Abstract
The ongoing discovery of disease-associated epitopes detected by CD8 T cells greatly facilitates peptide-based vaccine approaches and the construction of multimeric soluble recombinant proteins (e.g. tetramers) for isolation and enumeration of antigen-specific CD8 T cells. Related to these outcomes of epitope discovery is the recent demonstration that MHC class I/peptide complexes can be expressed as single chain trimers (SCTs) with peptide, β2m and heavy chain connected by linkers to form a single polypeptide chain. Studies using clinically relevant mouse models of human disease have shown that SCTs expressed by DNA vaccination are potent stimulators of cytotoxic T lymphocytes. Their vaccine efficacy has been attributed to the fact that SCTs contain a preprocessed and preloaded peptide that is stably displayed on the cell surface. Although SCTs of HLA class I/peptide complexes have been previously reported, they have not been characterized for biochemical stability or susceptibility to exogenous peptide binding. Here we demonstrate that human SCTs remain almost exclusively intact when expressed in cells and can incorporate a disulfide trap that dramatically excludes the binding of exogenous peptides. The mechanistic and practical applications of these findings for vaccine development and T cell isolation/enumeration are discussed.
CD8 T cells play a prominent role in immune defenses against viruses, intracellular bacteria, and tumors. To discriminate infected or malignant cells from normal cells, the CD8 T cell must detect disease-associated, processed peptide epitopes bound to self major histocompatibility complex class I (MHC-I)2 molecules. The feasibility of identifying disease-associated epitopes was first demonstrated by Thierry Boon and co-workers (1) using a mouse model of tumor rejection. Since then, many clinically relevant peptide epitopes have been identified for class I human leukocyte antigens (HLA-I) (2). A few examples include HLA-A2-restricted epitopes from human T-lymphotropic virus (HTLV) (3), influenza A (4), and melanoma (5). One clinically important HLA-B27-restricted epitope is the nucleoprotein sequence from influenza A (6). The identification of these and other epitopes opened the door for new peptide-based vaccination strategies to elicit CD8+ T cell responses to tumors or infections (7). Furthermore, knowing the epitope presented by different MHC-I molecules allowed for the isolation and enumeration of T cells specific for disease-associated epitopes (8, 9).
Relevant to the consequences of epitope discovery is the recent construction of MHC-I molecules as single chain trimers (SCTs) (10–12). SCT approaches have resulted in novel strategies for vaccination (13–16) and for the production of unique multivalent MHC-I reagents for isolating and enumerating antigen-specific CD8 T cells (17–19). The composition of the SCT is peptide – flexible linker – β2-microglobulin (β2m) – flexible linker – heavy chain (Fig. 1). Although SCT constructs have been reported for several different mouse and human MHC-I complexes, only the Kb/OVA SCT (Ovalbumin257–264-[G4S]3-β2m-[G4S]4-Kb) has been rigorously characterized (10, 18) in terms of (i) the biochemical integrity of the covalent SCT polypeptide and (ii) refractoriness of the SCT to exogenous peptide binding, which measures the binding strength of SCT-encoded peptide. Specifically, in initial studies, the Kb/OVA SCT was found to remain biochemically intact when expressed in cells because all three SCT components were linker-attached as detected in Western blots. Cellular proteases did not sever the antigenic peptide sequence from the SCT, making it unlikely that a cleaved peptide could rebind to the severed SCT or other endogenous Kb molecules. This important finding verified a necessary condition of the SCT approach: direct presentation of the covalently bound SCT peptide to bypass the normal inefficiencies of MHC-I processing, presentation, or cross-presentation (20, 21). Cross-presentation is a major pathway by which DNA vaccines elicit T cell responses, but the inefficiency of cross-presentation can be a contributing factor to relatively weak T cell responses after DNA vaccination. In addition, recent studies have documented the cross-presentation of synthetic peptides from recombinant MHC-I tetramers used to activate T cells (22, 23). Thus biochemical stability of the SCT is a requirement for improved DNA vaccination or MHC-I tetramer approaches.
FIGURE 1.
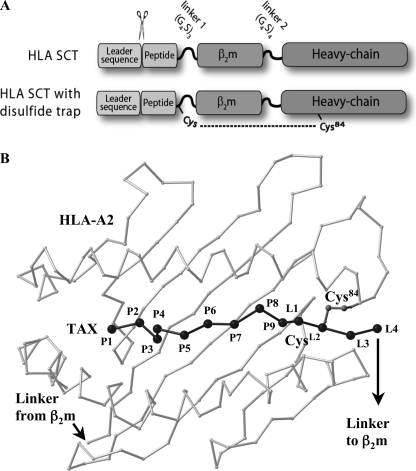
HLA-I SCT format and positioning of the disulfide trap. A, schematic of SCT format and cysteine positions for HLA-I dtSCTs. HLA-I SCTs were expressed in mammalian cells using the SCT format originally described by Yu et al. (10) for murine MHC-I. The signal peptide is precisely cleaved within the endoplasmic reticulum to reveal the antigenic sequence that folds into the HLA-I peptide binding groove. Flexible linkers fuse the subunits of the heterotrimer for presentation of a pre-processed CD8 T cell epitope from a single open-reading frame. The first SCT linker perturbs conserved MHC interactions that normally engage the peptide carboxyl group; however, this decrease in C-terminal anchoring was offset by the introduction of a disulfide bond, which locks the peptide securely into the HLA-I groove. B, model of a disulfide trap A2/TAX SCT. Presented is a Cα trace of the peptide binding platform of HLA-A2 (light gray) as well as the TAX peptide (black) in the conformation observed in complex with the A6 T cell receptor (56). The first four linker residue Cα positions (L1–L4), as well as the Cys side chains that form a disulfide bond between linker position 2 (CysL2) and Cys84 were taken from the superimposed crystal structure of the dtSCT of Kb/OVA (24).
Furthermore in these initial characterizations, the Kb/OVA SCT was found to have a significantly prolonged cell surface half-life compared with native Kb/OVA complexes or Kb loaded with endogenous peptides (18). Given that peptide occupancy determines the surface half-life of MHC-I, SCT were presumed to have superior peptide occupancy compared with native Kb/OVA. Unexpectedly, however, the Kb/OVA SCT was found to be more susceptible to exogenous peptide binding than Kb loaded with free ovalbumin peptide. The clear implication of these findings was that the superior surface stability of the SCT compared with native Kb/OVA was not due to continuous peptide binding, but rather the ability to rebind the linker-attached peptide. Subsequent crystallographic studies of the Kb/OVA SCT demonstrated that the linker extending from the OVA peptide moiety disrupted a highly conserved H-bonding network that normally serves to anchor peptide at the C terminus (24). Specifically, the heavy chain residue Tyr-84 forced the linker to protrude above from the peptide binding platform, thereby disrupting the H-bonding network of the peptide C terminus with the heavy chain. This provided a structural explanation for the increased susceptibility of the SCT to exogenous peptide binding.
To better accommodate linker extension and also improve C-terminal peptide anchoring, we recently engineered a disulfide trap into murine SCT molecules (dtSCTs) between position 84 of the MHC-I heavy chain and the second position on the SCT linker extending from the C terminus of the peptide (24, 25). Not only did the dtSCT constructs fold efficiently in cells, but the disulfide trap oxidized in a reliable manner and displayed a tenacious ability to prevent the binding of exogenous peptides. More importantly, these same properties were observed when dtSCT constructs incorporated peptides that were relatively poor MHC binders. Thus the dtSCT technology provided a mechanism to stably anchor a relative weak binding peptide into a single chain format. Because several known antigenic peptides are not particularly tight binders (26), incorporating traps could increase the general applications of SCTs as vaccines and diagnostic reagents. However, the proof of concept thus far for SCT biochemical stability, incorporation of disulfide traps, and exclusion of exogenous peptides has been almost exclusively with the Kb/OVA SCT.
