Abstract
Recently, association of celiac disease with common single-nucleotide polymorphism (SNP) variants in an extensive linkage-disequilibrium block of 480 kb containing the KIAA1109, Tenr, IL2, and IL21 genes has been demonstrated in three independent populations (rs6822844 Pcombined=1.3×10-14). The KIAA1109/Tenr/IL2/IL21 block corresponds to the Idd3 locus in the nonobese diabetic mouse model of type 1 diabetes (T1D). This block was recently found to be associated with T1D in a genomewide association study, although this finding lacks unequivocal confirmation. We therefore aimed to investigate whether the KIAA1109/Tenr/IL2/IL21 region is involved in susceptibility to multiple autoimmune diseases. We tested SNP rs6822844 for association with disease in 350 T1D-affected and 1,047 rheumatoid arthritis (RA)–affected Dutch patients and in 929 controls. We replicated the association with T1D (P=.0006; OR 0.64 [95% CI 0.50–0.83]), and revealed a similar novel association with RA (P=.0002; OR 0.72 [95% CI 0.61–0.86]). Our results replicate and extend the association found in the KIAA1109/Tenr/IL2/IL21 gene region with autoimmune diseases, implying that this locus is a general risk factor for multiple autoimmune diseases.
Type 1 diabetes (T1D [MIM 222100]), rheumatoid arthritis (RA [MIM 180300]), and celiac disease (CD [MIM 212750]) are common autoimmune diseases (AIDs), each affecting ∼1% of the general population. The manifestation and progression of AIDs depend on a combination of multiple genetic and environmental factors, yet the co-occurrence of these three different AIDs in families, or even within single patients, has been reported frequently.1–3 For instance, 6.2% of patients with T1D also have CD,4 and 2.8% of first-degree relatives of RA-affected probands have T1D.1 This overlap in etiology is most likely due to a shared genetic predisposition to autoimmunity. The well-known and major genetic risk factor for all three diseases are the human leukocyte antigen class II genes. Other risk factors in common that have been confirmed in various AIDs are the PTPN22 (MIM 600716) (in T1D and RA) and CTLA4 (MIM 123890) (in T1D and CD) genes, where the differences in overlap may point to differences in disease pathways.5
The region encompassing KIAA1109/Tenr/IL2/IL21 is contained in a large block (480 kb) of linkage disequilibrium (LD) recently reported to be a strong genetic factor involved in CD.6 This block is located on chromosome 4q27 and includes the IL2 (MIM 147680) and IL21 (MIM 605384) genes, which are both plausible functional candidate loci for AIDs. IL2 is a susceptibility gene in the nonobese diabetic (NOD) mouse model of T1D,7 and association of the IL2 receptor (IL2RA and CD25 [MIM 147730]) to T1D has been demonstrated in extended cohorts, highlighting the importance of the IL2 pathway in the predisposition to AIDs.8,9 Both IL2 and IL21 belong to the type 1 cytokine family, share a large degree of homology, and possess pleotropic functions in immune cells.10,11
Hence, the association of the KIAA1109/Tenr/IL2/IL21 gene region with T1D in mouse and with CD in human may reflect a general role for this locus in the etiology of autoimmunity. Given the comorbidity among CD, T1D, and RA, we sought to confirm this hypothesis by testing the most associated CD variant from the U.K. genomewide association (GWA) screen (rs6822844) in a Dutch T1D cohort and a Dutch RA cohort.
The patients with T1D were retrieved from the Kolibri T1D cohort, which includes 350 patients with juvenile-onset T1D (median age 8.7 years [range 1–17 years]). The cohort was collected consecutively after diagnosis by pediatricians in the southwestern part of the Netherlands between 1995 and 1999. The diagnosis was made according to International Society of Pediatric and Adolescent Diabetes and World Health Organization criteria. The RA group comprised two independent cohorts from Groningen (n=408) and Nijmegen (n=639). The characteristics of patients with RA in the Nijmegen cohort have been described elsewhere.12 All the patients were given diagnoses in accordance with American College of Rheumatology (ACR) criteria for RA,13 had a disease duration of >1 year, and had no history of using disease-modifying antirheumatic drugs or biological agents before presentation. The patients with RA in the Groningen cohort were recruited from the outpatient clinic of the Department of Rheumatology, University Medical Center Groningen (UMCG). All patients were given diagnoses in accordance with ACR criteria13 and had rheumatoid factor (RF)–positive and/or RF-erosive RA. Patients with T1D and RA were first compared with a panel of 929 unrelated Dutch controls who were genotyped and presented in the replication GWA study for CD.6 All the patients and controls gave their informed consent, and the medical ethics committees of the University Medical Center Utrecht, the UMCG, and the Radboud University Nijmegen Medical Center approved the respective original studies.
