Abstract
We previously discovered that mutations of the human FOXP2 gene cause a monogenic communication disorder, primarily characterized by difficulties in learning to make coordinated sequences of articulatory gestures that underlie speech. Affected people have deficits in expressive and receptive linguistic processing and display structural and/or functional abnormalities in cortical and subcortical brain regions. FOXP2 provides a unique window into neural processes involved in speech and language. In particular, its role as a transcription factor gene offers powerful functional genomic routes for dissecting critical neurogenetic mechanisms. Here, we employ chromatin immunoprecipitation coupled with promoter microarrays (ChIP-chip) to successfully identify genomic sites that are directly bound by FOXP2 protein in native chromatin of human neuron-like cells. We focus on a subset of downstream targets identified by this approach, showing that altered FOXP2 levels yield significant changes in expression in our cell-based models and that FOXP2 binds in a specific manner to consensus sites within the relevant promoters. Moreover, we demonstrate significant quantitative differences in target expression in embryonic brains of mutant mice, mediated by specific in vivo Foxp2-chromatin interactions. This work represents the first identification and in vivo verification of neural targets regulated by FOXP2. Our data indicate that FOXP2 has dual functionality, acting to either repress or activate gene expression at occupied promoters. The identified targets suggest roles in modulating synaptic plasticity, neurodevelopment, neurotransmission, and axon guidance and represent novel entry points into in vivo pathways that may be disturbed in speech and language disorders.
Neurodevelopmental disorders that disrupt language acquisition tend to be complex at the genetic level, potentially involving a large number of different susceptibility loci, such that identification of the relevant molecular pathways remains challenging.1,2 In earlier studies, we discovered that heterozygous mutations of the human FOXP2 gene (MIM 605317) are responsible for a rare monogenic communication disorder, primarily characterized by difficulties in learning to make the coordinated sequences of articulatory gestures that underlie speech (developmental verbal dyspraxia [MIM 602081]).3,4 The disorder also involves deficits in many aspects of linguistic processing, affecting both oral and written abilities, across expressive and receptive domains.5 To date, speech and language impairments have been documented in two different multigenerational families segregating missense and nonsense point mutations of FOXP2,3,4 as well as in several cases of gross chromosomal rearrangements (translocations and deletions) that disturb the integrity of the FOXP2 genomic locus in 7q31.3,6–8 People who are affected with Silver-Russell syndrome (MIM 180860), associated with uniparental disomy of the maternal copy of chromosome 7, can also display verbal dyspraxia, which appears to relate to reductions in FOXP2 expression.6
FOXP2 encodes a regulatory protein belonging to the forkhead-box (FOX) group of transcription factors.3 Members of this class of protein share a distinctive type of DNA-binding motif, the FOX domain, and are prominent regulators of eukaryotic gene expression, associated with a wide variety of cellular and developmental processes.9 FOX gene dysfunction has been implicated in a range of disease states, including developmental eye disorders, ovarian failure, immune deficiency, and carcinogenesis.10,11 Several FOX transcription factors are key players in CNS development; for example, Foxg1 regulates proliferation and differentiation of progenitor cells of the telencephalon,12 whereas Foxb1 is critical for normal development of diencephalon and midbrain.13 FOXP2 itself belongs to a functionally divergent subgroup of the FOX proteins, characterized by a shorter DNA-binding domain and the presence of other defining motifs, including glutamine-rich stretches, dimerization domains, and an acidic C-terminus.14
Much of our current knowledge of the neural correlates of FOXP2 disruption comes from intensive phenotypic studies of a single human family (the “KE” family) in which 15 people have verbal dyspraxia due to a missense mutation in the FOX domain.3 The mutation in this family is associated with bilateral abnormalities in gray-matter density, including significant decreases in the inferior frontal gyrus (including Broca’s area), caudate nucleus, and cerebellum and increases in the posterior temporal gyrus (including Wernicke’s area), angular gyrus, and putamen,15 as well as altered patterns of neural activity during linguistic processing.16 Intriguingly, the neural sites of structural and/or functional abnormalities in the KE family are concordant with regions of high FOXP2 expression in the developing human brain.17 We recently used human cell lines to demonstrate that the KE family's missense mutation and a nonsense mutation causing verbal dyspraxia in a second multiplex family4,18 dramatically interfered with transcription factor function.18
Overall, the combined findings from phenotypic evaluation, neuroimaging studies, expression analyses, and functional genetic assays suggest that a reduced dosage of functional FOXP2 has an impact on the development and function of a subset of distributed neural circuits, including those important for speech and language acquisition. Thus, the FOXP2 gene provides a unique molecular window into the neural basis of human communication.19 In particular, its role as a transcription factor, modulating the expression of target genes, offers elegant functional genomic routes for dissecting the associated neurogenetic pathways. However, at present, there are no neural targets of FOXP2 reported in the literature.
The aim of the present study was to discover downstream targets directly regulated by FOXP2 in neurons, by exploiting emerging strategies based on the chromatin-immunoprecipitation (ChIP) method. This is a powerful technique for studying protein-DNA interactions inside the nucleus under physiological conditions,20 allowing characterization of genomic sites bound by a protein of interest in the native chromatin of living cells. Here, we develop FOXP2 ChIP, couple it to high-throughput screening of microarrays (ChIP-chip), and identify occupied promoters in native chromatin of human neuron-like cells. We focus on a subset of targets uncovered via this approach, demonstrating that altered FOXP2 levels yield significant changes in their expression and that FOXP2 binds in a specific manner to consensus sites within the relevant promoters. Finally, we identify significant quantitative differences in target expression in the embryonic brains of mutant mice, mediated by specific in vivo Foxp2-chromatin interactions. This work, along with that of Spiteri et al.,21(in this issue) represents the first identification and validation of neural targets regulated by FOXP2 and suggests roles for this gene in modulating synaptic plasticity, neurodevelopment, neurotransmission, and axon guidance.
Material and Methods
Cell Culture and Reagents
SH-SY5Y cells were grown in Dulbecco's modified Eagle medium (DMEM):F12 media (Sigma), and HEK293T cells in DMEM media (Sigma). Media was supplemented with 10% fetal calf serum (Sigma), 2 mM l-glutamine (Sigma), and 2 mM penicillin/streptomycin (Sigma). Cells were grown at 37°C in the presence of 5% CO2. Stable SH-SY5Y cell lines overexpressing FOXP2 or nonexpressing controls were generated by transfection with pcDNA3.1/FOXP2 (isoform I–untagged) or the empty vector, respectively, by use of Lipofectamine Plus (Invitrogen) in accordance with the manufacturer's instructions. Cells were cultured in complete medium supplemented with 500 μg/ml G418 (Calbiochem) as a selective agent. Resistant single colonies were isolated 20 d after transfection and then were cultured and expanded independently in the presence of G418 (500 μg/ml). Quantitative RT-PCR (qRT-PCR) (see “qRT-PCR” section) and western blotting (performed as described elsewhere18) confirmed expression of recombinant FOXP2. A clone with a high and consistent level of expression was chosen for use in further experiments. Transient transfections of SH-SY5Y or HEK293T cells were performed using Transfast (Promega) or GeneJuice (Novagen) transfection reagents, respectively, in accordance with the manufacturer's instructions and were harvested 48 h after transfection. FOXP2 detection was performed using N-terminal (Santa Cruz Biotechnology) or C-terminal (Serotec) goat polyclonal antibodies.18
ChIP
SH-SY5Y cells stably expressing FOXP2 isoform I were cross-linked using 1% formaldehyde in cross-linking buffer (50 mM HEPES, 100 mM NaCl, 1 mM EDTA, and 0.5 mM ethylene glycol tetraacetic acid [EGTA]) at room temperature. Cells were incubated for 10 min in ice-cold ChIP lysis buffer (10 mM Tris, 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, and protease inhibitors) and were centrifuged at 10,000 g at 4°C for 5 min to pellet nuclei. Nuclei from 3×107 cells were resuspended in 1 ml Sonication Buffer (10 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitors) with 0.1 g of 212–300-μm glass beads (Sigma) before undergoing 10 rounds of 20-s sonication pulses at 65% power, with 2 min on ice between each round (with use of Bandelin SONOPULS HD2070 Ultrasonic Homogenisor and MS72 2-mm titanium tip with 200-μm SS amplitude). Cells were centrifuged at 10,000 g at 4°C for 5 min to remove glass beads and cell debris. Then, 1 μg of FOXP2 N-terminal antibody (Santa Cruz Biotechnology) was incubated with the sonicated supernatants in IP buffer (0.1 M Tris, 10 mM EDTA, 150 mM NaCl, 0.2 % Triton X-100, 1% phenylmethylsulfonyl fluoride, and protease inhibitors), rotating overnight at 4°C. Immune complexes were captured by addition of 5 μg sonicated salmon sperm DNA and 50 μl Protein G–sepharose beads, incubated for 3 h at 4°C. Protein was eluted from beads first by 1.5% SDS buffer (1.5% SDS, 1× TE [pH 7.5], and 30 mM NaCl) and then by 0.5% SDS buffer (0.5% SDS, 1× TE [pH 7.5], and 30 mM NaCl), with incubation of the beads with each in turn at room temperature for 15 min. Pooled supernatants were incubated at 65°C overnight to reverse cross-links. DNA was isolated via phenol-chloroform extraction followed by ethanol precipitation. Concentration and purity of the DNA was evaluated by spectrophotometry, and size was assessed via gel electrophoresis. Protein samples were extracted in parallel via precipitation with use of trichloroacetic acid (Sigma), and western blotting was used to confirm immunoprecipitation of the FOXP2 protein.
In vivo Foxp2 ChIP with use of embryonic brains from wild-type or homozygous mutant mice was performed according to the protocol described by Spiteri et al.21(in this issue) In each case, whole-brain tissue at embryonic day 16 (E16) was pooled from six mice. Mutant mice carry an early nonsense mutation in Foxp2, which leads to both nonsense-mediated RNA decay and protein instability, resulting in an absence of detectable Foxp2 protein, as confirmed using both N- and C-terminal antibodies (M.G., J.N., and S.E.F., unpublished data). Wild-type and mutant mice were littermates, to maximize the homogeneity of the genomic background. Despite a lack of Foxp2 protein, homozygous mutants show no gross anomalies in anatomy or brain development during embryogenesis. Postnatally, they display developmental delays and reduced cerebellar growth, dying ∼3–4 wk after birth for as-yet unknown reasons (M.G., J.N., and S.E.F., unpublished data). All animal studies were performed conforming to the regulatory standards of the U.K. Home Office, under Project Licence 30/2016.
Ligation-Mediated PCR Amplification of ChIP Products
Purified chromatin was amplified via ligation-mediated PCR in accordance with published protocols.22 Size and purity of DNA was assessed via spectrophotometry and gel electrophoresis.
Hybridization of Human FOXP2 ChIP Products to Promoter Microarrays
Two hundred nanograms of amplified immunoprecipitated chromatin or total input DNA was fluorescently labeled with Cy5 or Cy3, respectively, by use of random primers provided in the BioPrime DNA labeling system (Invitrogen), in accordance with the manufacturer's instructions. The labeling reaction was allowed to proceed for 16 h at 37°C, before purification by sodium-acetate precipitation. A total of 2 μg of labeled DNA was hybridized to high-density human promoter arrays (Aviva Biosystems).23 Three biological replicates were performed.
Microarray Data Analysis
Array images were scanned using the Axon GenePix 4000B. Data were retrieved, and initial quality control was performed using the GenePix Pro 6.0 software package (Molecular Devices). Microarray data analysis was performed using the Limma package for R.24,25 Print-tip loess normalization and background correction was performed within each array. Data were normalized between arrays by use of quantile normalization, and the median value was calculated from triplicate experiments for each probe, for use in further analyses. Probes that displayed statistically significant differences of abundance (P<.05) were ranked according to both fold change and P value. The P values were adjusted for multiple testing by use of the false-discovery–rate method in the p.adjust package in R.26 All microarray data can be found in the tab-delimited ASCII files of data sets 1, 2, 3, 4, and 5.
