Abstract
The Synechococcus sp. PCC 7002 genome encodes three genes, denoted cpcS-I, cpcU, cpcV, with sequence similarity to cpeS. CpcS-I copurified with His6-tagged (HT) CpcU as a heterodimer, CpcSU. When CpcSU was assayed for bilin lyase activity in vitro with phycocyanobilin (PCB) and apophycocyanin, the reaction product had an absorbance maximum of 622 nm and was highly fluorescent (λmax = 643 nm). In control reactions with PCB and apophycocyanin, the products had absorption maxima at 635 nm and very low fluorescence yields, indicating they contained the more oxidized mesobiliverdin (Arciero, D. M., Bryant, D. A., and Glazer, A. N. (1988) J. Biol. Chem. 263, 18343–18349). Tryptic peptide mapping showed that the CpcSU-dependent reaction product had one major PCB-containing peptide that contained the PCB binding site Cys-82. The CpcSU lyase was also tested with recombinant apoHT-allophycocyanin (aporHT-AP) and PCB in vitro. AporHT-AP formed an ApcA/ApcB heterodimer with an apparent mass of ∼27 kDa. When aporHT-AP was incubated with PCB and CpcSU, the product had an absorbance maximum of 614 nm and a fluorescence emission maximum at 636 nm, the expected maxima for monomeric holo-AP. When no enzyme or CpcS-I or CpcU was added alone, the products had absorbance maxima between 645 and 647 nm and were not fluorescent. When these reaction products were analyzed by gel electrophoresis and zinc-enhanced fluorescence emission, only the reaction products from CpcSU had PCB attached to both AP subunits. Therefore, CpcSU is the bilin lyase-responsible for attachment of PCB to Cys-82 of CpcB and Cys-81 of ApcA and ApcB.
Cyanobacteria are a morphologically and developmentally diverse group of prokaryotes. Their light-harvesting complexes, phycobilisomes (PBS),4 are very similar to those found in red algal chloroplasts but are quite distinct from the chlorophyll-based, light-harvesting protein complexes of higher plants (1–3). The PBS of the genetically amenable cyanobacterium Synechococcus sp. PCC 7002 are ideal objects for detailed characterization because they are among the simplest known PBS in structure and composition. These PBS contain only 12 polypeptides and are principally composed of only two phycobiliproteins (PBP): allophycocyanin (AP) and phycocyanin (PC) (3–12). Each of these major PBP is composed of two subunits, α and β, and each of these subunits carries at least one covalently attached phycocyanobilin (PCB) chromophore (1–3). The attachment of PCB to the polypeptide subunits occurs through thioether bonds to specific cysteine residues (1–3).
For some PBP it has been demonstrated that lyase enzymes are required for the attachment, isomerization, and detachment of the bilin chromophores from the cysteine residues of the PBP (13–21). For example, enzymes appear to be involved in the attachment of PCB to each of the three Cys attachment sites of PC. The products of two genes, cpcE and cpcF, which occur downstream of the cpcBA structural genes that encode the β and α subunits of PC, respectively, comprise a heterodimeric lyase that specifically attaches PCB to Cys-84 of α-PC (CpcA) (13–17). The PecEF and CpeYZ lyases are similar in sequence to CpcEF but are active on different substrate proteins (18–21). A completely different family of genes, first sequenced as part of an operon encoding PBS linker proteins for phycoerythrin in Fremyella diplosiphon (22), is involved in PCB attachment to the Cys-82 attachment site of β-PC as well as to Cys-153 of the same polypeptide (23). The Synechococcus sp. PCC 7002 genome encodes four members of this second lyase family (23–25). Three genes show highest sequence similarity to cpeS and have been named cpcS-I, cpcU, and cpcV. One gene is most similar to cpeT of F. diplosiphon and has been named cpcT (23–25). CpcT specifically attaches PCB to Cys-153 of β-PC in Synechococcus sp. PCC 7002 (24). In Nostoc sp. PCC 7120 it was recently shown that the product of open reading frame alr0617, which we shall refer to as CpcS-III based on phylogenetic analyses (25), is another member of this lyase family (26). Zhao et al. (26, 27) have shown that CpcS-III attaches PCB to Cys-82 on β-PC and β-phycoerythrocyanin as well as to several AP subunits in Nostoc sp. PCC 7120.
The PBS core linker, also known as LCM or ApcE, contains a PBP-like domain at its N terminus that is related in sequence to all other PBP but has the Cys residue for PCB attachment at a structurally distinct position (5, 10, 28, 29). ApcE may not require a lyase enzyme for bilin attachment. Zhao et al. (29) have shown that PCB addition to ApcE can occur in the absence of lyases in vitro, although it is currently not known if this also occurs in vivo. Moreover, PCB addition to ApcA (α-AP) may not require a lyase when the protein is synthesized in Escherichia coli cells that co-express apcA and the genes required to synthesize PCB from heme (30). When purified HT-ApcA and PCB were combined in vitro, PCB addition occurred, and the resulting product was reported to have an absorption spectrum that matched that of native holo-AP subunits (30). In some studies in which no requirement for lyases was reported, detergents or urea were added to modify the conformation of the PBP, the bilin, or both (29, 31).
We have used a combination of reverse genetics and biochemical methods to identify and characterize the PCB lyases of Synechococcus sp. PCC 7002. In an accompanying manuscript (25), we have presented strong evidence that null mutants for cpcS-I and cpcU define a heterodimeric PCB lyase that attaches PCB to Cys-82 of β-PC. The characterization of these mutants also suggested that CpcS-I and CpcU are required for PCB attachment to Cys-81 of the α and β subunits of AP (23, 25). In the studies presented here, the conclusions from these reverse genetics analyses are validated in vitro, and we show that CpcS-I and CpcU form a 1:1 heterodimer that attaches PCB to Cys-82 of β-PC. Neither CpcS-I nor CpcU alone is able to perform this lyase reaction. Additionally, we show that these two proteins are also required for correct attachment of PCB to the α and β subunits of AP in vitro.
EXPERIMENTAL PROCEDURES
Construction of Recombinant Expression Plasmids—The cpcS-I gene was amplified by PCR from Synechococcus sp. PCC 7002 chromosomal DNA using primers 7002cpcS5 (5′-AATTTTTCCATATGCAAAGCTTTGCGGATGCC-3′) and 7002cpcS3 (5′-TTGACTCGAGCAACACGGATATCTCTGTGGG-3′). The PCR product was cloned into pAED4 T7 expression vector using the restriction enzymes NdeI and XhoI (underlined in the primer sequences). The cpcU gene was amplified by PCR from Synechococcus sp. PCC 7002 chromosomal DNA using two primers (7002cpcU5, 5′-GTAACTGTTCATATGGATATCAATGCCTTTATCC-3′; 7002cpcU3, 5′-CTAAAAGCTTTCGTTAGTTACTGGCTTCAGCGG-3′). The PCR product was cloned into pBS150v vector using NdeI and HindIII (underlined in the primer sequences). The pBS150v vector includes a His6 tag for easy purification of the protein. The cpcV gene was amplified by PCR using primers 7002cpcV5 (5′-GCTCTTCGCATATGAATTTACTTGCGAC-3′) and 7002cpcV3 (5′-TTTAAGCTTACTAAAGACGCGTTTCTAAATACTGCGC-3′). After PCR amplification, the cpcV gene was cloned into vectors pAED4 and pBS150v using restriction enzymes NdeI and HindIII (underlined in the primer sequences). The cpcB and cpcA genes were cloned into pAED4 as described (24). The apcA and apcB genes were cloned in pET100 (24). The expression constructs were sequenced at the W. M. Keck Conservation and Molecular Genetics laboratory (University of New Orleans) to confirm that no unwanted mutations had been introduced.
