Abstract
Waardenburg syndrome (WS) is an auditory-pigmentary disorder that exhibits varying combinations of sensorineural hearing loss and abnormal pigmentation of the hair and skin. Depending on additional symptoms, WS is classified into four subtypes, WS1–WS4. Absence of additional features characterizes WS2. The association of facial dysmorphic features defines WS1 and WS3, whereas the association with Hirschsprung disease (aganglionic megacolon) characterizes WS4, also called “Waardenburg-Hirschsprung disease.” Mutations within the genes MITF and SNAI2 have been identified in WS2, whereas mutations of EDN3, EDNRB, and SOX10 have been observed in patients with WS4. However, not all cases are explained at the molecular level, which raises the possibility that other genes are involved or that some mutations within the known genes are not detected by commonly used genotyping methods. We used a combination of semiquantitative fluorescent multiplex polymerase chain reaction and fluorescent in situ hybridization to search for SOX10 heterozygous deletions. We describe the first characterization of SOX10 deletions in patients presenting with WS4. We also found SOX10 deletions in WS2 cases, making SOX10 a new gene of WS2. Interestingly, neurological phenotypes reminiscent of that observed in WS4 (PCWH syndrome [peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, WS, and Hirschsprung disease]) were observed in some WS2-affected patients with SOX10 deletions. This study further characterizes the molecular complexity and the close relationship that links the different subtypes of WS.
During development, the pluripotent neural-crest cells migrate from the neural tube throughout the embryo along several pathways and give rise to different cell types, including glia and neurons of the peripheral nervous system, enteric neurons and glia, some of the craniofacial skeletal tissue, and melanocytes of the skin and inner ear.1 Defects in neural-crest development are a significant cause of human disease. The term “neurocristopathies” collectively refers to these neural-crest disorders, one of which is Waardenburg syndrome (WS).2–4 The association of hearing loss and pigmentary abnormalities (e.g., heterochromia irides, white skin patches, and white forelock) characteristic of this syndrome results from an abnormal proliferation, survival, migration, or differentiation of neural-crest–derived melanocytes.3 Several subtypes of WS were defined on the basis of the presence of additional symptoms.
Type I WS (WS1 [MIM 193500]) refers to the first cases described by Waardenburg.5 Additional symptoms are dystopia canthorum and broad nasal root. Nearly all patients present with heterozygous mutations of PAX3 (encoding paired box gene 3 [PAX3], a member of the paired box family of transcription factors). Type III WS (WS3 [MIM 148820]), or Klein-Waardenburg syndrome, is an extreme presentation of type I WS, with hypoplasia of limb muscles, and is also caused by heterozygous or homozygous mutations of PAX3.3,6,7
Type II WS (WS2 [MIM 193510]) is characterized by deafness and pigmentation defects without additional features. Heterozygous mutations in the MITF gene (encoding microphthalmia-associated transcription factor [MITF], a basic helix-loop-helix transcription factor) have been identified in ∼15% of cases.3,8 Homozygous deletions of the SNAI2 gene (encoding snail homolog 2, a C2H2-type zinc-finger transcription factor) have also been described in two patients.9 Therefore, 85% of WS2 cases are still unexplained at the molecular level.
Type IV WS (WS4 [MIM 277580]), also called “Shah-Waardenburg syndrome” or “Waardenburg-Hirschsprung disease,” combines pigmentation defects, deafness, and Hirschsprung disease.10 Mutations in EDNRB, which encodes the endothelin B receptor (a G-protein–coupled transmembrane receptor), and EDN3, which encodes its ligand, endothelin-3, have been described. Homozygous (most frequently) or heterozygous mutations are found in probands with WS4, whereas heterozygous family members occasionally present with some of the features.4,11–17
Dominant mutations of SOX10 have also been identified in WS4.18 SOX10 is a key transcription factor of neural-crest development. It is crucial for the survival and maintenance of pluripotency of migrating neural-crest progenitors19,20 and also influences fate decisions and differentiation at later stages.21–25 SOX10 belongs to the SOX family of transcription factors and is closely related to SOX8 and SOX9, the latter of which is involved in campomelic dysplasia.21,23,25–27 All SOX proteins contain a DNA-binding motif known as the high-mobility group (HMG) domain. In addition, SOX10, like SOX8 and SOX9, contains a transactivation domain located in the C-terminal part of the protein and a dimerization domain immediately preceding the HMG domain.21,22,28,29 Functional studies revealed the importance of these domains for monomeric or dimeric DNA binding and transactivation of natural target genes.21,22 Among them are genes/factors crucial for the specification and differentiation of melanocytes or enteric nervous-system development, such as MITF/Mitf, TRYP2/Dct (encoding dopachrome tautomerase), tyrosinase, EDNRB, and the RET protooncogene.30–41 SOX10 targets also include genes important for glia development and identity, such as MPZ (encoding myelin protein zero [P0]), MBP (encoding myelin basic protein), and GJB1 (encoding the gap-junction protein connexin 32).29,42–46
The SOX10 mutations characterized so far are mostly truncating mutations—that is, nonsense and frameshift mutations and one splice mutation—which most often remove all or part of the transactivation domain.18,47–58 An insertion of 2 aa and an amino acid substitution in the HMG domain, as well as two mutations of the stop codon supposed to give rise to elongated SOX10 protein, have also been described.18,48,49,53 Unexpectedly, some of the patients with SOX10 mutations present with chronic intestinal pseudo-obstruction instead of Hirschsprung disease51,55 and/or with neurological features, either peripheral demyelinating neuropathy or central neuropathy or both, which leads to a syndrome called “PCWH” (peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, WS, and Hirschsprung disease).57 This more severe disease is mostly caused by mutations in the last coding exon of SOX10 and has been proposed to occur when the mutant mRNAs escape the nonsense-mediated mRNA decay (NMD) pathway.57
However, some WS4 cases remain unexplained at the molecular level, which suggests that other genes may be involved or that some mutations within the known genes are not detected by the methods commonly used for genotyping. Therefore, we used semiquantitative fluorescent multiplex PCR (QMF-PCR) to search for heterozygous SOX10 deletions. Here, we describe the first characterization of SOX10 deletions in patients presenting with WS4. In light of the phenotypic variability observed among patients with SOX10 point mutations, we also searched for SOX10 deletions in unexplained cases of WS2 and found several, making SOX10 a new gene of WS2.
Subjects and Methods
Subjects
We investigated a total of 30 patients presenting with the classic form of WS4 or PCWH; 29 were found to be negative for EDN3, EDNRB, and SOX10 point mutations, and 1 was found to be hemizygous for a SOX10 point variation within the course of this work. Our study also included 30 WS2-affected patients without MITF mutations. Clinical information and DNA samples were obtained with informed consent in accordance with French law for genetic testing. The main clinical findings in patients presenting with SOX10 gene deletions are summarized in table 1. Detailed clinical descriptions of the patients presenting with SOX10 gene deletions are as follows.
Table 1. .
Summary of Clinical Findings
| Phenotypea |
Molecular Defect |
||||||||||
| Patient | Sex | Age | WS Type | Deafness | Pigmentation Anomalies | HSCR | MR | Other(s) | Inheritance | Deletion Nomenclatureb | Deletion Size |
| 1 | M | 1 year | WS4 | Bilateral | Hair and skin hypopig. | Short | No | Bilateral cryptorchidism | De novo | c.697-740_1085del ins CCT | 1,128 bp del and 3-bp ins |
| 2 | M | 13 years | PCWH | Profound bilateral | Hair and skin hypopig., sapphire blue eyes | Short | Mild | Anosmia, hypermetropia, dental-enamel abnormalities | De novo | g.(17,738,296_17,740,110)_(17,794,727_17,801,789) del | 56–68 kb |
| 3 | M | 36 years | PCWH | Profound bilateral | Hair and skin hypopig., sapphire blue eyes | Short | Mild | Anosmia, cryptorchidism, hypogonadism | De novo | g.(17,712,505_17,716,229)_(17,929,647_17,933,832) del | 213–222 kb |
| 4c | M | 9 years | WS2 | Profound bilateral | Skin, irides, and retinal hypopig. | No | No | … | Maternal (mosaicism) | c.219_428+43del | 253 bp |
| 5 | M | 8 years | WS2 | Profound bilateral | Skin depigmentation, hypoplastic irides | No | No | … | Maternal | c.429-1112_697+396del | 1,777 bp |
| 6 | M | 8 years | WS2 | Profound bilateral | White frontal forelock, heterochromia irides | No | No | … | De novo | g.(16,173,196_16,489,188)_(17,790,847_17,791,697) del | 1.3–1.6 Mb |
| 7 | M | 23 years | WS2 | Bilateral | Skin hypopig., sapphire blue eyes | No | Severe | Short stature, pectus excavatum, autism | De novo | g.(17,357,039_17,432,022)_(18,006,412_18,254,748) del | 574–898 kb |
| 8 | F | 19 mo | WS2 | Yes | Skin hypopig., white forelock | No | Delayed | Thumb duplication, congenital heart disease | De novo | g.(Z83846_Z69042)_ (18,938,054_19,010,379) del | 5.5–6.1 Mb |
HSCR = Hirschsprung disease; MR = mental retardation; hypopig. = hypopigmentation.
The deletions are described in relation to the human genome reference sequence National Center for Biotechnology Information (NCBI) build 36.2 accession number NT_011520.11.
The brother of patient 4 presented with similar clinical symptoms.
Patient 1, a 1-year-old boy, was born at term to unrelated parents after unremarkable pregnancy and delivery. At age 48 h, he presented with clinical signs of meconium plug syndrome caused by short-segment Hirschsprung disease. Bilateral absence of responses to brain stem auditory-evoked potential strongly suggested bilateral deafness. He also had hair and skin hypopigmentation and bilateral cryptorchidism.
