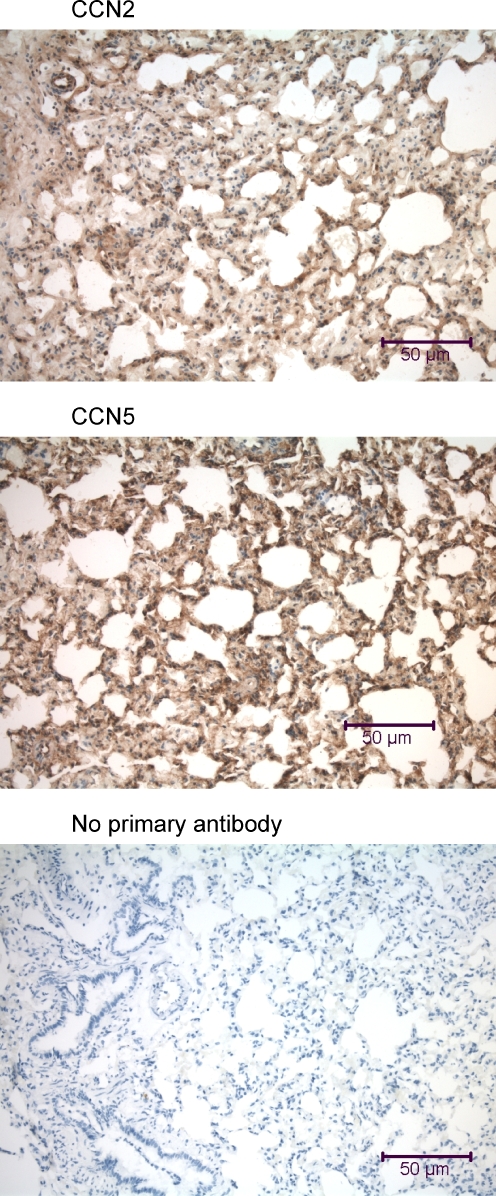
Immunohistochemical analysis of the mouse lung using anti-CCN2 and anti-CCN5. Lung samples were obtained from a mouse that was fixed by intracardial perfusion immediately post-mortem. In each panel is a transversely-oriented 7 μm lung tissue section that reveals a large bronchiole (BR) with a prominent lumenal columnar epithelium (EP) surrounded by a thin layer of circumferential smooth muscle (SM), large thin-walled pulmonary veins (PV), some small pulmonary arteries (PA), and numerous alveolar air sacs. All types of cells in the mouse lung demonstrate some nuclear localization of both CCN2 and CCN5
Fig. 4.
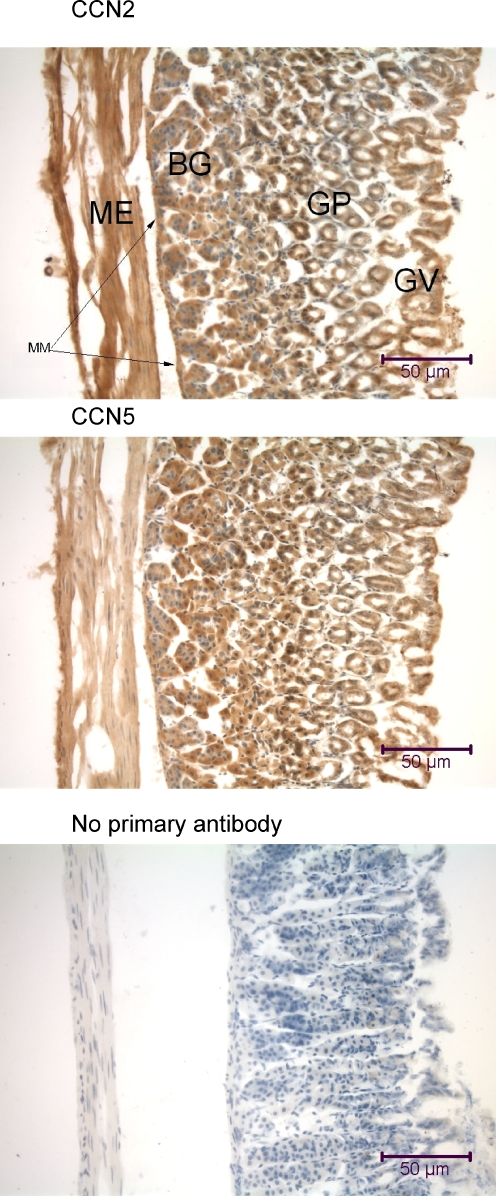
Immunohistochemical analysis of the mouse stomach wall using anti-CCN2 and anti-CCN5. The 7 μm transversely-oriented tissue sections reveal (from left to right) the muscularis externa (ME) and muscularis mucosae (MM) smooth muscle layers, secretory Brunner’s glands (BG), crypts of Lieberkuhn or gastric pits (GP), gastric villi (GV), and surface mucous cells. Cells at the base of the gastric pits demonstrate nuclear localization of both CCN2 and CCN5
Fig. 5.