Genotyping was performed using TaqMan technology. SNP genotyping assays for PCR were supplied by Applied Biosystems, as described elsewhere.6 The DNA samples were processed in 384-well plates. Each plate contained 8 negative controls and 16 genotyping controls (4 duplicates of four different CEPH samples). In the control group, the frequencies of all the SNPs were in Hardy-Weinberg equilibrium. Hardy-Weinberg equilibrium was tested by comparing the expected and observed genotypes in a 2×3 χ2 table. Odds ratios (ORs) were calculated, and the CIs were approximated using Woolf’s method with Haldane’s correction.14
The rs6822844 G→T SNP was tested in 350 patients with early-onset T1D and in 1,047 patients with RA. We saw a decrease in frequency of the rs6822844*T allele in both T1D (12.7%) and RA (14.1%) groups compared with controls (18.5%); this is similar to our earlier result for CD in the Dutch population (12.4%).6 The association was significant in both T1D (P=.0006; OR 0.64 [95% CI 0.50–0.83]) and RA (P=.0002; OR 0.72 [95% CI 0.61–0.86]) (table 1).
Table 1. .
Frequency of SNP rs6822844 in Controls (n=929), Patients with T1D (n=350), and Patients with RA (n=1,047)[Note]
| Patients with T1D |
Patients with RA |
||||||
| rs6822844 | Control Counts (%) | Counts (%) | OR (95% CI) | P | Counts (%) | OR (95% CI) | P |
| Allele G | 1,506 (81.5) | 585 (87.3) | 1 (ref) | … | 1,739 (85.9) | 1 (ref) | … |
| Allele T | 342 (18.5) | 85 (12.7) | .64 (.50–.83) | .0006 | 285 (14.1) | .72 (.61–.86) | .0002 |
| GG | 613 (66.3) | 258 (77.0) | 1 (ref) | … | 748 (73.9) | 1 (ref) | … |
| GT | 280 (30.3) | 69 (20.6) | .59 (.44–.80) | .0014 | 243 (24.0) | .71 (.58–.87) | .0009 |
| TT | 31 (3.4) | 8 (2.4) | .67 (.31–1.44) | … | 21 (2.1) | .56 (.32–.99) | … |
Note.— ref = Reference. The number of dropouts was 5 in controls, 14 in patients with T1D, and 35 in patients with RA. Reported P values were obtained with 1-df test for allelic effect and 2-df test for genotyping effect.
While we were performing this study, the results of a GWA study of T1D became available.15 One of the reported loci was on 4q27 and contained the KIAA1109/Tenr/IL2/IL21 gene region. A follow-up replication study indicated moderate association of SNPs in the KIAA1109/Tenr/IL2/IL21 gene region with T1D.8 We compared the SNPs from the T1D GWA study with the haplotype-tagging (ht) SNPs that we used for replicating the CD GWA results and observed that the most T1D-associated SNP (rs3136534 A→C) was in complete LD with one of the SNPs included in the CD screen (rs4505848 A→G) (r2=1; LOD 32.26 [HapMap data]), where the rs3136534*A allele corresponds to rs4505848*A and rs3136534*C corresponds to rs4505848*G. The haplotype structure and LD in the region were investigated using HapMap data16 with the Haploview application.17
We also genotyped the rs4505848 A→G SNP in our cohorts of patients with T1D and RA and found a slight increase of the rs4505848*G allele in patients with T1D (36.2%) and RA (37.1%) compared with controls (34.9%). Although the observed difference in allele frequency was not significant in either T1D (P=.53; OR 1.06 [95% CI 0.88–1.28]) or RA (P=.15; OR 1.10 [95% CI 0.97–1.26]), it did show a trend similar to that reported for the U.K. T1D-affected population (OR 1.11)8,15 (table 2).
