Abstract
We have conducted a multistage genomewide association study, using 1,620,742 single-nucleotide polymorphisms to systematically investigate the genetic factors influencing intrinsic skin pigmentation in a population of South Asian descent. Polymorphisms in three genes—SLC24A5, TYR, and SLC45A2—yielded highly significant replicated associations with skin-reflectance measurements, an indirect measure of melanin content in the skin. The associations detected in these three genes, in an additive manner, collectively account for a large fraction of the natural variation of skin pigmentation in a South Asian population. Our study is the first to interrogate polymorphisms across the genome, to find genetic determinants of the natural variation of skin pigmentation within a human population.
Humans possess an impressive range of skin pigmentation, both within and between populations. This diversity is highly correlated with geographical location, indicating that environmental factors as well as genetics strongly influence skin color. The predominant environmental variable affecting skin pigmentation is sunlight, and it is certain that skin pigments play an important role in both protecting DNA from the effects of UV irradiation1–4 and influencing the availability of UV radiation for the synthesis of necessary compounds, such as vitamin D.5–7 Epidemiological studies in humans show that skin pigmentation is a polygenic quantitative trait with high heritability,8–10 and, with the direct correlation between skin pigmentation and incident UV exposure, it has long been postulated that it is a trait under intense selective pressure.11–14 Because of its striking diversity and potential evolutionary importance as a highly selected trait, human skin pigmentation is of great scientific interest. Over the past 100 years, pigmentary mutants in model organisms and human pigmentation disorders have been the main source for the discovery of genes involved in skin color. More than 100 pigmentation genes have been identified in mouse alone, most with identified human orthologs, and at least 18 genes are currently listed in OMIM as being involved in human albinism.15,16 However, until very recently, few direct studies to identify the genes responsible for the normal range of human skin pigmentation have been reported13,17–26; in addition, all these studies have investigated the role of only known pigmentation-disorder genes in normal skin-color variation. Therefore, many questions about the loci responsible for the natural diversity of human skin pigmentation have remained unanswered: how many genes are involved, are the different gene effects additive, are the genes involved in skin pigmentation the same across different ethnic populations, and are there still-undiscovered pigmentation genes?
With the availability of the entire human genomic sequence in 2001,27,28 the identification of millions of SNPs across the genome,29–31 and the development of high-throughput genotyping technologies, the tools were available for investigation of the genetic components controlling human skin pigmentation with use of a high-density genomewide association study. In the present study, we applied a three-tiered methodology of quantitative pooled genotyping followed by individual genotyping of associated SNPs in original and replicate population sets, as has been successfully used elsewhere.32
In all, 1,620,742 SNPs across the genome were measured for allele-frequency differences between DNA pools made from the 20% tails of the skin-pigmentation distribution, as objectively measured by reflectance spectroscopy in a South Asian population. The top 30,000 SNPs (∼2%) with the largest allele-frequency differences between the pools were individually genotyped in the original population sample of 737 individuals, followed by individual genotyping of associated SNPs in an independent replicate sample of 231 individuals of South Asian ancestry. Despite confounding of phenotype-genotype associations by population stratification, we found three genes with replicated genomewide significance that together account for a large fraction of the skin-pigmentation variation in the South Asian population.
Material and Methods
Sample Selection
Approval for the study was obtained from ethics review boards in the United Kingdom. After a complete description of the study was given to the subjects, written informed consent was obtained. Recruitment of volunteers of South Asian descent was conducted at >50 different sites across the United Kingdom. For each qualifying subject, either both parents or all four grandparents were born in India, Pakistan, Bangladesh, or Sri Lanka. Reported ancestry was used to group subjects by region into eastern (mainly Bangladesh), northwestern (mainly Gujarat and Punjab), southern (mainly Sri Lanka), or mixed origin. Reported ancestry was defined for each individual from the grandparental information if known and from the parental information otherwise. If the country and state of birth of all four grandparents were the same, then the reported ancestry of the volunteer was defined similarly; this was also the case if there was missing information from one grandparent but the origin of the other three grandparents was the same. Furthermore, if information from both maternal and/or paternal grandparents was missing, then maternal and/or paternal information was used in its place. Subjects with disagreements about reported ancestry were categorized as “mixed.” Both males and females aged ⩾18 years were included. Volunteers were excluded if they reported consumption of oral dietary foods or supplements or the use of topical skin ointments on the measurement areas that are designed to change skin color. Other exclusion criteria included pigmentation disorders or any current skin disease of any type anywhere on the body.
Phenotype and DNA Collection
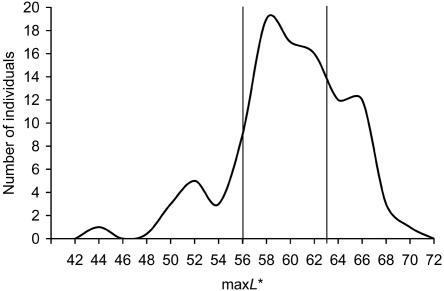
Skin pigmentation was measured with a Minolta chromameter with use of the Commission Internationale de l’Eclairage L*a*b* color system. The L* value, which measures skin reflectance, or lightness, ranges from 0 to 100, where 0 is the darkest and 100 is the lightest skin pigmentation. Skin reflectance was measured on three relatively hairless sites per arm—the sun-exposed lower dorsal forearm, the sun-protected inner volar forearm, and the sun-protected inner arm above the elbow—giving six measurements per volunteer. The highest of the six L* values was defined as maxL*. Skin chromameter measurements for 98 randomly selected individuals of South Asian ancestry living in the United Kingdom were used to estimate the distribution of maxL* within the South Asian population (fig. 1). Individuals were selected for the first phase of our association study by targeted recruitment in the upper and lower quintiles of maxL* values determined as described above. Targeted recruitment of individuals was conducted by prescreening volunteers for skin color by visual assessment. More than 2,800 volunteers were enrolled in the study and had their skin reflectance measured, with blood samples taken from >1,100 individuals for the isolation of DNA. Genomic DNA was extracted from whole blood with use of the Qiagen DNA isolation kit, in accordance with the manufacturer’s instructions. The sample recruited for the pooled genotyping phase of the study (cohort 1) consists of 395 individuals with maxL* values ⩽56 and 383 individuals with maxL* values ⩾63, who were classified as “L” for having low reflectance and as “H” for having high reflectance, respectively. For the replication population (cohort 2), 119 L and 116 H individuals were recruited.
Figure 1. .
Histogram of maximum L* values (maxL*) in volunteers of South Asian ancestry. The maxL* values were obtained from 98 randomly selected individuals of South Asian ancestry living in the United Kingdom. The maxL* values for the 20th and 80th percentiles of the distribution are indicated by vertical lines.
Population-Structure Analysis
The genetic ancestry of the individuals in cohort 1 was examined by individually genotyping them on a set of 312 SNPs, referred to as “genomic control” (GC) SNPs, which are spaced approximately uniformly across the human autosomes.33 Population structure within the study population was evaluated using the structure program34 with models having 1–4 ancestral clusters. For each model, subsets of the L and H individuals were selected so as to construct pools matched on mean cluster membership, as described elsewhere.32 Simple χ2 tests for association with skin reflectance were performed for these GC SNPs. Any residual inflation in test statistics was assessed in comparison with the experimental error of allele-frequency determination in pooled genotyping.32
For the association analysis of individual genotypes, both population cohorts used in this study were assessed jointly by use of principal-components analysis (PCA)35 of standardized genotypes on the GC SNPs. The presence of population structure increases the fraction of genotypic variance that is accounted for by the first few principal components. The genotypes and phenotypes are subsequently adjusted for these to project out the contributions of ancestry to the genotypes. To assess the significance of the eigenvalues corresponding to the first several principal components, the PCA was repeated for 100 permutations of the genotype data in which SNP genotypes were scrambled, so as to eliminate all SNP-SNP correlations.
Pooled Genotyping on Oligonucleotide Arrays
From cohort 1, 286 L individuals and 285 H individuals were used to construct two DNA pools, L and H, respectively, for the estimation of 1,620,742 SNP allele-frequency differences between the low- and high-reflectance groups, with the use of methods described elsewhere.32,36 For pooled genotyping, each DNA pool was amplified in quadruplicate with the use of 266,851 long-range PCRs; each PCR replicate was then pooled, labeled, hybridized to 228 high-density arrays, stained, and detected as described elsewhere.36
Determination of Pooled Allele-Frequency Estimates
Estimates of the pooled allele frequency, 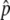 , were computed from fluorescence intensities on the arrays, as described elsewhere.32 For each SNP, we obtained eight independent measurements of
, were computed from fluorescence intensities on the arrays, as described elsewhere.32 For each SNP, we obtained eight independent measurements of 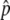 , four for each DNA pool. The allele-frequency difference between the L and H pools, Δ
, four for each DNA pool. The allele-frequency difference between the L and H pools, Δ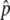 , was determined from the average for each pool. We also used an independently derived genomewide haplotype map35 to obtain a better estimate of the allele-frequency difference for some SNPs, using a method described elsewhere.32 In brief, within each haplotype block across the genome, linear regression was used to solve for frequency differences of the underlying common haplotype patterns from the measured estimates of Δ
, was determined from the average for each pool. We also used an independently derived genomewide haplotype map35 to obtain a better estimate of the allele-frequency difference for some SNPs, using a method described elsewhere.32 In brief, within each haplotype block across the genome, linear regression was used to solve for frequency differences of the underlying common haplotype patterns from the measured estimates of Δ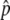 . A good quality of fit (P<.05 for the F test) was considered to be indicative of conformance between the genetic structure of the population in our study and the independently derived haplotype map. For such haplotype-conforming SNPs, the regression provided improved estimates of allele-frequency differences, called fitted Δ
. A good quality of fit (P<.05 for the F test) was considered to be indicative of conformance between the genetic structure of the population in our study and the independently derived haplotype map. For such haplotype-conforming SNPs, the regression provided improved estimates of allele-frequency differences, called fitted Δ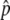 , by effectively averaging over redundant information within each haplotype block. These linear regressions within haplotype blocks also allowed for the elimination of some redundancy in the SNP set selected for subsequent individual genotyping.
, by effectively averaging over redundant information within each haplotype block. These linear regressions within haplotype blocks also allowed for the elimination of some redundancy in the SNP set selected for subsequent individual genotyping.
The selection of SNPs for further assessment with individual genotyping used a ranking by the magnitude of the estimated allele-frequency difference |Δ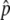 |. For haplotype-conforming SNPs, a lower threshold was employed on the fitted |Δ
|. For haplotype-conforming SNPs, a lower threshold was employed on the fitted |Δ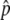 |, on the basis of the fact that estimates of allele-frequency differences are better for these SNPs. This approach to selecting SNPs from pooled genotype data for subsequent individual genotyping has been explained in detail elsewhere.32
|, on the basis of the fact that estimates of allele-frequency differences are better for these SNPs. This approach to selecting SNPs from pooled genotype data for subsequent individual genotyping has been explained in detail elsewhere.32
Individual Genotyping on Oligonucleotide Arrays
High-density oligonucleotide arrays for individual genotyping were designed as described elsewhere,30 with each SNP interrogated by 40 distinct 25-mer probes. DNA samples were amplified by short-range multiplex PCR and were labeled, hybridized to the arrays, stained, and detected as described elsewhere.33 The individual genotypes were determined by clustering measurements from multiple scans in the two-dimensional space defined by reference and alternate perfect-match trimmed mean intensities, as described elsewhere.37,38 Quality-control filters were applied to both the DNA samples and the SNPs. DNA samples with call rates <0.75 or that showed evidence of misidentification were excluded from analysis. SNPs genotyped in <0.8 of the DNA samples were excluded from further analysis. Deviations from Hardy-Weinberg equilibrium (HWE) are sometimes indicative of problems with genotype clustering; however, such deviations could also legitimately arise because of population structure or association with the trait of interest. Therefore, SNPs that did not follow HWE at a level of P<.001 were discarded in most cases, with exceptions for SNPs that were also statistically significant at a level of P⩽.001 on a simple trend test for association with skin reflectance corrected with GC.39 For these potentially associated SNPs, the clustering was visually inspected, and SNPs were excluded from further analysis if problems were detected.
Single-SNP Association Tests
Association tests were performed with logistic regression, with covariates to account for sex and population structure, with the use of likelihood-ratio tests to assess the significance of association with SNP genotypes. Population structure was represented by the first several principal components of the genotype matrix for the set of individually genotyped GC SNPs (see the “Population-Structure Analysis” section). Sex is a known confounder with skin pigmentation24 that was not explicitly matched for in selecting study participants; thus, it was also included as a covariate in logistic regression. The model for our primary single-SNP association test can be written as
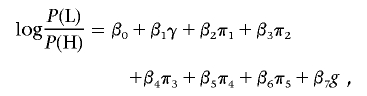 |
where P(H) is the likelihood of membership in the H group, P(L) is the likelihood of membership in the L group, γ is a factor denoting sex, π1–π5 are the first five principal components determined as described in the “Population-Structure Correction in the Association Analysis” section, and g is the SNP genotype of the individual, coded as g=0,1,2 on the basis of the count of an arbitrarily designated allele. This linear coding of genotypes corresponds to a multiplicative model of risk. A likelihood-ratio test was used to test for association of skin reflectance with genotype, comparing the model above with a null model without the genotype term,
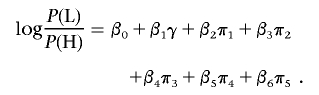 |
We tested for residual systematic inflation in the test statistics by applying this likelihood-ratio test to all samples taken together (cohorts 1 and 2) for the GC SNPs that were polymorphic in the study population and that passed our quality filters for individual genotyping. Other features of interest in the genotype data (dominance, epistasis, and independence of associations) were modeled by adding corresponding terms to the logistic-regression models.40
Independence of Associations
Cohort 2 samples were genotyped for 408 SNPs in the region on chromosome 15 between positions 45,524,265 and 48,470,992 (National Center for Biotechnology Information [NCBI] build 36), where many SNPs showed significant associations with the phenotype in cohort 1. To assess the independence of these SNP associations, an association analysis was performed conditioning on the genotype of the strongest associated SNP in this region, rs1426654. The conditional association for each other SNP in this region was assessed with a likelihood-ratio test that compared the logistic-regression model,
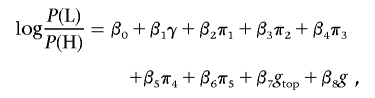 |
with the null model,
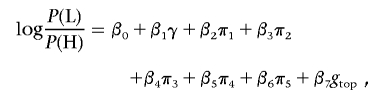 |
where g represents the genotypes of the SNP being assessed and gtop represents the genotypes of rs1426654. An analogous analysis with the roles of g and gtop reversed was also performed.
Dominance and Interactions
For the three primary SNPs that were found to be strongly associated with skin reflectance, dominance effects and epistasis were investigated by adding appropriate terms to the logistic-regression model. To test for dominance effects, genotypes of these SNPs were represented by two variables, gadd and gdom, and the genotypes AA, Aa, and aa (where A and a represent the two SNP alleles, arbitrarily designated) were coded as gadd=(-1,0,1) and gdom=(-0.5,0.5,-0.5).40 Thus, gdom represents deviations from a multiplicative risk model. A logistic-regression model was built on the cohort 2 samples, and a likelihood-ratio test was used to evaluate the significance of the association with gdom.
