Abstract
The insulin-responsive glucose transporter, GLUT4, is regulated in various physiologic states at the transcriptional level. When expressed in transgenic mice, the human GLUT4 promoter is governed by two cis-acting sequences: an MEF2 binding domain and Domain I, that function both as positive and negative regulators depending on the physiologic state. MEF2 proteins and GLUT4 enhancer factor (GEF) are known ligands for these cis-acting elements, but their mechanism of action is unclear. To begin to understand this important process, we have characterized GEF structural domains and its interactions with the MEF2A isoform. We find that the C terminus of GEF comprises its DNA-binding domain, but does not contribute to GEF homo-oligomerization. We also have found that GEF dimerizes with increased affinity to a hypophosphorylated form of MEF2A. Furthermore, we demonstrated that MEF2A binding to its cognate binding site can increase the DNA binding activity of GEF to Domain I, suggesting a novel mechanism for MEF2A transcriptional activation. Finally, we have demonstrated that the transcriptional co-repressor HDAC5 can interact with GEF in the absence of MEF2 proteins and specifically inhibit GLUT4 promoter activity. These findings lead to the hypothesis that GEF and the MEF2 proteins form a complex on the GLUT4 promoter that allows for recruitment of transcriptional co-regulators (repressors and/or activators) to control GLUT4 promoter activity.
GLUT4, the insulin-responsive glucose transporter, is expressed in heart, skeletal muscle, and adipose tissues, and is regulated at the transcriptional level in a variety of physiologic and pathologic states (1). As glucose transport is the rate-limiting step of glucose uptake (2), the ability to efficiently clear glucose from the blood after a meal depends on both a fully functional insulin-signaling cascade and the total number of transporters available for insertion into the cell membrane. Changes in GLUT4 abundance can then have profound effects on glucose homeostasis. As GLUT4 levels are decreased in diabetes mellitus (3–7), up-regulation of GLUT4 transcription can alleviate the pathologic hyperglycemia present in this disease (8–10).
The human GLUT4 promoter has putative binding sites for several transcriptional regulators, but two cis-acting regions specifically been have identified in vivo to impart correct tissue-specific and hormonal/metabolic regulation of GLUT4 expression: Domain I, bound by the novel human transcription factor GEF,2 and the MEF2 domain, bound by MEF2A and MEF2D in GLUT4-expressing tissues (11–13). Whereas each binding site is required for full promoter activity and correct transcriptional regulation, neither alone is sufficient to drive GLUT4 expression. Interestingly, we and others have shown that these elements play both a positive role in tissue-specific expression and a negative role under certain physiologic states such as insulin deficiency (14–16). Therefore, examination of the transcription factors that bind these regions and how they function to regulate GLUT4 promoter activity is required.
GEF was first identified using a yeast one-hybrid screen searching for human transcription factors that interacted specifically with Domain I (12). In transgenic mice, deletion of Domain I in a human GLUT4 (hGLUT4)-driven chloramphenicol acetyltransferase reporter prevented chloramphenicol acetyltransferase expression, demonstrating the necessity of this region for GLUT4 promoter activity. GEF mRNA has been detected in all GLUT4 expressing tissues, and Domain I DNA binding activity mirrors the tissue distribution of GEF (12). Utilizing a null-cell GLUT4 promoter luciferase assay, GEF increased GLUT4 promoter activity, but only significantly when co-expressed with MEF2A (11). As MEF2A and MEF2D both could form a complex with GEF, the interactions of these transcription factors likely play a role in regulating the GLUT4 promoter.
The MEF2 transcription factor family often control various promoters in conjunction with other transcription factors and transcriptional co-regulators (17, 18). MEF2A, and to a lesser degree MEF2D appear to regulate the human GLUT4 promoter in conjunction with GEF (11). Furthermore, MEF2 transcription factors can down-regulate transcription via interactions with the class II histone deacetylases (HDACs) isoforms 4, 5, 7, and 9 (19, 20). HDAC5 has been implicated in regulating the GLUT4 promoter, although whether this regulation is direct has not been tested. Constitutive localization of HDAC5 to the nucleus leads to a 3-fold decrease of GLUT4 in the heart (21); in contrast, decreased association of MEF2 and HDAC association has been observed in exercised skeletal muscle, correlating with an increase in GLUT4 promoter activity (22). We hypothesized that GEF may also interact with HDAC5 to contribute potentially to the down-regulation of GLUT4 promoter activity in states such as insulin deficiency.
Whereas GEF and MEF2A/D can regulate the GLUT4 promoter, little is known how these transcription factors function at the molecular level. As GEF bears little sequence homology with previously identified transcription factors, we have begun to identify mechanisms by which it might govern GLUT4 transcription. We have narrowed the DNA-binding domain of GEF to the C terminus. We found that whereas GEF can homooligomerize, it does not self-associate within the DNA-binding domain. Furthermore, we have demonstrated a novel function of MEF2A: an ability to increase the affinity of GEF for its cognate DNA sequence. We have determined that both GEF and MEF2 can each form a complex with HDAC5 that inhibits GLUT4 transcription. Finally, we show that HDAC5 associates with the GLUT4 promoter in vivo in a temporal manner that is consistent with GLUT4 expression. We conclude that the HDAC5 plays a significant role in the regulation of GLUT4 gene expression in cultured adipocytes.
EXPERIMENTAL PROCEDURES
Transfections and Nuclear and Whole Cell Extraction—MEF2A, MEF2D, HA-GEF-11, GEF-FL, FLAG-MEF2D, and HDAC5 were transiently transfected into COS-7 cells or 3T3-L1 pre-adipocytes using the FuGENE 6 Transfection Reagent (Roche). In short, 6 mg of total DNA was incubated with 12 ml of FuGENE 6 reagent in F-12 Ham's starvation media for 10 min at room temperature. Each transfection mixture was then added to an ∼80% confluent plate of COS-7 cells and incubated for 24 h at 37°C + 5% CO2. For Flag-MEF2D-GEF-HDAC5 co-transfections, Flag-MEF2D and HDAC5 plasmids were held constant, whereas GEF-encoding plasmid was increased. Empty vector (pcDNA3) was used to normalize total DNA added. Nuclear extracts were prepared using the NE-PER protein extraction kit (Pierce), with 1× Complete-mini EDTA-free Protease Inhibitors (Roche) added to prevent proteolysis during the extraction procedure. Whole cell extracts were prepared as described as in Ref. 23. Cells were initially washed in 1× PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4) and resuspended in ice-chilled whole cell extract buffer (1× PBS, 0.5% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1× Complete-mini EDTA-free protease inhibitors (Roche)) with vortexing. Extracts were sonicated for 10 s on ice and insoluble debris was pelleted by centrifugation in a microcentrifuge at 12,000 × g for 10 min at 4 °C. Total protein concentrations were determined with Coomassie Plus Protein Assay Reagent (Pierce).
