Abstract
Mutations in FOXP2, a member of the forkhead family of transcription factor genes, are the only known cause of developmental speech and language disorders in humans. To date, there are no known targets of human FOXP2 in the nervous system. The identification of FOXP2 targets in the developing human brain, therefore, provides a unique tool with which to explore the development of human language and speech. Here, we define FOXP2 targets in human basal ganglia (BG) and inferior frontal cortex (IFC) by use of chromatin immunoprecipitation followed by microarray analysis (ChIP-chip) and validate the functional regulation of targets in vitro. ChIP-chip identified 285 FOXP2 targets in fetal human brain; statistically significant overlap of targets in BG and IFC indicates a core set of 34 transcriptional targets of FOXP2. We identified targets specific to IFC or BG that were not observed in lung, suggesting important regional and tissue differences in FOXP2 activity. Many target genes are known to play critical roles in specific aspects of central nervous system patterning or development, such as neurite outgrowth, as well as plasticity. Subsets of the FOXP2 transcriptional targets are either under positive selection in humans or differentially expressed between human and chimpanzee brain. This is the first ChIP-chip study to use human brain tissue, making the FOXP2-target genes identified in these studies important to understanding the pathways regulating speech and language in the developing human brain. These data provide the first insight into the functional network of genes directly regulated by FOXP2 in human brain and by evolutionary comparisons, highlighting genes likely to be involved in the development of human higher-order cognitive processes.
Spoken language and written language are uniquely human traits with a significant but complex genetic component. As with other developmental processes, the study of rare Mendelian forms of language or speech disorders provides an efficient means to begin to understand the molecular basis of human speech and language.1 FOXP2 was identified as involved in speech and language when affected members of the “KE” family were found to carry a mutated allele, and an unrelated individual was found to carry a balanced translocation with a break in FOXP22 (causing speech-language disorder 1 [SPCH1 {MIM 602081}]). Its role in speech and language was reiterated when an additional mutation that caused a truncation of FOXP2 was identified in a family with speech and language deficits.3 Individuals with FOXP2 mutations have dominantly inherited verbal dyspraxia and linguistic and/or grammatical difficulties.2,4,5 Additionally, patients with FOXP2 mutations have demonstrated developmental abnormalities of the basal ganglia (BG) and inferior frontal cortex (IFC).6,7 FOXP2 expression overlaps with perisylvian frontal and temporal regions, including the inferior frontal gyrus (Broca’s region), but also extends more broadly to suggest a role in complex sensory motor integration involving auditory vocal learning,8,9 as well as mirror neuron system function, which is highly evolved in primates and disrupted in autism.10,11
FOXP2 belongs to a family of proteins that contain a forkhead DNA binding, or “winged helix,” domain, a region responsible for DNA binding that is found in transcription factors.2 FOXP2 also contains a transcriptional repression domain including a zinc-finger motif in the N-terminal region12 and has been shown to interact with the corepressor protein C-terminal binding protein 1.13 Foxp1, Foxp2, and Foxp4 have been demonstrated to form hetero- and homotypic dimers that are important for their transcriptional activity.13 The expression patterns of FOXP1 and FOXP2 are not identical8; therefore, in some cases, the proteins may be forming heterotypic dimers, whereas, in others, they may act alone.13 Foxp2, the mouse orthologue of FOXP2, has been proposed to be an important regulator of proximal versus distal epithelial differentiation in the lung, on the basis of in vitro repression of the mouse CC10 promoter and human SP-C promoter.12 Additionally, other members of the forkhead-box (FOX) family of transcription factors are well established as transcriptional repressors or activators involved in development.14–16 However, the role of FOXP2 in the brain and its transcriptional targets remain to be elucidated.
Previous analyses of amino acid changes in FOXP2 across species indicate that FOXP2 has undergone accelerated evolution in humans.17,18 The most parsimonious explanation for the observed acceleration is positive selection of FOXP2 in humans, since there is no evidence of an increase in the mutation rate or purifying selection. Thus, the study of FOXP2 provides a potentially powerful avenue for investigations into the molecular and physical adaptations that allowed for the development of speech and language in humans. Language is a complex trait and necessarily involves the interaction of many genes,1 some of which may have coevolved. Since FOXP2 is a transcription factor, identification of its transcriptional targets in the brain and the assessment of their evolution would provide an important advance by elucidating the molecular pathways involved and potential evidence of their adaptive evolution.
Here, using chromatin immunoprecipitation (ChIP) followed by hybridization of the precipitated DNA to human promoter arrays (hereafter, “ChIP-chip”), we have identified targets of FOXP2 in vivo in both the BG region and the IFC of the human fetal brain. A subset of ChIP-chip–identified targets were confirmed by ChIP–quantitative PCR, and the functional consequence of FOXP2 binding to target promoters was demonstrated by FOXP2 overexpression in vitro. This study, along with that by Vernes et al.,19(in this issue) provides the first insight into the direct functional targets of FOXP2 during human brain development, as well as a core set of genes for further exploration into the genetic basis of human speech and language.
Material and Methods
Antibody Production
A 15-mer amino acid sequence was chosen from the C-terminal region of FOXP2. A FOXP2 antibody was made against the 14-aa sequence EDLNGSLDHIDSNG (fig. 1) in the C-terminal region of FOXP2. The 14-aa peptide was chosen on the basis of its antigenicity (DNASTAR) and dissimilarity between family members FOXP1 and FOXP4 (fig. 1). Similarity of the amino acid sequence to other proteins was excluded by comparisons with family members FOXP1 and FOXP4, as well as by protein blast (blastp) analysis (BLAST). The peptide was conjugated to keyhole limpet hemocyanin, and rabbits were immunized with the peptide and complete Freund’s adjuvant (Sigma-Genosys). Antibody was purified on an affinity purification column with the peptide (Sigma-Genosys).
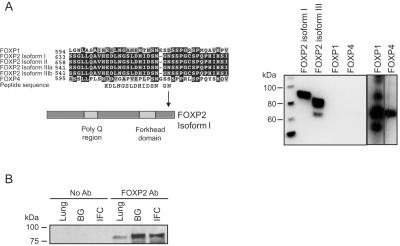
Figure 1. .
FOXP2 antibody detection of human brain and lung FOXP2 expression. A, Peptide used as an antigen to create the FOXP2 antibody aligned against the amino acid sequence of FOXP2 and family members FOXP1 and FOXP4. Protein from MRC5 cells transfected with expression vectors containing FOXP2 isoforms I and III and FOXP1 and FOXP4 transcripts were run on an SDS-PAGE gel, were transferred to PVDF membrane, and were hybridized with polyclonal anti-FOXP2 antibody (lanes 1–4). FOXP1 and FOXP4 were also hybridized with protein-specific antibodies (lanes 5 and 6) as a positive control for protein expression. B, FOXP2 isoform I protein immunoprecipitated from lung, BG, and IFC regions run on an SDS-PAGE gel, transferred to PVDF membrane, and hybridized with polyclonal anti-FOXP2 antibody (Ab).
Western-Blot Analysis
MRC5 or SH-SY5Y cells were lysed in hypotonic buffer (10 mM Tris-Cl, 10 mM KCl, 0.1 mM EDTA, and 0.1 mM ethylene glycol tetraacetic acid [EGTA]) and were sonicated briefly. Cell debris was removed by centrifugation. Protein was run on a 7.5% SDS-PAGE gel and was transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% milk in PBS-Tween for 2 h. The corresponding lanes were then incubated with FOXP2 in-house antibody (1:5,000 dilution), FOXP1 in-house antibody (1:10,000 dilution), FOXP4 ab17726 Abcam antibody (1:1,000 dilution), or FLAG F3165 Sigma antibody (1:10,000 dilution) in 5% milk in PBS-Tween overnight at 4°C. Blots were washed and incubated with 1:10,000 dilution of horseradish peroxidase–conjugated anti-rabbit or anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) and then were exposed to SuperSignal West Pico Chemiluminescent Substrate (Pierce) in accordance with the manufacturer’s directions.
Tissue Samples
Human tissue samples were obtained from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders under contracts N01-HD-4-3368 and N01-HD-4-3383 (lung = University of Maryland Brain Bank [UMB] number 1926 [18 gestational wk {GW}]; BG = UMB number 1664 [19 GW], UMB number 1888 [19 GW], and UMB number 1876 [20 GW]; IFC = UMB number 638 [16 GW], UMB number 899 [18 GW], and UMB number 1876 [20 GW]). FOXP2 targets in human fetal brain during midgestation (16–20 GW) were investigated because this time corresponds to the peak period of neuronal migration, differentiation, and cortical regionalization and is a time of high FOXP2 expression.8,9,20
ChIP
ChIP was performed as described elsewhere.21 For each experiment, 0.6 g of human tissue was used. Tissue was finely minced in PBS on ice, and the cross-linking reaction was subsequently performed for 15 min at room temperature. Nuclei were isolated, and DNA was sonicated in 1 ml of buffer (1 μM EDTA, 0.5 μM EGTA, and 10 μM Tris-HCl) to DNA fragments that were ∼0.2–1 kb in size. Then, 100 μl of Protein A beads were mixed with 10 μg of FOXP2 antibody overnight. Nuclear lysate was hybridized to the beads overnight. Beads were then washed five times with RIPA buffer (50 mM Hepes, 1 mM EDTA, 0.7% deoxycholic acid, 1% NP-40, and 0.5 M LiCl), and protein-DNA complexes were eluted (50 mM Tris, 10mM EDTA, and 1% SDS) at 65°C for 12 min. The cross-linking reaction was reversed at 65°C overnight (50 mM Tris, 10 mM EDTA, and 1% SDS). The ends of the DNA were blunted with T4 DNA polymerase (New England Biolabs) for 20 min at 12°C. DNA was ligated to annealed linkers (oJW102, GCGGTGACCCGGGAGATCTGAATTC, and oJW103, GAATTCAGATC) at 16°C for 16 h. ChIP and input DNA were amplified by PCR, with 1 mM deoxynucleotide triphosphates, 2 U Taq (Qiagen), and 1 μM oJW102. Cycling conditions were 1 cycle for 5 min at 72°C; 95°C for 2 min; 24 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min; and 1 cycle for 5 min at 72°C. For some ChIPs, a second and third round of PCR was performed to obtain enough DNA.
Labeling and Hybridization
A total of 200 ng of purified PCR product was labeled with Cy5 (ChIP DNA) and Cy3 (input DNA) with BioPrime DNA labeling system (Invitrogen) at 37°C for 16 h. Human promoter arrays (Aviva Systems Biology)22 were hybridized with 2 μg of each labeled ChIP and input DNA.
Microarray and Data Analysis
Arrays were scanned on a GenePix 4000B (Molecular Devices), and image analysis was performed with GenePix Pro 6.0. Three independent biological replicates were analyzed for BG and IFC; two independent biological replicates were analyzed for lung. Poor-quality spots were flagged by GenePix Pro 6.0. Background was determined on a spot-by-spot basis. Data analysis was performed using R with Bioconductor23 packages Limma and Marray. The quality of arrays was determined on the basis of signal plots before normalization. Spots whose signal intensities were twofold greater than the local background were considered present. Median normalization was performed. Data have been deposited into NCBI Gene Expression Omnibus (GEO) and can be accessed with accession number GSE8547.
Gene Ontology (GO) Analysis
Target genes with an M score >0.5 and a P value of >.05 were analyzed using DAVID.24,25 The Fisher exact test was used, with all array genes as background.
Stable and Transient-Overexpression Cell Lines
cDNA was transcribed from RNA isolated (Qiagen) from human fetal brain by use of the Superscript First-Strand Synthesis System (Invitrogen). FOXP2 was amplified via PCR with forward primer FOXP2-1F(BamHI)+ (5′-aaggatccatgatgcaggaatctgcgac-3′) and reverse primer FOXP2-1R(EcorI)+ (5′-ccgaattcttccagatcttcagataaaggc-3′). FOXP1 was amplified with forward primer Foxp1kozakF (5′-caccatgatgcaagaatctgggac-3′) and reverse primer Foxp1w/otagR (5′-tcactccatgtcctcgtttac-3′). A FOXP4 clone was obtained from Open Biosystems (MHS1010-9204774). Products were cloned into a pEF6/V5-His TOPO TA vector for transient expression in MRC5 cells. MRC5 cells were transiently transfected with linearized clones with Lipofectamine 2000 (Invitrogen) for 24 h. For stable transfection in SH-SY5Y cells, PCR products were cloned into the pCMV-Tag4A vector (Stratagene) with three C-terminal FLAG tags. SH-SY5Y cells were transfected with linearized clones either with the FOXP2 insert or without (vector only) by use of Lipofectamine 2000 (Invitrogen) and were selected for stably transfected cells with 1.428 mg/ml Geneticin (Invitrogen) for >5 d. The antibiotic was removed at least 48 h before harvesting for quantitative PCR experiments. Seven passages of each cell line were used as biological replicates.