In this report, we study SCTs made from several different clinically relevant HLA-I complexes. Our findings show that HLA-I SCTs fold efficiently in cells and, similar to the Kb/OVA prototype, maintain their covalent attachments in cells. Furthermore, HLA-I SCTs can be secured using disulfide trap technology, as measured by their remarkable capacity to exclude high affinity competitor peptides. In all cases, these HLA-I dtSCT molecules are recognized by human cytotoxic T cells specific for native MHC-I complexes. These observations validate the use of SCTs as vaccines for the treatment of human diseases or for the production of tetramers for isolating and enumerating human disease-specific T cells. These and other applications of HLA-I SCTs are discussed.
EXPERIMENTAL PROCEDURES
DNA Clones and Mutagenesis—PCR-generated inserts with appropriate DNA restriction sites flanking mature HLA-A0201 (A2) and -B2705 (B27) heavy chain sequences replaced the mature H2-Kb sequence of the previously constructed murine SCT (10) in the pIRESneo vector (Clontech Laboratories, Palo Alto, CA). The mature human β2m sequence was spliced into these constructs in place of the mouse β2m sequence. Thus, HLA-I SCTs retained the subunit order and linker lengths [G4S]3 and [G4S]4 found in the original SCT design (10) (Fig. 1). Synthetic DNA oligonucleotides (Integrated DNA Technologies, Coralville, IA) encoding peptide antigens were ligated into the SCT vectors at restriction sites specifically designed for expeditious shuttling of peptide sequences into the SCT construct, which for this study included the melanoma G280-9V peptide (27), the human T-lymphotropic virus TAX peptide (3), and the influenza A virus M158–66 peptide (4) for A2, as well as the influenza A virus NP383–391 peptide (6) for B27. As a control, a cDNA was produced for expression of β2m.[G4S]4.A2, with no covalently linked peptide at the N terminus. A single point mutation (R48Q) generated by site-directed mutagenesis (QuikChange II XL, Stratagene) was required to introduce the 64-3-7 epitope (28–32) into A2. Two point mutations were made in the G280-9V.β2m.A2 SCT and the FluM1.β2m.A2 SCT to create disulfide trap forms (25): a cysteine in the second position of the first linker and a cysteine at position 84 of the A2 heavy chain. DNA sequencing confirmed the correct sequence of all constructs.
Cell Lines and Transfections—HeLa cells (human cervical carcinoma) transfected with 64-3-7 epitope-tagged HLA-B27 have been described (32). Transfection of HeLa cells with cDNAs encoding A2 and B27 SCTs was performed using FuGene6 (Roche Applied Science, Indianapolis, IN). Stable transfectants were selected and maintained in 0.6–1.2 mg/ml Geneticin (Invitrogen, Grand Island, NY), identified by flow cytometry, and cloned by limiting dilution. All cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, UT), 2 mm l-glutamine, 0.1 mm nonessential amino acids, 1.25 mm HEPES, 1 mm sodium pyruvate, and 100 units/ml penicillin/streptomycin (all from the Tissue Culture Support Center, Washington University School of Medicine, St. Louis, MO).
Antibodies—mAb BB7.2 is specific for folded A2 molecules (33), and mAb ME-1 is specific for folded B27 (34); both of these were used for immunoprecipitation and flow cytometry. mAb HCA2 (a kind gift from H. Ploegh, Harvard Medical School, Boston, MA) recognizes unfolded A2 molecules (35) and was used for immunoblots. Open/peptide-empty conformers of human MHC-I molecules or SCT constructs were also detected by mAb 64-3-7 epitope-tagging (28–32). This antibody was also used for immunoprecipitations and immunoblots of epitope-tagged A2 and B27 and the corresponding SCT constructs.
Flow Cytometry—Flow cytometric analyses were performed using a FACSCalibur (BD Biosciences, San Jose, CA). Dead cells and debris were excluded from analysis on the basis of forward angle and side-scatter light gating. A minimum of 10,000 gated events was collected for analysis. Data were analyzed using CellQuest software (BD Biosciences). For surface staining, ∼5 × 105 cells per sample were incubated on ice in microtiter plates with culture supernatant from the appropriate hybridoma. After washing, PE-conjugated goat anti-mouse IgG (BD Pharmingen, San Diego, CA) was used to visualize MHC-I staining. In some experiments, flow cytometry followed incubation (1 × 106 cells/ml) with exogenous peptides TAX (A2-specific) (3) or MCMV (H2-Ld-specific) (36) at indicated concentrations.
Immunoprecipitations—Cells were lysed in PBS, 1.0% Nonidet P-40 (Sigma), 20 mm iodoacetamide, protease inhibitors, and a saturating concentration of precipitating mAb. After lysis for 30 min on ice, postnuclear lysates were incubated with protein A-Sepharose (Amersham Biosciences, Uppsala, Sweden) for 1 h. Beads were washed four times in PBS, 0.1% detergent, and bound proteins were eluted by boiling in 1× SDS-PAGE sample buffer. For nonreducing gels, β2-mercaptoethanol was excluded. For some samples, endoglycosidase H treatment followed immunoprecipitation. Bound proteins were eluted from protein A-Sepharose by boiling in 10 mm TrisCl, pH 6.8, 0.5% SDS, 1% β2-mercaptoethanol. Eluates were mixed with an equal volume of 100 mm sodium acetate, pH 5.4, and either digested (or mock-digested) at 37 °C for >1 h with 1 milliunit of endoglycosidase H (ICN Pharmaceuticals, Costa Mesa, CA) that was reconstituted in 50 mm sodium acetate, pH 5.4.
Immunoblotting—Immunoblotting was performed following SDS-PAGE separation of precipitated proteins and transfer to Immobilon P membranes (Millipore, Bedford, MA). Membranes were blocked (1 h to overnight) with PBS, 10% dried milk, 0.05% Tween 20. Primary Abs were added and incubated for 1 h, followed by washing in PBS, 0.05% Tween 20. As a second step, membranes were incubated for 1 h with biotin-conjugated anti-mouse Ig (Caltag Laboratories, San Francisco, CA). In some cases, biotin-conjugated mAb 64-3-7 was used, which obviated the second step. After washing, horseradish peroxidase-conjugated streptavidin (Zymed Laboratories) was added for 1 h, followed by three washes. Specific proteins were visualized by chemiluminescence using the ECL system (Amersham Biosciences, Boston, MA).
CTL Generation and Chromium Release CTL Assays—HeLa cell targets expressing A2 and B27 SCTs were tested against specific CTL lines. Protocols for the generation of these CTLs and chromium release assays to measure specific lysis have been described (37–41). The RR10 CTL was generated from a patient immunized with G280-9V pulsed DC as described (42). The CMV pp65-specific CTL line was generated from a CMV seropositive healthy donor using purified CD8+ T cells, peptide-pulsed irradiated autologous DC, and 10 units/ml IL-2 added on the second day of culture. Primed CD8+ T cells were repeatedly stimulated with antigen to generate CTL lines. Peptide competition assays were performed by incubating targets with competitor peptide at the indicated concentrations. On the following day, targets were trypsinized, washed twice, and used in a standard chromium release assay.
RESULTS
Design and Characterization of HLA-I SCTs—The SCT format encodes the MHC-I heterotrimer as a single polypeptide (Fig. 1), and the prototype SCT made from Kb/OVA has been well characterized in terms of assembly and expression in cells, biochemical stability, structure, T cell recognition, and incorporation of a disulfide trap to secure peptide (10, 18, 25). Although there are now several reports of SCTs made with HLA-I complexes (Table 1), these SCTs have not been characterized for biochemical stability, incorporation of disulfide traps, and exclusion of exogenous peptides. The functional ramifications of such investigations were the impetus for the current study.
TABLE 1.