GST-GEF Fusion Protein Purification—GST-GEF fusion proteins were expressed via plasmids maintained in Escherichia coli Bl-21 cells. Cultures expressing GST-GEF-FL were grown in the presence of 100 mm zinc acetate. Following sonication to disrupt bacterial membranes, one-step affinity purification was performed via incubation with glutathione-Sepharose (Amersham Biosciences), rocking for 1 h at 4°C. Bead-bound protein was washed 3 times in ice-cold TBS (20 mm Tris, pH 7.6, 137 mm NaCl) + 1% Nonidet P-40, then 3 times in TBS + 0.5 m NaCl, then 5 times in TBS. For GST fusion protein co-precipitations, proteins were stored at 4 °C in TBS + 5 mm dithiothreitol. For electrophoretic mobility shift assay (EMSA) analysis, proteins were eluted with TBS + 20 mm glutathione. Eluted fusion protein was concentrated using Amicon Centricon concentrators and dialyzed into storage buffer (10 mm HEPES, pH 7.4, 50 mm NaCl, 1 mm dithiothreitol).
Electrophoretic Mobility Shift Assays—EMSA analysis was performed as previously described (12). Radiolabeled probes were initially prepared by end-labeling oligonucleotides corresponding to the human GLUT4 Domain I binding site with excess [γ-32P]ATP (GE Healthcare) and T7 DNA kinase (Promega). Radiolabeled oligonucleotides were separated from unincorporated [γ-32P]ATP by centrifugation over Sepharose beads at 100 × g for 2 min. Purified GST-GEF fusion proteins were incubated with radiolabeled Domain I for 30 min in EMSA binding buffer (15 mm HEPES, pH 7.4, 40 mm KCl, 5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 0.4 mg/ml poly(dI-dC) (Sigma), 5% glycerol). DNA-protein complexes were resolved by non-denaturing polyacrylamide gel electrophoresis in 0.5× TBE at 4 °C, and visualized using autoradiography.
GST Fusion Protein Co-precipitations—For co-precipitations, equimolar amounts of GST fusion proteins (as determined by separation by SDS-PAGE and Coomassie staining) were mixed with nuclear or whole cell extracts in 1 ml of binding buffer (25 mm Tris-HCl, pH 7.5, 20 mm zinc acetate, 1 mm dithiothreitol, 1 mm MgCl2, 40 mm KCl) by rocking at 4 °C for 1 h. Protein complexes were washed 3 times in TBS + 0.5% Nonidet P-40 and twice in TBS. Proteins were eluted by boiling at 100 °C for 5 min in 2× Laemmli buffer (120 mm Tris, pH 6.8, 4% SDS, 20% glycerol, 140 mm β-mercaptoethanol) and analyzed by immunoblotting.
Biotinylated Oligonucleotide Precipitations—Whole cell extracts from COS-7 cells overexpressing GEF alone, MEF2A alone, or GEF and MEF2A together were incubated in EMSA binding buffer with 20 mm of one of the various biotinylated double-stranded oligonucleotides (Invitrogen) for 1 h at 4°C with gentle mixing: two repeats of the GLUT4 promoter MEF2 binding site (2XMEF2): 5′-GGGAGCTAAAAATAGCAGCGGGAGCTAAAAATAGCAGC-3′, Domain I (D): CTTGTCCCTCGGACCGGCTCCAGGAACCAA, a mutant Domain I as previously described in Ref. 12 (Dmut): 5′-CTTGTCCCTCGGGTTAACTCCAGGAACCAA-3′, or an internal deletion of the human GLUT4 promoter lacking the non-conserved region between Domain I and the MEF2 binding site (hG4): 5′-CTTGTCCCTCGGACCGGCTCCAGGAACCAAGGGCCCCCGCCTGGGACAGGCCGGGACTGACATTTGGAGGCTCCCAACGTGGGAGCTAAAAATAGCAGC-3′. Streptavidin-agarose (Fluka) was then added to precipitate protein-oligonucleotide complexes, and further mixing proceeded for 1 h at 4°C. The beads were collected by centrifugation at 500 × g for 5 min at 4 °C and washed three times in ice-cold 1× PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4). Protein was eluted in 2× Laemmli buffer with boiling at 100 °C for 5 min and analyzed by SDS-PAGE and immunoblotting.
Real Time Polymerase Chain Reaction—RT-PCR was utilized to determine the presence of HDAC5 mRNA in skeletal muscle and adipose tissue. RNA was isolated from respective frozen tissues as follows. Tissue was homogenized in tissue lysis buffer (5 m guanidinium isothiocyanate, 30 mm sodium acetate, pH 5.2, 10 mm EDTA, pH 8.0, 5% β-mercaptoethanol, 0.2% Sarkosyl). CsCl was then added to 3.3 m, and the homogenate was layered over a CsCl cushion (5.7 m CsCl, 30 mm sodium acetate, 10 mm EDTA, pH 5.5). Samples were centrifuged at 120,000 × g overnight at 20 °C. After removal of the supernatant, the RNA pellet was resuspended in HES buffer (20 mm HEPES, pH 7.4, 5 mm EDTA, pH 8.0, 1% SDS). An equal volume of 4:1 chloroform:butanol was added, the suspension was vortexed, and RNA was separated at 500 × g for 3 min. The aqueous layer containing the RNA was removed and RNA was precipitated with 0.1 volume of 3 m sodium acetate, pH 5.2, and 2.5 volumes of 95% ethanol. RNA was stored in precipitated form at–20 °C.
RT-PCR was performed using the Titan One-Tube RT-PCR System (Roche) according to the manufacturer's specifications. HDAC5-specific oligonucleotides were used to expand a region of HDAC5 cDNA from skeletal muscle and adipose tissues: HDAC5 upper, 5′-ATGAACTCTCCCAACGAGTC-3′; HDAC5 lower, 5′-CGGTGCTGGCGATGGCACTC-3′.