Functional Classification of Genes
The GOTree Machine (GOTM),27 part of WebGestalt (Web-based gene set analysis toolkit), was used to visualize gene-function relationships. This program queries the Genekey database incorporating the Locuslink, Ensembl, Swiss-Prot, HomoloGene, Unigene, Gene Ontology Consortium, and Affymetrix databases. Statistical significance of overrepresentation in the target gene list of 303 genes was calculated using the entire probe set as a reference data set, via a hypergeometric test, where significance is defined as P<.05. Functional annotation was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID).28
Pathway Network Analysis
Ingenuity pathway analysis software was used to identify interactions between target genes (Ingenuity Systems). All 303 enriched genes with a P value <.05 (table 1) were included in this analysis, and both direct and indirect interactions were considered. The full set of genes from the array was used as a reference data set.
Table 1. .
Genes Displaying Significant Enrichment (P<.05) in FOXP2 High-Throughput Location Analysis
| Gene | GenBank Accession Number | P |
| ABCG2 | NM_004827 | .027 |
| ACSL5 | NM_016234 | .0063 |
| ACSS2 | NM_018677 | .017 |
| ADAM28 | NM_021777 | .022 |
| ADMR | NM_007264 | .036 |
| AKAP6 | NM_004274 | .031 |
| ALDOA | NM_000034 | .0015 |
| ARL1 | NM_001177 | .026 |
| ARL4A | NM_005738 | .050 |
| ATF6 | NM_007348 | .0075 |
| ATP1A2 | NM_000702 | .040 |
| ATP1B4 | NM_012069 | .044 |
| BAIAP3 | NM_003933 | .019 |
| BUD31 | NM_003910 | .025 |
| C13orf24 | NM_006346 | .021 |
| C1orf38 | NM_004848 | .032 |
| C20orf24 | NM_018840 | .037 |
| CABP1 | NM_031205 | .037 |
| CACNG3 | NM_006539 | .0053 |
| CALCRL | NM_005795 | .015 |
| CCK | NM_000729 | .045 |
| CCKAR | NM_000730 | .035 |
| CCL19 | NM_006274 | .0088 |
| CCNK | NM_003858 | .012 |
| CD164 | NM_006016 | .014 |
| CD180 | NM_005582 | .0042 |
| CD5 | NM_014207 | .015 |
| CDH5 | NM_001795 | .0025 |
| CER1 | NM_005454 | .0019 |
| CGB | NM_033142 | .029 |
| CHST11 | NM_018413 | .035 |
| CILP | NM_003613 | .026 |
| CLEC10A | NM_182906 | .039 |
| CLNS1A | NM_001293 | .035 |
| CLPX | NM_006660 | .049 |
| CNBP | NM_003418 | .041 |
| COL8A1 | NM_001850 | .0060 |
| COL9A1 | NM_001851 | .047 |
| COPS5 | NM_006837 | .048 |
| COX11 | NM_004375 | .000099 |
| CRISP3 | NM_006061 | .012 |
| CRYBA4 | NM_001886 | .027 |
| CXCL2 | NM_002089 | .011 |
| CYB5B | NM_030579 | .032 |
| DCTN2 | NM_006400 | .010 |
| DGAT1 | NM_012079 | .0028 |
| DLG4 | NM_001365 | .027 |
| DLL3 | NM_016941 | .025 |
| DPAGT1 | NM_001382 | .034 |
| DUSP12 | NM_007240 | .020 |
| DYRK1B | NM_004714 | .0062 |
| EBI2 | NM_004951 | .00062 |
| EBP | NM_006579 | .016 |
| EIF3S10 | NM_003750 | .0019 |
| ELL2 | NM_012081 | .022 |
| ENTPD7 | NM_020354 | .042 |
| EPHX2 | NM_001979 | .013 |
| EPOR | NM_000121 | .012 |
| ERO1L | NM_014584 | .000051 |
| F8 | NM_019863 | .046 |
| FADD | NM_003824 | .014 |
| FBN1 | NM_000138 | .0051 |
| FLT1 | NM_002019 | .00037 |
| FMO4 | NM_002022 | .0038 |
| FOLR1 | NM_016725 | .038 |
| FRY | NM_023037 | .015 |
| FTH1 | NM_002032 | .010 |
| FTSJ2 | NM_013393 | .0055 |
| FUT2 | NM_000511 | .023 |
| GABBR1 | NM_001470 | .044 |
| GAS7 | NM_005890 | .048 |
| GBAS | NM_001483 | .041 |
| GDF5 | NM_000557 | .036 |
| GENX-3414 | NM_003943 | .032 |
| GGH | NM_003878 | .049 |
| GNAZ | NM_002073 | .0045 |
| GPR160 | NM_014373 | .031 |
| GPR17 | NM_005291 | .047 |
| GPR75 | NM_006794 | .019 |
| GRHPR | NM_012203 | .038 |
| GUCA1B | NM_002098 | .049 |
| HAPLN1 | NM_001884 | .0028 |
| HAS1 | NM_001523 | .0019 |
| HAT | NM_004262 | .014 |
| HIST1H2AG | NM_021064 | .021 |
| HNRPK | NM_002140 | .031 |
| HOXB6 | NM_156036 | .0023 |
| HRSP12 | NM_005836 | .0000062 |
| HSPA2 | NM_021979 | .018 |
| HSPB7 | NM_014424 | .0022 |
| HTRA2 | NM_012103 | .050 |
| HUWE1 | NM_031407 | .042 |
| HYAL2 | NM_003773 | .012 |
| IFI30 | NM_006332 | .00024 |
| IGLL1 | NM_020070 | .017 |
| IL18 | NM_001562 | .034 |
| IL1B | NM_000576 | .042 |
| IL4R | NM_000418 | .035 |
| ISLR | NM_005545 | .010 |
| ITPK1 | NM_014216 | .0071 |
| KCNB1 | NM_004975 | .041 |
| KCNE1L | NM_012282 | .0079 |
| Kifap3 | NM_014970 | .047 |
| KLK8 | NM_007196 | .035 |
| KRT17 | NM_000422 | .031 |
| LBR | NM_002296 | .0036 |
| LDHA | NM_005566 | .013 |
| LECT1 | NM_007015 | .0054 |
| LENEP | NM_018655 | .0085 |
| LILRA2 | NM_006866 | .043 |
| LILRB5 | NM_006840 | .043 |
| LILRP2 | NR_003061 | .017 |
| LIM2 | NM_030657 | .044 |
| LNPEP | NM_005575 | .00012 |
| LTB | NM_002341 | .012 |
| LTF | NM_002343 | .020 |
| LY6G6E | NM_024123 | .035 |
| LYPLA1 | NM_006330 | .033 |
| MAD2L2 | NM_006341 | .030 |
| MAEA | NM_005882 | .021 |
| MAPK14 | NM_001315 | .046 |
| MAPK7 | NM_139032 | .0042 |
| MAPK8IP1 | NM_005456 | .0021 |
| MAPRE3 | NM_012326 | .036 |
| MARK2 | NM_004954 | .019 |
| MDFI | NM_005586 | .023 |
| MEST | NM_002402 | .019 |
| MFGE8 | NM_005928 | .045 |
| MORF4L2 | NM_012286 | .027 |
| MOS | NM_005372 | .044 |
| MPO | NM_000250 | .026 |
| MPP3 | NM_001932 | .034 |
| MPV17 | NM_002437 | .043 |
| MYOT | NM_006790 | .0057 |
| NCOR2 | NM_006312 | .0013 |
| NDUFA2 | NM_002488 | .046 |
| NDUFA8 | NM_014222 | .030 |
| NEDD8 | NM_006156 | .039 |
| NEU2 | NM_005383 | .020 |
| NEUROD2 | NM_006160 | .010 |
| NEUROG1 | NM_006161 | .018 |
| NRTN | NM_004558 | .045 |
| NRXN3 | NM_004796 | .016 |
| NUDT1 | NM_002452 | .011 |
| NXF1 | NM_006362 | .019 |
| OPN1SW | NM_001708 | .0046 |
| ORC6L | NM_014321 | .025 |
| OXR1 | NM_018002 | .037 |
| P115 | NM_003715 | .043 |
| PAM | NM_000919 | .0084 |
| PAX1 | NM_006192 | .0013 |
| PAX3 | NM_000438 | .0084 |
| PCCA | NM_000282 | .015 |
| PCSK1 | NM_000439 | .029 |
| PCSK6 | NM_002570 | .013 |
| PCYT1B | NM_004845 | .042 |
| PDCD8 | NM_004208 | .026 |
| PDE1C | NM_005020 | .030 |
| PDE4C | NM_000923 | .045 |
| PDE6B | NM_000283 | .012 |
| PEX1 | NM_000466 | .013 |
| PEX16 | NM_057174 | .034 |
| PIGC | NM_153747 | .014 |
| PKP1 | NM_000299 | .036 |
| PLA2G4B | NM_005090 | .046 |
| PLA2R1 | NM_007366 | .049 |
| PLAUR | NM_002659 | .012 |
| PM5 | NM_014287 | .018 |
| PMF1 | NM_007221 | .024 |
| PNKP | NM_007254 | .030 |
| POLS | NM_006999 | .017 |
| POU4F3 | NM_002700 | .032 |
| PPP2R3A | NM_002718 | .0098 |
| PPP2R5D | NM_006245 | .037 |
| PRKAG3 | NM_017431 | .046 |
| PRSC | NM_006587 | .031 |
| PRSS12 | NM_003619 | .023 |
| PRSS22 | NM_022119 | .018 |
| PRSS8 | NM_002773 | .037 |
| PSEN2 | NM_000447 | .013 |
| PSMA3 | NM_002788 | .017 |
| PSMD1 | NM_002807 | .018 |
| PTCH2 | NM_003738 | .026 |
| PTGER1 | NM_000955 | .0026 |
| PTGER3 | NM_000957 | .021 |
| PTK9 | NM_198974 | .0091 |
| PYCR1 | NM_153824 | .012 |
| RAB18 | NM_021252 | .013 |
| RAB27A | NM_004580 | .035 |
| RAB5C | NM_004583 | .021 |
| RABGGTA | NM_004581 | .016 |
| RAD51AP1 | NM_006479 | .014 |
| RAI1 | NM_030665 | .015 |
| RALA | NM_005402 | .0031 |
| RALBP1 | NM_006788 | .0015 |
| RARB | NM_000965 | .040 |
| RBM4 | NM_002896 | .039 |
| RBP2 | NM_004164 | .046 |
| RCN2 | NM_002902 | .0051 |
| RECQL5 | NM_004259 | .033 |
| RFPL3 | NM_006604 | .010 |
| RGN | NM_004683 | .041 |
| RGS2 | NM_002923 | .047 |
| ROR2 | NM_004560 | .033 |
| RPL10 | NM_006013 | .023 |
| RPL22 | NM_000983 | .0040 |
| RPL28 | NM_000991 | .0098 |
| RQCD1 | NM_005444 | .035 |
| RRAGB | NM_016656 | .048 |
| RRP9 | NM_004704 | .011 |
| RYR3 | NM_001036 | .021 |
| S100A3 | NM_002960 | .028 |
| SAS10 | NM_020368 | .050 |
| SCGB2A2 | NM_002411 | .030 |
| SCRG1 | NM_007281 | .028 |
| SCRN1 | NM_014766 | .033 |
| SELENBP1 | NM_003944 | .030 |
| SEMA3B | NM_004636 | .0071 |
| SFRP4 | NM_003014 | .027 |
| SFRS11 | NM_004768 | .047 |
| SIRT3 | NM_012239 | .0053 |
| SLBP | NM_006527 | .032 |
| SLC17A3 | NM_006632 | .0093 |
| SLC20A1 | NM_005415 | .0047 |
| SLC22A14 | NM_004803 | .0099 |
| SLC22A3 | NM_021977 | .042 |
| SLC25A3 | NM_002635 | .0041 |
| SLC2A4 | NM_001042 | .0058 |
| SLC4A4 | NM_003759 | .036 |
| SLC4A8 | NM_001039960 | .0028 |
| SLC5A1 | NM_000343 | .025 |
| SLC5A6 | NM_021095 | .026 |
| SLIT1 | NM_003061 | .025 |
| SMAD3 | NM_005902 | .020 |
| SMARCB1 | NM_003073 | .0042 |
| SMC2 | NM_006444 | .0022 |
| SMCX | NM_004187 | .044 |
| SMS | NM_004595 | .038 |
| SNFT | NM_018664 | .00025 |
| SNRPG | NM_003096 | .020 |
| SNW1 | NM_012245 | .016 |
| SOCS1 | NM_003745 | .050 |
| SOD3 | NM_003102 | .00046 |
| SORBS3 | NM_005775 | .0047 |
| SOX15 | NM_006942 | .023 |
| SOX21 | NM_007084 | .031 |
| SPEG | XM_001131579 | .0024 |
| SPOCK1 | NM_004598 | .011 |
| SPOCK3 | NM_016950 | .018 |
| SRD5A2 | NM_000348 | .029 |
| SRPK1 | NM_003137 | .018 |
| SSX2IP | NM_014021 | .012 |
| ST3GAL5 | NM_003896 | .012 |
| STC1 | NM_003155 | .0098 |
| STC2 | NM_003714 | .042 |
| SYK | NM_003177 | .0022 |
| SYNJ1 | NM_203446 | .031 |
| TACSTD2 | NM_002353 | .020 |
| TAGLN | NM_003186 | .0034 |
