Abstract
Autophagy is an intracellular phenomenon in which a cell digests its own constituents. Autophagy is well conserved in nature from lower eukaryotes to mammals and has been attributed to disparate physiological events – including cell death, the mechanism of which is different from apoptosis. However, unlike in apoptosis, in which a family of cysteine proteases (caspases) and a number of other regulatory proteins have been identified and characterized, the mechanism of autophagic cell death remains unclear. In addition, the general mechanisms by which autophagy is initiated and modulated are just emerging, and several lines of evidence indicate that activated class I phosphatidylinositol 3-kinase and mammalian target of rapamycin (mTOR) inhibit autophagy, while class III phosphatidylinositol 3-kinase acts as a facilitator. Autophagy has been attributed to a number of cardiac disorders, such as ischemic cardiomyopathy, cardiac hypertrophy, hemochromatosis and myocardial aging. Induction of ventricular hypertrophy is associated with decreased autophagy, whereas it is enhanced during the regression of hypertrophy. Induction of acute cardiotoxicity by the anticancer drug anthracycline is also associated with massive cardiomyocyte loss due to autophagy (and apoptosis). Myocyte loss due to autophagy has also been reported during progression from compensated hypertrophy to heart failure in a pressure-overloaded model. Although the depth and dimension of the regulatory network that modulates autophagy in mammalian cells has yet to emerge, existing evidence suggests that it is an integral part of maintaining cellular metabolism, organelle homeostasis and redox equilibrium. Thus, it is a likely possibility that autophagy plays a crucial role in maintaining healthy myocytes in the myocardium.
Keywords: Autophagy, Cardiac myocytes, Cell death
Sustenance and propagation in every organism, whether it be unicellular or multicellular, requires a well-coordinated mechanism of cell proliferation and cell death. However, since the mid 1960s, with the advent of sophisticated tools in cellular and molecular biology, cell proliferation has received preferential attention, while the mechanisms of cell death and its importance in tissue and organismal homeostasis have only lately been appreciated (1). Certain cell types such as mature blood cells and epithelial cells, among others, are periodically derived from their progenitors, thus, their loss under certain pathological conditions is less deleterious. However, certain others, such as cardiac myocytes and neuronal cells, are largely nonrenewable and cell death in those organs is lethal. Therefore, an understanding of cell death in general, as well as in the context of the myocardium (and the brain) in particular, has been the subject of intense interest. Until the early 1970s, cell death was primarily described by microscopic observations and ‘necrosis’ was the common term used for all forms of cell death. Thereafter, the term ‘apoptosis’, a biochemically defined and genetically orchestrated phenomenon of cell death, was established (1,2). However, the precise demarcation between necrosis and apoptosis is still debated, and apoptotic cell death with certain features of necrosis has also been documented (3,4). Moreover, with the progress in our understanding of cell death, it has also emerged that, depending on the mechanisms of induction and execution, cell death can be of multiple types (5). In this context, another form of cell death, named autophagic cell death, has drawn considerable attention during the past several years (1). The present review primarily describes some recent developments in understanding autophagic cell death, with specific reference to the myocardium. Of note, autophagic cell death is only one among various consequences of a cellular phenomena called autophagy.
AUTOPHAGY AND CELLULAR HOMEOSTASIS
Autophagy (in Greek, it means ‘to eat oneself’) is an intracellular event in which a cell digests its own constituents. The phenomenon of autophagy is well conserved in nature and has been documented in yeast, Caenorhabditis elegans, Drosophila and in mammals (6–8). In lower eukaryotes, it is essentially involved in recycling-salvaging of cellular constituents under starvation, whereas in mammals, it has been attributed to disparate events such as: nutritional stress in the liver, to ensure an adequate supply of amino acids (9); response to developmental cues (10); cell differentiation (11); removal of excess organelles (12,13); oxidative stress (14); intracellular accumulation of aberrant proteins (15,16); protection against diseases such as cancer, muscular disorders and neurodegeneration; progression of certain other diseases such as Huntington, Alzheimer and Parkinson; and the prevention of infection (17). In whole, it is thus expected that induction of autophagy in mammalian cells is a highly evolved and coordinated event whereupon multiple signal inputs are integrated in a given cellular context followed by its execution. Autophagy may also occur under normal conditions, to remove long-lived proteins and excess organelles, and to maintain cellular homeostasis (12). Finally, autophagy has been attributed to cell death, the mechanism of which is different from apoptosis and, thus, it is called type II (while apoptosis is type I) programmed cell death. Overall, induction of autophagy and its consequences are varied, especially in mammalian cells (18).