Immunoprecipitations—HDAC5-GEF co-immunoprecipitations were carried out from whole cell extracts of COS-7 cells transiently transfected with GEF alone, HDAC5 alone, or GEF and HDAC5 co-transfected. 750 mg of whole cell extract was incubated with 2 mg of affinity-purified α-GEF-N antibody (Cocalico Biologicals) bound to Protein A/G PLUS-agarose (Santa Cruz Biotechnology) for 1 h at 4°C with gentle mixing. Samples were washed three times in fresh ice-chilled 1× PBS, and samples were eluted with boiling for 5 min in 2× Laemmli sample buffer and analyzed via SDS-PAGE gels and immunoblotting. FLAG-MEF2D-GEF-HDAC5 co-immunoprecipitations were performed from whole cell extracts prepared as described, but using 40 ml of EZview Red ANTI-FLAG M2 Affinity Gel (Sigma) prewashed in 1× PBS. Immunoprecipitations were incubated overnight at 4 °C with gentle mixing. Samples were washed five times in fresh ice-chilled whole cell extract buffer, and samples were eluted with boiling for 5 min in 2× Laemmli sample buffer.
Immunoblot Analysis—Denatured samples were separated by SDS-PAGE using 10% polyacrylamide gels. Proteins were then transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) overnight in transfer buffer (25 mm Tris, 190 mm glycine) at 0.2 Å and 4 °C. Membranes were blocked for 1 h at room temperature with Block Buffer (Li-Cor Biosciences). Membranes were then probed with rabbit a-GEF-N sera, rabbit polyclonal α-MEF2A (Santa Cruz Biotechnology), or goat polyclonal α-HDAC antibodies. For co-precipitations and oligonucleotide precipitations, GEF, MEF2A, and HDAC immunoreactive peptides were visualized following incubation with respective Alexa Fluor 680 goat anti-rabbit IgG or donkey anti-goat IgG secondary antibodies (Molecular Probes) using the Odyssey Infrared Imaging System (Li-Cor Biosciences). Following co-immunoprecipitations GEF immunoreactive peptides were visualized via incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (H+L) (Bio-Rad) and enhanced chemiluminescence.
3T3-L1 Adipocyte Differentiation and Electroporation—3T3-L1 were differentiated and transfected by electroporation as previous described (24). Luciferase assays were performed 24 h post-electroporation.
Luciferase Assays—3T3-L1 pre-adipocytes or day 6 adipocytes were transiently transfected with plasmids containing the –895-hGLUT4 promoter driving the firefly luciferase and pRLTK (Renilla luciferase driven by the TK promoter for transfection efficiency, Promega) along with plasmids encoding GEF, MEF2A, or HDAC5 as described in Fig. 4. 24 h post-transfection, luciferase assays were performed using the Dual Glo Luciferase Assay system (Promega) according to the manufacturer's specifications, with slight modification. All reactions were carried out at 25 °C. In short, cells plated in 6-well tissue culture dishes were resuspended by scraping into 1 ml of PBS. Cells were collected by centrifugation at 12,000 × g in a microcentrifuge for 20 s, and the supernatant was removed. Cell pellets were resuspended in 200 ml of F-12 starvation medium by vortexing. 200 ml of Dual Glo Luciferase substrate was added, and cells were further lysed by 2 passes in a 26-gauge needle. Reactions were incubated at room temperature for 10 min, and firefly luciferase relative light units/s were determined in a Femtomaster FB12 Luminometer (Zylux). 200 ml of Stop&Glo substrate and buffer were then added, and reactions were incubated at room temperature for 10 min. Renilla luciferase relative light units/s was then determined for each sample to control for transfection efficiency and general promoter activity.
FIGURE 4.
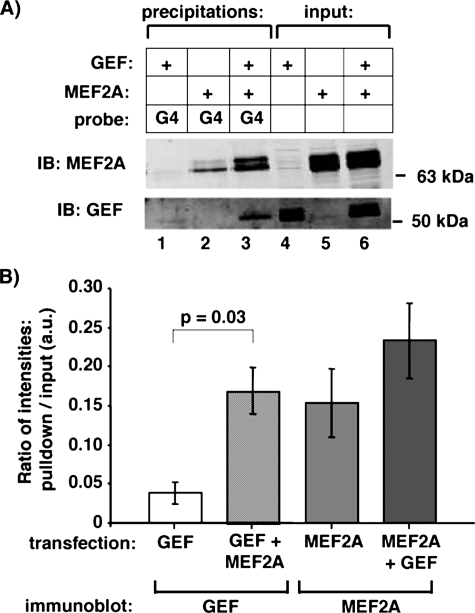
MEF2A increases the affinity of GEF for domain I. A, whole cell extracts of COS-7 cells transfected with GEF, MEF2A, or co-transfected with both were incubated with a biotinylated oligonucleotide corresponding to a portion of the hGLUT4 promoter (hG4, see Fig. 4 and “Experimental Procedures”). DNA-oligonucleotide complexes were precipitated with streptavidin-agarose, eluted by denaturation, and analyzed with anti-GEF and anti-MEF2A specific antibodies via immunoblot (IB). B, the ratio of band intensities of the precipitation lane versus the corresponding input lane from A were calculated for GEF alone (lanes 1 and 4), GEF + MEF2A (lanes 3 and 6), MEF2A (lanes 2 and 5), and MEF2A + GEF (lanes 3 and 6). Average and S.E. from three independent experiments are shown; a statistically significant increase in bound GEF was observed in the GEF + MEF2A co-transfection versus GEF alone (p = 0.03).
Chromatin Immunoprecipitation (ChIP) Assays—ChIP assays were performed using nuclear preparations from 3T3-L1 adipocytes using the ChIP-It express kit (Active Motifs) according to the manufacturer's specifications. The HDAC5 antibody used for ChIP assays was obtained from Santa Cruz Biotechnology, Inc. (San Diego, CA).
Statistical Analysis—Statistical significance of oligonucleotide precipitations and some luciferase activities was determined using Student's t test. Multiple experimental conditions were analyzed using one-way analysis of variance and Duncan's multiple range pairwise comparison to identify significant differences between conditions.
RESULTS
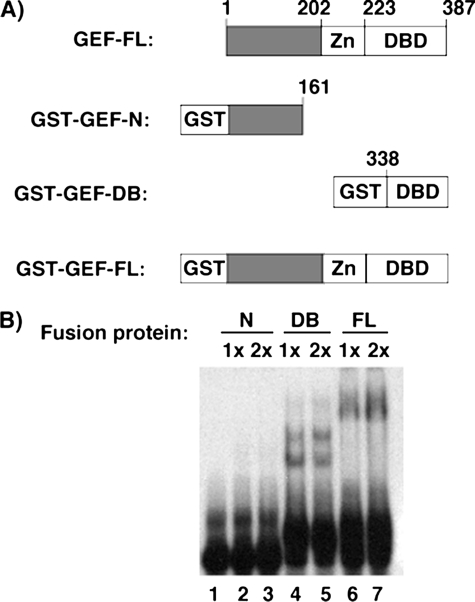
To better determine what regions of GEF are involved in protein-protein and protein-DNA interactions, we generated and purified various GST fusion products of GEF (Fig. 1). EMSAs were performed using a radiolabeled oligonucleotide corresponding to the human Domain I sequence and increasing amounts of GST-GEF-N, GST-GEF-DB, and GST-GEF-FL. Both full-length GEF and the C-terminal region of GEF were found to shift the radiolabeled probe, indicating that the DNA-binding domain falls within the final 49 amino acids.