Patient 2, a 13-year-old boy, was born at term to unrelated parents after a pregnancy complicated by gestational diabetes. He has two healthy sisters. He had delayed psychomotor development, hair and skin hypopigmentation, sapphire blue eyes, and short-segment Hirschsprung disease that required a surgical procedure at age 15 mo. Nine months later, he had implantation of a unilateral cochlear device for profound bilateral congenital deafness. In spite of good perceptual results, he had speech and sign-language impairment because of dyspraxia. He had mild mental retardation, anosmia, hypermetropia, and dental-enamel abnormalities.
Patient 3, a 36-year-old man, was born at term to unrelated parents after unremarkable pregnancy and delivery. In the neonatal period, he had axial hypotonia and delayed psychomotor development; he could hold his head up at age 1 year, sit alone at age 3 years, and walk at age 4–5 years. He had hair and skin hypopigmentation, sapphire blue eyes, short-segment Hirschsprung disease, and profound bilateral congenital deafness, for which he had implantation of a unilateral cochlear device at age 32 years. Temporal bone CT scan revealed a bilateral vestibular malformation and hypoplasia of the external and posterior semicircular canals. He also presented with anosmia, bilateral cryptorchidism, hypogonadotropic hypogonadism, and bone-age and pubertal growth delay. He now has mild mental retardation with marked abstraction difficulties.
Patient 4, a 9-year-old boy, was born at term to unrelated parents after unremarkable pregnancy and delivery. He began to walk at age 21 mo. A bilateral sensorineural hearing impairment was discovered at age 6 mo and progressed to profound deafness. A cochlear implantation was performed with success, in terms of his comprehension and language skills. Temporal bone CT scan revealed a symmetrical bilateral vestibular malformation with dilatation of the vestibule, hypoplasia, and dilatation of the external and posterior semicircular canals. He presented with hypopigmentation of the skin, irides, and retina. He had no history of constipation, and his neurological development was normal. His brother presented with similar clinical symptoms. No other cases of hearing defect or pigmentation anomalies were observed in the family.
Patient 5, an 8-year-old boy, was born at term to unrelated parents after unremarkable pregnancy and delivery. He began to walk at age 20 mo. A profound bilateral sensorineural hearing impairment was diagnosed at age 5 mo. Temporal bone CT scan did not reveal any malformation of the inner ears. He presented with skin depigmentation and hypoplastic irides. He had no history of constipation, and his neurological development was normal. His mother presented with a bilateral severe sensorineural prelingual hearing impairment. She was born with a frontal white forelock and heterochromia (i.e., one green iris and one hypoplastic iris).
Patient 6, an 8-year-old boy, is the third child of a nonconsanguineous couple from the French Caribbean islands. At age 9 mo, a severe bilateral sensorineural hearing impairment was diagnosed and progressed to bilateral profound deafness by age 8 years. He presented a white frontal forelock at birth and heterochromia irides (i.e., one black and one hypoplastic) but has no skin depigmentation. He began walking at age 17 mo and has no history of severe constipation or neurological anomaly.
Patient 7, a 23-year-old man, was born at term to unrelated parents after unremarkable pregnancy and delivery. He presented with a white frontal forelock (now disappeared), skin hypopigmentation, sapphire blue eyes, pectus excavatum, and statural growth on the 3rd percentile. He has no constipation problems. In addition, this patient has severe autism and developmental delay (he started to walk at age 3 years, is not toilet trained, and has little autonomy). Brain CT scan was normal. Bilateral sensorineural deafness was diagnosed at age 9 mo, and he had a cochlear implant until age 13 years. However, because of severe behavioral problems, hearing loss has not been evaluated since age 13 years.
Patient 8 was born at term to unrelated parents after unremarkable pregnancy and delivery. She presented with severe congenital heart disease associating double-outlet right ventricle, transposition of the great arteries, pulmonary atresia, patent ductus arteriosus, ventricular septal defect, and atrial septal defect. A white forelock and a duplication of the thumb on the left side were noted. At age 10 mo, she presented obvious evidence of deafness, medial flare of the eyebrows, strabismus, and general hypotonia. Brain magnetic resonance imaging scan at age 14 mo showed delayed myelination. Specific evaluation of the skin on Wood’s lamp revealed multiple hypopigmented areas. She had no history of constipation. Heart surgery had been partially successful, but episodes of unexplained bradychardia and peripheral cyanosis were observed, and the patient died at age 19 mo.
QMF-PCR
QMF-PCR has been shown to be a sensitive method for the detection of gene-dosage anomalies and has been successfully used in our laboratory to characterize deletions and duplications within several genes.59,60 We adapted the protocol described elsewhere,61,59 to screen for SOX10 gene deletions. In brief, the three coding exons of SOX10, exon 4 of POLR2F, and four regions located 5′ of SOX10 were amplified in two multiplex reactions. The beginning of exon 3 and the middle of the exon 5 coding sequences are not covered by the amplicons; however, a deletion restricted to one of these regions would have been found during the point-mutation screening. Noncoding regions of SOX10 were not studied. GenBank accession numbers, positions, sequences of the primers, and PCR product sizes are shown in table 2. In each set, two controls were used: DSCR1, located on chromosome 21, and F9, located on chromosome X (see reference genes in table 2). The reverse primers were labeled with the fluorescent phosphoramidite 6-FAM dye. Amplifications were performed in duplicates in 25-μl reactions with use of the QIAGEN Multiplex PCR kit (Qiagen), with 75 ng of genomic DNA, a mix of primers (concentration range 0.1–1 μM), and 5% dimethyl sulfoxide (DMSO). The reaction started with an initial denaturation for 15 min at 95°C, followed by 22 cycles at 95°C for 30 s, at 55°C (multiplex reaction mix 1) or 58°C (multiplex reaction mix 2) for 30 s, and at 72°C for 45 s with an increment of 3 s per cycle, and a final extension for 10 min at 72°C. Then, 3 μl of the purified PCR products were processed as described elsewhere.59 Two control DNA samples (male and female) were included in each experiment. Results were analyzed by superimposing fluorescent profiles of tested patients and controls and by calculating dosage quotient (DQ).59 DQ values <0.6 were considered as indicating potential deletions. Table 3 summarizes examples of DQ values obtained.
Table 2. .
QMF-PCR Primer Sequences[Note]
| Primer Sequence (5′→3′) |
||||||
| Primer | Forward | Reverse | Reaction Mix | PCR Product Size (bp) |
Gene Position | GenBank Accession Number |
| Reference: | ||||||
| DSCR1 | GCGACGAGGACGCATTCCAA | GTCCTTGTGCGATCACCACA | 1 and 2 | 238 | DSCR1 exon 4 | NC_000021.7 |
| F9 | AAATGATGCTGTTACTGTCTA | GAAGTTTCAGATACAGATTTTC | 1 and 2 | 214 | F9 exon 5 | NC_000023.9 |
| SOX10: | ||||||
| P5 | GGCCAGGCGAGCTGGGCAAGGTC | GAATCCACCCGAAGCTAGAG | 1 | 341 | SOX10 exon 3 | NC_000022.9 |
| P6 | GGAGTGCTCTGGCATTCACG | CTTGCCCCACCCTCAGCTCT | 1 | 366 | SOX10 exon 4 | NC_000022.9 |
| P7 | GGAAGTTCACGTGCGCCCAC | GCGGCAGGTACTGGTCCAAC | 1 | 286 | SOX10 exon 5 | NC_000022.9 |
| P8 | CCACACTACACCGACCAGCC | GGGTGGTGGCGACAGGGC | 1 | 326 | SOX10 exon 5 | NC_000022.9 |
| External markers: | ||||||
| P9 | GATGACGTTTCTAGGTGG | TGTTCCTAAGGATCTGTAGG | 1 | 309 | POLR2F exon 4 | NC_000022.9 |
| P1 | GAGGAGGGAGTTTGGGTGGGTGG | GCACAGGATGGGACGGGTTGAGA | 2 | 310 | −54694/−55003 | NC_000022.9 |
| P2 | TGGGAACAATGTCAACGTCG | CAGAAGGCCTCCTCCAATGA | 2 | 330 | −32490/−32819 | NC_000022.9 |
| P3 | TACCAGGGCGGGCTCCTGTA | GCCCCTGTTCCTTCCAGACTCCC | 2 | 363 | −14793/−15155 | NC_000022.9 |
| P4 | ACACGACGGCAGATGCTCGT | CCTCGGAGCCCAGTGAATTA | 2 | 335 | −2116/−2450 | NC_000022.9 |
Note.— GenBank accession numbers, positions, sequences, and expected PCR product sizes are given for primers P1–P9. Positions of primers P1–P4 are given with the A of the SOX10 ATG numbered as 1. NCBI build 36.2 was used.
Table 3. .
DQ Values from QMF-PCR Performed with Mix 1[Note]
|
SOX10 |
||||||
| DNASample | SOX10 5′ Region S4 (P4) | Exon 3 (P5) | Exon 4 (P6) | Exon 5 (P7) | Exon 5 (P8) | POLR2F Exon 4 (P9) |
| Subject 1 | 1.07 | 1.02 | 1.12 | 1.02 | 1.02 | 1.00 |
| Subject 2 | 1.02 | 1.08 | 1.03 | .97 | .93 | .94 |
| Patient 1 | 1.00 | 1.02 | .96 | .47 | 1.10 | .86 |
| Patient 2 | .46 | .50 | .46 | .51 | .49 | .45 |
| Patient 3 | .47 | .47 | .48 | .51 | .51 | .51 |
| Patient 4 | 1.20 | .58 | .90 | 1.17 | 1.20 | .83 |
| Patient 5 | .90 | 1.10 | .56 | 1.08 | .96 | .99 |
| Patient 6 | .52 | .54 | .48 | .52 | .56 | .43 |
| Patient 7 | .56 | .56 | .45 | .54 | .57 | .48 |
| Patient 8 | .51 | .46 | .53 | .50 | .49 | .50 |
Note.— The analysis is based on the comparison of the peak height from the tested DNA (two patients without deletion, called subjects 1 and 2, and patients 1–8) to control DNA samples. The copy-number change for each amplicon was calculated using the peak value normalized to the peak value of DSCR1 exon 4. DQ values <0.6 are indicated in bold.