Applications of HLA-I complexes constructed as SCTs
Several class I human leukocyte antigens (HLA-I) have been expressed either in cells or as recombinant proteins in single-chain trimer format (peptide-linker-β2-microglobulin-linker-heavy chain). Two of the most prevalent and successful applications have been DNA vaccination and flow cytometry. Asterisks denote SCTs verified in the current report.
| HLA-I allele | Peptide | Origin | Sequence | Mammalian cell surface expression | Immune receptor engagement | DNA vaccine | Recombinant multimers for flow cytometry | Refs. |
|---|---|---|---|---|---|---|---|---|
| A0201 | G280-9V | Melanoma | YLEPGPVTV | Yes (shown here) | TCR | • | *19 | |
| A0201 | TAX | HTLV | LLFGYPVYV | Yes (shown here) | TCR | • | *17 | |
| A0201 | M1 | Influenza A | GILGFVFTL | Yes | TCR | • | *17 | |
| A0201 | pp65 | CMV | NLVPMVATV | Yes | TCR | • | 17 | |
| A0201 | G209-2M | Melanoma | IMDQVPFSV | N.D.a | TCR | • | 19 | |
| A0201 | GLC280-288 | EBV | GLCTLVAML | N.D. | TCR | • | 19 | |
| A0201 | MAM-A2.1 | Mammaglobin (breast cancer) | LIYDSSLCDL | N.S.b | TCR | • | 13 | |
| A0201 | HBcAg C18-27 | Hepatitis B virus | FLPSDFFPSV | Yes | TCR | • | 16 | |
| HBcAg C107-115 | CLTFGRETV | |||||||
| A0201 | HIVgag | HIV | SLYNTVATL | Yes | TCR | • | 16 | |
| A0201 | Hmeso540-549 | Mesothelin (ovarian cancer) | KLLGPHVEGL | N.S. | TCR | • | 15 | |
| B2705 | NP383-391 | Influenza A | SRYWAIRTR | Yes (shown here) | TCR | * | ||
| HLA-E | Sequence found within signal peptide of many HLA-C alleles | VMAPRTLIL | Yes | NK | 53 | |||
N.D., not determined.
N.S., not shown but expected based on DNA vaccine efficacy.
Three different SCTs were used for this study, each with disease relevance. The first SCT was constructed with the peptide, G280-9V (YLEPGPVTV), bound to A2. This peptide is derived from the melanocyte-specific gp100 protein, which is expressed in most melanomas (27, 43). Adoptive transfer of cultured tumor-infiltrating lymphocytes specific for gp100 has been shown to cause tumor regression (5). The second SCT was constructed with the peptide, TAX (LLFGYPVYV) also bound to A2. This peptide is derived from p40tax protein, the major antigenic protein of the HTLV, which causes adult T cell leukemia (3, 44). The third SCT was constructed with the peptide, NP383–391, (SRYWAIRTR) bound to B27, and is a fragment of the influenza A nucleoprotein (6). These antigens were chosen for their predominant roles in CTL responses to melanoma, HTLV, or influenza, respectively. However, the HLA-B27 allele is also of interest because of its clear association with arthritic disease (45). SCTs were constructed for each of these MHC-I complexes to test the efficiency of cell surface expression, folding of the molecules (by monitoring conformational changes serologically), and T cell recognition.
Efficient Folding, Cell Surface Expression, and T Cell Recognition of HLA-I SCTs—HeLa cells were transfected with native HLA-I heavy chains or SCTs of the complexes A2/G280-9V, A2/TAX, or B27/NP383–391. The SCT constructs exhibited high level surface expression, comparable to the native HLA-I molecules, as detected in flow cytometry experiments by mAbs specific for the respective folded HLA-I molecules (Fig. 2A and shown later in Fig. 4). Thus these HLA-I SCTs clearly passed ER quality control, and linkers were accommodated such that the proper folding was detected by mAbs specific for the native, fully assembled HLA-I molecules. It is noteworthy that a construct similar to our G280-9V.β2m.A2 SCT was expressed as a recombinant protein (19); however, for certain applications such as DNA vaccination (46, 47), it was critical to verify cell surface expression in mammalian cells where the SCT faces competition with endogenous β2m, peptides, and molecular chaperones.
FIGURE 2.
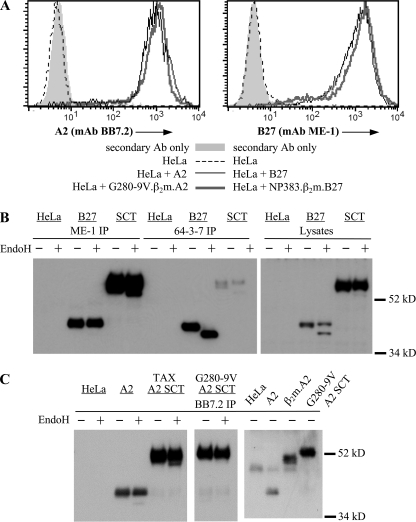
Efficient cell surface expression and biochemical stability of HLA-A2 and HLA-B27 SCTs. A, flow cytometry of stable HeLa transfectants stained with mAbs specific for either HLA-A2 (mAb BB7.2) or HLA-B27 (mAb ME-1). Surface staining is shown for SCTs (thick gray lines), native MHC-I molecules (thin black lines), untransfected HeLa cells (dotted lines), and for secondary reagent staining only (gray-filled histograms). B, HLA-B27 and the NP383–391.β2m.B27 SCT, both 64-3-7 epitope-tagged, were immunoprecipitated from HeLa cell lysates using either mAb ME-1, specific for folded B27, or mAb 64-3-7, specific for open MHC conformers. The immunoprecipitates were digested or mock-digested with endoglycosidase H (EndoH), as were postnuclear cell lysates. Immunoblots detected the molecular weights, folding, and the EndoH sensitivity of the transfected molecules. C, HeLa cells transfected with the indicated A2 constructs were lysed and immunoprecipitated using mAb BB7.2, specific for folded HLA-A2. Samples were digested or mock-digested with EndoH. Immunoblots using mAb HCA2 revealed the biochemical integrity and EndoH resistance of the A2 SCTs (left and middle panels). Densitometry using NIH Image revealed that 99% of the A2 SCTs remained intact, while 1% of the signal was found in a fragment at ∼40 kDa. The gel migration of the A2 SCT was also directly compared with that of a β2m.A2 covalently linked dimer (right panel) to demonstrate that the antigenic peptide and first SCT linker are not cleaved by cellular proteases.
FIGURE 4.
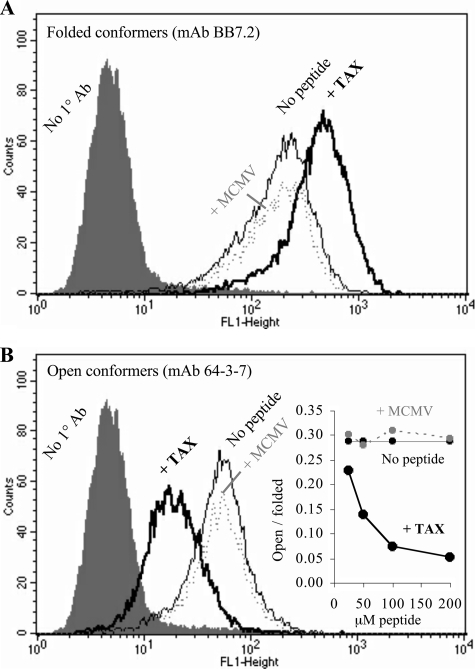
Detection of peptide-empty A2 conformers by means of the 64-3-7 epitope tag. HeLa cells expressing 64-3-7 epitope-tagged A2 molecules were stained with either (A) mAb BB7.2, which is specific for folded A2 conformers, or (B) mAb 64-3-7, which has been shown for several other MHC alleles to be specific for peptide-empty, or “open” MHC conformers. Addition of specific peptide TAX (thick black line) increased levels of BB7.2 and decreased reactivity with 64-3-7. Nonspecific peptide MCMV (gray dotted line) had no effect. No peptide (thin black line) and secondary-only controls (filled histogram) are also shown. The inset in B shows peptide dose-dependent changes in the ratio of open to folded MHC conformers for cells fed TAX peptide (thick black line), MCMV negative control peptide (dotted gray line), or no peptide (thin black line).
