Abstract
FOXO1, a member of the evolutionarily conserved forkhead family of transcription factors, regulates expression of a number of genes that play critical roles in cell cycle and apoptosis. A pivotal regulatory mechanism of FOXO is reversible phosphorylation, catalyzed by kinases and phosphatases. Phosphorylation of FOXO1 is associated with 14-3-3 binding and cytosolic localization, whereas dephosphorylated FOXO1 translocates to the nucleus and is transcriptionally active. Experiments were performed to identify the serine/threonine phosphatase that dephosphorylates FOXO1. PP2A inhibitors, okadaic acid and fostriecin, increased FOXO1 phosphorylation in vitro and in cells. Microcystin-agarose pull-downs suggested that a phosphatase binds to FOXO1, and PP2A catalytic subunit was identified in endogenous FOXO1 immunocomplexes, indicating that PP2A is a FOXO1 phosphatase. Purified PP2A interacted directly with FOXO1 and dephosphorylated FOXO1 in vitro. Silencing of PP2A protected FOXO1 from dephosphorylation and delayed FOXO1 nuclear translocation, confirming the physiologic role of PP2A in the regulation of FOXO1 function. Furthermore, inhibition of PP2A phosphatases rescued FOXO1-mediated cell death by regulating the level of the pro-apoptotic protein BIM. We conclude that PP2A is a physiologic phosphatase of FOXO1.
FOXO (Forkhead box, class O) proteins, an evolutionarily conserved subgroup of the winged helix or forkhead transcription factors family, regulate a wide variety of cellular responses, including cell cycle arrest, cell death, and protection from stress stimuli (1). In humans, four members are identified: FOXO1 (FKHR), FOXO3a (FKHRL1), and FOXO4 (AFX), and FOXO6; the first three were found at chromosomal translocations in tumors, suggesting roles in oncogenesis (2). Activation of FOXO proteins leads to G1 cell cycle arrest, apoptosis, and gluconeogenesis, through up-regulation of cyclin-dependent kinase inhibitor p27KIP1, apoptotic-related proteins such as FAS ligand, tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), BIM, insulin-like growth factor-binding protein-1 (IGFBP-1), and phosphoenolpyruvate carboxykinase (PEPCK), respectively (3–11). The main regulator of FOXO function is the phosphoinositide 3-kinase (PI3K)2 effector AKT/PKB (4, 12). FOXO transcription factors are also targets of several other kinases including serum and glucocorticoid-regulated kinase, casein kinase 1, and dual-specificity tyrosine-phosphorylated regulated kinase 1a (DYRK1a) (1, 13, 14).
In response to survival factors such as IL3 and insulin-like growth factor, AKT/PKB and the related kinase, Serum and glucocorticoid-regulated kinase (SGK), phosphorylate FOXO1 directly at Thr24, Ser256, and Ser319, inactivating FOXO1 by promoting nuclear export and inhibiting transcriptional function (15–17). Thr24 is located in a consensus 14-3-3-binding motif, and phosphorylation of Thr24 is required for 14-3-3 binding. Phosphorylation of Ser256 inhibits FOXO1 transactivation and is essential for its interaction with SCF ubiquitin ligase complex and subsequent degradation (18, 19). Ser319 phosphorylation generates a consensus site for casein kinase 1 phosphorylation at Ser322 and Ser325. The cluster of phosphorylation sites at Ser319, Ser322, and Ser325 is relevant to the apoptotic function of FOXO1 (14).
Physiologic importance of FOXO proteins is demonstrated in gene deletions studies. Knock-out of FOXO1 results in embryonic lethality due to incomplete vascular development (20). Recent publications of simultaneous Cre-mediated disruption of FOXO1, FOXO3, and FOXO4 revealed the collective tumor suppressor role of these genes and their essential function in the maintenance of hematopoietic stem cells (21–24). FOXO proteins accomplish these functions as active transcription factors in the dephosphorylated state.
Reversible phosphorylation of proteins, catalyzed by kinases and phosphatases, regulates many organismal and cellular processes including embryonic development, cell proliferation, and cell death (25, 26). Mammalian serine/threonine phosphatase families consist of the most abundant protein phosphatases PP1 and PP2A, Ca2+-dependent PP2B (calcineurin), Mg2+-dependent PP2C, other PP2A-like phosphatases PP4 (PPX) and PP6 (27, 28), as well as PP5, and PP7. PP2A holoenzyme is a heterotrimer consisting of a catalytic subunit C, a structural scaffolding subunit A, and a highly variable B regulatory subunit, giving rise to many different PP2A holoenzymes (29).
Because the phosphorylation state of FOXO1 plays a critical role in its physiological function, we sought to determine whether phosphatase(s) are involved in the activation of FOXO1. In this study, we used phosphatase inhibitors, coimmunoprecipitations, knockdowns, as well as functional studies to investigate the role of serine-threonine phosphatases in FOXO1-mediated apoptosis.
EXPERIMENTAL PROCEDURES
Reagents—Serine/threonine phosphatase inhibitors microcystin-LR, okadaic acid, fostriecin, and calyculin A, PI 3-kinase inhibitor wortmannin, AKT inhibitor SH5 and His-AKT1 were purchased from EMD Biosciences. The following antibodies were used: phospho-AKT, AKT, phospho-Thr24-FOXO1 and phospho-Ser256-FOXO1 (Cell signaling Technology); FOXO1 (H128 and N18), PP1 (E9), phospho-ERK (E4), 14-3-3 (K19 and H8) (Santa Cruz Biotechnology); PP2A catalytic subunit, BIM (BD Biosciences); PP2A A subunit (Zymed Laboratories); PP4 (Chemicon International); and PP6 (Sigma). PP2A and microcystin-agarose were purchased from Upstate. PP1 siRNA, PP2A siRNA, and scrambled siRNA were purchased from Santa Cruz Biotechnology. pGEX-mFOXO1 plasmid was kindly provided by Dr. Masahiko Negishi (National Institutes of Health, Research Triangle Park, NC). Dr. David Brautigan (University of Virginia, Charlottesville, VA) generously provided purified PP2A AC core enzyme.
Cell Line and Cultures—The FL5.12 cell line with doxycycline-inducible wild type FOXO1 (F14) and the FL5.12 cell line with the inducible FOXO1-AAA mutant (T24A, S256A, S319A) (A3) were kindly provided by Dr. David Plas (University of Cincinnati, Cincinnati, OH). Plasmids pcDNA3-Flag-FOXO1, pcDNA3-Flag-FOXO1-AAA mutant, and pcDNA3-GFP-FOXO1 were gifts from Dr. William R. Sellers (Harvard Medical School, Boston, MA). FL5.12 cells were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and IL3. FOXO1 expression was induced with 2 μg/ml doxycycline for 14 h. NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% calf serum. BOSC cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. In IL3 deprivation experiments, FL5.12 cells with inducible FOXO1 were washed with phosphate-buffered saline 3 times and re-cultured in medium lacking IL3.