Real-Time PCR
RNA was extracted using RNeasy Mini Kit (Qiagen) following the manufacturer's directions. DNA was removed by digestion with RNase-free DNase (Qiagen). A quantity of 1.2–5 μg was used in a reaction to synthesize cDNA with oligo dT primers. Quantitative real-time PCR was performed on an ABI 7900HT (Applied Biosystems) with SDS 2.1 software. The reaction mix contained iTaq SYBR Green Supermix (Bio-Rad) and 0.3 μM of each primer. Cycling conditions were 50°C for 2 min and 95°C for 3 min, followed by 45 cycles at 95°C for 15 s and 58°C for 45 s, and, finally, 95°C for 15 s, 60°C for 20 s, and 95°C for 15 s.
ChIP-PCR
ChIP was done as described above by use of anti-FOXP2 (rabbit polyclonal [Abcam]) or anti-FLAG (mouse monoclonal [Sigma]) antibodies. PCR was performed using either SYBR Green Supermix (Bio-Rad) or Taq (Qiagen) and the following primers: for ANK1, 5′-ccccctccttaggaaacaaa-3′ and 5′-agcccagagttggacatcag-3′; for CALCRL, 5′-tcactctttcccaccttgct-3′ and 5′-gaaacattgccaaactatatgagaa-3′; for CDH1, 5′-ctcgacacccgattcaaagt-3′ and 5′-gcgtgactttggtggaaaac-3′; for LBR, 5′-taaagctgggaggtgctgtc-3′ and 5′-ggctgctgtaggcttgagag-3′; for KCNJ15, 5′-ccagtaggcaaatccttcca-3′ and 5′-ggggatagaaattcgggtgt-3′; for PIR51, 5′-cagtccaagtgcccctatgt-3′ and 5′-ggaactacccacctcacagg-3′; for PPP2R1B, 5′-acaacagaaggcaccattcc-3′ and 5′-ccgctcagactcaaacttcc-3′; for TGM2, 5′-tggctgtgtcaggctgtatc-3′ and 5′-acacagagagcagacgcaga-3′; and, for TNNI1, 5′-tgctggtttcactcagttgg-3′ and 5′-aatgcacacaacaggcacat-3′.
Comparison of Gene-Expression Levels and Estimates of Protein-Sequence Divergence Rates between Humans and Chimpanzees
Gene-expression levels in human and chimpanzee cerebral cortex were determined by combining microarray data from three independent studies.26–28 To identify probes common to both species, megablast (BLAST) was used to align all probes from the Affymetrix HGU95Av2 microarray to the human genome (build 34) and the chimpanzee draft genome. Any probe without a perfect match in both species (∼1/4) was masked during the calculation of expression values (GCOSv1.2 [Affymetrix]). In addition, only probe sets with six or more matching probes were retained for subsequent analyses (n=11,768 of 12,625 sets). For each array, expression values were scaled to an average intensity of 200 (GCOSv1.2 [Affymetrix]). Two samples, “Hs3_MFG” and “Pt4_FP” from the study of Caceres et al.,26 were identified as outliers and were removed from the analysis. Technical replicates were averaged, followed by biological replicates (i.e., different cortical samples from the same individual). After averaging, there were 11 unique human and 8 unique chimpanzee individuals in the data set (two of the chimpanzees from these studies26–28 were identical). Quantile normalization26 was then performed, and data were log transformed. Gene-expression levels were compared between the species by use of a Bayesian t test via the “bayesreg” R package, with the settings betaFit = 1, winSize = 101, and conf = 10. Estimated rates of protein-sequence divergence between humans and chimpanzees were obtained from the study of Khaitovich et al.29
Results
The FOXP subfamily of transcription factors have relatively high homology among themselves, so it was important to generate an immunoreagent that would meet the specificity and efficiency requirements of ChIP. We produced a high-affinity FOXP2 polyclonal antibody on the basis of immunization with a relatively divergent region near the C-terminus (fig. 1). Since isoform III does not share the N-terminus with isoforms I and II, this C-terminal moiety provided the additional benefit of permitting detection of all FOX domain–containing isoforms of FOXP2. No cross-reactivity was detected for FOXP1 or FOXP4, and the antibody detected two major FOXP2 isoforms, designated isoform I and III (GenBank accession numbers NP_055306 and NP_683697 or NP_683698) as predicted (fig. 1). A protein of ∼80 kDa was immunoprecipitated from human fetal brain regions and lung (fig. 1). Immunohistochemistical staining of mouse brain by use of this antibody reflects previously described patterns of postmitotic neuronal staining in the cortex and BG (data not shown).20
Identification of Core FOXP2 Targets by In Vivo ChIP-Chip in Fetal Human Brain
Given previous data suggesting adaptive evolution of FOXP2 in humans17,18 and its involvement in human higher cognitive functions, we were interested in elucidating FOXP2 targets in the human brain. We performed ChIP-chip experiments on human fetal brain during midgestation, which corresponds to the peak period of neuronal migration, differentiation, and cortical regionalization and is a time of high FOXP2 expression.8,9,20 The two regions we chose, one cortical (IFC) and one subcortical (BG), are part of a parallel-distributed circuitry that, among other functions, is involved in language and speech,30,31 express high levels of FOXP2 during this period of development,8 and are sites of abnormalities in patients with FOXP2 mutations.32 Thus, FOXP2 targets identified within these regions during human brain development would be particularly germane to the understanding of the molecular circuitry involved in the development of these regions and their relationship to speech and language functions. Furthermore, the importance of this period for key aspects of the development of the human cerebral cortex is also highlighted by previous studies that demonstrate key elements of patterning—including brain asymmetry, a structural correlate of language—that occur during this time.31,33
We used rapidly frozen tissue stored at −80°C, with short postmortem intervals, to optimize the likelihood of detecting regions of DNA bound by FOXP2 (see the “Material and Methods” section). ChIP products from three independent replicates were hybridized onto Aviva Systems Biology cDNA promoter arrays containing ∼6,000 DNA fragments from potential regulatory regions, which were initially validated in the first genomewide ChIP-chip studies (see the “Material and Methods” section).22 Although this is not a whole-genome microarray, it has been validated in several important ChIP studies and provides a very solid cross-section of genes.34,35 Since no FOXP2 targets had been previously identified, we reasoned that this platform would provide a good cross-section of targets.
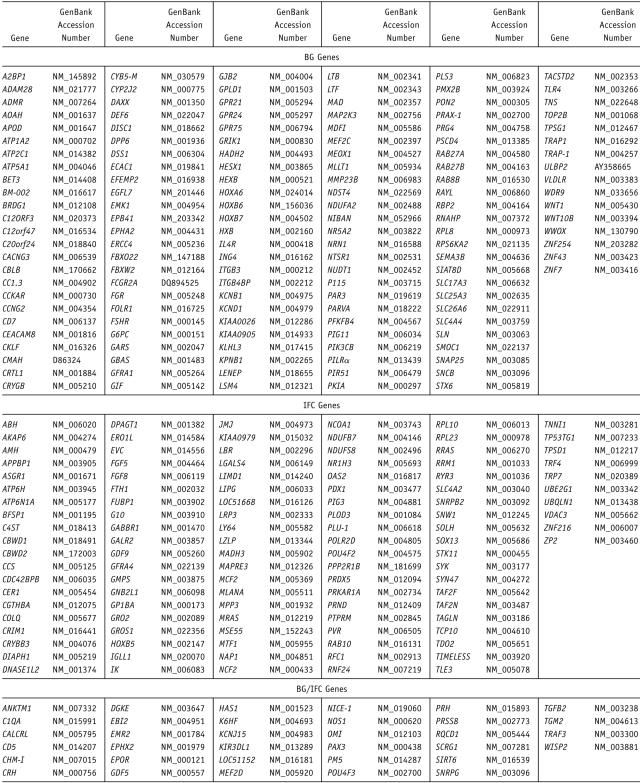
We identified 175 targets in BG and 144 targets in IFC, using conservative criteria (see the “Material and Methods” section and table 1). The overlap between the two regions was highly significant, with 24% of IFC genes overlapping with BG genes (hypergeometric probability p<6.767×10-22). An additional set of genes identified as regionally specific in either BG (141 genes) or IFC (110 genes) were identified (fig. 2). These genes may represent specific regional targets of FOXP2 regulation.
Table 1. .
Summary of Results for All Identified Target Genes[Note]
| Tissue Detected | BG | IFC | Lung | Evolutionary Significance | TATTT[A/G]T | CAAATT | AAAT | |||||||||||||
| Gene | GenBank Accession Number | Chromosome | BG | IFC | Lung | P | M Score | P | M Score | P | M Score | P | Ka/Ki | Ka/Ks | 5′→3′ | 3′→5′ | 5′→3′ | 3′→5′ | 5′→3′ | 3′→5′ |
| A2BP1 | NM_145892 | 16 | X | X | .0131 | .9143 | NA | NA | .0359 | .7260 | NA | NA | NA | 0 | 0 | 1 | 1 | 10 | 4 | |
| ABH | NM_006020 | 14 | X | NA | NA | .0150 | .5692 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 10 | 3 | ||
| ADAM28 | NM_021777 | 8 | X | .0079 | 1.0674 | NA | NA | NA | NA | .0395 | .7658 | .437 | 0 | 0 | 0 | 1 | 9 | 10 | ||
| ADMR | NM_007264 | 12 | X | .0387 | .6588 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| AKAP6 | NM_004274 | 14 | X | X | NA | NA | .0058 | .5468 | .0246 | .8147 | NA | NA | NA | 0 | 0 | 1 | 0 | 12 | 9 | |
| AKR7A2 | NM_003689 | 1 | X | NA | NA | NA | NA | .0319 | .7002 | .412 | .5579 | .4332 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| AMH | NM_000479 | 19 | X | NA | NA | .0031 | .5806 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | ||
| AMY2A | NM_000699 | 1 | X | NA | NA | NA | NA | .0386 | .7024 | NA | NA | NA | 1 | 1 | 1 | 0 | 7 | 12 | ||
| ANK1 | NM_000037 | 8 | X | NA | NA | NA | NA | .0175 | .6487 | NA | NA | NA | 0 | 0 | 1 | 0 | 3 | 1 | ||
| ANKTM1 | NM_007332 | 8 | X | X | .0460 | .7704 | .0026 | .7532 | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 4 | 3 | |
| AOAH | NM_001637 | 7 | X | .0016 | 1.0052 | NA | NA | NA | NA | .021 | .3038 | .7755 | 0 | 0 | 1 | 0 | 9 | 6 | ||
| AP1GBP1 | NM_007247 | 17 | X | NA | NA | NA | NA | .0243 | .5700 | NA | NA | NA | 1 | 1 | 0 | 1 | 11 | 10 | ||
| APOD | NM_001647 | 3 | X | .0498 | .6387 | NA | NA | NA | NA | .0001 | NA | NA | 1 | 0 | 0 | 0 | 7 | 14 | ||
| APPBP1 | NM_003905 | 16 | X | NA | NA | .0096 | .7178 | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 7 | 4 | ||
| ASGR1 | NM_001671 | 17 | X | NA | NA | .0099 | .6434 | NA | NA | .0235 | NA | NA | 0 | 0 | 0 | 0 | 3 | 3 | ||
| ATF6 | NM_007348 | 1 | X | NA | NA | NA | NA | .0071 | 1.0159 | NA | NA | NA | 0 | 1 | 1 | 2 | 13 | 12 | ||
| ATP1A2 | NM_000702 | 1 | X | X | .0473 | .6462 | NA | NA | .0281 | .6642 | NA | NA | NA | 0 | 0 | 2 | 0 | 2 | 2 | |
| ATP2C1 | NM_014382 | 3 | X | .0335 | .7018 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| ATP5A1 | NM_004046 | 18 | X | .0077 | .5099 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 6 | 5 | ||
| ATP6B1 | NM_001692 | 2 | X | NA | NA | NA | NA | .0395 | .5274 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 0 | ||
| ATP6H | NM_003945 | 5 | X | X | NA | NA | .0023 | .5945 | .0190 | .5686 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 2 | |
| ATP6N1A | NM_005177 | 17 | X | NA | NA | .0056 | .5537 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| ATP6S1 | NM_001183 | X | X | NA | NA | NA | NA | .0437 | .5208 | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 5 | ||
| BET3 | NM_014408 | 1 | X | X | .0149 | .5926 | NA | NA | .0315 | .5852 | NA | NA | NA | 0 | 0 | 0 | 1 | 4 | 3 | |
| BFSP1 | NM_001195 | 20 | X | NA | NA | .0153 | .5430 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 3 | ||
