Abstract
The stearoyl-CoA desaturase 1 (SCD1) catalyzes the synthesis of monounsaturated fatty acids. This enzyme is a critical control point regulating hepatic lipogenesis and lipid oxidation. Therefore SCD1 may be a potential therapeutic target in the treatment of obesity and metabolic syndrome. Regulation of SCD1 expression occurs primarily at the level of transcription. In the present study, we characterized the insulin response elements (IREs) and the insulin signaling pathway mediating the regulation of SCD1 gene transcription in liver. In chicken embryo hepatocytes (CEH) and HepG2 cells, insulin stimulates SCD1 promoter activity by 2.5 folds. This activation is mediated by two different IREs on the chicken promoter, one localized between −1,975 and −1,610 bp and one between −372 and −297 bp. The latter binds both NF-Y and SREBP-1 transcription factors in response to insulin. We also demonstrated that insulin induction of SCD1 gene expression and promoter activity is abolished by pre-incubation of cells with specific inhibitors of both PI3-kinase (LY294002) and mTor (Rapamycin) or by over-expression of a dominant negative mutant of PI3-kinase. The PI3-kinase and mTor pathway mediates the insulin response on both IREs. In summary, insulin activates SCD1 gene expression in liver via a signaling pathway that involves PI3-kinase and mTor and the downstream transcription factors NF-Y and SREBP-1. Sentence summary: Insulin regulates SCD1 gene expression via two different IREs. The most 3′ IRE is localized between −372 and −297 bp and binds the NF-Y and SREBP-1 transcription factors in response to insulin. PI3-kinase and mTor mediate the action of insulin on both IREs.
Keywords: SCD1, Transcription, Insulin, mTor, NF-Y, SREBP-1
Introduction
The stearoyl CoA Desaturase (SCD1) (EC 1.14.99.5) is a key enzyme involved in the synthesis of monounsaturated fatty acids. It catalyzes the introduction of a cis double bound in Δ9 position of de novo synthesized fatty acids mainly from palmitoyl- and stearoyl-CoA (Enoch et al. 1976; Kasturi and Joshi 1982). Its regulation is of physiological importance because changes in the enzyme activity will lead to changes in cell membrane fluidity leading to various metabolic diseases such as obesity and cancers (Ntambi 1995).
In liver, under a standard diet, SCD1 mRNA is weakly expressed while fat-free high carbohydrate diet dramatically increased it (Kaestner et al. 1989; Mihara 1990; Ntambi 1995; Thiede and Strittmatter 1985). This regulation is mainly attributed to a modulation of gene transcription (Ntambi 1992). It was demonstrated that in human (Bene et al. 2001; Zhang et al. 2001), chicken (Lefevre et al. 2001) and rodent (Enser 1979; Legrand and Bensadoun 1991; Ntambi 1999), SCD1 genes are under the positive control of insulin. The effect of insulin on SCD1 appears to be largely mediated by SREBP-1c (Shimomura et al. 1998) however a direct binding of SREBP-1 on the SCD1 promoter has never been clearly associated to an insulin response (Tabor et al. 1998, 1999; Waters et al. 1997). In chicken LMH hepatoma cells, it was showed that insulin stimulates SCD1 gene expression through an Insulin Response Element (IRE) localized in the first 372 pb of the chicken promoter (Lefevre et al. 2001).
Insulin controls gene transcription by modifying the binding of transcription factors on IREs or by regulating their transcriptional activities (Mounier and Posner 2006). Several IREs have been defined through which insulin up-regulates gene transcription. Among them, the sterol response element binds the transcription factors of the SREBP family, particularly SREBP-1c (Eberle et al. 2004). A binding of activator protein 1 (AP-1) has also been identified in the promoter regions of several genes up-regulated by insulin. A large number of insulin responsive genes also harbour a new type of IRE containing GC-rich regions which binds the ubiquitous transcription factor Sp1 (Samson and Wong 2002).
The binding of insulin to the insulin receptor tyrosine kinase (IRK) is followed by IRK autophosphorylation, activation, and the phosphorylation of IRSs on tyrosine residues. These phospho-tyrosine motifs bind adaptor proteins, such as the p85 regulatory subunit of PI3-kinase. (Alessi and Downes 1998; Yenush et al. 1998). Observations indicate that the activation of PI3-kinase is a major route through which insulin modulates gene transcription (Band and Posner 1997; Dickens et al. 1998; Sutherland et al. 1995, 1998). Several pathways have been implicated downstream of PI3-kinase such as the activation of Akt (Agati et al. 1998; Liao et al. 1998; Schmoll et al. 2000) which directly phosphorylates the transcription factors Foxo-1, HNF-3β and CREB (Brunet et al. 1999; Chakravarty et al. 2005; Ogg et al. 1997). We and others have also clearly identified a role for mTor, downstream of PI3-kinase, in mediating insulin effect on IGFBP-1 gene transcription (Band and Posner 1997; Mounier et al. 2006; Patel et al. 2002). Independently of PI3-kinase, insulin can also mediate its effect on gene expression through the activation of the MAPK pathway (Ayala et al. 2004; Keeton et al. 2003; Kotzka et al. 2004; Nadeau et al. 2004).
In the present study we showed, that in chick embryo hepatocytes in culture (CEH) and in HepG2 cells, insulin up-regulates SCD1 gene expression. The insulin action was mediated by 2 different IREs on the chicken promoter, one localized between −1,975 and −1,610 bp upstream of the CAP site and one between −372 bp and −297 bp. The most 3′ element recruited the NF-Y and SREBP-1 transcription factors in response to insulin. In addition, we demonstrated that upon insulin treatment of both CEH and HepG2 cells a PI3-kinase and mTor dependent pathway was activated mediating the insulin effect on chicken and human SCD1 promoters. Finally, we showed that this insulin-activated pathway targets the two different IREs on the chicken SCD1 promoter.
Materials and methods
Materials
Leghorn chickens were obtained from Couvoir Simentin (Mirabel, Canada). LY294002 and Rapamycin were from Calbiochem (EMD Biosciences, San Diego, CA). The GAPDH, Akt, anti-phospho-Akt (ser273), p70S6-kinase and anti-phospho-p70S6-kinase (Thr 389) antibodies were from Cell Signaling Technology, Inc. (Danvers, MA). The Tyr(P) (PY99), SREBP-1 and NF-Y antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The PI3-kinase p85 and IRS1 antibodies were from Upstate Biotechnology, Inc. (Lake Placid, NY). The rat SCD1 antibody was a generous gift of Dr. J. Ozols (Farmington, CT). Protein A-Sepharose was from Amersham Biosciences Inc. (Baie d’Urfé, Canada). The collagenase H and the Fugene-HD reagent were purchased from Roche Diagnostics (Laval, Canada). The Waymouth medium MD 705/1, MEM and the porcine insulin were obtained from Sigma. Lipofectin reagent was from Invitrogen Canada Inc. (Burlington, Canada). The 6 wells-cell + culture plates were purchased from Sarstedt (St. Laurent, Canada) and luciferase reagent from Promega (Madison WI). ATP was purchased from PerkinElmer Life Sciences (Wellesley, MA). The human SCD1 promoter construct was kindly provided by Drs S.M. Prouty and W. Samuels (NIH, Bethesda, MD). Most other reagents and chemicals were obtained from Sigma and were of the highest grade available.
Plasmid constructions
The SCD1-1 to the SCD1-6 DNA constructs were a generous gift of Dr. C. Diot (Lefevre et al. 2001). pGL2-Basic Vector and pGL2-Promoter Vector were used as negative and positive controls respectively (Promega, Ontario, Canada). The SCD1-7 promoter construct was generated by deleting a 75 bp fragment (−372 to −297 bp) from the SCD1-6 plasmid using the NruI and SmaI restriction enzymes (New England Biolabs, Ipswich, MA). The RSV β−galactosidase plasmid (pRSV-βGal) was used to evaluate transfection efficiency (Baillie et al. 1993). The dominant negative PI3-kinase construct (Δp85) contains a cDNA encoding the p85 regulatory subunit, whose p110 binding region was deleted (Kong et al. 2000). The 2156 bp of the avian SCD1 promoter sequence was published in Genbank under the number DQ645535.