Immunoprecipitation and Immunoblotting—Cell lysates for Western blotting were prepared in radioimmunoprecipitation assay (RIPA) buffer (100 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mm Tris-Cl, pH 8.0, and 1 mm EDTA). Immunoblotting of lysates with antibodies specific to phospho-Thr24 or phospho-Ser256 FOXO1 often interfered with subsequent detection of total FOXO1 using rabbit antibody, therefore, phospho-FOXO bands were standardized against actin on the same blots and total FOXO1 was detected in a separate loading of the same samples. For immunoprecipitation, cells were lysed in isotonic immunoprecipitation (IP) buffer (142.5 mm KCl, 5 mm MgCl2, 10 mm HEPES, and 0.1% Nonidet P-40) with protease inhibitors, incubated with FOXO1 antibody (N-18 or H128), precipitated with protein A and G Sepharose, fractionated by 12.5% SDS-PAGE, and transferred to polyvinylidene difluoride membrane. For immunoprecipitated FOXO1, blots were probed with phosphospecific antibody first, then re-probed with antibody against total FOXO1.
Cross-linking Assay—Whole cell extracts were prepared in IP buffer with a protease inhibitor mixture, incubated with cross-linker 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP, 2 mm) at 4 °C for 2 h. A final concentration of 20 mm Tris-Cl, pH 7.5, was added to quench the reaction for 15 min at room temperature. Thereafter, the immunoprecipitation assay was conducted as usual, except that IP buffer was replaced by RIPA buffer in the final washings.
In Vitro Dephosphorylation Assay—For FOXO1 dephosphorylation in whole cell lysates, cell extracts were prepared in phosphatase assay buffer A (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.25% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin), and incubated on ice for 30 min. Cell lysates were centrifuged at 12,000 × g for 10 min at 4 °C, and the supernatant was further incubated at 30 °C for 20 min with or without the addition of serine/threonine phosphatase inhibitors. For dephosphorylation of FOXO1 by PP2A, FOXO1 was immunoprecipitated from F14 cells and incubated in phosphatase assay buffer B (50 mm Tris-Cl, pH 8.5, 20 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Purified PP2A was added to immunoprecipitated FOXO1 and incubated at 30 °C for 20 min.
In Vitro Binding Assay—FOXO1 was immunoprecipitated from F14 cells. Purified PP2A was added to immunoprecipitated FOXO1 and incubated at 4 °C for 2 h. GST-mFOXO1 was expressed in Escherichia coli strain BL21 and purified with glutathione-Sepharose. Phosphorylated GST-pFOXO1 was made by adding purified His-AKT1 (Calbiochem) to GST-mFOXO1-glutathione-Sepharose in kinase buffer (50 mm Tris-Cl, pH 7.5, 1 mm dithiothreitol, 50 μm ATP, 100 mm NaCl, 20 mm MgCl2) at 30 °C for 30 min. For in vitro binding, purified PP2A was added to GST-Sepharose, GST-FOXO1-Sepharose, or GST-pFOXO1-Sepharose and incubated at 4 °C for 2 h with the cross-linker 3,3′-dithiobis(sulfosuccinimidylpropionate).
PP2A and PP1 Knock-down Assay—The F14 cell line was transfected with PP2A/C siRNA or PP1 siRNA using the Amaxa Nucleofector kit V. Transfection frequencies were over 70% as indicated by fluorescent control vector pmaxGFP. 24 h after transfection, FOXO1 expression was induced with doxycycline (2 μg/ml) for 16 h and collected at 40 h after transfection.
Nuclear Translocation Assay—NIH 3T3 cells were transfected with pcDNA3-GFP-FOXO1 expression plasmid. Twenty-four to 36 h after transfection, cells were pretreated or not with serine/threonine phosphatase inhibitors for 1 h and then serum deprived for 2 h with or without the continued presence of inhibitors. Using Lipofectamine 2000 transfection kit, BOSC cells were transfected with pcDNA3-GFP-FOXO1 and scrambled siRNA or pcDNA3-GFP-FOXO1 and PP2A siRNA. Red indicator siRNA was used to assess efficiency of siRNA delivery. Usually, the majority of the cells were transfected by siRNA. Twenty-four h after transfection, cells were treated with staurosporine (2 μm) for 1 h. Subcellular GFP localization was assessed by fluorescence microscopy.
Cell Survival Assay—After F14 and A3 cells were treated with doxycycline (2 μg/ml) for 12 h, fostriecin (3 μm) or calyculin A (5 nm) was added to the medium for 3 h in the presence of IL3, followed by IL3 deprivation. Cell death was measured by propidium iodide exclusion or annexin V-fluorescein isothiocyanate staining, analyzed by flow cytometry. BOSC cells transfected with GFP-FOXO1 were preteated with calyculin A (5 nm) for 1 h, then treated with 2 μm staurosporine for 6 h with or without the continued presence of calyculin A. One μg/ml Hoechst was added to the medium at 37 °C for 20 min.
RESULTS
PP1 and PP2A Inhibitors Prevent FOXO1 Dephosphorylation—To determine whether serine/threonine phosphatases are involved in FOXO1 dephosphorylation in cells, we treated cells with okadaic acid (OA), a cell-permeable inhibitor of PP1 and PP2A, and checked FOXO1 phosphorylation status. F14, the FL5.12 cell line with doxycycline-inducible FOXO1 (30), was treated with increasing concentrations of OA for 3 h and assayed for FOXO1 phosphorylation by Western blot analysis (Fig. 1A). Treatment of cells with OA resulted in increased phosphorylation of FOXO1 at both the Thr24 and Ser256 sites. The dose-dependent increase in FOXO1 phosphorylation by OA suggested the involvement of either PP1 or a phosphatase of the PP2A family in FOXO1 dephosphorylation.
FIGURE 1.