| BM-002 | NM_016617 | 13 | X | .0055 | .6497 | NA | NA | NA | NA | NA | NA | NA | 1 | 1 | 1 | 0 | 6 | 4 | ||
| BRDG1 | NM_012108 | 4 | X | X | .0356 | .7008 | NA | NA | .0185 | .8701 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| C12ORF2 | NM_007211 | 12 | X | NA | NA | NA | NA | .0128 | .6040 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| C12ORF3 | NM_020373 | 12 | X | .0220 | .6247 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 3 | 9 | ||
| C12orf47 | NM_016534 | 12 | X | .0064 | .8127 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 6 | 6 | ||
| C1QA | NM_015991 | 1 | X | X | X | .0099 | .7268 | .0025 | .6873 | .0265 | .6403 | NA | .4237 | .791 | NA | NA | NA | NA | NA | NA |
| C20orf24 | NM_018840 | 20 | X | X | .0171 | .6045 | NA | NA | .0223 | .6199 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 3 | |
| C4ST | NM_018413 | 12 | X | NA | NA | .0079 | .5435 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 4 | ||
| CA4 | NM_000717 | 17 | X | NA | NA | NA | NA | .0112 | .6899 | .2482 | .6937 | 1.5476 | NA | NA | NA | NA | NA | NA | ||
| CACNG3 | NM_006539 | 16 | X | X | .0164 | .9633 | NA | NA | .0128 | .9116 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | |
| CALCRL | NM_005795 | 2 | X | X | X | .0415 | 1.3025 | .0010 | .8368 | .0026 | .7912 | NA | NA | NA | 1 | 1 | 1 | 0 | 7 | 11 |
| CBLB | NM_170662 | 3 | X | .0479 | .5994 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| hCBWD1 | NM_018491 | 9 | X | NA | NA | .0021 | .6602 | NA | NA | NA | NA | NA | 0 | 1 | 0 | 1 | 12 | 5 | ||
| CBWD2 | NM_172003 | 2 | X | NA | NA | .0085 | .5472 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| CC1.3 | NM_004902 | 20 | X | .0218 | .6451 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 1 | 1 | 2 | 5 | ||
| CCK | NM_000729 | 3 | X | NA | NA | NA | NA | .0126 | .8671 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| CCKAR | NM_000730 | 4 | X | .0289 | .5102 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 5 | 2 | ||
| CCNG2 | NM_004354 | 4 | X | X | .0364 | .5585 | NA | NA | .0265 | .6804 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | |
| CCS | NM_005125 | 11 | X | NA | NA | .0046 | .5259 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| CD5 | NM_014207 | 11 | X | X | X | .0220 | .9352 | .0016 | .7112 | .0167 | .5848 | NA | NA | NA | 0 | 0 | 1 | 0 | 9 | 2 |
| CD7 | NM_006137 | 17 | X | .0494 | .6252 | NA | NA | NA | NA | NA | .7805 | 1.5375 | 0 | 0 | 0 | 0 | 1 | 2 | ||
| CDC42BPB | NM_006035 | 14 | X | NA | NA | .0024 | .7014 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| CDH5 | NM_001795 | 16 | X | NA | NA | NA | NA | .0031 | .7681 | NA | NA | NA | 0 | 0 | 0 | 0 | 5 | 4 | ||
| CEACAM8 | NM_001816 | 19 | X | .0132 | .6363 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| CER1 | NM_005454 | 9 | X | X | NA | NA | .0009 | .7446 | .0091 | .6421 | NA | .5913 | 1.7308 | 0 | 0 | 0 | 1 | 8 | 7 | |
| CGTHBA | NM_012075 | 16 | X | NA | NA | .0033 | .5614 | NA | NA | NA | NA | NA | 1 | 0 | 1 | 0 | 1 | 5 | ||
| CHM-I | NM_007015 | 13 | X | X | .0019 | .8620 | .0005 | 1.4095 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 4 | 2 | |
| CKLF | NM_016326 | 16 | X | .0034 | .6263 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 2 | 4 | ||
| CMAH | D86324 | 6 | X | .0232 | .6378 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 8 | 12 | ||
| COLQ | NM_005677 | 3 | X | X | NA | NA | .0063 | .5571 | .0124 | .7294 | NA | NA | NA | 0 | 1 | 0 | 0 | 8 | 8 | |
| CRH | NM_000756 | 8 | X | X | X | .0184 | .8799 | .0009 | .7169 | .0192 | .7540 | NA | NA | NA | 0 | 1 | 0 | 0 | 5 | 6 |
| CRIM1 | NM_016441 | 2 | X | NA | NA | .0024 | .5528 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | ||
| CRSP3 | NM_015979 | 6 | X | NA | NA | NA | NA | .0240 | .7035 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 5 | ||
| CRTL1 | NM_001884 | 5 | X | .0322 | .5414 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 0 | 1 | 4 | 5 | ||
| CRYBA4 | NM_001886 | 22 | X | NA | NA | NA | NA | .0285 | .5111 | NA | NA | NA | 0 | 0 | 1 | 0 | 5 | 5 | ||
| CRYBB3 | NM_004076 | 22 | X | NA | NA | .0062 | .5106 | NA | NA | .4505 | .6762 | .6374 | 0 | 1 | 0 | 0 | 4 | 2 | ||
| CRYGB | NM_005210 | 2 | X | X | .0366 | .5356 | NA | NA | .0208 | .5789 | NA | NA | NA | 0 | 0 | 0 | 1 | 6 | 8 | |
| CXADR | NM_001338 | 21 | X | NA | NA | NA | NA | .0263 | .6591 | .0001 | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| CYB5-M | NM_030579 | 16 | X | .0167 | .6880 | NA | NA | NA | NA | .0048 | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | ||
| CYP21A2 | NM_000500 | 6 | X | NA | NA | NA | NA | .0326 | .5541 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 4 | ||
| CYP2J2 | NM_000775 | 1 | X | .0332 | .5021 | NA | NA | NA | NA | .0001 | .4403 | .7101 | 1 | 0 | 0 | 0 | 7 | 3 | ||
| DAXX | NM_001350 | 6 | X | X | .0102 | .8786 | NA | NA | .0331 | .6247 | .0308 | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | |
| DCT | NM_001922 | 13 | X | NA | NA | NA | NA | .0220 | .6489 | .0331 | NA | NA | 0 | 0 | 0 | 1 | 9 | 5 | ||
| DEF6 | NM_022047 | 6 | X | .0206 | .8739 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| DGKE | NM_003647 | 17 | X | X | X | .0260 | .8173 | .0001 | .8459 | .0091 | .8624 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 |
| DIAPH1 | NM_005219 | 5 | X | NA | NA | .0054 | .5502 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| DISC1 | NM_018662 | 1 | X | .0215 | .6402 | NA | NA | NA | NA | NA | .8117 | .3802 | 0 | 0 | 0 | 0 | 3 | 1 | ||
| DNASE1L2 | NM_001374 | 16 | X | NA | NA | .0240 | .6027 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| DPAGT1 | NM_001382 | 11 | X | NA | NA | .0087 | .5263 | NA | NA | .1513 | .1004 | .5 | 0 | 0 | 0 | 2 | 2 | 4 | ||
| DPP6 | NM_001936 | 7 | X | .0440 | .6273 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 2 | 0 | 16 | 13 | ||
| DSS1 | NM_006304 | 7 | X | X | .0369 | .7390 | NA | NA | .0284 | .7568 | NA | NA | NA | 0 | 1 | 1 | 0 | 9 | 6 | |
| E2-EPF | NM_014501 | 17 | X | NA | NA | NA | NA | .0192 | .7032 | NA | NA | NA | 1 | 0 | 0 | 0 | 3 | 5 | ||
| EBI2 | NM_004951 | 13 | X | X | X | .0431 | 1.5721 | .0001 | .9837 | .0003 | 1.0850 | NA | NA | NA | 0 | 0 | 3 | 1 | 17 | 18 |
| ECAC1 | NM_019841 | 7 | X | .0285 | .5386 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 3 | 4 | ||
| EDF1 | NM_003792 | 9 | X | NA | NA | NA | NA | .0061 | .8091 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 4 | ||
| EFEMP2 | NM_016938 | 11 | X | X | .0436 | .5835 | NA | NA | .0171 | .6877 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | |
| EFNB3 | NM_001406 | 17 | X | NA | NA | NA | NA | .0158 | .6209 | .0365 | .3821 | .8846 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| EGFL7 | NM_201446 | 9 | X | X | NA | .9198 | NA | NA | .0137 | .9309 | NA | NA | NA | 0 | 0 | 1 | 0 | 3 | 0 | |
| EIF4EBP2 | NM_004096 | 10 | X | NA | NA | NA | NA | .0163 | .7665 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | ||
| EMK1 | NM_004954 | 11 | X | X | .0028 | .8569 | NA | NA | .0425 | .5757 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 3 | |
| EMR2 | NM_001784 | 19 | X | X | .0425 | .7391 | .0079 | .5144 | NA | NA | .4906 | .5098 | .6667 | 0 | 0 | 0 | 0 | 1 | 4 | |
| EPB41 | NM_203342 | 1 | X | .0354 | .6121 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 0 | 1 | 7 | 16 | ||
| EPHA2 | NM_004431 | 1 | X | .0379 | .5563 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | ||
| EPHX2 | NM_001979 | 8 | X | X | .0214 | .9185 | .0018 | .6189 | NA | NA | .0472 | NA | NA | 0 | 0 | 0 | 0 | 1 | 2 | |
| EPOR | NM_000121 | 19 | X | X | X | .0171 | .9258 | .0263 | .5590 | .0037 | 1.1961 | .0832 | .1999 | .5185 | 1 | 0 | 0 | 0 | 3 | 8 |
| ERCC4 | NM_005236 | 16 | X | .0115 | .5582 | NA | NA | NA | NA | .0439 | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | ||
| ERO1L | NM_014584 | 14 | X | NA | NA | .0021 | .7909 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 10 | 8 | ||
| ERO1Lβ | NM_019891 | 1 | X | NA | NA | NA | NA | .0187 | .5690 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| EVC | NM_014556 | 4 | X | X | NA | NA | .0016 | .5830 | .0181 | .6591 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 0 | |
| FAAH | NM_001441 | 1 | X | NA | NA | NA | NA | .0252 | .6041 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 3 | ||
| FACL3 | NM_004457 | 2 | X | NA | NA | NA | NA | .0496 | .5204 | NA | NA | NA | 1 | 0 | 1 | 1 | 5 | 21 | ||
| FBXO22 | NM_147188 | 15 | X | X | .0454 | .6194 | NA | NA | .0431 | .5989 | NA | NA | NA | 0 | 0 | 0 | 1 | 5 | 6 | |
| FBXW2 | NM_012164 | 9 | X | X | .0236 | .7398 | NA | NA | .0182 | .6760 | NA | NA | NA | 0 | 0 | 0 | 1 | 2 | 4 | |
| FCGR2A | DQ894525 | 1 | X | .0222 | .6197 | NA | NA | NA | NA | .0008 | .7127 | 2.8529 | 0 | 0 | 0 | 0 | 2 | 2 | ||
| FGF5 | NM_004464 | 4 | X | NA | NA | .0077 | .5140 | NA | NA | NA | NA | NA | 0 | 1 | 0 | 0 | 3 | 3 | ||
| FGF8 | NM_006119 | 10 | X | NA | NA | .0085 | .5008 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| FGR | NM_005248 | 1 | X | .0283 | .5766 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 5 | 1 | ||
| FNTB | NM_002028 | 14 | X | NA | NA | NA | NA | .0404 | .5170 | NA | NA | NA | 0 | 1 | 0 | 1 | 8 | 3 | ||
| FOLR1 | NM_016725 | 11 | X | .0338 | .7973 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 5 | ||
| FSHR | NM_000145 | 2 | X | X | .0278 | .7671 | NA | NA | .0349 | .6176 | .339 | .5082 | .7816 | 0 | 0 | 0 | 0 | 7 | 7 | |
| FTH1 | NM_002032 | 11 | X | X | NA | NA | .0004 | .7659 | .0173 | .5704 | NA | NA | NA | 1 | 0 | 0 | 1 | 11 | 12 | |
| FUBP1 | NM_003902 | 1 | X | X | NA | NA | .0009 | .7045 | .0213 | .6357 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| FY | NM_002036 | 1 | X | NA | NA | NA | NA | .0179 | .6230 | .0361 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| G10 | NM_003910 | 7 | X | NA | NA | .0057 | .8101 | NA | NA | .0119 | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| G6PC | NM_000151 | 17 | X | X | .0070 | .5722 | NA | NA | .0269 | .6607 | NA | NA | NA | 0 | 0 | 1 | 0 | 7 | 6 | |