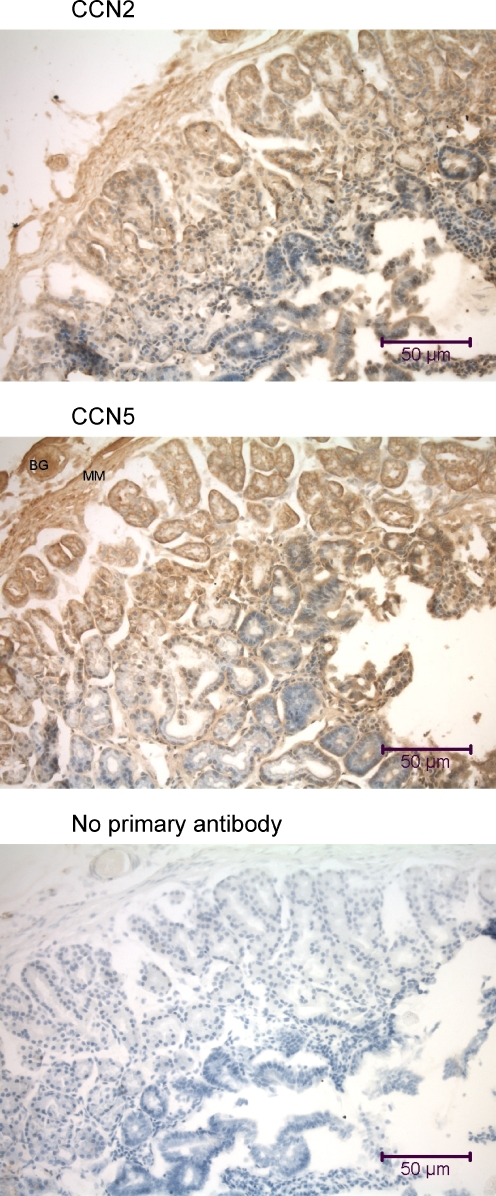
Immunohistochemical analysis of the mouse duodenum using anti-CCN2 and anti-CCN5. The tissue sections (7 μm oblique) include (from left to right) submucosal Brunner’s glands (BG), muscularis mucosae (MM; a smooth muscle layer), and the mucosa with numerous villi and pits. Some cells in the villi demonstrate nuclear localization of CCN2 and CCN5
Fig. 6.
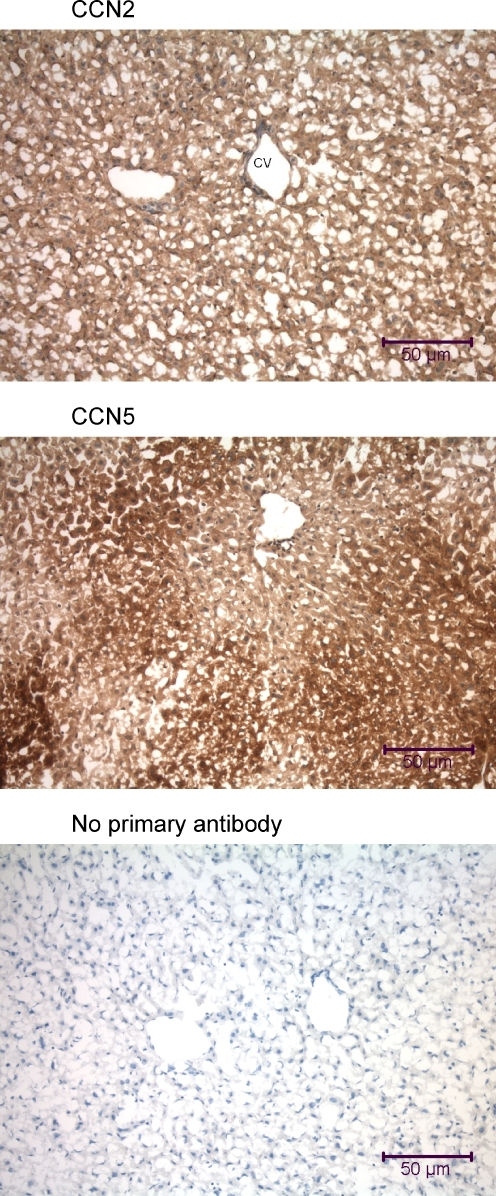
Immunohistochemical analysis of the mouse liver using anti-CCN2 and anti-CCN5. Liver samples were obtained from a mouse that was fixed by intracardial perfusion immediately post-mortem. The tissue sections (7 μm transverse) display cords of hepatocytes surrounded by sinusoids, large central veins (CV), and smaller portal vein branches
Fig. 7.
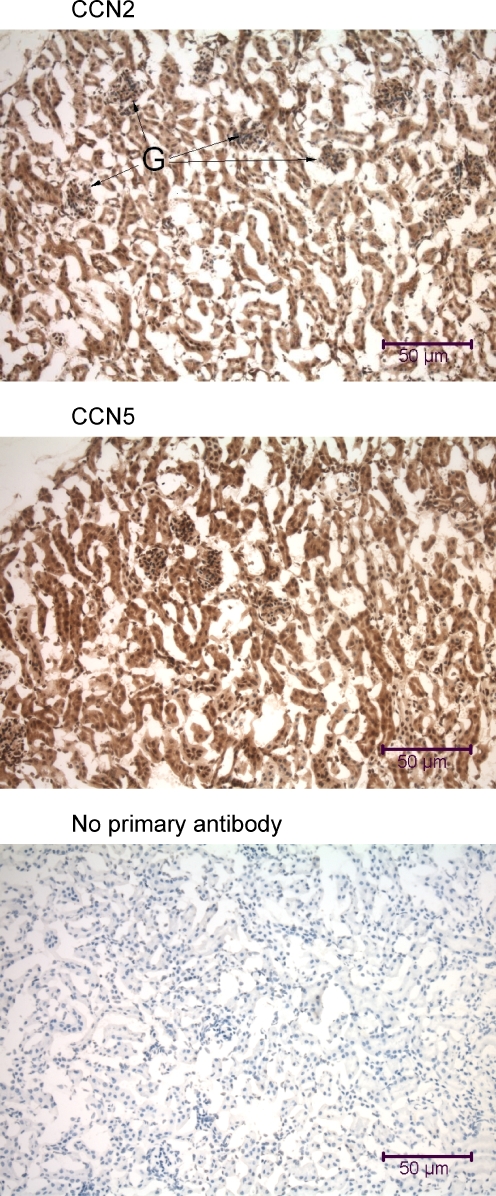
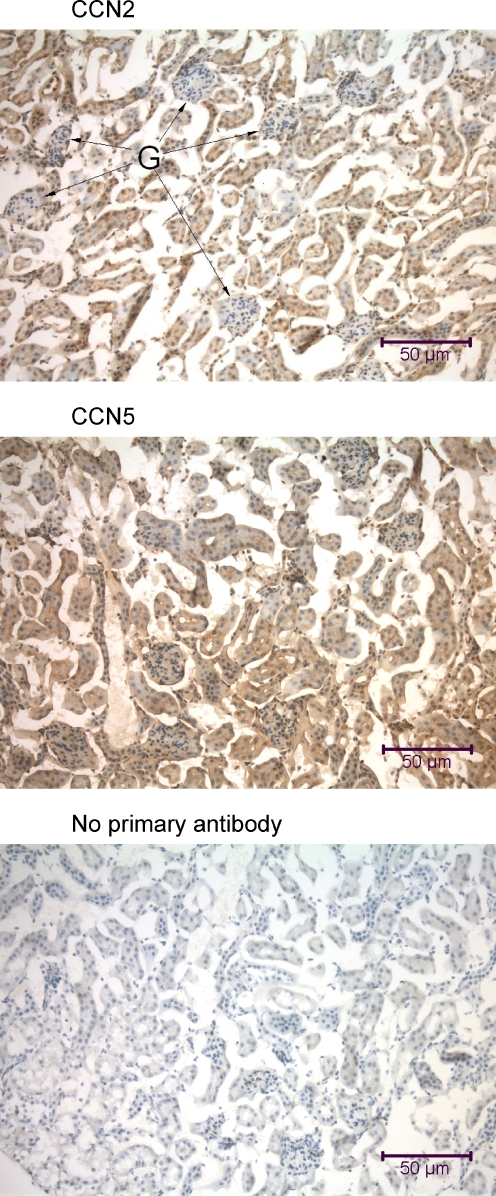