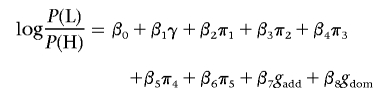 |
was compared with
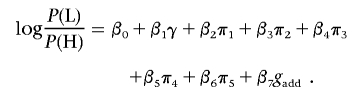 |
To test for interactions between pairs of SNPs, we constructed indicator variables of possible combinations of genotypes at the two loci. Denoting the alleles at SNP 1 by a and A and those at SNP 2 by b and B, the indicator variables are IaAbB, IaABB, IAAbB, and IAABB, where, for example, IaAbB=1 for an individual whose genotype is aA on SNP 1 and bB on SNP 2. These terms were added to a base model that has additive terms gadd,1, gadd,2, and gadd,3 for the three SNPs. The interaction between each pair of SNPs was then assessed using a likelihood-ratio test, comparing the model
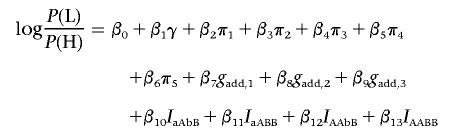 |
with
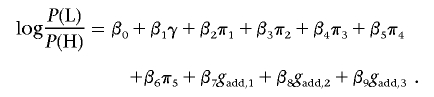 |
Results
Determination of the Phenotypic Groups
The population of South Asia, which, in this study, includes India, Pakistan, Bangladesh, and Sri Lanka, has a wide natural variation in skin pigmentation and consists of the most genetically diverse population outside of Africa,41 which makes it well suited for the investigation of the complex genetic trait of human skin pigmentation. Human skin color is largely determined by two biological components, melanin and hemoglobin, and one major environmental component, exposure to UV light.42 Melanin is the major determining factor in intrinsic skin pigmentation43; therefore, we were most interested in identifying genes that affect melanin. The melanin contribution to skin pigmentation can be quantitatively measured and distinguished from redness caused by hemoglobin and/or inflammation by reflectance spectrophotometry with use of the L* value from the L*a*b* color system.44 Therefore, the highest L* value (maxL*) from hairless, sun-protected sites on the arm was used to define intrinsic skin reflectance in this study, thereby controlling for confounding effects due to sun exposure and hair. To empirically determine the top 20% and bottom 20% of the skin-reflectance distribution in the South Asian population, 98 randomly selected individuals of South Asian ancestry living in the United Kingdom were recruited and had skin chromameter measurements taken. From the plot of the distribution of the maxL* values in this set (fig. 1), individuals with maxL* values ⩽56 were classified as belonging to the low-reflectance quintile (L) with the darkest skin pigmentation, and those with maxL* values ⩾63 were classified as belonging to the high-reflectance (H) quintile with the lightest skin pigmentation of the distribution. Targeted recruitment of individuals in these tails resulted in the collection of DNA from 395 and 383 subjects with low and high skin reflectance, respectively, for the pooled genotyping phase of the study (cohort 1).
Population Structure and Construction of Balanced Pools
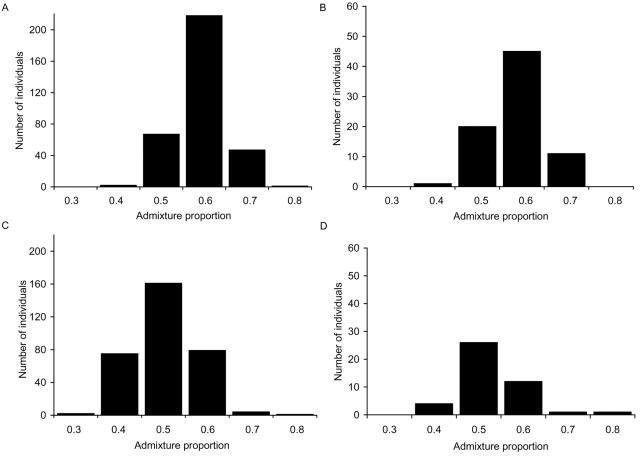
The genetic diversity within South Asian populations potentially introduces significant confounding in an association study because of population structure.45 To control for spurious associations due to systematic differences in ancestry between the L and H groups, we composed the two DNA pools to be used in the pooled genotyping with individuals who were matched for ancestry. The genetic ancestry of the 395 L and 383 H individuals was investigated by individually genotyping them for a set of 312 unlinked GC SNPs. Of the 312 GC SNPs, 294 yielded high-quality genotype data and were analyzed for population structure by use of the structure program,34 with the use of models with 1–4 clusters. The results yielded good evidence of more than one distinct genetic population cluster in our set of 778 individuals but yielded little support for more than two population clusters. The inferred cluster-membership values for a two-cluster model correlate with reported geographical ancestry. Individuals of southern (mainly Sri Lankan) and eastern (mainly Bangladeshi) origin have a similar distribution of admixture, which is distinct from individuals of northwestern (mainly Punjabi) origin (fig. 2).
Figure 2. .
Histogram of admixture proportions in cohort 1 for the geographical ancestry groups. Admixture proportions were determined for 778 individuals in cohort 1 with use of the structure program for a two-cluster model, with genotypes from 294 GC SNPs. Reported parental and grandparental ancestry was used to group individuals by region into (A) eastern (mainly Bangladesh), (B) southern (mainly Sri Lanka), (C) northwestern (mainly Gujarat and Punjab), or (D) mixed origin.
Using the same genotype data set of 294 GC SNPs and a method described elsewhere,32,33 we found significant population stratification between the tails of the skin-reflectance distribution. This result was not unexpected, since there was a clear bias in the recruitment of individuals for cohort 1 from the three geographical regions with respect to the reflectance tails (table 1). Under the null (one-cluster) model of no population structure, for which the L and H groups are assumed to be well matched, we observed a large excess of small P values in χ2 tests for association with skin color (table 2), and a global test for population stratification based on the sum of χ2 statistics46 was highly statistically significant (P<10-40). Using the inferred ancestry values obtained from the structure program with the two-cluster model, we composed the two phenotypic pools, using a subset of individuals such that the average proportions of genetic ancestry were similar between the groups and as many individuals as possible were retained. This matching required us to exclude ∼25% of the original individuals but reduced the population stratification considerably, as can be measured by a reduction in the sum of χ2 statistics versus the unmatched groups (table 2). Although the residual stratification in the matched groups is statistically significant (P=3.9×10-5), the effect of a sum statistic of 400 is comparable to additional experimental error of ∼1% in the pooled allele-frequency measurements. This error size is smaller than the actual level of experimental error. On the basis of these results, we constructed the DNA pools from 286 L and 285 H individuals from cohort 1. Recruitment of individuals for the replication population, cohort 2, was designed to reduce the population stratification bias with respect to skin reflectance, with the use of reported geographical ancestry as a marker for the genetic ancestry profiles we detected in cohort 1 (fig. 2).
Table 1. .
Cohort Distribution for Regional Ancestry and Skin Reflectance[Note]
| No. of Individualsin Reflectance Group |
||
| Cohort and Region | L | H |
| 1: | ||
| Northwestern | 75 | 247 |
| Eastern | 235 | 100 |
| Southern | 75 | 2 |
| Mixed | 10 | 34 |
| 2: | ||
| Northwestern | 61 | 62 |
| Eastern | 58 | 53 |
| Southern | 0 | 0 |
| Mixed | 0 |
1 |
| Total | 514 | 499 |
Note.— Self-reported ancestry was used to group subjects by region into eastern (Bangladesh), northwestern (Gujarat and Punjab), southern (Sri Lanka), or mixed origin.
Table 2. .
Measures of Population Stratification for Skin-Reflectance Pools of Different Compositions
| No. of SNPs with Small P Values in χ2 Tests for Associationb |
|||||||
| Pooling Strategy | L/Ha | P<.0001 | P<.001 | P<.01 | P<0.1 | χ2 Sum | Overall P |
| Expectedc | 395/383 | 0 | 0 | 3 | 29 | 294 | … |
| All of cohort 1 included | 395/383 | 5 | 9 | 24 | 86 | 740 | 2.9×10−40 |
| Matched by inferred ancestryd | 286/285 | 1 | 2 | 8 | 43 | 400 | 3.9×10−5 |
Number of individuals included in each of the skin-reflectance phenotypic pools.
χ2 tests for association were based on 294 GC SNPs.
The expected number of SNPs with low P values, based on the null hypothesis of no population structure in cohort 1.
Inferred ancestry values were obtained from the structure program, with the two-cluster model.
Pooled Genotyping Results
We attempted to estimate the allele-frequency difference, Δ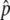 , between the L and H pools for 1,620,742 SNPs across the human genome. Individual measurements of
, between the L and H pools for 1,620,742 SNPs across the human genome. Individual measurements of 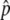 were excluded when we could identify specific experimental errors—saturated signal intensities, inconsistent hybridization patterns, or low signal-to-background ratios. We excluded SNPs for which more than one
were excluded when we could identify specific experimental errors—saturated signal intensities, inconsistent hybridization patterns, or low signal-to-background ratios. We excluded SNPs for which more than one 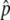 measurement per pool failed these quality-control criteria. SNPs for which the SE of Δ
measurement per pool failed these quality-control criteria. SNPs for which the SE of Δ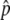 was >0.035 were also excluded, so that the set of SNPs having the largest absolute Δ
was >0.035 were also excluded, so that the set of SNPs having the largest absolute Δ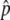 were not dominated by a subset of measurements with very high experimental variance. “Fitted Δ
were not dominated by a subset of measurements with very high experimental variance. “Fitted Δ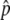 ” values, where information from Perlegen’s haplotype map is used to improve pooled allele-frequency estimates, were generated for a fraction of the SNPs, called “haplotype-conforming” SNPs. These fitted Δ
” values, where information from Perlegen’s haplotype map is used to improve pooled allele-frequency estimates, were generated for a fraction of the SNPs, called “haplotype-conforming” SNPs. These fitted Δ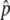 values provide more-accurate estimates than do the measured Δ
values provide more-accurate estimates than do the measured Δ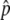 values of individual SNPs, by exploiting correlations among SNPs within haplotype blocks. After all the data-quality filters were applied, we had high-quality Δ
values of individual SNPs, by exploiting correlations among SNPs within haplotype blocks. After all the data-quality filters were applied, we had high-quality Δ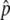 or fitted Δ
or fitted Δ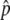 estimates for 1,502,205 SNPs.
estimates for 1,502,205 SNPs.
Individual Genotyping for Cohort 1
SNPs were ranked on the basis of absolute Δ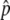 or fitted Δ
or fitted Δ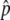 , from largest to smallest, and 30,000 were chosen for individual genotyping on the basis of the capacity of a high-density oligonucleotide array. Selected SNPs from the pooled genotyping results had a fitted
, from largest to smallest, and 30,000 were chosen for individual genotyping on the basis of the capacity of a high-density oligonucleotide array. Selected SNPs from the pooled genotyping results had a fitted 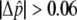 or a
or a 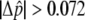 . A lower threshold was used for haplotype-conforming SNPs, because their consistency with the haplotype map provides additional evidence of allele-frequency differences at those positions. An additional set of 66 candidate SNPs was selected to provide denser coverage of variants in genes known to be involved in the pigmentation process (table 3), and 312 GC SNPs used to test for population structure were also included in the array. Short-range PCR assays were successfully designed for 30,045 SNPs, 98.9% of those selected for all categories, and were genotyped with the 778 individuals of cohort 1. After application of quality filters to the genotypes and the DNA samples, a total of 25,928 SNPs for 737 individuals, 363 L and 374 H, were used for the association analysis, including 292 polymorphic GC SNPs. These included individuals who had been excluded from the balanced pools constructed for cohort 1; for, unlike pooled genotyping, with individual genotypes we can explicitly model population structure in the association analysis, and, therefore, the inclusion of all individuals maximizes our power to detect associations.
. A lower threshold was used for haplotype-conforming SNPs, because their consistency with the haplotype map provides additional evidence of allele-frequency differences at those positions. An additional set of 66 candidate SNPs was selected to provide denser coverage of variants in genes known to be involved in the pigmentation process (table 3), and 312 GC SNPs used to test for population structure were also included in the array. Short-range PCR assays were successfully designed for 30,045 SNPs, 98.9% of those selected for all categories, and were genotyped with the 778 individuals of cohort 1. After application of quality filters to the genotypes and the DNA samples, a total of 25,928 SNPs for 737 individuals, 363 L and 374 H, were used for the association analysis, including 292 polymorphic GC SNPs. These included individuals who had been excluded from the balanced pools constructed for cohort 1; for, unlike pooled genotyping, with individual genotypes we can explicitly model population structure in the association analysis, and, therefore, the inclusion of all individuals maximizes our power to detect associations.
Table 3. .
Candidate SNPs
| Allele |
||||||
| SNPa | Chrb | Positionc | Gened | Functione | 1 | 2 |
| rs6058017 | 20 | 32320659 | ASIP | 3′ UTR | A | G |
| rs11551042 | 13 | 93887736 | DCT | Down | A | T |
| rs1540979 | 13 | 93888693 | DCT | Down | T | A |
| rs9516414 | 13 | 93893332 | DCT | Int | A | G |
| rs9524491 | 13 | 93893392 | DCT | Int | T | A |
| rs2892680 | 13 | 93893530 | DCT | Int | A | G |
| rs1407995 | 13 | 93894014 | DCT | Int | T | C |
| rs2296498 | 13 | 93894112 | DCT | Int | A | G |
| rs12876569 | 13 | 93916607 | DCT | Int | C | G |
| rs3212379 | 16 | 88512632 | MC1R | 5′ UTR | C | T |
| rs3212359 | 16 | 88512678 | MC1R | 5′ UTR | C | T |
| rs3212360 | 16 | 88512718 | MC1R | 5′ UTR | C | T |
| rs3212361 | 16 | 88512723 | MC1R | 5′ UTR | G | A |
| rs3212362 | 16 | 88512845 | MC1R | 5′ UTR | G | A |
| rs3212363 | 16 | 88512942 | MC1R | 5′ UTR | A | T |
| rs1805005 | 16 | 88513345 | MC1R | Nonsyn | G | T |
| rs1805006 | 16 | 88513419 | MC1R | Nonsyn | C | A |
| rs2228479 | 16 | 88513441 | MC1R | Nonsyn | G | A |
| rs2229617 | 16 | 88513477 | MC1R | Nonsyn | G | A |
| rs3212364 | 16 | 88513485 | MC1R | Syn | G | A |
| rs1805007 | 16 | 88513618 | MC1R | Nonsyn | C | T |
| rs1110400 | 16 | 88513631 | MC1R | Nonsyn | T | C |
| rs3212365 | 16 | 88513633 | MC1R | Nonsyn | G | C |
| rs1805008 | 16 | 88513645 | MC1R | Nonsyn | C | T |
| rs885479 | 16 | 88513655 | MC1R | Nonsyn | G | A |
| rs3212366 | 16 | 88513753 | MC1R | Nonsyn | T | C |
| rs1805009 | 16 | 88514047 | MC1R | Nonsyn | G | C |
| rs3212367 | 16 | 88514067 | MC1R | Syn | C | T |
| rs2228478 | 16 | 88514109 | MC1R | Syn | A | G |
| rs3212368 | 16 | 88514133 | MC1R | 3′ UTR | G | A |
| rs3212369 | 16 | 88514261 | MC1R | 3′ UTR | A | G |
| rs3212370 | 16 | 88514278 | MC1R | 3′ UTR | C | A |
| rs3212371 | 16 | 88514702 | MC1R | 3′ UTR | A | G |
| rs12592271 | 15 | 25763517 | OCA2 | Int | G | A |
| rs12592307 | 15 | 25763768 | OCA2 | Syn | G | A |
| rs17566952 | 15 | 25870480 | OCA2 | Int | G | C |
| rs7170989 | 15 | 25874003 | OCA2 | Int | T | C |
| rs11638265 | 15 | 25876168 | OCA2 | Int | C | T |
| rs12439067 | 15 | 25876220 | OCA2 | Int | G | T |
| ss69374775 | 15 | 25876236 | OCA2 | Int | G | A |
| rs1800411 | 15 | 25885516 | OCA2 | Syn | C | T |
| rs12910433 | 15 | 25902239 | OCA2 | Int | C | T |
| rs1037208 | 15 | 25904952 | OCA2 | Int | C | A |
| rs2290100 | 15 | 25946945 | OCA2 | Int | A | G |
| rs1052165 | 12 | 54637613 | SILV | Syn | C | T |
| ss69374774 | 5 | 33980486 | SLC45A2 | Down | A | C |
| rs250416 | 5 | 33983301 | SLC45A2 | Int | G | T |
| rs2278007 | 5 | 33987308 | SLC45A2 | Int | A | G |
| rs2287949 | 5 | 33990268 | SLC45A2 | Syn | T | C |
| rs26722 | 5 | 33999627 | SLC45A2 | Nonsyn | C | T |
| rs183671 | 5 | 33999967 | SLC45A2 | Int | A | C |
| rs4547091 | 11 | 88550469 | TYR | Up | C | T |
| rs1799989 | 11 | 88550571 | TYR | Up | C | A |
| rs1042602 | 11 | 88551344 | TYR | Nonsyn | C | A |
| rs12804012 | 11 | 88600438 | TYR | Int | G | A |
| rs3793975 | 11 | 88600836 | TYR | Int | C | T |
| rs1827430 | 11 | 88658088 | TYR | Int | A | G |
| rs28521275 | 11 | 88668617 | TYR | Down | G | A |
| rs2075508 | 9 | 12688363 | TYRP1 | Int | T | C |
| rs2762462 | 9 | 12689776 | TYRP1 | Int | T | C |
| rs2733832 | 9 | 12694725 | TYRP1 | Int | C | T |
| rs2733833 | 9 | 12695095 | TYRP1 | Int | T | G |
| rs2733834 | 9 | 12698910 | TYRP1 | Int | C | G |
| rs2762464 | 9 | 12699586 | TYRP1 | 3′ UTR | A | T |
| rs2382360 | 9 | 12700413 | TYRP1 | Int | T | C |
| ss69374773 | 9 | 12700473 | TYRP1 | Down | T | G |
rs or ss identifier in dbSNP.