| GABBR1 | NM_001470 | 6 | X | NA | NA | .0063 | .6116 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| GADD45G | NM_006705 | 9 | X | NA | NA | NA | NA | .0294 | .6485 | NA | NA | NA | 0 | 0 | 2 | 0 | 6 | 3 | ||
| GALR2 | NM_003857 | 17 | X | NA | NA | .0057 | .5536 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| GALR3 | NM_003614 | 22 | X | NA | NA | NA | NA | .0118 | .6806 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| GARS | NM_002047 | 7 | X | X | .0290 | .6988 | NA | NA | .0181 | .6901 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 9 | |
| GBAS | NM_001483 | 7 | X | X | .0275 | .8326 | NA | NA | .0254 | .5739 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | |
| GDF5 | NM_000557 | 20 | X | X | X | .0416 | .9516 | .0025 | .6273 | .0075 | .7943 | NA | NA | NA | 0 | 1 | 0 | 0 | 5 | 5 |
| GDF9 | NM_005260 | 5 | X | NA | NA | .0027 | .7455 | NA | NA | NA | NA | NA | 0 | 0 | 5 | 1 | 16 | 7 | ||
| GFRA1 | NM_005264 | 10 | X | X | .0128 | 1.0440 | NA | NA | .0438 | .5414 | .0287 | NA | NA | 0 | 1 | 0 | 0 | 7 | 3 | |
| GFRA4 | NM_022139 | 20 | X | NA | NA | .0027 | .7145 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| GIF | NM_005142 | 11 | X | X | .0410 | .7947 | NA | NA | .0152 | .9503 | NA | .4007 | .7541 | NA | NA | NA | NA | NA | NA | |
| GJB2 | NM_004004 | 13 | X | .0170 | .5045 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 5 | 5 | ||
| GLUD1 | NM_005271 | 10 | X | NA | NA | NA | NA | .0218 | .6532 | .0011 | NA | NA | 0 | 3 | 0 | 0 | 12 | 10 | ||
| GMPS | NM_003875 | 3 | X | NA | NA | .0079 | .5831 | NA | NA | NA | NA | NA | 1 | 0 | 1 | 2 | 9 | 16 | ||
| GNB2L1 | NM_006098 | 5 | X | NA | NA | .0099 | .5403 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 4 | 8 | ||
| GP1BA | NM_000173 | 17 | X | NA | NA | .0026 | .8359 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 2 | ||
| GPLD1 | NM_001503 | 6 | X | .0134 | .5954 | NA | NA | NA | NA | .009 | .5473 | .4088 | 0 | 0 | 1 | 0 | 12 | 7 | ||
| GPR21 | NM_005294 | 9 | X | .0315 | .6415 | NA | NA | NA | NA | NA | .4147 | 1.3871 | 0 | 0 | 1 | 0 | 7 | 8 | ||
| GPR24 | NM_005297 | 22 | X | .0383 | .6207 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | ||
| GPR75 | NM_006794 | 2 | X | .0069 | .5032 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 5 | ||
| GRIK1 | NM_000830 | 21 | X | X | .0270 | .8532 | NA | NA | .0181 | .7106 | .0439 | NA | NA | 0 | 0 | 0 | 1 | 6 | 4 | |
| GRO2 | NM_002089 | 4 | X | X | NA | NA | .0038 | .6167 | .0113 | .6158 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| GROS1 | NM_022356 | 1 | X | NA | NA | .0041 | .5843 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| HADH2 | NM_004493 | X | X | .0050 | .6023 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| HAS1 | NM_001523 | 19 | X | X | X | .0475 | 1.3249 | .0009 | .8220 | .0011 | .8959 | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 1 |
| HBQ1 | NM_005331 | 16 | X | NA | NA | NA | NA | .0235 | .5414 | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 5 | ||
| HESX1 | NM_003865 | 3 | X | X | .0415 | .5856 | NA | NA | .0482 | .6339 | .3485 | .5971 | 75 | 1 | 1 | 0 | 0 | 11 | 15 | |
| HEXB | NM_000521 | 5 | X | .0322 | .6266 | NA | NA | NA | NA | .2235 | .6457 | .9425 | 0 | 0 | 0 | 1 | 1 | 4 | ||
| HIVEP1 | NM_002114 | 6 | X | NA | NA | NA | NA | .0108 | .6495 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| HOXA6 | NM_024014 | 7 | X | .0271 | .5207 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | ||
| HOXB5 | NM_002147 | 17 | X | NA | NA | .0042 | .5741 | NA | NA | .0025 | NA | NA | 1 | 0 | 0 | 0 | 6 | 5 | ||
| HOXB6 | NM_156036 | 17 | X | X | .0230 | 1.1453 | NA | NA | .0198 | .5892 | NA | NA | NA | 1 | 0 | 0 | 0 | 1 | 6 | |
| HOXB7 | NM_004502 | 17 | X | .0426 | .5147 | NA | NA | NA | NA | .2016 | .4565 | .7049 | NA | NA | NA | NA | NA | NA | ||
| HSPB7 | NM_014424 | 1 | X | NA | NA | NA | NA | .0006 | .9824 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| HXB | NM_002160 | 9 | X | X | .0053 | .8143 | NA | NA | .0456 | .5818 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 3 | |
| IGLL1 | NM_020070 | 22 | X | X | NA | NA | .0004 | .7403 | .0062 | .7784 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | |
| IK | NM_006083 | 5 | X | NA | NA | .0063 | .5097 | NA | NA | 0 | NA | NA | 0 | 0 | 0 | 0 | 2 | 3 | ||
| IL4R | NM_000418 | 16 | X | X | .0397 | .6960 | NA | NA | .0196 | .7007 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | |
| ING4 | NM_016162 | 12 | X | X | .0435 | .8067 | NA | NA | .0355 | .8162 | NA | NA | NA | 1 | 0 | 0 | 0 | 6 | 3 | |
| INHA | NM_002191 | 2 | X | NA | NA | NA | NA | .0230 | .6319 | .0123 | .4267 | .6471 | 0 | 1 | 0 | 1 | 3 | 1 | ||
| ITGB3 | NM_000212 | 17 | X | .0362 | .5971 | NA | NA | NA | NA | .0164 | NA | NA | 0 | 0 | 0 | 1 | 2 | 1 | ||
| ITGB4BP | NM_002212 | 20 | X | X | .0280 | .6697 | NA | NA | .0048 | .7917 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | |
| JMJ | NM_004973 | 6 | X | NA | NA | .0043 | .5377 | NA | NA | NA | NA | NA | 1 | 0 | 1 | 0 | 4 | 11 | ||
| K6HF | NM_004693 | 12 | X | X | X | .0409 | .6749 | .0041 | .5061 | .0092 | .8341 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 |
| KCNB1 | NM_004975 | 20 | X | X | .0419 | .5127 | NA | NA | .0184 | .6952 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | |
| KCND1 | NM_004979 | X | X | .0037 | .7040 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| KCNJ15 | NM_004983 | 21 | X | X | .0310 | .7042 | .0020 | .5799 | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 9 | 10 | |
| KIAA0026 | NM_012286 | X | X | X | .0394 | .6612 | NA | NA | .0045 | .9468 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 5 | |
| KIAA0905 | NM_014933 | 4 | X | X | .0094 | .5448 | NA | NA | .0285 | .5473 | NA | NA | NA | 0 | 0 | 1 | 0 | 1 | 3 | |
| KIAA0979 | NM_015032 | 13 | X | NA | NA | .0038 | .5382 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| KIR3DL1 | NM_013289 | 19 | X | X | X | .0178 | .5990 | .0031 | .6378 | .0067 | .7822 | NA | NA | NA | 0 | 0 | 1 | 0 | 12 | 9 |
| KIT | NM_000222 | 4 | X | NA | NA | NA | NA | .0183 | .8021 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 2 | ||
| KLHL3 | NM_017415 | 5 | X | .0046 | .7050 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 0 | 1 | 1 | 9 | ||
| KPNB1 | NM_002265 | 17 | X | X | .0067 | .8313 | NA | NA | .0192 | .7568 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| LBR | NM_002296 | 1 | X | X | NA | NA | .0004 | .7878 | .0021 | .8348 | NA | .4127 | 1.0833 | 0 | 0 | 2 | 3 | 12 | 18 | |
| LENEP | NM_018655 | 1 | X | .0191 | .5764 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | ||
| LGALS4 | NM_006149 | 19 | X | NA | NA | .0025 | .6104 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 0 | 1 | ||
| LIMD1 | NM_014240 | 3 | X | NA | NA | .0043 | .5049 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 3 | 2 | ||
| LIPG | NM_006033 | 18 | X | NA | NA | .0121 | .5269 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 3 | 1 | ||
| LOC51152 | NM_016181 | 1 | X | X | X | .0111 | .8251 | .0040 | .5132 | .0165 | .6583 | NA | NA | NA | 1 | 0 | 0 | 0 | 1 | 2 |
| LOC51668 | NM_016126 | 1 | X | NA | NA | .0049 | .5406 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| LOC55829 | NM_018445 | 15 | X | NA | NA | NA | NA | .0414 | .5091 | NA | NA | NA | 0 | 1 | 0 | 0 | 4 | 3 | ||
| LRP3 | NM_002333 | 19 | X | X | NA | NA | .0037 | .6851 | .0164 | .7159 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| LSM4 | NM_012321 | 19 | X | X | .0441 | .5264 | NA | NA | .0133 | .6764 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 7 | |
| LTB | NM_002341 | 6 | X | X | .0256 | .8010 | NA | NA | .0019 | 1.0656 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 2 | |
| LTF | NM_002343 | 3 | X | .0176 | .5066 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | ||
| LY64 | NM_005582 | 5 | X | X | NA | NA | .0001 | .8310 | .0102 | .7571 | NA | NA | NA | 0 | 0 | 1 | 0 | 5 | 6 | |
| LY95 | NM_004828 | 6 | X | NA | NA | NA | NA | .0282 | .5141 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | ||
| LZLP | NM_013344 | 11 | X | NA | NA | .0027 | .5994 | NA | NA | NA | NA | NA | 0 | 1 | 0 | 1 | 9 | 7 | ||
| MAD | NM_002357 | 2 | X | .0412 | .6233 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| MADH3 | NM_005902 | 15 | X | X | NA | NA | .0003 | .7307 | .0254 | .7535 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | |
| MAP2K3 | NM_002756 | 17 | X | X | .0116 | .6834 | NA | NA | .0272 | .5976 | .0345 | NA | NA | NA | NA | NA | NA | NA | NA | |
| MAPRE3 | NM_012326 | 2 | X | X | NA | NA | .0039 | .5069 | .0177 | .7014 | NA | NA | NA | 0 | 0 | 0 | 1 | 3 | 1 | |
| MCF2 | NM_005369 | X | X | NA | NA | .0050 | .5946 | NA | NA | .0413 | NA | NA | 2 | 1 | 0 | 1 | 11 | 12 | ||
| MDFI | NM_005586 | 6 | X | .0266 | .5077 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| MEF2C | NM_002397 | 5 | X | X | .0212 | .7250 | NA | NA | .0267 | .8366 | NA | NA | NA | 0 | 1 | 1 | 0 | 23 | 15 | |
| MEF2D | NM_005920 | 1 | X | X | X | .0316 | .8039 | .0005 | .7094 | .0113 | .9174 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 |
| MEOX1 | NM_004527 | 17 | X | .0315 | .5022 | NA | NA | NA | NA | .0839 | .4824 | 52 | 0 | 1 | 0 | 2 | 9 | 10 | ||
| MEP1B | NM_005925 | 18 | X | NA | NA | NA | NA | .0209 | .6317 | NA | NA | NA | 0 | 1 | 0 | 1 | 11 | 11 | ||
| MEST | NM_177525 | 7 | X | NA | NA | NA | NA | .0168 | .6294 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| MFAP2 | NM_017459 | 1 | X | NA | NA | NA | NA | .0281 | .5131 | NA | NA | NA | 0 | 0 | 0 | 1 | 7 | 7 | ||
| MGST2 | NM_002413 | 4 | X | NA | NA | NA | NA | .0101 | .6587 | .0716 | .313 | .623 | 0 | 1 | 0 | 0 | 5 | 2 | ||
| MLANA | NM_005511 | 9 | X | NA | NA | .0072 | .5979 | NA | NA | NA | NA | NA | 1 | 0 | 1 | 0 | 11 | 10 | ||
| MLLT1 | NM_005934 | 19 | X | X | .0090 | .7103 | NA | NA | .0321 | .6642 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | |
| MMP23B | NM_006983 | 1 | X | X | .0423 | .7716 | NA | NA | .0143 | .8810 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 1 | |