Immunohistochemical analysis of the mouse kidney using anti-CCN2 and anti-CCN5. Kidney samples were obtained from a mouse that was fixed by intracardial perfusion immediately post-mortem. The tissue sections (7 μm parasagittal) are from the renal cortex and include renal corpuscles with Bowman’s capsules and glomeruli (G) interspersed within a dense network of convoluted tubules. Many of the cells in the mouse kidney demonstrate nuclear localization of CCN2 and CCN5
Fig. 11.
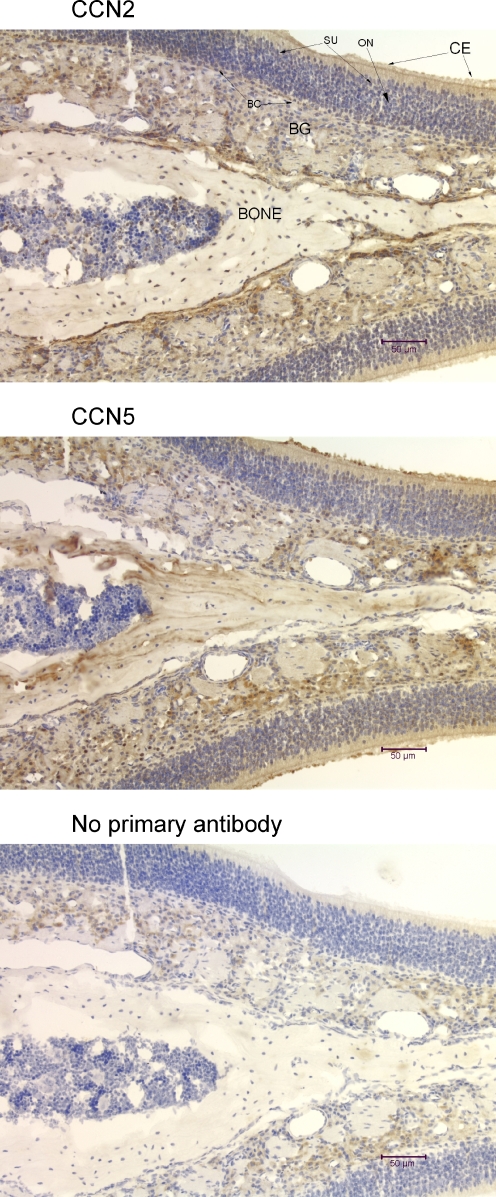
Immunohistochemical analysis of the mouse olfactory epithelium using anti-CCN2 and anti-CCN5. The tissue sections (7 μm, coronal) show all of the layers of the olfactory epithelium, with the ciliated columnar epithelium of the nasal mucosa on the outside (CE), and the sustentacular cell layer (SU), mature and immature bipolar olfactory neurons (ON), globose and horizontal basal cells (BC), the cells of Bowman’s glands (BG), and bone. Some cells from all regions of the mouse olfactory system demonstrate nuclear localization of CCN2 and CCN5
Table 1.