MECHANISMS OF AUTOPHAGY
In a common form of autophagy (ie, macroautophagy), a double membrane cytoplasmic vesicle, called the autophagosome, engulfs cellular constituents, including organelles. Thereafter, autophagosomes fuse with lysosomes, forming autolysosomes, followed by lysis (19). Autophagosomes may also deliver the sequestered cytoplasmic contents into a collecting vacuole called the amphisome, which then delivers the cargo to a lysosome, where sequestered components are digested (20). Although autophagy is largely a nonselective process, wherein a bulk of the cytoplasmic contents is sequestered into the autophagosome, under certain conditions it can be selective, leading to the removal of injured mitochondria, endoplasmic reticulum and excess peroxisome, among others (13). In another form of autophagy, called microautophagy, cytosolic vesicles bud into the lysosomal lumen by direct invagination followed by the degradation of both cytoplasmic components and the lysosomal membrane (21). In a third form of autophagy, called chaperone-mediated autophagy, only selective proteins directly cross the lysosomal membrane into the lumen, where they are rapidly degraded (22). While the mechanisms of macro-autophagy are well investigated, that of microautophagy and chaperone-mediated autophagy are less understood. Autophagy was first described in mammalian cells as a morphological phenomenon in the mid 1960s (23). However, its mechanism and relevance only recently emerged with the identification of a family of over 20 genes named autophagy-related genes (ATG) in yeast and their counterparts in mammals (8,24–28). Additional genes are also likely to be involved, especially in the early stages of autophagosome formation as well as its fusion with the vacuoles at a later stage (29). Noticeably, in yeast, ATG genes are involved in two other related but distinct intracellular events named the cytoplasm-to-vacuole targeting pathway and peroxisome degradation (pexophagy), thereby indicating that autophagy has evolved from an ancestral cellular event common to these pathways (30).
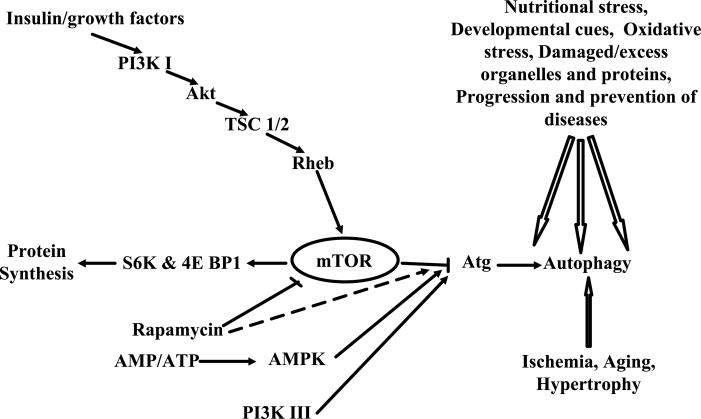
The precise mechanisms by which autophagy is initiated and modulated under various conditions are just emerging, and several lines of evidence indicate critical roles for the target of rapamycin (TOR) and phosphatidylinositol 3- (PI3) kinases. TOR proteins are members of a family termed phosphatidylinositol kinase-related kinases, which comprises a number of signalling kinases, namely, ataxia telangiectasia mutated kinase (ATM), ataxia telangiectasia- and Rad3-related kinase (ATR) and DNA-dependent protein kinase. In addition, despite its homology to lipid kinases, TOR is a serine/threonine protein kinase (31). Mammalian TOR (mTOR) (approximately 280 kD) consists of multiple regulatory and interacting domains (31,32). Insulin and other progrowth, prosurvival factors bind to their cognate receptors, resulting in the activation of the class I PI3-protein kinase B pathway, which, in turn, phosphorylates the tuberous sclerosis 1/2 complex. Phosphorylated tuberous sclerosis 1/2 then degrades, releasing the small GTPase Rheb (Ras homologue enriched in brain), which stimulates TOR activity (33,34). Activated mTOR phosphorylates downstream targets such as p70 S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), increasing protein synthesis (35). Studies performed with a number of mammalian cells demonstrate that amino acid depletion results in dephosphorylation of S6K1 and 4EBP1, whereas the addition of amino acids rescues such effects (31). Inactivation of mTOR by rapamycin induces autophagy, thereby assigning it an inhibitory role. Interestingly, it has recently been demonstrated that S6K activity is also required for autophagy (36). Because activated mTOR induces S6K, whereas it inhibits autophagy, the role of S6K in autophagy is yet to be understood. Furthermore, in Huntington disease, mTOR is sequestered in polyglutamine aggregates, impairing its kinase activity and leading to autophagy (37). Similarly, the inactivation of mTOR by the activation of p53 (by etoposide