| MPDU1 | NM_004870 | 17 | X | NA | NA | NA | NA | .0150 | .6057 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| MPP3 | NM_001932 | 17 | X | NA | NA | .0093 | .8638 | NA | NA | NA | NA | NA | 1 | 1 | 0 | 0 | 3 | 5 | ||
| MRAS | NM_012219 | 3 | X | X | NA | NA | .0008 | .8490 | .0113 | .7427 | .0137 | NA | NA | NA | NA | NA | NA | NA | NA | |
| MSE55 | NM_152243 | 22 | X | NA | NA | .0032 | .5731 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| MSLN | NM_013404 | 16 | X | NA | NA | NA | NA | .0220 | .6583 | NA | .5922 | .5 | 0 | 0 | 0 | 0 | 0 | 3 | ||
| MT1H | NM_005951 | 16 | X | NA | NA | NA | NA | .0439 | .5132 | NA | NA | NA | 0 | 0 | 1 | 0 | 1 | 1 | ||
| MT2A | NM_005953 | 16 | X | NA | NA | NA | NA | .0195 | .5782 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 2 | ||
| MTF1 | NM_005955 | 1 | X | X | NA | NA | .0127 | .5936 | .0201 | .5506 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 0 | |
| MTHFD2 | NM_006636 | 2 | X | NA | NA | NA | NA | .0143 | .5956 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| NAP1 | NM_004851 | 19 | X | NA | NA | .0052 | .5228 | NA | NA | NA | .7813 | .7522 | 0 | 1 | 0 | 0 | 6 | 2 | ||
| NCF2 | NM_000433 | 1 | X | NA | NA | .0038 | .5397 | NA | NA | .1293 | .4255 | .8846 | 0 | 0 | 0 | 1 | 3 | 8 | ||
| NCOA1 | NM_003743 | 2 | X | NA | NA | .0304 | .7087 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 10 | 7 | ||
| NDST4 | NM_022569 | 4 | X | .0038 | .8242 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 1 | 0 | 8 | 7 | ||
| NDUFA2 | NM_002488 | 5 | X | .0331 | .7012 | NA | NA | NA | NA | .2525 | 1.6458 | 1.2958 | 0 | 1 | 0 | 0 | 4 | 1 | ||
| NDUFB7 | NM_004146 | 19 | X | NA | NA | .0054 | .5247 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 2 | 8 | 5 | ||
| NDUFS8 | NM_002496 | 11 | X | NA | NA | .0016 | .8725 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| NIBAN | NM_052966 | 1 | X | .0286 | .6822 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 7 | ||
| NICE-1 | NM_019060 | 1 | X | X | .0153 | .8528 | .0075 | .5080 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | |
| NOS1 | NM_000620 | 12 | X | X | X | .0340 | 1.1315 | .0011 | .6678 | .0100 | .6303 | NA | NA | NA | 1 | 0 | 0 | 0 | 8 | 3 |
| NR1H3 | NM_005693 | 11 | X | NA | NA | .0031 | .5660 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 1 | ||
| NR5A2 | NM_003822 | 1 | X | .0081 | .7970 | NA | NA | NA | NA | NA | NA | NA | 1 | 1 | 0 | 1 | 5 | 14 | ||
| NRIP1 | NM_003489 | 21 | X | NA | NA | NA | NA | .0337 | .5853 | NA | NA | NA | 1 | 0 | 1 | 0 | 7 | 15 | ||
| NRN1 | NM_016588 | 6 | X | .0120 | .7757 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 5 | 4 | ||
| NTSR1 | NM_002531 | 20 | X | X | .0133 | .6634 | NA | NA | .0383 | .6024 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | |
| NUDT1 | NM_002452 | 7 | X | X | .0110 | .8431 | NA | NA | .0432 | .5571 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 2 | |
| OAS2 | NM_016817 | 12 | X | NA | NA | .0038 | .6665 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| OMI | NM_012103 | 2 | X | X | X | .0479 | .7194 | .0144 | .5025 | .0132 | 1.0094 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 |
| OPN1LW | NM_020061 | X | X | NA | NA | NA | NA | .0153 | .6269 | NA | NA | NA | 0 | 0 | 0 | 0 | 5 | 0 | ||
| OR1A1 | NM_014565 | 17 | X | NA | NA | NA | NA | .0210 | .6286 | NA | NA | NA | 3 | 1 | 0 | 0 | 9 | 13 | ||
| P115 | NM_003715 | 4 | X | .0140 | .5315 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 5 | ||
| PAR3 | NM_019619 | 10 | X | .0372 | .6575 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 0 | 2 | 10 | 7 | ||
| PARG | NM_003631 | 10 | X | NA | NA | NA | NA | .0275 | .5181 | .0064 | NA | NA | 0 | 0 | 0 | 0 | 4 | 0 | ||
| PARVA | NM_018222 | 11 | X | .0057 | .5302 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | ||
| PAX1 | NM_006192 | 2 | X | NA | NA | NA | NA | .0243 | .5289 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| PAX3 | NM_000438 | 11 | X | X | X | .0300 | 1.1898 | .0034 | .6882 | .0010 | .9766 | .0235 | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 |
| PDX1 | NM_003477 | 21 | X | X | NA | NA | .0035 | .5188 | .0234 | .6471 | NA | NA | NA | 0 | 1 | 0 | 0 | 2 | 1 | |
| PDXK | NM_003681 | 3 | X | NA | NA | NA | NA | .0187 | .6162 | .0132 | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PFKFB4 | NM_004567 | 19 | X | .0499 | .6651 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 0 | ||
| PGLYRP | NM_005091 | 11 | X | NA | NA | NA | NA | .0262 | .6950 | NA | 1.8051 | .5485 | 1 | 0 | 0 | 0 | 8 | 3 | ||
| PIG11 | NM_006034 | 2 | X | .0211 | .7178 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 0 | ||
| PIG3 | NM_004881 | 3 | X | NA | NA | .0127 | .5042 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 3 | ||
| PIK3CB | NM_006219 | 7 | X | .0151 | .5663 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 1 | 4 | 13 | 19 | ||
| PILRα | NM_013439 | 12 | X | X | .0358 | .6271 | NA | NA | .0246 | .6280 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | |
| PIR51 | NM_006479 | 4 | X | X | .0130 | .8437 | NA | NA | .0051 | .9570 | NA | .404 | 56 | 0 | 0 | 1 | 2 | 6 | 7 | |
| PKIA | NM_000297 | 8 | X | .0456 | .5862 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 2 | 0 | 10 | 5 | ||
| PLAA | NM_004253 | 9 | X | NA | NA | NA | NA | .0267 | .5389 | NA | NA | NA | 2 | 0 | 0 | 0 | 7 | 16 | ||
| PLOD3 | NM_001084 | 7 | X | NA | NA | .0243 | .5394 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 2 | ||
| PLS3 | NM_006823 | X | X | .0243 | .5527 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 3 | 4 | ||
| PLU-1 | NM_006618 | 1 | X | NA | NA | .0205 | .5359 | NA | NA | NA | NA | NA | 0 | 0 | 1 | 0 | 5 | 1 | ||
| PM5 | NM_014287 | 16 | X | X | X | .0227 | 1.3605 | .0012 | .7689 | .0009 | .9788 | NA | NA | NA | 0 | 0 | 1 | 0 | 4 | 3 |
| PMX2B | NM_003924 | 4 | X | .0172 | .7359 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 5 | 8 | ||
| PNOC | NM_006228 | 8 | X | NA | NA | NA | NA | .0089 | .7730 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 4 | ||
| PNR | NM_003967 | 6 | X | NA | NA | NA | NA | .0093 | .9334 | NA | NA | NA | 1 | 0 | 0 | 0 | 7 | 14 | ||
| POLM | NM_013284 | 7 | X | NA | NA | NA | NA | .0241 | .5581 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| POLR2D | NM_004805 | 2 | X | NA | NA | .0053 | .6817 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 6 | ||
| PON2 | NM_000305 | 7 | X | X | .0319 | .7761 | NA | NA | .0494 | .5680 | .0112 | NA | NA | 1 | 0 | 0 | 0 | 3 | 3 | |
| POU4F2 | NM_004575 | 4 | X | NA | NA | .0104 | .5192 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 2 | ||
| POU4F3 | NM_002700 | 5 | X | X | X | .0403 | .8517 | .0012 | .6869 | .0085 | .7370 | NA | NA | NA | 0 | 0 | 1 | 0 | 2 | 1 |
| PPIF | NM_005729 | 10 | X | NA | NA | NA | NA | .0430 | .5081 | NA | NA | NA | 0 | 0 | 1 | 0 | 2 | 2 | ||
| PPP2R1B | NM_181699 | 11 | X | X | NA | NA | .0076 | .5880 | .0300 | .5046 | .0366 | NA | NA | 1 | 0 | 0 | 0 | 4 | 2 | |
| PPP2R5C | NM_015512 | 3 | X | NA | NA | NA | NA | .0084 | .7154 | NA | NA | NA | 0 | 0 | 1 | 1 | 6 | 3 | ||
| PRAX-1 | NM_002700 | 17 | X | .0445 | .5316 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | ||
| PRDX5 | NM_012094 | 11 | X | NA | NA | .0200 | .6032 | NA | NA | NA | .4122 | .902 | 0 | 0 | 2 | 0 | 7 | 3 | ||
| PRG4 | NM_004758 | 17 | X | X | .0380 | .5144 | NA | NA | .0390 | .5280 | .1725 | 2.4455 | .5826 | NA | NA | NA | NA | NA | NA | |
| PRH | NM_015893 | 2 | X | X | X | .0202 | 1.0341 | .0009 | .7512 | .0094 | .9816 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| PRKAR1A | NM_002734 | 17 | X | NA | NA | .0056 | .5440 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 3 | 10 | ||
| PRND | NM_012409 | 20 | X | NA | NA | .0013 | .6788 | NA | NA | NA | .7955 | .4877 | 1 | 0 | 0 | 0 | 3 | 5 | ||
| PRSS12 | NM_003619 | 4 | X | NA | NA | NA | NA | .0078 | 1.0619 | NA | NA | NA | 0 | 0 | 0 | 0 | 11 | 14 | ||
| PRSS8 | NM_002773 | 16 | X | X | X | .0299 | .7276 | .0019 | .5939 | .0090 | .7619 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 |
| PSA | NM_021154 | 9 | X | NA | NA | NA | NA | .0299 | .5052 | NA | NA | NA | 0 | 1 | 0 | 0 | 4 | 0 | ||
| PSCD4 | NM_013385 | 22 | X | .0116 | .5300 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| PSG6 | NM_002782 | 19 | X | NA | NA | NA | NA | .0188 | .6806 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 0 | ||
| PSMB7 | NM_002799 | 9 | X | NA | NA | NA | NA | .0348 | .5884 | NA | NA | NA | 0 | 0 | 2 | 0 | 2 | 1 | ||
| PSME1 | NM_006263 | 14 | X | NA | NA | NA | NA | .0160 | .5836 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | ||
| PTGER1 | NM_000955 | 19 | X | NA | NA | NA | NA | .0024 | .8247 | NA | NA | NA | 0 | 1 | 0 | 0 | 1 | 2 | ||
| PTPRM | NM_002845 | 18 | X | NA | NA | .0130 | .5310 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 9 | ||
| PVR | NM_006505 | 19 | X | NA | NA | .0303 | .6698 | NA | NA | NA | .886 | .3417 | 0 | 0 | 0 | 0 | 1 | 1 | ||
| RAB10 | NM_016131 | 2 | X | NA | NA | .0018 | .5768 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 3 | ||
| RAB27A | NM_004580 | 15 | X | .0209 | .5453 | NA | NA | NA | NA | NA | NA | NA | 2 | 0 | 0 | 0 | 6 | 9 | ||
| RAB27B | NM_004163 | 18 | X | .0027 | .7780 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 5 | 3 | ||
| RAB33A | NM_004794 | X | X | NA | NA | NA | NA | .0330 | .5027 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| RAB5EP | NM_004703 | 17 | X | NA | NA | NA | NA | .0331 | .5354 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 5 | ||
| RAB8B | NM_016530 | 15 | X | .0071 | .6620 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 3 | ||
| RAYL | NM_006860 | 22 | X | .0114 | .5825 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 4 | ||
| RBP2 | NM_004164 | 3 | X | .0320 | .5133 | NA | NA | NA | NA | NA | .9006 | 106 | 0 | 0 | 0 | 0 | 5 | 2 | ||
| RECQL5 | NM_004259 | 17 | X | NA | NA | NA | NA | .0041 | .7924 | NA | NA | NA | 1 | 0 | 1 | 0 | 6 | 8 | ||
| RFC1 | NM_002913 | 4 | X | NA | NA | .0002 | .8516 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 4 | 4 | ||
| RNAHP | NM_007372 | 17 | X | .0270 | .7921 | NA | NA | NA | NA | NA | NA | NA | 2 | 0 | 0 | 3 | 11 | 15 | ||