Protein Overexpression and Purification—Expression plasmids were transformed into E. coli BL21 DE3 cells, and colonies were selected on Luria Bertani plates in the presence of ampicillin (100 μg ml–1) or spectinomycin (100 μg ml–1). For expression of cpcS-I, cpcU, and cpcV, cells from a 50-ml starter culture were added to 1 liter of Luria-Bertani medium with the appropriate antibiotic and grown for 4 h. Cells harboring plasmids encoding cpcS-I or cpcV were grown at 30 °C, whereas those encoding cpcU were grown at 37 °C. Production of proteins was induced by the addition of 0.5 mm isopropyl β-d-thiogalactoside. Cells were incubated with shaking for another 4 h before cells were harvested by centrifugation, and pellets were frozen at –20 °C until required. Purification of rCpcBA and aporHT-AP was performed as described (24).
To purify CpcS-I, cells were resuspended in 50 mm Tris-HCl, pH 8.0, and lysed by three passages through a chilled French pressure cell at 138 megapascals. The lysed cell suspension was centrifuged for 25 min at 17,000 × g to remove unbroken cells and inclusion bodies. The supernatant was brought to 40% saturation with ammonium sulfate and left at 4 °C overnight. After centrifugation at 17,000 × g for 20 min, the pellet containing the CpcS-I protein was resuspended in a small amount and dialyzed exhaustively against the same buffer to remove the ammonium sulfate. Aliquots (10 ml) of the CpcS-I (pI 4.79) solution were loaded onto a DEAE column (Whatman DE-52: 2.5 × 12.5 cm) that had been equilibrated with 50 mm Tris-HCl, pH 8.0, 1 mm NaN3 (buffer 1; sodium azide was added to prevent microbial growth during storage of the column/buffer) by using the BioLogic LP system at room temperature (Bio-Rad). The column was developed using the same procedure described for CpcT (24). Fractions containing CpcS-I were pooled and dialyzed against 50 mm Tris-HCl, pH 8.0. CpcS-I was concentrated using an Amicon YM-10 concentrator and stored in 2-ml aliquots at ×20 °C until required. The CpcS-I protein has a calculated molecular mass of 22,526.3 Da. Antibodies were raised against the CpcS-I protein present in inclusion bodies. Inclusion bodies were washed as described in Fairchild et al. (15), and CpcS-I was purified by preparative SDS-PAGE on a 15% (w/v) acrylamide gel. The band corresponding to CpcS-I was excised and minced, and protein was electroeluted from the gel slices using a procedure recommended by Sigma Genosys, who generated rabbit polyclonal antibodies to the protein.
Because CpcU is produced with an N-terminal His6 tag, it was purified by metal affinity chromatography using Ni-NTA resin (Qiagen, Valencia, CA). E. coli cells containing HTCpcU were resuspended in 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 50 mm KCl (buffer 0) and lysed by three passages through a chilled French pressure cell at 138 megapascals. After clarification of the extract by centrifugation at 17,000 × g for 25 min, the supernatant was added to 10 ml of Ni-NTA resin that had been washed with buffer 0. The resin and the extracts were incubated together for 15 min and then loaded into a column. The column was washed as described in Shen et al. (24), and HT-CpcU was eluted from the Ni-NTA resin by the addition of 20 ml buffer C (20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 50 mm KCl, 200 mm imidazole), dialyzed against Buffer 0, concentrated, and stored in 2-ml aliquots at 20 °C until required. The HT-CpcU protein has a calculated molecular mass of 23,471.71 Da. The HT-CpcV protein was purified in the same manner and had a molecular mass of 22,728.2 Da. CpcV (non-tagged; calculated molecular mass of 19,851.3 Da) was not purified; whole cell extracts containing CpcV were used in some enzyme assays.
Interaction Assays—A pulldown interaction assay was performed to determine whether HT-CpcU and CpcS-I interacts in vitro. An aliquot (50 μl) of E. coli whole-cell extract containing HT-CpcU was mixed with a whole-cell extract (50 μl) containing CpcS-I, and the solution was incubated on ice for 15 min. The Ni-NTA resin (150 μl) was pelleted by centrifugation at 3000 × g for 4 min. The supernatant was discarded, and 500 μl of buffer 0 was added. The resin was pelleted again under the same conditions, and the supernatant was discarded. The protein solution was then added to the washed resin and incubated for 30 min with gentle agitation. The mixture was then centrifuged for 5 min at 3000 × g. The supernatant was discarded, and 500 μl of buffer A1 was added, and the resin was pelleted by centrifugation for 5 min at 3000 × g. The same procedure was followed for buffer B and buffer A2 (as described in Shen et al. (24)). The bound proteins were eluted by the addition of buffer C (40 μl). An equal amount of 2× SDS-loading buffer was added to the sample, which was boiled and loaded on a 15% (w/v) acrylamide gel as described previously (24). For purification of large amounts of the CpcSU complex, whole cell extracts containing each subunit were incubated together on ice for 1 h, and then the complexes were purified using metal affinity chromatography in the same way as described above for HT-CpcU.
In Vitro PcyA Reactions—A plasmid for the expression of pcyA was kindly provided by Dr. J. C. Lagarias (32). This enzyme was overproduced as a glutathione S-transferase fusion and purified as described previously (32–34). PcyA reduces biliverdin in two sequential 2-electron reductions using reduced ferredoxin to produce PCB. rCpcBA (1 mg ml–1) reactions (total volume, 4 ml) were set up with one of the following additions; 200 μl of whole-cell extract from E. coli cells harboring pAED4 as the negative control (24), 200 μl of CpcS-I (1 mg ml–1), 600 μl of HT-CpcU (0.33 mg ml–1), or 400 μl of co-purified CpcSU (1 mg ml–1). The following were added to the reaction mixture: 50 mm HEPES buffer, pH 7.3, 1 mm MgCl2 6.5 mm glucose-6-phosphate, 1.6 mm NADP+, 1.1 units of glucose-6-phosphate dehydrogenase ml–1, 4.6 μm recombinant ferredoxin from Synechococcus sp. PCC 7002 (35) or spinach ferredoxin (Sigma), 0.025 units ml–1 of spinach (Sigma) or recombinant Synechococcus sp. PCC 7002 ferredoxin:NADP+ oxidoreductase (12, 35, 36), 10 μm bovine serum albumin, 5 μm biliverdin (Porphyrin Products, Logan, UT), and 10 μm PcyA (32–34). All reactions were incubated in a 30 °C water bath for 1 h in the dark. Another aliquot of biliverdin was added (for a final concentration of 10 μm), and the reactions were allowed to continue for 1 h at 30 °C. The reaction solutions were clarified by centrifugation at 14,000 × g for 10 min.