We also examined the level of steady-state folding of the HLA-I SCTs. To quantify the amount of open MHC-I conformers, the 64-3-7 epitope tag was introduced into both native B27 (92) and the NP383–391.β2m.B27 SCT (28–32). HeLa cells expressing these constructs were lysed and analyzed by immunoprecipitation and blotting (Fig. 2B, left panel). Almost all of the B27 SCT was folded (ME-1+) and mature (EndoH-resistant) at steady-state. Very few peptide-empty (64-3-7+) forms of the HLA-B27 SCT could be detected, indicating that these SCTs fold rapidly. In contrast, native B27 molecules had approximately equal amounts of folded and open conformers, and all of the open conformers were immature (Endo H-sensitive). Furthermore, these immunoblots confirmed that the covalent [G4S]3 and [G4S]4 linkers remained intact when the NP383–391.β2m.B27 SCT was expressed in cells, as indicated by the absence of proteolytic fragments in both immunoprecipitates (Fig. 2B, left panel) and lysates (Fig. 2B, right panel). Similarly, efficient maturation and intact covalent structure was also apparent in HeLa cells expressing either the TAX.β2m.A2 or G280-9V.β2m.A2 SCT (Fig. 2C, left panel). The vast majority of each SCT was EndoH-resistant, indicating rapid maturation, and each SCT was full-length, indicating the lack of proteolytic digestion.
Because one of the main purposes of expressing HLA-I complexes as SCTs was to prevent peptide dissociation, it was important to demonstrate that the SCT peptide moieties were not proteolytically cleaved and “re-presented” in the context of the HLA-I SCT binding groove. Upon immunoprecipitation and blotting, we found that the G280-9V.β2m.A2 SCT migrated more slowly than the β2m.A2 covalently linked dimer (Fig. 2C, right panel). The antigenic peptide and first linker conferred upon the SCT a higher molecular weight. This provided direct biochemical evidence of the intact nature of the A2 SCT.
We next verified that the HLA-I SCTS are recognized by cytotoxic T cells specific for these clinically relevant HLA-I complexes. Targets expressing the NP383–391.β2m.B27 SCT (Fig. 3A), TAX.β2m.A2 SCT (Fig. 3B), and G280-9V.β2m.A2 SCT (Fig. 6B) were lysed specifically by T cells originally stimulated by APC expressing the native HLA-I complexes. As expected, the SCTs were not recognized by T cells specific for other peptide antigens that bind to A2 (Figs. 3B and 6, C and D and data not shown). In summary, these clinically relevant A2 and B27 SCTs were highly expressed, intact, and recognized by T cells. In fact, each human MHC-I/peptide combination covalently linked in the SCT configuration (see Table 1) has engaged T cells specific for the corresponding native MHC-I complexes. This strongly argues that the SCT linkers are essentially transparent to T cells. Our previous work in the mouse system has further demonstrated that SCT linkers also allow for binding of the CD8 co-receptor (48) and NK receptor Ly49C (49). These cumulative findings validate the use of SCTs in basic immunological research as well as clinical studies of vaccines and immunotherapy.
FIGURE 3.
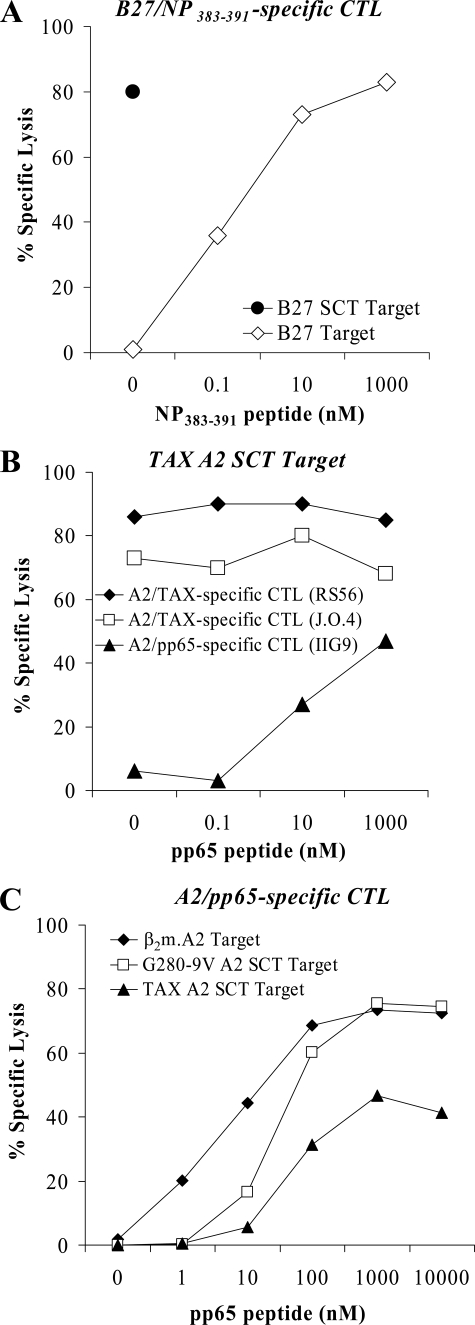
Efficient T cell recognition of HLA-A2 and HLA-B27 SCTs and susceptibility to peptide exchange. A, CTL recognition of the NP383–391.β2m.B27 SCT. Target cells transfected with the B27 SCT or with native B27 and pulsed with increasing concentrations of exogenous NP383–391 peptide were tested in a 51Cr release assay for recognition by B27/NP383–391-specific CTL effectors. The SCT was strongly recognized without the addition of exogenous peptide. B, CTL recognition of the TAX.β2m.A2 SCT and its susceptibility to peptide competition. Two distinct A2/TAX-specific CTL lines, but not an A2/pp65-specific CTL line, recognized the TAX.β2m.A2 SCT. However, when increasing concentrations of competitor peptide pp65 were added to the TAX.β2m.A2 SCT-expressing target cells, significant lysis was observed with the A2/pp65-specific CTL line. C, TAX.β2m.A2 SCT and G280-9V.β2m.A2 SCT are susceptible to peptide exchange albeit to different degrees. TAX.β2m.A2 SCT and G280-9V.β2m.A2 SCT-expressing targets were incubated with increasing concentrations of competitor pp65 peptide and tested in a 51Cr release assay using an A2/pp65-specific CTL line. Although both SCT are susceptible to peptide exchange, higher concentrations of exogenous pp65 peptide are required for TAX SCT displacement, a finding consistent with the tighter A2 binding of TAX versus G280-9V peptide (50).
FIGURE 6.
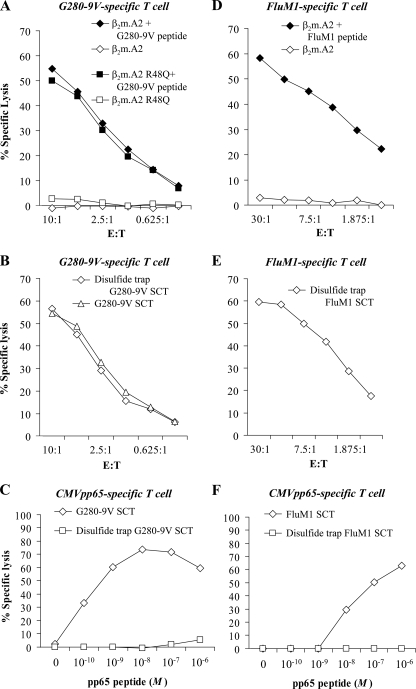
Disulfide trap HLA-I SCTs are refractory to binding of competitor peptide. A, CTLs specific for the A2/G280-9V recognized A2 molecules with or without the 64-3-7 epitope, demonstrating that the point mutation required for introducing this epitope did not affect peptide binding or T cell recognition. B, G280-9V.β2m.A2 SCT as well as the disulfide trap-engineered SCT were both strongly recognized by G280-9V/A2-specific CTL. C, increasing concentrations of the A2-binding CMVpp65 competitor peptide were incubated with target cells expressing the A2 SCT with or without the disulfide trap. While the SCT of original design readily accepted the competitor at 10–11 m, the disulfide trap SCT did not allow any significant access to the A2 binding groove, even with a 10,000-fold higher competitor peptide concentration. D–F, second SCT was engineered to include the disulfide trap and tested for T cell recognition and resistance to competitor peptide binding. A2/FluM1-specific T cells (D) strongly recognized the disulfide-trap version of the FluM1.β2m.A2 SCT (E), and A2/CMVpp65-specific T cells were unable to detect competitor peptide binding to the FluM1.β2m.A2 dtSCT (F).