Chr = chromosome.
Chromosome base position on NCBI build 36.2.
Gene symbol from Entrez Gene.
Int = intronic; nonsyn = nonsynonymous; syn = synonymous; up = within 10 kb of the transcriptional start site; down = within 10 kb of the transcriptional stop site.
Population-Structure Correction in the Association Analysis
Since we observed significant population stratification between the tails of the skin-reflectance distribution in this South Asian population, we assessed and adjusted for population structure in the association analysis, using the genotypes of our GC panel of 312 SNPs. Population structure was modeled jointly for both population cohorts used in this study, with use of a PCA of the standardized genotypes on the 292 polymorphic GC SNPs that passed our quality filters. The significance of the eigenvalues corresponding to the first several principal components was assessed by repeating the PCA on 100 scrambled realizations of the SNP genotype data. The largest three eigenvalues for the true data set were larger than the largest eigenvalue in any of the permuted data sets (P<.01); the largest five eigenvalues in the true data set were observed in fewer than half the permuted data sets. In addition, four of the top five principal components show significant association with reported ancestry regions in an analysis of variance (PC1, P<1.0×10-16; PC2, P=.19; PC3, P=2.9×10-3; PC4, P=2.1×10-3; PC5, P=5.9×10-8), indicating that these axes of variation likely reflect true features of population structure. We also investigated the relationship between skin reflectance (classified as “L” or “H”) and the top 10 principal components, using logistic regression with sex as a covariate. The first and fifth components showed significant associations (P=4.6×10-22 and P=1.7×10-3, respectively). On the basis of these findings, we adjusted all association analyses for the first five principal components in addition to sex. After this adjustment, we tested for evidence of remaining population structure bias in the association analysis, using the 292 polymorphic GC SNPs that passed quality filters. One SNP (rs8041414) was located in a region on chromosome 15 showing the strongest association with skin reflectance. The results for the remaining 291 SNPs did not reveal a significant inflation in single-SNP test statistics for association with skin reflectance (mean deviance 1.0232; P=.37). Thus, no further corrections for population structure were deemed necessary in association tests for other SNPs in cohort 1 or cohort 2.
Association Results for Cohort 1
Single-SNP tests for association with skin reflectance were performed with logistic regression, including sex and five principal components of the GC SNP genotypes as covariates. Likelihood-ratio tests were used to assess the significance of associations with SNP genotypes. In total, we tested 1,502,205 SNPs for association with skin pigmentation. Although there is likely to be some linkage disequilibrium among the SNPs within this population, it could not be evaluated from our pooled genotyping allele-frequency estimates. Therefore, we employed a conservative Bonferroni threshold for significance of association, α=0.05/1,502,205=3.3×10-8, and found 42 SNPs associated at this level of significance across the genome (table 4). Surprisingly, 39 of the 42 associated SNPs are all located within a single 2.4-Mb chromosomal region of 15q21.1-21.2, between chromosomal positions 45,774,265 and 48,220,992. The strongest SNP association for the entire genome is in this region, with an allele-frequency difference of 45% between the H and L reflectance groups, yielding a P value of 1.0×10−50. This SNP, rs1834640, is located 21 kb from the closest gene, SLC24A5. Of the three SNPs with genomewide significance located outside the chromosomal 15q region, two are located within the TYR gene on chromosome 11; one is in an intron (rs12295166), and the other is a nonsynonymous polymorphism (rs1042602 [p.S192Y]) and is the only SNP in this set of 42 associations that was selected for individual genotyping as a candidate SNP (table 4). The last associated SNP, rs16891982, is also a nonsynonymous polymorphism (p.L374F) in the gene SLC45A2 on chromosome 5.
Table 4. .
Associated SNPs Showing Genomewide Significance in Cohort 1
| Frequencyf |
|||||||||||
| SNPa | Categoryb | Chromosome | Positionc | Gene(s)d | Functione | Allele 1 | Allele 2 | L | H | P | OR (95% CI)g |
| rs1834640 | PG | 15 | 46179457 | … | … | A | G | .47 | .92 | 1.01×10−50 | .08 (.05–.12) |
| rs12913316 | PG | 15 | 46275146 | … | … | C | T | .33 | .05 | 4.95×10−32 | 8.9 (5.68–13.97) |
| rs11070627 | PG | 15 | 46258816 | MYEF2 | Up | A | T | .33 | .05 | 9.52×10−32 | 8.77 (5.6–13.75) |
| rs2924566 | PG | 15 | 46056053 | … | … | G | A | .55 | .25 | 4.17×10−23 | 3.86 (2.88–5.18) |
| rs4775730 | PG | 15 | 46087470 | … | … | T | C | .52 | .78 | 3.56×10−21 | .26 (.2–.36) |
| rs11637235 | PG | 15 | 46420445 | DUT | Int | C | T | .66 | .35 | 6.09×10−21 | 3.44 (2.6–4.54) |
| rs9788730 | PG | 15 | 46098702 | … | … | A | C | .65 | .89 | 3.87×10−19 | .23 (.17–.33) |
| rs10519170 | PG | 15 | 46473467 | … | … | A | G | .57 | .81 | 1.16×10−15 | .3 (.22–.41) |
| rs2965317 | PG | 15 | 46049012 | … | … | C | T | .33 | .13 | 2.44×10−15 | 3.65 (2.58–5.17) |
| rs2965318 | PG | 15 | 46051787 | … | … | T | G | .33 | .13 | 4.43×10−15 | 3.4 (2.44–4.73) |
| rs16960541 | PG | 15 | 46157395 | … | … | G | T | .83 | .96 | 1.70×10−14 | .16 (.1–.27) |
| rs1820489 | PG | 15 | 46472393 | … | … | C | T | .41 | .19 | 8.09×10−14 | 2.98 (2.2–4.05) |
| rs7164700 | PG | 15 | 46097633 | … | … | G | A | .8 | .95 | 1.04×10−13 | .22 (.14–.34) |
| rs16960682 | PG | 15 | 46306954 | SLC12A1 | Int | G | C | .84 | .97 | 1.14×10−13 | .16 (.09–.27) |
| rs2413890 | PG | 15 | 46313654 | SLC12A1 | Int | G | T | .45 | .23 | 1.07×10−11 | 2.55 (1.92–3.38) |
| rs16891982 | PG | 5 | 33987450 | SLC45A2 | Nonsyn | C | G | .97 | .83 | 3.21×10−11 | 4.86 (2.88–8.21) |
| rs16960451 | PG | 15 | 46069778 | … | … | C | T | .81 | .95 | 4.96×10−11 | .28 (.18–.42) |
| rs2924567 | PG | 15 | 46055778 | … | … | G | T | .82 | .67 | 1.74×10−10 | 2.54 (1.89–3.42) |
| ss69356377 | PG | 15 | 46157464 | … | … | A | G | .89 | .98 | 3.13×10−10 | .13 (.06–.27) |
| rs4775727 | PG | 15 | 46067503 | … | … | A | T | .84 | .95 | 3.91×10−10 | .25 (.16–.4) |
| rs2924572 | PG | 15 | 46039330 | … | … | T | A | .22 | .08 | 3.98×10−10 | 3.23 (2.18–4.77) |
| rs1042602 | CS | 11 | 88551344 | TYR | Nonsyn | C | A | .96 | .84 | 4.48×10−10 | 4.36 (2.64–7.2) |
| rs7170781 | PG | 15 | 48180287 | ATP8B4 | Int | C | G | .54 | .73 | 8.85×10−10 | .45 (.35–.59) |
| rs1869454 | PG | 15 | 46080741 | … | … | C | T | .84 | .95 | 1.33×10−9 | .26 (.17–.42) |
| rs16960450 | PG | 15 | 46069520 | … | … | C | T | .83 | .94 | 1.49×10−9 | .27 (.17–.42) |
| rs4774527 | PG | 15 | 46971973 | SHC4 | Int | G | A | .8 | .63 | 1.73×10−9 | 2.34 (1.76–3.12) |
| rs16960434 | PG | 15 | 46062007 | … | … | G | C | .83 | .94 | 1.77×10−9 | .27 (.17–.43) |
| rs1531916 | PG | 15 | 46313563 | SLC12A1 | Int | G | A | .35 | .18 | 2.69×10−9 | 2.4 (1.78–3.25) |
| rs504376 | PG | 15 | 45957669 | … | … | C | G | .59 | .4 | 2.96×10−9 | 2.21 (1.68–2.89) |
| rs16960453 | PG | 15 | 46070428 | … | … | C | G | .85 | .95 | 3.07×10−9 | .27 (.17–.43) |
| rs494230 | PG | 15 | 45903689 | … | … | T | C | .42 | .61 | 3.60×10−9 | .47 (.36–.61) |
| rs12295166 | PG | 11 | 88615805 | TYR | Int | T | C | .95 | .77 | 4.81×10−9 | 2.84 (1.93–4.17) |
| rs1843144 | PG | 15 | 46311814 | SLC12A1 | Int | G | C | .34 | .18 | 5.85×10−9 | 2.35 (1.74–3.17) |
| rs4774557 | PG | 15 | 48220992 | … | … | T | C | .55 | .73 | 6.47×10−9 | .47 (.36–.61) |
| rs1912640 | PG | 15 | 45774265 | … | … | T | C | .27 | .11 | 7.53×10−9 | 2.57 (1.84–3.61) |
| rs4775728 | PG | 15 | 46078857 | … | … | C | G | .87 | .95 | 9.04×10−9 | .24 (.15–.41) |
| ss69356376 | PG | 15 | 45998012 | … | … | G | A | .91 | .98 | 1.12×10−8 | .19 (.1–.35) |
| rs1872304 | PG | 15 | 46446080 | … | … | A | G | .45 | .28 | 1.95×10−8 | 2.16 (1.64–2.86) |
| rs11854994 | PG | 15 | 46986684 | SHC4, DUT | Int | G | A | .69 | .51 | 2.32×10−8 | 2.07 (1.59–2.69) |
| rs784416 | PG | 15 | 46800217 | … | … | G | C | .22 | .1 | 2.57×10−8 | 2.68 (1.86–3.86) |
| rs16961610 | PG | 15 | 46847375 | CEP152 | Int | G | T | .79 | .91 | 2.74×10−8 | .36 (.25–.53) |
| rs16960843 | PG | 15 | 46454117 | … | … | A | G | .85 | .97 | 3.28×10−8 | .27 (.17–.45) |
rs or ss identifier in dbSNP.
PG = SNP selection based on pooled genotyping results. CS = candidate SNP.
Chromosome base position in NCBI build 36.2.
Gene symbol from Entrez Gene.
Int = intronic; nonsyn = nonsynonymous; up = within 10 kb of the transcriptional start site.
Frequency of allele 1 in the reflectance groups of cohort 1.
Odds ratios (ORs) and 95% CIs for allele 1 are shown. The OR is the ratio of the likelihood of an individual being in the low-reflectance group to the likelihood of being in the high-reflectance group.
Replication of Associations in Cohort 2
All of the 42 genomewide-significant SNPs in cohort 1 were individually genotyped with an independent replicate population of 235 individuals (cohort 2). After quality filters were applied, genotypes of the 42 SNPs in 115 H and 116 L individuals were used in the association analysis, where single-SNP likelihood-ratio tests were performed in the same manner as were those for cohort 1. A majority of the associated SNPs, 32 (76%) of 42, yielded P values with a significance threshold of <.05 (table 5). These included the nonsynonymous polymorphisms in TYR and SLC45A2, as well as 30 SNPs in the 2.4-Mb region on chromosome 15. As was seen in cohort 1, the most significant association among these SNPs in cohort 2 was for rs1834640 on 15q21.1, with P=3.28×10-15 and a 36% allele-frequency difference between the high- and low-reflectance groups. Treating the two cohorts as a single population of 968 individuals in the association analysis, instead of independently, yielded different statistical conclusions for only four SNPs, the intronic TYR SNP rs12295166 and three chromosome 15 SNPs, which show genomewide significance in the joint analysis but failed to reach statistical significance by the previous analysis (table 5).
Table 5. .