Molecular Characterization of Rearrangements
When QMF-PCR revealed a short-size deletion (only one exon removed), the genomic region encompassing the deletion breakpoint was amplified either by classic or long-range PCR with the use of Dynazyme Ext DNA polymerase (Ozyme) or the Expand Long Template PCR system (Roche Diagnosis), respectively. The resulting PCR fragments were cloned into the TOPO-TA cloning kit dual promoter or the TOPO-XL PCR cloning kit (Invitrogen) and were sequenced. Bioinformatics analysis was performed to predict the functional consequences of intragenic deletions, with the use of Netgene2 (neural-network predictions of splice sites in humans) and HMMgene (gene-structure prediction) software.
In the case of whole SOX10 gene deletions, FISH was used to confirm the QMF-PCR results. Molecular cytogenetic studies were performed on chromosomes prepared from cultured fibroblasts (patient 8) or peripheral blood cells. Metaphase chromosomes were obtained according to standard techniques. FISH was performed as described elsewhere.62 PAC and BAC clones used in FISH experiments were provided by the BACPAC Resources Center (Children’s Hospital Oakland, Oakland, CA) or by The Wellcome Trust Sanger Institute (Cambridge, United Kingdom). Clones localized on chromosome 22q12-q13.2 (CTA-415G2, LL22NCO1-95B1, RP1-288L1, CTA-714B7, RP1-41P2, CTA-390B3, RP5-1177I5, RP1-37E16, RP3-466N1, RP5-1014D13, RP5-1039K5, CTA-228A9, CTA-447C4, RP1-5O6, RP3-434P1, RP1-319F24, RP3-508I1S, RP3-327516, CTA-150C2, RP4, 742C19, and LL22NC03-10C3) were directly labeled with Cy3. The control probes (RP1-41P2 and RP1127L4) were directly labeled with fluorescein isothiocyanate, and chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The specific signal intensity and its sublocalization along the chromosome axis were analyzed using a Leica fluorescence microscope equipped with the Visilog-6 program (Noesis). On the basis of FISH results, additional QMF-PCR primers were designed to delineate the extent of deletions. Positions and sequences of these additional primers are reported in table 4.
Table 4. .
Additional QMF-PCR Primers, Ordered Centromeric to Telomeric[Note]
| Primer Sequence (5′→3′) |
||
| Gene Position | Forward | Reverse |
| FBX07 exon 1 | CGCCTCAGCTACCCCTCAGC | CCGGCCCCAGCGTACCTGTA |
| TIMP3 exon 1 | CTGGAGGGGTAGCAGTTAGC | GATCGTTCAGATCCTTATAG |
| ISX exon 1 | GATCAACAACTGTCAGCTCC | CCTGCAGGTGGAGGCTGAGG |
| TOM1 exon 1 | CGGTAGCAGCAATGGACTTT | TCCTTACCTACTGGCTGTCA |
| APOL3 exon 4 | TTCAGGCATCTTCCTTGCAC | AAGTCACTAACACTCAGCTC |
| MYH9 intron 1 | AGGCGGCTGTCTCCACCGGG | GAGTGATTGTCTCAAAAGGC |
| CACNG2 exon 1 | CACACTCTCCCGAGAACCAG | CAAGTACCTTCTAGGCAGCA |
| LGALS2 exon 4 | GAGAGCCTGGCCTGGACACA | GAAAGAGGACATGTTGAACC |
| SH3BP1 exon 10 | TGTGACGGATGATTTCAGAT | ATGCCCTCAGAAAGCAGCAT |
| TRIOBP exon 1 | GGTGAAATTCCTCAGCTCTC | GTATCTTACTTGTGCAAGGG |
| MICALL1 exon 8 | CCCATGGATCACGCTGGTGC | TAGGGGTGATGCCGTACCAG |
| MICALL1 intron 9 | TATAATGTACATGCTGCTGT | GTACTGAGCCCAGGCAATCC |
| MICALL1 exons 11–12 | GACATCCATGGAGAGATGGA | GAGCTCGGACTCTCGCCGCA |
| MICALL1 exon 16 | GGCAGCGGTGATTCAGCCCT | ACTGAGATCCAAGGGCGCCT |
| C22ORF23 exon 4 | GATGCTTTGCCCCTACAGTG | GCTGAGGCTCAAGAGCATTG |
| C22ORF23 exon 3 | CAGAGGGCAGAAATTTTATT | CATGATGTCCATGATGTGGC |
| POLR2F exon 1 | CAGTCAGCGCATGCGCACTT | CAGACATTGCCCCTAGACGC |
| P2_3-4 −20540/−20829 | CAGAGGTAATCAAGTTAGCC | GTATTAGGATGGGTCGTAGG |
| P2_3-3 −21390/−21739 | CACCTGGCCAGGATGAGCTT | CCACAGTGAGCACTCAGGAA |
| P2_3-2 −23920/−24166 | TTGTAAAGCAACAGAGGAGC | CCTTCCGCAAAGACTTGCTG |
| P2_3-1 −31482/−31759 | CCAAAAGCAGAAATCTGGGA | GGCCTGAGGCTCCAGCTGAG |
| PLA2G6 exon 7 | GATGGGATGGGAGGCAGTCT | CCGTGGGTCAGCAGCACTAT |
| PLA2G6 exon 4 | GGCGGGAATGCTCTTGAGCT | CACCCTGGACAGCATAATGG |
| PLA2G6 intron 2 | GGCTTCAGGAGGGCAGAGCA | CCAGGGCTTTTAACATACCA |
| PLA2G6 exon 2 | CCTTCAGTGGCGTCACCAAC | GGTCCAAGAGTACAGTCTCC |
| C22ORF5 exon 9 | CTCTAGACCTGGGGCCTCCG | ATGCAGGCCTCTGCCACGAG |
| KDELR3 exon 1 | TGGACCATGAACGTGTTCCG | ATCATCTCGAGGGGTCCTCC |
| DDX17 exon 1 | TTGTGCAGTCGCTGGGAAGG | GATAGAGATCTGGGAGCGGG |
| CBX7 intron 1 | TCCTACCCCATCTGCCTTCT | CCTGGAGCCTCCCAAAGGGC |
| PDGFB exon 7 | AGCCTGTGGCTTGGAGTGGC | GTGGGAGCTCAGATCCCACC |
Chromosome Segregation Analysis
Seven chromosome 22 microsatellites (D22S420, D22S539, D22S315, D22S280, D22S283, D22S423, and D22S274) were analyzed in patient 3 and his parents, with use of the linkage mapping set (Applied Biosystems) in accordance with the manufacturer’s instructions.
SOX10 Point-Mutation Screening in Patients with WS2
In the absence of a full description of the 5′ UTR noncoding exon(s) of SOX10, we used, for convenience, the exon-numbering system used elsewhere—that is, noncoding exons 1 and 2, the ATG codon in exon 3, and the stop codon in exon 5.18 Three sets of primers were used to amplify the SOX10 gene fragments covering coding exons 3–5 and intron-exon boundaries (first set: exon 3, forward 5′-ACCCACCTAGAGTCTGGCATG-3′ and reverse 5′-CTCGGCTACCCTGAATCCAC-3′, 733-bp PCR product; second set: exon 4, forward 5′-CCACAAATCATAGGGCACAG-3′ and reverse 5′-TAGAGTCCAGGGTCTCATTG-3′, 523-bp PCR product; third set: exon 5, forward 5′-CCTGCCTCTAACCTGCTTCC-3′ and reverse 5′-ACCTCCTTCTCCTCTGTCCA-3′, 997-bp PCR product). The PCR covering the exon 3 region contained 10% DMSO. Resulting PCR products were sequenced using a 16-capillary ABI Prism sequencer and the Terminator Cycle Sequencing kit. Additional internal sequencing primers were used for exon 3 (forward 5′-GCGAGCTGGGCAAGGTCAAG-3′ and reverse 5′-TCGCCGTCCTGCTGCTCCTT-3′) and for exon 5 (forward 5′-GGATGCCAAAGCCCAGGTGA-3′ and reverse 5′-GTAGGCGATCTGTGAGGTGG-3′).
Plasmids, Cell Culture, Transfection, and Reporter Assays
The pECE-SOX10, pECE-SOX10-E189X, pECE-SOX10-Y313X, pECE-SOX10-482ins6, pECE-PAX3, pECE-EGR2, pGL3-MITFdel1718, and pGL3-Cx32 vectors were described elsewhere.30,43 The p.Val92Leu mutation (named “V92L” for convenience) was introduced within the pECE-SOX10 construct by site-directed mutagenesis with use of the QuikChange Mutagenesis Kit (Stratagene). Luciferase assays and immunofluorescence experiments were performed 24 h after transfection of HeLa cells, as described elsewhere.30,43,63 The SOX10 antibody used for immunofluorescence experiments was described elsewhere.64 The P0 construct was kindly provided by M. Wegner.29
Results
Identification of SOX10 Deletions in Patients Presenting with WS4 or PCWH
In our experience, ∼20%–40% of affected individuals with the WS4 or PCWH phenotypes have no mutations within SOX10, EDN3, or EDNRB identifiable by conventional genetic analysis with the use of DNA sequencing of PCR-amplified gene segments. Since this technique does not detect heterozygous deletions, we decided to search for SOX10 deletions or rearrangements by QMF-PCR. Our study included 29 patients presenting with the classic form of WS4 or PCWH, previously found to be negative for SOX10, EDN3, and EDNRB point mutations. We first analyzed the three coding exons of SOX10, part of the POLR2F downstream gene, and sequences located up to 50 kb upstream of SOX10 (S1–S4) (see fig. 1A). One of them, S1, was described elsewhere as a SOX10 enhancer65 (see table 2 for sequences of primers P1–P9).