Rationale for the Introduction of a Disulfide Trap: HLA-I SCTs Are Susceptible to Peptide Exchange—Studies of Kb/OVA have shown that the SCT format is highly effective at preventing the binding of exogenous peptides (10, 18). However, in the presence of high concentrations of exogenous peptide, the OVA peptide moiety was displaced from the SCT groove. Furthermore, SCTs formed with lower affinity binding OVA peptide analogues were found to be more susceptible to peptide displacement. Thus it was of considerable interest to determine whether SCT made with HLA-I heavy chains and antigenic peptides were refractory to exogenous peptide binding.
We first measured the ability of the TAX.β2m.A2 SCT to exclude binding of another A2-restricted peptide, CMV pp65. Increasing concentrations of this competitor peptide were incubated with TAX.β2m.A2 SCT-expressing targets, which were then subjected to a cytotoxicity assay using TAX- or pp65-specific CTLs (Fig. 3B). The addition of competitor peptide did not significantly decrease the ability of TAX-specific T cells to recognize the SCT; however, pp65-specific T cells did recognize the SCT to a certain extent after incubation with pp65 peptide. Because the target cells themselves did not express A2 endogenously (not shown), it was clear that the pp65 peptide was accessing the peptide binding groove of the A2 SCT. Furthermore, a comparison of TAX.β2m.A2 SCT and G280-9V.β2m.A2 SCT ability to exclude exogenous peptide using the same approach indicated that the G280-9V.β2m.A2 SCT was considerably more susceptible than TAX.β2m.A2 SCT to binding exogenous pp65 peptide (Fig. 3C). This finding is consistent with the predicted t½ stabilities of these peptides bound to A2 (G280-9V, 20.4 min; CMV pp65, 159.9 min; and TAX, 2496.1 min) (50). Thus the weaker binding peptide in the SCT is more displaceable. This is of particular importance in designing SCT as vaccines as most tumor antigens are poor MHC binders.
To alleviate the potential for the SCT to bind exogenous peptides, we recently engineered a disulfide trap into the OVA.mβ2m.Kb SCT (25). Two cysteine residues were introduced by site-directed mutagenesis creating a strategically placed disulfide bond to anchor the antigenic peptide moiety of the SCT to the peptide binding groove (Fig. 1). One cysteine was positioned in the first SCT linker approximately where the C terminus of a noncovalently bound peptide normally resides. The second cysteine was introduced at position 84 of the MHC-I heavy chain, where normally a tyrosine residue interacts with the C terminus of a noncovalently bound peptide. The disulfide bond formed properly in cells expressing the OVA.mβ2m.Kb SCT and effectively prevented the release of OVA under stringent conditions that included high concentrations of Kb-binding competitor peptides. Because of its ability to trap the peptide into the SCT groove, we designated this engineered covalent bond a “disulfide trap.” Here we report the first introduction of functional disulfide traps in HLA-I SCTs including both G280-9V.β2m.A2 and FluM1.β2m.A2. A model of the disulfide trap in an HLA-I complex is shown in Fig. 1.
Serological and Biochemical Analyses of Disulfide Trap HLA-I SCT—The peptide binding state of MHC-I molecules can be monitored serologically using the aforementioned mAb 64-3-7. This mAb detects a peptide-empty or “open” conformation of MHC-I heavy chains that bear the appropriate epitope for this reagent (28–32). The conserved sequence recognized by 64-3-7 is naturally found in H2-Ld, but a single point mutation (R48Q) was all that was required to recreate the 64-3-7 epitope in HLA-A2. We tested the functionality of the 64-3-7 epitope in HeLa cells expressing the β2m.A2 dimer with the intent to use this approach later to measure the open conformers of A2 SCTs with and without the disulfide trap.
HeLa cells expressing 64-3-7 epitope-tagged β2m.A2 were incubated overnight with the A2-binding TAX peptide, control peptide, or no peptide. Specific peptide induced expression of folded A2 conformers was detected by BB7.2 (Fig. 4A), and reduced expression of open A2 conformers was detected by 64-3-7 (Fig. 4B). With increasing amounts of specific peptide, the ratio of open to folded conformers was markedly reduced (Fig. 4, inset). Thus 64-3-7 reliably detected the loss of open A2 conformers after addition of specific peptide. This validated the use of the 64-3-7 epitope in A2 SCTs to measure serologically the effects of a disulfide trap.
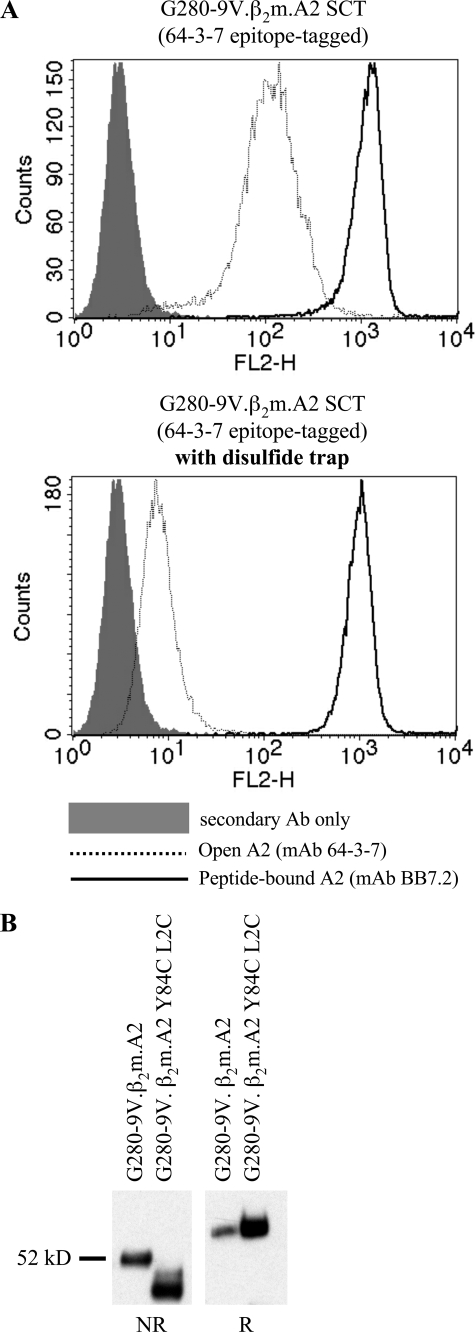
Using flow cytometry, we compared cell surface levels of open and closed conformers of 64-3-7 epitope-tagged G280-9V.β2m.A2 SCT either with or without the disulfide trap (Fig. 5A). Like the parent molecule, the dtSCT was fully capable of high levels of surface expression. However, introducing the disulfide trap resulted in a >10-fold decrease in the number of cell surface open conformers. This finding indicated that the disulfide trap prevented dissociation of the linker attached peptide and thus lowered steady-state detection of SCT molecules with an open peptide binding groove.
FIGURE 5.
Introduction of a disulfide trap into the HLA-A2 SCT. A, cells expressing the G280-9V.β2m.A2 SCT (upper panel) or the same construct engineered with the disulfide trap (lower panel) were stained with antibodies specific for folded A2 (BB7.2, thin black line) or open A2 conformers (64-3-7, thin dotted line, see Fig. 3). Negative control staining is also shown (gray histogram). Addition of the disulfide trap resulted in a dramatic decrease in open conformers (∼10-fold) detected at the cell surface. B, biochemical confirmation of cysteine residue oxidation. The same cell lines were lysed and A2 SCTs were immunoprecipitated and subjected to SDS-PAGE under non-reducing (NR) or reducing (R) conditions. The additional disulfide bond contracts the radius of gyration of the denatured molecule, allowing it to migrate faster through the gel matrix.
To directly test whether the disulfide trap was formed, a biochemical approach was taken. Considering the placement of the engineered disulfide bond (Fig. 1), the radius of gyration of the denatured SCT molecule should be significantly shortened during nonreducing SDS-PAGE, which would result in faster migration through polyacrylamide. This was indeed the case (Fig. 5B). All of the disulfide trap SCT material migrated more quickly through the gel matrix than non-trapped parental G280-9V.β2m.A2 SCT, while both migrated equivalently under reducing conditions. Thus, the disulfide trap is in place with cysteines properly oxidized, and the approach is compatible with thioreductase activities present in the endoplasmic reticulum.