treatment) also induces autophagy (38). Autophagy is an ATP-dependent process, and the depletion of ATP is likely to suppress it. In addition, a fall in ATP level is concurrent with an increase in the AMP to ATP ratio, resulting in the activation of AMP-activated protein kinase. Accordingly, high levels of AMP and activated AMP-activated protein kinase suppress autophagy (38). Nevertheless, the induction of autophagy following ATP depletion has also been reported (39). Accumulating data suggest that PI3 kinase also plays a critical role in modulating autophagy (40). Noticeably, while class III PI3 kinase acts as a mediator of autophagy, class I PI3 kinase antagonizes it. Among three mammalian PI3 kinases, class III kinase is the most ancient, and is conserved from yeasts to humans (Vps34 and hVps34, respectively). Genetic studies in yeast have shown that Vps34, its regulatory subunit Vps15, and two other associated proteins, namely, Vps14 and Vps30/Atg6, are all required for autophagy (41). 3-Methyladenine, an inhibitor of class III PI3 kinase, inhibits autophagy in mammalian cells and phosphatidylinositol-3-phosphate (PtdIns-3-P) can reverse this effect (42). Beclin 1, the human orthologue of yeast autophagy gene Apg6/Vps30, promotes autophagy in mammalian cells (8,42). Although Beclin 1 was initially identified as an interacting partner of antiapoptotic protein Bcl-2, subsequent coimmunoprecipitation assays identified it as a partner of mammalian class III PI3 kinase hVps34 (43). Overall, it appears that class III PI3 kinase and its product PtdIns-3-P are required for autophagy. Available evidence suggests that Vps34 regulates autophagy at an early phase of elongation but the specific role of PtdIns-3-P in autophagy is yet to be understood (44). Contrary to class III PI3 kinase, class I PI3 kinase inhibits autophagy, presumably by the activation of Akt, which, in turn, activates TOR as its downstream target. In agreement, while autophagy is induced by PI3 phosphatase PTEN (phosphatase and tensin homologue), it is inhibited by insulin, an inducer of class IA PI3 kinase/Akt and also by constitutively active Akt (45,46). In addition to the above mentioned kinases, monomeric G proteins (Rab7 and Rab24) and heterotrimeric G proteins (Gi3) have also been attributed to autophagy. It appears that while monomeric G proteins modulate the recruitment of cytosolic proteins into autophagic vesicles, Gi3 and its associated proteins modulate the flow of membranes from the exocytic pathway to autophagic pathways (47,48). Although genetic analysis in yeast has significantly enhanced our understanding of mechanisms of autophagy, the precise details are still emerging, especially in mammalian cells (Figure 1). Autophagy is initiated by the formation of a C-shaped double membrane structure in the cytoplasm; thereafter, both ends grow and finally close to form the autophagosome. The source of the membrane structure of the autophagosome remains a subject of intense debate (49). Two ubiquitin-like conjugation systems, namely, Atg12-Atg5 and Atg8-phosphatidylethanolamine, are involved in the biogenesis of the autophagic vesicles and a substantial number of Atg genes are dedicated to their formation. They are also conserved in higher eukaryotes, thereby highlighting their critical role in autophagy (50,51). The Atg12-Atg5 conjugate noncovalently binds another protein, Atg16 (the mouse counterpart is Atg16L), forming a multimeric structure that associates with a small crescent-shaped vesicle, initiating the formation of autophagosomes while dissociating from the mature one (52). Yeast Atg8 is a unique ubiquitin-like protein whose target of modification is a phospholipid (ie, phosphatidylethanolamine) (53). Three mammalian homologues of Atg8 (golgi-associated ATPase enhancer of 16 kDa [GATE-16], gamma-aminobutyric acid receptor-associated protein [GABARAP] and microtubule-associated protein 1 light chain 3 beta [MAP1LC3]), have been identified, which also conjugate with phospholipids and are localized in the autophagosome (54). Furthermore, a number of variants of MAP1LC3 have recently been