Cell culture and transfection procedure
Chick embryo hepatocytes (CEH) were isolated from livers of 19-day-old chick embryos (Goodridge 1973) (protocol no. 500 approved by the University animal care comity). 2.5 × 106 cells were plated in 6-wells-cell + culture plates and cultured at 40°C under 5% CO2 in serum-free Waymouth medium MD 705/1 containing streptomycin (100 μg/ml) and penicillin G (60 μg/ml). After 6 h, cells were transfected with either 1.5 μg of the SCD1–1 construct or 1.5 μg of the constructs containing the different SCD1 5′ deletions and 0.5 μg of pRSV-βGal (Baillie et al. 1993). Briefly, cells were incubated with the DNA/Lipofectin mixture for 16–24 h. Thereafter, the medium was replaced with fresh medium with or without addition of 50 nM insulin and the incubation was continued for an additional 24 h. For the experiences implicating the use of kinases inhibitors, CEH were stimulated 24 h with 50 nM insulin after 30 min of preincubation with either vehicle (DMSO 0,5%), 50 μM LY294002 or 200 nM Rapamycin. The HepG2 cells were used to test the activity of the human SCD1 promoter. Cells were cultured in MEM media containing 4 mM glutamine and transfected with DNA constructs using Fugene HD (Roche Diagnostics) as previously described (Fillion-Forté and Mounier 2007). HepG2 cells were also used to overexpress the dominant negative p85. Cells were transfected with 1.5 μg of the Δp85 construct and in presence or not of either 1.5 μg of the SCD1–1 construct or the pBluescript vector (Kong et al. 2000). After 24 h, the medium was changed to a medium containing 50 nM of insulin for an additional 24 h.
Effects of kinases inhibitors on CEH viability were evaluated by flow-cytometry with PI staining using FACScan (Sakurai et al. 1999).
Analysis of cell extract
CEH and HepG2 cells were lysed at room temperature in 500 μl of CAT Elisa lysis buffer (Roche Diagnostics, Laval, Canada) and protein concentration (Bradford 1976), luciferase (de Wet et al. 1987) and β-galactosidase (Sambrook and Russell 2000) activities were measured by the indicated methods. The results were expressed as luciferase activity per milligram of soluble protein and then normalized for transfection efficiency using the β-galactosidase activity. Transcriptional rates were expressed as a percentage of the standardized activity measured in the untreated cells.
Gel electrophoretic mobility shift assay
Nuclear extracts were prepared from HepG2 cells incubated for 24 h with 100 nM insulin (Andrews and Faller 1991). Double-stranded oligonucleotides were labelled with [γ-32P]ATP using the T4 polynucleotide kinase. 6 μg of nuclear proteins were mixed with 12 μl of binding buffer containing 20,000 cpm of 32P-labeled probe, 2 μg of poly(dI-dC), 0.01% Nonidet P-40, 0.8 μg of bovine serum albumin, 5% (v/v) glycerol, and 5 μg of salmon sperm DNA. The reaction was incubated for 15 min at room temperature. Competition experiments were performed using a molar excess of unlabelled wild-type oligonucleotide. The sequence of the mutated probes 1 and 2 were respectively: GCGAACCATCACAGACGGCAGCTTCGAGGCAGG; TCCCGCAGTTCGACGGCTCG. Antibody experiments used the same incubation conditions except that 2 μl of either SREBP-1 or NF-Y antibodies (2 μg) were incubated with the reaction mixture for an additional 15 min at room temperature. The reaction mixtures were then subjected to electrophoresis on a 6% polyacrylamide gel at 150 V in 25 mM Tris-HCl, 0.19 M glycine, 1 mM EDTA. Gels were dried and visualized by autoradiography using the phospho-imager system (Molecular imager FX, Biorad, Mississauga, Canada).
Western blot analysis
After treatment with the test agents for the time and the concentration indicated in the figure legends, CEH or HepG2 cells were rinsed twice with ice-cold phosphate-buffered saline (pH 7.4) and solubilized with lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10 mM sodium pyrophosphate, 100 mM sodium fluoride, 1.5 mM MgCl2, 1 mM EGTA, 200 μM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10% glycerol, and 1% Triton X-100). Cell lysates were clarified by centrifugation at 10,000 × g for 20 min at 4°C, and protein concentrations in the resulting supernatants were determined using the BioRad protein assay (BioRad, Mississauga, Canada). Twenty micrograms of proteins from cell lysates were mixed with 4 μl of 3× Laemmli sample buffer (2% SDS, 2% β-mercaptoethanol, 10% V/V glycerol and 50 mg/l bromophenol blue in 0.1 M Tris-HCl buffer, pH 6.8), heated at 100°C for 5 min, subjected to SDS–PAGE and then transferred to Immobilon-P membranes for immunoblotting. Membranes were incubated for 1 h in blocking buffer (1X TBS, 0,1% Tween-20: TBST) containing 5% BSA and then overnight in the same buffer with addition of antibodies raised against, SCD1 (1/6,000), GAPDH (1/2,500), PI3-kinase p85 (1/4,000), Akt (1/2,000), phospho-Akt (1/2,000), p70-S6 kinase (1/1,000), phospho-p70-S6 kinase (1:1,000) or IRS-1 (1/2,000). After three successive washes in 1X TBST, membranes were incubated for 1 h at room temperature in 1X TBS in presence of an anti Rabbit IgG binds to the horseradish peroxidase (1:2,000). Signals were revealed using the ECL plus Western blotting detection reagent according to the manufacturer instructions (GE Healthcare, Baie d’Urfé, Canada). The appropriate bands were quantified using the phospho-imager system (Molecular imager FX, Biorad, Mississauga, Canada).
PI3-kinase activity assay
Lysates (300 μg of protein) from insulin-treated (100 nM insulin for 5 min) or non-treated HepG2 cells over-expressing or not the dominant negative p85, were immunoprecipitated in the presence of protein A-Sepharose, using a specific an anti-phosphotyrosine antibody. Immunoprecipitates were extensively washed, and the protein A-Sepharose pellets were resuspended in 50 μl of kinase assay buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EGTA) containing 0.5 mg/ml L-α-phosphatidylinositol (Avanti Polar Lipids, Inc., Alabaster, AL) and assayed for PI3-kinase activity as described previously (Band and Posner 1997).
RNA analysis
Total RNA was extracted from CEH as previously described (Chomczynski and Sacchi 1987), UV-quantified and diluted in 15 μl of DEPC-treated water at a concentration of 1 μg/μl. Reverse transcription (RT) was performed using the Omniscript enzyme kit of Qiagen (Montreal, Canada) and Oligo-dT (Roche Diagnostics, Laval, Canada) for 1 h at 37°C with an inactivation step at 93°C during 5 min. qPCRs were performed using the QuantiTect SYBR Green PCR Kit from Qiagen (Montreal, Canada) and the LightCycler device (Roche Diagnostics, Laval, Canada). The reactions were realized with 10% of the cDNA and 0.5 μM of both forward and reverse specifics primers for 50 cycles to ensure exponential amplification phase. The HPRT-1 gene was used as reference. The relative quantification was then performed using the RelQuant software (Roche Diagnostics, Laval, Canada). For the SCD1 gene, primers were defined on avian sequences: GCCTTCCAGAATGACATCTATG (sense) and AAATCACTCAGGTCCAGCTTC (antisense) and for the HPRT1 gene primers were defined on human sequences: ATGACCTCTCAACCTTGACTGG (sense) and GGCCACTTTCACCATCTTTG (antisense).