For reactions containing aporHT-AP (in a 1.5 ml volume), aporHT-AP (600 μl of 2.0 mg ml–1 solution) was added to one of the following: 50 μl of pAED4 E. coli control extract, 50 μl of CpcS-I (1 mg ml–1), 150 μl of HT-CpcU (0.33 mg ml–1), or both CpcS-I and HT-CpcU (50 and 150 μl, respectively; i.e. not copurified). These reactions contained the same concentrations of buffers, PcyA, and other additions as specified above for the rCpcBA reactions.
Fluorescence emission spectra were acquired with a PerkinElmer Life Sciences fluorometer with the slits set at 10 nm (excitation and emission), and the excitation wavelength was set to 590 nm. The absorbance spectra were measured on a Lambda 35, dual-beam UV-visible spectrophotometer (PerkinElmer Life Sciences). Zinc-enhanced bilin fluorescence of proteins was performed after SDS-PAGE using the FX imaging system to excite the bilin chromophores at 532 nm and a 555-nm long-pass filter (Bio-Rad) (24).
Tryptic Digestion of Phycocyanin—The products of the four rCpcBA lyase reactions described above and holo-PC (100 μl of a 4 mg ml–1 solution), which had been purified as described from Synechococcus sp. PCC 7002 (37), were purified by ion-exchange chromatography on separate DEAE columns (1 × 3 cm) that had been equilibrated with 5 mm sodium phosphate, pH 7.0. The colored products were eluted with 100 mm sodium phosphate buffer, pH 7.0. The samples were exhaustively dialyzed against 50 mm sodium phosphate buffer, pH 7.0, 1 mm 2-mercaptoethanol and then concentrated by ultrafiltration over an Amicon YM10 membrane (Millipore, Billerica, MA). The resulting samples were analyzed by SDS-PAGE. Concentrated reaction samples were diluted 1:4 and were titrated to pH 2.0 with 1 n HCl. These solutions were incubated for 45 min in the dark at room temperature to allow proteins to unfold. Tryptic digestion was performed as described (38), and peptides were purified for HPLC analysis as described (24). The peptide samples were stored in the dark at –20 °C until required for HPLC analysis.
High Performance Liquid Chromatography—Tryptic peptides were separated on a C18 reverse-phase HPLC column (5 μm × 10 mm × 250 mm; Waters Corp., Milford, MA) using a Waters HPLC equipped with a 600E pump and a photodiode array detector. The peptide separation was performed as described (38) using 0.1 m phosphate buffer, pH 2.1, and 100% acetonitrile. Bilin peptides were monitored at 600 nm. For size exclusion chromatography, a Bio-Sil SEC250 column (Bio-Rad; 300 × 7.8 mm) equipped with a guard column of the same material (80 × 7.8 mm) was used in conjunction with the Waters HPLC pump and detector. The temperature for all runs was 22 °C. The liquid phase was 50 mm sodium phosphate, pH 7.0, and was delivered at a rate of 0.8 ml min–1. Samples (200 μl) were injected onto the column that was equilibrated in the phosphate buffer. A standard curve was established using molecular weight standards from Bio-Rad (thyroglobulin, 670 kDa; bovine γ-globulin, 158 kDa; chicken ovalbumin, 44 kDa; horse myoglobin, 17 kDa; vitamin B12, 1.35 kDa). Samples were collected from various peaks, and the proteins present were precipitated by the addition of an equal volume of 20% (w/v) trichloroacetic acid before SDS-PAGE.
SDS-PAGE and Immunoblotting—SDS-PAGE, immunoblotting with rabbit antibodies against Synechococcus sp. PCC 7002 CpcS-I, and determination of zinc-enhanced fluorescence of proteins were performed as previously described (24, 25).
RESULTS
Purification of CpcS-I, HT-CpcU, and HT-CpcV—The genome of the unicellular marine cyanobacterium Synechococcus sp. PCC 7002 encodes three paralogs, denoted cpcS-I, cpcU, and cpcV, of the cpeS gene of F. diplosiphon (22). In the accompanying paper (25), a reverse genetics approach provided strong evidence that two of these genes, cpcS-I and cpcU, encode the subunits of a heterodimeric PCB lyase specific for the Cys-82 position of β-PC and Cys-81 of the α and β subunits of AP. To test directly whether these proteins are lyases, each gene was amplified by PCR and cloned into expression vectors as described under “Experimental Procedures.” The cpcU gene was expressed as a fusion protein with a His6 tag at its N terminus, whereas the cpcS-I gene was expressed with no affinity tag. The cpcV gene was expressed with and without a His6 tag. After induction with isopropyl 1-thio-β-d-galactopyranoside, the recombinant proteins were readily detectable among the soluble proteins of E. coli cell extracts as shown in supplemental Fig. 1 (the arrows indicate the polypeptides corresponding to each protein). The apparent molecular masses on SDS-PAGE were consistent with the calculated molecular masses and ranged from 19 kDa (CpcV) to 23 kDa (HT-CpcU; see “Experimental Procedures”). CpcS-I, HT-CpcU, and HT-CpcV were purified as described above.