Results for Associated SNPs in Cohort 2
| Frequencye |
|||||||||
| SNPa | Chromosome | Positionb | Gene(s)c | Functiond | L | H | P | OR (95% CI)f | Joint Cohort Pg |
| rs1834640 | 15 | 46179457 | … | … | .51 | .87 | 3.28×10−15 | .01 (.00–.04) | 3.39×10−64 |
| rs11070627 | 15 | 46258816 | MYEF2 | Up | .31 | .06 | 1.49×10−10 | 54.67 (13.08–228.52) | 6.63×10−41 |
| rs12913316 | 15 | 46275146 | … | … | .31 | .06 | 2.30×10−10 | 46.44 (11.57–186.35) | 3.18×10−41 |
| rs16960682 | 15 | 46306954 | SLC12A1 | Int | .85 | .97 | 2.32×10−6 | .02 (.00–.12) | 1.46×10−19 |
| rs4775730 | 15 | 46087470 | … | … | .52 | .77 | 4.40×10−6 | .14 (.06–.34) | 2.10×10−25 |
| rs2924566 | 15 | 46056053 | … | … | .51 | .25 | 6.25×10−6 | 6.84 (2.84–16.47) | 7.65×10−27 |
| rs9788730 | 15 | 46098702 | … | … | .68 | .88 | 7.30×10−6 | .09 (.03–.27) | 6.84×10−24 |
| rs11854994 | 15 | 46986684 | SHC4, DUT | Int | .72 | .48 | 1.36×10−5 | 6.41 (2.67–15.39) | 2.65×10−13 |
| rs4774527 | 15 | 46971973 | SHC4 | Int | .84 | .63 | 1.76×10−5 | 8.11 (2.96–22.2) | 4.09×10−14 |
| rs7164700 | 15 | 46097633 | … | … | .8 | .94 | 1.81×10−5 | .05 (.01–.23) | 3.07×10−18 |
| rs504376 | 15 | 45957669 | … | … | .58 | .38 | 5.90×10−5 | 5.77 (2.36–14.09) | 3.68×10−12 |
| rs16960541 | 15 | 46157395 | … | … | .87 | .97 | 4.21×10−4 | .05 (.01–.32) | 1.93×10−16 |
| rs2924567 | 15 | 46055778 | … | … | .8 | .64 | 9.62×10−4 | 4.7 (1.83–12.05) | 1.63×10−12 |
| rs11637235 | 15 | 46420445 | DUT | Int | .59 | .42 | 1.00×10−3 | 4.17 (1.72–10.06) | 2.53×10−22 |
| rs2965318 | 15 | 46051787 | … | … | .31 | .16 | 2.10×10−3 | 4.91 (1.73–13.9) | 1.65×10−16 |
| ss69356377 | 15 | 46157464 | … | … | .9 | .97 | 2.35×10−3 | .06 (.01–.45) | 1.73×10−13 |
| rs2965317 | 15 | 46049012 | … | … | .3 | .15 | 3.50×10−3 | 4.79 (1.63–14.09) | 1.97×10−16 |
| rs16891982 | 5 | 33987450 | SLC45A2 | Nonsyn | .94 | .85 | 4.56×10−3 | 7.37 (1.76–30.96) | 5.02×10−13 |
| rs784416 | 15 | 46800217 | … | … | .21 | .11 | 4.83×10−3 | 4.66 (1.56–13.93) | 5.05×10−11 |
| rs10519170 | 15 | 46473467 | … | … | .64 | .76 | 6.84×10−3 | .28 (.11–.72) | 1.03×10−16 |
| rs1820489 | 15 | 46472393 | … | … | .35 | .24 | 6.98×10−3 | 3.59 (1.39–9.29) | 3.71×10−15 |
| rs2924572 | 15 | 46039330 | … | … | .19 | .09 | 1.09×10−2 | 5.02 (1.41–17.91) | 1.85×10−11 |
| rs1869454 | 15 | 46080741 | … | … | .88 | .95 | 1.22×10−2 | .14 (.03–.68) | 6.05×10−11 |
| rs16961610 | 15 | 46847375 | CEP152 | Int | .81 | .89 | 1.32×10−2 | .25 (.08–.76) | 2.80×10−10 |
| rs16960450 | 15 | 46069520 | … | … | .87 | .95 | 1.98×10−2 | .16 (.03–.79) | 1.00×10−10 |
| rs1042602 | 11 | 88551344 | TYR | Nonsyn | .94 | .87 | 2.05×10−2 | 5.05 (1.23–20.74) | 6.54×10−11 |
| rs16960453 | 15 | 46070428 | … | … | .88 | .95 | 2.38×10−2 | .16 (.03–.82) | 2.86×10−10 |
| rs4775727 | 15 | 46067503 | … | … | .87 | .94 | 2.59×10−2 | .18 (.04–.84) | 5.38×10−11 |
| rs16960434 | 15 | 46062007 | … | … | .87 | .94 | 2.88×10−2 | .18 (.04–.87) | 2.78×10−10 |
| rs4775728 | 15 | 46078857 | … | … | .88 | .95 | 2.98×10−2 | .17 (.03–.88) | 1.23×10−9 |
| rs16960451 | 15 | 46069778 | … | … | .86 | .94 | 3.04×10−2 | .24 (.06–.92) | 9.01×10−12 |
| ss69356376 | 15 | 45998012 | … | … | .91 | .96 | 4.65×10−2 | .17 (.03–1.01) | 5.93×10−9 |
| rs12295166 | 11 | 88615805 | TYR | Int | .89 | .81 | 8.26×10−2 | 2.25 (.89–5.68) | 1.94×10−9 |
| rs494230 | 15 | 45903689 | … | … | .49 | .6 | 8.44×10−2 | .49 (.22–1.11) | 6.24×10−10 |
| rs2413890 | 15 | 46313654 | SLC12A1 | Int | .4 | .31 | 1.85×10−1 | 1.77 (.76–4.14) | 6.55×10−11 |
| rs1912640 | 15 | 45774265 | … | … | .17 | .19 | 3.47×10−1 | .58 (.18–1.83) | 4.64×10−6 |
| rs1843144 | 15 | 46311814 | SLC12A1 | Int | .3 | .24 | 4.23×10−1 | 1.44 (.59–3.49) | 1.26×10−7 |
| rs4774557 | 15 | 48220992 | … | … | .61 | .62 | 4.79×10−1 | .74 (.33–1.69) | 6.20×10−8 |
| rs7170781 | 15 | 48180287 | ATP8B4 | Int | .61 | .63 | 5.04×10−1 | .76 (.34–1.71) | 1.12×10−8 |
| rs1872304 | 15 | 46446080 | … | … | .38 | .36 | 5.60×10−1 | 1.28 (.55–2.99) | 3.89×10−7 |
| rs1531916 | 15 | 46313563 | SLC12A1 | Int | .29 | .24 | 6.71×10−1 | 1.22 (.49–3.02) | 1.67×10−7 |
| rs16960843 | 15 | 46454117 | … | … | .9 | .91 | 9.46×10−1 | .95 (.24–3.79) | 1.55×10−6 |
rs or ss identifier in dbSNP.
Chromosome base position on NCBI build 36.2.
Gene symbol from Entrez Gene.
Int = intronic; nonsyn = nonsynonymous; up = within 10 kb of the transcriptional start site.
Frequency of allele 1 in the reflectance groups of cohort 2.
Odds ratios (ORs) and 95% CIs for allele 1 are shown. The OR is the ratio of the likelihood of an individual being in the low-reflectance group to the likelihood of being in the high-reflectance group.
P value for the combined analysis of both cohorts.
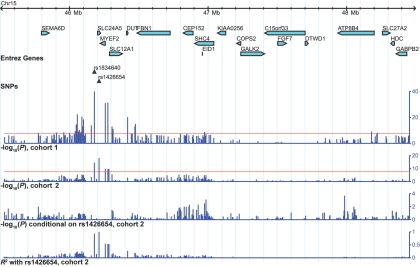
Independent Associations on Chromosome 15 in the Vicinity of SLC24A5
To determine whether the large number of SNP associations observed within the 15q21.1-21.2 region were due to the presence of multiple independently associated SNPs or to one association having a large effect, we individually genotyped a dense set of SNPs within this region for cohort 2. The range for SNP selection was extended by 0.25 Mb in both directions from the boundaries established by significant cohort 1 associations in the region—that is, from positions 45,524,265 to 48,470,992 on chromosome 15. A total of 408 SNPs were successfully assayed, and association test results for this set of SNPs revealed several other significant associations across the region (table 6 and fig. 3). The most significant association in this region was observed for rs1426654, a nonsynonymous polymorphism (p.A111T) in SLC24A5, with P=1.06×10-18 and a 39% allele-frequency difference between the high- and low-reflectance groups. This SNP is in strong linkage disequilibrium with the most significant SNP from the genomewide scan, rs1834640 (R2=0.93), indicating that the two probably represent a single associated locus.
Table 6. .
Association Test Results for a 2.4-Mb Region on Chromosome 15 for Cohort 2
| Frequencye |
|||||||
| SNPa | Positionb | Genec | Functiond | L | H | P | Conditional Pf |
| rs1426654 | 46213776 | SLC24A5 | Nonsyn | .51 | .90 | 1.06×10−18 | … |
| rs12440932 | 47985162 | ATP8B4 | Int | .96 | .90 | 3.01×10−3 | 2.20×10−4 |
| rs11854994 | 46986684 | SHC4 | Int | .72 | .48 | 1.36×10−5 | 8.65×10−4 |
| rs4774527 | 46971973 | SHC4 | Int | .84 | .63 | 1.76×10−5 | 3.49×10−3 |
| rs17465874 | 46866258 | CEP152 | Int | .90 | .95 | 1.42×10−2 | 4.77×10−3 |
| rs17467239 | 46930524 | SHC4 | Int | .75 | .51 | 2.39×10−5 | 5.57×10−3 |
| rs8041414 | 46843962 | CEP152 | Int | .56 | .77 | 4.15×10−6 | 8.92×10−3 |
| rs784411 | 46827089 | CEP152 | Int | .56 | .76 | 1.07×10−5 | 9.05×10−3 |
| rs7176696 | 46861195 | CEP152 | Int | .57 | .78 | 1.48×10−5 | 9.30×10−3 |
| rs1912640 | 45774265 | … | … | .17 | .19 | 3.47×10−1 | 9.61×10−3 |
| rs4143620 | 48055351 | ATP8B4 | Int | .95 | .89 | 1.46×10−2 | 1.07×10−2 |
| rs16961455 | 46799115 | … | … | .88 | .94 | 6.90×10−3 | 1.31×10−2 |
| rs2289179 | 46861831 | CEP152 | Int | .78 | .90 | 3.13×10−3 | 1.49×10−2 |
| rs17463995 | 46791064 | … | … | .61 | .40 | 6.99×10−6 | 1.54×10−2 |
| rs1797225 | 45556171 | … | … | .14 | .10 | 1.81×10−1 | 1.84×10−2 |
| rs8023809 | 47984948 | ATP8B4 | Int | .38 | .24 | 3.30×10−3 | 2.32×10−2 |
| rs8039142 | 48059752 | ATP8B4 | Int | .47 | .63 | 9.04×10−4 | 2.43×10−2 |
| rs7182710 | 46892226 | CEP152 | Up | .74 | .55 | 1.04×10−4 | 2.51×10−2 |
| rs12914304 | 46800835 | … | … | .66 | .46 | 5.85×10−4 | 3.23×10−2 |
| rs4775777 | 46918903 | SHC4 | Int | .73 | .56 | 4.75×10−4 | 3.23×10−2 |
| rs16960682 | 46306954 | SLC12A1 | Int | .85 | .97 | 2.32×10−6 | 3.27×10−2 |
| rs2304546 | 46890536 | CEP152 | Up | .70 | .84 | 1.87×10−4 | 3.63×10−2 |
| rs937171 | 47982041 | ATP8B4 | Int | .48 | .34 | 2.00×10−3 | 4.15×10−2 |
| rs854151 | 48330011 | HDC | Int | .80 | .72 | 7.05×10−2 | 4.21×10−2 |
| rs10519188 | 46842454 | CEP152 | Int | .83 | .90 | 1.82×10−2 | 4.22×10−2 |
| rs16961645 | 46862521 | CEP152 | Int | .83 | .90 | 1.82×10−2 | 4.22×10−2 |
| rs934741 | 46994194 | SHC4 | Int | .23 | .19 | 9.30×10−2 | 4.97×10−2 |
| rs2413980 | 48008650 | ATP8B4 | Int | .84 | .78 | 2.26×10−1 | 5.03×10−2 |
| rs10519251 | 48056965 | ATP8B4 | Int | .91 | .84 | 7.89×10−3 | 5.24×10−2 |
| rs7175415 | 46777881 | … | … | .72 | .81 | 5.29×10−3 | 5.25×10−2 |
| rs16961470 | 46804316 | … | … | .83 | .90 | 2.63×10−2 | 5.30×10−2 |
| rs2413890 | 46313654 | SLC12A1 | Int | .40 | .31 | 1.85×10−1 | 5.49×10−2 |
| rs421853 | 45531623 | … | … | .23 | .20 | 2.58×10−1 | 6.00×10−2 |
| rs7174374 | 46778158 | … | … | .34 | .19 | 1.12×10−3 | 6.37×10−2 |
| rs12906304 | 48359690 | GABPB2 | Int | .86 | .84 | 2.81×10−1 | 6.58×10−2 |
| rs16962583 | 47600407 | C15orf33 | Int | .92 | .89 | 1.09×10−1 | 7.23×10−2 |
| rs627566 | 47839646 | … | … | .66 | .61 | 2.58×10−2 | 7.28×10−2 |
| rs1036477 | 46702218 | FBN1 | Int | .27 | .09 | 1.96×10−5 | 7.34×10−2 |
| rs1224660 | 45788054 | SEMA6D | Up | .20 | .13 | 3.32×10−1 | 8.33×10−2 |
| rs1496917 | 45529635 | … | … | .79 | .69 | 1.12×10−2 | 8.49×10−2 |
| rs11070739 | 48043671 | ATP8B4 | Int | .61 | .73 | 5.93×10−3 | 8.68×10−2 |
| rs785016 | 45922856 | … | … | .35 | .53 | 1.10×10−4 | 8.81×10−2 |
| rs12923 | 48357328 | GABPB2 | 3′ UTR | .86 | .85 | 3.63×10−1 | 8.82×10−2 |
| rs17393761 | 47164183 | … | … | .78 | .83 | 1.13×10−1 | 9.28×10−2 |
| rs16960451 | 46069778 | … | … | .86 | .94 | 3.04×10−2 | 9.29×10−2 |
| rs5020564 | 46451845 | … | … | .78 | .72 | 5.21×10−1 | 9.39×10−2 |
| rs1656602 | 45581286 | … | … | .23 | .18 | 2.26×10−1 | 9.49×10−2 |
| rs7164451 | 47001348 | SHC4 | Int | .24 | .36 | 1.57×10−2 | 9.93×10−2 |
| rs1484556 | 47829263 | … | … | .83 | .83 | 6.16×10−1 | 1.05×10−1 |
| rs8034382 | 48059377 | ATP8B4 | Int | .75 | .66 | 1.05×10−1 | 1.06×10−1 |
| rs16960434 | 46062007 | … | … | .87 | .94 | 2.88×10−2 | 1.14×10−1 |
| rs9806310 | 45650255 | … | … | .95 | .94 | 4.89×10−1 | 1.15×10−1 |
| rs2452524 | 48013605 | ATP8B4 | Nonsyn | .47 | .36 | 2.98×10−2 | 1.16×10−1 |
| rs1035704 | 45615618 | … | … | .22 | .17 | 3.23×10−1 | 1.19×10−1 |
| rs12594966 | 46107263 | … | … | .90 | .97 | 1.50×10−3 | 1.19×10−1 |
| rs8031403 | 48062303 | ATP8B4 | Int | .42 | .49 | 1.08×10−1 | 1.21×10−1 |
| rs7164700 | 46097633 | … | … | .80 | .94 | 1.81×10−5 | 1.28×10−1 |
| rs9788730 | 46098702 | … | … | .68 | .88 | 7.30×10−6 | 1.30×10−1 |
| rs4775727 | 46067503 | … | … | .87 | .94 | 2.59×10−2 | 1.35×10−1 |
| rs7174453 | 46804645 | … | … | .94 | .99 | 4.67×10−2 | 1.35×10−1 |
| rs9806753 | 46953709 | EID1 | Up | .37 | .27 | 1.40×10−2 | 1.39×10−1 |
| rs12914876 | 47225707 | COPS2 | Int | .91 | .94 | 4.11×10−1 | 1.40×10−1 |
| rs10519132 | 45661803 | … | … | .78 | .66 | 3.22×10−2 | 1.41×10−1 |