| TCEB1 | NM_005648 | .020 |
| TCF12 | NM_207037 | .0020 |
| TDO2 | NM_005651 | .0046 |
| TG | NM_003235 | .0088 |
| TGM2 | NM_004613 | .014 |
| THBS1 | NM_003246 | .041 |
| THPO | NM_000460 | .028 |
| TIAF1 | NM_004740 | .033 |
| TIAM1 | NM_003253 | .0013 |
| TITF1 | NM_003317 | .024 |
| TJP2 | NM_004817 | .026 |
| TLL2 | NM_012465 | .0015 |
| TMEM4 | NM_014255 | .043 |
| TNFRSF14 | NM_003820 | .032 |
| TNFSF9 | NM_003811 | .038 |
| TNNI1 | NM_003281 | .0038 |
| TNS1 | NM_022648 | .034 |
| TOPBP1 | NM_007027 | .022 |
| TP53AP1 | NM_007233 | .0051 |
| TPM2 | NM_003289 | .013 |
| TPO | NM_000547 | .0064 |
| TPP2 | NM_003291 | .0028 |
| TPSD1 | NM_012217 | .032 |
| TRAF1 | NM_005658 | .0057 |
| TREH | NM_007180 | .030 |
| TRIM21 | NM_003141 | .043 |
| TRIP13 | NM_004237 | .026 |
| TRPA1 | NM_007332 | .0057 |
| TUBB2B | NM_178012 | .0057 |
| TXNIP | NM_006472 | .043 |
| UBE2D2 | NM_181838 | .045 |
| UBE2G1 | NM_003342 | .036 |
| UBN1 | NM_016936 | .030 |
| VAMP2 | NM_014232 | .0074 |
| VASP | NM_003370 | .049 |
| VLDLR | NM_003383 | .026 |
| YRDC | NM_024640 | .038 |
| ZFP95 | NM_014569 | .039 |
| ZNF259 | NM_003904 | .036 |
| ZNF331 | NM_018555 | .030 |
| ZNF384 | NM_133476 | .032 |
| ZNF74 | NM_003426 | .030 |
| ZNF76 | NM_003427 | .039 |
Motif Analysis
The top 100 statistically significant probe sequences were assessed for the presence of known FOX family binding sites by use of the Emboss FUZZNUC nucleic acid–pattern search tool. Predicted sites for FOXP2 binding were based on previously published consensus binding sites for FOX family members or recently reported sequences bound by FOXP2. Sites were defined as follows: for FOXP2, (A)ATTTG(T) (i.e., AATTTG or ATTTGT)29,30; for FOXP, TATTTRT14; and, for FOX, TRTTKRY,31 where R = A or G, K = T or G, and Y = T or C. When a site fell into two classes of motifs, only the most specific level of classification was used—that is, sites were preferentially categorized as FOXP2, FOXP, or FOX consensus sites, in that order. Significance was calculated using χ2 tests comparing frequencies to counts of predicted sites in permuted probe sequences from the top 100 statistically significant probe sequences. When potential sites for hetero- or homodimerization were considered, only nonoverlapping sites conforming to the exact consensus that lay within 100 bp of each other were included.
Sequences were assessed for the presence of overrepresented motifs by use of the Multiple Em for Motif Elucidation (MEME) and/or Motif Alignment and Search Tool (MAST) programs.32,33 Top-scoring matrices were investigated for matches to any known binding-site matrices by use of the TRANSFAC database. Sequences were investigated for coincident motifs that could potentially interact with the function of FOX family binding sites. A total of 542 binding-site matrices from TRANSFAC were used to query the enriched probe sequence set (303 genes) (table 1), as well as the set of sequences from the whole array as a reference data set. Statistical significance (P<.05) was assessed using Student's t test, to determine the overrepresentation of the binding sites present in the enriched probe data set compared with the whole array data set.
qRT-PCR
RNA was extracted from cells or tissue harvested in TRIzol reagent by use of the QIAGEN RNeasy kit. Human cell-based experiments exploited SH-SY5Y cells transfected with either FOXP2 or empty control vectors. For stable transfectants, multiple independent passages of a single clone were used (see “Cell Culture and Reagents” section), whereas transient transfectants involved separately transfected clones. For the latter, cells were harvested 48 h after transfection for RT-PCR analyses. In vivo mouse experiments used dissected brain tissue from E16 embryos, including mutant mice lacking Foxp2 protein and wild-type littermate controls. Reverse transcription was performed with Superscript III reverse transcriptase (Invitrogen) in accordance with the manufacturer's instructions. In brief, 1 μg RNA was primed via incubation with 100 ng of random primers and 0.8 mM deoxynucleotide triphosphates (dNTPs) at 65°C for 5 min and then on ice for 1 min. Reverse transcription was mediated via the addition of 200 U of Superscript III Reverse Transcriptase (Invitrogen) and 20 U of Superase-In RNase Inhibitor (Ambion), in the presence of first-strand buffer (Invitrogen).
Primers specific for candidate genes and for the control housekeeping gene GAPDH/Gapdh (glyceraldehyde 3-phosphate dehydrogenase) were designed using PrimerBank.34 Human and mouse primers were as given in table 2. PCRs used SYBR Green supermix (Bio-Rad), including 0.2 μM each of forward and reverse primers and 1 μl of cDNA template prepared as described above. Quantitative PCRs were performed on the iQ5 thermal cycler real-time PCR detection system (Bio-Rad) in accordance with the manufacturer's instructions. Reaction conditions were (1) 95°C for 3 min, (2) 95°C for 15 s, (3) 60°C for 30 s, and (4) 72°C for 30 s, then (5) repeat from step 2 for 49 cycles, followed by (6) 95°C for 1 min, and (7) 55°C for 30 s. Melting-curve analysis was performed to assess the specificity of the amplification. Data analysis was performed using iCycler software (Bio-Rad). Quantification was calculated using the comparative CT method.35 Fold changes are reported in response to FOXP2 expression (transient or stable) compared with cells transfected with an empty vector, following normalization to an internal control, the GAPDH housekeeping gene. Reported fold changes for in vitro experiments are the mean of comparisons between seven (stable) or six (transient) cDNA preparations. In vivo fold changes are reported as the mean of comparisons between cDNA preparations from five wild-type and five knockout littermates. Data are expressed as mean±SEM. Statistical significance was assessed using unpaired t tests (two-tailed).
Table 2. .
qRT-PCR Primers
| Primer(5′→3′) |
||
| Gene | Forward | Reverse |
| SLC17A3 | GCCCTCGTCTTACATTTCTGC | AGGAATCATTGAGCTGGGATTG |
| CALCRL | AAGACCCCATTCAACAAGCAG | CCAGTTTCCATCTTGGTCACAG |
| LNPEP | TTCACCAATGATCGGCTTCAG | CTCCATCTCATGCTCACCAAG |
| HSPB7 | GAGCATGTTTTCCGATGACTTTG | GGTGACAATGATGTCTTCAGGTG |
| PSEN2 | GAGGATGGAGAGAACACTGCC | CCACTACAGACATAGCGGTCAG |
| COX11 | GCGTTCCTTTCTGTGGCTG | CCACCTCAGTCCTCTCTCG |
| PM5 | GGTGGCTTCGTCAAGTCGG | GGGCACAGTCTGTCTGGTAT |
| RCN2 | TTGGACTCAGATGGCTTTCTCA | TCCCAAGTCACAGTATCATCACT |
| CD164 | CCCTCCCCTTCTACAACTTCC | TGAGGTTGGAGTCACAGTGTTAT |
| CER1 | CCTGCCTCTAGGAAAGACCAC | TGGCACTGCGACAAACAGA |
| SLC22A14 | GTTTGCTGACCACTTCGTGTT | CCATTGGGTGCTTGGGGTAT |
| ERO1L | GGCTGGGGATTCTTGTTTGG | AGTAACCACTAACCTGGCAGA |
| MAPK8IP1 | TGTGCGACTAGAGGCCACT | AGGGTCTGGATCGGAGCTG |
| SYK | TGGTCAGCGGGTGGAATAATC | GGGCTCTCGTACACCTCTG |
| GAPDH | CAGTCCATGCCATCACTGC | TTCGTTGTCATACCAGGAAATG |
| Slc17a3a | CCTCAATAGCTCTACTGAACAGC | AGGACTCCAGTTATACACAGGG |
| Cer1a | ATCACCTCTACTGGAGGAAGC | GGTCTCCCAGTGTACTTCGTG |
| Foxp2a | AAGCAGCTTGCCTTTGCTAAG | GGATTGAATGTATGTGTGGCTGA |
| Gapdha | TGACGTGCCGCCTGGAGAAAC | CCGGCATCGAAGGTGGAAGAGT |
Mouse gene.
qRT-PCR experiments with Slc17a3 in wild-type versus mutant mice indicated loss of repression in E16 brains and were followed up with independent replications in separate litters by use of standard RT-PCR. Samples were prepared and reactions performed in the same manner as qRT-PCR experiments, except that PCR cycle number was varied depending on primers: 30 cycles for Foxp2 and Gapdh and 40 cycles for Slc17a3. In this case, PCR products were run on 2% agarose gels in the presence of ethidium bromide to allow visualization of DNA.
Electrophoretic Mobility Shift Assays (EMSAs) for DNA Binding
HEK293 nuclear extracts (transfected with FOXP2 or an empty vector control) and purified FOXP2 protein (ΔQ-rich: amino acids 262–715) expressed in a bacterial system were prepared as described in our earlier study.18 HEK293 cells yield higher FOXP2 overexpression than do SH-SY5Y cells, facilitating clean EMSAs. Probes were designed as 20–28-nt oligomers. A consensus probe, based on a previously described FOXP1 consensus binding sequence, was used as a positive control.14 Oligonucleotide labeling and binding reactions were performed as described elsewhere.18 When an unlabeled competitor probe was used to confirm specificity of DNA binding, it was added in 5-fold or 10-fold excess and was incubated at room temperature for 15 min before addition of labeled probe. Supershift assays were performed via preincubation of an N-terminal FOXP2 antibody (Santa Cruz) with nuclear lysates, for 15 min at room temperature before the binding reaction. Protein-DNA interactions were resolved on a 5% polyacrylamide Tris/borate/EDTA gel.