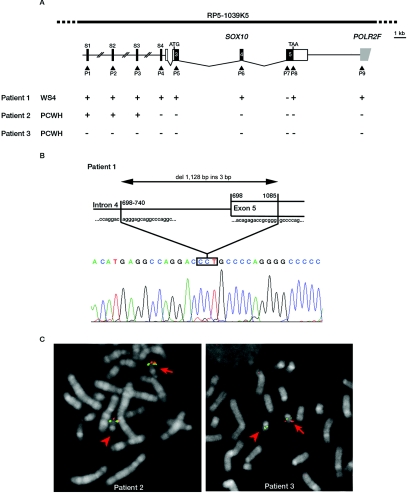
Figure 1. .
SOX10 deletions in patients presenting with the classic form of WS4 or PCWH. A, Schematic representation of the three deletions identified by QMF-PCR. The scheme on the top indicates the SOX10 gene structure (an approximate scale is shown on the right). SOX10 coding sequence (exons 3–5) is indicated with black boxes, and noncoding exonic sequence is indicated with white boxes. Arrowheads indicate the position of the segments analyzed by QMF-PCR (P1–P9; see table 2 for corresponding primer sequences). They include the downstream adjacent POLR2F gene (gray box) and four short regions located up to 50 kb upstream of SOX10, indicated by dark lines and labeled S1–S4. QMF-PCR results for patients 1–3 are indicated: + = not deleted; − = deleted. The phenotypes are indicated on the left. B, Schematic representation (top) and electropherogram (bottom) of the deletion breakpoint region of patient 1. The size of the deletion and the nucleotidic localization of the breakpoint are indicated on the diagram. The 3 inserted nt are boxed. C, Hybridization pattern of patients 2 and 3 with use of BAC clone RP5-1039K5 encompassing the SOX10 locus (indicated at the top of panel A). The BAC clone RP5-1039K5 is shown in red, and the control probe (RP1-41P2) is shown in green. The normal chromosome 22 is indicated by an arrow, and the deleted chromosome 22 is indicated by an arrowhead.
We identified two heterozygous deletions (patients 1 and 2; see the “Subjects” section and table 1 for clinical descriptions). The first, found in a patient presenting with a classic WS4 phenotype, removes part of exon 5 (fig. 1A, patient 1). PCR amplification with the use of primers located in intron 4 and exon 5 and cloning and sequencing of the resulting products revealed a complex rearrangement combining a 1,128-bp deletion, encompassing 740 bp of intron 4 and 388 bp of exon 5, and a 3-bp insertion (fig. 1B). The other deletion, found in a patient with PCWH, includes the whole SOX10 gene, POLR2F, and the upstream S4 sequence (fig. 1A, patient 2). FISH analysis with use of a BAC encompassing the whole region (clone RP5-1039K5) (fig. 1A) confirmed the QMF-PCR results. Indeed, we observed a significantly decreased signal on one of the chromosomes 22 in all the metaphases (fig. 1C, patient 2), showing that the deletion removes only part of the RP5-1039K5 probe (S1–S3 are not deleted) (see fig. 1A). In each of the two cases, the analysis of parent DNA by QMF-PCR revealed the de novo occurrence of the deletion (table 1).
Detection of a SOX10 Deletion in a Patient with PCWH with p.Val92Leu Variation
Sequencing of the SOX10 coding exons and intron-exon boundaries in newly recruited patients led us to identify a new point mutation (nucleotide substitution c. 274G→C in exon 3) that predicts the replacement of a valine by a leucine at codon 92 (p.Val92Leu) in a patient presenting with PCWH (patient 3) (see the “Subjects” section and table 1 for clinical description). This amino acid substitution affects a residue located within a region directly preceding the HMG domain that is well conserved not only between SOX10 proteins across evolution but also among the SOX E members, SOX8 and SOX9 (fig. 2A).28 Surprisingly, this variation was observed at the homozygous state and therefore contrasts with the heterozygous state of all the mutations identified to date. The parents were not consanguineous and did not present any feature of the disease, except for early graying hair in the mother. We found the mutation at the heterozygous state in the father’s DNA only. The analysis of seven chromosome 22 microsatellites excluded a chromosome segregation abnormality and the possibility of a chromosome 22 paternal isodisomy (data not shown).
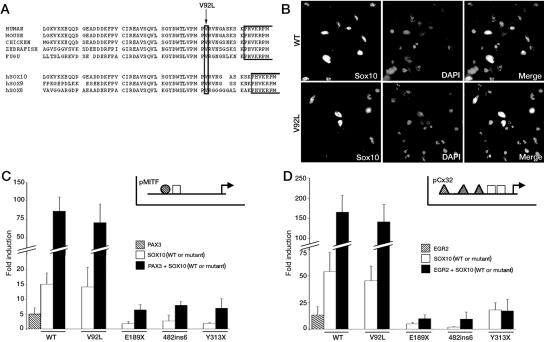
Figure 2. .
Functional consequences of the p.Val92Leu (V92L) mutation. A, Amino acid sequence comparison of the region immediately preceding the HMG domain of SOX10 proteins across evolution and of SOX subgroup E proteins (human sequences). Gaps are indicated by blanks. The first part of the HMG domain is boxed, and the amino acid substitution V92L from patient 3 is indicated by an arrow. SOX10 sequence reference numbers: human (accession number NP_008872.1), mouse (XP_128139.4), chicken (NP_990123.1), fugu (NP_001072112.1), zebrafish (NP_571950.1), human SOX8 (NP_055402.2), and human SOX9 (NP_000337.1). B, Subcellular localization of wild-type SOX10 and p.Val92Leu mutant. HeLa cells were transfected with pECE-SOX10 (WT) or mutant (V92L) constructs for 24 h, were fixed, and were immunostained for SOX10. Cultures were counterstained with DAPI. C and D, Transactivation capacities of SOX10 proteins. The MITF promoter (pMITF) (C) or the GJB1 (pCx32) promoter (D) luciferase reporters were transfected in Hela cells in combination with wild-type (WT) or mutant SOX10 proteins (V92L, E189X, 482ins6, and Y313X), and/or PAX3 (C) or EGR2 (D). Reporter-gene activations are presented as fold induction relative to the empty expression vector (pECE). Results represent the mean±SEM from six experiments, each performed in duplicate. Insets in C and D show a schematic representation of relative localization of SOX10 (white boxes), PAX3 (striped circles) or EGR2 (striped triangles) binding sites on MITF or GJB1 (Cx32) promoters.
To explain these data, we searched for a deletion or rearrangement of SOX10 that would result in a hemizygous p.Val92Leu mutation. QMF-PCR experiments indeed unraveled a deletion that removes the whole SOX10 gene, along with the upstream and downstream sequences tested (fig. 1A, patient 3). FISH analysis with use of the BAC clone RP5-1039K5 confirmed the presence of a deletion encompassing the whole region (fig. 1C, patient 3, showing presence of only one signal on the normal chromosome 22). Analysis of parent samples established the de novo occurrence of the deletion (table 1).
These results suggested that the phenotype of patient 3 is related to this large deletion, but we could not exclude the possibility that the p.Val92Leu variation contributes to the phenotype. To test this possibility, we introduced the mutation into the SOX10 cDNA and analyzed its functional consequences in vitro. Immunofluorescence experiments on HeLa cells transiently transfected with wild-type or p.Val92Leu constructs revealed correct nuclear localization of the mutant protein (fig. 2B).
The region affected by the mutation was previously implicated in DNA-dependent dimerization, both in SOX10 and SOX9.29,42,45,66 Moreover, a SOX9 mutation located in the same region (p.Ala76Glu) was shown to selectively abrogate DNA-dependent dimerization and to thus interfere with promoter activation via natural target sites that require binding of SOX9 as dimers.66 We therefore analyzed the transactivation potential of the p.Val92Leu mutant on two promoters previously shown to contain monomeric or dimeric SOX10 binding sites—MITF and Cx32, respectively. Indeed, SOX10, in synergy with PAX3, regulates MITF expression by directly binding to its promoter as monomers.30,33 SOX10, on the other hand, regulates GJB1 (encoding connexin 32 [Cx32]) expression by directly binding to its promoter on a dimeric configuration and in synergy with its cofactor EGR243,67 (insets in fig. 2C and 2D). Cotransfection of either promoter with wild-type or p.Val92Leu SOX10 mutant and/or SOX10 cofactors (PAX3 and EGR2) revealed that normal and mutant SOX10 have similar transactivation capacities on these promoters, alone or in synergy with the cofactors (fig. 2C and 2D). In contrast, three previously identified SOX10 mutations (p.Glu189X, c.482ins6, and p.Tyr313X, named “E189X,” “482ins6,” and “Y313X,” respectively, in fig. 2C and 2D) failed to transactivate these reporter constructs, as described elsewhere.30,43,68 We also performed similar experiments, using another SOX10-responsive promoter known to contain SOX10 dimeric binding sites, P0, and found no difference between wild-type and p.Val92Leu mutant transactivation capacities (data not shown). Therefore, the p.Val92Leu variation does not seem to affect SOX10 function, at least in vitro, which argues in favor of the deletion as the cause of the phenotype observed in this patient.