Results
Insulin stimulates the avian SCD1 promoter activity in chick embryo hepatocytes (CEH)
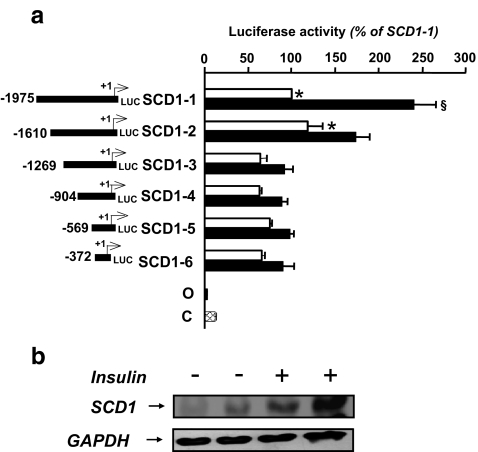
Previous studies have demonstrated that insulin stimulates SCD1 gene expression in various species. However, the molecular mechanism underlying insulin action remains to be delineated. In the present study, we have used the chick embryo hepatocytes transfected with the avian promoter to characterize this response. The choice of this model was justified by the fact that in birds like in human, lipogenesis is mainly hepatic. In addition this primary cell culture model has been extensively used to study lipogenic gene expression and was therefore well characterized (Hillgartner et al. 1995). As described under “Materials and methods”, cells were transiently transfected with luciferase promoter constructs (SCD1-1 to SCD1-6, Fig. 1a containing different 5′ flanking regions of the chicken SCD1 gene promoter (Lefevre et al. 2001). All the constructs presented a high level of luciferase activity compared to the SV40 promoter (left panel, C). When the fragment localized between −1,975 and −1,610 bp was deleted, a significant decrease in the basal SCD1 promoter activity was observed suggesting the presence of a positive transcriptional element in this region.
Fig. 1.
Effect of insulin on cells transfected with constructs containing different deletions of the 5′-flanking region of the SCD1 gene linked to luciferase. a Chick embryo hepatocytes were isolated and incubated in serum free media for 6 h before transfection. Cells were then transiently transfected using lipofectine (10 μg/plate), SCD1–1 (1.5 μg/plate or an equimolar amount of the other constructs) and pRSV-βGal (0.5 μg/plate) as described under “Materials and methods”. After removing the transfection medium, the hepatocytes were incubated for an additional 24 h in either serum-free Waymouth medium (white bars) or in the same medium containing 100 nM insulin (black bars)—Left section, reporter gene DNA constructs used in this experiment. The number at the left of each construct indicates the 5′-end of the fragment in nucleotides relative to the major start site for transcription. +1 indicates the site of initiation of transcription. For all constructs, the 3′-end is at +125 bp. O: pGL2 basic vector (negative control); C: pGL2 basic vector containing the SV40 early promoter (positive control). Right section, values represented luciferase activities normalized by β-galactosidase activities per milligram of soluble protein. Luciferase activities are expressed as a percentage of the activity measured with the SCD1–1 construct in non stimulated cells. Asterisk p < 0.05 comparing non insulin-stimulated activities of SCD1–3, SCD1–4, SCD1–5 and SCD1–6 versus SCD1–1 and SCD1–2. Section sign p < 0.05 comparing the insulin effect on SCD1–1 versus the insulin effect on either SCD1–2, SCD1–3, SCD1–4, SCD1–5 or SD1–6. Results are the means ± S.D. of at least five experiments, each one using an independent batch of isolated hepatocytes. b HepG2 cells were treated (+) or not (−) for 10 min with 100 nM insulin. Cells were lysed, and extracted proteins were resolved on SDS–PAGE and immunoblotted with either an anti-SCD1 antibody (top panel) or an anti-GAPDH antibody (bottom panel)
We subsequently evaluated the effect of insulin on the SCD1 promoter activity. As shown in Fig. 1a, insulin increased SCD1-1 luciferase activity by about 2.5 folds. However, insulin activated SCD1 mRNA and protein levels by more than 5 folds (Figs. 1b, 5a and b). This suggested that additional 5′ sequences may be required to mediate the full insulin action or that insulin acted at a post-transcriptional level. In order to localize the insulin response element(s) (IREs), we subsequently tested the effect of insulin on various 5′ deletions. Deletion of the −1,975 and −1,610 fragment decreased the effect of insulin to 1.5 fold. A similar level of activation is measured when cells were transfected with all the other SCD1 constructs (viz. SCD1–3 to SCD1–6). These experiments suggested that at least two different IREs mediate the action of insulin on the SCD1 promoter, one element localized between −1,975 and −1,610 bp and a second in the first 372 bp of the avian promoter.
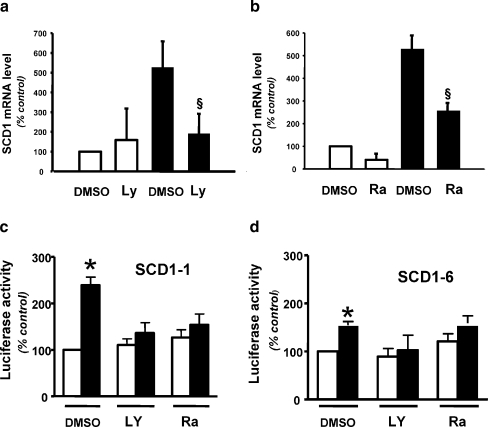
Fig. 5.
Effect of LY294002 and rapamycin on insulin-induced SCD1 gene expression. a and b Chick embryo hepatocytes were cultured for 24 h with (black bars) or without insulin (white bars). Prior to incubation with the hormone, cells were treated for 30 min with either vehicle (0.5% DMSO), 50 μM Ly290042 (Ly) or 200 nM rapamycin (Ra). Total RNA were subsequently extracted as described under “Materials and methods” and SCD1 mRNA levels were evaluated by qPCR using the HPRT-1 gene as standard. Results were expressed as a percentage of SCD1 mRNA level expressed in basal condition in absence of inhibitor. Bars indicate standard deviation (S.D.). Section sign p < 0.05 inhibitor-treated versus non-treated cells. Chick embryo hepatocytes were transfected with SCD1–1 (C) or SCD1–6 (D) and treated (black bars) or not (white bars) with insulin as described in the legend of Fig. 1 with preincubation with either vehicle (0.5% DMSO), 50 μM LY294002 (Ly) or 200 nM rapamycin (Ra). Values represented luciferase activities normalized by β-galactosidase activity per milligram of soluble protein. Results were expressed as a percentage of the activity measured in non-stimulated cells transfected with the corresponding construct and incubated with vehicle only. They are the mean of at least three separate experiments; bars indicate standard deviation (S.D.). Asterisk p < 0.05 insulin-treated versus non-treated cells
Characterization of the most 3′ IRE
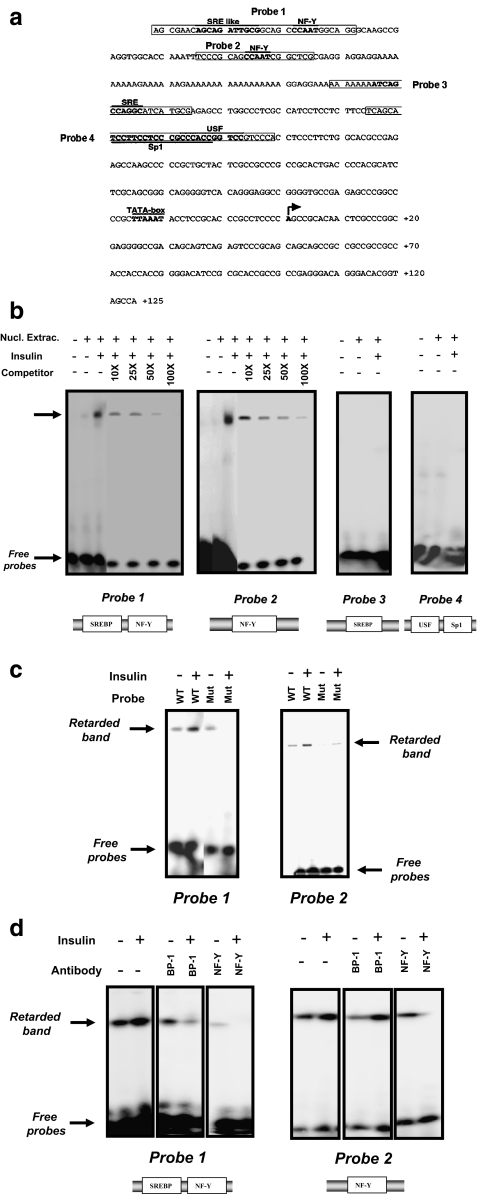
We next focused on the IRE localized on the SCD1–6 construct. This 372 bp fragment contains several consensus binding sites for transcription factors required for insulin-mediated activation of transcription in other genes (Fig. 2a) (Mounier and Posner 2006). To demonstrate DNA-binding of nuclear factor(s) to the putative cis-element(s) previously identified, EMSA experiments were conducted using 32P-labeled double-stranded oligonucleotides corresponding to different parts of the 372 bp sequence (probes 1–4, Fig. 2a). When radiolabeled probes were incubated with nuclear extracts prepared from insulin-stimulated HepG2 cells, DNA-protein complexes were observed only with the probes 1 and 2 (Fig. 2b). The specificity of DNA binding on probes 1 and 2 was confirmed by competition with increasing concentration of non-radioactive probes. Finally, we performed EMSA with mutated probes 1 and 2 and have showed that the DNA binding of insulin-treated nuclear extracts was strongly inhibited (Fig. 2c).