| rs10519193 | 46950245 | EID1 | Up | .40 | .30 | 1.89×10−2 | 1.44×10−1 |
| rs7177445 | 46489749 | FBN1 | Exon | .08 | .15 | 7.62×10−2 | 1.47×10−1 |
| rs1426200 | 46885308 | CEP152 | Int | .08 | .06 | 4.07×10−1 | 1.59×10−1 |
| rs16960453 | 46070428 | … | … | .88 | .95 | 2.38×10−2 | 1.59×10−1 |
| rs4775728 | 46078857 | … | … | .88 | .95 | 2.98×10−2 | 1.60×10−1 |
| rs11857760 | 45778132 | … | … | .53 | .45 | 1.00×10−1 | 1.62×10−1 |
| rs16960450 | 46069520 | … | … | .87 | .95 | 1.98×10−2 | 1.63×10−1 |
| rs10519145 | 45885244 | … | … | .80 | .74 | 3.65×10−1 | 1.67×10−1 |
| rs16961388 | 46757556 | … | … | .91 | .96 | 3.98×10−2 | 1.70×10−1 |
| rs1869454 | 46080741 | … | … | .88 | .95 | 1.22×10−2 | 1.71×10−1 |
| rs1453854 | 46016892 | … | … | .69 | .64 | 3.53×10−1 | 1.73×10−1 |
| rs16963635 | 48407932 | GABPB2 | Int | .90 | .97 | 1.14×10−2 | 1.83×10−1 |
| rs12439630 | 45822483 | SEMA6D | Int | .72 | .62 | 9.47×10−2 | 1.87×10−1 |
| ss69356377 | 46157464 | … | … | .90 | .97 | 2.35×10−3 | 1.91×10−1 |
| rs1531916 | 46313563 | SLC12A1 | Int | .29 | .24 | 6.71×10−1 | 1.95×10−1 |
| rs1390870 | 45541604 | … | … | .78 | .69 | 4.48×10−2 | 1.98×10−1 |
| rs1224662 | 45789839 | SEMA6D | Up | .23 | .14 | 1.06×10−1 | 1.99×10−1 |
| rs16962923 | 47882435 | … | … | .83 | .86 | 1.21×10−1 | 2.00×10−1 |
| rs2413914 | 46891800 | CEP152 | Up | .08 | .07 | 6.93×10−1 | 2.07×10−1 |
| rs16963151 | 48066954 | ATP8B4 | Nonsyn | .80 | .75 | 6.39×10−1 | 2.09×10−1 |
| rs504376 | 45957669 | … | … | .58 | .38 | 5.90×10−5 | 2.11×10−1 |
| rs16959669 | 45612375 | … | … | .95 | .95 | 7.16×10−1 | 2.12×10−1 |
| rs16960071 | 45846061 | SEMA6D | Int | .92 | .92 | 9.57×10−1 | 2.12×10−1 |
| rs1820489 | 46472393 | … | … | .35 | .24 | 6.98×10−3 | 2.16×10−1 |
| rs1974961 | 46938695 | SHC4 | Int | .09 | .10 | 9.62×10−1 | 2.17×10−1 |
| rs530734 | 45908604 | … | … | .93 | .95 | 9.45×10−1 | 2.25×10−1 |
| rs16962243 | 47350024 | GALK2 | Int | .80 | .75 | 3.36×10−2 | 2.26×10−1 |
| rs854158 | 48340369 | HDC | Int | .78 | .71 | 1.36×10−1 | 2.29×10−1 |
| rs2059474 | 45595646 | … | … | .82 | .72 | 2.77×10−2 | 2.30×10−1 |
| rs955845 | 45776451 | … | … | .79 | .75 | 3.20×10−1 | 2.35×10−1 |
| rs11634585 | 48443741 | GABPB2 | Up | .24 | .25 | 6.12×10−1 | 2.37×10−1 |
| rs4259993 | 47011664 | SHC4 | Int | .87 | .83 | 3.84×10−1 | 2.39×10−1 |
| rs16960541 | 46157395 | … | … | .87 | .97 | 4.21×10−4 | 2.40×10−1 |
| rs2279842 | 46782678 | … | … | .20 | .16 | 3.01×10−1 | 2.44×10−1 |
| rs1045688 | 45852779 | SEMA6D | 3′ UTR | .74 | .66 | 1.60×10−1 | 2.48×10−1 |
| rs16961560 | 46835997 | CEP152 | Nonsyn | .93 | .92 | 1.39×10−1 | 2.48×10−1 |
| rs16963682 | 48448856 | … | … | .91 | .90 | 5.01×10−1 | 2.52×10−1 |
| rs1453857 | 46116200 | … | … | .53 | .49 | 8.71×10−1 | 2.53×10−1 |
| rs4775783 | 46963495 | EID1 | Down | .60 | .76 | 1.37×10−3 | 2.54×10−1 |
| rs17466389 | 46894932 | CEP152 | Up | .84 | .86 | 8.61×10−1 | 2.55×10−1 |
| rs12442429 | 48071636 | ATP8B4 | Int | .82 | .79 | 7.58×10−1 | 2.56×10−1 |
| rs628501 | 45814620 | SEMA6D | Int | .72 | .63 | 1.47×10−1 | 2.67×10−1 |
| rs10519170 | 46473467 | … | … | .64 | .76 | 6.84×10−3 | 2.68×10−1 |
| rs12913008 | 47980304 | ATP8B4 | Int | .68 | .61 | 1.83×10−1 | 2.68×10−1 |
| rs4775785 | 46979618 | SHC4 | Int | .60 | .76 | 2.21×10−3 | 2.70×10−1 |
| rs7169897 | 46939299 | SHC4 | Int | .71 | .82 | 4.95×10−3 | 2.73×10−1 |
| rs11070711 | 47703708 | C15orf33 | Up | .06 | .06 | 6.60×10−1 | 2.81×10−1 |
| rs479062 | 47877548 | … | … | .62 | .63 | 2.01×10−1 | 2.83×10−1 |
| rs10851472 | 47005151 | SHC4 | Int | .55 | .65 | 1.39×10−1 | 2.86×10−1 |
| rs7162626 | 46731907 | FBN1 | Up | .54 | .35 | 3.55×10−4 | 2.91×10−1 |
| rs16960139 | 45871972 | … | … | .95 | .97 | 9.23×10−1 | 2.92×10−1 |
| rs1025760 | 45855645 | SEMA6D | Down | .88 | .94 | 1.28×10−1 | 2.98×10−1 |
| rs2278167 | 48285140 | SLC27A2 | Int | .69 | .59 | 6.14×10−2 | 3.01×10−1 |
| rs12914000 | 45694010 | … | … | .93 | .93 | 9.69×10−1 | 3.04×10−1 |
| rs1435752 | 45702304 | … | … | .66 | .80 | 3.17×10−3 | 3.07×10−1 |
| rs7175546 | 46511988 | FBN1 | Int | .86 | .92 | 3.51×10−2 | 3.11×10−1 |
| rs1843144 | 46311814 | SLC12A1 | Int | .30 | .24 | 4.23×10−1 | 3.19×10−1 |
| rs2413922 | 47009173 | SHC4 | Int | .74 | .61 | 1.63×10−2 | 3.19×10−1 |
| rs17399591 | 47500513 | C15orf33 | Int | .98 | .98 | 7.92×10−1 | 3.19×10−1 |
| rs596942 | 45869443 | … | … | .88 | .94 | 1.50×10−1 | 3.20×10−1 |
| rs925104 | 47703947 | C15orf33 | Up | .75 | .81 | 1.67×10−1 | 3.21×10−1 |
| rs10851470 | 46757572 | … | … | .18 | .13 | 9.41×10−2 | 3.22×10−1 |
| rs4774504 | 45827510 | SEMA6D | Int | .15 | .07 | 7.09×10−2 | 3.22×10−1 |
| rs11638981 | 47108821 | KIAA0256 | Int | .75 | .65 | 1.61×10−1 | 3.30×10−1 |
| rs1968825 | 46837115 | CEP152 | Int | .22 | .13 | 6.16×10−3 | 3.30×10−1 |
| rs11854557 | 47360252 | GALK2 | Int | .79 | .83 | 3.27×10−1 | 3.31×10−1 |
| rs1872304 | 46446080 | … | … | .38 | .36 | 5.60×10−1 | 3.34×10−1 |
| rs7162426 | 46974966 | SHC4 | Int | .73 | .84 | 3.20×10−3 | 3.34×10−1 |
| rs12898878 | 47025722 | SHC4 | Int | .61 | .47 | 1.69×10−2 | 3.39×10−1 |
| rs17472989 | 47211658 | COPS2 | Int | .80 | .83 | 3.56×10−1 | 3.46×10−1 |
| rs8036322 | 48054416 | ATP8B4 | Int | .21 | .21 | 7.35×10−1 | 3.48×10−1 |
| rs11639262 | 48186938 | ATP8B4 | Int | .92 | .88 | 2.54×10−1 | 3.53×10−1 |
| rs2413996 | 48060962 | ATP8B4 | Int | .56 | .65 | 1.57×10−1 | 3.53×10−1 |
| rs16960843 | 46454117 | … | … | .90 | .91 | 9.46×10−1 | 3.53×1010−1 |
| rs494230 | 45903689 | … | … | .49 | .60 | 8.44×10−2 | 3.54×10−1 |
| rs12593937 | 48051824 | ATP8B4 | Int | .20 | .21 | 6.98×10−1 | 3.55×10−1 |
| rs647903 | 45877708 | … | … | .33 | .42 | 5.43×10−2 | 3.55×10−1 |
| rs17383671 | 46986669 | SHC4 | Int | .80 | .88 | 1.53×10−1 | 3.57×10−1 |
| rs4491452 | 47015113 | SHC4 | Int | .17 | .12 | 1.26×10−1 | 3.64×10−1 |
| rs963031 | 45901235 | … | … | .89 | .91 | 8.85×10−1 | 3.75×10−1 |
| rs17479589 | 47540824 | FGF7 | Int | .77 | .81 | 2.44×10−1 | 3.77×10−1 |
| rs1369637 | 45732119 | … | … | .22 | .32 | 4.40×10−2 | 3.79×10−1 |
| rs586118 | 45837068 | SEMA6D | Int | .40 | .46 | 3.18×10−1 | 3.79×10−1 |
| rs3743286 | 45853084 | SEMA6D | 3′ UTR | .73 | .67 | 2.80×10−1 | 3.80×10−1 |
| rs669653 | 45944278 | … | … | .71 | .86 | 1.67×10−5 | 3.80×10−1 |
| rs17431095 | 48448741 | … | … | .77 | .74 | 7.90×10−1 | 3.80×10−1 |
| rs2924572 | 46039330 | … | … | .19 | .09 | 1.09×10−2 | 3.84×10−1 |
| rs1561483 | 45856780 | SEMA6D | Down | .73 | .66 | 2.39×10−1 | 3.87×10−1 |
| rs1699400 | 46834784 | CEP152 | Int | .78 | .87 | 8.09×10−3 | 3.91×10−1 |
| rs935001 | 47141674 | … | … | .75 | .72 | 1.31×10−1 | 3.94×10−1 |
| rs17388803 | 45814496 | SEMA6D | Int | .93 | .91 | 5.10×10−1 | 3.94×10−1 |
| rs586799 | 45812214 | … | … | .76 | .70 | 2.03×10−1 | 3.99×10−1 |
| rs16959595 | 45582548 | … | … | .89 | .94 | 1.13×10−1 | 4.04×10−1 |
| rs12438387 | 45768317 | … | … | .30 | .32 | 7.80×10−1 | 4.12×10−1 |
| rs688475 | 45818759 | SEMA6D | Int | .76 | .70 | 1.77×10−1 | 4.12×10−1 |
| rs3784296 | 48356641 | GABPB2 | Down | .57 | .49 | 8.74×10−2 | 4.15×10−1 |
| rs8029889 | 48338192 | HDC | Int | .75 | .78 | 9.91×10−1 | 4.15×10−1 |
| rs12898855 | 45801857 | SEMA6D | Int | .74 | .69 | 2.70×10−1 | 4.16×10−1 |
| rs6493312 | 46329848 | SLC12A1 | Int | .88 | .90 | 5.61×10−1 | 4.16×10−1 |
| rs784416 | 46800217 | … | … | .21 | .11 | 4.83×10−3 | 4.18×10−1 |
| rs17384124 | 47005799 | SHC4 | Int | .97 | .95 | 8.36×10−1 | 4.18×10−1 |
| rs2555470 | 46534732 | FBN1 | Int | .66 | .74 | 1.86×10−1 | 4.24×10−1 |
| rs4482220 | 46889123 | CEP152 | Int | .81 | .81 | 7.47×10−1 | 4.24×10−1 |
| rs16960351 | 46001269 | … | … | .87 | .94 | 6.30×10−3 | 4.28×10−1 |
| rs12440824 | 46012263 | … | … | .80 | .90 | 1.02×10−2 | 4.34×10−1 |
| rs4775854 | 48201197 | ATP8B4 | Up | .73 | .78 | 5.14×10−1 | 4.35×10−1 |
| rs16962047 | 47174522 | … | … | .94 | .95 | 6.40×10−1 | 4.37×10−1 |
| rs10519262 | 48219786 | … | … | .73 | .76 | 6.73×10−1 | 4.41×10−1 |
| rs76739 | 45829438 | SEMA6D | Int | .72 | .68 | 3.19×10−1 | 4.42×10−1 |
| rs12903325 | 48140569 | ATP8B4 | Int | .75 | .74 | 6.10×10−1 | 4.44×10−1 |
| rs17374298 | 45581770 | … | … | .77 | .69 | 2.00×10−1 | 4.44×10−1 |
| rs4775692 | 45599347 | … | … | .89 | .94 | 1.29×10−1 | 4.45×10−1 |
| rs11634375 | 47536840 | FGF7 | Int | .46 | .46 | 9.32×10−1 | 4.47×10−1 |
| rs12591300 | 47492033 | C15orf33 | Int | .75 | .79 | 2.62×10−1 | 4.47×10−1 |
| rs12441775 | 46637949 | FBN1 | Int | .53 | .34 | 4.93×10−4 | 4.52×10−1 |
| rs1610098 | 45593304 | … | … | .63 | .62 | 5.54×10−1 | 4.52×10−1 |
| rs7178146 | 47792109 | … | … | .73 | .78 | 2.07×10−1 | 4.55×10−1 |
| rs11070743 | 48137976 | ATP8B4 | Int | .44 | .43 | 9.98×10−1 | 4.56×10−1 |
| rs2899440 | 47980269 | ATP8B4 | Int | .46 | .49 | 3.23×10−1 | 4.59×10−1 |
| rs11634811 | 45783028 | … | … | .74 | .61 | 9.56×10−2 | 4.61×10−1 |
| rs537052 | 45844911 | SEMA6D | Int | .39 | .40 | 9.09×10−1 | 4.61×10−1 |
| rs11633714 | 47929321 | ATP8B4 | Down | .86 | .85 | 8.66×10−1 | 4.63×10−1 |
| rs17486446 | 47928420 | ATP8B4 | Down | .86 | .85 | 8.66×10−1 | 4.63×10−1 |
| rs11637901 | 48426740 | … | … | .59 | .54 | 1.85×10−1 | 4.74×10−1 |
| rs11855823 | 48407007 | GABPB2 | Int | .59 | .54 | 1.85×10−1 | 4.74×10−1 |
| rs2081628 | 48379835 | GABPB2 | Int | .59 | .54 | 1.85×10−1 | 4.74×10−1 |
| rs8041517 | 48055189 | ATP8B4 | Int | .38 | .46 | 1.79×10−1 | 4.74×10−1 |
| rs17508371 | 48378086 | GABPB2 | Int | .60 | .54 | 2.02×10−1 | 4.75×10−1 |
| rs11634947 | 47784508 | … | … | .76 | .80 | 2.73×10−1 | 4.82×10−1 |
| rs3809485 | 45838748 | SEMA6D | Int | .38 | .40 | 3.60×10−1 | 4.83×10−1 |
| rs1007662 | 47002601 | SHC4 | Int | .72 | .75 | 1.84×10−1 | 4.86×10−1 |
| rs4774517 | 46546583 | FBN1 | Int | .66 | .74 | 1.76×10−1 | 4.86×10−1 |
| rs662968 | 47795735 | … | … | .46 | .47 | 3.92×10−1 | 4.86×10−1 |
| rs7171359 | 46544752 | FBN1 | Int | .66 | .74 | 1.76×10−1 | 4.86×10−1 |
| rs16961049 | 46586202 | FBN1 | Int | .92 | .98 | 3.12×10−3 | 4.89×10−1 |
| rs16960244 | 45941874 | … | … | .71 | .86 | 5.57×10−5 | 4.95×10−1 |
| rs16960510 | 46109866 | … | … | .96 | .96 | 6.40×10−1 | 4.97×10−1 |
| rs11632411 | 48224343 | … | … | .85 | .84 | 5.79×10−1 | 5.02×10−1 |
| rs10519249 | 48055662 | ATP8B4 | Int | .38 | .46 | 1.89×10−1 | 5.07×10−1 |
| rs784405 | 46819991 | CEP152 | Down | .13 | .12 | 8.92×10−1 | 5.08×10−1 |
| rs8030581 | 47180201 | … | … | .11 | .03 | 1.52×10−2 | 5.08×10−1 |
| rs16961937 | 47033530 | SHC4 | Int | .96 | .96 | 9.84×10−1 | 5.09×10−1 |
| rs559561 | 47803588 | … | … | .60 | .59 | 2.41×10−1 | 5.12×10−1 |
| rs17487348 | 47951100 | ATP8B4 | Int | .78 | .71 | 1.47×10−1 | 5.13×10−1 |
| rs10519208 | 47175541 | … | … | .71 | .68 | 1.32×10−1 | 5.14×10−1 |
| rs10519225 | 47508070 | FGF7 | Int | .75 | .79 | 3.74×10−1 | 5.19×10−1 |