Molecular Characterization of Large Deletions in Patients with PCWH
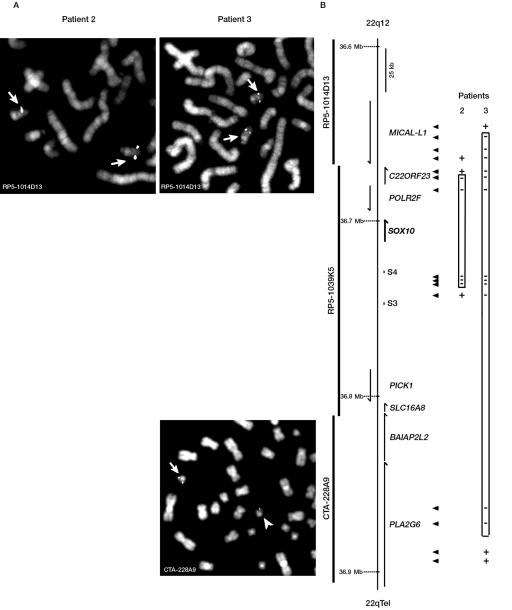
To define the boundaries of the deletions encompassing the SOX10 gene found in patients 2 and 3, we extended our analysis to adjacent regions, using two complementary strategies: FISH and QMF-PCR. In the case of patient 2, we chose additional sets of QMF-PCR primers localized between upstream S3 and S4 sequences, which allowed us to map the telomeric border of the deletion to a region located 24–31 kb upstream of the SOX10 start codon (fig. 3B, patient 2). In parallel, FISH experiments with the probe RP5-1014D13 (covering regions proximal to POLR2F, including MICAL-L1) (see fig. 3) showed a distinct signal on both chromosomes 22, positioning the centromeric border of the deletion between POLR2F and MICAL-L1 (fig. 3A). We chose additional sets of QMF-PCR primers within the genes located in the region and localized the end of the deletion between exons 3 and 4 of the C22ORF23 sequence (see fig. 3B and table 4 for corresponding primers). The whole deletion therefore encompasses 56–68 kb including, in addition to SOX10, POLR2F and part of C22ORF23.
Figure 3. .
Determination of the extent of large deletions in patients with PCWH. Schematic representation of the deletions in patients 2 and 3 was determined by FISH (A) or QMF-PCR (B). A, Representative FISH results. The names of the BACs used for hybridization are indicated on each panel. B, Scheme with the names, positions, and orientation of genes located proximal or distal of SOX10. An approximate scale is shown. BACs used for FISH experiments are shown on the left, and segments analyzed by QMF-PCR are indicated by arrowheads (see table 4 for corresponding primer sequences). QMF-PCR results of patients 2 and 3 are indicated next to the corresponding fragments: + = not deleted; − = deleted.
In the case of patient 3, we performed FISH analysis, using RP5-1014D13 on one side and CTA-228A9 on the other. This allowed us to localize the centromeric border of the deletion between RP5-1014D13 (that shows hybridization on both chromosomes 22) and RP5-1039K5 (that shows only one signal on normal chromosome 22) (figs. 1C and 3A). The telomeric border was localized within CTA-228A9 (we observed a significantly decreased signal on one of the chromosomes 22 in all the metaphases) (fig. 3A). On the basis of these results, we repeated a series of multiplex PCR, using primers located within C22ORF23 or MICAL-L1 on one side and in PLA2G6 on the other, and were able to finally determine the deletion boundaries between intron 9 and exon 8 of MICAL-L1 and between exon 4 and intron 2 of PLA2G6 (see fig. 3B and table 4 for corresponding primers). The whole deletion therefore encompasses 213–222 kb, including part of the MICAL-L1 gene, the C22ORF23, POLR2F, SOX10, PICK1, SLC16A8, and BAIAP2L2 genes, and part of the PLA2G6 gene.
Identification of SOX10 Deletions in Patients with WS2
WS4-affected patients with SOX10 point mutations or deletions exhibit a large variability and an incomplete penetrance of each feature (i.e., fully blue irides, patchy blue irides, or normal eyes; large, small, or no depigmented skin patches; short- or long-segment Hirschsprung disease or chronic intestinal pseudo-obstruction, etc.). Interestingly, we previously described a c.1076delGA mutation in a patient with a classic form of WS418 that was inherited from a mother presenting with deafness and white forelock only. We also described a p.Ser135Thr (S135T) mutation in a patient presenting with a peculiar phenotype named “Yemenite deaf-blind hypopigmentation syndrome” without any intestinal dysfunction.48 These two phenotypes, which are reminiscent of that observed in patients with WS2, prompted us to search for SOX10 deletions in unexplained cases of WS2. Screening of 30 cases (previously found to be negative for MITF) by means of QMF-PCR allowed us to identify five different SOX10 deletions (in patients 4–8) (see the “Subjects” section and table 1 for clinical descriptions).
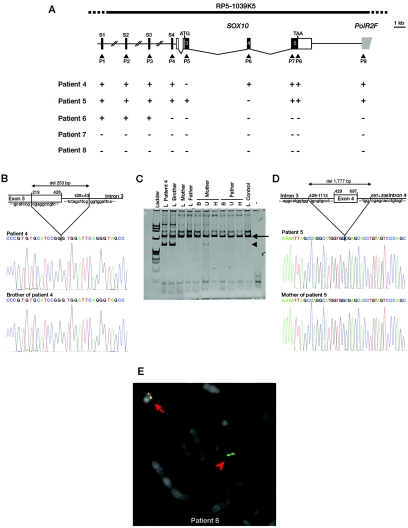
The first encompasses exon 3 (fig. 4A, patient 4). PCR amplification with use of sets of primers located in exon 3 and intron 3, followed by cloning and sequencing of the PCR products, revealed a 253-bp deletion that removed 210 bp of exon 3 and 43 bp of intron 3 (fig. 4B). Interestingly, the patient’s brother, who presented with similar WS2 features, also carries the deletion (fig. 4B). However, it was not found in the parents' DNA, suggesting germline mosaicism. We analyzed hair roots and buccal and uroepithelial cell samples of both parents by PCR amplification and found the band specific to the deleted allele in the mother’s uroepithelial cell sample, showing the existence of somatic mosaicism (fig. 4C and table 1).
Figure 4. .
SOX10 deletions in patients presenting with WS2. A, Schematic representation of the five deletions identified by QMF-PCR. The top scheme indicates the SOX10 gene structure and QMF-PCR experiment results (+ = not deleted; − = deleted), as described in figure 1. B and D, Schematic representation (top) and electropherograms (bottom) of the breakpoint deletion region of patient 4 (B) and patient 5 (D) and their relatives. The sizes of the deletions and the breakpoints are indicated on the scheme. C, PCR covering exon 3 and intron-exon boundaries, with the use of DNA from patient 4, his brother, his mother, and his father, extracted from leukocytes (L), buccal cells (B), uroepithelial cells (U), and hair roots (H). The normal band is indicated by an arrow, and the mutant allele is indicated by an arrowhead. Note the faint mutant band in the mother’s uroepithelial cells. E, Representative FISH result obtained for patient 8 with probe RP5-1039K5 encompassing the SOX10 locus (indicated on the top scheme of panel A). The BAC clone RP5-1039K5 is shown in red, and the control probe (RP1-127L4) is shown in green. The normal chromosome 22 is indicated by an arrow, and the deleted chromosome 22 is indicated by an arrowhead.
The second deletion encompasses 1,777 bp and removes the whole exon 4, 1,112 bp of intron 3, and 396 bp of intron 4 (fig. 4A and 4D, patient 5). The father’s DNA was not available. However, analysis of the mother’s DNA by QMF-PCR and PCR sequencing techniques revealed that the deletion is maternally inherited, in agreement with the observation that the mother also presents with WS2 (fig. 4D and table 1).
The other three deletions included the whole SOX10 gene, as well as POLR2F and the upstream S4 sequence (fig. 4A, patients 6, 7, and 8). Additionally, sequences S1–S3 were deleted in patients 7 and 8. FISH analysis with use of the BAC clone RP5-1039K5 confirmed the presence of a deletion encompassing the whole region analyzed in patient 8 (fig. 4A and 4E). Analysis of the parents’ samples revealed the de novo occurrence of the deletions in these three patients (table 1).
Molecular Characterization of Large Deletions in Patients Presenting with WS2
To define the boundaries of the three deletions encompassing the whole SOX10 gene, we extended our analyses to adjacent regions. In the absence of material with which to perform FISH experiments in patients 6 and 7, we determined the boundaries of the deletion by repeating many sets of QMF-PCR (fig. 5B and data not shown). In the case of patient 6, additional sets of QMF-PCR primers between upstream S3 and S4 sequences allowed us to map the telomeric border of the deletion to a region located 20–21 kb upstream of the SOX10 start codon. The centromeric border was localized between genes CACNG2 and MYH9, therefore delimiting a deletion of 1.3–1.6 Mb that removes at least 42 genes, including SOX10 and TRIOBP (fig. 5B).
Figure 5. .