Table 2. .
Frequency of SNP rs4505848 in Controls (n=929), Patients with T1D (n=350), and Patients with RA (n=1,047)[Note]
| Patients with T1D |
Patients with RA |
||||||
| rs4505848 | Control Counts (%) | Counts (%) | OR (95% CI) | P | Counts (%) | OR (95% CI) | P |
| Allele A | 1,180 (65.1) | 417 (63.8) | 1 (ref) | … | 1,270 (62.9) | 1 (ref) | … |
| Allele G | 632 (34.9) | 237 (36.2) | 1.06 (.88–1.28) | .53 | 750 (37.1) | 1.10 (.97–1.26) | .15 |
| AA | 380 (41.9) | 144 (44.0) | 1 (ref) | … | 396 (39.2) | 1 (ref) | … |
| AG | 420 (46.4) | 129 (39.4) | .81 (.62–1.07) | .03 | 478 (47.3) | 1.09 (.90–1.32) | .34 |
| GG | 106 (11.7) | 54 (16.5) | 1.35 (.93–1.97) | … | 136 (13.5) | 1.23 (.92–1.64) | … |
Note.— ref = Reference. The number of dropouts was 23 in controls, 23 in patients with T1D, and 37 in patients with RA. Reported P values were obtained with 1-df test for allelic effect and 2-df test for genotyping effect.
We further investigated the haplotypes of the KIAA1109/Tenr/IL2/IL21 gene region by typing three additional ht SNPs: rs11732095, rs4492018, and rs1398553. The combined five SNPs tag seven haplotypes of the block in which rs6822844 is located. These haplotypes have a combined frequency of >98%. Interestingly, rs6822844 is a perfect proxy for the AATGG haplotype, which is the most associated haplotype in all three diseases with haplotype frequencies of ∼13% in cases versus 18.6% in controls (table 3).
Table 3. .
Haplotype Analysis of SNPs rs4505848, rs11732095, rs6822844, rs4492018, and rs1398553[Note]
| Patients with RA |
Patients with T1D |
Patients with CD |
||||||||
| Haplotype | Control Counts (%) | Counts (%) | OR (95% CI) | P | Counts (%) | OR (95% CI) | P | Counts (%) | OR (95% CI) | P |
| GAGGA | 536 (29.1) | 636 (31.2) | 1 | Ref | 192 (29.0) | 1 | Ref | 312 (31.1) | 1 | Ref |
| AAGAG | 409 (22.2) | 433 (21.2) | .89 (.75–1.07) | .21 | 161 (24.3) | .91 (.71–1.16) | .45 | 216 (21.5) | .91 (.73–1.13) | .38 |
| AATGG | 343 (18.6) | 283 (13.9) | .70 (.57–.85) | .00025 | 87 (13.2) | .71 (.53–.95) | .018 | 124 (12.4) | .62 (.49–.80) | .00016 |
| AAGGG | 213 (11.6) | 293 (14.3) | 1.16 (.94–1.43) | .17 | 81 (12.2) | 1.07 (.79–1.44) | .70 | 150 (15.0) | 1.21 (.94–1.56) | .14 |
| AGGGG | 154 (8.3) | 176 (8.6) | 1.04 (.81–1.33) | .76 | 49 (7.5) | .90 (.63–1.29) | .52 | 91 (9.0) | 1.02 (.76–1.37) | .92 |
| GAGGG | 101 (5.4) | 108 (5.3) | .90 (.67–1.21) | .49 | 41 (6.2) | 1.15 (.77–1.70) | .54 | 61 (6.1) | 1.04 (.74–1.47) | .84 |
| AAGGA | 67 (3.6) | 71 (3.5) | .89 (.63–1.27) | .53 | 43 (6.5) | 1.72 (1.13–2.62) | .01 | 34 (3.4) | .88 (.57–1.36) | .54 |
Note.— Combined frequency of haplotypes presented is >98%. Ref = reference.