Fig. 2.
Analysis of DNA binding activity of the nuclear proteins from insulin-treated cells on the chicken SCD1 promoter- a Analysis of DNA binding activity of the nuclear proteins from insulin-treated cells on the chicken SCD1 promoter- a Sequence of the first 372 bp of the chicken SCD1 promoter (Genebank no. DQ645535). Consensus binding sites for transcription factors are indicated in bold. Squares indicate the different probes used in the EMSA experiments. b 6 μg of nuclear extracts prepared from untreated (−) or insulin stimulated (+) HepG2 cells were incubated with 4 different 32P-labeled double-stranded fragment of the SCD1 promoter (probes 1–4). Competition with unlabelled probe was performed using molar excess of the double-stranded probe 1 or probe 2. Putative binding sites for transcription factors are indicated on the bottom of each gel. The major DNA-protein complex is designated by the arrow (retarded band). A representative autoradiography of three separate experiments is depicted. c 6 μg of nuclear extracts prepared from untreated (−) or insulin stimulated (+) HepG2 cells were incubated with either a wild type (WT) or mutated (Mut) 32P-labeled double-stranded probe 1 or 2. d 6 μg of nuclear extracts prepared from untreated (−) or insulin stimulated (+) HepG2 cells were incubated with either probe 1 or 2. IgGs (1 μg each) were added after mixing labelled probe and nuclear extract; they were anti-SREBP-1 (BP-1) or anti-NF-Y (NF-Y). Putative binding sites for transcription factors characterized in each probes are indicated. A representative autoradiography of three separate experiments is depicted
In order to identify the trans-acting factors involved in the insulin-activation of SCD1 gene expression, supershift EMSA experiments were conducted using antibodies raised against the known transcription factors binding to the serum response element (SRE) and the NF-Y motifs (SREBP-1 and NF-Y). The retarded band observed with probe 1 and insulin-treated nuclear extracts, disappeared in presence of either SREBP-1 or NF-Y antibody while with probe 2, the retarded band disappeared only in presence of the NF-Y antibody (Fig. 2d). This indicated that both SREBP-1 and NF-Y transcription factors were recruited on the SCD1 promoter upon insulin treatment of the cells.
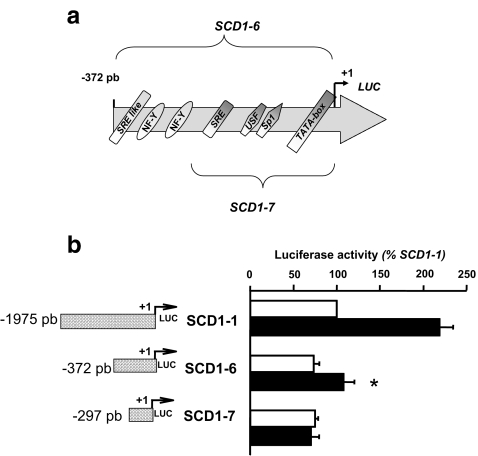
The functionality of this region was subsequently validated by transfection experiments in hepatic cells. Deletion of the SRE like and NF-Y motifs (SCD1–7, Fig. 3a) totally abolished the insulin effect on the SCD1 promoter activity (Fig. 3b). When combined, these data suggested that the −372 to −297 bp fragment of the chicken SCD1 promoter contains an IRE that binds SREBP-1 and NF-Y transcription factors in response to insulin.
Fig. 3.
Effect of insulin on cells transfected with a construct lacking the Insulin Response Element 2. a Schematic representation of SCD1–6 and SCD1–7 constructs. Consensus binding sites for transcription factors are indicated. b Chick embryo hepatocytes were transfected with the SCD1–1, the SCD1–6 or the SCD1–7 construct and treated (black bars) or not (white bars) with 100 nM insulin as described in the legend of Fig. 1. Left section, reporter gene DNA constructs used in this experiment. Right section, bars represented luciferase activities normalized by β-galactosidase activities per milligram of soluble protein. Luciferase activities were expressed as a percentage of the activity measured in the respective non insulin treated sample. *p < 0.05 comparing insulin-treated cells transfected with SCD1–6 versus insulin-treated cells transfected with SCD1–7. Results are the means ± S.D. of at least three experiments
Role of PI3-kinase and mTor on the insulin-induced SCD1 gene transcription
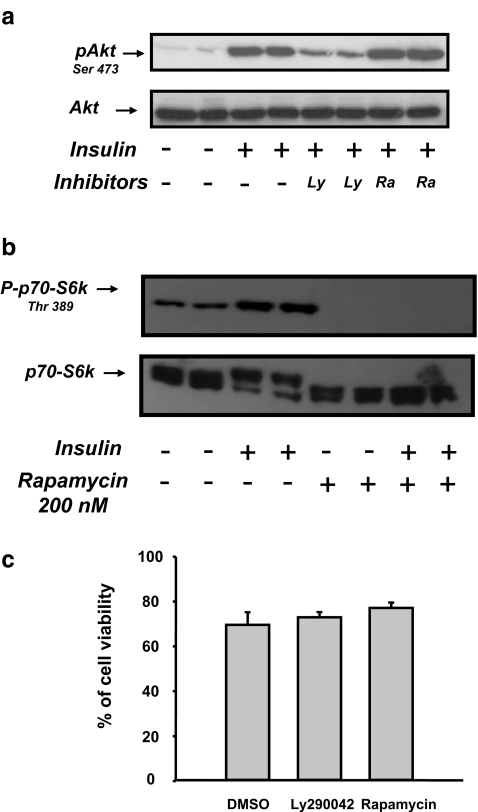
We next characterized the signaling pathway mediating the insulin effect on the SCD1 promoter. As described under “Materials and methods,” CEH were pre-treated for 30 min with specific inhibitor of either PI3-kinase (50 μM LY294002) or mTor (200 nM rapamycin) and subsequently stimulated for 10 min with 100 nM insulin. As shown in Fig. 4a insulin stimulated Akt phosphorylation on the serine 473 residue in CEH. This stimulation was abolished by the presence of LY290042 while presence of rapamycin had no effect. The effect of rapamycin was evaluated by measuring the level of p70-S6 kinase phosphorylation on residue Thr389 in response to insulin. As shown in Fig. 4b, insulin stimulated p70-S6 kinase phosphorylation in CEH while incubation with 200 nM of rapamycin totally abolished the insulin effect. Presence of either PI3-kinase or mTor inhibitor did not affected CEH viability in culture as evaluated by PI staining (Fig. 4c).