Semiquantitative PCR
Chromatin isolated during ChIP from mutant and wild-type mouse brains was amplified using a semiquantitative PCR technique. PCR amplification was performed using 10 ng of template chromatin from each sample, in the presence of 1× Reaction Buffer (Applied Biosystems), 0.2 mM dNTPs, 2.5 mM MgCl2, 0.375 μM forward and reverse primers, and 0.75 U AmpliTaq Gold DNA polymerase (Applied Biosystems). A touchdown PCR protocol was used as follows: (1) 95°C for 5 min, (2) 95°C for 30 s, (3) 68°C for 30 s and −0.5°C per second until temperature reaches 55°C, and (4) 72°C for 2.5 min, then (5) repeat from step 2 for 14 cycles, followed by (6) 95°C for 30 s, (7) 55°C for 30 s, and (8) 72°C for 3 min, and then (9) repeat from step 6 for 19 cycles, followed by (10) 72°C for 10 min. PCR products were run on 2% agarose gels in the presence of ethidium bromide to allow visualization of DNA. Reactions were performed in triplicate. Primers were as follows: for Slc17a3, forward 5′-TGGTAACATGTGCAAATTGAGA-3′ and reverse 5′-AATGGACAAAACCCACAAGC-3′; for Cer1, forward 5′-GCTGTGACCAACAGCTTCAA-3′ and reverse 5′-AGATGTCTGCCACAGCACAG-3′; for Psen2, forward 5′-CCAGCTTTCCTTGAGCTGAC-3′ and reverse 5′-ACAAATGGCACATCGACAGA-3′; and, for β-actin, forward 5′-AGGGTACCACCGGAAAAGTC-3′ and reverse 5′-CCCCAAAGGCTGAGAAGTTA-3′.
Results
Isolation of Promoters That Are Bound by FOXP2 in a Human Neuronal Cell Model
To shed new light on the roles of FOXP2 in normal and abnormal brain development, we set out to discover direct neural targets of this transcription factor via ChIP-based strategies in an appropriate human cell-line. SH-SY5Y cells have been exploited commonly for many years as a standard model for investigating human neuronal function36 and were used effectively in a previous investigation to explore properties of etiological FOXP2 mutations implicated in speech-language disorder.18 For the present study, we employed stable transfection to generate SH-SY5Y cells displaying constitutive expression of the 715-aa isoform of FOXP2 (isoform I) (hereafter, “SH-FOXP2 cells”), in parallel with control cells transfected with an empty vector (see the “Materials and Methods” section). Stable expression in SH-FOXP2 cells was confirmed at the mRNA level by use of qRT-PCR and at the protein level via western-blotting experiments with polyclonal antibodies recognizing epitopes at the N- and C-termini of FOXP2 isoform I.18 Both antibodies were specific to FOXP2 and did not show cross-reactivity to other highly similar FOX proteins, such as FOXP1 and FOXP4.
FOXP2-specific ChIP was developed using the antibody recognizing the N-terminus of isoform I. SH-FOXP2 cells were chemically cross-linked, nuclei were isolated and treated with sonication to shear DNA, and FOXP2-bound DNA was enriched via immunoprecipitation. After reversal of cross-links, the resulting DNA fragments were purified and amplified by ligation-mediated PCR. Initial characterization of pulled-down fragments via shotgun sequencing, followed by PCR-based comparisons with ChIP with a no-antibody control, indicated that the FOXP2 ChIP protocol was successful in yielding enrichment of bound sites (data not shown).
We went on to perform location analysis of FOXP2 binding sites in SH-FOXP2 cells via ChIP-chip,20 by cohybridizing differentially labeled ChIP-enriched and nonenriched (total input) DNA to human promoter arrays (fig. 1). The experiments involved high-density microarrays (Aviva Biosystems) including PCR products from almost 5,000 well-characterized promoters. The DNA fragments are an average of ∼900 bp in size and typically span sequence from ∼650 bp upstream to ∼250 bp downstream of each transcription start site (see the work of Li et al.23 for full description of array characteristics). These arrays were designed in earlier studies for the specific purpose of high-throughput location analysis and have been used successfully to identify binding sites of other important transcription factors.23
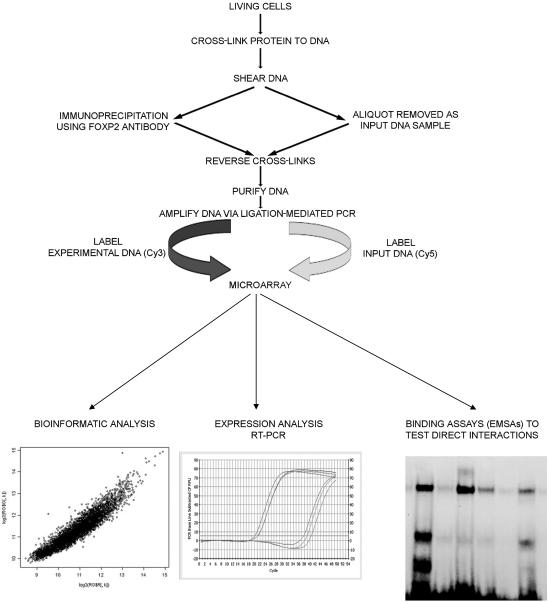
Figure 1. .
Overview of ChIP-chip technique. Living cells are cross-linked by treatment with formaldehyde to preserve interactions between transcription factors and target DNA. Following cell lysis, genomic DNA is sheared into fragments of ∼0.5–1 kb. At this stage, an aliquot is removed to be used as “input” reference DNA. Chromatin fragments are immunoprecipitated using an antibody directed to the N-terminus of FOXP2. Cross-links are heat reversed in both the immunoprecipitated and input samples. After DNA purification and labeling with fluorescent dyes, samples are applied to microarrays containing fragments from thousands of human promoters. Genes whose promoters show positive fold changes can then be investigated using in silico analysis. Functional validation via qRT-PCR expression analysis and DNA binding assays is performed on a gene-by-gene basis.
ChIP-chip experiments in SH-FOXP2 cells identified 303 DNA fragments that were consistently enriched in immunoprecipitated material across three independent biological replicates (P<.05; see table 1). We performed detailed bioinformatic analyses of the 100 most significantly enriched promoters—these encompassed a range of gene ontology (GO) categories but included many genes relevant to the brain, in particular those involved in neurotransmitter release, G-protein–coupled receptor signaling, the Notch and Wnt signaling pathways, ion transport and/or binding, and nervous-system development (table 3). Empirical analyses of statistical significance (permutation-based simulations; see the “Material and Methods” section) indicated that this subset of promoters includes a large excess of positive findings beyond those expected to occur by chance (false-discovery rate ∼22%).
Table 3. .
GO Characterization of the 100 Most Significantly Enriched Genes in FOXP2 High-Throughput Location Analysis[Note]
| GO Level 1 and Child GO Categories |
Genes |
| Signal transducer: | |
| Cell communication | STC1,a KCNE1L, TIAM1,a TRAF1,a,b ATF6,a,c EPOR,a CD180,a,d CD5,a PLAUR,a,e ITPK1,b,c,e,f RALBP1, and RAB18c,f |
| Synaptic transmission/synapse | VAMP2, LNPEP,b,d TDO2,b RALA,a,c,d MAPK8IP1,a,e DCTN2, and MAPK7c,e |
| Notch signaling pathway | NCOR2,a,c SNW1,a and PSEN2a |
| Wnt signaling pathway | PM5, CER1,a and SFRP4 |
| G-protein–coupled receptor signaling | CXCL2,a,d CCL19,a,d TGM2,b,c,d,e EBI2,d GNAZ,c,f PTGER1, CALCRL, and OPN1SWd |
| Development: | HOXB6c and MAEA |
| Nervous system development | NEUROD2,c SPOCK1,b and PAX3c |
| Organ development/morphogenesis | TCF12,c LECT1, CD164,d,g SYK,a,c,e,d,g DGAT1,e PAX1,c FLT1,c,e,g TAGLN,a and SPEGc,e |
| Binding: | |
| Ion binding | PYCR1,c,f RFPL3,a RCN2,a DUSP12,c,f TPO,d SOD3, and ACSL5 |
| Nucleic acid/nucleotide binding | SNFT,a RPL22, RPL28, EIF3S10, RAD51AP1,a,d TOPBP1,a LBR,a PCCA, TUBB2B,f PEX1,f FMO4,d and SMC2a,f |
| Protein binding | TNNI1, SMARCB1, ALDOA, HSPB7,d PSMD1, and C13orf24 |
| Transporter activity: | SLC2A4a and ERO1L |
| Ion transporter | COX11,b SLC17A3,b CACNG3,b FTH1,a SLC20A1,g TRPA1, SLC25A3, SLC4A8, and SLC22A14 |
| Catalytic activity: | IFI30d and LDHAa |
| Transferase | FTSJ2, PIGC, FUT2, ST3GAL5, and PTK9a |
| Hydrolase | HYAL2, TLL2, PSMA3,a and TPP2 |
| Cell adhesion | SSX2IP,a HAS1,e SORBS3,a HAPLN1, and HRSP12f |
| Cellular process: | |
| Response to stimulus | IGLL1 |
Note.— Genes classified as being in additional categories are indicated with the following footnotes.
Protein binding.
Ion binding.
Nucleic acid/nucleotide binding.
Response to stimulus.
Transferase.
Hydrolase.
Cell communication.
Identification of Biological Themes Associated with FOXP2 Binding
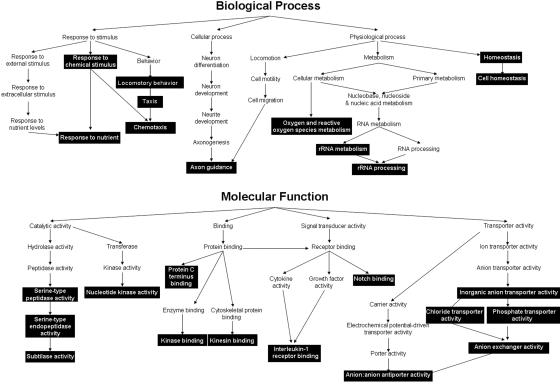
We performed in silico analyses of our set of significantly enriched target promoters using WebGestalt.27 These tools help to identify the GO categories and biological themes that characterize a set of genes and that are significantly overrepresented in the data set compared with an unbiased reference set (in this case, all the genes represented on the promoter arrays). We used this program to build GOTrees, graphical representations of the relationships between the overrepresented gene categories. GO categories are divided into three separate classifications: cellular compartment, biological process, and molecular function.37 With regard to cellular compartment, a particularly large proportion of the putative FOXP2 targets identified by ChIP-chip encode membrane-bound proteins, significantly more than would be expected by chance (data not shown). Notably, biological processes related to homeostasis, locomotory behavior, and axon guidance were significantly overrepresented in our data set, as well as molecular functions, such as ion-transporter activities, kinase binding and/or activity, and Notch binding (fig. 2).
Figure 2. .
GO clustering of genes enriched in FOXP2 high-throughput location analysis. GOTrees demonstrate GO classifications that were overrepresented in the target list of 303 genes and relationships between categories, with regard to molecular function and biological process. Significantly overrepresented categories (P<.05) are highlighted in black.
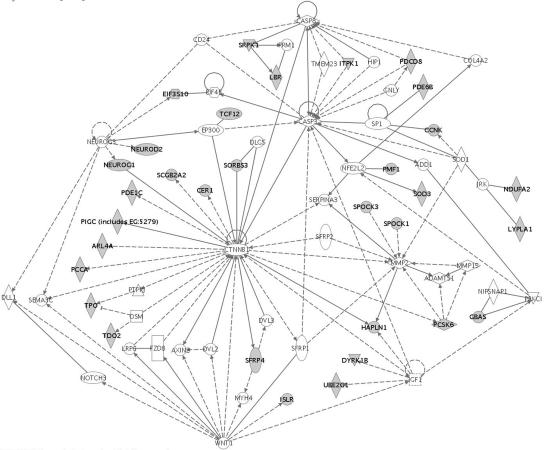
To further clarify the functional networks related to FOXP2 binding, we searched for connectivity between putative target genes, using Ingenuity pathway-analysis software. This software exploits a highly comprehensive curated database established from the literature to determine direct and indirect interactions between genes of interest, thereby identifying canonical pathways associated with a particular set of genes. Several networks were generated (fig. 3 and data not shown), representing functional relationships implicated in development (including embryonic and nervous-system development), neurogenesis, cell death, cell migration, and transcription. Figure 3 shows one such network that centers largely on the Wnt/β-catenin and IGF-1 signaling pathways.
Figure 3. .