Summary of mouse tissue CCN5 and CCN2 expression
| Tissue | CCN2 | CCN5 |
|---|---|---|
| Heart | Cytoplasmic throughout ventricular and atrial myocardia and valves | Cytoplasmic throughout ventricular and atrial myocardia and valves |
| Lung | Cytoplasmic in all cell types; many nuclei | Cytoplasmic in all cell types; many nuclei |
| Stomach | Cytoplasmic—all cell types; nuclear-gastric pit cells | Cytoplasmic—all cell types; nuclear-gastric pit cells |
| Duodenum | Cytoplasmic—all cell types; some nuclei in villi | Cytoplasmic—all cell types; some nuclei in villi |
| Liver | Cytoplasmic in all cell types | Cytoplasmic in all cell types |
| Kidney | Cytoplasmic in all cell types; many nuclei in all cell types | Cytoplasmic in all cell types; many nuclei in all cell types |
| Ovary | Cytoplasmic in all cell types; many nuclei among all cell types | Cytoplasmic in all cell types; many nuclei among all cell types |
| Pancreas | Cytoplasmic in all cell types | Cytoplasmic in all cell types |
| Thymus | Cytoplasmic in all cells; nuclear in most cells | Cytoplasmic in all cells; nuclear in most cells |
| Olfactory | Cytoplasmic in most cell types; nuclear some cell types | Cytoplasmic in most cell types; nuclear some cell types |
| Uterus | Cytoplasmic in all cell types; nuclear in some stroma cells | Cytoplasmic in all cell types; nuclear in some stroma cells |
| Fallopian tube | Cytoplasmic in all cell types; nuclear in some stroma cells | Cytoplasmic in all cell types; nuclear in some stroma cells |
| Skeletal muscle | Cytoplasmic in all cells | Cytoplasmic in all cells |
| Spleen | Cytoplasmic in all cells | Cytoplasmic in all cells |
Fig. 3.
Immunohistochemical analysis of the rat lung using anti-CCN2 and anti-CCN5. Each panel includes a transversely-oriented 7 μm lung tissue section that reveals a portion of the alveolar network. As in the mouse lung (Fig. 2), some cells in the rat lung demonstrate nuclear localization of both CCN2 and CCN5
Fig. 8.
Immunohistochemical analysis of the rat kidney using anti-CCN2 and anti-CCN5. The tissue sections (7 μm parasagittal) are from the renal cortex and include renal corpuscles with Bowman’s capsules and glomeruli (G) interspersed within a dense network of convoluted tubules. Many of the cells in the rat kidney, particularly those from the convoluted tubules, demonstrate nuclear localization of CCN2 and CCN5
Fig. 9.
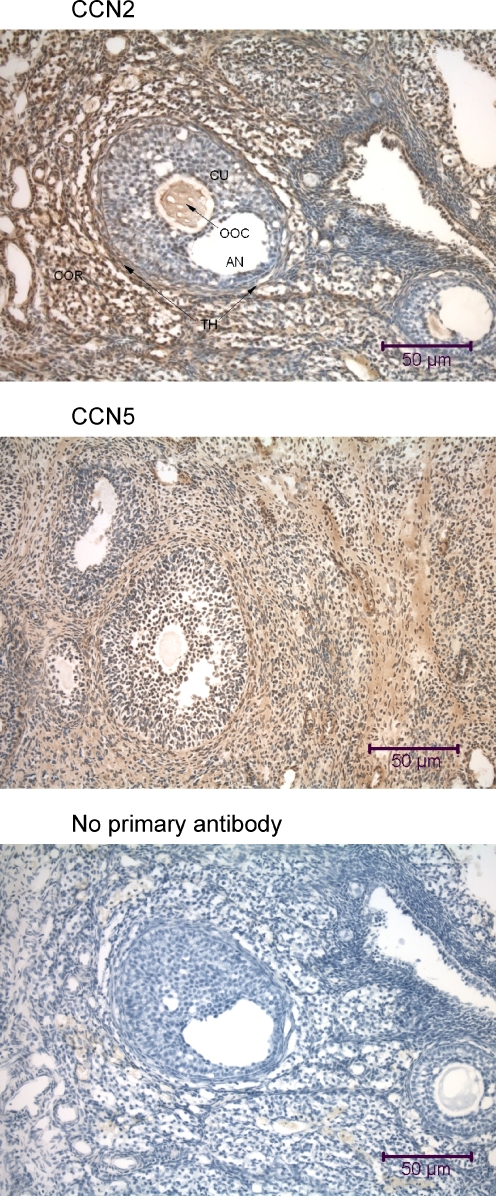
Immunohistochemical analysis of the rat ovary using anti-CCN2 and anti-CCN5. The tissue sections (7 μm transverse) reveal the ovarian cortex (COR), and demonstrate numerous small primordial follicles, and several follicles of later maturational stages embedded within cortical stroma cells. The largest follicles in the middle of each panel are Graafian follicles, with an antrum (AN) and an oocyte (OOC) surrounded by numerous cumulus follicle cells, and an outside layer of thecal cells (TH). Many cells in the rat ovary, particularly the cumulus cells (CU), demonstrate nuclear localization of CCN2 and CCN5. Both proteins are also present in the oocyte, surrounded by the clear zona pellucida
Fig. 12.
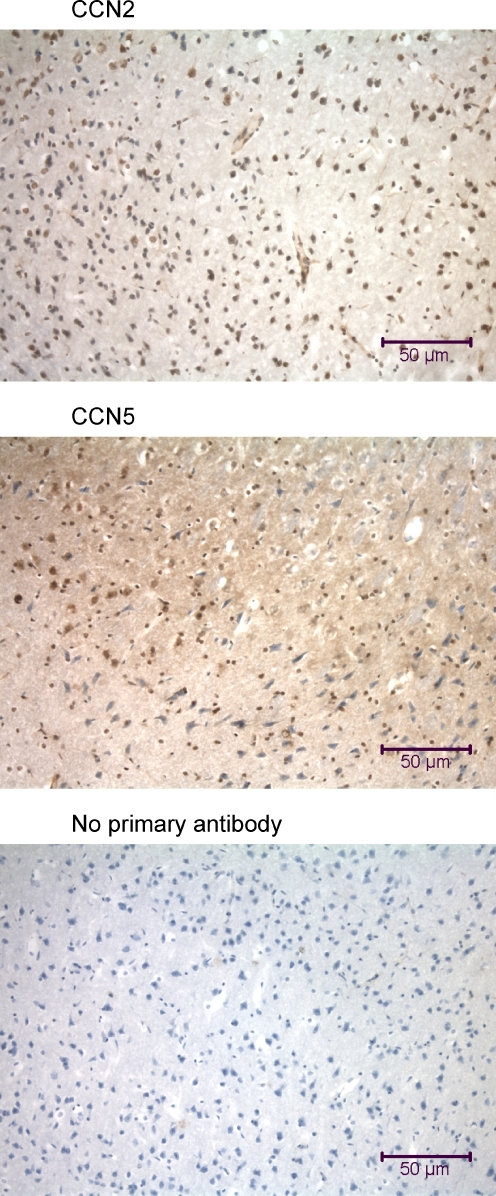
Immunohistochemical analysis of the rat brain using anti-CCN2 and anti-CCN5. The tissue sections (7 μm parasagittal) were made from the cerebral cortex of the rat brain. Many cells of the rat cerebrum demonstrate nuclear localization of CCN2 and CCN5
Table 2.