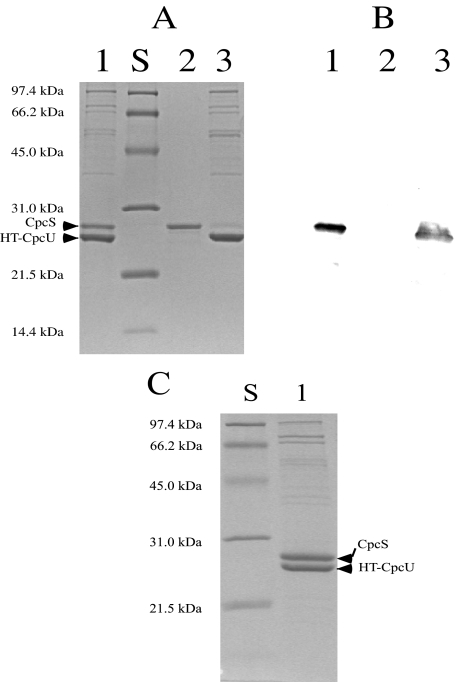
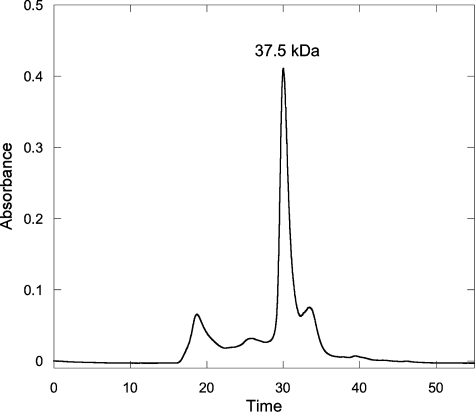
Lyase Subunit Interactions—Some PCB lyases, such as CpcE/CpcF and PecE/PecF (13–20), are heterodimers, and therefore, it was important to learn whether CpcS-I and CpcU interact to form a heterodimer. To determine whether these proteins can form a stable complex, purified CpcS-I (Fig. 1A, lane 2) and purified HT-CpcU (Fig. 1A, lane 3) were mixed as described, and HT-CpcU was affinity-purified using the Ni-NTA resin in a pulldown experiment. As shown in Fig. 1A, lane 1, a protein with the electrophoretic mobility of CpcS-I copurified with HT-CpcU. To verify the identity of the protein that copurified with HT-CpcU, an immunoblot was prepared from an identical gel and probed with antibodies raised against CpcS-I. The antibodies to CpcS-I did not cross-react with HT-CpcU (Fig. 1B, lane 2) but recognized both purified CpcS-I (Fig. 1B, lane 1) and CpcS-I that copurified with HT-CpcU in the pulldown experiment (Fig. 1B, lane 3). When individual E. coli whole-cell extracts containing CpcS-I and HT-CpcU were combined and allowed to interact for 1 h on ice before purification by Ni-NTA chromatography, CpcS-I and HT-CpcU could be co-purified and were present in an approximate 1:1 stoichiometry as judged by Coomassie Blue staining after SDS-PAGE (Fig. 1C, lane 1). To determine the molecular weight of this complex, the copurified, recombinant CpcS-I and HT-CpcU (CpcSU) complex was subjected to size exclusion chromatography (Fig. 2). As judged by SDS-PAGE, both CpcS-I and HT-CpcU were present in all fractions (data not shown). The molecular weight of the most abundant component was calculated to be 37,500, and the latest eluting (∼33 min peak) corresponded to single-subunit monomers with molecular weights of ∼20,000–25,000. The combined Mr of HT-CpcU (23,475) and CpcS-I (22,526) is ∼46,000. Therefore, these results indicated that CpcS-I and HT-CpcU form a 1:1 heterodimer. Unless stated otherwise, the CpcSU heterodimer complex was used for all subsequent experiments.
FIGURE 1.
SDS-PAGE and immunoblot analysis of CpcS-I and HT-CpcU. Panel A shows a Coomassie Blue-stained SDS-polyacrylamide gel that was loaded with the products of the interaction between HT-CpcU and CpcS-I after elution from metal chelation chromatography. Lane 1, purified HT-CpcU and CpcS-I complex; lane 2, molecular mass standards (S) whose sizes are given to the left; lane 3, purified CpcS-I; lane 4, purified HT-CpcU. Panel B shows an immunoblot of another part of the gel shown in panel A that had been probed with anti-CpcS-I antibodies. Lane 1 contained purified CpcS-I; lane 2 contains HT-CpcU; lane 3 contains the eluate from metal chelation chromatography that contains both CpcS-I and HT-CpcU. Panel C shows the results from SDS-PAGE analysis of the product of a large scale copurification of HT-CpcU and CpcS-I complex. E. coli extracts containing the individual proteins were combined as described, and CpcS-I was copurified with HT-CpcU with a 1:1 stoichiometry as shown in lane 1. Lane S shows molecular mass standards whose masses are indicated at the left.
FIGURE 2.
Size exclusion chromatography of CpcSU. Approximately 200 μl of CpcSU (∼1 mg ml–1) was injected onto the size exclusion column, and the protein absorbance was monitored at 280 nm. The apparent molecular mass of the complex, calculated after chromatography of molecular mass standards (see under “Experimental Procedures”), is shown above the major peak.
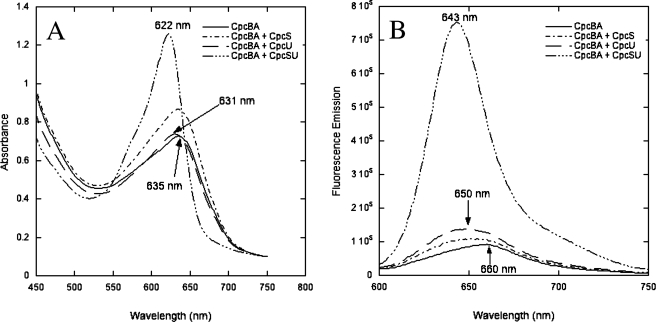
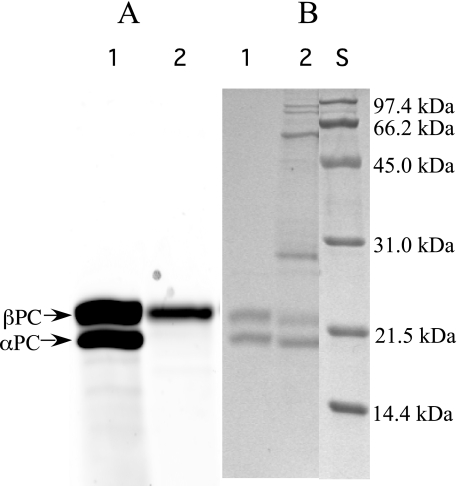
Bilin Addition Assays with ApoPC—As described under “Experimental Procedures,” recombinant CpcB and CpcA (rCpcBA) were co-purified from E. coli and used for PCB addition assays (with PcyA-generated PCB in situ) with CpcS-I, HT-CpcU, CpcSU, or a control extract from E. coli cells harboring an empty vector (no lyase). In Fig. 3A, the absorption spectra of these reaction products are shown. The absorption maximum for the no-lyase control (PCB and rCpcBA only) was 635 nm. This product likely represents covalently bound mesobiliverdin (MBV), a more oxidized bilin product that occurs when no lyases are present (24, 38). PCB attachment reactions with CpcS-I or the HT-CpcU produced reaction products with absorption maxima at 631 nm, which is also probably MBV. However, when the CpcSU heterodimer was present, the absorption maximum of the rCpcBA product was 622 nm, which is very similar to the maximum expected for PCB bound at Cys-82 on β-PC (39). The same results were achieved when purified PCB replaced PcyA-generated PCB in addition reactions with rCpcBA (see supplemental Fig. 2). The fluorescence emission spectra of these reaction products are shown in Fig. 3B. The product of the CpcSU reaction was extremely fluorescent and had an emission maximum at 643 nm, whereas the control reaction had a small fluorescence yield with a maximum at ∼660 nm. The products of reactions with CpcS-I or HT-CpcU alone also had low fluorescence yields with maxima near 650 nm. The PCB chromophore bound to Cys-82 of β-PC should be highly fluorescent at ∼644–650 nm (39–41), whereas MBV is much less fluorescent and has an emission maximum at 660–668 nm (38). The absorbance and fluorescence emission spectra of the reaction product obtained from CpcSU are both consistent with the presence of PCB at the Cys-82 site on CpcB. However, because both apoCpcA (one PCB addition site at Cys-84) and apoCpcB (two PCB addition sites, Cys-82 and Cys-153) are present in the reaction mixture, it was necessary to determine experimentally which site(s) carried the PCB chromophore(s). An aliquot of each reaction was separated by SDS-PAGE, and the zinc-enhanced fluorescence of PCB-bound peptides (Fig. 4A) was recorded; subsequently, the gel was stained with Coomassie Blue (Fig. 4B). Fig. 4A shows the bilin fluorescence of holo-PC purified from Synechococcus sp. PCC 7002 (lane 1) and of the CpcSU-dependent, rCpcBA reaction product (lane 2), showing only one fluorescent band corresponding to β-PC. The bilin-linked CpcB subunit of this reaction (Fig. 4B, lane 2) had an electrophoretic mobility that was slightly faster than that of holo-β-PC. This finding is consistent with the presence of a single PCB chromophore (588 Da) rather than the two PCB chromophores of holo-β-PC (Fig. 4B, lane 1).