| rs12439479 | 47522135 | FGF7 | Int | .76 | .72 | 1.20×10−1 | 5.34×10−1 |
| rs1042078 | 46490165 | FBN1 | Exon | .34 | .26 | 1.69×10−1 | 5.39×10−1 |
| rs25458 | 46584599 | FBN1 | Exon | .76 | .79 | 3.76×10−1 | 5.40×10−1 |
| rs7164052 | 46779873 | … | … | .90 | .88 | 9.72×10−1 | 5.44×10−1 |
| rs7162180 | 48286546 | SLC27A2 | Int | .56 | .59 | 5.34×10−1 | 5.45×10−1 |
| rs16961610 | 46847375 | CEP152 | Int | .81 | .89 | 1.32×10−2 | 5.49×10−1 |
| rs1369636 | 45732174 | … | … | .82 | .89 | 1.41×10−1 | 5.51×10−1 |
| rs3743281 | 45844250 | SEMA6D | Syn | .73 | .67 | 4.14×10−1 | 5.56×10−1 |
| rs16961671 | 46887207 | CEP152 | Int | .81 | .88 | 1.23×10−2 | 5.56×10−1 |
| rs8036777 | 48208710 | ATP8B4 | Up | .43 | .44 | 7.72×10−1 | 5.57×10−1 |
| rs17352842 | 46481503 | FBN1 | Down | .86 | .93 | 3.55×10−1 | 5.57×10−1 |
| rs4494483 | 47939566 | ATP8B4 | 3′ UTR | .82 | .78 | 3.58×10−1 | 5.58×10−1 |
| rs12594698 | 46362248 | SLC12A1 | Int | .69 | .74 | 2.25×10−1 | 5.63×10−1 |
| rs3985863 | 45820660 | SEMA6D | Int | .89 | .81 | 3.34×10−2 | 5.63×10−1 |
| rs1224659 | 45787001 | … | … | .60 | .59 | 7.95×10−1 | 5.66×10−1 |
| rs11539517 | 47704633 | C15orf33 | Up | .85 | .82 | 7.64×10−1 | 5.68×10−1 |
| rs2163095 | 47429179 | C15orf33 | Int | .53 | .59 | 2.27×10−1 | 5.69×10−1 |
| rs17314486 | 45557607 | … | … | .88 | .84 | 5.86×10−1 | 5.69×10−1 |
| rs11854679 | 46344865 | SLC12A1 | Int | .69 | .75 | 2.03×10−1 | 5.79×10−1 |
| rs4143837 | 48193475 | ATP8B4 | Int | .54 | .61 | 6.58×10−2 | 5.81×10−1 |
| rs1435763 | 45647928 | … | … | .18 | .26 | 9.56×10−2 | 5.82×10−1 |
| rs2078139 | 48447126 | … | … | .59 | .65 | 5.08×10−1 | 5.83×10−1 |
| rs2924566 | 46056053 | … | … | .51 | .25 | 6.25×10−6 | 5.89×10−1 |
| rs12442472 | 46191894 | SLC24A5 | Up | .91 | .97 | 1.09×10−2 | 5.90×10−1 |
| rs8029928 | 45821317 | SEMA6D | Int | .68 | .74 | 1.46×10−1 | 5.90×10−1 |
| rs6493364 | 47432702 | C15orf33 | Int | .72 | .69 | 1.54×10−1 | 5.92×10−1 |
| rs16960195 | 45913521 | … | … | .93 | .92 | 8.48×10−1 | 5.93×10−1 |
| rs1023683 | 47494739 | FGF7 | Up | .85 | .82 | 9.41×10−1 | 5.98×10−1 |
| rs8024406 | 45871559 | … | … | .77 | .84 | 1.70×10−1 | 6.00×10−1 |
| rs16961125 | 46628336 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs16961144 | 46637064 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs16961172 | 46651769 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs16961186 | 46660907 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs16961202 | 46664308 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs16961232 | 46682983 | FBN1 | Int | .92 | .98 | 1.05×10−3 | 6.02×10−1 |
| rs11637235 | 46420445 | DUT | Int | .59 | .42 | 1.12×10−3 | 6.04×10−1 |
| rs934740 | 46994089 | SHC4 | Int | .15 | .12 | 5.46×10−1 | 6.06×10−1 |
| rs10519252 | 48070953 | ATP8B4 | Int | .57 | .60 | 5.93×10−1 | 6.08×10−1 |
| rs2924567 | 46055778 | … | … | .80 | .64 | 9.62×10−4 | 6.17×10−1 |
| rs17382306 | 46923318 | SHC4 | Int | .84 | .88 | 4.25×10−1 | 6.20×10−1 |
| rs12907752 | 45890486 | … | … | .77 | .82 | 2.34×10−1 | 6.22×10−1 |
| rs11632303 | 47317613 | GALK2 | Int | .86 | .83 | 9.10×10−1 | 6.23×10−1 |
| rs1059852 | 47938555 | ATP8B4 | 3′ UTR | .86 | .84 | 9.91×10−1 | 6.25×10−1 |
| rs2414021 | 48198648 | ATP8B4 | 5′ UTR | .73 | .71 | 8.00×10−1 | 6.26×10−1 |
| rs16963010 | 47957733 | ATP8B4 | Int | .76 | .81 | 3.65×10−1 | 6.30×10−1 |
| rs17402569 | 47702698 | DTWD1 | Int | .87 | .83 | 7.18×10−1 | 6.33×10−1 |
| rs2965317 | 46049012 | … | … | .30 | .15 | 3.50×10−3 | 6.44×10−1 |
| rs16960346 | 45999405 | … | … | .81 | .90 | 2.95×10−2 | 6.46×10−1 |
| rs4775730 | 46087470 | … | … | .52 | .77 | 4.40×10−6 | 6.49×10−1 |
| rs616614 | 45889302 | … | … | .25 | .31 | 1.89×10−1 | 6.51×10−1 |
| rs2414049 | 48453732 | … | … | .27 | .30 | 5.62×10−1 | 6.52×10−1 |
| rs4580097 | 48102545 | ATP8B4 | Int | .41 | .44 | 3.39×10−1 | 6.62×10−1 |
| rs363830 | 46507944 | FBN1 | Exon | .93 | .97 | 1.53×10−1 | 6.64×10−1 |
| rs2289181 | 46877467 | CEP152 | Nonsyn | .81 | .88 | 1.83×10−2 | 6.65×10−1 |
| rs2899453 | 48154202 | ATP8B4 | Int | .44 | .43 | 9.20×10−1 | 6.66×10−1 |
| rs17478785 | 47527596 | FGF7 | Int | .76 | .79 | 5.75×10−1 | 6.67×10−1 |
| rs1059850 | 47938821 | ATP8B4 | 3′ UTR | .77 | .75 | 8.52×10−1 | 6.71×10−1 |
| rs11635140 | 46569855 | FBN1 | Int | .44 | .62 | 1.13×10−3 | 6.74×10−1 |
| rs17376095 | 45620983 | … | … | .90 | .84 | 3.26×10−1 | 6.77×10−1 |
| rs16963076 | 47999573 | ATP8B4 | Int | .91 | .88 | 5.79×10−1 | 6.78×10−1 |
| rs4592603 | 47018806 | SHC4 | Int | .26 | .18 | 1.59×10−1 | 6.79×10−1 |
| rs2965318 | 46051787 | … | … | .31 | .16 | 2.10×10−3 | 6.80×10−1 |
| rs16959719 | 45671245 | … | … | .89 | .94 | 9.34×10−2 | 6.83×10−1 |
| rs10519233 | 47693662 | DTWD1 | Up | .87 | .82 | 6.41×10−1 | 6.91×10−1 |
| rs17483139 | 47791804 | … | … | .87 | .82 | 6.41×10−1 | 6.91×10−1 |
| rs17361868 | 46584438 | FBN1 | Int | .88 | .92 | 1.40×10−1 | 6.98×10−1 |
| rs607289 | 45862845 | SEMA6D | Down | .37 | .38 | 8.72×10−1 | 7.01×10−1 |
| rs17480434 | 47600635 | C15orf33 | Int | .87 | .82 | 6.17×10−1 | 7.01×10−1 |
| rs1993150 | 45854810 | SEMA6D | Down | .53 | .56 | 7.30×10−1 | 7.05×10−1 |
| rs2009833 | 47980845 | ATP8B4 | Int | .48 | .53 | 1.76×10−1 | 7.06×10−1 |
| rs688709 | 45870675 | … | … | .79 | .71 | 9.31×10−2 | 7.09×10−1 |
| rs11070627 | 46258816 | MYEF2 | Up | .31 | .06 | 1.49×10−10 | 7.12×10−1 |
| rs11631496 | 46863210 | CEP152 | Int | .88 | .88 | 8.60×10−1 | 7.12×10−1 |
| rs12913316 | 46275146 | … | … | .31 | .06 | 2.30×10−10 | 7.12×10−1 |
| rs1320052 | 46282800 | SLC12A1 | Up | .31 | .06 | 2.30×10−10 | 7.12×10−1 |
| rs6493311 | 46326879 | SLC12A1 | Syn | .57 | .67 | 6.92×10−2 | 7.23×10−1 |
| rs1834640 | 46179457 | … | … | .51 | .87 | 3.28×10−15 | 7.25×10−1 |
| rs16959955 | 45789992 | SEMA6D | Up | .86 | .87 | 5.77×10−1 | 7.30×10−1 |
| rs34912622 | 46882494 | CEP152 | Int | .97 | .96 | 3.73×10−1 | 7.31×10−1 |
| rs363836 | 46510176 | FBN1 | Exon | .93 | .97 | 3.67×10−1 | 7.33×10−1 |
| rs12593270 | 45534632 | … | … | .64 | .64 | 9.21×10−1 | 7.40×10−1 |
| rs11636795 | 47463394 | C15orf33 | Int | .24 | .27 | 1.58×10−1 | 7.41×10−1 |
| rs1912643 | 45777558 | … | … | .83 | .78 | 2.70×10−1 | 7.41×10−1 |
| rs17394420 | 47208108 | COPS2 | Int | .85 | .81 | 8.78×10−1 | 7.41×10−1 |
| rs16963644 | 48425969 | … | … | .89 | .89 | 9.23×10−1 | 7.43×10−1 |
| rs11070672 | 47048808 | SHC4 | Up | .38 | .40 | 6.87×10−1 | 7.53×10−1 |
| rs8039653 | 46359927 | SLC12A1 | Int | .70 | .76 | 1.53×10−1 | 7.60×10−1 |
| rs16960880 | 46481074 | FBN1 | Down | .88 | .93 | 1.85×10−1 | 7.70×10−1 |
| rs938043 | 45696805 | … | … | .82 | .74 | 2.03×10−1 | 7.77×10−1 |
| rs532598 | 45845363 | SEMA6D | Down | .64 | .72 | 2.16×10−1 | 7.81×10−1 |
| rs17476940 | 47412076 | GALK2 | Down | .80 | .76 | 9.78×10−1 | 7.84×10−1 |
| rs16961174 | 46652169 | FBN1 | Int | .93 | .98 | 2.84×10−3 | 7.85×10−1 |
| rs751467 | 46034246 | … | … | .29 | .41 | 4.74×10−2 | 7.89×10−1 |
| rs585451 | 47795017 | … | … | .78 | .78 | 9.03×10−1 | 7.90×10−1 |
| rs10519257 | 48176213 | ATP8B4 | Int | .75 | .75 | 7.73×10−1 | 7.93×10−1 |
| rs479173 | 45978990 | … | … | .81 | .89 | 5.82×10−3 | 7.93×10−1 |
| rs8040116 | 47369881 | GALK2 | Int | .75 | .76 | 6.77×10−1 | 7.93×10−1 |
| rs17470994 | 47091617 | KIAA0256 | Int | .95 | .94 | 8.79×10−1 | 7.93×10−1 |
| rs16963141 | 48061447 | ATP8B4 | Int | .89 | .87 | 3.43×10−1 | 7.99×10−1 |
| rs10519150 | 45926447 | … | … | .85 | .89 | 2.76×10−1 | 8.03×10−1 |
| rs17404262 | 47754911 | … | … | .87 | .82 | 4.95×10−1 | 8.04×10−1 |
| rs4774545 | 47847312 | … | … | .93 | .93 | 7.21×10−1 | 8.05×10−1 |
| rs17499601 | 48214938 | … | … | .88 | .85 | 6.29×10−1 | 8.05×10−1 |
| rs10519174 | 46521916 | FBN1 | Int | .53 | .37 | 3.09×10−3 | 8.08×10−1 |
| rs16962914 | 47880927 | … | … | .95 | .96 | 6.01×10−1 | 8.08×10−1 |
| rs16961274 | 46712407 | FBN1 | Int | .94 | .98 | 3.37×10−2 | 8.10×10−1 |
| rs671291 | 45925000 | … | … | .68 | .78 | 7.81×10−2 | 8.11×10−1 |
| rs733220 | 46170229 | … | … | .93 | .95 | 8.52×10−1 | 8.11×10−1 |
| rs755067 | 47955564 | ATP8B4 | Int | .52 | .56 | 6.03×10−1 | 8.12×10−1 |
| rs4375601 | 47959139 | ATP8B4 | Int | .77 | .77 | 7.16×10−1 | 8.15×10−1 |
| rs16960538 | 46156777 | … | … | .94 | .97 | 7.52×10−1 | 8.15×10−1 |
| rs16960540 | 46156866 | … | … | .94 | .97 | 7.52×10−1 | 8.15×10−1 |
| rs17423970 | 46089356 | … | … | .80 | .65 | 1.52×10−3 | 8.21×10−1 |
| rs4544192 | 47939482 | ATP8B4 | 3′ UTR | .83 | .80 | 5.81×10−1 | 8.23×10−1 |
| ss69356384 | 48416695 | GABPB2 | Int | .75 | .71 | 3.99×10−1 | 8.24×10−1 |
| rs6493426 | 48375850 | GABPB2 | Int | .75 | .71 | 3.94×10−1 | 8.25×10−1 |
| rs16961323 | 46746374 | … | … | .85 | .88 | 1.57×10−1 | 8.25×10−1 |
| rs8041979 | 47951146 | ATP8B4 | Int | .82 | .80 | 6.25×10−1 | 8.26×10−1 |
| rs16962871 | 47838116 | … | … | .75 | .79 | 3.01×10−1 | 8.29×10−1 |
| rs11636875 | 47040769 | … | … | .67 | .59 | 1.90×10−1 | 8.35×10−1 |
| rs893160 | 46737569 | … | … | .19 | .18 | 8.44×10−1 | 8.36×10−1 |
| rs491996 | 45965538 | … | … | .67 | .82 | 1.39×10−4 | 8.37×10−1 |
| rs17499691 | 48216033 | … | … | .89 | .87 | 7.31×10−1 | 8.39×10−1 |
| rs16963067 | 47996550 | ATP8B4 | Int | .91 | .88 | 3.37×10−1 | 8.40×10−1 |
| rs16963180 | 48084344 | ATP8B4 | Int | .97 | .96 | 6.30×10−1 | 8.43×10−1 |
| rs11853852 | 48199132 | ATP8B4 | Up | .55 | .52 | 4.17×10−1 | 8.44×10−1 |
| rs12912380 | 45734583 | … | … | .94 | .94 | 6.52×10−1 | 8.47×10−1 |
| rs1365508 | 48288276 | SLC27A2 | Int | .70 | .69 | 5.26×10−1 | 8.47×10−1 |
| rs10519258 | 48187342 | ATP8B4 | Int | .95 | .94 | 3.26×10−1 | 8.50×10−1 |
| rs7170781 | 48180287 | ATP8B4 | Int | .61 | .63 | 5.04×10−1 | 8.50×10−1 |
| rs8036806 | 46926246 | SHC4 | Int | .96 | .96 | 6.58×10−1 | 8.54×10−1 |
| rs12915792 | 47225618 | COPS2 | Int | .26 | .24 | 5.80×10−1 | 8.56×10−1 |
| rs2067885 | 48009237 | ATP8B4 | Int | .83 | .83 | 4.96×10−1 | 8.59×10−1 |
| rs16960438 | 46062139 | … | … | .87 | .93 | 1.17×10−1 | 8.59×10−1 |
| rs8039548 | 45888247 | … | … | .84 | .88 | 1.64×10−1 | 8.60×10−1 |
| rs16960516 | 46116834 | … | … | .90 | .83 | 5.49×10−2 | 8.63×10−1 |
| rs4494482 | 47939458 | ATP8B4 | 3′ UTR | .84 | .81 | 5.95×10−1 | 8.65×10−1 |
| rs2934177 | 46000839 | … | … | .03 | .02 | 7.78×10−1 | 8.66×10−1 |
| rs16960341 | 45997010 | … | … | .93 | .94 | 5.23×10−1 | 8.68×10−1 |
| rs17509458 | 48452654 | … | … | .74 | .70 | 4.02×10−1 | 8.68×10−1 |
| rs16963311 | 48192467 | ATP8B4 | Int | .88 | .88 | 7.08×10−1 | 8.70×10−1 |
| rs1656631 | 45623301 | … | … | .70 | .67 | 6.34×10−1 | 8.71×10−1 |
| rs12324326 | 45935427 | … | … | .82 | .88 | 1.79×10−1 | 8.73×10−1 |
| rs16959550 | 45549747 | … | … | .91 | .95 | 2.28×10−1 | 8.73×10−1 |
| rs4426299 | 48000077 | ATP8B4 | Int | .91 | .88 | 3.55×10−1 | 8.74×10−1 |