Interestingly, a reported RA association with PTPN22-1858T, another general autoimmune risk factor, was preferentially observed in the subgroup of patients with RF-positive RA18; similarly, linkage to the PTPN22 region is also limited to families with RF-positive RA.19 We investigated whether the association in our RA group was dependent on the presence of RF. Information about RF was available for 776 individuals genotyped for rs6822844, of whom 664 had RF-positive RA. We did not observe a significant difference in association between the RF-positive and RF-negative groups, although the protective effect in the RF-positive group was moderately stronger than in the RF-negative group (OR 0.71 [95% CI 0.59–0.87] and OR 0.89 [95% CI 0.61–1.29], respectively) (table 4).
Table 4. .
Frequency of SNP rs6822844 in Patients with RF-Positive (n=664) and RF-Negative (n=112) RA Compared with Controls[Note]
| RF Positive |
RF Negative |
||||||
| rs6822844 | Control Counts (%) | Counts (%) | OR (95% CI) | P | Counts (%) | OR (95% CI) | P |
| Allele G | 1,506 (81.5) | 1,143 (86.1) | 1 (ref) | … | 187 (83.5) | 1 (ref) | … |
| Allele T | 342 (18.5) | 185 (13.9) | .71 (.59–.87) | .0006 | 37 (16.5) | .89 (.61–1.29) | .47 |
| GG | 613 (66.3) | 491 (73.9) | 1 (ref) | … | 79 (70.5) | 1 (ref) | … |
| GT | 280 (30.3) | 161 (24.2) | .72 (.57–.90) | .0028 | 29 (25.9) | .82 (.52–1.28) | .63 |
| TT | 31 (3.4) | 12 (1.8) | .51 (.26–.99) | … | 4 (3.6) | 1.20 (.43–3.31) | … |
Note.— ref = Reference.
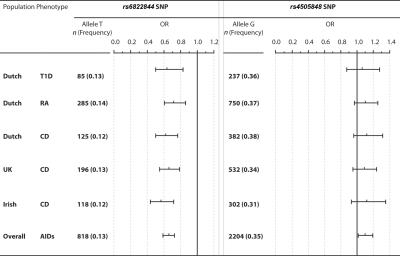
To estimate overall association of the studied SNPs with AIDs in the Dutch population, we then pooled all the Dutch patients with T1D, RA, and CD into one group (Dutch AIDs), which was compared with controls. This group was then included in a meta-analysis with recently published data from a U.K. and Irish CD genome-scan study presented elsewhere,6 with use of a random-effect model. There was no significant heterogeneity among the studies. In total, 6,236 patient chromosomes and 5,812 control chromosomes were included in the meta-analysis. Overall, a meta-analysis of our findings showed a consistently lower frequency of the rs6822844*T allele (random model test for an overall effect, P<.00001). It yielded a 1.5-fold decrease (95% CI 1.36–1.69) in risk of AIDs in carriers (OR 0.67 [95% CI 0.59–0.74]) (fig. 1).
Figure 1. .
The frequency of rs6822844 and rs4505848 alleles in Dutch AIDs cohorts and controls and in a meta-analysis of Dutch, U.K., and Irish AIDs cohorts. Dutch AIDs cohorts include T1D (n=670), CD (n=1,012), and RA (n=2,024). Data for the U.K. and Irish cohorts were extracted from the work of van Heel et al.6 n = Number of chromosomes. ORs and the corresponding 95% CIs were estimated using a random-effect model. Vertical lines represent no effect (i.e., disease risk = 1).