| RNF24 | NM_007219 | 20 | X | NA | NA | .0130 | .5324 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 3 | ||
| RPL10 | NM_006013 | X | X | X | NA | NA | .0015 | .7964 | .0199 | .7086 | .026 | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | |
| RPL23 | NM_000978 | 17 | X | NA | NA | .0120 | .5285 | NA | NA | NA | NA | NA | 0 | 1 | 0 | 0 | 3 | 0 | ||
| RPL8 | NM_000973 | 8 | X | .0106 | .5005 | NA | NA | NA | NA | .0136 | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | ||
| RPS6KA2 | NM_021135 | 6 | X | .0413 | .6324 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| RQCD1 | NM_005444 | 2 | X | X | X | .0385 | .7866 | .0003 | .7407 | .0057 | 1.0475 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 5 |
| RRAS | NM_006270 | 19 | X | NA | NA | .0118 | .5014 | NA | NA | .0468 | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| RRM1 | NM_001033 | 11 | X | NA | NA | .0124 | .6071 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| RYR3 | NM_001036 | 15 | X | NA | NA | .0045 | .5486 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| S100A10 | NM_002966 | 1 | X | NA | NA | NA | NA | .0333 | .5531 | .0219 | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| SCRG1 | NM_007281 | 4 | X | X | X | .0245 | .8622 | .0037 | .6126 | .0050 | .8163 | .0001 | NA | NA | NA | NA | NA | NA | NA | NA |
| SDHA | NM_004168 | 5 | X | NA | NA | NA | NA | .0463 | .5104 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 3 | ||
| SEMA3B | NM_004636 | 3 | X | X | .0248 | .7193 | NA | NA | .0272 | .5989 | .001 | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | |
| SIAT8D | NM_005668 | 5 | X | X | .0308 | .6569 | NA | NA | .0182 | .7220 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | |
| SIRT6 | NM_016539 | 19 | X | X | X | .0455 | 1.1561 | .0007 | .7775 | .0151 | .5832 | NA | NA | NA | 0 | 0 | 0 | 1 | 3 | 4 |
| SKD1 | NM_004869 | 18 | X | NA | NA | NA | NA | .0226 | .6079 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | ||
| SLC13A2 | NM_003984 | 17 | X | NA | NA | NA | NA | .0427 | .5468 | .299 | .4976 | .1465 | 0 | 0 | 0 | 1 | 1 | 1 | ||
| SLC17A3 | NM_006632 | 6 | X | .0071 | .8426 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 1 | 0 | 7 | 5 | ||
| SLC25A3 | NM_002635 | 12 | X | X | .0052 | .6005 | NA | NA | .0012 | 1.3485 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 2 | |
| SLC26A6 | NM_022911 | 3 | X | X | .0113 | .5554 | NA | NA | .0442 | .5924 | NA | .4473 | .6154 | 0 | 0 | 0 | 0 | 1 | 0 | |
| SLC28A1 | NM_004213 | 15 | X | NA | NA | NA | NA | .0297 | .7211 | .0178 | NA | NA | 0 | 0 | 0 | 1 | 2 | 3 | ||
| SLC4A2 | NM_003040 | 7 | X | NA | NA | .0075 | .5017 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 0 | 4 | ||
| SLC4A4 | NM_003759 | 4 | X | .0381 | .6320 | NA | NA | NA | NA | .0023 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| SLC4A8 | NM_004858 | 12 | X | NA | NA | NA | NA | .0064 | .6813 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 4 | ||
| SLN | NM_003063 | 11 | X | .0409 | .5860 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 1 | 0 | 10 | 8 | ||
| SMOC1 | NM_022137 | 14 | X | .0162 | .5248 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | ||
| SMTN | NM_006932 | 22 | X | NA | NA | NA | NA | .0157 | .6823 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| SNAP25 | NM_003085 | 5 | X | X | .0495 | .6919 | NA | NA | .0182 | .7532 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| SNCB | NM_003096 | 5 | X | X | .0389 | .7511 | NA | NA | .0341 | .5934 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | |
| SNRPB2 | NM_003092 | 20 | X | NA | NA | .0053 | .5351 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 5 | ||
| SNRPG | NM_003096 | 2 | X | X | X | .0259 | .7083 | .0097 | .5847 | .0072 | .8152 | .0008 | NA | NA | 2 | 0 | 0 | 0 | 3 | 7 |
| SNW1 | NM_012245 | 14 | X | NA | NA | .0062 | .6774 | NA | NA | .0095 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| SOLH | NM_005632 | 16 | X | NA | NA | .0050 | .5447 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| SOX13 | NM_005686 | 1 | X | NA | NA | .0033 | .5509 | NA | NA | NA | NA | NA | 0 | 1 | 0 | 0 | 3 | 3 | ||
| STAC | NM_003149 | 3 | X | NA | NA | NA | NA | .0117 | .9971 | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| STK11 | NM_000455 | 19 | X | NA | NA | .0496 | .5056 | NA | NA | .002 | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| STX6 | NM_005819 | 1 | X | X | .0476 | .5889 | NA | NA | .0102 | .9135 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 5 | |
| SYK | NM_003177 | 9 | X | X | NA | NA | .0052 | .6308 | .0008 | 1.0833 | .0001 | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | |
| SYN47 | NM_004272 | 5 | X | X | NA | NA | .0005 | .7356 | .0087 | .7914 | NA | NA | NA | 0 | 1 | 1 | 0 | 9 | 5 | |
| TACSTD2 | NM_002353 | 1 | X | .0184 | .6988 | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | 1 | 1 | 8 | 6 | ||
| TACTILE | NM_005816 | 3 | X | NA | NA | NA | NA | .0450 | .5909 | NA | NA | NA | 0 | 0 | 1 | 1 | 14 | 10 | ||
| TAF1C | NM_005679 | 16 | X | NA | NA | NA | NA | .0329 | .5503 | .0000008 | .4367 | .6475 | 2 | 0 | 0 | 0 | 3 | 7 | ||
| TAF2F | NM_005642 | 5 | X | NA | NA | .0108 | .5514 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 4 | ||
| TAF2N | NM_003487 | 17 | X | X | NA | NA | .0078 | .5738 | .0222 | .5442 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| TAGLN | NM_003186 | 11 | X | X | NA | NA | .0002 | .7463 | .0002 | 1.2027 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | |
| TCP10 | NM_004610 | 6 | X | NA | NA | .0195 | .5170 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 2 | 6 | 5 | ||
| TDO2 | NM_005651 | 4 | X | X | NA | NA | .0004 | .7506 | .0022 | .8348 | NA | NA | NA | 1 | 0 | 1 | 3 | 10 | 16 | |
| TGFB2 | NM_003238 | 1 | X | X | .0351 | .9198 | .0054 | .5532 | NA | NA | .003 | NA | NA | 0 | 0 | 0 | 0 | 4 | 6 | |
| TGM2 | NM_004613 | 20 | X | X | X | .0302 | .8491 | .0057 | .5918 | .0135 | .6604 | NA | NA | NA | 0 | 0 | 0 | 1 | 0 | 2 |
| TIMELESS | NM_003920 | 12 | X | NA | NA | .0046 | .6596 | NA | NA | NA | NA | NA | 1 | 0 | 0 | 0 | 5 | 6 | ||
| TLE3 | NM_005078 | 15 | X | X | NA | NA | .0046 | .5051 | .0366 | .5070 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 3 | |
| TLR4 | NM_003266 | 9 | X | .0403 | .6244 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| TNNI1 | NM_003281 | 1 | X | NA | NA | .0140 | .5406 | NA | NA | NA | NA | NA | 0 | 0 | 1 | 1 | 3 | 6 | ||
| TNS | NM_022648 | 2 | X | X | .0319 | .6754 | NA | NA | .0231 | .6409 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 3 | |
| TOM1 | NM_005488 | 22 | X | NA | NA | NA | NA | .0357 | .5146 | NA | NA | NA | 0 | 0 | 0 | 1 | 4 | 3 | ||
| TOP2B | NM_001068 | 3 | X | .0387 | .5029 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 0 | ||
| TP53TG1 | NM_007233 | 7 | X | NA | NA | .0041 | 1.2632 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| TPSD1 | NM_012217 | 16 | X | NA | NA | .0050 | .5075 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 0 | ||
| TPSG1 | NM_012467 | 16 | X | X | .0204 | .5671 | NA | NA | .0351 | .5145 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 8 | |
| TRAF3 | NM_003300 | 14 | X | X | X | .0226 | .9777 | .0276 | .6833 | .0061 | .9782 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| TRAP1 | NM_016292 | 16 | X | .0063 | .7397 | NA | NA | NA | NA | .0016 | NA | NA | 0 | 0 | 0 | 0 | 1 | 1 | ||
| TRAP-1 | NM_004257 | 2 | X | X | .0096 | .7332 | NA | NA | .0149 | .7419 | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 6 | |
| TRF4 | NM_006999 | 5 | X | NA | NA | .0046 | .5055 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 6 | 9 | ||
| TRP7 | NM_020389 | 5 | X | NA | NA | .0227 | .5889 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||
| TTK | NM_003318 | 6 | X | NA | NA | NA | NA | .0437 | .5624 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 3 | ||
| UBE2G1 | NM_003342 | 17 | X | X | NA | NA | .0007 | .6810 | .0064 | .7475 | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| UBQLN1 | NM_013438 | 9 | X | NA | NA | .0043 | .5068 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 3 | ||
| UK114 | NM_005836 | 8 | X | NA | NA | NA | NA | .0291 | .5440 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | ||
| ULBP2 | AY358665 | 6 | X | .0237 | .6179 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 1 | ||
| VDAC3 | NM_005662 | 8 | X | NA | NA | .0068 | .5347 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 4 | 0 | ||
| VDR | NM_000376 | 12 | X | NA | NA | NA | NA | .0096 | .7698 | NA | NA | NA | 0 | 0 | 0 | 0 | 3 | 0 | ||
| VLDLR | NM_003383 | 9 | X | X | .0190 | .8386 | NA | NA | .0051 | .9363 | .0207 | NA | NA | 0 | 0 | 0 | 0 | 0 | 1 | |
| WDR9 | NM_033656 | 21 | X | X | .0081 | .7508 | NA | NA | .0191 | .5976 | NA | NA | NA | 0 | 0 | 0 | 0 | 7 | 17 | |
| WISP2 | NM_003881 | 20 | X | X | .0255 | .7554 | .0006 | .6905 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | |
| WNT1 | NM_005430 | 12 | X | .0372 | .5091 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| WNT10B | NM_003394 | 12 | X | .0376 | .5500 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | ||
| WWOX | NM_130790 | 16 | X | .0435 | .7051 | NA | NA | NA | NA | NA | NA | NA | 1 | 0 | 0 | 1 | 6 | 7 | ||
| ZNF216 | NM_006007 | 9 | X | X | NA | NA | .0034 | .6026 | .0306 | .5161 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 1 | |
| ZNF236 | NM_007345 | 18 | X | NA | NA | NA | NA | .0056 | .7160 | NA | NA | NA | 0 | 0 | 0 | 0 | 1 | 8 | ||
| ZNF254 | NM_203282 | 19 | X | .0193 | .5085 | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 14 | 19 | ||
| ZNF264 | NM_003417 | 19 | X | NA | NA | NA | NA | .0246 | .6508 | NA | NA | NA | 0 | 0 | 0 | 0 | 2 | 2 | ||
| ZNF43 | NM_003423 | 19 | X | .0377 | .5609 | NA | NA | NA | NA | .103 | .3964 | 1.5405 | 0 | 0 | 0 | 1 | 16 | 12 | ||
| ZNF7 | NM_003416 | 8 | X | X | .0103 | 1.1337 | NA | NA | .0319 | .6670 | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | |
| ZP2 | NM_003460 | 16 | X | NA | NA | .0099 | .5085 | NA | NA | NA | NA | NA | 0 | 0 | 0 | 1 | 5 | 5 | ||
Note.— NA = not available.