Fig. 4.
Effect of LY294002 and rapamycin on insulin responses in chick embryo hepatocytes. a Chick embryo hepatocytes were treated (+) or not (−) for 10 min with 100 nM insulin after 30 min of preincubation with either vehicle (−), 50 μM LY290042 (Ly) or 200 nM rapamycin (Ra). Cells were lysed, and extracted proteins were resolved on 8% SDS–PAGE and immunoblotted with either an anti-Akt antibody (lower panel) or an antibody that recognized the phosphorylated isoform of Akt (top panel). The autoradiographs are representatives of three different experiments b Chick embryo hepatocytes were treated (+) or not (−) for 10 min with 100 nM insulin after 30 min of preincubation with 200 nM rapamycin. Cells were lysed, and extracted proteins were resolved on SDS–PAGE and immunoblotted with either an anti-p70-S6 kinase antibody (lower panel) or an antibody that recognized the phosphorylated isoform of p70-S6 kinase (top panel). The autoradiographs are representatives of three different experiments. c Chick embryo hepatocytes were treated with either 50 μM LY290042 or 200 nM rapamycin for 30 min. Cell viability was subsequently evaluated by PI staining using FAScan. Bars represent the percentage of living cells measured in at least two different wells
The effects of both inhibitors were subsequently evaluated on the insulin-induced SCD1 gene expression. Pre-incubation of CEH with LY294002 totally prevented the effect of insulin on SCD1 mRNA expression (Fig. 5a while presence of rapamycin inhibited it by more than 50% (Fig. 5b). In order to validate our observations at the transcriptional level, the SCD1–1 construct which contains the two IREs and the SCD1–6 construct which contains the most 3′ element were transfected in CEH. LY294002 and rapamycin also abolished the insulin effect on both SCD1–1 and SCD1–6 transcriptional activity (Fig. 5c and d respectively). Taken together, our data suggested that both PI3-kinase and mTor mediated the insulin action of the SCD1 gene expression. The total abolition of the insulin effect on transcription in presence of one of these inhibitors probably indicated that both kinases worked on the same pathway.
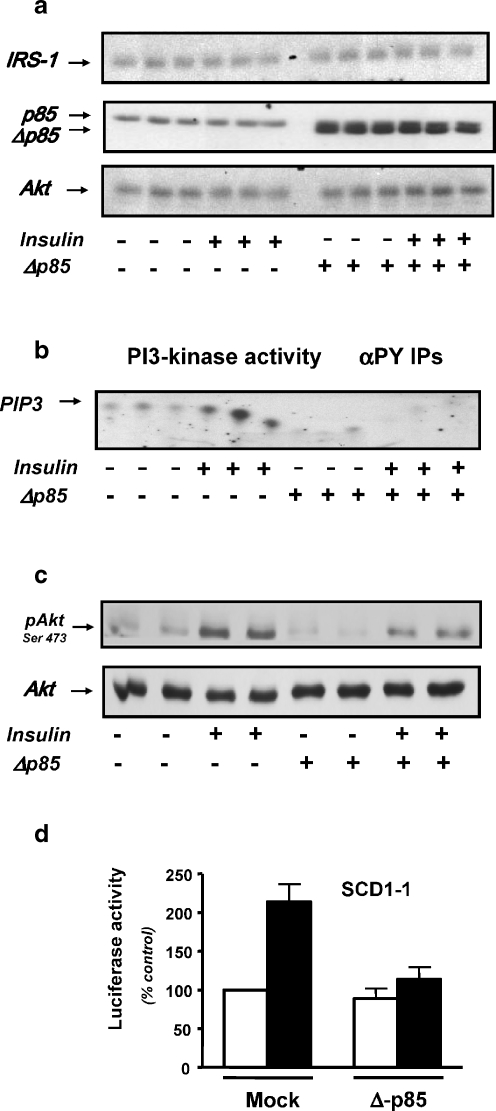
In order to confirm the role of PI3-kinase in mediating the effect of insulin on SCD1 gene expression, we have co-transfected the SCD1–1 plasmid with a dominant negative (DN) mutant of PI3-kinase (Δp85) (Kong et al. 2000) in the human hepatocarcinoma cell line HepG2. This cell line was used instead of CEH in order to attain a high level of Δp85 expression. Figure 6a shows that in our experimental conditions, Δp85 was specifically over-expressed while the levels of IRS1 and Akt expression, upstream and downstream effectors of PI3-kinase respectively, were unaffected. Over-expression of the Δp85 strongly inhibited the insulin-induced PI3-kinase activity (Fig. 6b) and Akt phosphorylation on serine 473 residue (Fig. 6c). Co-transfection of the Δp85 construct with the SCD1–1 plasmid abolished the insulin effect on the SCD1 promoter activity (Fig. 6d). In conclusion, the effects of both LY294002 and Δp85 on the SCD1 promoter activity confirmed the role of PI3-kinase in mediating the insulin action on the regulation of SCD1 gene expression.
Fig. 6.
Effect of the dominant-negative p85 mutant on PI3- kinase, Akt and SCD1 promoter activities in response to insulin in HepG2 cells. HepG2 cells were treated (+) or not (−) for 5 min with 100 nM insulin in presence (+) or absence (−) of Δp85 over-expression. a Cells were lysed, and extracted proteins were resolved on 7% SDS–PAGE then immunoblotted with either an anti-IRS-1 (top panel), an anti-p85 antibody (middle panel) or and anti-Akt antibody (bottom panel). A representative autoradiograph is presented. b The HepG2 cell lysates were immunoprecipitated with αPY antibody and the immunoprecipitates were analyzed for PI3-kinase activity as described under “Materials and methods”. A representative autoradiograph is presented. PIP3 indicates the product of the PI3-kinase reaction. c Cells were lysed, and extracted proteins were resolved on SDS–PAGE then immunoblotted with either an anti-Akt antibody (lower panel) or an antibody that recognized the phosphorylated isoform of Akt (top panel). A representative autoradiograph is presented. d HepG2 cells were transfected with either Bluescript (Mock) or with the dominant negative p85 (Δp85) construct in combination with the SCD1–1 construct as described under “Materials and methods”. After 24 h, medium was removed and replaced by the same medium in presence (black bars) or absence (white bars) of 100 nM insulin. Values represented luciferase activities normalized by β-galactosidase activities per milligram of soluble protein. Results were expressed as a percentage of the activity measured in non-stimulated cells transfected with the SCD1–1 construct and the Bluescript. They are the mean of at least three separate experiments; bars indicate standard deviation (S.D.)
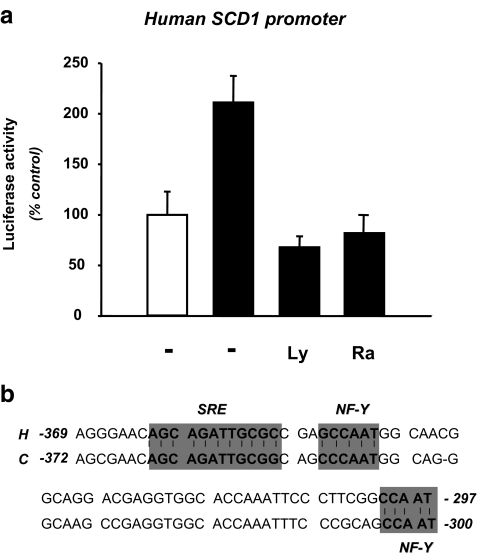
We next evaluated the role of this insulin-signaling pathway on the human SCD1 promoter. Therefore, the HepG2 cells were transfected with a luciferase reporter construct containing about 4 kb of the human SCD1 promoter (kindly provided by Drs Samuel and Prouty, Bethesda, MD). As demonstrated for the chicken promoter, insulin stimulated the human SCD1 promoter activity by about 2 folds (Fig. 7a. Comparison (Fig. 7b) between the proximal human and chicken promoter sequences revealed the presence of conserved consensus binding sites for transcription factors previously identified as mediators of the insulin effect on the chicken promoter (Figs. 2 and 3). Presence of both LY20042 and rapamycin also abrogated the insulin effect on the human SCD1 promoter expression.