FIGURE 1.
The C terminus of GEF contains the DNA-binding domain. A, a schematic representation of GST-GEF fusion proteins utilized in EMSA analysis. Residue numbers are shown for full-length (FL) and truncated versions of GEF. B, eluted GST-GEF fusion proteins were analyzed by EMSA to detect binding to the Domain I oligonucleotide. Increasing amounts of fusion proteins were mixed with radiolabeled Domain I oligonucleotides, separated over a non-denaturing polyacrylamide, and analyzed by autoradiography.
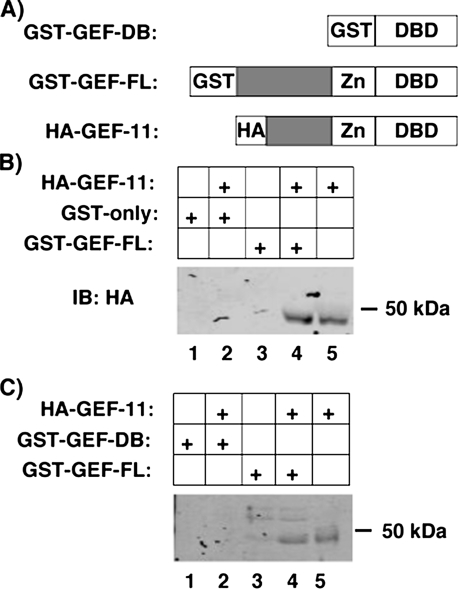
As GEF has previously been demonstrated to hetero-oligomerize with both MEF2A and MEF2D (11), we examined whether GEF can self-associate. Various GST-GEF fusion proteins were incubated with nuclear extracts from COS-7 cells transiently expressing HA-tagged GEF-11 (Fig. 2). HA-GEF-11 could be co-precipitated by GST-GEF-FL but not by GST-GEF-DB. We conclude that GEF can homo-oligomerize, and that the domain governing self-association falls between amino acids 68 and 338, which does not include the DNA-binding domain.
FIGURE 2.
Homo-oligomerization of GEF. GST-GEF can homo-oligomerize, but not within the DNA-binding domain (DB). A, schematic representation of GST- and HA-tagged GEF utilized in co-precipitations. GEF-11 comprises residues 68–387 of GEF-FL. B, nuclear extracts from COS-7 cells transiently expressing HA-GEF-11 were incubated with various GST-GEF fusion proteins bound to glutathione-Sepharose beads. Protein complexes were washed, eluted by denaturation, and analyzed by SDS-PAGE and immunoblotting for HA. FL, full-length.
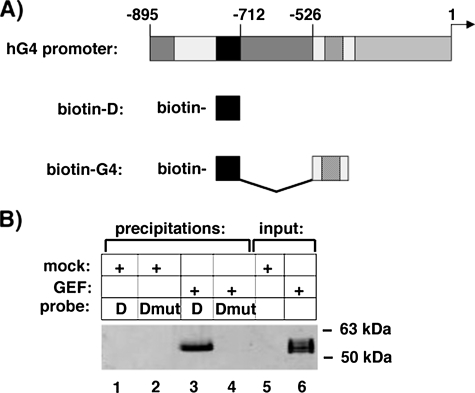
Knowing that GEF both homo- and hetero-oligomerizes, we hypothesized that a GEF-MEF2A complex might have altered DNA binding characteristics. To better examine the isoforms of proteins bound to DNA, our laboratory has developed a biotinylated oligonucleotide precipitation assay (16), in which biotinylated oligonucleotides corresponding to regions of the GLUT4 promoter are incubated with nuclear or whole cell extracts of interest. To first determine whether GEF could be precipitated using this methodology, we incubated nuclear extracts of COS-7 cells transiently transfected with either empty vector or plasmid expressing full-length GEF with biotinylated oligonucleotides corresponding to wild type Domain I or a mutant form of Domain I previously shown to not interact with GEF (12). GEF was specifically precipitated by Domain I, but not by mutant Domain I (Fig. 3). As the short Domain I oligonucleotide does not include an MEF2 binding site, we decided to expand the DNA to include a larger portion of the GLUT4 promoter. Previous deletion analysis of the GLUT4 promoter had determined that the non-conserved region between bases –712 and –526 of the human GLUT4 promoter were not required for full tissue-specific and metabolic/hormonal control of GLUT4 expression (12). We therefore designed a biotinylated oligonucleotide that recapitulated this promoter containing both Domain I and MEF2-binding sites linked together within a 99-base pair stretch (biotin-hG4). The biotin-hG4 oligonucleotide was able to precipitate GEF and MEF2A alone from whole cell extracts of transiently transfected COS-7 cells (Fig. 4). Surprisingly, there was an apparent increase in GEF DNA binding activity when MEF2A was co-transfected, when compared with the single transcription factors alone. When corrected for input levels of the transcription factors, there was a statistically significant increase in GEF precipitated by the biotin-hG4 oligonucleotide in the presence of MEF2A. Whereas there was an apparent trend for increased MEF2A DNA binding activity in the presence of GEF, the increase did not reach statistical significance.
FIGURE 3.
Biotinylated domain I oligonucleotide can specifically precipitate GEF. A, schematic of the human GLUT4 promoter and the biotinylated oligonucleotides designed as described under “Experimental Procedures.” B, nuclear extracts from COS-7 cells transiently transfected with empty vector (mock) or a vector expressing GEF (GEF) were incubated with biotinylated oligonucleotides corresponding to Domain I (D) or a mutant Domain I unable to bind GEF (Dmut). DNA-oligonucleotide complexes were precipitated with streptavidin-agarose, eluted by denaturation, and analyzed by SDS-PAGE and immunoblot for GEF and compared with 10% of input lysates (lanes 5 and 6).