In this study, we have identified the 4q27 KIAA1109/Tenr/IL2/IL21 gene region as a general autoimmune locus, since we have shown genetic association with three AIDs—namely, CD, T1D, and RA. These results were corroborated by the recent report of a GWA study of T1D.15 The KIAA1109/Tenr/IL2/IL21 block is characterized by extremely strong LD that shows a very similar pattern in the different populations studied (HapMap CEU, Dutch, U.K., and Irish).6 Four SNPs, all in strong LD with each other, tag the same haplotype and show strong association with CD in multiple populations. Other common SNPs located in the KIAA1109/Tenr/IL2/IL21 gene region have shown strong association with T1D in a recent GWA study.15 However, a large replication study of T1D in a U.K. population reported only moderate association with rs3136534, the most associated SNP in the study.8 rs4505848 is a perfect proxy for rs3136534 (r2=1), and it was included in both the CD GWA study and the replication analysis. In the three CD-affected populations tested, this proxy SNP was related to only a moderate increase in the frequency of the G allele compared with controls (P value is nonsignificant in Dutch, U.K., and Irish cohorts).6 When we investigated the T1D proxy SNP rs4505848 in our AIDs cohort, we did not detect significant associations with either T1D or RA, although there was a similar increase in frequency of the G allele in both groups (fig. 1). Our haplotype analysis showed that a single haplotype shows consistent association with the majority of the decreased risk for AID. The rs6822844*T allele differentiates this haplotype perfectly from the others, indicating that, in our data set, this allele is the best proxy for the unknown disease variant. Further investigation of this region, including comprehensive detection and testing of all variants, is required to pinpoint the underlying disease variant.
The 4q27-associated block contains four genes: KIAA1109, Tenr, IL2, and IL2. The extensive LD within this block means that none of these genes could be excluded by genetic methods from being the causal one. The two genes IL2 and IL21 are both plausible functional candidates as genetic modifiers of autoimmunity. The importance of the IL2 pathway in T1D is underpinned by recent findings that have shown that regulatory variants in the IL2 gene modify the predisposition to organ-specific AID in NOD mice.7 The IL2 receptor (CD25) is also a proven T1D-susceptibility locus that has recently been reported to be associated with RA.15 These considerations highlight the importance of the IL2 pathway in autoimmunity.
IL21 is involved in both cell-mediated and humoral responses and has a pleiotropic effect on a variety of immune and nonimmune cells.10 A crucial role of IL21 in autoimmune pancreatic B-cell destruction was demonstrated in NOD mice,20 and a contribution from IL21 to the progression of lupus in mouse models has also been reported.21,22 IL21 has a crucial role in regulation of antibody production. The role of IL21 in eliminating autoreactive B cells through apoptosis suggests that dysregulation of IL21-IL21 receptor signaling might contribute to the development of antibody-mediated AIDs.10
In conclusion, we have now shown association of T1D and RA with the KIAA1109/Tenr/IL2/IL21 region. Summarizing our findings with previous data about the association of 4q27 with CD and T1D leads us to conclude that the KIAA1109/Tenr/IL2/IL21 region is a general autoimmune risk locus.
Acknowledgments
We thank all patients and families for participating in the study. This study was supported by grants from the Celiac Disease Consortium, an Innovative Cluster approved by the Netherlands Genomics Initiative, and was partially funded by Dutch Government grant BSIK03009, Netherlands Organization for Scientific Research grant 918.66.620, European Union STREP KP6 grant 036383, the Dutch Diabetes Research Foundation grant 97.137, the Netherlands Organization for Health Research and Development (ZonMW), the Juvenile Diabetes Research Foundation International grant 2001.10.004, and the Wellcome Trust Clinician Scientist Fellowship grant GR068094MA (to D.A.v.H.). We are indebted to Erik Toonen for collecting DNA from patients with RA. We are grateful to Jaap Fransen for his analysis of the clinical characteristics of the patients with RA. We thank all the people from the Department of Rheumatology in Nijmegen who were involved in setting up and monitoring the database of patients with RA. We thank Harry van Someren for his role as database manager and Jackie Senior for critically reading the manuscript.