PP2A inhibitors block FOXO1 dephosphorylation in vitro and in cells. A, FOXO1 inducible cell line F14 was treated with the indicated doses of okadaic acid (OA) for 3 h, and analyzed by Western blotting. The lysates were run on triplicate Western blots and each blot was probed for either Thr(P)24, or Ser(P)256, or total FOXO1. One of two experiments is shown. B, F14 lysates were treated with or without phosphatase inhibitors microcystin-LR (2 μm), OA (1 μm), fostriecin (FST) (10 nm), and analyzed by Western blotting. Phospho-Thr24 and phospho-Ser256 were probed on separate blots. This time, the same blots were reprobed for total FOXO1. One of two experiments is shown. C, F14 cells were IL3 starved for 3 h, stimulated with IL3 for 30 min to achieve maximal phosphorylation of FOXO1, then washed and seeded into IL3-free medium for 3 h in the presence of the indicated inhibitors: AKT inhibitor SH5 (10 μm), or PI3K inhibitor wortmannin (wort) (100 nm) and OA (250 nm) or FST (10 μm). Cell lysates were run on triplicate Western blots and each blot was probed for either Thr(P)24 or Ser(P)256, or total FOXO1. This experiment is done in part or in total 9 times. D, F14 cells were IL3 starved for 3 h, then re-stimulated with IL3 for 30 min in the absence (+IL3) or presence of 10 μm AKT inhibitor SH5 (+SH5 +IL3) or 100 nm PI3K inhibitor wortmannin (+wort +IL3). Cell lysates were immunoblotted for phospho-AKT. One of 3 experiments is shown. E, NIH 3T3 cells transfected with GFP-FOXO1 were serum starved for 10 or 30 min. Lysates were run in parallel, and Western blotted for either Thr(P)24 or total FOXO1. ACTIN was used as loading control for each blot. One of two experiments is shown.
To distinguish between the action of PP1 or PP2A, serine/threonine phosphatase inhibitors were used in in vitro phosphatase assays of FOXO1 dephosphorylation. Both Thr(P)24 and Ser(P)256 were dephosphorylated in F14 lysates after incubation for 20 min at 30 °C (Fig. 1B, lanes 1 and 2). FOXO1 dephosphorylation was effectively prevented by PP1 and PP2A inhibitors microcystin-LR, okadaic acid, and fostriecin (Fig. 1B, lanes 3–5). Fostriecin is a highly selective inhibitor of PP2A and PP2A-like phosphatases and is 10,000 to 40,000 times more potent against PP2A than PP1 (31). The concentration of fostriecin used in this experiment, 10 nm, was far lower than the IC50 for PP1, which suggested that the phosphatase acting on FOXO1 in cell-free extracts is PP2A or a PP2A-like phosphatase.
Increased FOXO1 Phosphorylation by PP2A Inhibitors Is Due to Inhibition of Phosphatases, Not Activation of Upstream Kinases—In cells, increased phosphorylation of FOXO1 can result from the inhibition of FOXO1 phosphatases or the activation of FOXO1 kinases. To rule out the possibility that maintenance of FOXO1 phosphorylation by phosphatase inhibitors was the result of activation of upstream kinases, we combined inhibitors of AKT/PKB or PI 3-kinase with phosphatase inhibitors. Because FOXO1 is a downstream substrate in the PI3K-AKT signaling pathway, we used the AKT inhibitor SH5 and PI3K inhibitor wortmannin together with okadaic acid or fostriecin. After removal of IL3 from the culture medium, FOXO1 was dephosphorylated at Thr24 and Ser256 (Fig. 1C, compare lane 1 to lanes 5 and 9, lane 10 to lanes 14 and 18). The phosphatase inhibitors okadaic acid, fostriecin, and calyculin A all prevented the dephosphorylation at Thr24 and Ser256 when used alone or when AKT and PI3K were also inactivated by SH5 or wortmannin, respectively (compare lanes 2 and 4, 6 and 8, 11 and 13, 15, and 17). These results suggested that increased FOXO1 phosphorylation was due to direct inhibition of phosphatase(s) acting on FOXO1 rather than increased kinase activity of AKT or PI3K. Inhibition of AKT phosphorylation upon IL3 stimulation confirmed that the kinase inhibitors SH5 and wortmannin were functional at the doses used (Fig. 1D).
Activation of phosphatase would be expected to lead to rapid dephosphorylation of substrate, whereas inhibition of kinases would result in a gradual decrease of already phosphorylated substrate over time. In NIH 3T3 cells, dephosphorylation of at least the Thr(P)24 site was observed as early as 10 min after serum withdrawal (Fig. 1E), supporting the hypothesis that an enzyme was activated, which directly dephosphorylated FOXO1.
PP2A Binds Directly to FOXO1 Regardless of 14-3-3 and Phosphorylation Status—The inhibitor experiments above suggested that PP2A or a PP2A-like phosphatase might directly dephosphorylate FOXO1, which would be strongly supported by demonstration of interaction of FOXO1 and PP2A. Microcystin-agarose binds PP1 and PP2A from cell lysates efficiently. Both FOXO1 wild type and AAA mutant proteins were captured by microcystin-agarose from lysates of FL5.12 cells with inducible FOXO1, suggesting that FOXO1 bound to either PP1 or PP2A pulled down by microcystin (Fig. 2A). To determine whether the phosphatase binding to FOXO1 was PP1 or PP2A, lysates of NIH 3T3 cells transiently transfected with FLAG-FOXO1wt, FLAG-FOXO1-AAA, or GFP-FOXO1 were incubated with FLAG beads, eluted with FLAG peptide, then the eluates were mixed with microcystin beads. FLAG-FOXO1wt and mutant, but not GFP-FOXO1, were pulled down by FLAG beads, and microcystin-agarose captured the FLAG-FOXO1wt and AAA mutant from the FLAG eluates. Western blotting of the microcystin pull-down revealed the presence of the catalytic subunit of PP2A (PP2A/C), but not PP1, indicating that the phosphatase in the FOXO1 immunocomplex was PP2A (Fig. 2B).
FIGURE 2.
PP2A associates with FOXO1. A, lysates from FL5.12 cells expressing vector alone, inducible wild type FOXO1, or inducible AAA mutant FOXO1 were incubated with microcystin-agarose at 4 °C for 2 h. Microcystin-agarose pull-downs were immunoblotted for Thr(P)24 and total FOXO1. B, FOXO1 was pulled down with FLAG-agarose from NIH 3T3 cells transfected with GFP-FOXO1, FLAG-FOXO1wt, or FLAG-FOXO1-AAA mutant, and eluted with FLAG peptide. Eluates from the FLAG-agarose were incubated with microcystin-agarose at 4 °C for 2 h, which was centrifuged and immunoblotted for FOXO1, PP2A/C, or PP1. Two percent of the initial wild type FOXO1 lysate was loaded as “input.” C, recombinant GST, GST-FOXO1, or GST-pFOXO1 on glutathione beads were incubated with FL5.12 cell lysate, respun, washed, and immunoblotted. FL5.12 lysate was loaded as input. D, FOXO1 was IP from F14 cells in the presence (+R18) or absence (–R18) of R18 peptide and immunoblotted. IP in the absence of antibody (Beads + lysate) or in the absence of lysate (Beads + Ab) served as negative controls. E, FOXO1 was IP from F14, K562, or NIH 3T3 cell lines and immunoblotted. IP of F14 lysate with rabbit IgG was used as negative control. F14, K562, and NIH 3T3 lysates were loaded separately as input. F, FOXO1 was immunoprecipitated from the F14 or FOXO1-AAA inducible cell line, washed extensively with RIPA buffer, then incubated with purified PP2A AC core enzyme for 2 h at 4°C, respun, and immunoblotted. Cell total lysates were loaded as input. Sepharose beads alone were incubated with PP2A as control. G, recombinant GST, GST-FOXO1, or GST-pFOXO1 on glutathione-Sepharose was incubated with purified PP2A AC core enzyme for 2 h at 4 °C and immunoblotted. All experiments in this figure were done at least three times, except E, which was done twice.