Summary of rat tissue CCN5 and CCN2 expression
| Tissue | CCN2 | CCN5 |
|---|---|---|
| Heart | Cytoplasmic throughout ventricular and atrial myocardia and valves | Cytoplasmic throughout ventricular and atrial myocardia and valves |
| Lung | Cytoplasmic in all cell types; many nuclei | Cytoplasmic in all cell types; many nuclei |
| Liver | Cytoplasmic in all cell types | Cytoplasmic in all cell types |
| Kidney | Cytoplasmic in all cell types; many nuclei in all cell types | Cytoplasmic in all cell types; many nuclei in all cell types |
| Ovary | Cytoplasmic in all cell types; many nuclei among all cell types | Cytoplasmic in all cell types; many nuclei among all cell types |
| Brain | Cytoplasmic in all cells; nuclear in most cells | Cytoplasmic in all cells; nuclear in most cells |
| Uterus | Cytoplasmic in all cell types; nuclear in some stroma cells | Cytoplasmic in all cell types; nuclear in some stroma cells |
| Fallopian tube | Cytoplasmic in all cell types; nuclear in some stroma cells | Cytoplasmic in all cell types; nuclear in some stroma cells |
| Skeletal muscle | Cytoplasmic in all cells | Cytoplasmic in all cells |
| Aorta | Cytoplasmic in all cell types | Cytoplasmic in all cell types |
| Spleen | Cytoplasmic in all cells | Cytoplasmic in all cells |
Cardiovascular and pulmonary systems
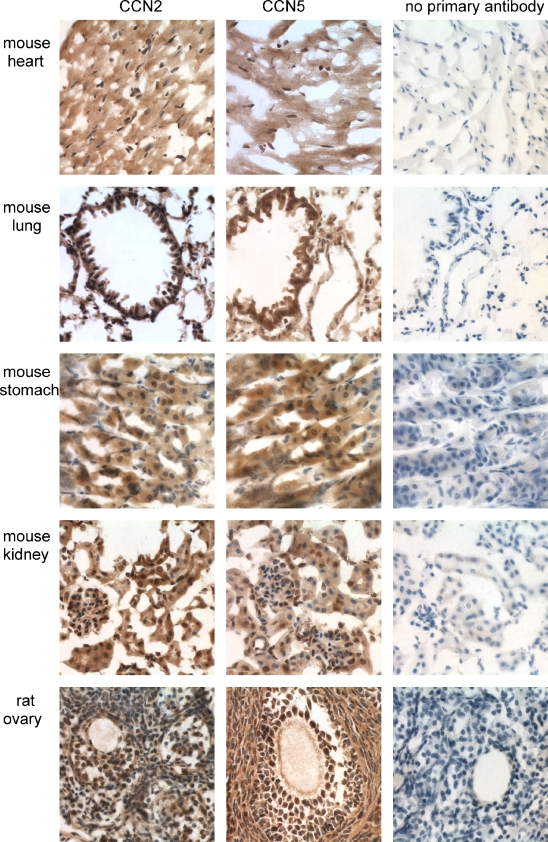
We have previously demonstrated high levels of CCN5 in cardiovascular cells. High levels of CCN5 protein were detected in rat arterial smooth muscle and lumenal epithelium and in cultured aortic smooth muscle cells (Lake et al. 2003). Immunohistochemical analysis of mouse heart tissue revealed similar patterns of both CCN2 and CCN5 protein distribution (Fig. 1). Both proteins were found throughout the cytoplasm of the ventricular myocardium. Both CCN2 and CCN5 were found in the atria and in the valves (data not shown). To our surprise, most nuclei in the myocardium displayed nuclear localization of both CCN2 and CCN5 (Fig. 13). Rat heart sections revealed the same pattern of CCN2 and CCN5 as that of the mouse (data not shown). In pectoralis major muscle and several other skeletal muscles, we detected a similar pattern of widespread CCN2 and CCN5 protein distribution (data not shown).
Both CCN2 and CCN5 displayed similar ubiquitous distribution patterns in both mouse lung (Fig. 2) and rat lung (Fig. 3). Both proteins were observed in the cytoplasm of alveolar and bronchiolar epithelial cells, endothelial cells, and smooth muscle cells, and also in many nuclei of all lung cell types (Fig. 13). We have also detected high levels of CCN2 and CCN5 on the surface and in the nuclei of cultured smooth muscle cells from mouse, rat, and human bronchial trachealis muscle (data not shown).
Gastrointestinal system
Several tissues from the mouse and rat gastrointestinal system were analyzed for the presence of CCN2 and CCN5, including the stomach, duodenum, and liver (Figs. 4, 6). All cells in mouse stomach wall (fundus portion) displayed both CCN2 and CCN5 in the cytoplasm (Fig. 4). Both proteins were observed in many nuclei in gastric pit cells in the deep zone of the mucosa (Figs. 4, 13).