Network analysis of target genes. Interaction networks were generated using Ingenuity pathway analysis. The figure shows one example of an important functional pathway identified via this approach, which centers on Wnt/β-catenin and IGF-1 signaling pathways. Genes shaded in gray were identified as significantly (P<.05) enriched in ChIP-chip analyses; others were brought into the pathway on the basis of relationships within the Ingenuity knowledge database. Solid and dotted lines represent direct and indirect interactions, respectively. Node shapes represent the class of molecule—vertical diamonds represent enzymes, horizontal diamonds represent peptidases, ovals represent transcription factors, squares represent cytokines, triangles represent kinases, hexagons represent translation factors, and circles represent other classes. Further information can be found at the Ingenuity Systems Web site.
Bioinformatic Characterization of Binding Sites in Enriched Promoters
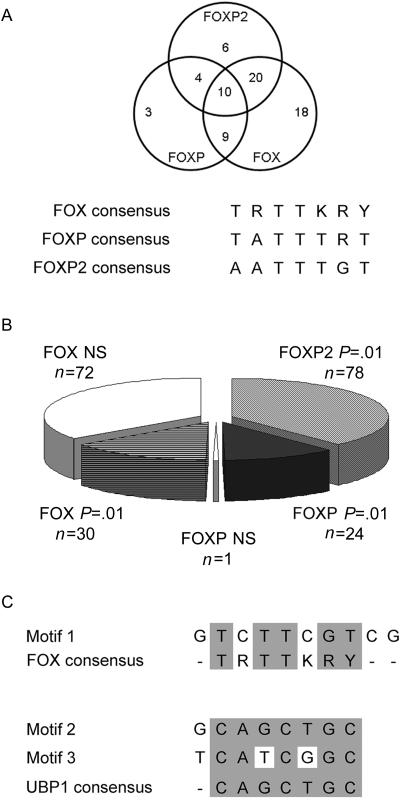
The set of 100 enriched target promoters were investigated in silico for the presence of FOXP2 consensus binding sites. This analysis focused specifically on the ∼1 kb of each promoter represented on the Aviva arrays. As shown in figure 4A, we searched for three categories of binding sites, which had different levels of specificity: (i) experimentally determined sites shown to be bound by FOXP2 in structural studies ((A)ATTTG(T)29,30), (ii) a consensus site determined in vitro for the closely related family member FOXP1 (TATTTRT),14 which can also be bound by FOXP2,18 and (iii) the general FOX family consensus sequence (TRTTKRY), to which most FOX proteins are able to bind.31 We refer to these three categories of sites as FOXP2, FOXP, and FOX, respectively. (Note that the FOXP consensus sequence also conforms to the general FOX family consensus, but each site was classified only once, on the basis of the category that gave the most specific match.)
Figure 4. .
In silico motif analysis of sequences enriched in FOXP2 ChIP-chip. A, Assessment of the presence of known FOX (TRTTKRY), FOXP (TRTTTRY), or FOXP2 ([A]ATTTG[T]) binding sites in the promoter regions of the genes in table 3. The Venn diagram represents the number of promoter sequences containing at least one of these binding sites, including overlaps where more than one class of site is present in the same sequence. Of the promoters, 70 contained at least one perfect FOXP2, FOXP, or FOX binding site, with most (61%) containing multiple classes of consensus sites. All promoters featured at least one FOXP2 binding site that contained a single mismatch. B, Number and significance of binding sites. The total number of binding sites identified in all sequences is represented, partitioned according to the site class and the statistical significance of identifying each site within the data set. NS = not significant. C, Results of unbiased motif analysis. Overrepresented motifs identified from the 100 target sequences analyzed included motif 1, which closely resembled the FOX family consensus binding site, and motifs 2 and 3, which were close or exact matches to the UBP1 (also known as “LBP-1a”) consensus binding site.
Figure 4A displays the numbers of target promoters containing consensus binding sites for each category, including overlap where multiple binding sites from alternative classes are present in the same sequence. Of note, the majority of the enriched promoters (70 of 100) contain at least one exact match to the consensus site for FOXP2, FOXP, or FOX, and a substantial number of these (43) include multiple sites from the different categories. In addition, multiple sites from the same category were often present within a single sequence. For example, 78 perfect FOXP2 consensus binding sites were identified within 40 of the top 100 enriched sequences. Using simulations, we determined that the probability of finding each of these FOXP2 consensus sites by chance was <0.01 (see the “Material and Methods” section). The numbers of sites and the statistical significance for all consensus binding sites that we identified are given in figure 4B. When criteria were relaxed to allow a single mismatch at any position, all sequences displayed the presence of a FOXP2 binding site. Unlike most other forkhead proteins, FOXP2 is thought to require dimerization (either with itself or with other members of the FOXP subfamily) to bind target DNA.14,38 We searched the putative target sequences for regions that could provide an opportunity for binding of FOXP dimers, focusing only on perfect consensus matches that were within 100 bp of each other. We thereby identified 29 independent compound sites involving the same category of binding site, as well as 37 compound sites involving different categories.
We next investigated the potential for other transcription factors to regulate these target genes. We determined whether there was overrepresentation of other known transcription factor binding sites, since this could suggest a direct or indirect role in regulation of gene expression by FOXP2 (e.g., as part of a transcriptional complex). We used two approaches to query the TRANSFAC database. First, we searched all 303 (significantly enriched) sequences for the presence of known TRANSFAC binding-site matrices and determined significant overrepresentation on the basis of the frequency of these sites in all sequences represented on the array. A total of 28 consensus motifs, recognized by 21 different transcription factors, were identified as being significantly overrepresented (described in table 4). Consensus binding sites for transcription factors known to play key roles in neuronal plasticity and CNS development—such as CREB and its related family member, ATF, as well as SP1 and PAX5—were found significantly more often in the target list than would be expected on the basis of the sequences represented on the array. Second, we independently used an unbiased approach to identify the presence of significantly overrepresented sequence motifs in our data set by useof the MEME algorithm.32,33 The resulting matrices were investigated for matches to known binding sites in the TRANSFAC database. Three of the overrepresented matrices showed the closest consensus to known binding sites (fig. 4C). Notably, motif 1 represented a close match to the FOX family consensus binding site, and motifs 2 and 3 were near or exact matches for the upstream binding protein 1 (UBP1; also known as “LBP-1a”) transcription factor binding site. UBP1 is a CCAAT-binding protein that is able to act in a heterodimer with its closely related family member, LBP-1c.39 The LBP-1c binding site was also found to be significantly overrepresented in the target gene sequences (table 4). These data raise the possibility that there may be some functional interaction between FOXP2 and the transcription factors detailed above (and in table 4)—that is, via direct interactions or indirectly as part of a larger complex.
Table 4. .
Significantly Overrepresented Co-Occurring Transcription Factor Binding Motifs
| Name (Description) and TRANSFAC Matrix |
Consensus Sequencea | P |
| AP-2 (activator protein 2): | ||
| AP2_Q6 | MKCCCSCNGGCG | <.01 |
| AP-4 (activator protein 4): | ||
| AP4_01 | WGARYCAGCTGYGGNCNK | <.01 |
| AP4_Q6 | CWCAGCTGGN | <.01 |
| ATF1 (activating transcription factor): | ||
| ATF_01 | CNSTGACGTNNNYC | <.01 |
| CAAT (CCAAT box): | ||
| CAAT_01 | NNNRRCCAATSA | <.01 |
| TFCP2 (transcription factor CP2 [also known as “LBP-1c”]): | ||
| CP2_01 | GCHCDAMCCAG | <.01 |
| CREB (cAMP-response element-binding protein): | ||
| CREB_02 | NNGNTGACGTNN | <.01 |
| CREB_Q2 | NSTGACGTAANN | <.01 |
| CREB_Q4 | NSTGACGTMANN | <.01 |
| CREBP1 (CRE-binding protein 1): | ||
| CREBP1_Q2 | VGTGACGTMACN | <.01 |
| E47: | ||
| E47_01 | VSNGCAGGTGKNCNN | <.01 |
| GATA1 (GATA-binding factor 1): | ||
| GATA1_01 | SNNGATNNNN | <.01 |
| GATA2 (GATA-binding factor 2): | ||
| GATA2_01 | NNNGATRNNN | <.01 |
| GC box (GC box elements): | ||
| GC_01 | NRGGGGCGGGGCNK | <.01 |
| Ik-2 (Ikaros 2): | ||
| IK2_01 | NNNTGGGAWNNC | <.01 |
| Lmo2 complex (complex of Lmo2 bound to Tal-1, E2A proteins, and GATA-1, half-site 1): | ||
| LMO2COM_01 | CNNCAGGTGBNN | <.05 |
| MZF1: | ||
| MZF1_01 | NGNGGGGA | <.01 |
| MZF1_02 | KNNNKAGGGGNAA | <.01 |
| NF1 (nuclear factor 1): | ||
| NF1_Q6 | NNTTGGCNNNNNNCCNNN | <.01 |
| NFY (nuclear factor Y [Y-box binding factor]): | ||
| NFY_01 | NNNRRCCAATSRGNNN | <.05 |
| NFY_C | NCTGATTGGYTASY | <.01 |
| NFY_Q6 | TRRCCAATSRN | <.01 |
| Pax-4 (Pax-4 binding sites): | ||
| PAX4_01 | NGNVGTCANGCGTGNNSNNYN | <.01 |
| PAX5 (B-cell specific activating protein): | ||
| PAX5_01 | BCNNNRNGCANBGNTGNRTAGCSGCHNB | <.01 |
| RREB-1 (Ras-responsive element binding protein 1): | ||
| RREB1_01 | CCCCAAACMMCCCC | <.01 |
| Sp1 (stimulating protein 1): | ||
| SP1_01 | GGGGCGGGGT | <.01 |
| SP1_Q6 | NGGGGGCGGGGYN | <.01 |
| USF (upstream stimulating factor): | ||
| USF_Q6_01 | GYCACGTGNC | <.01 |
International Union of Biochemistry ambiguity codes: R = A or G; Y = C or T; K = G or T; M = A or C; S = C or G; W = A or T; V = A, C, or G; B = C, G, or T; H = A, C, or T; D = A, G, or T; and N = A, G, C, or T.
Significant Alteration of ChIP Target Expression Due to Increased FOXP2 Expression
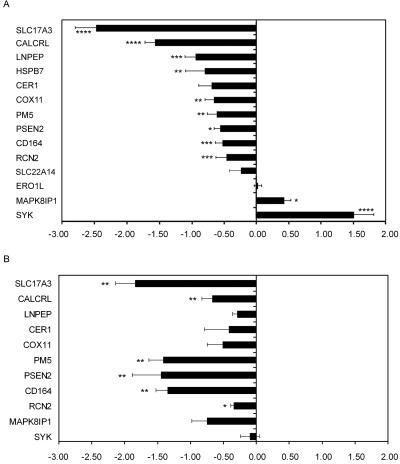
Our ChIP-chip experiments successfully identified promoter sites that are enriched for FOXP2 binding in the native chromatin of living SH-SY5Y cells. We went on to obtain functional verification for a subset of genes from table 3, using multiple independent approaches (figs. 5 and 6). First, we directly assessed the evidence of a functional link between FOXP2 and selected ChIP targets, by asking whether presence or absence of FOXP2 protein led to significant changes in expression of the gene in question. These experiments exploited our SH-SY5Y cell lines that had been stably transfected with either FOXP2 or an empty vector control and involved quantitative assessments of ChIP-target mRNA expression on a gene-by-gene basis with use of qRT-PCR (fig. 5). We thereby identified robust and significant differences in ChIP-target expression. Of the 14 ChIP targets investigated in figure 5A, 11 genes displayed significant effects, across replicate experiments. In line with previous studies suggesting that FOXP proteins act as repressors,14,18,38 10 of the targets tested (SLC17A3, CALCRL, LNPEP, HSPB7, CER1, COX11, PM5, PSEN2, CD164, and RCN2) showed reduced expression in the presence of FOXP2. However, two genes (MAPK8IP and SYK) displayed significant increases in transcription in response to FOXP2 upregulation in our stable cell lines, suggesting that the protein may also act as an activator for a minority of direct targets. Targets that showed altered gene expression were also assessed in multiple additional biological replicates involving transient transfections of SH-SY5Y cells. As shown in figure 5B, several of the targets that were functionally validated in the stable cell models showed similarly significant effects during multiple independent transient transfections.
Figure 5. .