FIGURE 3.
Absorbance and fluorescence emission spectra of in vitro bilin addition reactions with CpcS-I and HT-CpcU and rCpcBA. A, absorbance spectra of reactions with rCpcBA alone (control, solid line) and rCpcBA with CpcS-I alone (dash dots), HT-CpcU alone (long dashes), or CpcS-I and HT-CpcU copurified (long dash triple dot). The absorption maxima of some reaction products are indicated. B, fluorescence emission spectra of the same reactions with excitation at 590 nm (10-nm slit widths). The fluorescence emission maximum for rCpcBA with CpcS-I/HT-CpcU was at 643 nm, whereas all products had emission maxima between 650 and 660 nm.
FIGURE 4.
SDS-PAGE analysis of bilin addition reaction of rCpcBA products from CpcSU reactions. A, zinc-enhanced fluorescence of holo-PC purified from Synechococcus sp. PCC 7002 (lane 1) and the CpcSU-dependent rCpcBA reaction product (lane 2). The identity of each fluorescing polypeptide is indicated at the left. B, the same SDS-PAGE gel after staining with Coomassie Blue. The masses of molecular mass standards (S) are indicated to the right in kDa. The same gel was used in both panels, but a different procedure and instrument, each using a different magnification, was used to acquire the two images.
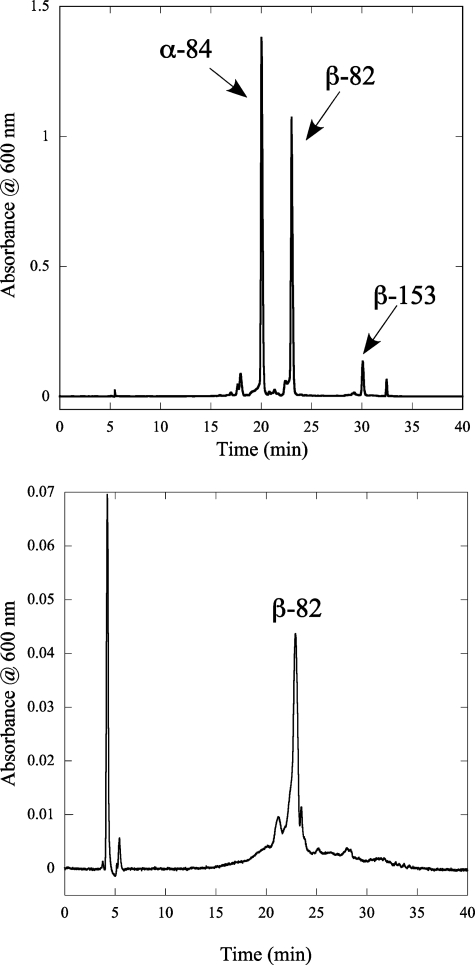
Identification of the PCB Addition Site on β-PC—Because β-PC has two PCB chromophore binding sites, one at Cys-82 and one at Cys-153, tryptic mapping of the CpcSU-dependent, rCpcBA reaction product was performed to identify which Cys residue(s) carried PCB. The rCpcBA product of the CpcSU addition reaction and the purified holo-PC from Synechococcus sp. PCC 7002 were digested with trypsin. The products of the tryptic digestions were analyzed by HPLC chromatography on a C18 reverse-phase column. The elution profile of the bilin peptides (recorded at 600 nm) is shown in Fig. 5. The top chromatogram shows the elution profile of tryptic digestion of holo-PC. There are three main peaks in the profile, which correspond to the chromopeptides containing the α-Cys-84, β-Cys-82, andβ-Cys-153 residues, at which PCB is covalently bound in holo-PC. The peptide containing α-Cys-84 eluted first at 20.1 min and was followed by the β-Cys-82 peptide at 23.1 min and the β-Cys-153 peptide at 30.1 min. This elution order of the peptides is consistent with the previously reported results from Arciero et al. (38). It is also consistent with the calculated masses of the tryptic peptides, which are: α-Cys-84 = 1251 Da, β-Cys-82 = 1323 Da, and β-Cys-153 = 4075 Da (24). The lower chromatogram shows the elution profile for the tryptic peptides from rCpcBA that had been incubated with PCB and CpcSU. Only one PCB-containing tryptic peptide, with a retention time of 23.0 min, was observed. This retention time identifies this peptide as the one that includes the β-Cys-82 binding site. This result consequently demonstrates that CpcSU is a bilin lyase that specifically attaches PCB to Cys-82 of the β-PC subunit.
FIGURE 5.
Reverse-phase HPLC separation of tryptic peptides with bound bilins. The top chromatogram shows the elution pattern of tryptic peptides containing bilins (as determined by absorbance at 600 nm) derived from wild-type Synechococcus sp. PCC 7002 PC. Arrows identify the three bilin-containing peptides by the cysteinyl residue that carries the PCB chromophore. The bottom chromatogram shows the retention times for the PCB peptides derived from CpcSU-dependent rCpcBA product. Only one PCB-linked peptide, with a retention time at 23.0 min and corresponding to the Cys-82 site of β-PC, is observed.
Bilin Addition to ApoAP—We next tested whether CpcS-I, CpcU, or CpcSU was involved in bilin attachment to the α and β subunits of AP (aporHT-AP). We presumed that aporHT-AP would purify as an (αβ) heterodimer (i.e. the “monomer”). To test this assumption experimentally, size-exclusion chromatography of aporHT-AP was performed, and the majority of the protein eluted as a complex with a Mr of ∼27,000, which suggested that the recombinant protein was forming an αβ protomer (monomer; the calculated Mr is 38,600) (supplemental Fig. 3). When both CpcS-I and HT-CpcU are present in the PCB addition reaction, the absorbance maximum of the product was 614 nm, which is consistent with that expected for holo-monomeric AP (Fig. 6A) (42). When CpcS-I, CpcU, or only aporHT-AP (control) was incubated with PCB, the absorbance maximum was 645–647 nm. However, only the CpcSU-dependent rHT-AP reaction product was fluorescent and had the expected emission maximum at 636 nm. The other reaction products, including the negative control, were nearly non-fluorescent and had only very low fluorescence emission at 650 nm, consistent with the presence of MBV. Native holo-AP is isolated as a trimer (α3β3), and in this form, its absorbance is red-shifted to 650 nm (1–3). The holo-monomeric AP has an absorption spectrum that is more similar to that of the individual α or β subunits, which have absorption maxima near 614 nm (43). Therefore, it was important to determine whether bilin addition had taken place on both subunits or on only one of the two subunits.