Cells of the mouse duodenum revealed both CCN2 and CCN5 in patterns similar to that observed in the stomach (Fig. 5). CCN2 and CCN5 protein were found in the cytoplasm of cells of the Brunner’s glands, muscularis mucosae, the crypts, and the villi, and in many nuclei of cells in Brunner’s glands and villi.
Both mouse liver (Fig. 6) and rat liver (data not shown) displayed ubiquitous distribution of both CCN2 and CCN5 in the cytoplasm of all cell types, including hepatocytes and endothelial cells.
Urogenital system
The tissues of the urogenital system in both mouse and rat revealed similar patterns of ubiquitous CCN2 and CCN5 distribution (Figs. 7, 8, 9). In the kidneys of both mice and rats, both proteins were present in the cytoplasm and many nuclei of all cell types present, including distal and proximal tubular cells, glomerular endothelial cells, and mesangial cells (Figs. 7, 8).
The rat ovary (Fig. 9) and mouse ovary (data not shown) revealed both CCN2 and CCN5 in all cell types, including stromal cells, thecal cells, granulosa cells, and oocytes. Most nuclei in the ovary demonstrated both CCN2 and CCN5 (Figs. 9, 13).
Endocrine system
Both CCN2 and CCN5 were present in other endocrine glands and in immune tissues. In the mouse pancreas, CCN2 and CCN5 were expressed throughout the cytoplasm of exocrine cells (data not shown). No nuclear staining for CCN2 or CCN5 was observed in the pancreas.
The mouse thymus revealed CCN2 and CCN5 protein throughout both the medulla and cortex (Fig. 10). Both proteins were present in the cytoplasm and most nuclei of lymphocytes., and most lymphocyte nuclei had high levels of staining for both CCN2 and CCN5.
Nervous system and ectodermally-derived tissues
CCN2 and CCN5 were found in ectodermally-derived tissues of the mouse and rat, including the olfactory epithelium and brain (Figs. 11, 12). In the mouse nasal cavity, CCN2 and CCN5 were present in the cytoplasm and some nuclei of all cell types of the olfactory epithelium and lamina propria (Fig. 11).
CCN2 and CCN5 proteins were found in throughout the cytoplasm of cells of the cerebrum of the rat brain, and in many nuclei (Figs. 12, 13). The proportion of nuclei that had nuclear localization of CCN2 and CCN5 varied widely according to the region of the brain (data not shown).
Summary
CCN5 and CCN2 proteins were detected in all organs and tissues examined in both mouse and rat. Immunohistochemistry results for both mouse and rat tissues varied in distribution and intensity among the different cell types in each tissue tested (Tables 1, 2). Both proteins were most commonly observed in the cytoplasm, varying in intensity from low levels (e.g., spleen) to very high levels (e.g., stomach). The lowest levels of CCN2 and CCN5 among all the tissues analyzed were observed in the spleen (data not shown). In many tissues, such as heart, lung, stomach, duodenum, kidney, ovary, thymus, olfactory epithelium, brain, and 14-day gestation stage mouse brain, nuclear staining was observed for both CCN2 and CCN5 (Tables 1, 2; Fig. 13). In almost all tissues, the intensity and distribution of CCN5 resembled that of CCN2 (Tables 1, 2).
Discussion
We expected that CCN5 protein would be present mostly in smooth muscle, glandular epithelium, and endothelial cells, and not in other tissues. Instead, CCN5 was found widely distributed throughout all tissues of the adult mouse and rat. Although we expected to find CCN2 mostly in endothelial cells and connective tissue, and not in other tissues, it was also found widely distributed throughout all rodent tissues analyzed. Since the biological effects of CCN2 and CCN5 have been demonstrated to be opposite in many assays, we expected that the distributions of both proteins would be largely non-overlapping and complementary. Instead, both proteins had very similar patterns of tissue distribution, with only small differences in levels of expression. Because we have not measured the affinity constants of either antibody used, we cannot interpret the significance of immunohistochemical staining differences. We expected that CCN2 and CCN5 would localize only in the cytoplasm or on cell surfaces. We were surprised to find that both proteins were frequently present in the nuclei of many tissues (Fig. 13).
We have previously demonstrated the presence of CCN5 protein in rat uterine smooth muscle and endometrial glandular epithelium (Mason et al. 2004) and in rat arterial smooth muscle (Lake et al. 2003). CCN5 has also been described in human fetal bone (Kumar et al. 1999); human pancreas (Dhar et al. 2007); and breast ductal and lobular epithelium (Banerjee et al. 2003). CCN5 protein and mRNA has also been detected in numerous cultured cells and tissue specimens using Western and Northern blots, respectively (summarized in Gray and Castellot 2004). The results of some screening studies were extremely variable and inconsistent, with an excess of negative findings in some studies. For example, in one early survey of CCN5 mRNA expression using Northern blots, no CCN5 mRNA was detected in most organs and tissues of the rat and mouse (Zhang et al. 1998). It is possible that CCN5 and CCN2 proteins have a long half-life in adult tissues, and could have been translated from very low levels of mRNA, or mRNA present during an earlier stage of organogenesis. It is unlikely that CCN5 is made in specific cells and then transported via the blood to other tissues, since we have been unable to detect it in blood plasma. It is also possible that some posttranslational modifications of CCN5 protein might mask the epitopes detected by either antibody used in this study, leading to undetected CCN5 proteins in some cell types. Until the present studies, no comprehensive analysis for CCN5 protein distribution in adult vertebrate tissues has been reported.