To test whether FOXO1 can bind to PP2A in cell-free extracts, we incubated recombinant GST-FOXO1 or GST-pFOXO1 (in vitro phosphorylated by AKT) with FL5.12 lysates. PP2A catalytic subunit was captured by GST-FOXO1 or GST-pFOXO1, but not GST (Fig. 2C). The presence of 14-3-3 in the GST-pFOXO1 complex confirmed that GST-pFOXO1 was indeed phosphorylated. The presence of both PP2A/C and 14-3-3 in the GST-pFOXO1 pull down strongly suggested that PP2A/C binds both phosphorylated and dephosphorylated FOXO1 and that binding was unaffected by 14-3-3. Other PP2A-like phosphatases, PP4 and PP6, were not detected in the GST-FOXO1 complexes, confirming that PP2A, but not other phosphatases, were involved in FOXO1 dephosphorylation.
PP2A Coimmunoprecipitates with FOXO1—To directly visualize PP2A/C in the FOXO1 complex, wild type or AAA mutant FOXO1 were immunoprecipitated and Western blotted for PP2A/C. The PP2A catalytic subunit coimmunoprecipitated with FOXO1 from the F14 cell lysates (Fig. 2D). R18, a peptide that displaces 14-3-3 from its substrates by interacting with basic residues in the amphipathic groove of 14-3-3 (32), did not significantly affect the interaction of FOXO1 and PP2A (Fig. 2D, lanes 3 and 4), indicating that the presence of 14-3-3 did not inhibit PP2A binding to FOXO1. However, when 14-3-3 was present, phosphorylation at Thr24 and Ser256 was protected from PP2A action. When 14-3-3 was displaced by R18, Thr(P)24 was greatly diminished and Ser(P)256 became undetectable (Fig. 2D, compare Thr(P)24 and Ser(P)256 bands in lanes 3 and 4). Similar PP2A association with FOXO1wt and the AAA mutant indicated that binding of PP2A was not regulated by the phosphorylation status of the AKT sites on FOXO1. PP1 was never detected in any of the FOXO1 immunocomplexes.
To verify that PP2A will bind to endogenous FOXO1, we immunoprecipitated endogenous FOXO1 from the human erythroleukemia cell line K562 and NIH 3T3 cells. Both PP2A/C and 14-3-3 were readily detected in endogenous FOXO1 complexes, as well as in complexes of induced FOXO1 in F14 cells, supporting the physiologic relevance of the FOXO1 and PP2A interaction (Fig. 2E).
FOXO1 Interacts Directly with PP2A—We tested whether the PP2A AC core dimer interacted directly with FOXO1. Immunoprecipitated FOXO1 extensively washed with detergent buffer to remove associated proteins was incubated with purified AC dimer. The absence of 14-3-3 in the complex confirmed that the known associated protein was successfully removed by detergent washing. Immunoblotting suggested that PP2A/C bound to FOXO1 directly (Fig. 2F). Direct binding of PP2A to FOXO1 was further tested by incubating bacterially produced GST-FOXO1 or GST-pFOXO1 with purified PP2A AC dimer (Fig. 2G). PP2A/C bound to both recombinant GST-FOXO1 and GST-pFOXO1, but not GST. PP2A/A was also detected in the complex. The experiments in Fig. 2 demonstrated conclusively that a PP2A enzyme binds both phosphorylated and dephosphorylated FOXO1 directly.
PP2A Dephosphorylates FOXO1—Next, we tested whether PP2A has enzymatic activity against FOXO1. Immunoprecipitated FOXO1 was incubated with purified PP2A AC dimer in an in vitro phosphatase assay. PP2A dephosphorylated both Thr(P)24 and Ser(P)256 in a dose-dependent manner (Fig. 3A). To confirm that PP2A is a physiologic phosphatase of FOXO1, PP2A siRNA was nucleofected into F14 cells and FOXO1 phosphorylation was assayed by Western analysis. The PP2A/C level was significantly decreased in cells nucleofected with PP2A siRNA, and phosphorylation of Thr(P)24 and Ser(P)256 was concomitantly increased (Fig. 3B, left). PP1 siRNA had no significant effect on FOXO1 dephosphorylation (Fig. 3B, right). These results are consistent with PP2A being the major phosphatase for FOXO1.
FIGURE 3.
PP2A dephosphorylates FOXO1. A, immunoprecipitated FOXO1 from F14 cells was treated with the indicated units of purified PP2A enzyme at 30 °C for 20 min, and loaded twice for Western analysis. The blots were first probed for phospho-FOXO1, then for total FOXO1. This experiment was repeated 6 times. B, F14 cells were nucleofected with PP2A siRNA or PP1 siRNA and immunoblotted. Phospho-Thr24, phospho-Ser256, and total FOXO1 were analyzed in separate loadings, using ACTIN as loading control. Grouped panels were from the same blot. This experiment was repeated 3 times.
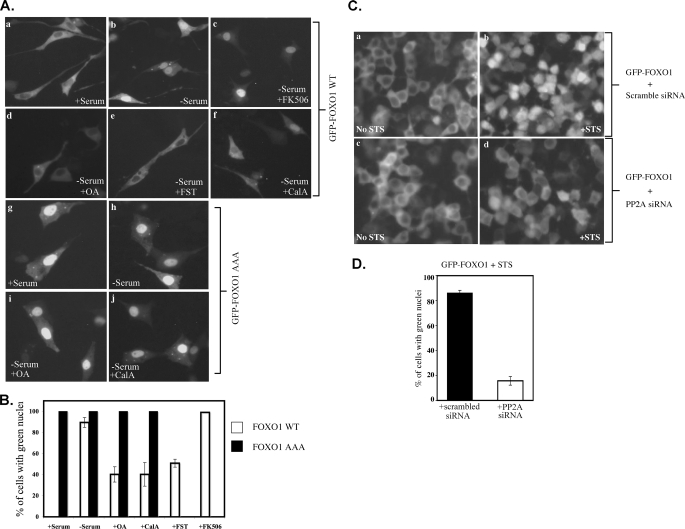
Inhibition of PP2A Prevents Nuclear Translocation of FOXO1—Phosphorylation of Ser256 is important for FOXO1 transactivation by inhibiting nuclear import through suppression of a nuclear localization signal. To address the physiologic function of FOXO1 dephosphorylation, we asked whether GFP-FOXO1 translocation is affected by inhibition of phosphatases. In the presence of serum, GFP-FOXO1 was distributed diffusely in the cytoplasm and excluded from the nucleus. After serum withdrawal, GFP fluorescence translocated into the nucleus (Fig. 4A, panels a and b). However, when the serine/threonine phosphatase inhibitor OA, calyculin A, or fostriecin were added in the absence of serum, GFP-FOXO1 remained cytoplasmic in a significant number of the cells (Fig. 4A, panels d–f). Nuclear translocation was decreased ∼50% by these PP1 and PP2A phosphatase inhibitors (Figs. 4B). In contrast, FK506, which inhibits PP2B, had no effect on FOXO1 nuclear translocation (Fig. 4, A, panel c, and B). The PP1 and PP2A inhibitors acted on the physiologic phosphorylation sites of FOXO1, because they failed to affect the localization of the GFP-FOXO1-AAA mutant (Fig. 4, A panels g–j, and B).