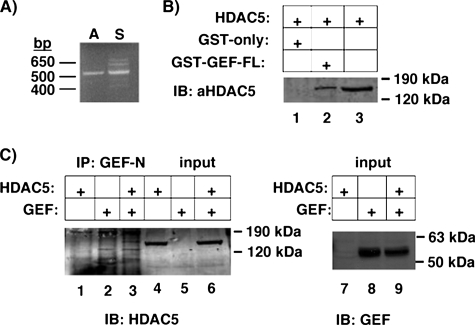
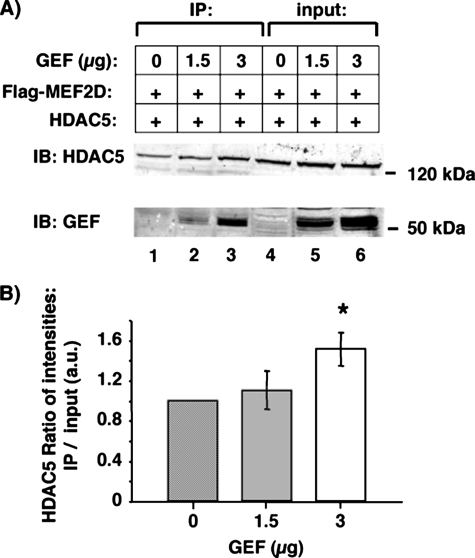
MEF2 proteins are known to regulate transcription via several co-activators and co-repressors, in particular HDAC5. To determine whether HDAC5 can interact with GEF as well as the MEF2 proteins to regulate GLUT4 promoter activity, we first established if HDAC5 is expressed in GLUT4-expressing tissues. As HDAC5 expression has been demonstrated in heart and skeletal muscle, we determined if HDAC5 is expressed in adipose tissue. HDAC5 mRNA was detected by RT-PCR in both adipose tissue and skeletal muscle (Fig. 5). The ability of GEF and HDAC5 to form a complex in vitro was tested by co- and immunoprecipitations. GST-GEF-FL was able to co-precipitate HDAC5 from whole cell extracts of COS-7 cells overexpressing the transcriptional repressor. Furthermore, GEF antibodies could specifically co-immunoprecipitate GEF and HDAC5. As GEF and HDAC5 could interact in the absence of any MEF2 proteins, we then hypothesized that GEF and MEF2D together might co-precipitate more HDAC5 than MEF2D alone. To test this, whole cell extracts from COS-7 cells expressing FLAG-MEF2D, HDAC5, and increasing amounts of GEF were incubated with anti-FLAG-agarose (Fig. 6). Co-immunoprecipitated HDAC5 and GEF were analyzed by SDS-PAGE and immunoblot, and compared with input lysates. A statistically significant increase of HDAC5 co-immunoprecipitated by FLAG-MEF2D in the presence of GEF was observed. Therefore, HDAC5 can form a multimeric complex with both GEF and the MEF2 transcription factors.
FIGURE 5.
GEF can interact with HDAC5. A, cDNA from a portion of HDAC5 was expanded using RT-PCR from mouse adipose (A) or skeletal muscle (S) RNA and specific primers corresponding to an approximate 500-bp region of HDAC5. B, whole cell extracts from COS-7 cells transiently expressing HDAC were incubated with bacterially expressed GST alone or GST-GEF-FL bound to glutathione-Sepharose. Protein complexes were washed, eluted by denaturation, and analyzed by SDS-PAGE and immunoblotting (IB) for HDAC5. C, proteins from COS-7 whole cell extracts from cells overexpressing GEF alone, HDAC5 alone, or GEF and HDAC5 together were immunoprecipitated (IP) using affinity purified anti-GEF polyclonal antibodies and Protein A/G PLUS-agarose. Immunoprecipitates were eluted by denaturation and analyzed by SDS-PAGE and immunoblot for HDAC5 (lanes 1–3). Input (3%) was loaded as a comparison for HDAC levels (lanes 4–6) or GEF (lanes 7–9). FL, full-length.
FIGURE 6.
GEF can increase the association of HDAC5 with FLAG-MEF2D. A, whole cell extracts of COS-7 transiently transfected with plasmids encoding FLAG-MEF2D, HDAC5, and increasing amounts of GEF were co-immunoprecipitated (IP) with anti-FLAG-agarose. Immunoprecipitated HDAC5 and GEF (lanes 1–3) were analyzed by SDS-PAGE and immunoblot (IB) and compared with 3% total input (lanes 4–6). B, the ratio of intensities of the HDAC5 bands from IP and input lanes were calculated in the absence (0 mg, lanes 1 and 4) and presence (1.5 mg, lanes 2 and 5; 3 mg, lanes 3 and 6) of GEF. The -fold increase of the HDAC5 IP/input ratio from 0 mg of GEF was statistically significant when comparing 0 to 3 mg of GEF (p < 0.05).
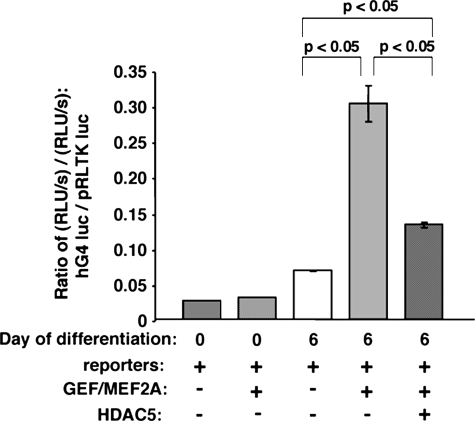
We next determined if HDAC5 could down-regulate the human GLUT4 promoter in 3T3-L1 adipocytes. We transiently transfected undifferentiated pre-adipocytes with a firefly luciferase reporter driven by the human GLUT4 promoter, as well as a plasmid encoding Renilla luciferase under the control of the TK promoter as a control for transfection efficiency and general promoter activity. Whereas firefly and Renilla luciferase activities were detected, we found that GEF and MEF2A could not activate the GLUT4 promoter in 3T3-L1 pre-adipocytes (Fig. 7), indicating that pre-adipocytes have an as yet undescribed mechanism for inhibiting GLUT4 gene transcription. Six days post-differentiation, however, GEF and MEF2A were able to significantly activate the human GLUT4 promoter in 3T3-L1 adipocytes. Furthermore, HDAC5 was able to specifically down-regulate the human GLUT4 promoter in the presence of MEF2A and GEF.
FIGURE 7.
HDAC5 can down-regulate the GLUT4 promoter in the presence of GEF and MEF2A. 3T3-L1 pre-adipocytes or adipocytes differentiated to day 6 were transiently transfected with luciferase reporter plasmids–895-hG4-luc (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), as well as plasmids encoding GEF, MEF2A, and HDAC5 (+ indicates inclusion in transfection). Luciferase assays were performed after 24 h; specific activation of the –895-hG4 promoter was calculated from the ratio of firefly to Renilla luciferase activities. GEF and MEF2A did not increase hGLUT4 promoter activity in pre-adipocytes. GEF and MEF2A statistically significantly increased–895-hG4 promoter activity in day 6 adipocytes; HDAC5 significantly decreased promoter activity in the presence of GEF and MEF2A in these cells. (Day 0 preadipocytes, n = 2; day 6 adipocytes, n = 3, shown are mean ± S.E.).