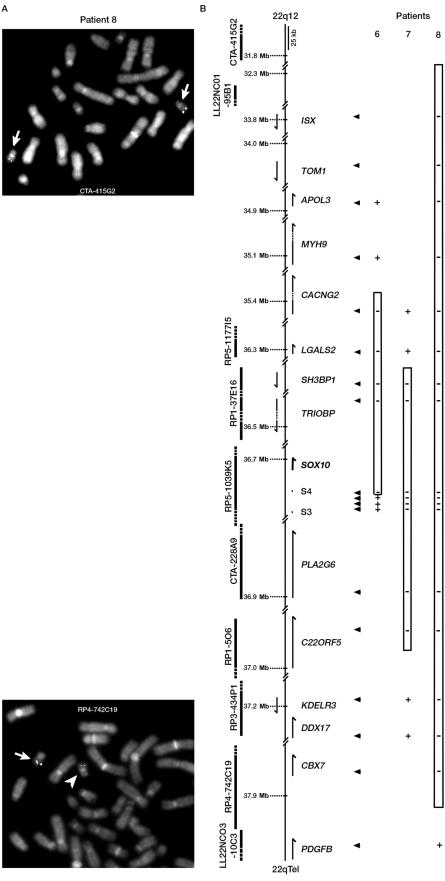
Determination of the extent of large deletions in WS2. Schematic representation of the deletions in patients 6, 7, and 8 was determined by FISH (A) or QMF-PCR (B), as described in figure 3. Only the first nondeleted or partially deleted BACs are shown in panel A.
Patient 7′s deletion removes 574–898 kb. Indeed, boundaries were located between LGALS2 and SH3BP on one side and C22ORF5 and KDELR3 on the other side. The deletion therefore encompasses 23 genes, including SOX10, TRIOBP, and PLA2G6 (fig. 5B).
In the case of patient 8, FISH analysis was performed using several probes, including CTA-415G2, LL22NC01-95B1, RP1-288L1, CTA-714B7, RP5-1177I5, RP1-37E16, CTA-228A9, RP1-5O6, RP3-434P1, CTA-150C2, and RP4-742C19. The telomeric border was localized within the RP4-742C19 BAC (we observed a significantly decreased signal on one of the chromosomes 22 in all the metaphases) (fig. 5A), and the last centromeric deleted BAC was LL22NCO1-95B1 (fig. 5B and data not shown). QMF- PCR with the use of primers within some of the genes located in the region confirmed the results, therefore delimiting a deletion of 5.5–6.1 Mb encompassing 102 different genes, including SOX10, LARGE , RASD2, RBM9, MYH9, CACNG2, TRIOBP, PLA2G6, KCNJ4, and NPTXR (fig. 5B and data not shown).
The identification of five SOX10 deletions in the 30 cases studied revealed that SOX10 is a new gene of WS2 and thus prompted us to screen for SOX10 point mutations. However, we failed to detect any mutation by DNA sequencing of the three SOX10 coding exons. Taken together, these results make SOX10 the third gene discovered to be involved in WS2, with an estimated frequency (15%) similar to that of MITF.
Discussion
Considering our panel of patients presenting with WS4 (classic forms of WS4 and PCWH), we estimated that the phenotype is caused, for 20%–30% of patients, by EDN3 or EDNRB point mutations and, for 40%–50% of patients, by SOX10 point mutations. In patients with WS2, mutations in the MITF gene were identified in ∼15% of cases. Thus, 20%–40% of WS4 cases and 85% of WS2 cases remained unexplained at the molecular level, raising the possibility that other genes are involved or that some mutations within the known genes are not detected by the methods commonly used for genotyping. In this study, we describe the first characterization of SOX10 deletions in patients presenting with WS4 and WS2. We found three deletions among 30 WS4 cases. Taking into account the fact that we tested only DNA samples of patients negative for SOX10, EDN3, and EDNRB point mutations, we estimate that SOX10 deletions are involved in ∼5% of all cases of WS4. We also found five deletions among 30 WS2 cases. Since we tested only patients negative for MITF mutations, we can estimate that SOX10 deletions account for ∼15% of all WS2 cases. These results extend the spectrum of SOX10 mutations found in patients with WS4 and make SOX10 the third gene discovered to be involved in WS2, with an estimated frequency similar to that of MITF. In terms of molecular diagnosis, SOX10 deletions should now be searched for when no SOX10 point mutations (PCWH) or no SOX10, EDN3, and EDNRB point mutations (classic form of WS4) are found. More importantly, SOX10 deletions should be considered for first-step analysis in WS2, as well as MITF mutations.
Full characterization of the eight deletions by PCR and sequencing, or by a combination of QMF-PCR and FISH, revealed eight different deletion events ranging from the deletion of a single exon of SOX10 to the deletion of up to 6 Mb around the SOX10 locus (table 1). Both intragenic and full SOX10 deletions were observed in either WS2 or WS4. Intragenic deletions involved different exons. The observation of sequences surrounding the breakpoints of the three intragenic deletions revealed no clear mechanism except for in patient 5, in whom the deletion occurred between two hexanucleotide repeats, tggtgg, retaining one copy. Bioinformatics analysis with the use of Netgene2 and HMMgene software was performed to predict the functional consequences of these deletions. In the case of patient 1, the rearrangement was predicted to activate a cryptic splice-acceptor site within exon 5, located downstream of the stop codon (780 nt after the usual acceptor site; estimated frequency of 97%). The predicted protein would lack the last 206 aa and would be replaced by 27 unrelated aa, thereby removing the transactivation domain. In the case of patient 4, the deletion removed the exon 3–intron 3 boundary. In silico analysis revealed that this deletion could result in translation of a short intronic sequence with a nearby immediate stop codon (5 aa downstream), with the resulting protein devoid of all functional domains. Alternatively, the use of a cryptic splice-donor site (estimated frequency 55% or 88%, depending on the program used), located 55 nt downstream of the missing donor site, could produce a protein lacking 69 aa, removing the dimerization domain and half of the HMG domain but leaving the transactivation domain intact. In the case of patient 5, the Netgene2 software predicted that the deletion could lead to exon 4 skipping, therefore resulting in a frameshift and a truncated protein of 189 aa with 47 unrelated aa in the carboxyterminal part. Half of the HMG domain and the following domains would be removed. Unfortunately, there is no SOX10-expressing tissue easily accessible by noninvasive methods in patients, which precludes analysis of the protein produced in vivo.
Most of the SOX10 disease-associated point mutations identified so far, regardless of whether they cause WS4 or PCWH, result in premature termination codons. An explanation for the presence or absence of a neurological phenotype (i.e., central or peripheral) that characterizes the PCWH syndrome has been hypothesized to be related to the NMD process.57 Truncating mutations located in the first coding exons (exons 3 and 4) activate the NMD RNA surveillance pathway, leading to haploinsufficiency and classic forms of WS4. On the other hand, truncating mutations located in the last coding exon (exon 5) escape NMD, leading to translation of an abnormal SOX10 protein with a dominant negative effect and therefore resulting in the more severe PCWH phenotype. Considering the published18,47,49,50,52–58 and our unpublished cases with SOX10 point mutations, we observed that the length of intestinal aganglionosis may also fit the NMD hypothesis, since all the point mutations associated with the long-segment Hirschsprung disease are located in exon 5. Accordingly, full deletions of SOX10 would be expected to cause classic forms of WS4 with short-segment Hirschsprung disease as a result of haploinsufficiency. All three patients indeed present with a short form; however, two of them have mild PCWH syndrome, an observation that does not match the NMD hypothesis (table 1).
We wondered whether the PCWH syndrome might also result from other mechanisms, at least in some of the patients. Interestingly, one of the PCWH-affected patients with a SOX10 deletion (patient 3) also carries a SOX10 valine→leucine substitution at the hemizygous state. This variation, which could worsen the phenotype of the patient, is located in a highly conserved region crucial for the mediation of DNA-dependent dimerization. We thus reasoned that the p.Val92Leu variation, although not a drastic substitution, might interfere with the formation of SOX10 dimers and thus specifically hamper promoter activation via natural target sites that require binding of SOX10 dimers. We therefore compared wild-type and p.Val92Leu SOX10 transactivation capacities on MITF and GJB1 (Cx32) promoters containing monomeric or dimeric binding sites, respectively, and found no differences. These results suggest that the p.Val92Leu variation does not affect SOX10 function, at least in vitro, arguing in favor of the deletion as the major or sole cause of the PCWH phenotype observed. A contiguous gene syndrome may also account for the neurological features. The deletion identified in patient 3 encompasses the PLA2G6 gene known to be involved in a recessive neurological defect (infantile neuroaxonal dystrophy 1, neurodegeneration with brain iron accumulation, and Karak syndrome).69 However, this gene is not deleted in patient 2, who has a mild PCWH syndrome resembling that of patient 3, suggesting that the heterozygous PLA2G6 deletion is not involved in the phenotypic expression of the disease.
In WS2, as in WS4, the largest deletions are found in patients presenting with additional symptoms, suggesting that some features may be influenced by the presence of other genes. Indeed, several genes removed by one or more of the deletions have a well-established neurological role (i.e., LARGE, RASD2, RBM9, CACNG2, KCNJ4, and NPTXR). Two genes involved in human hearing loss are also located within some of the deleted regions: TRIOBP (recessive nonsyndromic deafness [DFNB28]70) and MYH9 (Epstein and Fechtner syndromes and dominant progressive isolated deafness [DFNA17]71,72). However, when comparing all eight patients, we observed no clear correlation between the deletion of one of these genes and the presence of neurological features or the deafness phenotype. In fact, as for SOX10 point mutations, SOX10 deletions by themselves might be sufficient to explain the phenotypes observed. On the other hand, the autism of patient 7 and the cardiac defects of patient 8 have not previously been linked to SOX10 mutations and may result from the deletion of additional genes. We found no evidence of a known gene removed by the deletion in patient 8, which could explain the cardiopathy, but this deletion encompasses 102 genes, and they are not all functionally characterized. In autism, recurrent deletions of chromosome 22q13 have been characterized,73 including a frequent breakpoint within the SHANK3 gene.74 However, this region is distal to the deletion found in patient 7. The question of whether the association with autism in patient 7 is fortuitous or results from the deletion remains unanswered.