FIGURE 4.
Inhibition of PP2A prevents GFP-FOXO1 nuclear translocation. A, NIH 3T3 cells transfected with GFP-FOXO1wt or GFP-FOXO1-AAA mutants were pretreated for 1 h with 100 nm OA, 3 μm FST, or 5 nm calyculin A (CalA), or 5 μm FK506, or vehicle. Serum was then withdrawn for 2 h in the continued presence of phosphatase inhibitors or vehicle. Representative photographs from 3 different experiments are shown. B, cells with green nuclei in A are graphed as percent of total GFP+ cells. C, BOSC cells transfected with GFP-FOXO1 and scrambled siRNA or PP2A siRNA were treated or not with 2 μm STS for 1 h. D, percentages of GFP-positive cells with green nuclei after STS treatment (panels b and d in C) are graphed. For B and D, data are mean ± S.D. of triplicate assays from 3 separate experiments where error bars are present. At least 100 cells were counted for each condition.
To determine whether decreased FOXO1 nuclear translocation by phosphatase inhibitors was specifically due to inhibition of PP2A, PP2A siRNA was co-transfected with GFP-FOXO1. Induction of cell death by staurosporine (STS) led to translocation of GFP-FOXO1 into the nucleus in the majority of cells within 1 h. In cells with PP2A knockdown, GFP-FOXO1 remained largely cytoplasmic at the same time point, whereas scrambled siRNA had no effect on GFP-FOXO1 translocation (Fig. 4, C and D). The delay in FOXO1 nuclear translocation associated with PP2A knockdown linked PP2A to a physiologic FOXO1 function.
Inhibition of Serine/Threonine Phosphatase Prevents BIM Up-regulation by FOXO1 and Rescues FOXO1-induced Cell Death—BIM, a pro-apoptotic BCL2 family member, is up-regulated by FOXO in hematopoietic cell death (5, 33). Because serine/threonine phosphatase inhibitors prevented dephosphorylation and nuclear translocation of FOXO1, we tested whether BIM expression was altered when FOXO1 dephosphorylation was inhibited. After IL3 withdrawal from F14 cells, FOXO1 was dephosphorylated and the BIM level increased at the times tested, 1.5 and 3 h. Calyculin A prevented FOXO1 dephosphorylation after IL3 withdrawal and BIM expression remained unchanged (Fig. 5A). This result demonstrated that inhibition of FOXO1 dephosphorylation abrogated a known transcriptional function of FOXO1.
FIGURE 5.
Phosphatase inhibitors block BIM up-regulation by FOXO1 and rescue FOXO1-induced cell death. A, IL3 was withdrawn from F14 cells for the times indicated without or with 5 nm CalA, and immunoblotted for BIM and Ser(P)256. Total FOXO1 was assessed by a separate loading. ACTIN was used as loading control. B, F14 cells or FL5.12 cells with inducible FOXO1-AAA mutant were pretreated with 3 μm FST or 5 nm CalA or vehicle, then IL3 was withdrawn for 11 h in the presence of inhibitors or vehicle. Cell viability was assayed by PI exclusion. Data are mean ± S.D. of triplicate assays of 3 experiments. The viability differences between IL3-deprived cultures in the presence and absence of inhibitors have p values of <0.05 by analysis of variance. C, F14 cells or FL5.12 cells with inducible FOXO1-AAA mutant (A3) were induced and cultured in medium with or without IL3 for 22 h, incubated with annexin V-fluorescein isothiocyanate (FITC), and analyzed by flow cytometry. Percentages in the figures represent the mean from three independent experiments. Lysates of F14 and A3 cells cultured in the presence of IL3 were Western blotted for BIM. FL5.12 cells with empty vector were used as control (Con). D, BOSC cells transfected with GFP-FOXO1 (over 90% transfection efficiency) were preteated with Cal A (5 nm) for 1 h, then treated with 2μm STS for 6 h with or without the continued presence of CalA. Apoptosis was assessed by visualizing nuclei with Hoechst stain. E, number of apoptotic nuclei over total nuclei counted was graphed. Data are mean ± S.D. of triplicate assays from three separate experiments. At least 150 cells were counted for each condition.
To assess the physiologic relevance of FOXO1 phosphatase in apoptosis, we examined whether the inhibition of FOXO1 dephosphorylation enhanced cell survival in the absence of survival factors. We chose to use the PP1 and PP2A inhibitor calyculin A for its cell permeability and low toxicity, and fostriecin for its PP2A selectivity. The rate of death of F14 cells treated with calyculin A or fostriecin and deprived of IL3 was significantly decreased compared with cells treated with vehicle alone (Fig. 5B, left panel). Calyculin A and fostriecin had no effect on IL3 deprivation-induced death in cells expressing the AAA mutant of FOXO1, demonstrating that the survival effect was mediated through the AKT phosphorylation sites of FOXO1 (Fig. 5B, right panel).
Cell death of F14 cells and cells expressing the FOXO1-AAA mutant A3 was further compared using annexin V binding to specifically assay for apoptosis. In the presence of IL-3, 24% of cells expressing transcriptionally active FOXO1-AAA mutant were annexin V positive, compared with 5% annexin V positivity in the FOXO1 wild type culture (Fig. 5C, left panels). Upon IL-3 withdrawal, annexin V positivity in the wild type FOXO1 culture increased to 54%, similar to the 66% seen in the culture expressing the FOXO1-AAA mutant (Fig. 5C, right panels). This is consistent with data in Fig. 1 that FOXO1 dephosphorylation is mostly complete after 3 h of IL-3 deprivation, after which, wild type FOXO1 and FOXO1-AAA are functionally equivalent. Therefore, a difference in apoptosis would be expected when wild type FOXO1 is phosphorylated, i.e. in the presence of IL-3, but less so after wild type FOXO1 is dephosphorylated, i.e. after 3 h of IL-3 deprivation. We further demonstrated that BIM was up-regulated in FOXO1-AAA cells even in the presence of IL-3, consistent with transcriptional activity of unphosphorylated FOXO1 (Fig. 5C, bottom panel). Thus, FOXO1-mediated cell death in IL-3 withdrawal was an apoptotic process involving BIM up-regulation. In cells expressing FOXO1-AAA, BIM up-regulation was constitutive, leading to baseline apoptosis detectable by annexin V binding, an early apoptotic event, but not by PI exclusion, a late and less sensitive marker of apoptosis.