| rs17499726 | 48216520 | … | … | .95 | .94 | 3.90×10−1 | 8.75×10−1 |
| rs11854358 | 47007749 | SHC4 | Int | .30 | .31 | 8.34×10−1 | 8.85×10−1 |
| rs12591825 | 48098427 | ATP8B4 | Int | .25 | .28 | 3.34×10−1 | 8.89×10−1 |
| rs12909202 | 48400026 | GABPB2 | Int | .75 | .71 | 3.70×10−1 | 8.89×10−1 |
| rs16960493 | 46096739 | … | … | .91 | .94 | 5.24×10−1 | 8.92×10−1 |
| rs9806163 | 46591131 | FBN1 | Int | .65 | .74 | 9.19×10−2 | 8.98×10−1 |
| rs8030172 | 45924790 | … | … | .63 | .56 | 7.81×10−2 | 9.00×10−1 |
| rs2413959 | 47547717 | FGF7 | Int | .72 | .70 | 3.81×10−1 | 9.02×10−1 |
| rs11855291 | 45858167 | SEMA6D | Down | .76 | .80 | 4.81×10−1 | 9.03×10−1 |
| rs4775708 | 45824941 | SEMA6D | Int | .84 | .84 | 8.31×10−1 | 9.05×10−1 |
| rs2114438 | 46487221 | FBN1 | Down | .48 | .34 | 1.27×10−2 | 9.07×10−1 |
| rs4774557 | 48220992 | … | … | .61 | .62 | 4.79×10−1 | 9.07×10−1 |
| rs1669832 | 45897944 | … | … | .69 | .63 | 3.18×10−1 | 9.08×10−1 |
| rs12593807 | 46334019 | SLC12A1 | Int | .72 | .78 | 1.12×10−1 | 9.10×10−1 |
| rs8031680 | 47524988 | C15orf33 | Int | .06 | .06 | 6.59×10−1 | 9.13×10−1 |
| rs692339 | 47791940 | … | … | .04 | .04 | 5.79×10−1 | 9.14×10−1 |
| rs769136 | 47799924 | … | … | .70 | .75 | 1.51×10−1 | 9.14×10−1 |
| rs12594956 | 48409126 | GABPB2 | Int | .49 | .50 | 9.27×10−1 | 9.16×10−1 |
| rs16960904 | 46494063 | FBN1 | Int | .93 | .98 | 4.26×10−2 | 9.16×10−1 |
| rs6493427 | 48376049 | GABPB2 | Int | .47 | .48 | 9.67×10−1 | 9.22×10−1 |
| rs1559677 | 45525355 | … | … | .44 | .51 | 2.40×10−1 | 9.23×10−1 |
| rs8025221 | 48148835 | ATP8B4 | Int | .84 | .83 | 9.37×10−1 | 9.23×10−1 |
| rs1114003 | 47336143 | GALK2 | Int | .94 | .94 | 8.63×10−1 | 9.26×10−1 |
| rs568215 | 45850368 | SEMA6D | Syn | .21 | .30 | 3.62×10−2 | 9.26×10−1 |
| rs477077 | 45884409 | … | … | .44 | .47 | 5.75×10−1 | 9.27×10−1 |
| rs860526 | 48342303 | HDC | Int | .52 | .48 | 2.25×10−1 | 9.27×10−1 |
| rs1506524 | 46676280 | FBN1 | Int | .68 | .77 | 1.04×10−1 | 9.30×10−1 |
| rs16960979 | 46551641 | FBN1 | Int | .92 | .97 | 2.17×10−2 | 9.30×10−1 |
| rs17416784 | 48355447 | GABPB2 | Down | .97 | .96 | 5.49×10−1 | 9.34×10−1 |
| rs12912519 | 48220812 | … | … | .84 | .80 | 4.12×10−1 | 9.37×10−1 |
| rs9788716 | 45926940 | … | … | .83 | .88 | 2.14×10−1 | 9.41×10−1 |
| rs1797311 | 48392109 | GABPB2 | Int | .54 | .53 | 9.98×10−1 | 9.44×10−1 |
| rs8031017 | 46470746 | … | … | .38 | .30 | 6.23×10−2 | 9.49×10−1 |
| rs16961806 | 46966198 | EID1 | Down | .82 | .88 | 3.62×10−2 | 9.52×10−1 |
| ss69356376 | 45998012 | … | … | .91 | .96 | 4.65×10−2 | 9.53×10−1 |
| rs854160 | 48343002 | HDC | Int | .47 | .45 | 5.12×10−1 | 9.54×10−1 |
| rs501916 | 45840521 | SEMA6D | Syn | .57 | .58 | 9.55×10−1 | 9.55×10−1 |
| rs16960326 | 45984783 | … | … | .94 | .98 | 1.07×10−2 | 9.56×10−1 |
| rs12591556 | 46558253 | FBN1 | Int | .86 | .94 | 8.60×10−3 | 9.57×10−1 |
| rs17415911 | 48284586 | SLC27A2 | Int | .84 | .87 | 7.48×10−1 | 9.58×10−1 |
| rs11070741 | 48098440 | ATP8B4 | Int | .22 | .24 | 4.34×10−1 | 9.61×10−1 |
| rs8041014 | 48085001 | ATP8B4 | Int | .22 | .24 | 4.34×10−1 | 9.61×10−1 |
| rs1224606 | 45933110 | … | … | .86 | .91 | 4.22×10−1 | 9.66×10−1 |
| rs16959519 | 45528224 | … | … | .81 | .88 | 1.84×10−1 | 9.69×10−1 |
| rs597543 | 45930092 | … | … | .19 | .27 | 2.88×10−1 | 9.72×10−1 |
| rs16963207 | 48090156 | ATP8B4 | Int | .84 | .85 | 8.58×10−1 | 9.83×10−1 |
| rs7342574 | 48217475 | … | … | .58 | .59 | 4.44×10−1 | 9.90×10−1 |
| rs652281 | 45826380 | SEMA6D | Int | .83 | .91 | 1.94×10−2 | 9.92×10−1 |
| rs1866501 | 45839887 | SEMA6D | Syn | .83 | .91 | 1.76×10−2 | 9.93×10−1 |
| rs3743279 | 45843511 | SEMA6D | Nonsyn | .83 | .91 | 1.76×10−2 | 9.93×10−1 |
| rs8036851 | 45830750 | SEMA6D | Int | .83 | .91 | 1.76×10−2 | 9.93×10−1 |
| rs1797316 | 48273679 | SLC27A2 | Int | .09 | .10 | 5.98×10−1 | 9.94×10−1 |
| rs677207 | 45970045 | … | … | .70 | .82 | 6.41×10−4 | 9.97×10−1 |
| rs16959999 | 45816306 | … | … | .83 | .91 | 1.87×10−2 | 9.98×10−1 |
rs or ss identifier in dbSNP.
Chromosome base position on NCBI build 36.2.
Gene symbol from Entrez Gene.
Int = intronic; nonsyn = nonsynonymous; syn = synonymous; up = within 10 kb of the transcriptional start site; down = within 10 kb of the transcriptional stop site.
Frequency of allele 1 in the reflectance groups of cohort 2.
P value conditional on rs1426654.
Figure 3. .
Association signals, linkage disequilibrium, and genes in the region 45,524,265–48,470,992 on chromosome 15q21. Negative log10 association P values (found by logistic regression and likelihood-ratio tests) are displayed for cohorts 1 and 2 and for an analysis conditioned on rs1426654 in cohort 2. The R2 value between each SNP genotyped in the region and the primary associated SNP rs1426654 is also displayed. The SNPs rs1834640 and rs1426654 show the strongest associations in cohort 1 and cohort 2, respectively. Red horizontal lines are drawn at the Bonferroni-corrected significance threshold for the genomewide scan (α=0.05/1,502,205=3.3×10-8) for cohort 1 and 2 associations. In the association analysis conditional on rs1426654, no SNPs met a significance level that would correct for the number of SNPs in the region (α=0.05/407=1.23×10-4). This figure was made using the Generic Genome Browser.47 Chr = chromosome.
Furthermore, by performing an association analysis conditional on the genotypes of rs1426654 in cohort 2, we observed that none of the other SNPs rise above the Bonferroni threshold for significance of conditional association in the region (α=0.05/407=1.23×10-4) (fig. 3 and table 6). When the conditional analysis was performed in reverse, by evaluation of the association of rs1426654 in cohort 2 conditional on each of the other 407 SNPs in this region in turn, the rs1426654 association was found to be significant at the genomewide level (P<3.3×10-8) for all SNPs but rs1834640, with which it is in the strongest linkage disequilibrium (P=.003 for association of skin reflectance with rs1426654 conditional on rs1834640; P=.73 for association with rs1834640 conditional on rs1426654). Thus, the data are consistent with the existence of a single strong primary association at rs1426654 in the SLC24A5 gene, and the multiplicity of apparent single-SNP associations in this region can be explained by the pattern of linkage disequilibrium with this SNP (fig. 3). Because of the lack of genomewide individual genotyping data for a South Asian population, we are unable to compare the extent of linkage disequilibrium in this region with that in the rest of the genome. Of the 407 SNPs genotyped in this region, only rs1834640 is in high linkage disequilibrium with rs1426654 (R2>0.9), and 3 more SNPs are in moderate linkage disequilibrium (R2>0.5) (fig. 3). Although we cannot rule out other associations of small effect within this chromosomal region, the evidence points strongly to a single association of large effect size, located at or in strong disequilibrium with the nonsynonymous SNP rs1426654 within SLC24A5.
Dominance and Interactions among Three Nonsynonymous Polymorphisms
The single-SNP association analyses with cohorts 1 and 2 identified three nonsynonymous polymorphisms in three genes—rs1426654 (p.A111T) in SLC24A5, rs16891982 (p.L374F) in SLC45A2, and rs1042602 (p.S192Y) in TYR—that show genomewide significance for association with skin pigmentation, on the basis of a multiplicative model of risk. Under a simple additive allele-risk model, the squared correlation (R2) between the dichotomously defined skin-reflectance trait and the genotypes is directly related to the fraction of the variance of skin reflectance accounted for by each SNP. The primary associated SNP, rs1426654 in SLC24A5, has the largest effect on skin reflectance of the three SNPs, with R2=0.33, compared with R2=0.036 for rs16891982 in SLC45A2 and R2=0.025 for rs1042602 in TYR.
We examined whether there is evidence within the cohort 2 data of deviations from a multiplicative model of risk, such as a dominance effect or interactions among the loci. We have very good power to detect dominant or recessive inheritance for rs1426654, which has a high minor-allele frequency (49% in the L group and 10% in the H group) and a very large effect on skin reflectance. However, our power to detect deviations from additive inheritance for the other two SNP associations is much lower because of the smaller size of their effect. Likelihood-ratio tests were used to assess the significance of an added dominance term, but none of the tests showed significant deviations from multiplicative risk (P=.58 for rs1426654, P=.20 for rs16891982, and P=.91 for rs1042602), which provides no evidence to support dominant or recessive inheritance for any of the three SNPs.
We next explored whether the three SNPs collectively explain more of the trait than does the combination of their single-SNP associations. Interactions were modeled within the framework of logistic regressions by adding indicator variables corresponding to specific genotype combinations for pairs of SNPs. We tested each of the three pairs separately and found no evidence for pairwise interactions (rs1426654-rs16891982, P=.77; rs1426654-rs1042602, P=.38; rs16891982-rs1042602, P=.12). Therefore, our data are most consistent with a simple model for skin pigmentation comprising additive contributions from alleles at the three associated nonsynonymous polymorphisms. Because our study design used the tails of the skin-reflectance distribution, we cannot directly determine the overall fraction of the trait variance in the population that is accounted for by the associated SNPs. Nevertheless, it is evident that these three markers collectively account for a large fraction of the wide variability of skin pigmentation within South Asians.
Discussion
This is the first genomewide association study, to our knowledge, to investigate the genetic determinants of normal skin pigmentation within a human population. With the use of objective quantitative measurements of melanin content, our study clearly identifies three loci—TYR, SLC45A2, and SLC24A5—that contribute to the natural variation in skin pigmentation within a South Asian population. Within each of these genes, we found polymorphisms that met genomewide significance on association tests, with the associations replicated in a second South Asian cohort. The contributions of these polymorphisms to skin pigmentation were found to be independent and additive across genes, with no evidence of dominant or recessive effects at any of these loci. Collectively, these three genes account for a large fraction of the wide, naturally occurring variation in skin pigmentation in South Asians.