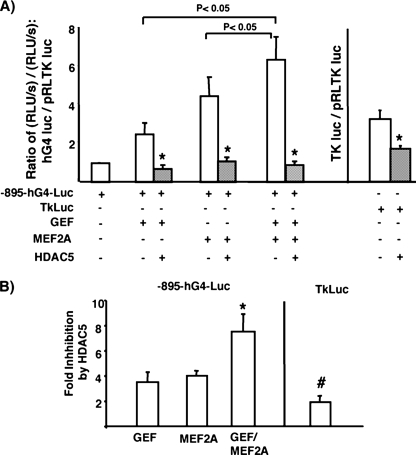
Because it is known that HDAC5 down-regulates MEF2A-dependent transcriptional activity (20, 25), we next determined if the HDAC5 was inhibiting GEF-dependent transcriptional activation. To this end, we transiently transfected fully differentiated adipocytes with the human GLUT4 promoter/luciferase reporter without or with GEF, MEF2A, or both together in the absence or presence of HDAC5 (Fig. 8A). Similar to transactivation in COS cells, both GEF and MEF2A contributed to GLUT4 promoter activity in fully differentiated adipocytes. Cotransfection with HDAC5 strongly inhibited both GEF and MEF2A-dependent transcriptional activation both separately and in combination. Calculation of the -fold inhibition by HDAC5 revealed that HDAC5 inhibited GEF-dependent activity 4-fold and MEF2A-dependent activity 4-fold. When both GEF and MEF2A were used in combination to transactivate the GLUT4 promoter, HDAC5 selectively inhibited transcriptional activity 8-fold, suggesting that both GEF and MEF2A-containing HDAC5 complexes were each contributing separately to the overall inhibition of GLUT4 transcriptional activity.
FIGURE 8.
HDAC5 specifically inhibits both GEF and MEF2A-dependent transcription of the GLUT4 promoter. A, 3T3-L1 adipocytes differentiated to day 6 were transiently transfected with luciferase reporter plasmids–895-hG4-luc (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), as well as plasmids encoding GEF, MEF2A, and HDAC5 (+ indicates inclusion in transfection). Luciferase assays were performed after 24 h; specific activation of the –895-hG4 promoter, or the TK promoter were calculated from the ratio of firefly to Renilla luciferase activities. Luciferase activity from activation of the –895-hG4 promoter transactivated by GEF, MEF2A, or both together was significantly greater than basal activation (p < 0.05) using one-way analysis of variance. The combination of GEF and MEF2A together significantly activated the promoter to a greater extent than either factor alone (using a Duncan's multiple range pairwise comparison). The asterisk indicates a significant inhibition of luciferase activity by the addition of HDAC5 (p < 0.05) using a Student's t test. B, the -fold inhibition was calculated for each case by dividing the ratio of expression without HDAC5 or the expression in the presence of HDAC5. The -fold inhibition was significantly increased when the promoter was transactivated by both GEF and MEF2A together (p < 0.05) using one-way analysis of variance and pairwise comparison. The nonspecific inhibition of TK-luc by HDAC5 was significantly lower than inhibition of –895-hG4-luc in each condition. Data shown are mean ± S.E. of n = 3 independent experiments performed in duplicate.
To determine whether the inhibition was specific for the GLUT4 promoter, we compared nonspecific inhibition of a control plasmid where the firefly luciferase was driven by the TK-promoter (the same promoter driving the Renilla luciferase control for transfection efficiency). The ratio of firefly luciferase to Renilla luciferase activity in this control was 2-fold lower in the presence of HDAC5 (Fig. 8, A and B). This -fold reduction was significantly lower than the -fold reductions observed with transactivation of the GLUT4 reporter with GEF and/or MEF2A (Fig. 8B).
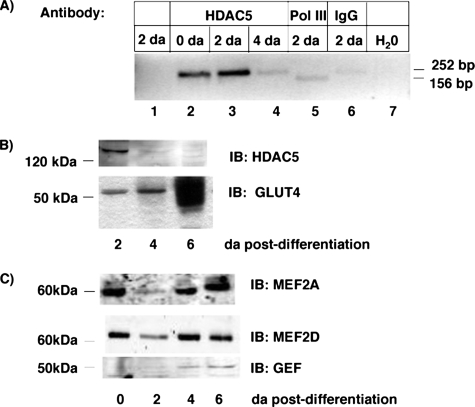
The physiologic significance of this work requires evidence that the HDAC5 complex can form on the GLUT4 promoter in a manner consistent with the level of expression of GLUT4. To begin to test the model, we used chromatin precipitation to determine whether HDAC5 forms a complex with the Glut4 promoter. HDAC5 immunoprecipitation of day 0 and day 2 adipocytes significantly enriched the DNA sequences containing Domain I- and MEF2-binding sites of the mouse Glut4 promoter (Fig. 9A, compare lanes 1, 2, and 3). By day 4 post-differentiation, HDAC5 immunoprecipitation did not significantly enrich Glut4 promoter sequences more than nonspecific IgG antibody (Fig. 9A, compare lanes 4 and 6). Suggesting that HDAC5 is not associated with the Glut4 promoter by 4 days post-differentiation in 3T3-L1 adipocytes.
FIGURE 9.
HDAC5 specifically associates with the mouse Glut4 promoter in a differentiation-dependent manner. A, chromatin immunoprecipitation assays were performed using HDAC5 antibody, RNA polymerase III (Pol III), or non-immune IgG antibody. Lane 1 shows the PCR performed on the input DNA not subjected to immunoprecipitation. Lane 7 shows the PCR performed with water. Lane 5 is the positive control for the PCR using immunoprecipitation with RNA polymerase III and a positive control primer set for mouse. B, nuclear extracts and cytoplasmic extracts were prepared from 3T3-L1 adipocytes harvested at days 2, 4, and 6 post-differentiation. The nuclear extracts were blotted for HDAC5, and cytoplasmic extracts blotted for GLUT4. Blots shown are representative of three independent experiments. C, nuclear extracts from adipocytes harvested at days 0, 2, 4, and 6 post-differentiation were blotted for MEF2A, MEF2D, and GEF. Results shown are representative of two independent experiments.