FIGURE 6.
Absorbance and fluorescence emission spectra of in vitro bilin addition reactions with aporHT-AP (HT-ApcA/ApcB). A, absorbance spectra of the control reaction (solid line), the reaction containing CpcS-I (medium dashes), HT-CpcU (short dashes), or both CpcSU (long dash triple dots). B, shows the room temperature, fluorescence emission spectra of the same reactions upon excitation at 590 nm with 10-nm slit widths. The maxima for some absorbance or fluorescence emission of some of the products are indicated.
Aliquots of each reaction were analyzed by SDS-PAGE, and proteins were tested for the presence of bilins using zinc-enhanced fluorescence (Fig. 7B). After recording the fluorescence emission, the gel was stained with Coomassie Blue (Fig. 7A). A bilin was attached to ApcB but not to HT-ApcA in the products from the control reaction with PCB and no enzyme (Fig. 7A, lane 1). The addition product to ApcB in the absence of an added enzyme is not the same as native AP, however, as evidenced by the absorbance and fluorescence emission spectra of this control reaction (Fig. 6, A and B). The ApcB product of this non-enzymatic attachment could either be MBV or PCB, which is not rigidly held in a stretched conformation. To determine which was the case, the products from a control reaction were dissolved in 8 m urea, pH 2 (see supplemental Fig. 4). The spectrum of the protonated bilin in the absence of any protein interactions was identical to that of PCB, which has absorption maxima at 359 and 664 nm under these conditions. MBV would have produced absorption maxima in the visible that were shifted 20 nm further to the red relative to PCB (31, 38, 44). Therefore, PCB can bind to ApcB in the absence of any lyase, but the resulting PCB product was not bound in a stretched conformation and did not have the absorption properties expected for native ApcB. Only when both CpcS-I and HTCpcU (Fig. 7B, lane 4), were present was there significant PCB addition to both AP subunits. The absorbance spectrum of this CpcSU-dependent rHT-AP product had a slight shoulder at 650 nm (Fig. 6A), which is probably attributable to the formation of some trimeric AP, which as noted above has an absorption maximum at 650 nm and a fluorescence emission maximum at ∼663 nm. The fluorescence emission spectrum of this reaction product was rather broad and had significant emission beyond 650 nm. Given the markedly different properties of the addition product for CpcSU, we conclude that CpcSU is also responsible for attachment of PCB to Cys-81 on both AP subunits in vitro.
FIGURE 7.
SDS-PAGE of products after in vitro bilin addition reactions. A, analysis of PCB addition to aporHT-AP (HT-ApcA/ApcB) by SDS-PAGE and Coomassie Blue staining. B, the same gel showing the zinc-enhanced fluorescence of PCB attached to the proteins in lanes 1–4. The legend at the top identifies which lyase subunit(s) was present in the samples using the +/– system. The HT-ApcA and ApcB proteins are identified at the left, and the sizes of molecular mass markers (lane S) are indicated in kDa at the right.
Bilin Addition Assays with CpcV—CpcV exhibited very little activity when proteins in whole-cell extracts of E. coli were added to rCpcBA and PCB to assay for PCB lyase activity. When CpcV was added alone or in combination with CpcS-I or CpcU (supplemental Fig. 5), a small amount of fluorescence from rCpcBA was often detected from such PCB addition assays. However, when compared with the activity obtained with CpcSU, the amount of this activity was consistently very low (supplemental Fig. 5). When CpcV alone or CpcV together with CpcS-I or CpcU from whole-cell extracts of E. coli were added to rCpcBA and PCB, the resulting products had absorption maxima near 600 nm and fluorescence emission maxima near 620 nm (supplemental Fig. 5). Because strains with null mutations in cpcV had no phenotype unless this mutation was combined with mutations in cpcS-I and cpcU (25), CpcV does not play a major role in PCB attachment to PBP.
DISCUSSION
The results presented here clearly show that both CpcS-I and CpcU are required to attach PCB to Cys-82 on β-PC, Cys-81 on α-AP, and probably to Cys-81 on β-AP. The CpcS-I and CpcU orthologs from Synechocystis sp. PCC 6803 are similarly required for PCB addition to β-PC (45). CpcS-I and HT-CpcU copurified as a stable heterodimer. Therefore, some cyanobacteria require a heterodimeric lyase for PCB attachment to the Cys-82 position on β-PC and both AP subunits. This result differs from those obtained by Zhao et al. (26, 27), who recently showed that only the product of open reading frame alr0617, denoted CpeS by these authors, of Nostoc sp. PCC 7120 is required for PCB addition to Cys-82 of β-PC and β-phycoerythrocyanin. It is presently unclear why two lyase subunits are required for some cyanobacteria but not for others. One obvious possibility is that the Alr0617 lyase is a homodimer, whereas a gene duplication event has occurred in some cyanobacteria, including Synechococcus sp. PCC 7002 and Synechocystis sp. PCC 6803. Phylogenetic analyses (see Fig. 1 of Ref. 25) show that Alr0617 belongs to a phylogenetically distinct subgroup of the CpcS family, which we have named CpcS-III. Whether organisms with CpcS-III arose by the loss of CpcU or organisms with CpcS-I gained CpcU by a gene duplication event, is presently unclear.
The in vitro biochemical results presented here are completely consistent with in vivo results obtained from the analysis of cpcS-I, cpcU, and cpcS-I and cpcU mutants (25). These mutants had severely reduced levels of PBP, and some of the PC that these mutants accumulated were missing the PCB at Cys-82 of β-PC. We also observed that when PBS were subjected to mass spectrometry, some of the PC present had the mass expected for wild-type β-PC, whereas some of the β-PC had a mass consistent with one missing PCB, and a compound with the mass of free PCB was observed at 587.5 Da (25). Our interpretation of these results is that, in the absence of CpcS-I or CpcU, PCB can associate with β-PC in the Cys-82 binding pocket non-covalently, and it slowly becomes covalently bound to Cys-82. However, only that fraction of the protein in which the chromophore has the requisite R stereochemistry at C-31 of the PCB is likely to be stably incorporated into PBS and avoid degradation.
Our results also show that a lyase is required for bilin addition to AP subunits. While the results described here were being prepared for publication, Zhao et al. (27) also reported that CpcS-III (Alr0617) attaches PCB to AP subunits in Nostoc sp. PCC 7120. Although we observed that some bilin addition took place on ApcB in the absence of enzymes, this product did not have the expected absorbance of holo-β-AP, probably because the attached bilin assumed a cyclic conformation rather than its normal stretched conformation (46–49). One of us (W. M. S.) had previously observed that bilin addition can take place to recombinant Nostoc sp. PCC 7120 apoAP subunits in vitro without enzymes. However, like the results shown in this paper, these products had red-shifted absorbance maxima when compared with native, holo-AP subunits; the products were not highly fluorescent like the native, holo-AP subunits, and when denatured, the spectra of these products were consistent with those for bound PCB.5 Similar results were also observed for PCB addition to CpcA in vitro. When the detergent Triton X-100 was added, the product in the absence of enzymes was PCB and not MBV, but its absorbance properties were altered, presumably due to the chromophore conformation and interaction (or lack of interaction) with CpcA (31). Recently, Hu et al. (30) showed that bilin addition to ApcA could take place without enzymes either in vitro or in E. coli cells. The ApcA product they obtained appeared to be similar to native, holo-ApcA.