Strikingly, our primary associated SNP in SLC24A5 may explain >30% of the variance of the dichotomously defined skin reflectance in our study population. As a result of the interplay between the strength of this single-SNP association and linkage disequilibrium with SNPs in its vicinity, several other significant associations were observed in a 2.4-Mb region on chromosome 15q21. Although we cannot completely exclude the possibility that multiple real independent associations exist in this chromosomal 15 region, our conditional analysis shows unambiguously that the association signals on all SNPs in the region are far less significant than those for the primary associated SNP in SLC24A5.
This is also one of the first genomewide association studies to focus on a South Asian population. This population is known to be highly genetically diverse, and, because ancestry is known to be highly confounded with skin pigmentation, the treatment of population structure was crucial to our association analysis. Our analysis of population structure was based on the individual genotypes of 968 individuals with 292 unlinked SNPs. Notwithstanding the limitations imposed by the size of this data set on the detection of structure,48 the results for these 292 SNPs indicated that we were able to adequately correct for stratification. It is encouraging that, despite the use of a study population that is genetically diverse and a trait that is strongly confounded with ancestry, we were able to detect three true SNP associations.
At the time of this study, mutations in two of these genes, TYR and SLC45A2, were known to be involved in different forms of oculocutaneous albinism (OCA1 [MIM 203100] and OCA4 [MIM 606574]),49,50 yet their contribution to the normal range of skin pigmentation between and within populations was just beginning to be studied. The remaining locus, SLC24A5, which displays the largest genetic effect on pigmentation status in the South Asian population studied here, was an unknown pigmentation gene during our study. The SLC24A5 gene has since been shown to play an important role in pigmentation, because a mutation in this gene was identified as the genetic basis of the hypopigmentation phenotype in golden zebrafish.22 During the past few years, several candidate-gene admixture studies on a cohort of African Americans and African Caribbeans has shown that the same three nonsynonymous polymorphisms that we found in our South Asian cohort—rs1042602 in TYR, rs16891982 in SLC45A2, and rs1426654 in SLC24A5—are significantly associated with melanin content measured by reflectometry.13,21,22,24 When taken together with the results of our study, the evidence is compelling that these three loci are a large component of the genetic influence of constitutive pigmentation in these diverse populations.
The importance of these three particular SNPs—rs1042602 in TYR, rs16891982 in SLC45A2, and rs1426654 in SLC24A5—to skin-pigmentation variation within other populations is likely to be highly variable across the globe. All three of the SNPs are essentially monomorphic in African and East Asian populations and thus are unlikely to contribute significantly to skin-pigmentation variation within those groups.13,25,29,30 For populations of European ancestry, the contributions of these three SNPs to intrapopulation skin-pigmentation variation are quite distinct from each other. The SLC24A5 SNP rs1426654 is nearly fixed for one allele in all European groups.11,13,22,29,30,51 In comparison, the TYR SNP rs1042602 is highly polymorphic in all European populations, but, despite its variability, previous studies found that this SNP was not significantly associated with skin-reflectance measurements in a white population.13,21,24,29 In contrast to the TYR SNP, the SLC45A2 SNP rs16891982 displays a gradient of increasing minor-allele frequency in European populations from north to south13,26,52 and has been significantly associated with subjectively measured skin color in a population of European ancestry.19 It is important to distinguish the involvement of these three particular SNPs in skin-pigmentation variation within populations from the potential contribution of the three loci (TYR, SLC24A4, and SLC45A2). For example, two functional promoter variants and another nonsynonymous polymorphism in SLC45A2, rs26722 (p.E272K), have also been significantly associated with subjectively measured skin color in a population of European ancestry.20 Whereas the promoter variants in SLC45A2 were not included in our study, rs26722 was assayed in the present study but showed weaker evidence of association with skin-reflectance measurements in our South Asian population (joint cohort association P=.02), as compared with the significantly associated rs16891982 SNP.
Other genes have been implicated either directly or indirectly with pigmentation-related traits in previous work. Several reported associations are based on the examination of markers in pigmentation candidate genes for allele-frequency differences between populations with known skin-pigmentation differences11–13,51 or are associations with a different qualitative measure of pigmentation, such as the Fitzpatrick skin type and subjective classification of skin color by visual assessment,53–55 all of which make direct comparisons with our results difficult. In contrast, other associations reported for skin pigmentation used quantitative measures of reflectance similar to those in our study17,18,21,23,24; therefore, our results can be compared directly with those studies. A modest association has been found between skin pigmentation from reflectance measurements for SNP rs1800404 within the gene OCA221 in an African American population (P=.01). Although rs1800404 was not assayed in our study, the closest OCA2 SNP that was included in our study, rs1037208, located 4,414 bp from rs1800404, yielded the most significant joint cohort association P value (P=5.6×10-5) of several OCA2 SNPs that we assayed with individual genotyping. On the basis of International HapMap data for the CEU sample (Utah residents with ancestry from northern and western Europe),29 these two SNPs are in modest linkage disequilibrium with one another (R2=0.35). Although we cannot establish beyond a doubt that these two SNPs correspond to a single locus in our population, it is quite plausible that they are highly correlated in South Asians; therefore, our results may support the prior association of OCA2 and skin pigmentation. Another study found an association with skin reflectance in a Tibetan population under an epistatic model (P=.01) for a pair of SNPs in OCA2 and MC1R17—rs12910433 and a nonsynonymous polymorphism, rs2228479 (p.V92M), respectively. Both these SNPs were individually genotyped in cohort 1 in our study, and, whereas the OCA2 SNP rs12910433 showed no evidence of association (P=.3), the MC1R SNP rs2228479 had the lowest P value (P=6.8×10-4) of several MC1R SNPs that were individually genotyped. There is no evidence of interactions between these two SNPs in our study population (P=.91). Lastly, one SNP in the 3′ UTR of the ASIP gene, rs6058017, has been associated with darker skin color in female African Americans18 (P<.001). This SNP was included in our pooled study but showed no allele-frequency difference between the low- and high-reflectance groups (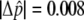 ), and attempts to individually genotype this SNP in cohort 1 were unsuccessful. Since the previous association showed significant association with only female, but not male, skin pigmentation, our pooled genotyping strategy that used mixed sexes would not have the power to find such an association.
), and attempts to individually genotype this SNP in cohort 1 were unsuccessful. Since the previous association showed significant association with only female, but not male, skin pigmentation, our pooled genotyping strategy that used mixed sexes would not have the power to find such an association.
It is important to note that the application of a very strict Bonferroni correction of 1,502,205 independent tests, combined with the pooled genotyping design, greatly reduces the power of this study to detect real SNP associations with small effects on skin pigmentation; thus, it is possible that loci other than TYR, SLC45A2, and SLC24A5 may affect skin-pigmentation variation within the South Asian population. Furthermore, as was mentioned earlier in this discussion, several studies comparing allele frequencies of candidate skin-pigmentation SNPs across multiple global populations present evidence supporting an independent genetic mechanism for the lighter skin pigmentation of East Asians that is distinct from genetic factors influencing lighter pigmentation in Europeans.12,13,25 Therefore, it is likely that loci other than the three identified in this study influence skin pigmentation in other global populations.
Acknowledgments
We thank Cathryn Lewis, C. V. Natraj, Geoffrey Probert, Pushker Sona, Ian Scott, Michael Barratt, Simon Alaluf, and Nicholas Holmberg, for useful discussions. We thank all Perlegen Sciences employees who provided technical assistance and scientific discussion for this project and manuscript. We also thank the volunteers who donated their blood and time, without which this study would not have been possible.
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Entrez Gene, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OCA1 and OCA4)
References
- 1.Armstrong BK, Kricker A, English DR (1997) Sun exposure and skin cancer. Australas J Dermatol 38:S1–S6 10.1111/j.1440-0960.1997.tb01115.x [DOI] [PubMed] [Google Scholar]
- 2.Palmer JS, Duffy DL, Box NF, Aitken JF, O’Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA (2000) Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet 66:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees JL (2004) The genetics of sun sensitivity in humans. Am J Hum Genet 75:739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins AH (1991) Biological perspectives on human pigmentation. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 5.Branda RF, Eaton JW (1978) Skin color and nutrient photolysis: an evolutionary hypothesis. Science 201:625–626 10.1126/science.675247 [DOI] [PubMed] [Google Scholar]
- 6.Chaplin G (2004) Geographic distribution of environmental factors influencing human skin coloration. Am J Phys Anthropol 125:292–302 10.1002/ajpa.10263 [DOI] [PubMed] [Google Scholar]
- 7.Murray FG (1934) Pigmentation, sunlight, and nutritional disease. Am Anthropol 36:438–445 10.1525/aa.1934.36.3.02a00100 [DOI] [Google Scholar]
- 8.Clark P, Stark AE, Walsh RJ, Jardine R, Martin NG (1981) A twin study of skin reflectance. Ann Hum Biol 8:529–541 10.1080/03014468100005371 [DOI] [PubMed] [Google Scholar]
- 9.Frisancho AR, Wainwright R, Way A (1981) Heritability and components of phenotypic expression in skin reflectance of Mestizos from the Peruvian lowlands. Am J Phys Anthropol 55:203–208 10.1002/ajpa.1330550207 [DOI] [PubMed] [Google Scholar]
- 10.Harrison GA, Owen JJ (1964) Studies on the inheritance of human skin colour. Ann Hum Genet 28:27–37 10.1111/j.1469-1809.1964.tb00457.x [DOI] [PubMed] [Google Scholar]
- 11.Izagirre N, Garcia I, Junquera C, de la Rua C, Alonso S (2006) A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol 23:1697–1706 10.1093/molbev/msl030 [DOI] [PubMed] [Google Scholar]
- 12.Myles S, Somel M, Tang K, Kelso J, Stoneking M (2007) Identifying genes underlying skin pigmentation differences among human populations. Hum Genet 120:613–621 10.1007/s00439-006-0256-4 [DOI] [PubMed] [Google Scholar]
- 13.Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD (2006) Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol 24:710–722 10.1093/molbev/msl203 [DOI] [PubMed] [Google Scholar]
- 14.Relethford JH (1997) Hemispheric difference in human skin color. Am J Phys Anthropol 104:449–457 [DOI] [PubMed] [Google Scholar]
- 15.Bennett DC, Lamoreux ML (2003) The color loci of mice—a genetic century. Pigment Cell Res 16:333–344 10.1034/j.1600-0749.2003.00067.x [DOI] [PubMed] [Google Scholar]
- 16.Sturm RA, Teasdale RD, Box NF (2001) Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene 277:49–62 10.1016/S0378-1119(01)00694-1 [DOI] [PubMed] [Google Scholar]
- 17.Akey JM, Wang H, Xiong M, Wu H, Liu W, Shriver MD, Jin L (2001) Interaction between the melanocortin-1 receptor and P genes contributes to inter-individual variation in skin pigmentation phenotypes in a Tibetan population. Hum Genet 108:516–520 10.1007/s004390100524 [DOI] [PubMed] [Google Scholar]
- 18.Bonilla C, Boxill LA, Donald SA, Williams T, Sylvester N, Parra EJ, Dios S, Norton HL, Shriver MD, Kittles RA (2005) The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum Genet 116:402–406 10.1007/s00439-004-1251-2 [DOI] [PubMed] [Google Scholar]
- 19.Graf J, Hodgson R, van Daal A (2005) Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat 25:278–284 10.1002/humu.20143 [DOI] [PubMed] [Google Scholar]
- 20.Graf J, Voisey J, Hughes I, van Daal A (2007) Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat 28:710–717 10.1002/humu.20504 [DOI] [PubMed] [Google Scholar]
- 21.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al (2005) SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310:1782–1786 10.1126/science.1116238 [DOI] [PubMed] [Google Scholar]
- 23.Naysmith L, Waterston K, Ha T, Flanagan N, Bisset Y, Ray A, Wakamatsu K, Ito S, Rees JL (2004) Quantitative measures of the effect of the melanocortin 1 receptor on human pigmentary status. J Invest Dermatol 122:423–428 10.1046/j.0022-202X.2004.22221.x [DOI] [PubMed] [Google Scholar]
- 24.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, et al (2003) Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112:387–399 [DOI] [PubMed] [Google Scholar]
- 25.Soejima M, Koda Y (2007) Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med 121:36–39 10.1007/s00414-006-0112-z [DOI] [PubMed] [Google Scholar]
- 26.Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, Dashnyam B, Jin F, Lucotte G, Chattopadhyay PK, et al (2006) Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet 70:802–811 10.1111/j.1469-1809.2006.00261.x [DOI] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 28.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al (2001) The sequence of the human genome. Science 291:1304–1351 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- 29.International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 10.1126/science.1105436 [DOI] [PubMed] [Google Scholar]
- 31.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- 32.Hinds DA, Seymour AB, Durham K, Banerjee P, Ballinger DG, Milos PM, Cox DR, Thompson JF, Frazer KA (2004) Application of pooled genotyping to scan candidate regions for association with HDL cholesterol levels. Hum Genomics 1:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG (2004) Matching strategies for genetic association studies in structured populations. Am J Hum Genet 74:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 36.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, et al (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 10.1126/science.1065573 [DOI] [PubMed] [Google Scholar]
- 37.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, et al (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 16:24–35 10.1093/hmg/ddl441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al (2007) Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 16:36–49 10.1093/hmg/ddl438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 10.1111/j.0006-341X.1999.00997.x [DOI] [PubMed] [Google Scholar]
- 40.Cordell HJ, Clayton DG (2002) A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet 70:124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumder PP (1998) People of India: biological diversity and affinities. Evol Anthropol 6:100–110 [DOI] [Google Scholar]
- 42.Barsh GS (2003) What controls variation in human skin color? PLoS Biol 1:E27 10.1371/journal.pbio.0000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rees JL (2003) Genetics of hair and skin color. Annu Rev Genet 37:67–90 10.1146/annurev.genet.37.110801.143233 [DOI] [PubMed] [Google Scholar]
- 44.Alaluf S, Atkins D, Barrett K, Blount M, Carter N, Heath A (2002) The impact of epidermal melanin on objective measurements of human skin colour. Pigment Cell Res 15:119–126 10.1034/j.1600-0749.2002.1o072.x [DOI] [PubMed] [Google Scholar]
- 45.Sengupta S, Farheen S, Mukherjee N, Dey B, Mukhopadhyay B, Sil SK, Prabhakaran N, Ramesh A, Edwin D, Usha Rani MV, et al (2004) DNA sequence variation and haplotype structure of the ICAM1 and TNF genes in 12 ethnic groups of India reveal patterns of importance in designing association studies. Ann Hum Genet 68:574–587 10.1046/j.1529-8817.2003.00126.x [DOI] [PubMed] [Google Scholar]
- 46.Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al (2002) The Generic Genome Browser: a building block for a model organism system database. Genome Res 12:1599–1610 10.1101/gr.403602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giebel LB, Strunk KM, King RA, Hanifin JM, Spritz RA (1990) A frequent tyrosinase gene mutation in classic, tyrosinase-negative (type IA) oculocutaneous albinism. Proc Natl Acad Sci USA 87:3255–3258 10.1073/pnas.87.9.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH (2001) Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 69:981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M (2007) Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet 71:354–369 10.1111/j.1469-1809.2006.00341.x [DOI] [PubMed] [Google Scholar]
- 52.Nakayama K, Fukamachi S, Kimura H, Koda Y, Soemantri A, Ishida T (2002) Distinctive distribution of AIM1 polymorphism among major human populations with different skin color. J Hum Genet 47:92–94 10.1007/s100380200007 [DOI] [PubMed] [Google Scholar]
- 53.Duffy DL, Montgomery GW, Chen W, Zhao ZZ, Le L, James MR, Hayward NK, Martin NG, Sturm RA (2007) A three–single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am J Hum Genet 80:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, Birch-Machin MA, Rees JL (2000) Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet 9:2531–2537 10.1093/hmg/9.17.2531 [DOI] [PubMed] [Google Scholar]
- 55.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ (1995) Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 11:328–330 10.1038/ng1195-328 [DOI] [PubMed] [Google Scholar]