Fig. 7.
Effect of LY294002 and rapamycin on insulin-mediated activation of the human SCD1 promoter activity in HepG2 cells. a HepG2 cells were transfected with the human SCD1 promoter construct and treated (black bars) or not (white bars) with insulin as described in the legend of Fig. 1. Prior to incubation with the hormone, cells were treated for 30 min with either vehicle (0.5% DMSO), 50 μM LY294002 or 200 nM rapamycin. Results are expressed as previously described in Fig. 5. b Sequence alignment between the human (AF320307) and chicken SCD1 promoters (DQ645535). Conserved consensus binding sites for the two key transcription factors are shaded. SRE: Sterol regulatory element. NF-Y: Nuclear factor Y
In conclusion, our data demonstrated that insulin-activation of both PI3-kinase and mTor were necessary and sufficient to mediate the action of insulin on both chicken and human SCD1 promoters. The abrogation of the insulin effect on SCD1 gene transcription in presence of only one of the inhibitor suggested that PI3-kinase and mTor probably worked on the same signaling pathway. In addition, our data suggested that at least 2 different IREs were present on the avian SCD1 promoter. The most 3′ IRE localized between −372 and −297 bp bound the SREBP-1 and NF-Y transcription factors in response to insulin. Finally we also showed that both IRES are targeted by the insulin-induced PI3-kinase/mTor dependent pathway.
Discussion
In the present study, we have demonstrated that insulin stimulates SCD1 gene transcription by 2.5 folds in both chick embryo hepatocytes and HepG2 cells (Figs. 1, 5 and 7). SCD1 gene transcription is stimulated by insulin to a similar level in LMH cells (Lefevre et al. 2001) and mouse liver (Waters and Ntambi 1994) while enzyme activity in cultured avian liver explants is induced by 4 fold (Joshi and Aranda 1979). In our study, we showed that in hepatic cells, insulin induced SCD1 mRNA and protein expressions by more than 5 folds (Figs. 1b, 5a and b). This suggested that additional sequences localized further upstream on the chicken promoter was necessary to mediate the full insulin response on SCD1 gene expression. It also suggested that insulin acts at a post-transcriptional level. Taken together, our data suggested that insulin regulates SCD1 gene expression in liver by modulating the level of SCD1 gene transcription but also by influencing the level of the mRNA and protein expression by a still unknown mechanism.
Using 5′ deletions constructs, we delineated two areas that mediate the insulin action on the avian SCD1 promoter. One area is localized between −1,975 and −1,610 bp (IRE1) and the second element between −372 and −297 bp (IRE2). A previous study performed in LMH cells had already suggested the presence of an insulin response sequence in the same region (viz. −372/+125) (Lefevre et al. 2001). Analysis of the avian SCD1 promoter sequence revealed the presence of several consensus binding sites for transcription factors known to mediate the insulin action on gene transcription (Mounier and Posner 2006) (Fig. 8). A consensus binding site for USF, known to mediate the positive action of insulin on FAS gene expression (Griffin and Sul 2004) has been localized in the most 5′ sequences containing the IRE1. In the same region, a consensus binding site for SREBP-1, also known to mediate the positive action of insulin on both Acetyl CoA carboxylase (Magana et al. 1997) and FAS genes expressions (Porstmann et al. 2005) has been identified.
Fig. 8.
Schematic representation of the regulation of SCD1 gene transcription in hepatic cells. Schematic representation of the regulation of SCD1 gene transcription in hepatic cells. The top part of the figure represents the signaling pathway implicated in the insulin regulation of SCD1 gene transcription. The large grey arrow represents the promoter where the different 5′-deletion constructs are indicated. The lower part represents the transcription factors implicated in the regulation of SCD1 gene transcription in liver in response to activation of the indicated signaling pathway
We characterized the IRE2 on the chicken promoter. This IRE2 is localized between 372 and −297 bp and binds SREBP-1 and NF-Y in response to insulin. Deletion of this DNA region abolished the insulin effect on SCD1 promoter activity. These transcription factors were previously described as positive mediators of insulin action on gene transcription (for review, (Mounier and Posner 2006)). Of great interest, the presence of these sites appears to be conserved between the chicken and human SCD1 promoters (Fig. 7b).
Using a specific pharmaceutical inhibitor for PI3-kinase as well as a dominant negative construct for p85 (Kong et al. 2000), we have clearly demonstrated that activation of PI3-kinase by insulin was necessary and sufficient to mediate the hormonal effect on the SCD1 promoter activity (Figs. 5, 6, and 7). A similar implication of PI3-kinase in mediating the positive action of insulin was previously described for the FAS, SREBP-1 and glucokinase genes (Matsumoto et al. 2002; Wang and Sul 1998). In addition, PI3-kinase was implicated in the insulin down-regulation of gene expression for the PEPCK, G6Pase and IGFBP-1 genes (Band and Posner 1997; Dickens et al. 1998; Mounier et al. 2006; Sutherland et al. 1995, 1998). Our observations are in agreement with the fact that activation of a PI3-kinase dependent pathway is the major route through which insulin modulates gene transcription.
Most of the studies looking at downstream PI3-kinase effectors have implicated Akt as a key mediator of insulin action on gene transcription. Activation of Akt mediates the insulin effect on PEPCK and G6Pase expression (Agati et al. 1998; Liao et al. 1998; Schmoll et al. 2000) by directly phosphorylating the transcription factors Foxo-1, HNF-3β and CREB (Brunet et al. 1999; Chakravarty et al. 2005; Ogg et al. 1997). Akt was also implicated in the insulin-induced FAS transcription (Wang and Sul 1998). In our study, we did not excluded a role of the Akt kinase in mediating the action of insulin on SCD1 gene transcription but we showed that it probably acted upstream of the kinase mTor as rapamycin abolished the effect of insulin on SCD1 promoter activity (Figs. 4 and 7).
As mentioned above, our data demonstrated that mTor mediated the insulin response on both chicken and human SCD1 gene transcription (Figs. 5 and 7). mTor was described as a nutrient sensor pathway and associated, like SCD1 with the development of obesity and several metabolic diseases (for review (Marshall 2006)). Of great interest, it was previously showed that PI3-kinase and mTor mediate the effect of PDGF on SCD1 gene expression in fibroblasts with evidences for the implication of the transcription factor SREBP-1 (Demoulin et al. 2004). In addition, growing evidences implicated the mTor kinase in modulating expression of several other different genes. We and others have previously demonstrated that insulin inhibition of IGFBP-1 gene expression required the activation of the mTor kinase in liver (Band and Posner 1997; Mounier et al. 2006; Patel et al. 2002). Furthermore, mTor was implicated in the phosphorylation of the transcription factors STAT1 and STAT3 leading to activation of STAT-dependent gene transcription (Kristof et al. 2003). mTor also plays a critical role in adipogenesis by directly modulating the activity of the transcription factor PPARγ (Kim and Chen 2004). It was also clearly established that ribosome biogenesis was controlled at the level of rDNA transcription by mTor through activation of the transcription factors UBF and TIF1A (Hannan et al. 2003; Mayer et al. 2004). Finally, as demonstrated by microarray analyses using rapamycin-treated mammalian cells, mTor signalling was implicated in the control of transcription for many genes, particularly those involved in the metabolic and biosynthetic pathways (Peng et al. 2002). In agreement with our data, Peng’s study showed that in rapamycin-treated cells, the level of SCD1 gene expression is inhibited compared to the level measured in non-treated cells.