We hypothesized that the decreased association of HDAC5 with the Glut4 promoter may have arisen from decreased HDAC5 protein in nuclear extracts. To test this, we measured the abundance of HDAC5 protein in nuclear extracts from the days 2, 4, and 6 3T3-L1 adipocytes (Fig. 9B). HDAC5 protein was highest in Day 2 extracts and significantly reduced in days 4 and 6. This was inversely correlated with the expression of GLUT4 protein measured in those adipocytes (Fig. 9B). This is consistent with a role for HDAC5 in regulation of Glut4 transcription as a function of differentiation.
We examined the developmental pattern of expression of GEF, MEF2A, and MEF2D as a function of time post-differentiation (Fig. 9C). Expression of the MEF2 isoforms was relatively constant over the time course measured, whereas GEF increased about 2-fold at days 4 and 6 post-differentiation. The appearance of the MEF2 and GEF transcription factors was not as tightly correlated with the appearance of GLUT4 protein as was the disappearance of HDAC5 protein. This suggests that the release of HDAC5-mediated repression may play a greater role in regulation of Glut4 transcription.
DISCUSSION
Correct tissue-specific and hormonal/metabolic regulation of human GLUT4 promoter in transgenic mice requires both of the cis-acting regulatory domains within the –895-bp promoter: Domain I and the MEF2 domain. Whereas the MEF2-binding proteins have been studied in depth in their regulation of muscle and neuronal promoters, little is known about the proteins binding to Domain I. Previous work to identify proteins that bound the human GLUT4 promoter within Domain I identified a novel protein called GEF (12). GEF forms a protein complex with MEF2 proteins suggesting a mechanism of action by which Domain I and MEF2 work together to regulate the GLUT4 promoter (11). In this paper we demonstrate in vitro that GEF can self-associate suggesting that it may form a dimer or higher order oligomer in vivo (Fig. 2). Transcription factors, including the MEF2 family, often homo-dimerize in or near the DNA-binding domains. In contrast to MEF2, the dimerization domain of GEF requires peptide sequence between amino acids 68 and 338 that lies outside the DNA-binding domain of the protein. This is not surprising because MEF2-dependent gene promoters often function through formation of MEF2 protein heterodimers with other transcription factors, including members of the MyoD family of basic helix-loop-helix proteins (for review, see Ref. 18).
The formation of protein complexes was carried out in the absence of DNA. For these protein complexes to regulate GLUT4 promoter function, we hypothesized that GEF and MEF2A and/or MEF2D form a large multimeric complex with DNA. To test this, we examined how the isoforms of the proteins that interact specifically with their cognate DNA-binding domains. Using a transcriptionally competent oligonucleotide carrying both GEF and MEF2A/D-binding domains (12), we performed GEF/MEF2 affinity precipitations from nuclear extracts (Fig. 4). Whereas both GEF and MEF2A could be precipitated alone from whole cell extracts of transiently transfected COS-7 cells, we were surprised to see an increase in affinity of GEF for DNA in the presence of MEF2A. Whereas a similar trend for MEF2A was observed, its increase in DNA binding did not reach statistical significance. We therefore conclude that MEF2A can increase the binding affinity of GEF. This, coupled with previous data demonstrating that MEF2A and GEF can physically interact (11), led us to conclude that these proteins form a multimeric complex on DNA to regulate the GLUT4 promoter. This cooperative functioning of MEF2 proteins and other transcription factors has been observed in other systems. For example, regulation of the atrial natiuretic factor promoter by the transcription factor HAND1 requires both HAND1 and MEF2 DNA binding activities (26).
The cis-acting elements that are required for tissue-specific expression of the GLUT4 gene also function to modulate the level of GLUT4 gene expression in a variety of physiologic states (14–16). Therefore, we hypothesized that additional factors must be required to modulate GLUT4 expression up and down to meet physiologic needs of the tissues. In this report we demonstrated that the class II histone deacetylase (HDAC) 5 isoform specifically down-regulated both MEF2 and GEF-dependent gene transcription. This is consistent with MEF2 and GEF forming a multimeric complex with HDAC5 to regulate the human GLUT4 promoter.
HDAC5 was implicated in regulation of the GLUT4 promoter in human skeletal muscle (22) and constitutive localization of HDACs to the nucleus of heart tissue leads to decreased GLUT4 levels (21). We now show that the regulation of Glut4 in adipocytes may also be regulated by HDAC5. Interestingly, HDAC5 mRNA was detected in mature 3T3-L1 adipocytes (Fig. 4), however, Western blot analysis indicated that the HDAC5 protein decreased as a function of differentiation state. Recently, it has been shown that HDAC5 is regulated post-translationally in skeletal muscle (27). Our results would be consistent with a post-translation regulation of HDAC5 in differentiating adipocytes as well.
Our time course of HDAC5 expression is inversely correlated with the appearance of GLUT4, strongly suggesting a physiologic relevance to the in vitro experiments. Further support for the model was provided through ChIP analysis of HDAC5 association with the Glut4 promoter (Fig. 9). HDAC5 antibody specifically immunoprecipitated Glut4 promoter DNA in nuclear extracts from day 0 through day 2 post-differentiation. By day 4 post-differentiation, HDAC5 antibody did not significantly immunoprecipitate Glut4 promoter DNA, consistent with the decreased protein concentration of HDAC5 in nuclear extracts as a function of differentiation (Fig. 5). The association of HDAC5 with the Glut4 promoter preadipocytes and early in differentiation may be the molecular mechanism responsible for the lack of Glut4 transcription in cells at this age. The increased Glut4 promoter activity as a function of differentiation may result from the release of transcriptional inhibition. This is further supported by our observation that the MEF2 and GEF transcription factors are expressed early in differentiation and to some extent in preadipocytes, yet Glut4 transcription is strongly inhibited (Fig. 9C). Our observation that the GLUT4 promoter cannot be transactivated by overexpression of MEF2A and GEF in preadipocytes is consistent with such a mechanism.
Our working hypothesis now is that the MEF2 proteins and GEF form a regulatory platform in binding their cognate DNA sequences on the GLUT4 promoter, where they serve to attract co-regulators such as the HDACs to affect GLUT4 transcription. It is notable that both the up-regulation and the down-regulation of GLUT4 promoter activity in vivo and in vitro have been shown to require both of the cis-acting sequences (15, 16, 28). Furthermore, simply overexpressing MEF2 proteins in vivo did not rescue the loss of GLUT4 expression in diabetes mellitus, even though MEF2A and/or MEF2D expression had decreased (29, 30). This correlative data suggests that multimeric complexes involving both GEF interacting with Domain I and MEF2A/D interacting with the MEF2 domain, along with tertiary binding partners, can regulate the GLUT4 promoter.