As in the case of CCN5, no systematic studies of CCN2 protein expression in adult tissues have been reported. Except for a few studies of normal tissue samples, most CCN2 protein expression data are from studies of cultured cells and tissues in pathological states. CCN2 protein has been found in normal human bronchiolar epithelium (Chien et al. 2006), chondrocytes of mouse and rat cartilage (Fukunaga et al. 2003), and in all normal uterine cell types in the mouse in patterns identical to our results in the rat uterus with both CCN2 and CCN5 (Surveyor et al. 1998). CCN2 mRNA has been localized using in situ hybridization methods in muring ovarian granulosa cells in preantral follicles (Harlow et al. 2002), in visceral epithelial cells of the rat kidney (Ito et al. 2001), and in porcine uterine smooth muscle, glandular epithelium, stromal, endothelial, and luminal epithelial cells (Moussad and Brigstock 2000).
The adult tissue CCN2 and CCN5 protein expression distribution patterns are similar to those observed in tissues of late gestation mouse embryos (Malmquist et al. 2007). The present immunohistochemistry results have been further corroborated by Western blot analyses of mouse and rat tissue protein lysates (manuscript in preparation), and in immunofluorescent microscopic analysis of cultured cells prepared from many of the tissue types analyzed (manuscript in preparation).
CCN2 and CCN3 proteins have been observed in the nucleus in some cells (Wahab et al. 2001; Planque et al. 2006). Nuclear localization of CCN2 and CCN5 has also been observed in cultured rat aortic, uterine, and tracheal smooth muscle cells using immunofluorescent staining and confocal microscopy methods (manuscript in preparation). In both CCN2 and CCN3, the nuclear localization signal is encoded by the CT domain. Experiments are underway to determine which portion of CCN5 is necessary and sufficient for nuclear localization in smooth muscle cells. Because CCN5 does not have a CT domain, its nuclear localization signal will be different from that of CCN2 and CCN3.
It has been suggested that CCN proteins in the nucleus might interact with transcriptional regulatory proteins (Planque 2006). Nuclear localization of CCN2 and CCN5 suggests the possibility of a direct mode of CCN protein signaling that is separate from, or complementary to, matricellular receptor-based mechanisms of regulating cell signaling pathways and cell cycle control proteins.
Widespread expression of both CCN5 and CCN2 strongly suggests that both proteins have numerous functions, most of which are not characterized. Since both CCN2 and CCN5 are expressed in similar patterns, they could be coordinately regulated by the same ligands and intracellular signaling pathways in many cell types. Since CCN5 and CCN2 frequently co-localize, they might modulate the effects of each other at the physical level by competing for or interfering with the same receptors. A balance of pro-proliferative CCN proteins (such as CCN2) and anti-proliferative CCN proteins (such as CCN5) might be necessary in many cells and tissues to achieve homeostasis, another potential example of a natural “yin-yang” effect. Because its gene is non-redundant, CCN5 might be necessary for all cell types, in contrast to the structural redundancy of the other four-domain CCN proteins. Further evidence for a vital role of CCN5 in vertebrates is the observed lethal effect caused by changes in CCN5 dosage during early embryonic development (data not shown).
Acknowledgments
Supported by grant numbers NIH HL49973 and NIH HD046251. We thank Dr. James Schwob for the mouse olfactory epithelium sections. We thank Ellis Johnson, Josh Russo, Joan Lemire, Ron Myers, Kristina Cvitanovic, Cassie Brougham, and Lan Wei for useful discussions and advice. We thank Po Kwok Tse for expert technical assistance.
Competing interests None declared
Footnotes
Contributions of authors: All the authors were involved in experimental design, data interpretation, and editing of the manuscript. MG provided intellectual direction, performed most of the experiments, and wrote the manuscript. JAM provided some of the tissue specimens as well as technical assistance. MS and MB provided technical assistance. JJC discovered CCN5 mRNA and protein, provided intellectual direction and oversight to the project, and financial support.
References
- Babic AM, Chen CC, Lau LF (1999) Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin αvβ3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol Cell Biol 19:2958–2966 [DOI] [PMC free article] [PubMed]
- Banerjee S, Saxena N, Sengupta K, Tawfik O, Mayo MS, Banerjee SK (2003) WISP-2 gene in human breast cancer: estrogen and progesterone inducible expression and regulation of tumor cell proliferation. Neoplasia 5:63–73 [DOI] [PMC free article] [PubMed]
- Bradham DM, Igarashi A, Potter RL, Grotendorst GR (1991) Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol 114:1285–1294 [DOI] [PMC free article] [PubMed]
- Brigstock DR (2003) The CCN family: a new stimulus package. J Endocrinol 178:169–175 [DOI] [PubMed]
- Brigstock DR, Lau L, Perbal B (2005) CCN workshop: report on the third international workshop on the CCN family of genes. J Clin Pathol 58:463–465 [DOI] [PMC free article] [PubMed]
- Chien W, Yin D, Gui D, Mori A, Frank JM, Said J, Kusuanco D, Marchevsky A, McKenna R, Koeffler HP (2006) Suppression of cell proliferation and signaling transduction by connective tissue growth factor in non-small cell lung cancer cells. Mol Cancer Res 4:1–8 [DOI] [PubMed]