Functional validation of FOXP2 target by expression assays. qRT-PCR was performed for cDNA prepared from stable SH-SY5Y cell lines (A) or transiently transfected SH-SY5Y cells (B). Expression changes (X-axes) are given as mean of log2 expression ratios of cells transfected with FOXP2 compared with cells transfected with an empty vector control and are normalized for equal expression of the internal control, GAPDH. Values are the mean of comparisons of seven independent cDNA preparations for stable cell lines and of six independent cDNA preparations for transient cell lines. Error bars indicate ±SEM. P values were calculated using a two-tailed unpaired t test. Four asterisks (****) indicate P<.0001, three asterisks (***) indicate P<.001, two asterisks (**) indicate P<.01, and one asterisk (*) indicates P<.05. Most expression changes observed in stable cell lines were verified in the transiently transfected cells. HSPB7 expression could not be detected in transient cell lines; thus, expression changes could not be assessed in this system.
Figure 6. .
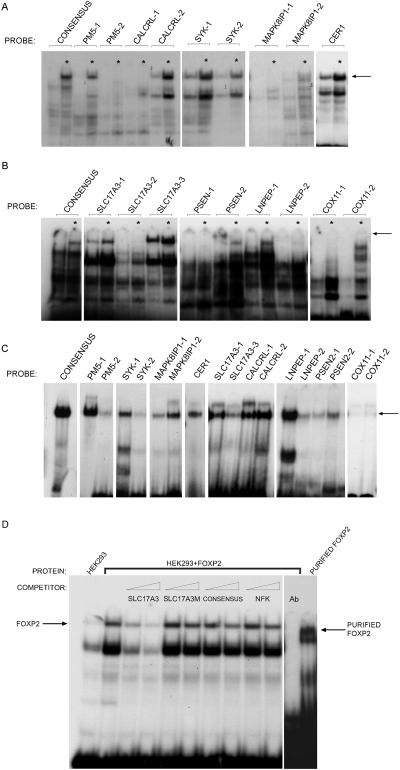
Functional validation of FOXP2 targets by DNA binding. EMSAs were used to determine direct interactions between FOXP2 and the promoter regions of targets already tested in expression assays. Probes were designed on the basis of the region represented on the promoter array and the presence of FOX(P) consensus binding sites (table 5). A and B, FOXP2 binding to target probes, first established using nuclei of HEK293T cells. Radiolabeled probes were incubated with either HEK293T nuclei sham transfected with an empty vector (HEK293 cells express a low level of endogenous FOXP2) or FOXP2 transfected nuclei (indicated by an asterisk [*]). The position of the shift caused by interaction between FOXP2 and the radiolabeled probe is indicated (arrow). C, Binding to target probes confirmed via EMSAs with purified FOXP2 protein, which gave results that paralleled those seen in HEK293T nuclei. In panels A–C, the binding of FOXP2 to a known consensus binding site (CONSENSUS) is shown as a positive control. D, More-detailed analysis undertaken for SLC17A3-1 probe. Binding assays were performed in the presence of competition from unlabeled probes. FOXP2 binding to the labeled probe was efficiently impaired via competition with unlabeled competitor probe (SLC17A3) in 5-fold or 10-fold excess, displaying the specificity of the interaction. Mutation of the core binding site resulted in a lack of competition by unlabeled probes (SLC17A3M), indicating the importance of the bases for FOXP2 binding. A consensus FOXP2 binding site showed slight competition, whereas an irrelevant promoter sequence (NFK) was unable to compete in binding at 5-fold or 10-fold excess. Addition of an antibody directed to FOXP2 (Ab) disrupted complex formation. This confirmed that the identity of the protein causing gel retardation is FOXP2, a result that was further verified using purified FOXP2 protein.
Direct Sequence-Specific Binding of FOXP2 to Promoters of Validated Targets
In the next stage of functional verification, we aimed to identify the particular sites within each target promoter that are directly bound by FOXP2 and to assess the specificity of the protein-DNA interaction. For this step, we focused on nine of the ChIP targets that had already shown evidence of up- or down-regulation in response to FOXP2 in stable SH-SY5Y cell lines (see previous paragraph) and evaluated binding by use of EMSAs as described elsewhere.18 For each validated target gene, the sequences originally represented on the promoter array were searched in silico to identify the closest matches to FOXP2, FOXP, and FOX consensus binding sites (see above for definitions of each category). Oligonucleotide probes spanning these regions (table 5) were used in EMSAs with nuclei from human cells overexpressing FOXP2 (fig. 6A and 6B), and FOXP2-specificity was further confirmed using purified protein (fig. 6C). Concordant results were seen for the nuclear extracts and purified protein, with the most robust and consistent binding seen for probes within SLC17A3, CALCRL, LNPEP, PSEN2, PM5, CER1, and SYK promoters. We went on to perform more-detailed analyses of the protein-DNA interactions for the SLC17A3 probe (fig. 6D), demonstrating that binding could be disrupted by competition with an excess competitor probe but not if the competitor carried a mutation in the putative core binding site. Subtle competition by the known FOXP2 binding site could be observed. Binding was not disrupted by competition with an irrelevant binding site—recognized by NFK but not expected to be recognized by FOXP2. Moreover, addition of antibodies directed to FOXP2 disrupted the formation of the protein-DNA complexes.
Table 5. .
Probes Used for EMSA DNA Binding Assays
| Class of Binding Sited |
||||||
| Gene or Transcript |
Probe Sequencea | Positionb (bp) | Boundc | FOXP2 | FOXP | FOX |
| Consensus | agcttTATTTATgttgttttgtat | … | No | − | − | − |
| PM5-1 | ctttaagAATTTGTgtaagc | −643 | Yes | + | − | − |
| PM5-2 | gacgggagATTTTGTtgtg | −360 | No | + | − | − |
| CALCRL-1 | cacagATTTGTtagattttttttc | −436 | No | + | − | − |
| CALCRL-2 | aataTATTTATtctaagtag | 232 | Yes | − | + | − |
| SYK-1 | caaagccTTTTGTaataattaaag | −565 | Yes | + | − | − |
| SYK-2 | ggtttAATTTATTTGgttgtgg | −616 | No | ++e | − | − |
| MAPK8IP1-1 | gcctcccAATTTCaggtgag | 255 | No | + | − | − |
| MAPK8IP1-2 | tgcTATTGTCccatttcacag | −499 | Yes | − | − | + |
| CER1 | gaaggatgttAATTTTtttg | −603 | Yes | + | − | − |
| CER1M | gaaggatgttAACCCTtttg | … | No | − | − | − |
| SLC17A3-1 | gaataTAGTTATctCTTTGTcc | −626 | Yes | + | + | − |
| SLC17A3-1M | gaataTAGGGATctCGGGGTcc | … | No | − | − | − |
| SLC17A3-2 | ccATTTCTtaaccTATGTATgat | −602 | No | + | + | − |
| SLC17A3-3 | ttttgtatAATTTGagaa | −489 | Yes | + | − | − |
| PSEN2-1 | gggcgTTTTGTtcttcttctc | −193 | No | + | − | − |
| PSEN2-2 | ccccTGTTTATTGCcttaataag | 224 | Yes | − | − | ++e |
| LNPEP-1 | tctgtgttTATTTATggtctg | −416 | Yes | − | + | − |
| LNPEP-2 | ATTTATggtcTGTTTCTggaatg | −407 | No | + | − | + |
| COX11-1 | cgggtaAGTTTGcgtt | −136 | No | + | − | − |
| COX11-2 | tcgcgagATTTGAcctctcg | −10 | No | + | − | − |
| NFK | ccgggggtgatttcactccccg | … | No | − | − | − |
Core binding sites are represented in capital letters. Bases in bold font indicate deviation from or mutation of the consensus binding site.
Position relative to transcriptional start site.
Ability to be bound by FOXP2 during EMSA.
A plus sign (+) indicates the presence of the consensus binding site, and a minus sign (−) indicates the absence of the site.
Indicates two overlapping sites.
Promoter Binding and Target-Gene Repression In Vivo in the Embryonic Mouse Brain
We next aimed to demonstrate that validated targets from functional genomic analyses of neuron-like cell-based models have biological significance in the CNS in vivo. We performed in vivo Foxp2 ChIP, using mouse brains at E16, a developmental time point showing high neural expression of Foxp2.17 The isolated chromatin could then be used to assess Foxp2 occupancy at a promoter of interest via a semiquantitative PCR approach. Crucially, we were able to compare data from wild-type mouse brains with those obtained from equivalent experiments in homozygous mutant mice that lack detectable Foxp2 protein, providing us with an ideal negative control (see the “Material and Methods” section). Primers were designed to amplify promoter regions of the mouse orthologues of SLC17A3, CER1, and PSEN2, three genes that had been rigorously tested in our cell-based models. PCR amplification of Foxp2 ChIP material from E16 brains indicated robust enrichments of regions of Slc17a3 and Cer1 promoters in chromatin isolated from wild-type mice, compared with that isolated from mutant littermates lacking Foxp2 protein (fig. 7A). Enrichment was not observed for the Psen2 promoter or β-actin negative control promoter region.40
Figure 7. .
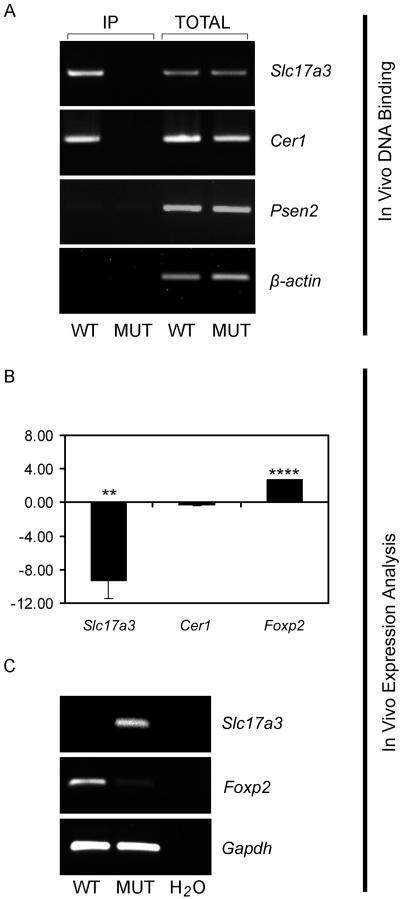
Foxp2 binding to target gene and regulation of target-gene expression in vivo in embryonic mouse brain. A, Analysis of in vivo promoter occupancy. FOXP2 ChIP was performed using whole brains at E16 from either wild-type mice (WT) or homozygous mutant littermates (MUT) that lack Foxp2 protein as a consequence of an exon 7 nonsense mutation (see the “Material and Methods” section). DNA was PCR amplified using primers directed toward Slc17a3, Cer1, Psen2, or β-actin control promoter regions. The promoters of Slc17a3 and Cer1 were specifically enriched in ChIP samples isolated from wild-type brains compared with those from mutant brains. Gels are representative of results from triplicate experiments. IP represents the immunoprecipitated sample. B, Expression of Slc17a3 showing an inverse relationship with Foxp2 expression in E16 mouse brains. RT-PCR was performed with cDNA generated from five wild-type versus five mutant whole-brain samples. Note that reduced expression of Foxp2 in mutant mice is the result of nonsense-mediated RNA decay; in addition, the potential truncated product is highly unstable, leading to a lack of detectable Foxp2 protein (M.G., J.N., and S.E.F., unpublished data). Expression changes are represented as mean of log2 expression ratios for comparisons between wild-type and mutant mice normalized for equal expression of the internal control, Gapdh. Statistical significance was calculated using a two-tailed unpaired t test. Two asterisks (**) indicate P<.01; four asterisks (****) indicate P<.0001. C, Confirmation of in vivo expression changes in independent mouse samples. Standard RT-PCR with use of samples prepared from an independent pair of wild-type and mutant mouse brains shows a clear loss of repression of Slc17a3 in the mutant.