The molecular mechanism of mTor action on SCD1 gene transcription remains however to be characterized. We do not know if mTor directly phosphorylates the insulin-targeted transcription factors, or if its action is mediated by a downstream kinase like the p70-S6kinase. Amino acids analysis of SREBP-1 and NF-Y revealed the presence of putative mTOR and p70-S6kinase phosphorylation sites (Xue et al. 2005). In addition, these putative phosphorylation sites appear to be conserved in both human and chicken transcription factors (data not shown). mTor may also regulate the transcription of SCD1 by modulating the level of expression of various transcription factors. In fact, a microarray analysis showed that in cells treated with rapamycin, SREBP-1 gene expression was inhibited (Peng et al. 2002 60). This hypothesis remains however to be demonstrated and is currently under investigation.
In conclusion, in hepatocytes, insulin regulates SCD1 gene expression by activating its level of transcription and possibly by modulating the mRNA stability. Insulin partially mediates its action acting on at least two different IREs. The most 3′ IRE is localized between −372 and −297 bp and binds both SREBP-1 and NF-Y transcription factors in response to insulin. Insulin regulation of SCD1 gene transcription is mediated by activation of a PI3-kinase and mTOR dependent signaling pathway targeting the two different IREs. Taken together, our present findings builds the link between the SCD1 gene expression and an insulin-stimulated signaling pathway both implicated in the development of several metabolic diseases such as obesity.
Acknowledgments
We wish to thank Dr. B.I. Posner and V. Dumas for their technical assistance on the measure of PI3-kinase activity. We also thank Dr. C. Diot for its generous gift of the SCD1 promoter constructs. This work was supported by a Quebec Heart and Stroke foundation grant.
Footnotes
Mauvoisin and Rocque contributed equally to this work.
References
- Agati JM, Yeagley D, Quinn PG (1998) Assessment of the roles of mitogen-activated protein kinase, phosphatidylinositol 3-kinase, protein kinase b, and protein kinase c in insulin inhibition of camp-induced phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem 273:18751–18759 [DOI] [PubMed]
- Alessi DR, Downes CP (1998) The role of pi 3-kinase in insulin action. Biochim Biophys Acta 1436:151–164 [DOI] [PubMed]
- Andrews NC, Faller DV (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19:2499 [DOI] [PMC free article] [PubMed]
- Ayala JE, Boustead JN, Chapman SC, Svitek CA, Oeser JK, Robey RB, O’Brien RM (2004) Insulin-mediated activation of activator protein-1 through the mitogen-activated protein kinase pathway stimulates collagenase-1 gene transcription in the mes 13 mesangial cell line. J Mol Endocrinol 33:263–280 [DOI] [PubMed]
- Baillie RA, Klautky SA, Goodridge AG (1993) Transient transfection of chick embryo hepatocytes. J Nutr Biochem 4:431–439 [DOI]
- Band CJ, Posner BI (1997) Phosphatidylinositol 3″-kinase and p70s6k are required for insulin but not bisperoxovanadium 1,10-phenanthroline (bpv(phen)) inhibition of insulin-like growth factor binding protein gene expression. Evidence for mek-independent activation of mitogen-activated protein kinase by bpv(phen). J Biol Chem 272:138–145 [DOI] [PubMed]
- Bene H, Lasky D, Ntambi JM (2001) Cloning and characterization of the human stearoyl-coa desaturase gene promoter: Transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochem Biophys Res Commun 284:1194–1198 [DOI] [PubMed]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 96:857–868 [DOI] [PubMed]
- Chakravarty K, Hanson RW, Cassuto H, Reshef L (2005) Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-c. Crit Rev Biochem Mol Biol 40:129–154 [DOI] [PubMed]
- Chomczynski P, Sacchi N (1987) Single-step method of rna isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed]
- Demoulin JB, Ericsson J, Kallin A, Rorsman C, Ronnstrand L, Heldin CH (2004) Platelet-derived growth factor stimulates membrane lipid synthesis through activation of phosphatidylinositol 3-kinase and sterol regulatory element-binding proteins. J Biol Chem 279:35392–35402 [DOI] [PubMed]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S (1987) Firefly luciferase gene: Structure and expression in mammalian cells. Mol Cell Biol 7:725–737 [DOI] [PMC free article] [PubMed]
- Dickens M, Svitek CA, Culbert AA, O’Brien RM, Tavare JM (1998) Central role for phosphatidylinositide 3-kinase in the repression of glucose-6-phosphatase gene transcription by insulin. J Biol Chem 273:20144–20149 [DOI] [PubMed]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F (2004) Srebp transcription factors: master regulators of lipid homeostasis. Biochimie 86:839–848 [DOI] [PubMed]
- Enoch HG, Catala A, Strittmatter P (1976) Mechanism of rat liver microsomal stearyl-coa desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem 251:5095–5103 [PubMed]
- Enser M (1979) The role of insulin in the regulation of stearic acid desaturase activity in liver and adipose tissue from obese-hyperglycaemic (ob/ob) and lean mice. Biochem J 180:551–558 [DOI] [PMC free article] [PubMed]
- Fillion-Forté V, Mounier C (2007) Optimization of transfection efficiency in chick embryo hepatocytes and hepg2 cells. Biochemica 1:13–15
- Goodridge AG (1973) Regulation of fatty acid synthesis in isolated hepatocytes. Evidence for a physiological role for long chain fatty acyl coenzyme a and citrate. J Biol Chem 248:4318–4326 [PubMed]
- Griffin MJ, Sul HS (2004) Insulin regulation of fatty acid synthase gene transcription: roles of usf and srebp-1c. IUBMB Life 56:595–600 [DOI] [PubMed]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD (2003) Mtor-dependent regulation of ribosomal gene transcription requires s6k1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor ubf. Mol Cell Biol 23:8862–8877 [DOI] [PMC free article] [PubMed]
- Hillgartner FB, Salati LM, Goodridge AG (1995) Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev 75:47–76 [DOI] [PubMed]
- Joshi VC, Aranda LP (1979) Hormonal regulation of the terminal enzyme of microsomal stearoyl coenzyme a desaturase in cultured avian liver explants. Role of insulin. J Biol Chem 254:11779–11782 [PubMed]
- Kaestner KH, Ntambi JM, Kelly TJ, Lane MD (1989) Differentiation-induced gene expression in 3t3-l1 preadipocytes. A second differentially expressed gene encoding stearoyl-coa desaturase. J Biol Chem 264:14755–14761 [PubMed]
- Kasturi R, Joshi VC (1982) Hormonal regulation of stearoyl coenzyme a desaturase activity and lipogenesis during adipose conversion of 3t3-l1 cells. J Biol Chem 257:12224–12230 [PubMed]
- Keeton AB, Bortoff KD, Bennett WL, Franklin JL, Venable DY, Messina JL (2003) Insulin-regulated expression of egr-1 and krox20: dependence on erk1/2 and interaction with p38 and pi3-kinase pathways. Endocrinology 144:5402–5410 [DOI] [PubMed]
- Kim JE, Chen J (2004) Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes 53:2748–2756 [DOI] [PubMed]
- Kong M, Mounier C, Wu J, Posner BI (2000) Epidermal growth factor-induced phosphatidylinositol 3-kinase activation and DNA synthesis. Identification of grb2-associated binder 2 as the major mediator in rat hepatocytes. J Biol Chem 275:36035–36042 [DOI] [PubMed]