reported, which are used as markers for autophagy (55). A third protein, Atg1, has also been characterized as a regulator of autophagosome formation. However, unlike Atg8 and Atg12-Atg5, Atg1 it is likely to be involved in the downstream events rather than in initiation. Atg1 is a serine/threonine protein kinase that interacts with a number of proteins and presumably acts as a switch between autophagy and vesicular trafficking (56). For example, in nutrient-rich conditions, Atg13 is hyperphosphorylated (mTOR dependent) and, thus, does not interact with Atg1. In conditions of starvation, Atg13 is partially dephosphorylated and interacts with Atg1, triggering autophagy (56,57). Another protein, Atg17, also interacts with Atg1, and plays a critical role in determining the extent of autophagy induced by Atg1 (58). Nevertheless, the exact mechanisms that integrate the functions of all ATGs are yet to be understood (56).
Figure 1).
Various regulatory pathways involved in the modulation of autophagy are shown. The inductive signals are shown by arrows, whereas inhibitory signals are shown as lines with blocked termini. The general stimulatory effect of rapamycin is shown by a broken arrow. The general inductive signals stimulating autophagy are shown on the top right, whereas those specific to the myocardium are shown in the bottom right. 4E BP1 Eukaryotic translation initiation factor 4E binding protein 1; AMPK AMP-activated protein kinase; Atg Autophagy-related genes; PI3K Phosphatidylinositol 3-kinase; Rheb Ras homologue enriched in brain; S6K p70 S6 kinase; TSC 1/2 Tuberous sclerosis 1/2
AUTOPHAGIC CELL DEATH
Despite a large number of studies that have observed autophagy in dying cells, whether it is an independent mechanism of cell death has been the subject of intense debate (18,59). Because autophagy is induced under conditions such as limited supply of nutrients and tropic factors, as well as during cellular remodelling (eg, organ development) and the removal of damaged organelles, it has been argued that autophagy is primarily a process of limited destruction aimed toward survival (60). Nevertheless, with the progress of our understanding of the biochemical, cell biological, molecular and the genetic basis of autophagy, it is becoming more and more apparent that, at least under certain conditions, autophagy may be directed toward programmed cell death, which is different from apoptosis and, thus, termed type II programmed cell death (whereas apoptosis is type I) (18). While the hallmarks of apoptosis are membrane blebbing, chromatin condensation, and fragmentation of the nucleus and the cytoplasm into apoptotic bodies, autophagic cell death is characterized by cell shrinkage and the formation of multiple autophagic vacuoles (18,61). Experimental evidence supporting autophagy as an independent mechanism of cell death has come from studies on mouse embryonic fibroblasts deficient in the proapoptotic proteins Bcl2-associated X protein (Bax) and BCL2-antagonist/killer 1 (Bak). These cells, when exposed to certain cytotoxic agents (such as etoposide and staurosporine [but not other agents such as tumour necrosis factor]), undergo autophagy followed by cell death. Furthermore, suppression of autophagy by 3-methyladenine or by silencing Atg5 and Atg6 prevents cell death, thereby demonstrating that autophagy is an alternative but independent form of programmed cell death (18,62). Autophagic cell death has also been documented in Purkinje cells with the lurcher mutation in the delta 2 glutamate receptor (63). However, unlike in apoptosis, where a family of cysteine proteases (caspases) and a number of other regulatory proteins (eg, cytochrome c, direct IAP binding protein with low pI [DIABLO], apoptosis inducing factor [AIF] and Bax) have been identified and extensively characterized, the precise mechanism by which autophagic cell death is executed remains unclear. Furthermore, while a number of studies have suggested that autophagy and apoptosis are mechanistically interconnected (64,65), a number of others have shown that autophagy is a distinct form of cell death (66,67).