T Cell Recognition of the Disulfide Trap HLA-I SCT and Exclusion of Competitor Peptides—To determine whether the disulfide-trapped SCT retains its native conformation for antigen presentation, a CTL line specific for the A2/G280-9V complex was used (Fig. 6A). This T cell line did not recognize untransfected HeLa target cells, even when incubated with the G280-9V peptide (not shown). In the presence of the G280-9V peptide, this T cell line recognized targets expressing the β2m.A2 dimer with or without the 64-3-7 tag (Fig. 6A). This latter finding was important because it established that the 64-3-7 tag itself did not interfere with T cell recognition or peptide binding. This finding is consistent with the location of the epitope tag and incorporation of this tag into other human and mouse MHC-I heavy chains. As shown in Fig. 6B, T cell recognition of the G280-9V.β2m.A2 SCT with or without the disulfide trap was strikingly similar. Therefore, this molecular approach to securing peptide into the SCT preserves both the positioning of the peptide in the A2 groove as well as the MHC-I/peptide antigenic structure presented to the TCR.
To test whether the disulfide trap functionally retains the antigenic peptide of the SCT, a T cell assay was used to monitor the intrusion of competitor peptides into the SCT peptide binding groove. For these assays, a CTL line specific for A2/CMVpp65 was used to monitor exogenous peptide binding by the G280-9V.β2m.A2 SCT (Fig. 6C). As shown, the pp65 competitor readily gained access to the SCT groove lacking a trap, because the HeLa G280-9V.β2m.A2 SCT targets were lysed even in the presence of low concentrations of pp65 peptide. Remarkably, introduction of the disulfide trap into the G280-9V.β2m.A2 SCT results in a molecule resistant to the binding of exogenous pp65 (Fig. 6C). No significant level of CTL recognition of pp65 could be detected even with 10,000 times the concentration of competitor peptide that would induce lysis of cells expressing the original SCT. To extend these remarkable findings to a second HLA-I complex, we also introduced a disulfide trap into the A2/FluM1 complex in the SCT format and characterized this dtSCT using similar assays. As with the A2/G280-9V complex, the engineered trap was reliably formed when the A2/FluM1 dtSCT was expressed in cells, as determined in biochemical and serological comparisons (not shown). Furthermore, this dtSCT was detected by a T cell line specific for FluM1 comparable to M1 peptide-fed targets expressing native A2 (Fig. 6, D and E). Finally and most importantly, the FluM1.β2m.A2 dtSCT was found to be completely refractory to exogenous peptide binding using T cells specific for the CMVpp65 peptide (Fig. 6F). By contrast the FluM1.β2m.A2 SCT lacking the trap displayed significant CMVpp65 peptide binding at peptide concentrations greater than 10–9 m (Fig. 6F). Interestingly, higher amounts of competitor CMVpp65 peptide were required to displace M1 from the SCT lacking the trap, compared with the amount of competitor required to displace the G280-9V sequence from the SCT lacking the trap. This is consistent with the predicted stability (t½) for the influenza M1 peptide binding to A2, which is 550.9 min compared with 20.4 min for the G280-9V peptide (50). Thus even SCTs constructed with relatively high affinity peptides (A2/TAX or A2/M1) are displaceable, accentuating the importance of the disulfide trap for engineering an HLA-I complex entirely refractory to exogenous peptide binding.
DISCUSSION
MHC class I SCT approaches are of great potential importance because some immunodominant antigenic epitopes of clinical relevance are relatively poor binders to their MHC restriction element. This apparent paradox can be accounted for by other factors beyond MHC binding (26), which are also heavily involved in determining epitope immunodominance, such as peptide abundance (as determined in part by processing efficiency) and T cell repertoire. However, these factors being equal, peptides that bind better are more effective at eliciting T cell responses in vivo (51). The primary reason for this is that tight binding peptides preferentially compete against an extensive pool of endogenous epitopes. Furthermore, MHC-I molecules with tight binding peptides are displayed at the cell surface longer than MHC-I complexed with weak binding peptides (29). SCTs with disulfide traps are highly resistant to competitor peptide binding (Fig. 6) and are stably expressed at the cell surface (Figs. 2A and 5A). These properties have significant implications for DNA vaccines, recombinant MHC-I multimers, and a diversity of other MHC-I applications.
DNA vaccination, the delivery of bacterial plasmid DNA encoding antigen directly into the individual being vaccinated, is a promising technique which is under intense study for eventual use in humans. However, a current challenge of DNA vaccination for eliciting CD8 T cell responses is to increase potency (47). Reduced potency is in large part due to the inefficiency of getting DNA encoded epitopes into the MHC-I antigen presentation pathway and can be dependent upon cross-presentation (52). Reflecting its pre-processed and pre-peptide loaded status, SCT-based DNA vaccination has shown great promise using animal model systems. For example, an HLA-A2 SCT was constructed with an immunodominant epitope (MamA2.1) derived from the mammaglobin-A protein that is expressed in 80% of breast cancers, but not normal tissue. Jaramillo et al. (13) vaccinated HLA-A2+/human CD8+ double transgenic mice with a plasmid encoding an SCT consisting of the MamA2.1 peptide.β2m.A2. SCT-vaccinated mice produced a robust, peptide-specific CTL response that specifically killed A2+ mammaglobin+ breast cancer cell lines. Two similar studies used mice expressing a chimeric transgene with the HLA-A2 α1/α2 domains and the α3 domain of a mouse MHC-I to improve CD8 interaction (15, 16). Zhang et al. (16) showed that vaccination with DNA encoding an A2 SCT incorporating an antigenic epitope of the core protein of hepatitis B virus induced a functional epitope-specific CTL response. Furthermore, Hung et al. (15) constructed an A2 SCT incorporating an antigenic epitope of human mesothelin that is highly expressed in a majority of ovarian cancer cells and expressed at a low level in normal cells. DNA vaccination with an A2/mesothelin SCT generated a strong epitope-specific CTL response that prevented the growth of an A2+ mesothelin+ tumor cell line. These combined studies establish the general applicability and effectiveness of SCT vaccination approaches in mouse models.
In the most mechanistically insightful SCT vaccine study thus far, Huang et al. (14) tested the efficacy of a DNA vaccine using a B6 mouse model for rejection of a syngeneic tumor cell line transformed by the E6 protein of human papillomavirus E6 protein. Mice were vaccinated with DNA encoding (i) an SCT encoding an E6 immunodominant epitope.β2m.Kb, (ii) a control SCT encoding OVA.β2m.Kb, or (iii) intact E6 protein. As expected, only vaccination with DNA encoding the SCT incorporating the E6 epitope and not OVA yielded peptide-specific CD8 T cells. More importantly, the response in mice vaccinated with DNA encoding the SCT with the E6 epitope yielded >40 fold more peptide-specific T cells than mice vaccinated with DNA encoding intact E6 protein. In vivo efficacy was furthermore demonstrated by the observation that 100% of the mice vaccinated with DNA encoding the SCT with the E6 peptide were protected against a lethal challenge of the E6-transformed syngeneic tumor. By contrast, mice vaccinated with DNA encoding the control SCT or intact E6 grew tumors. This superiority of the SCT was attributed to the fact that it bypassed the antigen processing and lead to stable presentation of the E6 epitope.
The implication of the current study on vaccine applications is that SCTs made with HLA-I heavy chains are biochemically stable. This finding strongly supports the conclusion that SCT vaccine efficacy likely results, at least in part, from its direct presentation. It is important to note that nascent SCTs assemble and fold more rapidly than native MHC-I complexes. This was demonstrated here by the observation that HLA-I SCTs are detected by Western blots in a predominantly folded conformation (Fig. 2, B and C). Rapid assembly/folding are important features of the SCT because it dissuades the binding of competitor peptides in the ER. Furthermore the chaperone-independent, preassembled nature of the SCT prevents several of the known immune evasion strategies of viruses and tumors that block antigen presentation to CD8+ CTL. All of these SCT properties have the potential to positively impact on the effectiveness of SCT vaccines.