The WS subtypes were initially clinically defined. It appears, however, that this classification does not reflect the molecular mechanisms. Indeed, an overlap between WS1 and WS3 has already been reported, and we now report an overlap between WS2 and WS4. In developmental syndromes, incomplete penetrance of some features is commonly observed. Incomplete penetrance of Hirschsprung disease is described both in isolated and in syndromic forms of the disease, and it may result from different mechanisms, including genetic modifiers.75,76 In the case of WS, incomplete penetrance of Hirschsprung disease could explain the overlap between WS2 and WS4. However, with regard to this hypothesis, it is surprising to not find SOX10 point mutations in WS2. To our knowledge, no genetic disorder has been described to result exclusively from deletions of the causative gene. It is possible that the penetrance of one feature varies between deletions and truncating point mutations, as shown, for example, in Von Hippel–Lindau disease.77 In our case, incomplete phenotypic penetrance may be explained by tissue-specific compensation of the loss of SOX10 by other SOX proteins having partly redundant function, such as SOX8 and SOX9. Interestingly, in mice that expressed SOX8 instead of SOX10, the enteric defect was partially or totally rescued in homozygotes or heterozygotes, respectively, whereas the pigmentation defect was not.78 As a result, SOX10 haploinsufficiency may be compensated by SOX8 or SOX9 during enteric nervous-system development, explaining the low penetrance of Hirschsprung disease associated with SOX10 deletions (leading to WS2 or WS4). In contrast, the presence of a truncated SOX10 protein may impair SOX8 or SOX9 function, resulting in a fully penetrant enteric phenotype (leading to WS4 only). However, it is possible that screening larger numbers of patients with WS2 will result in the identification of SOX10 point mutations.
An increasing diversity of clinical features is reported in WS; some patients with WS4 present with pseudo-obstruction instead of Hirschsprung disease, and others present with myelination defects of the peripheral nervous system and CNS (i.e., PCWH syndrome). In this study, patients 7 and 8 presented with WS2 and central and peripheral neurological features typical of PCWH—formally, a new syndrome indicating a continuum from isolated WS2 to severe PCWH. On the basis of our observation of SOX10 deletions in WS2, it appears possible that EDN3 and EDNRB mutations also play a role in WS2. Indeed, in a subset of WS4-affected families with EDN3 or EDNRB point mutations, heterozygous relatives present with a diversity of features that may occasionally recall WS2. It therefore appears necessary to undertake a more complete molecular analysis (including not only point mutations but also deletions) of all the genes involved in WS2 or WS4 (i.e., MITF, EDN3, and EDNRB). These comprehensive studies are necessary to fully document the molecular complexity and close relationship that link the different subtypes of WS and to reappraise their current clinical classification.
Acknowledgments
This work was supported by INSERM and Agence Nationale de la Recherche grant ANR-05-MRAR-008-01.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BACPAC Resources, http://bacpac.chori.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers NC_000021.7, NC_000023.9, and NC_000022.9)
- HMMgene, http://www.cbs.dtu.dk/services/HMMgene/ (for gene-structure prediction)
- Netgene2, http://www.cbs.dtu.dk/services/NetGene2/ (for neural-network predictions of splice sites in human DNA)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WS1–WS4) [PubMed]
- Wellcome Trust Sanger Institute, http://www.sanger.ac.uk/
References
- 1.Le Douarin NM, Kalcheim C (1999) The neural crest. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2.Bolande RP (1974) The neurocristopathies: a unifying concept of disease arising in neural crest maldevelopment. Hum Pathol 5:409–429 10.1016/S0046-8177(74)80021-3 [DOI] [PubMed] [Google Scholar]
- 3.Read AP, Newton VE (1997) Waardenburg syndrome. J Med Genet 34:656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amiel J, Lyonnet S (2001) Hirschsprung disease, associated syndromes, and genetics: a review. J Med Genet 38:729–739 10.1136/jmg.38.11.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waardenburg PJ (1951) A new syndrome combining developmental anomalies of the eyelids, eyebrows and nose root with pigmentary defects of the iris and head hair and with congenital deafness. Am J Hum Genet 3:195–253 [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin CT, Hoth CF, Amos JA, da-Silva EO, Milunsky A (1992) An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature 355:637–638 10.1038/355637a0 [DOI] [PubMed] [Google Scholar]
- 7.Tassabehji M, Read AP, Newton VE, Harris R, Balling R, Gruss P, Strachan T (1992) Waardenburg’s syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 355:635–636 10.1038/355635a0 [DOI] [PubMed] [Google Scholar]
- 8.Tassabehji M, Newton VE, Read AP (1994) Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet 8:251–255 10.1038/ng1194-251 [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I (2002) SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet 11:3231–3236 10.1093/hmg/11.25.3231 [DOI] [PubMed] [Google Scholar]
- 10.Shah KN, Dalal SJ, Desai MP, Sheth PN, Joshi NC, Ambani LM (1981) White forelock, pigmentary disorder of irides, and long segment Hirschsprung disease: possible variant of Waardenburg syndrome. J Pediatr 99:432–435 10.1016/S0022-3476(81)80339-3 [DOI] [PubMed] [Google Scholar]
- 11.Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, Chakravarti A (1994) A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell 79:1257–1266 10.1016/0092-8674(94)90016-7 [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti A (1996) Endothelin receptor-mediated signaling in Hirschsprung disease. Hum Mol Genet 5:303–307 [PubMed] [Google Scholar]
- 13.Edery P, Attie T, Amiel J, Pelet A, Eng C, Hofstra RM, Martelli H, Bidaud C, Munnich A, Lyonnet S (1996) Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome). Nat Genet 12:442–444 10.1038/ng0496-442 [DOI] [PubMed] [Google Scholar]
- 14.Hofstra RM, Osinga J, Tan-Sindhunata G, Wu Y, Kamsteeg EJ, Stulp RP, van Ravenswaaij-Arts C, Majoor-Krakauer D, Angrist M, Chakravarti A, et al (1996) A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome). Nat Genet 12:445–447 10.1038/ng0496-445 [DOI] [PubMed] [Google Scholar]
- 15.McCallion AS, Chakravarti A (2001) EDNRB/EDN3 and Hirschsprung disease type II. Pigment Cell Res 14:161–169 10.1034/j.1600-0749.2001.140305.x [DOI] [PubMed] [Google Scholar]
- 16.Pingault V, Bondurand N, Lemort N, Sancandi M, Ceccherini I, Hugot JP, Jouk PS, Goossens M (2001) A heterozygous endothelin 3 mutation in Waardenburg-Hirschsprung disease: is there a dosage effect of EDN3/EDNRB gene mutations on neurocristopathy phenotypes? J Med Genet 38:205–209 10.1136/jmg.38.3.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks AS, Oostra BA, Hofstra RM (2005) Studying the genetics of Hirschsprung’s disease: unraveling an oligogenic disorder. Clin Genet 67:6–14 10.1111/j.1399-0004.2004.00319.x [DOI] [PubMed] [Google Scholar]
- 18.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prehu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, et al (1998) SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet 18:171–173 10.1038/ng0298-171 [DOI] [PubMed] [Google Scholar]
- 19.Kapur RP (1999) Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatr Dev Pathol 2:559–569 10.1007/s100249900162 [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Lo L, Dormand E, Anderson DJ (2003) SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 38:17–31 10.1016/S0896-6273(03)00163-6 [DOI] [PubMed] [Google Scholar]
- 21.Wegner M (1999) From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res 27:1409–1420 10.1093/nar/27.6.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollaaghababa R, Pavan WJ (2003) The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene 22:3024–3034 10.1038/sj.onc.1206442 [DOI] [PubMed] [Google Scholar]
- 23.Hong CS, Saint-Jeannet JP (2005) Sox proteins and neural crest development. Semin Cell Dev Biol 16:694–703 10.1016/j.semcdb.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 24.Wegner M, Stolt CC (2005) From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci 28:583–588 10.1016/j.tins.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 25.Kelsh RN (2006) Sorting out Sox10 functions in neural crest development. Bioessays 28:788–798 10.1002/bies.20445 [DOI] [PubMed] [Google Scholar]
- 26.Foster JW, Dominguez-Steglich MA, Guioli S, Kowk G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525–530 10.1038/372525a0 [DOI] [PubMed] [Google Scholar]
- 27.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79:1111–1120 10.1016/0092-8674(94)90041-8 [DOI] [PubMed] [Google Scholar]
- 28.Bowles J, Schepers G, Koopman P (2000) Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol 227:239–255 10.1006/dbio.2000.9883 [DOI] [PubMed] [Google Scholar]
- 29.Peirano RI, Goerich DE, Riethmacher D, Wegner M (2000) Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol Cell Biol 20:3198–3209 10.1128/MCB.20.9.3198-3209.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Caignec CL, Wegner M, Goossens M (2000) Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 9:1907–1917 10.1093/hmg/9.13.1907 [DOI] [PubMed] [Google Scholar]
- 31.Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA (2000) Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest 106:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR (2000) Direct regulation of the microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem 275:37978–37983 10.1074/jbc.M003816200 [DOI] [PubMed] [Google Scholar]