AUTOPHAGY IN THE MYOCARDIUM
Emerging evidence suggests that the myocardium has regenerative potential because cardiac myocytes can be replenished by stem cells (68). However, natural replacement of cardiac myocytes is a slow and ineffective process, and thus, myocytes are largely nonrenewable (69,70). Therefore, the maintenance of healthy myocytes and prevention of their loss under pathological conditions has been a subject of intense therapeutic interest. Interestingly, the incidence of autophagy was described in the myocardium long before its implications were known (71). With progress in our understanding of the importance of autophagy in cellular homeostasis, only recently has it drawn sizable attention from cardiac biologists (72). Because they are long-living cells, cardiac myocytes (and neurons) are susceptible to age-related changes, which are further aggravated by other related conditions such as hypertension and atherosclerosis. With aging, oxidized cross-linked proteinaceous materials accumulate within the cells, and mitochondria and lysosomes become severely affected (73,74). In aging cells, mitochondria undergo structural alterations leading to decreased efficiency, while lysosomes accumulate lipofuscin, a brown, granular pigment that consists of cross-linked lipids and proteins produced during lysosomal digestion (75). While proliferating cells can remove those ‘biological wastes’ by cell division, cardiac myocytes cannot do so because they are terminally differentiated, and they maintain cellular homeostasis only by the activation of degrading pathways, namely, macroautophagy, microautophagy and chaperone-mediated autophagy (75). Thus, in essence, the accumulation of defective mitochondria and lysosomes in aged myocytes is a reflection of inefficient autophagy (73). In fact, the accumulation of lipofuscin in the lysosomes is primarily due to sustained autophagic activities in those long-living cells. Senescent myocytes are characterized by the presence of giant mitochondria arising due to oxidative damage followed by inefficient mitochondrial DNA repair and inept autophagy (76,77). Accumulating evidence also suggests that certain mutations in the mitochondial DNA may accelerate their fission, resulting in the replacement of normal mitochondria by the mutated one, which is less efficient in energy metabolism, and thus, less susceptible to oxidative damage and less vulnerable to autophagy (78). In agreement with this, the treatment of cultured cardiac myocytes with 3-methyladenosine, an inhibitor of autophagy, results in the accumulation of small normal mitochondria with a concomitant but moderate increase in the giant forms, thereby indicating that autophagy is more effective in removing small normal mitochondria (79). It is hypothesized that autophagocytosis of large mitochondria requires more energy, and thus, is less efficient (72). Inefficient mitochondrial autophagy may also be due to decreased lysosomal activity arising from the accumulation of lipofuscin with increasing age. When lytic enzymes are directed toward lipofuscin-filled lysosomes, they are trapped in it, thereby compromising their autophagic potential (80). Taken together, inefficient or a lack of autophagy plays a critical role in the development of aging-related disorders in the myocardium, and any therapeutic intervention that enhances autophagy may contribute toward increased cardiac performance (72). Such a proposition is corroborated by improvement in cardiac performance under calorie restriction. Autophagy has also been attributed to a number of other cardiac disorders such as ischemic cardiomyopathy, cardiac hypertrophy and hemochromatosis. The induction of ventricular hypertrophy (an anabolic process induced by hypertension, aortic constriction and adrenergic stress, among others) is associated with decreased autophagy (81), whereas it is enhanced during the regression of hypertrophy (82). Lysosomal storage disorders such as Fabry, Pompe and Danon diseases are characterized by the accumulation of undigested materials in the lysosomes due to defects in genes encoding lysosomal enzymes and other proteins. However, compared with the age-related accumulation of lipofuscin, the accumulation in lysosomal storage diseases is more extensive and has an early onset. One common manifestation of lysosomal storage diseases is cardiomyopathy. Although the mechanism by which lysosomal storage diseases induce cardiomyopathy is not yet known, a possible contribution from inefficient autophagy (due to lysosomal dysfunction), resulting in the accumulation of defective mitochondria, has been proposed (72,83). The induction of acute cardiotoxicity by the anticancer drug anthracycline is also associated with massive cardiomyocyte loss due to autophagy and apoptosis (84). A single cycle of