Figure 2. .
Transcriptional targets of FOXP2 identified by ChIP-chip assay
FOXP2 Targets in Lung versus Brain
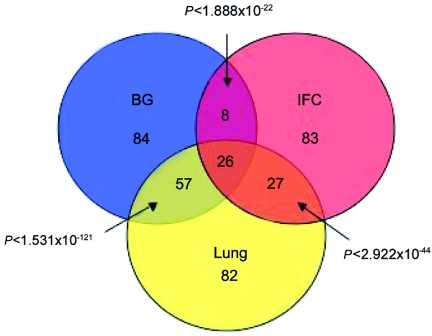
Murine Foxp2 has been shown to repress the promoter of the human lung-epithelial specific gene SP-C in vitro.12 Since Foxp2 has been reported as a transcriptional repressor that is expressed in the developing lung of mice,12 we were interested to see, which, if any, of the identified targets were potentially more-specific targets in the developing brain relative to lung, a non-CNS tissue expressing high levels of FOXP2 during development. Furthermore, no studies of targets of FOXP2 have been published from any human tissue, so even overlapping targets would be of general interest and would serve as a core set of non–tissue-specific FOXP2-regulated genes. We performed ChIP-chip in human fetal lung at 18 GW and identified 192 targets (table 1). There was 47% and 37% overlap between genes identified in lung and BG and in lung and IFC, respectively (fig. 3), providing further confidence in these genes as robust FOXP2 targets. Subtraction of the lung-enriched genes from the CNS data sets yielded 84 BG-specific genes and 83 IFC-specific genes (fig. 3 and table 1). In addition, there were eight targets found in both BG and IFC that were not enriched in lung. These highest-confidence brain-enriched targets include FGF8, which is a key effector of cortical patterning in mammals,36 and HOXB5 and HOXB7, members of the homeobox family of transcription factors, many of which are already known to be involved in CNS patterning.37,38
Figure 3. .
Distribution of FOXP2 targets identified among tissue regions. Shown are overlapping and tissue-specific targets of the 175 BG, 144 IFC, and 192 lung target genes among the three experiments. The P values based on the hypergeometric distribution show highly statistically significant overlap between the tissues.
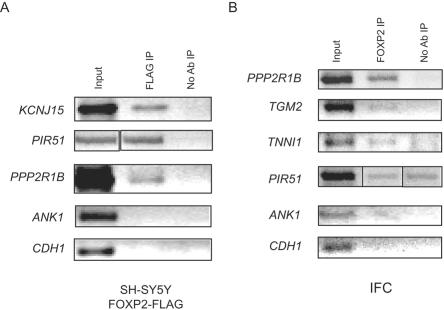
To provide independent validation of the array results, we checked a cross-section of the putative FOXP2 targets by ChIP-PCR, as is now standard.34,35,39,40 We used a second commercial antibody to FOXP2 to validate brain tissue and an antibody against the FLAG epitope in a neuronal cell line. For the cell-line confirmation, the neuronal cell line SH-SY5Y was stably transfected with FOXP2 isoform I with three C-terminal FLAG tags (see the “Material and Methods” section). Using either real-time quantitative or semiquantitative PCR, we assessed seven nervous-system targets in vitro and were able to confirm enrichment of FOXP2 occupancy at the promoters of all these genes (fig. 4 and data not shown). In contrast, the promoter of one lung target tested, ANK1, was not pulled down in these neuronal cells and served as a negative control; even though the level of expression of ANK1 changed in SY5Y cells with overexpressed FOXP2, it is likely a result of indirect regulation by FOXP2 and not direct binding. Although tissue was a limiting factor, we were able to test a subset of FOXP2 CNS targets in fetal brain tissue by ChIP-PCR and confirmed three of five promoters examined, providing a secondary level of confirmation for these targets (fig. 4 and data not shown).
Figure 4. .
Confirmation of identified FOXP2 targets by ChIP-PCR from SH-SY5Y cells overexpressing FLAG-tagged FOXP2 (A) or IFC (B). A, Promoters of three FOXP2 targets showing enrichment of FLAG-tagged FOXP2 at their promoters compared with control immunoprecipitations (IPs). No enrichment occurred at a lung-specific promoter, ANK1, or in a gene that was not a FOXP2 target, CDH1. B, Promoters of three FOXP2 targets showing enrichment of endogenous FOXP2 compared with control IPs. Also shown is an example of an IFC target, PIR51, that did not show confirmation by ChIP-PCR. No enrichment is seen when primers are used for the ANK1 promoter or CDH1 gene. No Ab = no addition of anti-FOXP2 antibody.
In Vitro Functional Validation of FOXP2 Targets
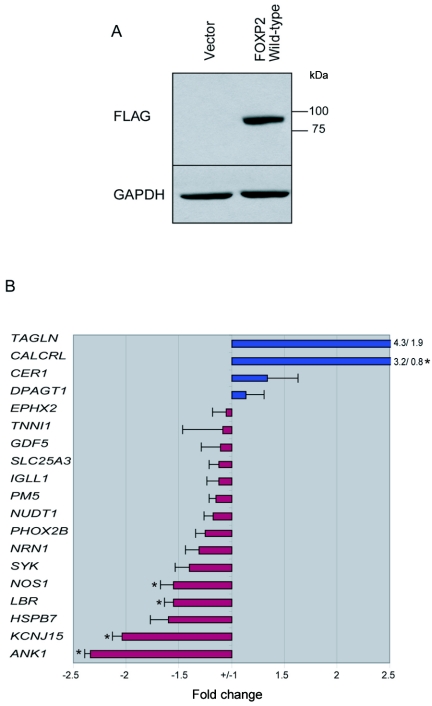
To provide some functional evidence of target regulation, we assessed the effect of FOXP2 overexpression in a neuronal cell culture system on identified FOXP2 targets. SH-SY5Y cells were stably transfected with FOXP2 isoform I with three C-terminal FLAG tags, and a population of stably transfected cells was used for the experiments at several different passages (fig. 5). Cells stably transfected with empty plasmid were used as a baseline control for comparison. Immunoblotting of the two cell lines for both FOXP2 and FLAG revealed robust expression of FLAG-tagged FOXP2 in the cell line transfected with FOXP2, compared with no expression in the cells transduced with the empty vector (fig. 5A and data not shown). This cell line provides an appropriate vehicle for overexpression studies, since FOXP1 is natively expressed in these cells, whereas FOXP2 is not detectable.4
Figure 5. .
FOXP2 overexpression confirming functional regulation of targets by FOXP2. A, Cell lysate from SH-SY5Y cells transfected with empty vector or FOXP2 isoform I, run on an SDS-PAGE gel and transferred to PVDF membrane. The membrane was hybridized with anti-FLAG antibody and anti-GAPDH antibody. B, Nineteen genes tested by qRT-PCR. The average of seven replicates and SEMs are indicated. Genes with a down-regulation in expression are shown in red, whereas those with a positive change in gene expression are shown in blue. Genes with significant difference in expression between control and FOXP2-overexpressing cells are indicated with an asterisk (*) (P<.05, by Student’s t test).
The majority of target genes investigated were chosen randomly, although a few were selected on the basis of their enrichment in all tissues studied (NOS1, CALCRL, PM5, and GDF5). Real-time quantitative RT-PCR (qRT-PCR) for 19 genes was performed on seven biological replicates of both control cells and cells overexpressing FOXP2. qRT-PCR analysis demonstrated that the majority of FOXP2 target genes identified (11 of 19) had at least a 25% change in expression with FOXP2 isoform I overexpression (fig. 5). Three genes, TAGLN, CALCRL, and CER1, showed up-regulation after overexpression of FOXP2, whereas DPAGT1 showed a trend toward increased expression (fig. 5). To test the reproducibility of the results, two genes, TNN1 and NUDT1, were examined using two different sets of primers in a blinded fashion. Both primer sets gave very similar results for each gene, indicating high specificity for the assay (data not shown). By use of a two-tailed paired t test, five of the genes had statistically significant fold changes between control and FOXP2-overexpressing cells. These genes were CALCRL, NOS1, LBR, KCNJ15, and ANK1. Accordingly, all these genes had changes >1.5 fold and were contained within the top-seven most changed genes. Interestingly, two of these genes, NOS1 and CALCRL, represent two of the genes enriched in all tissues by ChIP-chip and, as mentioned above, were selected a priori for this reason. Thus, we can conclude that the levels of at least 26% of the genes identified as targets of FOXP2 can be altered by manipulating expression of FOXP2 in a neuronal cell line. These data, confirming slightly more than 25% of targets examined in vitro, are consistent with those of other published studies, in which 20%–35% of the targets are typically confirmed in this manner.39–43
FOXP2 Binding Sites Sequence in Candidate Genes
We next determined whether the promoter regions of candidate genes present on the Aviva array contain putative FOXP2 binding sites. Since double-stranded PCR products were spotted onto the array, the binding site could lie in either strand, so we examined both strands. We inspected the sequences of identified candidate targets for the FOXP2 binding site CAAATT or the core FOXP2 binding site AAAT.44 The sequence from 323 of 367 potential targets identified by ChIP-chip was examined for binding sites, and 95% (307 genes) were found to contain at least one AAAT core FOXP2 binding site, whereas 106 contained at least one CAAATT binding site (table 1). Since the FOXP1 binding site (TATTT[A/G]T) has been shown to be a possible FOXP2 binding site,4,45 we also searched for the presence of a FOXP1 binding site. A total of 82 genes were found to have at least one FOXP1 binding site (table 1). All genes tested by overexpression of FOXP2 were found to have at least one copy of the core-binding site (AAAT), and most had multiple sites. HSPB7, CER1, CALCRL, ANK1, LBR, and KCNJ15, which showed directional changes in expression following overexpression of FOXP2, were also found to have at least one CAAATT binding site. Additionally, NOS1, which showed a large, statistically significant decrease in expression, and CALCRL, which showed a significant increase in expression after FOXP2 overexpression, as well as GDF5, were found to have a FOXP1 binding site present. Comparing this enrichment of FOXP2 sites within the target genes identified on the array with an equivalent number of random promoter sequences from Ref Seq genes showed a statistically significant enrichment for all sequences identified in BG as a group (P<.05, by χ2 with Yates correction), which was more significant for the more stringent canonical CAAATT site in BG targets (P=.007, by χ2). The targets identified in IFC and lung showed the same trend as BG but did not reach significance.
Functional Annotation
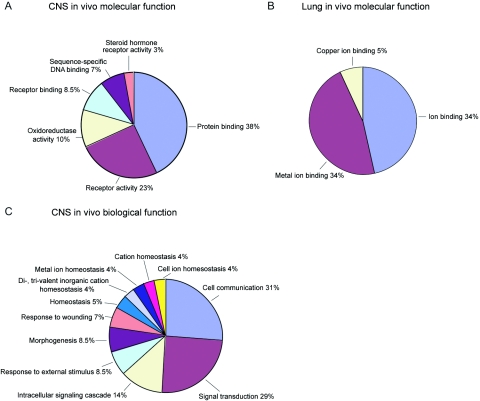
To investigate whether FOXP2 target genes belong to functional categories that provide insight into the role of FOXP2, we performed GO and pathway analysis on target genes, using the DAVID Bioinformatics Resources.24 Gene targets identified in BG or IFC were grouped together as “CNS-specific” genes and were compared with and contrasted to “lung-specific” genes. We focused on GO categories of level three or higher, containing at least three genes and having a P value ⩽.05 by Fisher’s exact test. CNS-specific genes were significantly enriched in six categories of molecular function, and the lung genes were enriched in three categories that did not overlap with those found in the CNS gene set (fig. 6). The CNS-specific genes were enriched for 11 biological-function GO categories, whereas the lung data set did not have any significant categories grouped according to biological function.
Figure 6. .
GO categories of in vivo targets, revealing tissue specificity of target pathways. FOXP2 target gene lists from either the CNS (BG and IFC) or lung were analyzed for significantly enriched GO categories by use of DAVID Bioinformatics Resources. Categories were considered significantly enriched if at least three genes were in one category with a P value ⩽.05. Significant CNS targets with a known molecular function fall into six categories (A), whereas lung targets can be classified into three categories (B), none of which overlap with the CNS results. Significant CNS targets with a known biological function are grouped into 11 categories (C). There were no significant biological function categories for lung-specific targets.