It is important to note that incorporation of the disulfide trap into the SCT appears to further increase the kinetics of assembly and folding, likely reflecting the improved C-terminal peptide anchoring (25). Furthermore, the disulfide trap incorporated into two different HLA-I complexes forms reliably and ablates exogenous peptide binding using a highly sensitive T cell readout. Despite the additional complexity of the cysteine engineering, these HLA-I dtSCT molecules were highly expressed at the cell surface and strongly recognized by specific T cells. Thus incorporation of a trap into human SCTs could improve their effectiveness as DNA vaccines, particularly for relatively weak binding epitopes.
Recombinant MHC-I multimers (e.g. tetramers) have gained widespread use as staining reagents for enumerating or isolating disease-related CD8 T cells. However in many cases, these reagents are difficult to construct because of weak binding peptides, or are unstable because of the loss of peptide (9). Peptide dissociation from MHC-I multimers is also a significant problem for in vitro or in vivo approaches to activate T cells recognizing a specific epitope. SCT approaches will likely be beneficial for overcoming the instability of MHC-I multimers resulting from peptide dissociation. Using an SCT construction scheme very similar to ours, Greten et al. (17) showed that the TAX.β2m.A2 and FluM1.β2m.A2 constructs could be expressed as Ig fusion proteins and used to stain cells. More recently, another group linked an A2 SCT with a highly immunogenic peptide to an antibody single chain variable domain specific for a tumor-associated antigen (19). Tumor cells expressing this antigen could be coated with the bi-functional recombinant protein and were lysed by CTL specific for the MHC-I SCT. This immunotherapeutic technique could potentially be used to target tumors cells in vivo that have down-regulated MHC-I expression. Such novel applications of recombinant MHC-I, as well as more established applications (e.g. MHC-I tetramers for antigen-specific T cell enumeration and isolation) are limited by susceptibility to loss of peptide both in vitro and in vivo. The utilization of disulfide-trapped MHC-I promises to expand and improve both diagnostic and therapeutic applications of multivalent MHC-I reagents by mitigating problems of peptide dissociation. In fact, for recombinant applications, the disulfide trap obviates the single chain trimer MHC-I format. In this regard, we have successfully incorporated a disulfide trap into recombinant Kb/OVA without SCT linkers by introducing cysteines into the recombinant heavy chain protein and synthetic peptide. These disulfide trap MHC-I tetramers stained T cells specific for Kb/OVA as well as NK cells expressing the cognate Ly49 NK inhibitory receptor (24). Thus the disulfide trap preserves MHC-I antigen specificity and is a promising option where the loss of peptide is a critical limitation.
SCTs have also been used for several novel applications (other than vaccines and multivalent reagents) where incorporation of a trap may be efficacious. For example, Crew et al. (53) applied the SCT technology to block human NK cell detection of a pig xenograft. More specifically, an SCT based on human HLA-E, a non-classical or MHC class Ib molecule loaded with an MHC class Ia signal peptide was expressed on porcine cells and shown to inhibit NK cells killing as mediated by the CD94/NKG2A inhibitory receptor. In another study, SCTs were used to demonstrate the importance of size-based molecular segregation for immune synapse formation (54). Along these same lines, a future application of SCTs should be to definitively test the importance of non-cognate co-receptor interactions in immune synapses (55). Finally SCTs have been expressed as transgenes and used to define MHC-I-mediated licensing during NK cell development (49). Each of these novel applications and many other likely ones, require SCTs to be stably expressed and highly resistant to exogenous peptide binding. Thus for all future SCT applications incorporation of disulfide traps should be a consideration.
Acknowledgments
We thank Nancy Myers and Michelle Becker-Hapak for providing expert technical support, for which the authors express sincere appreciation.
This work was supported by National Institutes of Health Grants AI055849 (to T. H. H.) and AI027568 (to J. M. C.), as well as Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Grant U54-AI057160 (to T. H. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MHC-I, major histocompatibility complex class I; HLA-I, human leukocyte antigen class I; A2, HLA allele A*0201; B27, HLA allele B*2705; β2m, β2-microglobulin; SCT, MHC class I single-chain trimer; dtSCT, disulfide trap single chain trimer; CTL, cytotoxic T lymphocyte; OVA, ovalbumin257–264 peptide; EndoH, endoglycosidase H; IP, immunoprecipitation; NK, natural killer; t½, surface half-life; mAb, monoclonal antibody; PBS, phosphate-buffered saline; CMV, cytomegalovirus; TCR, T cell receptor; HTLV, human T-lymphotropic virus; EBV, Epstein-Barr virus.
References
- 1.Lurquin, C., Van Pel, A., Mariame, B., De Plaen, E., Szikora, J. P., Janssens, C., Reddehase, M. J., Lejeune, J., and Boon, T. (1989) Cell 58 293–303 [DOI] [PubMed] [Google Scholar]
- 2.Peters, B., Sidney, J., Bourne, P., Bui, H. H., Buus, S., Doh, G., Fleri, W., Kronenberg, M., Kubo, R., Lund, O., Nemazee, D., Ponomarenko, J. V., Sathiamurthy, M., Schoenberger, S., Stewart, S., Surko, P., Way, S., Wilson, S., and Sette, A. (2005) PLoS. Biol. 3 e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannagi, M., Shida, H., Igarashi, H., Kuruma, K., Murai, H., Aono, Y., Maruyama, I., Osame, M., Hattori, T., Inoko, H., and Harada, S. (1992) J. Virol. 66 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison, J., Elvin, J., Latron, F., Gotch, F., Moots, R., Strominger, J. L., and McMichael, A. (1992) Eur. J. Immunol. 22 903–907 [DOI] [PubMed] [Google Scholar]
- 5.Kawakami, Y., Eliyahu, S., Jennings, C., Sakaguchi, K., Kang, X., Southwood, S., Robbins, P. F., Sette, A., Appella, E., and Rosenberg, S. A. (1995) J. Immunol. 154 3961–3968 [PubMed] [Google Scholar]
- 6.Huet, S., Nixon, D. F., Rothbard, J. B., Townsend, A., Ellis, S. A., and McMichael, A. J. (1990) Int. Immunol. 2 311–316 [DOI] [PubMed] [Google Scholar]
- 7.Purcell, A. W., McCluskey, J., and Rossjohn, J. (2007) Nat. Rev. Drug Discov. 6 404–414 [DOI] [PubMed] [Google Scholar]
- 8.Altman, J. D., Moss, P. A. H., Goulder, P. J. R., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J., and Davis, M. M. (1996) Science 274 94–96 [DOI] [PubMed] [Google Scholar]
- 9.Altman, J. D. (2004) Methods Cell Biol. 75 433–452 [DOI] [PubMed] [Google Scholar]
- 10.Yu, Y. Y., Netuschil, N., Lybarger, L., Connolly, J. M., and Hansen, T. H. (2002) J. Immunol. 168 3145–3149 [DOI] [PubMed] [Google Scholar]
- 11.Primeau, T., Myers, N. B., Yu, Y. Y., Lybarger, L., Wang, X., Truscott, S. M., Hansen, T. H., and Connolly, J. M. (2005) Immunol. Res. 32 109–122 [DOI] [PubMed] [Google Scholar]