The strongest evidence supporting the necessity of enzymes for PCB addition to AP subunits comes from the analysis of cpcS-I cpcU, cpcS-I cpcU cpcV, and cpcS-I cpcU cpcT mutants (25). ApcB levels in these three mutants, as quantitated from immunoblots, were 27, 13, and 7% of ApcB levels in wild-type cells, respectively. In contrast, a cpcBAC mutant had ≥68% of wild-type ApcB levels (25). If these lyase subunits were only required for the addition of PCB to Cys-82 on β-PC, then one would expect little or no effect on the AP content of cells. Clearly, there is a major defect in AP biogenesis in the CpcS-I and cpcU mutants, suggesting that lyases are normally required for PCB addition to the AP subunits in vivo.
Similar to the results of Zhao et al. (26, 27) for CpcS-III, we found no evidence that CpcS-I and CpcU could perform PCB transfer reactions from PC to aporHT-AP (see supplemental Fig. 6). Therefore, this class of lyases appears to differ from the CpcE/CpcF (13–17, 51, 52) and PecE/PecF (18–20) class of lyases, which might have a role in the repair of damaged PBP and in the removal of bilins during PBP degradation as well as their role in biogenesis. The β-82 chromophore has the important role of acting as the acceptor/fluorescing PCB within a trimer of PC (1, 50). If this β-82 PCB becomes damaged, there would be serious consequences to energy transfer. This chromophore is buried more deeply within the trimer interior and, therefore, might be less accessible for removal or replacement than the Cys-84 PCB of the α-PC. This could be one reason why the CpcSU lyase apparently is not involved in bilin repair or replacement and, thus, why it might never have evolved the capacity for the transferase reaction.
We have been unable to determine a function for CpcV to date. No obvious PCB attachment reaction could be demonstrated, and CpcV does not appear to play an important role in PBP degradation/turnover, since there was no difference in PBP degradation for the cpcV mutant and the wild type under nutrient starvation conditions.6 Interestingly, Synechocystis sp. PCC 6803 does not have a cpcV ortholog, and this gene is only found in some of the cyanobacterial genomes sequenced thus far (25). Because CpcV apparently does not play a critical role in PBP biogenesis or turnover, it is possible that this divergent, lyase-like protein is involved in PCB attachment to a protein other than a PBP.
In conclusion, we have shown here that CpcS-I and CpcU form a heterodimeric lyase that attaches PCB to Cys-82 on β-PC and to Cys-81 of the α and β subunits of AP. Together with the previously described CpcE/CpcF heterodimeric PCB lyases for Cys-84 on α-PC (13–17) and CpcT for Cys-153 of β-PC (24) it appears that all of the lyases required for PBP biogenesis in Synechococcus sp. PCC 7002 have been identified.
Supplementary Material
Acknowledgments
The W. M. Keck Foundation provided support for equipment utilized for this study, which is located in the Keck Conservation and Molecular Genetics laboratory at University of New Orleans. We thank Zvjezdana Begovic, Leon Harrison, Jr., Ariane Fletcher, and Alan Williams for technical support. We thank J. Clark Lagarias (University of California-Davis) for providing the Nostoc sp. PCC 7120 pcyA expression clone.
This research was supported in part by National Science Foundation Grants MCB-0133441 (to W. M. S.) and MCB-0077586 and MCB-0519743 (to D. A. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1–6.
Footnotes
The abbreviations used are: PBS, phycobilisome; AP, allophycocyanin; HPLC, high performance liquid chromatography; HT, His6-tagged; MBV, mesobiliverdin; Ni-NTA, nickel-nitrilotriacetic acid; PBP, phycobiliprotein; PC, phycocyanin; PCB, phycocyanobilin; rHT-AP, recombinant His6-tagged-ApcA/ApcB; rCpcBA, recombinant apoCpcB/CpcA; CpcSU, recombinant CpcS-I/HT-CpcU.
P. Fung, W. M. Schluchter, and A. N. Glazer, unpublished results.
G. Shen, W. M. Schluchter, and D. A. Bryant, unpublished results.
References
- 1.Glazer, A. N. (1989) J. Biol. Chem. 264 1–4 [PubMed] [Google Scholar]
- 2.Sidler, W. A. (1994) in The Molecular Biology of Cyanobacteria (Bryant, D. A., ed) pp. 139–216, Kluwer Academic Press, Dordrecht, The Netherlands
- 3.Bryant, D. A. (1991) in The Photosynthetic Apparatus: Molecular Biology and Operation. (Bogarad, L., and Vasil, I. K., eds) pp. 255–298, Academic Press, Inc., New York
- 4.de Lorimier, R., Bryant, D. A., Porter, R. D., Liu, W.-Y., Jay, E., and Stevens, S. E., Jr. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 7946–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, D. A. (1988) in Light-Energy Transduction in Photosynthesis: Higher Plant and Bacterial Models (Stevens, S. E., Jr., and Bryant, D. A., eds) pp. 62–90, American Society of Plant Physiologists, Rockville, MD
- 6.Bryant, D. A., de Lorimier, R., Guglielmi, G., and Stevens, S. E., Jr. (1990) Arch. Microbiol. 153 550–560 [DOI] [PubMed] [Google Scholar]
- 7.de Lorimier, R., Guglielmi, G., Bryant, D. A., and Stevens, S. E., Jr. (1990) Arch. Microbiol. 153 541–549 [DOI] [PubMed] [Google Scholar]
- 8.de Lorimier, R., Bryant, D. A., and Stevens, S. E., Jr. (1990) Biochim. Biophys. Acta 1019 29–41 [DOI] [PubMed] [Google Scholar]
- 9.Maxson, P., Sauer, K., Bryant, D. A., and Glazer, A. N. (1989) Biochim. Biophys. Acta 974 40–51 [DOI] [PubMed] [Google Scholar]
- 10.Gindt, Y. M., Zhou, J., Bryant, D. A., and Sauer, K. (1992) J. Photochem. Photobiol. B. 15 75–89 [DOI] [PubMed] [Google Scholar]
- 11.Gindt, Y. M., Zhou, J., Bryant, D. A., and Sauer, K. (1994) Biochim. Biophys. Acta 1186 153–162 [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Lojero, C., Pérez-Gómez, B., Shen, G., Schluchter, W. M., and Bryant, D. A. (2003) Biochemistry 42 13800–13811 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, J., Gasparich, G. E., Stirewalt, V. L., de Lorimier, R., and Bryant, D. A. (1992) J. Biol. Chem. 267 16138–16145 [PubMed] [Google Scholar]