To further demonstrate that cell death mediated by FOXO1 was apoptosis and rescuable by phosphatase inhibition, we assayed for apoptotic nuclear morphology by Hoechst staining in FOXO1 expressing cells in response to another apoptotic stimulus. Treatment by STS caused ∼20% of the cells to display fragmented and condensed nuclear morphology (Fig. 5D). In the presence of calyculin A, the percent of cells with such morphology was reduced to ∼5% (Fig. 5E), consistent with rescue of apoptosis.
These data showed that serine/threonine phosphatase inhibitors prevented FOXO1-mediated BIM up-regulation and rescued cells from apoptosis. We conclude that PP2A plays a physiologic role in activating FOXO1 transcriptional function in the regulation of apoptosis.
DISCUSSION
FOXO activity can be regulated through multiple post-translational modifications, including phosphorylation, acetylation, and ubiquitination (8, 34). Reversible phosphorylation plays a crucial role in FOXO1 subcellular localization, which in turn affects its transcriptional function (35, 36). In this study, we examined the effect of a panel of PP1 or PP2A inhibitors, as well as PP1 and PP2A siRNAs, on the regulation of FOXO1 phosphorylation and subsequent effects on FOXO1 nuclear translocation and cell death. We focused on FOXO1 sites phosphorylated by AKT, as these are important in the regulation of apoptosis.
PP1 and PP2A inhibitors microcystin, okadaic acid, calyculin A, and the PP2A-selective inhibitor fostriecin all prevented FOXO1 dephosphorylation (Fig. 1, A and B). The concentration of fostriecin used in these experiments was effective for PP2A inhibition but insufficient to inhibit PP1, suggesting that PP2A or a PP2A-like enzyme was involved in FOXO1 dephosphorylation. We excluded the possibility that increased phosphorylation was due to phosphatase inhibitors activating upstream kinases by concurrent inhibition of AKT and PI3K (Fig. 1C). The combination of data using cell permeable inhibitors suggested that PP1 or an enzyme of the PP2A family dephosphorylated FOXO1. In addition, dephosphorylation of FOXO1 upon growth factor withdrawal was rapid, consistent with phosphatase activation (Fig. 1E).
Using microcystin-agarose in conjunction with FOXO1 immunoprecipitations, we were able to identify PP2A, not PP1 or other PP2A-like enzymes such as PP4 or PP6, as the phosphatase interacting with FOXO1 (Fig. 2). We demonstrated coimmunoprecipitation of endogenous FOXO1 and PP2A in two different cell lines in addition to FL5.12, confirming the physiologic nature of the interaction (Fig. 2E). Identification of PP2A as a FOXO1 phosphatase was supported by the ability of purified PP2A AC dimer to bind and dephosphorylate FOXO1 in vitro (Fig. 3A). Knock-down of PP2A catalytic subunit further confirmed the role of PP2A in the regulation of FOXO1 phosphorylation (Fig. 3B).
The functional relevance of PP2A activity in FOXO1 regulation was demonstrated by a significant decrease in nuclear translocation of FOXO1 and lack of BIM up-regulation when PP2A was inhibited in cell death conditions (Figs. 4 and 5). These findings correlated functionally with decreased apoptosis (Fig. 5). Nuclear translocation of FOXO1 was examined using two stimuli, serum withdrawal and staurosporine treatment (Fig. 4), and PP2A was inhibited by pharmacologic inhibitors or siRNA knockdown. FOXO1 nuclear translocation was not completely prevented at the inhibitor doses we used, but toxicity precluded the use of higher doses. PP2A siRNA was more effective in preventing FOXO1 nuclear translocation in the short term, and had the added advantage of specificity over inhibitors. Enhancement of survival in IL-3 withdrawal or staurosporine treatment by PP2A inhibitors was partial, as would be expected, because transcriptional activation of FOXO1 is not the only mechanism of apoptosis in response to these stimuli (Fig. 5). These experiments provided evidence that PP2A dephosphorylates FOXO1 to activate its pro-apoptotic transcriptional function.
FOXO transcription factors induce not only cell death genes, but also genes regulating cell cycle arrest and glucose metabolism. It was demonstrated recently that in the absence of FOXO 1, 3, and 4, more hematopoietic stem cells were in cycle than wild type controls (22), consistent with the function of dephosphorylated FOXOs in transcribing cell cycle arrest genes such as p27. We were unable to measure the effect of inhibition of FOXO1 dephosphorylation on cell proliferation in our cell systems, because prolonged PP2A inhibition, which would be necessary for cell proliferation experiments, led to cell death. This is not surprising given the vast number of PP2A targets. We were able to observe rescue of FOXO1-mediated cell death by PP2A inhibition, most likely because our experiments were all complete in 24 h, or less than one cell cycle doubling time, before inhibitor toxicities became obvious.
We observed specific binding between FOXO1 and the PP2A catalytic subunit. The interaction between FOXO1 and PP2A exhibited distinct characteristics. First, FOXO1 phosphorylation status does not appear to significantly affect PP2A binding. The PP2A catalytic subunit binds to both phosphorylated wild type FOXO1 and phosphorylation-defective mutant (Fig. 2). Second, the presence of 14-3-3 does not appear to prevent FOXO1 and PP2A interaction. 14-3-3 proteins are known to bind and protect phosphorylated proteins from phosphatases. In our previous studies with pro-apoptotic BAD, we showed that 14-3-3 and PP2A binding to BAD protein were competitive, and dissociation of 14-3-3 from BAD was a prerequisite for BAD dephosphorylation (37). We investigated if there was a similar mechanism in the interaction between FOXO1 and PP2A, and the data demonstrated that PP2A did not compete with 14-3-3 for FOXO1 binding. Instead, PP2A associated with FOXO1 regardless of the presence of 14-3-3, but PP2A dephosphorylated FOXO1 only when 14-3-3 was displaced, confirming the phospho-protective role of 14-3-3 (Fig. 2). Similar to the situation reported for BAD and CDC25, 14-3-3 dissociation is a key regulatory event in the dephosphorylation of FOXO1 (37, 38). Third, we found FOXO1 interaction with PP2A catalytic subunit by several approaches. The catalytic and structural subunits of PP2A form the AC core dimer, which interacts with one of the diverse regulatory B subunits to form a PP2A holoenzyme (29). Recent structural data suggest that substrate binding by PP2A is likely mediated by contacts with both the regulatory subunit and the catalytic subunit (39–41). Other studies have shown that another protein such as α4 can bind and displace the PP2A catalytic subunit C from the AC core dimer and any of the variable B subunits, enhancing PP2A catalytic subunit activity and altering its substrate specificity (42–44). We did not detect α4 in FOXO1 complexes (data not shown), providing no evidence for this mode of PP2A regulation in the case of FOXO1. The regulatory subunit of the FOXO1 PP2A awaits discovery.