- Kotzka J, Lehr S, Roth G, Avci H, Knebel B, Muller-Wieland D (2004) Insulin-activated erk-mitogen-activated protein kinases phosphorylate sterol regulatory element-binding protein-2 at serine residues 432 and 455 in vivo. J Biol Chem 279:22404–22411 [DOI] [PubMed]
- Kristof AS, Marks-Konczalik J, Billings E, Moss J (2003) Stimulation of signal transducer and activator of transcription-1 (stat1)-dependent gene transcription by lipopolysaccharide and interferon-gamma is regulated by mammalian target of rapamycin. J Biol Chem 278:33637–33644 [DOI] [PubMed]
- Lefevre P, Tripon E, Plumelet C, Douaire M, Diot C (2001) Effects of polyunsaturated fatty acids and clofibrate on chicken stearoyl-coa desaturase 1 gene expression. Biochem Biophys Res Commun 280:25–31 [DOI] [PubMed]
- Legrand P, Bensadoun A (1991) Stearyl-coa desaturase activity in cultured rat hepatocytes. Biochim Biophys Acta 1086:89–94 [DOI] [PubMed]
- Liao J, Barthel A, Nakatani K, Roth RA (1998) Activation of protein kinase b/akt is sufficient to repress the glucocorticoid and camp induction of phosphoenolpyruvate carboxykinase gene. J Biol Chem 273:27320–27324 [DOI] [PubMed]
- Magana MM, Lin SS, Dooley KA, Osborne TF (1997) Sterol regulation of acetyl coenzyme a carboxylase promoter requires two interdependent binding sites for sterol regulatory element binding proteins. J Lipid Res 38:1630–1638 [PubMed]
- Marshall S (2006) Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci STKE 2006:re7 [DOI] [PubMed]
- Matsumoto M, Ogawa W, Teshigawara K, Inoue H, Miyake K, Sakaue H, Kasuga M (2002) Role of the insulin receptor substrate 1 and phosphatidylinositol 3-kinase signaling pathway in insulin-induced expression of sterol regulatory element binding protein 1c and glucokinase genes in rat hepatocytes. Diabetes 51:1672–1680 [DOI] [PubMed]
- Mayer C, Zhao J, Yuan X, Grummt I (2004) Mtor-dependent activation of the transcription factor tif-ia links rrna synthesis to nutrient availability. Genes Dev 18:423–434 [DOI] [PMC free article] [PubMed]
- Mihara K (1990) Structure and regulation of rat liver microsomal stearoyl-coa desaturase gene. J Biochem (Tokyo) 108:1022–1029 [DOI] [PubMed]
- Mounier C, Posner BI (2006) Transcriptional regulation by insulin: from the receptor to the gene. Can J Physiol Pharmacol. CJPP485 [DOI] [PubMed]
- Mounier C, Dumas V, Posner BI (2006) Regulation of hepatic insulin-like growth factor-binding protein-1 gene expression by insulin: central role for mammalian target of rapamycin independent of forkhead box o proteins. Endocrinology 147:2383–2391 [DOI] [PubMed]
- Nadeau KJ, Leitner JW, Gurerich I, Draznin B (2004) Insulin regulation of sterol regulatory element-binding protein-1 expression in l–6 muscle cells and 3t3 l1 adipocytes. J Biol Chem 279:34380–34387 [DOI] [PubMed]
- Ntambi JM (1992) Dietary regulation of stearoyl-coa desaturase 1 gene expression in mouse liver. J Biol Chem 267:10925–10930 [PubMed]
- Ntambi JM (1995) The regulation of stearoyl-coa desaturase (scd). Prog Lipid Res 34:139–150 [DOI] [PubMed]
- Ntambi JM (1999) Regulation of stearoyl-coa desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res 40:1549–1558 [PubMed]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G (1997) The fork head transcription factor daf-16 transduces insulin-like metabolic and longevity signals in c. Elegans Nature 389:994–999 [DOI] [PubMed]
- Patel S, Lochhead PA, Rena G, Fumagalli S, Pende M, Kozma SC, Thomas G, Sutherland C (2002) Insulin regulation of insulin-like growth factor-binding protein-1 gene expression is dependent on the mammalian target of rapamycin, but independent of ribosomal s6 kinase activity. J Biol Chem 277:9889–9895 [DOI] [PubMed]
- Peng T, Golub TR, Sabatini DM (2002) The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol 22:5575–5584 [DOI] [PMC free article] [PubMed]
- Porstmann T, Griffiths B, Chung YL, Delpuech O, Griffiths JR, Downward J, Schulze A (2005) Pkb/akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of srebp. Oncogene [DOI] [PubMed]
- Sakurai H, Mitsuhashi N, Tamaki Y, Akimoto T, Murata O, Kitamoto Y, Maebayashi K, Ishikawa H, Hayakawa K, Niibe H (1999) Interaction between low dose-rate irradiation, mild hyperthermia and low-dose caffeine in a human lung cancer cell line. Int J Radiat Biol 75:739–745 [DOI] [PubMed]
- Sambrook J, Russell DW (2000) Molecular cloning: A laboratory manual. Cold Spring Harbor, New-York
- Samson SL, Wong NC (2002) Role of sp1 in insulin regulation of gene expression. J Mol Endocrinol 29:265–279 [DOI] [PubMed]
- Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG (2000) Regulation of glucose-6-phosphatase gene expression by protein kinase balpha and the forkhead transcription factor fkhr. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem 275:36324–36333 [DOI] [PubMed]
- Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD (1998) Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J Biol Chem 273:35299–35306 [DOI] [PubMed]
- Sutherland C, O’Brien RM, Granner DK (1995) Phosphatidylinositol 3-kinase, but not p70/p85 ribosomal s6 protein kinase, is required for the regulation of phosphoenolpyruvate carboxykinase (pepck) gene expression by insulin. Dissociation of signaling pathways for insulin and phorbol ester regulation of pepck gene expression. J Biol Chem 270:15501–15506 [DOI] [PubMed]
- Sutherland C, Waltner-Law M, Gnudi L, Kahn BB, Granner DK (1998) Activation of the ras mitogen-activated protein kinase-ribosomal protein kinase pathway is not required for the repression of phosphoenolpyruvate carboxykinase gene transcription by insulin. J Biol Chem 273:3198–3204 [DOI] [PubMed]
- Tabor DE, Kim JB, Spiegelman BM, Edwards PA (1998) Transcriptional activation of the stearoyl-coa desaturase 2 gene by sterol regulatory element-binding protein/adipocyte determination and differentiation factor 1. J Biol Chem 273:22052–22058 [DOI] [PubMed]
- Tabor DE, Kim JB, Spiegelman BM, Edwards PA (1999) Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-coa desaturase 1 and 2. J Biol Chem 274:20603–20610 [DOI] [PubMed]
- Thiede MA, Strittmatter P (1985) The induction and characterization of rat liver stearyl-coa desaturase mrna. J Biol Chem 260:14459–14463 [PubMed]
- Wang D, Sul HS (1998) Insulin stimulation of the fatty acid synthase promoter is mediated by the phosphatidylinositol 3-kinase pathway. Involvement of protein kinase b/akt. J Biol Chem 273:25420–25426 [DOI] [PubMed]
- Waters KM, Ntambi JM (1994) Insulin and dietary fructose induce stearoyl-coa desaturase 1 gene expression of diabetic mice. J Biol Chem 269:27773–27777 [PubMed]
- Waters KM, Miller CW, Ntambi JM (1997) Localization of a polyunsaturated fatty acid response region in stearoyl-coa desaturase gene 1. Biochim Biophys Acta 1349:33–42 [DOI] [PubMed]
- Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X (2005) Gps: A comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res 33:W184–187 [DOI] [PMC free article] [PubMed]
- Yenush L, Zanella C, Uchida T, Bernal D, White MF (1998) The pleckstrin homology and phosphotyrosine binding domains of insulin receptor substrate 1 mediate inhibition of apoptosis by insulin. Mol Cell Biol 18:6784–6794 [DOI] [PMC free article] [PubMed]
- Zhang L, Ge L, Tran T, Stenn K, Prouty SM (2001) Isolation and characterization of the human stearoyl-coa desaturase gene promoter: requirement of a conserved ccaat cis-element. Biochem J 357:183–193 [DOI] [PMC free article] [PubMed]