Web Resources
The URLs for data presented herein are as follows:
- HapMap, http://www.hapmap.org/ (for information about SNPs and LD in population controls)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for T1D, RA, CD, PTPN22, CTLA4, IL2, IL21, and CD25)
References
- 1.Lin JP, Cash JM, Doyle SZ, Peden S, Kanik K, Amos CI, Bale SJ, Wilder RL (1998) Familial clustering of rheumatoid arthritis with other autoimmune diseases. Hum Genet 103:475–482 10.1007/s004390050853 [DOI] [PubMed] [Google Scholar]
- 2.Levin L, Tomer Y (2003) The etiology of autoimmune diabetes and thyroiditis: evidence for common genetic susceptibility. Autoimmun Rev 2:377–386 10.1016/S1568-9972(03)00080-6 [DOI] [PubMed] [Google Scholar]
- 3.Tait KF, Marshall T, Berman J, Carr-Smith J, Rowe B, Todd JA, Bain SC, Barnett AH, Gough SC (2004) Clustering of autoimmune disease in parents of siblings from the Type 1 diabetes Warren repository. Diabet Med 21:358–362 10.1111/j.1464-5491.2004.01162.x [DOI] [PubMed] [Google Scholar]
- 4.Barera G, Bonfanti R, Viscardi M, Bazzigaluppi E, Calori G, Meschi F, Bianchi C, Chiumello G (2002) Occurrence of celiac disease after onset of type 1 diabetes: a 6-year prospective longitudinal study. Pediatrics 109:833–838 10.1542/peds.109.5.833 [DOI] [PubMed] [Google Scholar]
- 5.Brand O, Gough S, Heward J (2005) HLA, CTLA-4 and PTPN22: the shared genetic master-key to autoimmunity? Expert Rev Mol Med 7:1–15 10.1017/S1462399405009981 [DOI] [PubMed] [Google Scholar]
- 6.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, et al (2007) A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet 39:827–829 10.1038/ng2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, et al (2007) Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 39:329–337 10.1038/ng1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39:857–864 10.1038/ng2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME, et al (2005) Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet 76:773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard WJ, Spolski R (2005) Interleukin-21: a modulator of lymphoid proliferation, apoptosis and differentiation. Nat Rev Immunol 5:688–698 10.1038/nri1688 [DOI] [PubMed] [Google Scholar]
- 11.Brandt K, Singh PB, Bulfone-Paus S, Ruckert R (2007) Interleukin-21: a new modulator of immunity, infection, and cancer. Cytokine Growth Factor Rev 18:223–232 10.1016/j.cytogfr.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 12.Welsing PM, van Riel PL (2004) The Nijmegen inception cohort of early rheumatoid arthritis. J Rheumatol Suppl 69:14–21 [PubMed] [Google Scholar]
- 13.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31:315–324 10.1002/art.1780310302 [DOI] [PubMed] [Google Scholar]
- 14.Haldane JB (1956) The estimation and significance of the logarithm of a ratio of frequencies. Ann Hum Genet 20:309–311 10.1111/j.1469-1809.1955.tb01285.x [DOI] [PubMed] [Google Scholar]
- 15.Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 18.Lee AT, Li W, Liew A, Bombardier C, Weisman M, Massarotti EM, Kent J, Wolfe F, Begovich AB, Gregersen PK (2005) The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun 6:129–133 10.1038/sj.gene.6364159 [DOI] [PubMed] [Google Scholar]
- 19.Michou L, Lasbleiz S, Rat AC, Migliorini P, Balsa A, Westhovens R, Barrera P, Alves H, Pierlot C, Glikmans E, et al (2007) Linkage proof for PTPN22, a rheumatoid arthritis susceptibility gene and a human autoimmunity gene. Proc Natl Acad Sci USA 104:1649–1654 10.1073/pnas.0610250104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King C, Ilic A, Koelsch K, Sarvetnick N (2004) Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell 117:265–277 10.1016/S0092-8674(04)00335-6 [DOI] [PubMed] [Google Scholar]
- 21.Herber D, Brown TP, Liang S, Young DA, Collins M, Dunussi-Joannopoulos K (2007) IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J Immunol 178:3822–3830 [DOI] [PubMed] [Google Scholar]
- 22.Ozaki K, Spolski R, Ettinger R, Kim HP, Wang G, Qi CF, Hwu P, Shaffer DJ, Akilesh S, Roopenian DC, et al (2004) Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol 173:5361–5371 [DOI] [PubMed] [Google Scholar]