The physiologic importance of FOXO1 has been demonstrated by embryonic lethality of the knock-out (20), as well as by recent publications reporting lymphomas and hemangiomas in mice in which FOXO1 was postnatally deleted along with FOXO3a and FOXO4 (21, 22). The mammalian FOXO factors, FOXO1, FOXO3a, and FOXO4, have different expression patterns in different cell types, but they share a common DNA recognition sequence and all three can regulate BIM expression and apoptosis (45–47). These three FOXO factors also share conserved phosphorylation sites, including the AKT sites, which are Thr24, Ser256, and Ser319 for FOXO1; Thr32, Ser253, and Ser315 for FOXO3; and Thr28, Ser193, and Ser258 for FOXO4. Interaction of PP2A with FOXO3a in hepatocyte lipoapoptosis was recently reported, revealing a role of PP2A in FOXO3a dephosphorylation at Thr32 upon stimulation by free fatty acids (48). Here, we showed the activity of PP2A on both the Thr24 and Ser256 sites of FOXO1 (equivalent to Thr32 and Thr253 of FOXO3a) in response to distinct stimuli, suggesting that PP2A dephosphorylation of AKT sites may be a common mechanism of activating FOXO factors.
It is well known that PP2A dephosphorylates and inactivates AKT. More recently, Trotman et al. (49) showed that AKT and PP2A are co-recruited into PML nuclear bodies and that the inability to recruit PP2A to AKT in PML deficiency resulted in FOXO3a inactivation. Our experiments were designed to examine dephosphorylation of already phosphorylated FOXO1, in that the dephosphorylation was rapid upon death stimulus (10 min, shown in Fig. 1E) and unlikely to be solely due to indirect activation of AKT. Inhibition of PP2A is expected to lead to increased AKT activity, however, we observed inhibition of FOXO1 dephosphorylation even when AKT was inhibited, indicating that an enzyme in addition to AKT was activated. It would make sense that the cell uses PP2A to both inhibit AKT and dephosphorylate FOXO to achieve activation of FOXO. It would be very interesting to see if the same PP2A holoenzyme dephosphorylates both AKT and FOXO. In addition, a new publication indicates that activation of FOXO can ultimately lead to suppression of PP2A and enhancement of AKT activity (50). This feedback loop to terminate FOXO activity may be more pertinent to glucose or cell cycle regulatory pathways in which the cell needs to stay alive, and less relevant in apoptosis regulation in which the goal is to remove the cell.
The activity of FOXO1 as a transcription factor requires that it be dephosphorylated. We demonstrated here that PP2A is a physiologic phosphatase of FOXO1. Dephosphorylation of the AKT sites on FOXO1 by PP2A is required for FOXO1 nuclear translocation and transactivation, thus PP2A is a key regulator of the pro-apoptotic function of FOXO1.
Acknowledgments
We are grateful to Dr. Brian E. Wadzinski (Vanderbilt University) for helpful advice and Dr. Charles Lin (Vanderbilt University) for the use of microscopy equipment.
This work was supported by National Institutes of Health Grant R01CA92498 and a grant from Hope St. Kids Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PI3K, phosphatidylinositol 3-kinase; IL, interleukin; siRNA, small interfering RNA; CalA, calyculin A; FST, fostriecin; OA, okadaic acid; STS, staurosporine; IP, immunoprecipitation; GFP, green fluorescent protein; GST, glutathione S-transferase.
References
- 1.Tran, H., Brunet, A., Griffith, E. C., and Greenberg, M. E. (2003) Sci. STKE 2003 RE5. [DOI] [PubMed] [Google Scholar]
- 2.Arden, K. C. (2006) Exp. Gerontol. 41 709–717 [DOI] [PubMed] [Google Scholar]
- 3.Medema, R. H., Kops, G. J., Bos, J. L., and Burgering, B. M. (2000) Nature 404 782–787 [DOI] [PubMed] [Google Scholar]
- 4.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J., and Greenberg, M. E. (1999) Cell 96 857–868 [DOI] [PubMed] [Google Scholar]
- 5.Dijkers, P. F., Medema, R. H., Lammers, J. W., Koenderman, L., and Coffer, P. J. (2000) Curr. Biol. 10 1201–1204 [DOI] [PubMed] [Google Scholar]
- 6.Accili, D., and Arden, K. C. (2004) Cell 117 421–426 [DOI] [PubMed] [Google Scholar]
- 7.Birkenkamp, K. U., and Coffer, P. J. (2003) Biochem. Soc. Trans. 31 292–297 [DOI] [PubMed] [Google Scholar]
- 8.Lam, E. W., Francis, R. E., and Petkovic, M. (2006) Biochem. Soc. Trans. 34 722–726 [DOI] [PubMed] [Google Scholar]
- 9.Modur, V., Nagarajan, R., Evers, B. M., and Milbrandt, J. (2002) J. Biol. Chem. 277 47928–47937 [DOI] [PubMed] [Google Scholar]
- 10.Nakae, J., Biggs, W. H., 3rd, Kitamura, T., Cavenee, W. K., Wright, C. V., Arden, K. C., and Accili, D. (2002) Nat. Genet. 32 245–253 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, W., Patil, S., Chauhan, B., Guo, S., Powell, D. R., Le, J., Klotsas, A., Matika, R., Xiao, X., Franks, R., Heidenreich, K. A., Sajan, M. P., Farese, R. V., Stolz, D. B., Tso, P., Koo, S. H., Montminy, M., and Unterman, T. G. (2006) J. Biol. Chem. 281 10105–10117 [DOI] [PubMed] [Google Scholar]
- 12.Tang, E. D., Nunez, G., Barr, F. G., and Guan, K. L. (1999) J. Biol. Chem. 274 16741–16746 [DOI] [PubMed] [Google Scholar]