ischemia-reperfusion induces autophagic cell death, whereas urocortin represses it by suppressing Beclin 1 expression via the PI3 kinase-Akt pathway (85). In a recent study, Kostin et al (86) elegantly demonstrated that in failing human heart, a loss of myocytes occurs due to a concurrent incidence of oncosis, apoptosis and autophagy. Furthermore, autophagic cell death is associated with an extensive accumulation of ubiquitin-protein complexes. Myocyte loss due to autophagy has also been reported during the progression from compensated hypertrophy to heart failure in a pressure-overloaded model (87). In chronically ischemic pig myocardium, autophagy is induced with a decrease in apoptosis, presumably by a homeostatic mechanism that suppresses apoptosis and stimulates autophagy, thereby providing cardioprotection (88).
CONCLUSION
Cellular and organismal well-being is maintained by multiple regulatory arms in coordination with various checks and balances. While a plethora of growth stimulatory factors may induce cell proliferation, it is also restrained by multiple check points, ensuring healthy growth. Similarly, intracellular stress is handled by the unfolded protein response and endoplasmic reticulum stress, proteasomal pathways and redox enzymes. Finally, when intracellular hazards become too much to cope with, discrete biochemical and cellular programs are initiated, leading to the systematic elimination of unhealthy cells. In this regard, autophagy can be perceived as a unique mechanism of maintaining cellular homeostasis. Although the depth and dimension of the regulatory network that modulates autophagy in mammalian cells is yet to emerge, existing evidence suggests that it is an integral part of cellular metabolism, organelle homeostasis and redox equilibrium. In this context, it is likely that autophagy may play a crucial role in maintaining healthy myocytes in the myocardium. Therefore, an enhanced understanding of regulatory points that connect the autophagic network with that of other intracellular pathways will open new avenues for cardiovascular therapy. It is thus expected that the coming years will see a quantum increase in the understanding of autophagy in the context of myocardium.
REFERENCES
- 1.Edinger AL, Thompson CB. Death by design: Apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–9. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papucci L, Formigli L, Schiavone N, et al. Apoptosis shifts to necrosis via intermediate types of cell death by a mechanism depending on c-myc and bcl-2 expression. Cell Tissue Res. 2004;316:197–209. doi: 10.1007/s00441-004-0872-z. [DOI] [PubMed] [Google Scholar]
- 4.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 5.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–16. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;19(173):963–74. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takacs-Vellai K, Vellai T, Puoti A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C elegans. Curr Biol. 2005;23(15):1513–7. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 9.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–82. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 10.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Fader CM, Savina A, Sanchez D, Colombo MI. Exosome secretion and red cell maturation: Exploring molecular components involved in the docking and fusion of multivesicular bodies in K562 cells. Blood Cells Mol Dis. 2005;35:153–7. doi: 10.1016/j.bcmd.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Iwata J, Ezaki J, Komatsu M, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–41. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 13.Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: A question of fate? Cell Death Differ. 2005;12(Suppl 2):1484–9. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- 14.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–62. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 15.Shibata M, Lu T, Furuya T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 16.Kamimoto T, Shoji S, Hidvegi T, et al. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–76. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–41. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 18.Tsujimoto Y, Shimizu S. Another way to die: Autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–34. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 19.Yorimitsu T, Klionsky DJ. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–52. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stromhaug PE, Seglen PO. Evidence for acidity of prelysosomal autophagic/endocytic vacuoles (amphisomes) Biochem J. 1993;291:115–21. doi: 10.1042/bj2910115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uttenweiler A, Schwarz H, Mayer A. Microautophagic vacuole invagination requires calmodulin in a Ca2+-independent function. J Biol Chem. 2005;280:33289–97. doi: 10.1074/jbc.M506086200. [DOI] [PubMed] [Google Scholar]