- Cicha I, Yilmaz A, Klein M, Raithel D, Brigstock DR, Daniel WG, Goppelt-Struebe M, Garlichs CD (2005) Connective tissue growth factor is overexpressed in complicated atherosclerotic plaques and induces mononuclear cell chemotaxis in vitro. Arterioscler Thromb Vasc Biol 25:1008–1013 [DOI] [PubMed]
- Delmolino LM, Stearns NA, Castellot JJ Jr (1997) Heparin induces a member of the CCN family which has characteristics of a growth arrest gene. Mol Biol Cell 8:287a 9190208
- Delmolino LM, Stearns NA, Castellot JJ Jr (2001) COP-1, a member of the CCN family, is a heparin-induced growth arrest specific gene in vascular smooth muscle cells. J Cell Physiol 188:45–55 [DOI] [PubMed]
- Dhar G, Mehta S, Banerjee S, Gardner A, McCarty BM, Mathur SC, Campbell DR, Kambhampati S, Banerjee SK (2007) Loss of WISP-2/CCN5 signaling in human pancreatic cancer: a potential mechanism for epithelial/mesenchymal-transition. Cancer Lett Epub (March) [DOI] [PubMed]
- Friedrichsen S, Heuer H, Christ S, Cuthill D, Bauer K, Raivich G (2005) Gene expression of connective tissue growth factor in adult mouse. Growth Factors 23:43–53 [DOI] [PubMed]
- Fukunaga T, Yamashiro T, Oya S, Takeshita N, Takigawa M, Takano-Yamamoto T (2003) Connective tissue growth factor mRNA expression pattern in cartilages is associated with their type I collagen expression. Bone 33:911–918 [DOI] [PubMed]
- Gao R, Ball DK, Perbal B, Brigstock DR (2004) Connective tissue growth factor induces c-fos gene activation and cell proliferation through p44/42 MAP kinase in primary rat hepatic stellate cells. J Hepatol 40:431–438 [DOI] [PubMed]
- Gray MR, Castellot JJ (2004) Function and regulation of CCN5, (Ch. 12). In: Perbal B (ed) CCN Proteins: a new Family of Cell Growth and Differentiation Regulators. Imperial College Press, London
- Harlow CR, Davidson L, Burns KH, Yan C, Matzuk MM, Hillier SG (2002) FSH and TGF-β superfamily members regulate granulosa cell connective tissue growth factor gene expression in vitro and in vivo. Endocrinol 143:3316–3325 [DOI] [PubMed]
- Ito Y, Goldschmeding R, Bende RJ, Claessen N, Chand MA, Kleij L, Rabelink TJ, Weening JJ, Aten J (2001) Kinetics of connective tissue growth factor expression during experimental proliferative glomerulonephritis. J Am Soc Nephrol 12:472–484 [DOI] [PubMed]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM (2003) Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791 [DOI] [PMC free article] [PubMed]
- Kumar S, Hand AT, Connor JR, Dodds RA, Ryan PJ, Trill JJ, Fisher SM, Nuttall ME, Lipshutz DB, Zou C, Hwang SM, Votta BJ, James IE, Rieman DJ, Gowen M, Lee JC (1999) Identification and cloning of a connective tissue growth factor-like cDNA from human osteoblasts encoding a novel regulator of osteoblast functions. J Biol Chem 274:17123–17131 [DOI] [PubMed]
- Lake AC, Bialik A, Walsh K, Castellot JJ Jr (2003) CCN5 is a growth arrest-specific gene that regulates smooth muscle cell proliferation and motility. Am J Pathol 162:219–231 [DOI] [PMC free article] [PubMed]
- Leask A (2004) Transcriptional profiling of the scleroderma fibroblast reveals a potential role for connective tissue growth factor (CTGF) in pathological fibrosis. Keio J Med 53:74–77 [DOI] [PubMed]
- Malmquist JA, Gray MR, Oliveira BE, Koch M, Castellot JJ (2007) CCN5 expression in mammals. I. Embryonic and fetal tissues. Cell communication and signaling (submitted as an accompanying paper) [DOI] [PMC free article] [PubMed]
- Mason HR, Grove-Strawser D, Rubin BS, Nowak RA, Castellot JJ Jr (2004) Estrogen induces CCN5 expression in the rat uterus in vivo. Endocrinol 145:976–982 [DOI] [PubMed]
- Moussad EEA, Brigstock DR (2000) Connective tissue growth factor: what’s in a name. Mol Genet Metab 71:276–292 [DOI] [PubMed]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, Watanabe C, Cohen RL, Melhem MF, Finley GG, Quirke P, Goddard AD, Hillan KJ, Gurney AL, Botstein D, Levine AJ (1998) WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA 95:14717–14722 [DOI] [PMC free article] [PubMed]
- Perbal B (2004) CCN proteins: multifunctional signalling regulators. Lancet 363:62–64 [DOI] [PubMed]
- Planque N (2006) Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun Signal 4:7–25 [DOI] [PMC free article] [PubMed]
- Planque N, Chang LL, Saule S, Bleau A-M, Perbal B (2006) Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem 99:105–116 [DOI] [PubMed]
- Rachfal AW, Luquette MH, Brigstock DR (2004) Expression of connective tissue growth factor (CCN2) in desmoplastic small round cell tumor. J Clin Path 57:422–425 [DOI] [PMC free article] [PubMed]
- Surveyor GA, Wilson AK, Brigstock DR (1998) Localization of connective tissue growth factor during the period of embryo implantation in the mouse. Biol Reprod 59:1207–1213 [DOI] [PubMed]
- Takigawa M, Nakanishi T, Kubota S, Nishida T (2003) Role of CTGF/HCS24/ecogenin in skeletal growth control. J Cell Physiol 194:256–266 [DOI] [PubMed]
- Tong ZY, Brigstock DR (2006) Intrinsic biological activity of the thrombospondin structural homology repeat in connective tissue growth factor. J Endocrinol 188:R1–8 [DOI] [PubMed]
- Ueberham U, Ueberham E, Gruschka H, Arendt T (2003) Connective tissue growth factor in Alzheimer’s disease. Neurosci 116:1–6 [DOI] [PubMed]
- Wahab NA, Brinkman H, Mason RM (2001) Uptake and intracellular transport of the connective tissue growth factor: a potential mode of action. J Biochem 359:89–97 [DOI] [PMC free article] [PubMed]
- Zhang R, Averboukh L, Zhu W, Zhang H, Jo H, Dempsey PJ, Coffey RJ, Pardee AB Liang P (1998) Identification of rCop-1, a new member of the CCN protein family, as a negative regulator for cell transformation. Mol Cell Biol 18:6131–6141 [DOI] [PMC free article] [PubMed]