- 14.Swanson, R. V., Zhou, J., Leary, J. A., Williams, T., de Lorimier, R., Bryant, D. A., and Glazer, A. N. (1992) J. Biol. Chem. 267 16146–16154 [PubMed] [Google Scholar]
- 15.Fairchild, C. D., Zhao, J., Zhou, J., Colson, S. E., Bryant, D. A., and Glazer, A. N. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, J. (1992) Mutational Analysis of the Genes Encoding Phycobilisome Components in the Cyanobacterium Synechococcus sp. PCC 7002. Ph.D. thesis, The Pennsylvania State University, University Park, PA
- 17.Fairchild, C. D., and Glazer, A. N. (1994) J. Biol. Chem. 269 8686–8694 [PubMed] [Google Scholar]
- 18.Jung, L. J., Chan, C. F., and Glazer, A. N. (1995) J. Biol. Chem. 270 12877–12884 [DOI] [PubMed] [Google Scholar]
- 19.Zhao, K. H., Deng, M. G., Zheng, M., Zhou, M., Parbel, A., Storf, M., Meyer, M., Strohmann, B., and Scheer, H. (2000) FEBS Lett. 469 9–13 [DOI] [PubMed] [Google Scholar]
- 20.Zhao, K. H., Wu, D., Zhou, M., Zhang, L., Bohm, S., Bubenzer, C., and Scheer, H. (2005) Biochemistry 44 8126–8137 [DOI] [PubMed] [Google Scholar]
- 21.Kahn, K., Mazel, D., Houmard, J., Tandeau de Marsac, N., and Schaefer, M. R. (1997) J. Bacteriol. 179 998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cobley, J. G., Clark, A. C., Weerasurya, S., Queseda, F. A., Xiao, J. Y., Bandrapali, N., D'Silva, I., Thounaojam, M., Oda, J. F., Sumiyoshi, T., and Chu, M. H. (2002) Mol. Microbiol. 44 1517–1531 [DOI] [PubMed] [Google Scholar]
- 23.Shen, G., Saunee, N. A., Gallo, E., Begovic, Z., Schluchter, W. M., and Bryant, D. A. (2004) in Photosynthesis 2004 Light-Harvesting Systems Workshop, August 26–29, 2004 (Niederman, R. A., Blankenship, R. E., Frank, H., Robert, B., and van Grondelle, R., eds) pp. 14–15, Saint Adele, Quebec, Canada
- 24.Shen, G., Saunée, N. A., Williams, S. R., Gallo, E. F., Schluchter, W. M., and Bryant, D. A. (2006) J. Biol. Chem. 281 17768–17778 [DOI] [PubMed] [Google Scholar]
- 25.Shen, G., Schluchter, W. M., and Bryant, D. A. (2008) J. Biol. Chem. 283 7503–7512 [DOI] [PubMed] [Google Scholar]
- 26.Zhao, K. H., Su, P., Li, J., Tu, J. M., Zhou, M., Bubenzer, C., and Scheer, H. (2006) J. Biol. Chem. 281 8573–8581 [DOI] [PubMed] [Google Scholar]
- 27.Zhao, K.-H., Su, P., Tu, M.-T., Wang, X., Liu, H., Ploscher, M., Eichacker, L., Yang, B., Zhou, M., and Scheer, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14300–14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capuano, V., Braux, A. S., Tandeau de Marsac, N., and Houmard, J. (1991) J. Biol. Chem. 266 7239–7247 [PubMed] [Google Scholar]
- 29.Zhao, K. H., Su, P., Bohm, S., Song, B., Zhou, M., Bubenzer, C., and Scheer, H. (2005) Biochim. Biophys. Acta 1706 81–87 [DOI] [PubMed] [Google Scholar]
- 30.Hu, I. C., Lee, T. R., Lin, H. F., Chiueh, C. C., and Lyu, P. C. (2006) Biochemistry 45 7092–7099 [DOI] [PubMed] [Google Scholar]
- 31.Zhao, K. H., Zhu, J. P., Song, B., Zhou, M., Storf, M., Bohm, S., Bubenzer, C., and Scheer, H. (2004) Biochim. Biophys. Acta 1657 131–145 [DOI] [PubMed] [Google Scholar]
- 32.Frankenberg, N., and Lagarias, J. C. (2003) J. Biol. Chem. 278 9219–9226 [DOI] [PubMed] [Google Scholar]
- 33.Frankenberg, N., Mukougawa, K. K., and Lagarias, J. C. (2001) Plant Cell 13 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yakota, A., and Lagarias, J. C. (2001) Plant Cell 13 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schluchter, W. M. (1994) The Characterization of Photosystem I and Ferredoxin-NADP+ Oxidoreductase in the Cyanobacterium Synechococcus sp. PCC 7002. Ph.D. thesis, The Pennsylvania State University, University Park, PA
- 36.Schluchter, W. M., and Bryant, D. A. (1992) Biochemistry 31 3092–3102 [DOI] [PubMed] [Google Scholar]
- 37.Glazer, A. N. (1988) Methods Enzymol. 167 291–303 [DOI] [PubMed] [Google Scholar]
- 38.Arciero, D. M., Bryant, D. A., and Glazer, A. N. (1988) J. Biol. Chem. 263 18343–18349 [PubMed] [Google Scholar]
- 39.Debreczeny, M. P., Sauer, K., Zhou, J., and Bryant, D. A. (1993) J. Phys. Chem. 97 9852–9862 [Google Scholar]
- 40.Sauer, K., Scheer, H., and Sauer, P. (1987) Photochem. Photobiol. 46 427–440 [Google Scholar]
- 41.Mimuro, M., Füglistaller, P., Rümbeli, R., and Zuber, H. (1986) Biochim. Biophys. Acta 848 155–166 [Google Scholar]
- 42.Gysi, J. R., and Zuber, H. (1979) Biochem. J. 181 577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck, W. F., and Sauer, K. (1992) J. Phys. Chem. 96 4658–4666 [Google Scholar]
- 44.Glazer, A. N., and Fang, S. (1973) J. Biol. Chem. 248 659–662 [PubMed] [Google Scholar]
- 45.Miller, C. A. (2007) Identification and Characterization of Enzymes Involved in Post-translational Modifications of Phycobiliproteins in the Cyanobacterium Synechocystis sp. PCC 6803. M.Sc. thesis, University of New Orleans, New Orleans, LA
- 46.Brejc, K., Ficner, R., Huber, R., and Steinbacher, S. (1995) J. Mol. Biol. 249 424–440 [DOI] [PubMed] [Google Scholar]
- 47.Reuter, W., Wiegand, G., Huber, R., and Than, M. E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1363–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schirmer, T., Bode, W., and Huber, R. (1987) J. Mol. Biol. 196 677–695 [DOI] [PubMed] [Google Scholar]
- 49.Schirmer, T., Huber, R., Schneider, M., Bode, W., Miller, M., and Hackert, M. L. (1986) J. Mol. Biol. 188 651–676 [DOI] [PubMed] [Google Scholar]
- 50.Schirmer, T., and Vincent, M. G. (1987) Biochim. Biophys. Acta 893 379–385 [Google Scholar]
- 51.Dolganov, N., and Grossman, A. R. (1999) J. Bacteriol. 181 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, H., and Sherman, L. A. (2002) Arch. Microbiol. 178 256–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.