- 22.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–35. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 23.Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33:437–49. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond EM, Brunet CL, Johnson GD, et al. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–5. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- 25.Meiling-Wesse K, Bratsika F, Thumm M. ATG23, a novel gene required for maturation of proaminopeptidase I, but not for autophagy. FEMS Yeast Res. 2004;4:459–65. doi: 10.1016/S1567-1356(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Cregg JM, Dunn WA, Jr, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 27.Kawamata T, Kamada Y, Suzuki K, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338:1884–9. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 28.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–6. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 29.Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:2189–204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–9. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 32.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Levine AJ, Feng Z, Mak TW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–75. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 34.Inoki K, Guan KL. Complexity of the TOR signaling network. Trends Cell Biol. 2006;16:206–12. doi: 10.1016/j.tcb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: Implications for cancer biology. Mol Cell. 2003;12:271–80. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 38.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005 doi: 10.1038/sj.cdd.4401697. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 40.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. (Erratum in 2000;275:12360) [DOI] [PubMed] [Google Scholar]
- 41.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes – Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–39. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 43.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–70. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 44.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–14. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 45.Gu Y, Wang C, Cohen A. Effect of IGF-1 on the balance between autophagy of dysfunctional mitochondria and apoptosis. FEBS Lett. 2004;577:357–60. doi: 10.1016/j.febslet.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 46.Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–6. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 47.Pattingre S, Petiot A, Codogno P. Analyses of Galpha-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy. Methods Enzymol. 2004;390:17–31. doi: 10.1016/S0076-6879(04)90002-X. [DOI] [PubMed] [Google Scholar]
- 48.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 49.Juhasz G, Neufeld TP. Autophagy: A forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–9. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 52.Mizushima N, Kuma A, Kobayashi Y, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 53.Amar N, Lustig G, Ichimura Y, Ohsumi Y, Elazar Z. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep. 2006;7:635–42. doi: 10.1038/sj.embor.7400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281:3017–24. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 55.Wu J, Dang Y, Su W, et al. Molecular cloning and characterization of rat LC3A and LC3B – Two novel markers of autophagosome. Biochem Biophys Res Commun. 2006;339:437–42. doi: 10.1016/j.bbrc.2005.10.211. [DOI] [PubMed] [Google Scholar]
- 56.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–8. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 57.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 58.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eskelinen EL. Doctor Jekyll and Mister Hyde: Autophagy can promote both cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1468–72. doi: 10.1038/sj.cdd.4401721. [DOI] [PubMed] [Google Scholar]