- 13.Van Der Heide, L. P., Hoekman, M. F., and Smidt, M. P. (2004) Biochem. J. 380 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rena, G., Woods, Y. L., Prescott, A. R., Peggie, M., Unterman, T. G., Williams, M. R., and Cohen, P. (2002) EMBO J. 21 2263–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A., and Greenberg, M. E. (2001) Mol. Cell. Biol. 21 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rena, G., Prescott, A. R., Guo, S., Cohen, P., and Unterman, T. G. (2001) Biochem. J. 354 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao, X., Gan, L., Pan, H., Kan, D., Majeski, M., Adam, S. A., and Unterman, T. G. (2004) Biochem. J. 378 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki, M., Jiang, H., and Vogt, P. K. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13613–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuzaki, H., Daitoku, H., Hatta, M., Tanaka, K., and Fukamizu, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11285–11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosaka, T., Biggs, W. H., 3rd, Tieu, D., Boyer, A. D., Varki, N. M., Cavenee, W. K., and Arden, K. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik, J. H., Kollipara, R., Chu, G., Ji, H., Xiao, Y., Ding, Z., Miao, L., Tothova, Z., Horner, J. W., Carrasco, D. R., Jiang, S., Gilliland, D. G., Chin, L., Wong, W. H., Castrillon, D. H., and DePinho, R. A. (2007) Cell 128 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tothova, Z., Kollipara, R., Huntly, B. J., Lee, B. H., Castrillon, D. H., Cullen, D. E., McDowell, E. P., Lazo-Kallanian, S., Williams, I. R., Sears, C., Armstrong, S. A., Passegue, E., DePinho, R. A., and Gilliland, D. G. (2007) Cell 128 325–339 [DOI] [PubMed] [Google Scholar]
- 23.Coffer, P. J., and Burgering, B. M. (2007) Nat. Cell. Biol. 9 251–253 [DOI] [PubMed] [Google Scholar]
- 24.Arden, K. C. (2007) Cell 128 235–237 [DOI] [PubMed] [Google Scholar]
- 25.Gallego, M., and Virshup, D. M. (2005) Curr. Opin. Cell Biol. 17 197–202 [DOI] [PubMed] [Google Scholar]
- 26.Ostman, A., Hellberg, C., and Bohmer, F. D. (2006) Nat. Rev. Cancer 6 307–320 [DOI] [PubMed] [Google Scholar]
- 27.Klumpp, S., and Krieglstein, J. (2002) Curr. Opin. Pharmacol. 2 458–462 [DOI] [PubMed] [Google Scholar]
- 28.Moorhead, G. B., Trinkle-Mulcahy, L., and Ulke-Lemee, A. (2007) Nat. Rev. Mol. Cell Biol. 8 234–244 [DOI] [PubMed] [Google Scholar]
- 29.Janssens, V., Goris, J., and Van Hoof, C. (2005) Curr. Opin. Genet. Dev. 15 34–41 [DOI] [PubMed] [Google Scholar]
- 30.Plas, D. R., and Thompson, C. B. (2003) J. Biol. Chem. 278 12361–12366 [DOI] [PubMed] [Google Scholar]
- 31.Walsh, A. H., Cheng, A., and Honkanen, R. E. (1997) FEBS Lett. 416 230–234 [DOI] [PubMed] [Google Scholar]
- 32.Wang, B., Yang, H., Liu, Y. C., Jelinek, T., Zhang, L., Ruoslahti, E., and Fu, H. (1999) Biochemistry 38 12499–12504 [DOI] [PubMed] [Google Scholar]
- 33.Stahl, M., Dijkers, P. F., Kops, G. J., Lens, S. M., Coffer, P. J., Burgering, B. M., and Medema, R. H. (2002) J. Immunol. 168 5024–5031 [DOI] [PubMed] [Google Scholar]
- 34.Vogt, P. K., Jiang, H., and Aoki, M. (2005) Cell Cycle 4 908–913 [DOI] [PubMed] [Google Scholar]
- 35.Greer, E. L., and Brunet, A. (2005) Oncogene 24 7410–7425 [DOI] [PubMed] [Google Scholar]
- 36.Burgering, B. M., and Kops, G. J. (2002) Trends Biochem. Sci. 27 352–360 [DOI] [PubMed] [Google Scholar]
- 37.Chiang, C. W., Kanies, C., Kim, K. W., Fang, W. B., Parkhurst, C., Xie, M., Henry, T., and Yang, E. (2003) Mol. Cell Biol. 23 6350–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolis, S. S., Perry, J. A., Forester, C. M., Nutt, L. K., Guo, Y., Jardim, M. J., Thomenius, M. J., Freel, C. D., Darbandi, R., Ahn, J. H., Arroyo, J. D., Wang, X. F., Shenolikar, S., Nairn, A. C., Dunphy, W. G., Hahn, W. C., Virshup, D. M., and Kornbluth, S. (2006) Cell 127 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing, Y., Xu, Y., Chen, Y., Jeffrey, P. D., Chao, Y., Lin, Z., Li, Z., Strack, S., Stock, J. B., and Shi, Y. (2006) Cell 127 341–353 [DOI] [PubMed] [Google Scholar]
- 40.Xu, Y., Xing, Y., Chen, Y., Chao, Y., Lin, Z., Fan, E., Yu, J. W., Strack, S., Jeffrey, P. D., and Shi, Y. (2006) Cell 127 1239–1251 [DOI] [PubMed] [Google Scholar]
- 41.Mumby, M. (2007) ACS Chem. Biol. 2 99–103 [DOI] [PubMed] [Google Scholar]
- 42.Murata, K., Wu, J., and Brautigan, D. L. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, J., Peterson, R. T., and Schreiber, S. L. (1998) Biochem. Biophys. Res. Commun. 247 827–832 [DOI] [PubMed] [Google Scholar]
- 44.Kong, M., Fox, C. J., Mu, J., Solt, L., Xu, A., Cinalli, R. M., Birnbaum, M. J., Lindsten, T., and Thompson, C. B. (2004) Science 306 695–698 [DOI] [PubMed] [Google Scholar]
- 45.Furuyama, T., Nakazawa, T., Nakano, I., and Mori, N. (2000) Biochem. J. 349 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilley, J., Coffer, P. J., and Ham, J. (2003) J. Cell Biol. 162 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urbich, C., Knau, A., Fichtlscherer, S., Walter, D. H., Bruhl, T., Potente, M., Hofmann, W. K., de Vos, S., Zeiher, A. M., and Dimmeler, S. (2005) FASEB J. 19 974–976 [DOI] [PubMed] [Google Scholar]
- 48.Barreyro, F. J., Kobayashi, S., Bronk, S. F., Werneburg, N. W., Malhi, H., and Gores, G. J. (2007) J. Biol. Chem. 282 27141–27154 [DOI] [PubMed] [Google Scholar]
- 49.Trotman, L. C., Alimonti, A., Scaglioni, P. P., Koutcher, J. A., Cordon-Cardo, C., and Pandolfi, P. P. (2006) Nature 441 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni, Y. G., Wang, N., Cao, D. J., Sachan, N., Morris, D. J., Gerard, R. D., Kuro, O. M., Rothermel, B. A., and Hill, J. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20517–20522 [DOI] [PMC free article] [PubMed] [Google Scholar]