- 12.Hansen, T. H., and Lybarger, L. (2006) Cancer Immunol. Immunother. 55 235–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaramillo, A., Narayanan, K., Campbell, L. G., Benshoff, N. D., Lybarger, L., Hansen, T. H., Fleming, T. P., Dietz, J. R., and Mohanakumar, T. (2004) Breast Cancer Res. Treat. 88 29–41 [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. H., Peng, S., He, L., Tsai, Y. C., Boyd, D. A., Hansen, T. H., Wu, T. C., and Hung, C. F. (2005) Gene Ther. 12 1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung, C. F., Calizo, R., Tsai, Y. C., He, L., and Wu, T. C. (2007) Vaccine 25 127–135 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Y., Li, S., Shan, M., Pan, X., Zhuang, K., He, L., Gould, K., and Tien, P. (2007) Immunology 121 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greten, T. F., Korangy, F., Neumann, G., Wedemeyer, H., Schlote, K., Heller, A., Scheffer, S., Pardoll, D. M., Garbe, A. I., Schneck, J. P., and Manns, M. P. (2002) J. Immunol. Methods 271 125–135 [DOI] [PubMed] [Google Scholar]
- 18.Lybarger, L., Yu, Y. Y., Miley, M. J., Fremont, D. H., Myers, N., Primeau, T., Truscott, S. M., Connolly, J. M., and Hansen, T. H. (2003) J. Biol. Chem. 278 27105–27111 [DOI] [PubMed] [Google Scholar]
- 19.Oved, K., Lev, A., Noy, R., Segal, D., and Reiter, Y. (2005) Cancer Immunol. Immunother. 54 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Princiotta, M. F., Finzi, D., Qian, S. B., Gibbs, J., Schuchmann, S., Buttgereit, F., Bennink, J. R., and Yewdell, J. W. (2003) Immunity 18 343–354 [DOI] [PubMed] [Google Scholar]
- 21.Yewdell, J. W., Reits, E., and Neefjes, J. (2003) Nat. Rev. Immunol. 3 952–961 [DOI] [PubMed] [Google Scholar]
- 22.Ge, Q., Stone, J. D., Thompson, M. T., Cochran, J. R., Rushe, M., Eisen, H. N., Chen, J., and Stern, L. J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13729–13734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schott, E., Bertho, N., Ge, Q., Maurice, M. M., and Ploegh, H. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13735–13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitaksov, V., Truscott, S. M., Lybarger, L., Connolly, J. M., Hansen, T. H., and Fremont, D. H. (2007) Chem. Biol. 14 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truscott, S. M., Lybarger, L., Martinko, J. M., Mitaksov, V. E., Kranz, D. M., Connolly, J. M., Fremont, D. H., and Hansen, T. H. (2007) J. Immunol. 178 6280–6289 [DOI] [PubMed] [Google Scholar]
- 26.Yewdell, J. W., and Bennink, J. R. (1999) Annu. Rev. Immunol. 17 51–88 [DOI] [PubMed] [Google Scholar]
- 27.Parkhurst, M. R., Salgaller, M. L., Southwood, S., Robbins, P. F., Sette, A., Rosenberg, S. A., and Kawakami, Y. (1996) J. Immunol. 157 2539–2548 [PubMed] [Google Scholar]
- 28.Lie, W. R., Myers, N. B., Connolly, J. M., Gorka, J., Lee, D. R., and Hansen, T. H. (1991) J. Exp. Med. 173 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, J. D., Myers, N. B., Gorka, J., and Hansen, T. H. (1993) J. Exp. Med. 178 2035–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, Y. Y., Myers, N. B., Hilbert, C. M., Harris, M. R., Balendiran, G. K., and Hansen, T. H. (1999) Int. Immunol. 11 1897–1906 [DOI] [PubMed] [Google Scholar]
- 31.Myers, N. B., Harris, M. R., Connolly, J. M., Lybarger, L., Yu, Y. Y., and Hansen, T. H. (2000) J. Immunol. 165 5656–5663 [DOI] [PubMed] [Google Scholar]
- 32.Harris, M. R., Lybarger, L., Myers, N. B., Hilbert, C., Solheim, J. C., Hansen, T. H., and Yu, Y. Y. (2001) Int. Immunol. 13 1275–1282 [DOI] [PubMed] [Google Scholar]
- 33.Brodsky, F. M., Parham, P., Barnstable, C. J., Crumpton, M. J., and Bodmer, W. F. (1979) Immunol. Rev. 47 3–61 [DOI] [PubMed] [Google Scholar]
- 34.Ellis, S. A., Taylor, C., and McMichael, A. (1982) Hum. Immunol. 5 49–59 [DOI] [PubMed] [Google Scholar]
- 35.Stam, N. J., Vroom, T. M., Peters, P. J., Pastoors, E. B., and Ploegh, H. L. (1990) Int. Immunol. 2 113–125 [DOI] [PubMed] [Google Scholar]
- 36.Reddehase, M. J., Rothbard, J. B., and Koszinowski, U. H. (1989) Nature 337 651–653 [DOI] [PubMed] [Google Scholar]
- 37.Carreno, B. M., Anderson, R. W., Coligan, J. E., and Biddison, W. E. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3420–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utz, U., Banks, D., Jacobson, S., and Biddison, W. E. (1996) J. Virol. 70 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biddison, W. E., Turner, R. V., Gagnon, S. J., Lev, A., Cohen, C. J., and Reiter, Y. (2003) J. Immunol. 171 3064–3074 [DOI] [PubMed] [Google Scholar]
- 40.Wang, Z., Turner, R., Baker, B. M., and Biddison, W. E. (2002) J. Immunol. 169 3146–3154 [DOI] [PubMed] [Google Scholar]
- 41.Carreno, B. M., Turner, R. V., Biddison, W. E., and Coligan, J. E. (1992) J. Immunol. 148 894–899 [PubMed] [Google Scholar]
- 42.Linette, G. P., Zhang, D., Hodi, F. S., Jonasch, E. P., Longerich, S., Stowell, C. P., Webb, I. J., Daley, H., Soiffer, R. J., Cheung, A. M., Eapen, S. G., Fee, S. V., Rubin, K. M., Sober, A. J., and Haluska, F. G. (2005) Clin. Cancer Res. 11 7692–7699 [DOI] [PubMed] [Google Scholar]
- 43.Kawakami, Y., Eliyahu, S., Delgado, C. H., Robbins, P. F., Sakaguchi, K., Appella, E., Yannelli, J. R., Adema, G. J., Miki, T., and Rosenberg, S. A. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6458–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poiesz, B. J., Ruscetti, F. W., Gazdar, A. F., Bunn, P. A., Minna, J. D., and Gallo, R. C. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 7415–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granfors, K., Marker-Hermann, E., de Keyser, F., Khan, M. A., Veys, E. M., and Yu, D. T. (2002) Arthritis Rheum. 46 606–613 [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, F. K., Ottensmeier, C. H., Johnson, P., Zhu, D., Buchan, S. L., McCann, K. J., Roddick, J. S., King, A. T., McNicholl, F., Savelyeva, N., and Rice, J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14646–14652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly, J. J., Wahren, B., and Liu, M. A. (2005) J. Immunol. 175 633–639 [DOI] [PubMed] [Google Scholar]
- 48.Truscott, S. M. (2007) Engineering and Applications of Single Chain Trimers of Major Histocompatibility Complex Class I Molecules, PhD Thesis, Washington University, St. Louis, MO
- 49.Kim, S., Poursine-Laurent, J., Truscott, S. M., Lybarger, L., Song, Y. J., Yang, L., French, A. R., Sunwoo, J. B., Lemieux, S., Hansen, T. H., and Yokoyama, W. M. (2005) Nature 436 709–713 [DOI] [PubMed] [Google Scholar]
- 50.Parker, K. C., Bednarek, M. A., and Coligan, J. E. (1994) J. Immunol. 152 163–175 [PubMed] [Google Scholar]
- 51.Lazarski, C. A., Chaves, F. A., Jenks, S. A., Wu, S., Richards, K. A., Weaver, J. M., and Sant, A. J. (2005) Immunity 23 29–40 [DOI] [PubMed] [Google Scholar]
- 52.Lauterbach, H., Gruber, A., Ried, C., Cheminay, C., and Brocker, T. (2006) J. Immunol. 176 4600–4607 [DOI] [PubMed] [Google Scholar]
- 53.Crew, M. D., Cannon, M. J., Phanavanh, B., and Garcia-Borges, C. N. (2005) Mol. Immunol. 42 1205–1214 [DOI] [PubMed] [Google Scholar]
- 54.Choudhuri, K., Wiseman, D., Brown, M. H., Gould, K., and van der Merwe, P. A. (2005) Nature 436 578–582 [DOI] [PubMed] [Google Scholar]
- 55.Yachi, P. P., Ampudia, J., Gascoigne, N. R., and Zal, T. (2005) Nat. Immunol. 6 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garboczi, D. N., Ghosh, P., Utz, U., Fan, Q. R., Biddison, W. E., and Wiley, D. C. (2006) Nature 384 134–141 [DOI] [PubMed] [Google Scholar]