Finally, we investigated whether the in vivo binding of Foxp2 to Slc17a3 and Cer1 promoters in E16 mouse brain has detectable effects on target-gene expression in the tissue in question. Our RT-PCR–based expression analyses detected Slc17a3 expression in the brains of E16 mouse embryos but at very low levels. Intriguingly, we found that mutant mice lacking Foxp2 protein demonstrate a substantial and highly significant increase in Slc17a3 expression in E16 brain tissue (fig. 7B and 7C). Only minor differences in overall Cer1 expression levels were seen between wild-type and mutant mouse brains. The Slc17a3 expression data are notably concordant with our earlier human cell-based findings, which showed that human FOXP2 severely down-regulates SLC17A3 expression in SH-SY5Y systems (c.f. fig. 5). The corresponding in vivo findings indicate that Foxp2 normally binds to the Slc17a3 promoter in the embryonic mouse brain, acting to repress its transcription, and that absence of Foxp2 in mutants leads to a dramatic loss of repression.
Discussion
Binding of FOXP2 to Promoters of Genes Involved in Cell Communication, Development, and Ion Transport in Living Neuron-Like Cells
It is well established that FOXP2 disruptions are correlated with abnormalities in acquisition of speech and language skills.3,4,6–8 However, the precise functions of this gene at molecular, cellular, and developmental levels have remained largely elusive in the 6 years following its discovery. In the present study, we have exploited a powerful functional genomics approach to gain entry into FOXP2-related networks and to begin to bridge the gaps between genes, brain, and behavior. To examine these pathways, we assessed the binding of FOXP2 to native chromatin in living neuron-like cells cultured in the laboratory and simultaneously screened thousands of promoters through a microarray-based approach. In this way, we were able to demonstrate significant enrichment during ChIP-chip experiments for promoters from a range of genes involved in diverse biological functions, including (but not limited to) synaptic transmission, cell signaling, ion transport, and development. As for other transcription factors, FOXP2 may be involved in the regulation of a large number of different genes; on the basis of the empirical analyses of significance in the array experiments, we would expect a minimum of 1.5% of promoters in the human genome (i.e., at least several hundred genes) to be occupied by FOXP2 in our cell-based models. This estimate is consistent with in vivo human data obtained from human fetal brain and lung in an accompanying study by Spiteri et al.,21(in this issue) which, despite the use of a different FOXP2 antibody, identified a highly significant overlapping list of targets, providing a large degree of independent confirmation of targets identified here (discussed further below).
Several key biological themes and functional pathways emerged when FOXP2-enriched targets from neuron-like cells were analyzed for gene-gene connectivity or overrepresentation of GO classes. For example, in terms of biological process, we observed an excess of genes contributing to axon guidance and homeostasis, whereas molecular functions such as anion transporter activity, notch binding, and kinase binding and/or activity were also significantly overrepresented. Moreover, in silico analyses of direct and indirect interactions between FOXP2-enriched targets highlighted a number of interesting networks, most notably one that was associated with Wnt/β-catenin and IGF-1 signaling pathways (fig. 3). Intriguingly, genes represented in this particular network have been implicated in differentiation of neurons and neuroglia, neuronal cell death, cell adhesion, and control of transcription. In addition, abnormalities in IGF-related pathways are thought to contribute to Silver-Russell syndrome, a disorder that is sometimes associated with reductions in FOXP2 expression.6
One advantage of assessing gene-gene interaction networks is that characterization can be based not only on the functions of target genes but also on those of interacting genes. Thus, targets can be clustered around functional “nodes” that may not be included in the target list itself, as shown for CTNNB1 (β-catenin) in figure 3. Of note, many of the networks uncovered by Ingenuity pathway analysis were similarly highlighted during GO analysis, including Wnt/Notch signaling and development. Wnt signaling plays a major highly conserved role in brain patterning in vertebrates and invertebrates, including humans.41,42 Given the role of Wnt genes in patterning the forebrain in mammals, targets in this pathway are attractive candidates for mediating structural and functional alterations in the brains of people carrying FOXP2 mutations.
Furthermore, we observed significant overrepresentation of a number of transcription factor binding sites in the enriched target sequences (P<.05). This included binding sites for transcription factors involved in processes such as neurodevelopment, Wnt signaling, and plasticity (CREB, SP1, PAX5, and E47/TCF3),43–48 as well as genes implicated in disease states, such as Alzheimer disease (LBP-1c).49 The co-occurrence of these binding sites highlights the potential interplay of FOXP2 with key pathways involved in neural development and function.
After identifying promoters bound by FOXP2 in cell-based models of neural function, we went on to show, in these same cells, that quantitative expression levels of putative target genes could be significantly altered by presence or absence of FOXP2. Until now, FOXP2 has been assumed to act as a transcriptional repressor, largely on the basis of experiments involving expression from artificial or viral promoters in vitro.14,18,38 Indeed, the majority of the direct targets validated in the current investigation showed significant reductions in mRNA expression as a consequence of increased FOXP2 levels, consistent with the repressor hypothesis. However, a minority showed up-regulation in response to increases in FOXP2, suggesting that, although this regulatory factor usually represses transcription, it is also able to switch from repressor to activator under certain circumstances. This is consistent with findings from a number of FOX family members, including the closely related FOXP3 protein, which have the capacity to mediate both repression and activation of direct transcriptional targets.40,50,51 The switch from repressor to activator may be dependent on binding-site affinity,52 cofactors that interact with FOXP2,53 or posttranslational modification status. Interaction with cofactors is particularly pertinent when considering FOXP2 regulation of gene expression, given that it requires dimerization for efficient DNA binding. Thus, the expression levels of known binding partners, such as FOXP family members,14,38 CTBP1,38 NFAT,30 or other as-yet unidentified interactors, could affect not only the affinity for the target DNA but also the switch from repressor to activator.
Overlap of FOXP2 Targets in Cultured Neuron-Like Cells and Those Identified in Human Fetal Brain
The present report demonstrates the power of extrapolating from cell-based models to gain insights into brain function. By using state-of-the-art functional genomics in cultured neuron-like cells, we uncovered regulatory mechanisms that we could subsequently confirm in vivo in the developing mouse brain (see below). Moreover, in a separate investigation performed in parallel to our own, Spiteri et al. applied ChIP-chip with FOXP2 in vivo by using human fetal brain tissue.21(in this issue) A between-study comparison of the lists of enriched promoters, obtained using equivalent statistical thresholds (P<.05; see table 1), reveals a remarkable degree of overlap in targets suggested by cell-based and in vivo methods: 29% of putative target promoters identified by Spiteri et al. in the basal ganglia (BG) and 30% of those found in the inferior frontal cortex (IFC) were independently highlighted in SH-SY5Y cells in the present study, representing 14%–19% of our potential targets (table 1). The probabilities of obtaining this extent of overlap by chance are highly significant (P<10-24 [BG/SH-SY5Y] and P<10-22 [IFC/SH-SY5Y]). The concordance between these data sets is particularly striking when it is considered that the cell-based and in vivo ChIP-chip studies employed antibodies recognizing distinct epitopes at opposite termini of the FOXP2 protein and that these antibodies have the potential to recognize different sets of alternatively spliced isoforms.
Like us, Spiteri et al.21(in this issue) were able to confirm regulation of ChIP targets by FOXP2 with use of cell-based models and similarly found that this transcription factor can act as both a repressor and an activator. Interestingly, although some targets that were commonly tested showed equivalent expression changes (e.g., HSPB7 and PM5) in the independent studies, others (e.g., CALCRL) were significantly activated in one study but were significantly repressed in the other. We retested CALCRL expression levels with use of the primers detailed by Spiteri et al. (unpublished data) to determine whether differences between studies were caused by detection of alternative transcripts. However, in our cells, these primers detected expression changes that were entirely consistent with our previous experiments (data not shown and fig. 5). Instead, the differing effects between studies may relate to the specific cellular models that were used. Although both investigations employed SH-SY5Y cells, the present study assessed expression in replicates of a stably transfected cell line generated from a single progenitor. This clonal line provided a homogenous population in which every cell expressed a consistent and relatively low level of FOXP2 over successive passages. By comparison, the study by Spiteri et al. used a more heterogeneous cell population. These findings may again indicate that the relative levels of FOXP2 and its interacting cofactors affect the protein’s ability to act as an activator or repressor and provide further evidence of the complexity of regulatory mechanisms mediated by the FOXP subgroup.40,51
Identification by Human ChIP-Chip of Conserved Pathways Regulated by Foxp2 in Vivo in Embryonic Mouse Brain
It is thought that orthologues of the FOXP2 gene play highly conserved roles in development and/or function of sensorimotor-related circuits in many vertebrates, on the basis of similarities in sequence and neural expression patterns across a range of distantly related species.19,54 As such, studies of animal model systems can offer important clues to FOXP2 function at multiple levels, in ways that are not possible when humans are investigated.55 In particular, there is a high degree of sequence conservation between human FOXP2 and its mouse orthologue (∼99.4% identity at the amino acid sequence level), and there is notably concordant expression in corresponding brain structures, with consistent patterns of sublocalization.17 We recently generated mutant mice that carry an early stop codon, making them effectively null for Foxp2 (M.G., J.N., and S.E.F., unpublished data). The availability of homozygous mutant animals that completely lack Foxp2 protein provides a valuable system for establishing whether targets identified in cell-based studies also show promoter occupancy and/or mRNA regulation by Foxp2 in vivo in the CNS.
Thus, in the present study, we followed up human ChIP-chip by performing Foxp2 ChIP in embryonic brain tissue (E16) from wild-type and mutant mice and assessed enrichment of promoter regions for three of our most promising targets. We were able to demonstrate clear in vivo promoter occupancy for Slc17a3 and Cer1 but not for Psen2. These data show interesting parallels with independent in vivo ChIP data from human fetal brain obtained by Spiteri et al., by which SLC17A3 and CER1 were identified as potential targets in human BG and IFC, respectively, and binding to the PSEN2 promoter was not noted in either region. We went on to test whether promoter occupancy of Slc17a3 and Cer1 was correlated with significant alterations in regulation of each target, by comparing quantitative expression levels in E16 brains of wild-type and mutant mice. Cer1 demonstrated only a minor difference, but Slc17a3 showed a highly significant loss of repression in absence of Foxp2 protein.
Genes like SLC17A3/Slc17a3 that show promoter occupancy in vivo in the developing brains of humans and mice, as well as FOXP2/Foxp2-mediated expression differences in both human neuron-like cells and mouse brain tissue, clearly represent confirmed targets in both species. Thus, further studies of such targets with use of mouse mutants will yield critical insights into conserved neural pathways and how they are disturbed by disruption of FOXP2/Foxp2. For example, at this stage, there is little information on the function of SLC17A3, other than that it is thought to act as a sodium/phosphate cotransporter and is highly expressed in the kidney and liver.56,57 We have uncovered low levels of Slc17a3 expression in embryonic brain, which is consistent with online data from the Cancer Genome Anatomy Project (CGAP)–EST project.58 Our findings may indicate a potential, as-yet undetermined role for this gene in neural pathways regulated by FOXP2, which could be important for speech and language development. Alternatively, loss of repression of SLC17A3 may lead to inappropriate expression in the brains of affected individuals, interfering with the normal development and function of FOXP2-related circuits.
It is worth emphasizing that, given the complex spatiotemporal patterns of FOXP2/Foxp2 neural expression,17 a target identified in human neuron-like cells may still represent a real conserved target even if it does not show validation of binding and/or regulation in E16 whole mouse brain. FOXP2/Foxp2 is expressed in tightly regulated subsets of neurons in several structures, including the striatum, thalamus, deep layers of the cortex, and Purkinje cells of the cerebellum.17 Regulatory effects in subpopulations of neurons may be diluted out in studies of whole brain. More crucially, different sets of targets are likely to be regulated by Foxp2 in distinct subpopulations and/or at different developmental stages, depending partly on the presence or absence of cofactors (such as Foxp1 or Foxp4). In relation to this, it is interesting to note that Cer1, although bound by Foxp2 in vivo in E16 whole-brain samples, showed only minor changes in overall expression in equivalent samples from mutant mice. In addition, recent ChIP-chip studies, including those investigating FOXP3 binding,40,51 have highlighted disparities between transcription factor promoter occupancy and transcriptional regulation.59 In some cases, target-gene expression could be validated under altered cellular conditions, where the relevant transcriptional cofactors were present.59
Finally, it is reasonable to expect that a subset of true FOXP2 targets will be human specific and hence impossible to validate or characterize in animal models. Indeed, evidence that FOXP2 was subject to positive selection in the human lineage suggests that the relevant mechanisms may have undergone modifications in our species, possibly in relation to their recruitment toward speech and language capacities.60,61 Future studies could address the important issue of species-specific targets, by using in vivo neural ChIP-chip in different species to facilitate a direct comparison of genomewide promoter occupancy in the brains of humans, nonhuman primates, and rodents.