Whereas we have not yet identified any co-activators, several transcriptional regulators have been implicated in either MEF2-driven transcription broadly or the GLUT4 promoter specifically. Identification of proteins that can interact with both MEF2A/D and GEF might give an indication of what co-regulators and upstream signaling cascades control GLUT4 promoter activity.
In summary, we have identified new molecular mechanisms by which the GLUT4 promoter can be regulated. Now recognizing that GEF and the MEF2 proteins may regulate GLUT4 transcription through complex protein-protein interactions, further work can be done to identify other proteins that can bind this complex in vivo. Understanding the signaling cascades that may regulate GEF and MEF2 localization and DNA binding might allow for new pharmacological interventions to control GLUT4 expression, allowing for increased insulin sensitivity and alleviation of diabetic symptoms.
This work was supported by National Institutes of Health Grant DK062341 and a predoctoral fellowship (to D. P. S.) from the American Diabetes Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GEF, GLUT4 enhancer factor; ChIP, chromatin immunoprecipitation; GLUT4, insulin responsive glucose transporter 4; MEF2, myocyte enhancer factor 2; HDAC, histone deactylase; TK, thymidine kinase; PBS, phosphate-buffered saline; GST, glutathione S-transferase; EMSA, electrophoretic mobility shift assay; HA, hemagglutinin; RT, reverse transcriptase.
References
- 1.Charron, M. J., Katz, E. B., and Olson, A. L. (1999) J. Biol. Chem. 274 3253–3256 [DOI] [PubMed] [Google Scholar]
- 2.Ziel, F. H., Venkatesan, N., and Davidson, M. B. (1988) Diabetes 37 885–890 [DOI] [PubMed] [Google Scholar]
- 3.Berger, J., Biswas, C., Vicario, P. P., Strout, H. V., Saperstein, R., and Pilch, P. F. (1989) Nature 340 70–72 [DOI] [PubMed] [Google Scholar]
- 4.Garvey, W. T., Huecksteadt, T. P., and Birnbaum, M. J. (1989) Science 245 60–63 [DOI] [PubMed] [Google Scholar]
- 5.Kahn, B. B., Simpson, I. A., and Cushman, S. W. (1988) J. Clin. Investig. 82 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha, M. K., Raineri-Maldonado, C., Buchanan, C., Pories, W. J., Carter-Su, C., Pilch, P. F., and Caro, J. F. (1991) Diabetes 40 472–477 [DOI] [PubMed] [Google Scholar]
- 7.Sivitz, W. I., DeSautel, S. L., Kayano, T., Bell, G. I., and Pessin, J. E. (1989) Nature 340 72–74 [DOI] [PubMed] [Google Scholar]
- 8.Gibbs, E. M., Stock, J. L., McCoid, S. C., Stukenbrok, H. A., Pessin, J. E., Stevenson, R. W., Milici, A. J., and McNeish, J. D. (1995) J. Clin. Investig. 95 1512–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brozinick, J. T., McCoid, S. C., Reynolds, T. H., Nardone, N. A., Hargrove, D. M., Stevenson, R. W., Cushman, S. W., and Gibbs, E. M. (2001) Diabetes 50 593–600 [DOI] [PubMed] [Google Scholar]
- 10.Christ-Roberts, C. Y., Pratipanawatr, T., Pratipanawatr, W., Berria, R., Belfort, R., Kashyap, S., and Mandarino, L. J. (2004) Metabolism 53 1233–1242 [DOI] [PubMed] [Google Scholar]
- 11.Knight, J. B., Eyster, C. A., Griesel, B. A., and Olson, A. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshel, K. M., Knight, J. B., Cao, K. T., Thai, M. V., and Olson, A. L. (2000) J. Biol. Chem. 275 23666–23673 [DOI] [PubMed] [Google Scholar]
- 13.Thai, M. V., Guruswamy, S., Cao, K. T., Pessin, J. E., and Olson, A. L. (1998) J. Biol. Chem. 273 14285–14292 [DOI] [PubMed] [Google Scholar]
- 14.Holmes, B. F., Lang, D. B., Birnbaum, M. J., Mu, J., and Dohm, G. L. (2004) Am. J. Physiol. 287 E739–E743 [DOI] [PubMed] [Google Scholar]
- 15.Olson, A. L., and Pessin, J. E. (1995) J. Biol. Chem. 270 23491–23495 [DOI] [PubMed] [Google Scholar]
- 16.Sparling, D. P., Griesel, B. A., and Olson, A. L. (2007) Am. J. Physiol. 292 E1149–E1156 [DOI] [PubMed] [Google Scholar]
- 17.Black, B. L., Molkentin, J. D., and Olson, E. N. (1998) Mol. Cell. Biol. 18 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black, B. L., and Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14 167–196 [DOI] [PubMed] [Google Scholar]
- 19.Han, A., He, J., Wu, Y., Liu, J. O., and Chen, L. (2005) J. Mol. Biol. 345 91–102 [DOI] [PubMed] [Google Scholar]
- 20.Lemercier, C., Verdel, A., Galloo, B., Curtet, S., Brocard, M. P., and Khochbin, S. (2000) J. Biol. Chem. 275 15594–15599 [DOI] [PubMed] [Google Scholar]
- 21.Czubryt, M. P., McAnally, J., Fishman, G. I., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee, S., and Hargreaves, M. (2004) Diabetes 53 1208–1215 [DOI] [PubMed] [Google Scholar]
- 23.Lu, J., McKinsey, T. A., Nicol, R. L., and Olson, E. N. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyster, C. A., Duggins, Q. S., and Olson, A. L. (2005) J. Biol. Chem. 280 17978–17985 [DOI] [PubMed] [Google Scholar]
- 25.Lu, J., McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2000) Mol. Cell 6 233–244 [DOI] [PubMed] [Google Scholar]
- 26.Morin, S., Pozzulo, G., Robitaille, L., Cross, J., and Nemer, M. (2005) J. Biol. Chem. 280 2272–32278 [DOI] [PubMed] [Google Scholar]
- 27.Potthoff, M. J., Wu, H., Arnold, M. A., Shelton, J. M., Backs, J., McAnally, J., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2007) J. Clin. Investig. 117 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLean, P. S., Zheng, D., Jones, J. P., Olson, A. L., and Dohm, G. L. (2002) Biochem. Biophys. Res. Commun. 292 409–414 [DOI] [PubMed] [Google Scholar]
- 29.Mora, S., and Pessin, J. E. (2000) J. Biol. Chem. 275 16323–16328 [DOI] [PubMed] [Google Scholar]
- 30.Mora, S., Yang, C., Ryder, J. W., Boeglin, D., and Pessin, J. E. (2001) Endocrinology 142 1999–2004 [DOI] [PubMed] [Google Scholar]