- 60.Levine B, Yuan J. Autophagy in cell death: An innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roach HI, Aigner T, Kouri JB. Chondroptosis: A variant of apoptotic cell death in chondrocytes? Apoptosis. 2004;9:265–77. doi: 10.1023/b:appt.0000025803.17498.26. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–8. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 63.Dusart I, Guenet JL, Sotelo C. Purkinje cell death: Differences between developmental cell death and neurodegenerative death in mutant mice. Cerebellum. 2006;5:163–73. doi: 10.1080/14734220600699373. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez-Polo RA, Boya P, Pauleau AL, et al. The apoptosis/autophagy paradox: Autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 65.Sadasivan S, Waghray A, Larner SF, Dunn WA, Jr, Hayes RL, Wang KK. Amino acid starvation induced autophagic cell death in PC-12 cells: Evidence for activation of caspase-3 but not calpain-1. Apoptosis. 2006 doi: 10.1007/s10495-006-7690-6. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 66.Guillon-Munos A, van Bemmelen MX, Clarke PG. Role of phosphoinositide 3-kinase in the autophagic death of serum-deprived PC12 cells. Apoptosis. 2005;10:1031–41. doi: 10.1007/s10495-005-0741-6. [DOI] [PubMed] [Google Scholar]
- 67.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;19(22):463–75. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 68.Urbanek K, Rota M, Cascapera S, et al. Cardiac stem cells possess growth factor-receptor systems that after activation regenerate the infarcted myocardium, improving ventricular function and long-term survival. Circ Res. 2005;97:663–73. doi: 10.1161/01.RES.0000183733.53101.11. (Erratum in 2006;98:e27) [DOI] [PubMed] [Google Scholar]
- 69.Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature. 2006;441:1097–9. doi: 10.1038/nature04961. [DOI] [PubMed] [Google Scholar]
- 70.MacLellan WR, Schneider MD. Death by design. Programmed cell death in cardiovascular biology and disease. Circ Res. 1997;81:137–44. doi: 10.1161/01.res.81.2.137. [DOI] [PubMed] [Google Scholar]
- 71.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. Am J Pathol. 1980;98:425–44. [PMC free article] [PubMed] [Google Scholar]
- 72.Terman A, Brunk UT. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc Res. 2005;68:355–65. doi: 10.1016/j.cardiores.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 73.Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem Biol Interact. 2006 doi: 10.1016/j.cbi.2006.04.013. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 74.Grune T, Merker K, Jung T, Sitte N, Davies KJ. Protein oxidation and degradation during postmitotic senescence. Free Radic Biol Med. 2005;39:1208–15. doi: 10.1016/j.freeradbiomed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 75.Terman A, Brunk UT. Oxidative stress, accumulation of biological ‘garbage’, and aging. Antioxid Redox Signal. 2006;8:197–204. doi: 10.1089/ars.2006.8.197. [DOI] [PubMed] [Google Scholar]
- 76.Coleman R, Silbermann M, Gershon D, Reznick AZ. Giant mitochondria in the myocardium of aging and endurance-trained mice. Gerontology. 1987;33:34–9. doi: 10.1159/000212851. [DOI] [PubMed] [Google Scholar]
- 77.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–52. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Nekhaeva E, Bodyak ND, Kraytsberg Y, et al. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci USA. 2002;99:5521–6. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terman A, Dalen H, Eaton JW, Neuzil J, Brunk UT. Aging of cardiac myocytes in culture: Oxidative stress, lipofuscin accumulation, and mitochondrial turnover. Ann NY Acad Sci. 2004;1019:70–7. doi: 10.1196/annals.1297.015. [DOI] [PubMed] [Google Scholar]
- 80.Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol. 2004;36:2376–91. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 81.Dammrich J, Pfeifer U. Cardiac hypertrophy in rats after supravalvular aortic constriction. II. Inhibition of cellular autophagy in hypertrophying cardiomyocytes. Virchows Arch B Cell Pathol Incl Mol Pathol. 1983;43:287–307. doi: 10.1007/BF02932962. [DOI] [PubMed] [Google Scholar]
- 82.Frenzel H, Schwartzkopff B, Rettig B, Vogelsang H. Morphologic criteria of progression and regression of cardiac hypertrophy. J Cardiovasc Pharmacol. 1987;10(Suppl 6):S20–8. [PubMed] [Google Scholar]
- 83.Verity MA. Infantile Pompe’s disease, lipid storage, and partial carnitine deficiency. Muscle Nerve. 1991;14:435–40. doi: 10.1002/mus.880140509. [DOI] [PubMed] [Google Scholar]
- 84.Semenov DE, Lushnikova EL, Nepomnyashchikh LM. Anthracycline-induced cardiomyopathy is manifested in decreased protein synthesis, impaired intracellular regeneration, and non-necrotic death of cardiomyocytes. Bull Exp Biol Med. 2001;131:505–10. doi: 10.1023/a:1017956922385. [DOI] [PubMed] [Google Scholar]
- 85.Valentim L, Laurence KM, Townsend PA, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–52. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 86.Kostin S, Pool L, Elsasser A, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–24. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 87.Hein S, Arnon E, Kostin S, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: Structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–91. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 88.Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA. 2005;102:13807–12. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]