- 33.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ (2000) Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet 107:1–6 10.1007/s004390050001 [DOI] [PubMed] [Google Scholar]
- 34.Verastegui C, Bille K, Ortonne JP, Ballotti R (2000) Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem 275:30757–30760 10.1074/jbc.C000445200 [DOI] [PubMed] [Google Scholar]
- 35.Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN (2003) Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 130:2809–2818 10.1242/dev.00461 [DOI] [PubMed] [Google Scholar]
- 36.Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ (2004) Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res 17:352–362 10.1111/j.1600-0749.2004.00154.x [DOI] [PubMed] [Google Scholar]
- 37.Ludwig A, Rehberg S, Wegner M (2004) Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett 556:236–244 10.1016/S0014-5793(03)01446-7 [DOI] [PubMed] [Google Scholar]
- 38.Zhu L, Lee HO, Jordan CS, Cantrell VA, Southard-Smith EM, Shin MK (2004) Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat Genet 36:732–737 10.1038/ng1371 [DOI] [PubMed] [Google Scholar]
- 39.Wegner M (2005) Secrets to a healthy Sox life: lessons for melanocytes. Pigment Cell Res 18:74–85 10.1111/j.1600-0749.2005.00218.x [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama S, Takeda K, Shibahara S (2006) SOX10, in combination with Sp1, regulates the endothelin receptor type B gene in human melanocyte lineage cells. FEBS J 273:1805–1820 10.1111/j.1742-4658.2006.05200.x [DOI] [PubMed] [Google Scholar]
- 41.Murisier F, Guichard S, Beermann F (2007) The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res 20:173–184 10.1111/j.1600-0749.2007.00368.x [DOI] [PubMed] [Google Scholar]
- 42.Peirano RI, Wegner M (2000) The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res 28:3047–3055 10.1093/nar/28.16.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M (2001) Human connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet 10:2783–2795 10.1093/hmg/10.24.2783 [DOI] [PubMed] [Google Scholar]
- 44.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M (2001) The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev 15:66–78 10.1101/gad.186601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlierf B, Ludwig A, Klenovsek K, Wegner M (2002) Cooperative binding of Sox10 to DNA: requirements and consequences. Nucleic Acids Res 30:5509–5516 10.1093/nar/gkf690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16:165–170 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Touraine RL, Attie-Bitach T, Pelet A, Auge J, Pingault V, Amiel J, Goossens M, Delezoide AL, Razavi F, Munnich A, et al (1998) Expression of SOX10 in human embryo and fetal brain accounts for a neurological phenotype in Waardenburg type 4 spectrum. Am J Hum Genet 63:A174 [Google Scholar]
- 48.Bondurand N, Kuhlbrodt K, Pingault V, Enderich J, Sajus M, Tommerup N, Warburg M, Hennekam RC, Read AP, Wegner M, et al (1999) A molecular analysis of the Yemenite deaf-blind hypopigmentation syndrome: SOX10 dysfunction causes different neurocristopathies. Hum Mol Genet 8:1785–1789 10.1093/hmg/8.9.1785 [DOI] [PubMed] [Google Scholar]
- 49.Inoue K, Tanabe Y, Lupski JR (1999) Myelin deficiencies in both the central and the peripheral nervous systems associated with a SOX10 mutation. Ann Neurol 46:313–318 [DOI] [PubMed] [Google Scholar]
- 50.Southard-Smith EM, Angrist M, Ellison JS, Agarwala R, Baxevanis AD, Chakravarti A, Pavan WJ (1999) The Sox10Dom mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res 9:215–225 [PubMed] [Google Scholar]
- 51.Pingault V, Guiochon-Mantel A, Bondurand N, Faure C, Lacroix C, Lyonnet S, Goossens M, Landrieu P (2000) Peripheral neuropathy with hypomyelination, chronic intestinal pseudo-obstruction and deafness: a developmental “neural crest syndrome” related to a SOX10 mutation. Ann Neurol 48:671–676 [DOI] [PubMed] [Google Scholar]
- 52.Touraine RL, Attie-Bitach T, Manceau E, Korsch E, Sarda P, Pingault V, Encha-Razavi F, Pelet A, Auge J, Nivelon-Chevallier A, et al (2000) Neurological phenotype in Waardenburg syndrome type 4 correlates with novel SOX10 truncating mutations and expression in developing brain. Am J Hum Genet 66:1496–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sham MH, Lui VC, Chen BL, Fu M, Tam PK (2001) Novel mutations of SOX10 suggest a dominant negative role in Waardenburg-Shah syndrome. J Med Genet 38:E30 10.1136/jmg.38.9.e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue K, Shilo K, Boerkoel CF, Crowe C, Sawady J, Lupski JR, Agamanolis DP (2002) Congenital hypomyelinating neuropathy, central dysmyelination, and Waardenburg-Hirschsprung disease: phenotypes linked by SOX10 mutation. Ann Neurol 52:836–842 10.1002/ana.10404 [DOI] [PubMed] [Google Scholar]
- 55.Pingault V, Girard M, Bondurand N, Dorkins H, Van Maldergem L, Mowat D, Shimotake T, Verma I, Baumann C, Goossens M (2002) SOX10 mutations in chronic intestinal pseudo-obstruction suggest a complex physiopathological mechanism. Hum Genet 111:198–206 10.1007/s00439-002-0765-8 [DOI] [PubMed] [Google Scholar]
- 56.Toki F, Suzuki N, Inoue K, Suzuki M, Hirakata K, Nagai K, Kuroiwa M, Lupski JR, Tsuchida Y (2003) Intestinal aganglionosis associated with the Waardenburg syndrome: report of two cases and review of the literature. Pediatr Surg Int 19:725–728 10.1007/s00383-003-1057-7 [DOI] [PubMed] [Google Scholar]
- 57.Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, et al (2004) Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet 36:361–369 10.1038/ng1322 [DOI] [PubMed] [Google Scholar]
- 58.Verheij JB, Sival DA, van der Hoeven JH, Vos YJ, Meiners LC, Brouwer OF, van Essen AJ (2006) Shah-Waardenburg syndrome and PCWH associated with SOX10 mutations: a case report and review of the literature. Eur J Paediatr Neurol 10:11–17 10.1016/j.ejpn.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 59.Niel F, Martin J, Dastot-Le Moal F, Costes B, Boissier B, Delattre V, Goossens M, Girodon E (2004) Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet 41:e118 10.1136/jmg.2004.022400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M (2007) ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat 28:313–321 10.1002/humu.20452 [DOI] [PubMed] [Google Scholar]
- 61.Yau SC, Bobrow M, Mathew CG, Abbs SJ (1996) Accurate diagnosis of carriers of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet 33:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 10.1073/pnas.83.9.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Girard M, Goossens M (2006) Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett 580:1635–1641 10.1016/j.febslet.2006.02.011 [DOI] [PubMed] [Google Scholar]
- 64.Maka M, Stolt CC, Wegner M (2005) Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev Biol 277:155–169 10.1016/j.ydbio.2004.09.014 [DOI] [PubMed] [Google Scholar]
- 65.Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A (2006) Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet 15:259–271 10.1093/hmg/ddi442 [DOI] [PubMed] [Google Scholar]
- 66.Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G (2003) Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum Mol Genet 12:1439–1447 10.1093/hmg/ddg158 [DOI] [PubMed] [Google Scholar]
- 67.Houlden H, Girard M, Cockerell C, Ingram D, Wood NW, Goossens M, Walker RW, Reilly MM (2004) Connexin 32 promoter P2 mutations: a mechanism of peripheral nerve dysfunction. Ann Neurol 56:730–734 10.1002/ana.20267 [DOI] [PubMed] [Google Scholar]
- 68.Lang D, Epstein JA (2003) Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet 12:937–945 10.1093/hmg/ddg107 [DOI] [PubMed] [Google Scholar]
- 69.Morgan NV, Westaway SK, Morton JE, Gregory A, Gissen P, Sonek S, Cangul H, Coryell J, Canham N, Nardocci N, et al (2006) PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat Genet 38:752–754 10.1038/ng1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shahin H, Walsh T, Sobe T, Abu Sa’ed J, Abu Rayan A, Lynch ED, Lee MK, Avraham KB, King MC, Kanaan M (2006) Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet 78:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN (2000) Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet 67:1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, Denison JC, Gregory MC, White JG, Barker DF, et al (2001) Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet 69:1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phelan MC, Rogers RC, Saul RA, Stapleton GA, Sweet K, McDermid H, Shaw SR, Claytor J, Willis J, Kelly DP (2001) 22q13 deletion syndrome. Am J Med Genet 101:91–99 [DOI] [PubMed] [Google Scholar]
- 74.Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O (2006) Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet 43:822–828 10.1136/jmg.2005.038604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Attie T, Pelet A, Edery P, Eng C, Mulligan LM, Amiel J, Boutrand L, Beldjord C, Nihoul-Fekete C, Munnich A, et al (1995) Diversity of RET proto-oncogene mutations in familial and sporadic Hirschsprung disease. Hum Mol Genet 4:1381–1386 10.1093/hmg/4.8.1381 [DOI] [PubMed] [Google Scholar]
- 76.de Pontual L, Pelet A, Clement-Ziza M, Trochet D, Antonarakis SE, Attie-Bitach T, Beales PL, Blouin JL, Dastot-Le Moal F, Dollfus H, et al (2007) Epistatic interactions with a common hypomorphic RET allele in syndromic Hirschsprung disease. Hum Mutat 28:790–796 10.1002/humu.20517 [DOI] [PubMed] [Google Scholar]
- 77.Wong WT, Agron E, Coleman HR, Reed GF, Csaky K, Peterson J, Glenn G, Linehan WM, Albert P, Chew EY (2007) Genotype-phenotype correlation in von Hippel-Lindau disease with retinal angiomatosis. Arch Ophthalmol 125:239–245 10.1001/archopht.125.2.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kellerer S, Schreiner S, Stolt CC, Scholz S, Bosl MR, Wegner M (2006) Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development 133:2875–2886 10.1242/dev.02477 [DOI] [PubMed] [Google Scholar]