Concluding Remarks
In the present study, we have exploited high-throughput location analysis of binding sites to identify direct neural targets regulated by FOXP2, the first gene to have been implicated in a human speech and language disorder. Moreover, we have provided compelling evidence at multiple levels that our findings are relevant to in vivo biological function. First, the promoters that we identified contained an excess of sequences conforming to previously determined consensus sites for FOXP2 binding. Second, targets included significant overrepresentations of a subset of GO categories, suggesting potential roles in modulating synaptic plasticity, neurodevelopment, neurotransmission, and neurite outgrowth. Third, quantitative expression levels of target genes isolated by ChIP were significantly altered by presence or absence of FOXP2 in human neuron-like cells, in both stable and transiently transfected systems. Fourth, EMSAs demonstrated that FOXP2 binds directly and specifically to particular sites within validated target promoters. Fifth, targets identified in human neuron-like cells showed substantial and significant overlap with those suggested by in vivo ChIP in human fetal brain tissue in an independent study that exploited a different FOXP2 antibody.21(in this issue) Finally, by studying ChIP targets in mutant mice, we have obtained in vivo validation of both binding and functional regulation in the embryonic brain.
Future research will assess the regulation of ChIP targets during embryogenesis and in postnatal brain function by use of relevant model systems, including rodents and song birds.55 With regard to human studies, a subset of neural targets of FOXP2 may represent novel candidates for involvement in language-related phenotypes. Such genes can be screened for genetic association and/or point mutations in children affected with disorders like developmental verbal dyspraxia, autism, and specific language impairment. Thus, this work, which represents the first isolation of direct neural targets regulated by FOXP2, demonstrates how functional genomics may yield exciting new insights into pathways that go awry in speech and language disorders.
Supplementary Material
Acknowledgments
We are grateful to Julian Knight, Olivia Osborn, and Sally Ashe for helpful discussions; Louise Bird for supplying bacterially expressed protein; and Svante Pääbo and Wolfgang Enard for assistance with mouse models. This study was supported by a Project Grant from the Wellcome Trust, United Kingdom, and a Pilot Grant Award from Autism Speaks, United States (both awarded to S.E.F.). S.C.V. is a Christopher Welch Biological Sciences Scholar and was supported in part by the Clarendon Fund, Oxford, United Kingdom. J.N. was supported by a Marie Curie Intra-European Fellowship. This research was supported in part by National Institutes of Health (NIH) grant 5R21MH075028. E.S. was supported by NIH grant T32GM008243. S.E.F. is a Royal Society Research Fellow.
Web Resources
The URLs for data presented herein are as follows:
- CGAP, http://cgap.nci.nih.gov/
- DAVID, http://niaid.abcc.ncifcrf.gov/
- FUZZNUC, http://bioweb.pasteur.fr/seqanal/interfaces/fuzznuc.html
- Ingenuity Systems, http://www.ingenuity.com/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FOXP2, developmental verbal dyspraxia, and Silver-Russell syndrome)
- WebGestalt, http://bioinfo.vanderbilt.edu/webgestalt/
References
- 1.Fisher SE, Lai CS, Monaco AP (2003) Deciphering the genetic basis of speech and language disorders. Annu Rev Neurosci 26:57–80 10.1146/annurev.neuro.26.041002.131144 [DOI] [PubMed] [Google Scholar]
- 2.Wassink TH, Brzustowicz LM, Bartlett CW, Szatmari P (2004) The search for autism disease genes. Ment Retard Dev Disabil Res Rev 10:272–283 10.1002/mrdd.20041 [DOI] [PubMed] [Google Scholar]
- 3.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- 4.MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, et al (2005) Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet 76:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins KE, Dronkers NF, Vargha-Khadem F (2002) Behavioural analysis of an inherited speech and language disorder: comparison with acquired aphasia. Brain 125:452–464 10.1093/brain/awf058 [DOI] [PubMed] [Google Scholar]
- 6.Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, Finucane B, Hartung D, Innes M, Kerem B, Nowaczyk MJ, et al (2006) Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet 79:965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shriberg LD, Ballard KJ, Tomblin JB, Duffy JR, Odell KH, Williams CA (2006) Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. J Speech Lang Hear Res 49:500–525 10.1044/1092-4388(2006/038) [DOI] [PubMed] [Google Scholar]
- 8.Zeesman S, Nowaczyk MJ, Teshima I, Roberts W, Cardy JO, Brian J, Senman L, Feuk L, Osborne LR, Scherer SW (2006) Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am J Med Genet A 140:509–514 [DOI] [PubMed] [Google Scholar]
- 9.Carlsson P, Mahlapuu M (2002) Forkhead transcription factors: key players in development and metabolism. Dev Biol 250:1–23 10.1006/dbio.2002.0780 [DOI] [PubMed] [Google Scholar]
- 10.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS (2003) Fox’s in development and disease. Trends Genet 19:339–344 10.1016/S0168-9525(03)00111-2 [DOI] [PubMed] [Google Scholar]
- 11.Katoh M, Katoh M (2004) Human FOX gene family (review). Int J Oncol 25:1495–1500 [PubMed] [Google Scholar]
- 12.Hanashima C, Li SC, Shen L, Lai E, Fishell G (2004) Foxg1 suppresses early cortical cell fate. Science 303:56–59 10.1126/science.1090674 [DOI] [PubMed] [Google Scholar]
- 13.Wehr R, Mansouri A, de Maeyer T, Gruss P (1997) Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development 124:4447–4456 [DOI] [PubMed] [Google Scholar]
- 14.Wang B, Lin D, Li C, Tucker P (2003) Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem 278:24259–24268 10.1074/jbc.M207174200 [DOI] [PubMed] [Google Scholar]
- 15.Watkins KE, Vargha-Khadem F, Ashburner J, Passingham RE, Connelly A, Friston KJ, Frackowiak RS, Mishkin M, Gadian DG (2002) MRI analysis of an inherited speech and language disorder: structural brain abnormalities. Brain 125:465–478 10.1093/brain/awf057 [DOI] [PubMed] [Google Scholar]
- 16.Liegeois F, Baldeweg T, Connelly A, Gadian DG, Mishkin M, Vargha-Khadem F (2003) Language fMRI abnormalities associated with FOXP2 gene mutation. Nat Neurosci 6:1230–1237 10.1038/nn1138 [DOI] [PubMed] [Google Scholar]
- 17.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ (2003) FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain 126:2455–2462 10.1093/brain/awg247 [DOI] [PubMed] [Google Scholar]
- 18.Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE (2006) Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet 15:3154–3167 10.1093/hmg/ddl392 [DOI] [PubMed] [Google Scholar]
- 19.Fisher SE, Marcus GF (2006) The eloquent ape: genes, brains and the evolution of language. Nat Rev Genetics 7:9–20 10.1038/nrg1747 [DOI] [PubMed] [Google Scholar]
- 20.Kim TH, Ren B (2006) Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet 7:81–102 10.1146/annurev.genom.7.080505.115634 [DOI] [PubMed] [Google Scholar]
- 21.Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH (2007) Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet 81:1144–1157 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberley MJ, Farnham PJ (2003) Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol 371:577–596 [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B (2003) A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA 100:8164–8169 10.1073/pnas.1332764100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31:265–273 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3 [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300 [Google Scholar]
- 27.Zhang B, Schmoyer D, Kirov S, Snoddy J (2004) GOTree Machine (GOTM): a Web-based platform for interpreting sets of interesting genes using gene ontology hierarchies. BMC Bioinformatics 5:16 10.1186/1471-2105-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 29.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L (2006) Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14:159–166 10.1016/j.str.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al (2006) FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126:375–387 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- 31.Overdier DG, Porcella A, Costa RH (1994) The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol 14:2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36 [PubMed] [Google Scholar]
- 33.Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54 10.1093/bioinformatics/14.1.48 [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Seed B (2003) A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 31:e154 10.1093/nar/gng154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 36.Pahlman S, Mamaeva S, Meyerson G, Mattsson ME, Bjelfman C, Ortoft E, Hammerling U (1990) Human neuroblastoma cells in culture: a model for neuronal cell differentiation and function. Acta Physiol Scand Suppl 592:25–37 [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Weidenfeld J, Morrisey EE (2004) Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24:809–822 10.1128/MCB.24.2.809-822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon JB, Li G, Roeder RG (1994) Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol 14:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, Macisaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445:931–935 10.1038/nature05478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH (2004) Wnt genes define distinct boundaries in the developing human brain: implications for human forebrain patterning. J Comp Neurol 474:276–288 10.1002/cne.20112 [DOI] [PubMed] [Google Scholar]
- 42.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ (2006) Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience 142:1119–1131 10.1016/j.neuroscience.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 43.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M (1994) Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901–912 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- 44.Rudolph D, Tafuri A, Gass P, Hammerling GJ, Arnold B, Schutz G (1998) Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc Natl Acad Sci USA 95:4481–4486 10.1073/pnas.95.8.4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lonze BE, Riccio A, Cohen S, Ginty DD (2002) Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron 34:371–385 10.1016/S0896-6273(02)00686-4 [DOI] [PubMed] [Google Scholar]
- 46.Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER (2002) Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 34:447–462 10.1016/S0896-6273(02)00684-0 [DOI] [PubMed] [Google Scholar]
- 47.Breslin MB, Zhu M, Lan MS (2003) NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem 278:38991–38997 10.1074/jbc.M306795200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ 3rd, Kandel ER, Duff K, et al (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42:23–36 10.1016/S0896-6273(04)00182-5 [DOI] [PubMed] [Google Scholar]
- 49.Bertram L, Parkinson M, McQueen MB, Mullin K, Hsiao M, Menon R, Moscarillo TJ, Blacker D, Tanzi RE (2005) Further evidence for LBP-1c/CP2/LSF association in Alzheimer’s disease families. J Med Genet 42:857–862 10.1136/jmg.2004.024596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF (2001) Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem 276:37672–37679 10.1074/jbc.M104521200 [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY (2007) Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445:936–940 10.1038/nature05563 [DOI] [PubMed] [Google Scholar]
- 52.Bird AJ, Blankman E, Stillman DJ, Eide DJ, Winge DR (2004) The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J 23:1123–1132 10.1038/sj.emboj.7600122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Washburn BK, Esposito RE (2001) Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol Cell Biol 21:2057–2069 10.1128/MCB.21.6.2057-2069.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scharff C, Haesler S (2005) An evolutionary perspective on FoxP2: strictly for the birds? Curr Opin Neurobiol 15:694–703 10.1016/j.conb.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 55.White SA, Fisher SE, Geschwind DH, Scharff C, Holy TE (2006) Singing mice, songbirds, and more: models for FOXP2 function and dysfunction in human speech and language. J Neurosci 26:10376–10379 10.1523/JNEUROSCI.3379-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishibashi K, Matsuzaki T, Takata K, Imai M (2003) Identification of a new member of type I Na/phosphate co-transporter in the rat kidney. Nephron Physiol 94:10–18 10.1159/000071070 [DOI] [PubMed] [Google Scholar]
- 57.Reimer RJ, Edwards RH (2004) Organic anion transport is the primary function of the SLC17/type I phosphate transporter family. Pflugers Arch 447:629–635 10.1007/s00424-003-1087-y [DOI] [PubMed] [Google Scholar]
- 58.Strausberg RL (2001) The Cancer Genome Anatomy Project: new resources for reading the molecular signatures of cancer. J Pathol 195:31–40 [DOI] [PubMed] [Google Scholar]
- 59.Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS (2006) T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem 281:11992–12000 10.1074/jbc.M513613200 [DOI] [PubMed] [Google Scholar]
- 60.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S (2002) Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418:869–872 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Webb DM, Podlaha O (2002) Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics 162:1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.