Several intriguing CNS-enriched GO categories were identified: morphogenesis (TIMELESS, WNT1, SOX13,HOXB5, and FGF8), intracellular signaling cascades (CDC42BPB, GABBR1, CCKAR, RP26KA2, and RRAS), and cation homeostasis (GALR2, RYR3, and CCKAR). Focusing this analysis on those genes contained within significantly enriched GO categories in the CNS allowed us to uncover FOXP2 targets with potential roles in neural development and to strengthen the hypothesis of FOXP2 as a crucial player in signaling cascades regulating this critical epoch. Salient examples of FOXP2 targets previously shown to be important for CNS development models include WNT146,47 and RPS6KA2, also known as RSK3, which is highly expressed in the cortical primordium.48 Mutations in a related family member of RSK3, RSK2, lead to Coffin-Lowry syndrome (MIM 303600), which is associated with cognitive abnormalities.49
Another critical pathway downstream of FOXP2 in the IFC appears to be neurite outgrowth and axonal morphology, including calcium-mediated growth cone dynamics (e.g., GALR2, POU4F2, RRAS, and RYR3).50–54 Further support for the role of FOXP2 transcriptional targets in dynamic regulation of neuronal structure was obtained using Ingenuity pathway-analysis software to analyze the 34 core CNS targets identified in both BG and IFG. Ingenuity identified several functions of neuronal activity significantly enriched, including branching of dendrites (NOS1 and CRH), mobilization of calcium (CALCRL, CD5, and PRLH), quantity of calcium (EPOR and CRH), and learning (CRH and EPOR) (data not shown). These data also suggest a possible function for FOXP2 signaling cascades in activity-based (e.g., long-term potentiation) modeling of neural connections, in addition to its role in development.
Possible Positive Selection of Several FOXP2 Targets in Humans
FOXP2 is clearly involved in multiple functions not related to human higher cognitive functions, including sensorimotor integration and vocal learning in birds8,55 and lung development outside the CNS.12 It would therefore be useful to identify a list of targets that most likely contribute to the development of higher cognitive specializations.11 We hypothesized that, given the genetic complexity of a highly advantageous trait such as human language, positive selection may be working on FOXP2 target proteins in addition to FOXP2 itself.17,18 We reasoned that identification of FOXP2 target genes potentially under positive selection will enrich for those target genes more likely to be involved in language development. We analyzed our data with respect to published estimates of protein sequence divergence (Ka/Ki and Ka/Ks) for 1,168 genes with available data.29
Ka measures the rate of nonsynonymous nucleotide substitutions, Ki measures the rate of nucleotide substitutions in interspersed repeats within a 250-kb window centered around each gene, and Ks measures the rate of synonymous substitutions. Low Ka/Ki or Ka/Ks values suggest strong purifying selection (deleterious mutations), whereas elevated Ka/Ki values suggest accelerated evolution via positive selection, or relaxation of constraint.29 Typically, a value >1.0 is indicative of acceleration.56 Fourteen genes identified as FOXP2 targets in either BG, IFC, or lung had Ka/Ks or Ka/Ki >1.0 (table 2). As a whole, these genes are potential key FOXP2 targets that, by virtue of their sequence divergence, show evidence of accelerated evolution. Remarkably, this list includes genes such as NDUFA2, which is part of the electron transport chain, a pathway identified to be under potential adaptive evolution in humans by comparative gene-expression studies in brain.57,58 It also includes a number of other genes implicated in vertebrate forebrain patterning, including HESX1 and CER1. Remarkably, four of the FOXP2 targets, HESX5, MEOX1, PIR51, and RBP2, have Ka/Ks values >50.29 Genes with evidence of accelerated evolution, which is likely to be indicative of positive selection in humans, comprise a key cohort potentially related to human cognitive specializations integrated by the BG and IFC, including speech and language.
Table 2. .
Fourteen Genes with Ka/Ki or Ka/Ks >1.0[Note]
| Positive Selection |
|||
| Gene | GenBank Accession Number | Ka/Ki | Ka/Ks |
| CA4 | NM_000717 | .6937 | 1.5476 |
| CD7 | NM_006137 | .7805 | 1.5375 |
| CER1 | NM_005454 | .5913 | 1.7308 |
| FCGR2A | DQ894525 | .7127 | 2.8529 |
| GPR21 | NM_005294 | NA | 1.3871 |
| HESX1 | NM_003865 | .5971 | 75 |
| LBR | NM_002296 | NA | 1.0833 |
| MEOX1 | NM_004527 | NA | 52 |
| NDUFA2 | NM_002488 | 1.6458 | 1.2958 |
| PGLYRP | NM_005091 | 1.8051 | .5485 |
| PIR51 | NM_006479 | NA | 56 |
| PRG4 | NM_004758 | 2.4455 | .5826 |
| RBP2 | NM_004164 | .9006 | 106 |
| ZNF43 | NM_003423 | NA | 1.5405 |
Note.— Estimated rates of protein-sequence divergence between humans and chimpanzees were obtained from the study by Khaitovich et al.29 NA = not available.
Differential Expression of Several FOXP2 Targets between Chimpanzee and Human Brain
Although measures of protein-sequence divergence measure one key metric of function relevant to evolution, they do not consider additional changes in promoter or other regions that might alter gene expression, another dimension relevant to brain evolution.58,59 This is especially important because FOXP2 is a transcriptional regulator, and one might expect its targets to exhibit differential expression between evolutionarily divergent species. Therefore, we performed a meta-analysis of primate-human gene-expression data, to identify genes differentially expressed in the brains of humans and nonhuman primates, using three published data sets26–28 to insure identification of the most robust changes in expression (see the “Material and Methods” section). This was essential since each study was comprised of only a few individuals. We identified 47 FOXP2 target genes that were differentially expressed between human and chimpanzee brains, including one of the genes whose coding region was also under positive selection, FCGR2 (table 3).
Table 3. .
Summary of Evolutionary Data[Note]
| Positive Selection | ||||
| Gene | GenBank Accession Number | Differential Expression P Value | Ka/Ki | Ka/Ks |
| ADAM28 | NM_021777 | .0395 | .7658 | NA |
| AKR7A2 | NM_003689 | NA | .5579 | NA |
| AOAH | NM_001637 | .021 | NA | .7755 |
| APOD | NM_001647 | .0001 | NA | NA |
| ASGR1 | NM_001671 | .0235 | NA | NA |
| C1QA | NM_015991 | NA | NA | .791 |
| CA4 | NM_000717 | NA | .6937 | 1.5476 |
| CD7 | NM_006137 | NA | .7805 | 1.5375 |
| CER1 | NM_005454 | NA | .5913 | 1.7308 |
| CRYBB3 | NM_004076 | NA | .6762 | .6374 |
| CXADR | NM_001338 | .0001 | NA | NA |
| CYB5-M | NM_030579 | .0048 | NA | NA |
| CYP2J2 | NM_000775 | .0001 | NA | .7101 |
| DAXX | NM_001350 | .0308 | NA | NA |
| DCT | NM_001922 | .0331 | NA | NA |
| DISC1 | NM_018662 | NA | .8117 | NA |
| DPAGT1 | NM_001382 | NA | NA | .5 |
| EFNB3 | NM_001406 | .0365 | NA | .8846 |
| EMR2 | NM_001784 | NA | .5098 | .6667 |
| EPHX2 | NM_001979 | .0472 | NA | NA |
| EPOR | NM_000121 | NA | NA | .5185 |
| ERCC4 | NM_005236 | .0439 | NA | NA |
| FCGR2A | DQ894525 | .0008 | .7127 | 2.8529 |
| FSHR | NM_000145 | NA | .5082 | .7816 |
| FY | NM_002036 | .0361 | NA | NA |
| G10 | NM_003910 | .0119 | NA | NA |
| GFRA1 | NM_005264 | .0287 | NA | NA |
| GIF | NM_005142 | NA | NA | .7541 |
| GLUD1 | NM_005271 | .0011 | NA | NA |
| GPLD1 | NM_001503 | .009 | .5473 | NA |
| GPR21 | NM_005294 | NA | NA | 1.3871 |
| GRIK1 | NM_000830 | .0439 | NA | NA |
| HESX1 | NM_003865 | NA | .5971 | 75 |
| HEXB | NM_000521 | NA | .6457 | .9425 |
| HOXB5 | NM_002147 | .0025 | NA | NA |
| HOXB7 | NM_004502 | NA | NA | .7049 |
| IK | NM_006083 | 0 | NA | NA |
| INHA | NM_002191 | .0123 | NA | .6471 |
| ITGB3 | NM_000212 | .0164 | NA | NA |
| LBR | NM_002296 | NA | NA | 1.0833 |
| MAP2K3 | NM_002756 | .0345 | NA | NA |
| MCF2 | NM_005369 | .0413 | NA | NA |
| MEOX1 | NM_004527 | NA | NA | 52 |
| MGST2 | NM_002413 | NA | NA | .623 |
| MRAS | NM_012219 | .0137 | NA | NA |
| MSLN | NM_013404 | NA | .5922 | .5 |
| NAP1 | NM_004851 | NA | .7813 | .7522 |
| NCF2 | NM_000433 | NA | NA | .8846 |
| NDUFA2 | NM_002488 | NA | 1.6458 | 1.2958 |
| PARG | NM_003631 | .0064 | NA | NA |
| PAX3 | NM_000438 | .0235 | NA | NA |
| PDXK | NM_003681 | .0132 | NA | NA |
| PGLYRP | NM_005091 | NA | 1.8051 | .5485 |
| PIR51 | NM_006479 | NA | NA | 56 |
| PON2 | NM_000305 | .0112 | NA | NA |
| PPP2R1B | NM_181699 | .0366 | NA | NA |
| PRDX5 | NM_012094 | NA | NA | .902 |
| PRG4 | NM_004758 | NA | 2.4455 | .5826 |
| PRND | NM_012409 | NA | .7955 | NA |
| PVR | NM_006505 | NA | .886 | NA |
| RBP2 | NM_004164 | NA | .9006 | 106 |
| RPL10 | NM_006013 | .026 | NA | NA |
| RPL8 | NM_000973 | .0136 | NA | NA |
| RRAS | NM_006270 | .0468 | NA | NA |
| S100A10 | NM_002966 | .0219 | NA | NA |
| SCRG1 | NM_007281 | .0001 | NA | NA |
| SEMA3B | NM_004636 | .001 | NA | NA |
| SLC13A2 | NM_003984 | NA | NA | NA |
| SLC26A6 | NM_022911 | NA | NA | .6154 |
| SLC28A1 | NM_004213 | .0178 | NA | NA |
| SLC4A4 | NM_003759 | .0023 | NA | NA |
| SNRPG | NM_003096 | .0008 | NA | NA |
| SNW1 | NM_012245 | .0095 | NA | NA |
| STK11 | NM_000455 | .002 | NA | NA |
| SYK | NM_003177 | .0001 | NA | NA |
| TAF1C | NM_005679 | 0 | NA | .6475 |
| TGFB2 | NM_003238 | .003 | NA | NA |
| TRAP1 | NM_016292 | .0016 | NA | NA |
| VLDLR | NM_003383 | .0207 | NA | NA |
| ZNF43 | NM_003423 | NA | NA | 1.5405 |
Note.— NA = not available.
Among FOXP2 in vivo targets showing primate-human differential expression were a number of genes encoding transcription factors involved in neural development, including PAX3 and HOXB5, and a number of other genes encoding known CNS patterning and guidance molecules, such as EPHX2, ITGB3, and SEMA3B.60 Several genes related to neural transmission, including neurotransmitter receptor genes GFRA1 and GRIK1, were also identified among this cohort. When combined with the analysis of sequence evolution, these data suggest a potential role of specific genes in human cognition by identifying genes with differential expression at the mRNA level or that are potentially under positive selection in humans.
Discussion
Here, we identify FOXP2 targets in vivo for the first time, providing an initial transcriptional network downstream of this gene known to be involved in human higher cognition and vocal-motor learning. We used previously published, well-validated ChIP-chip methods21,22 and, in addition, performed extensive functional validation. Although the majority of genes were repressed by FOXP2, we were able to identify a subset of genes that appear to be activated by FOXP2 at the transcriptional level, indicating that FOXP2 can act as a transcriptional activator in some contexts. Further, we show that some of the genes show evidence of accelerated evolution and thus may be under positive selection in humans, or they are differentially expressed between chimpanzee and human brain, providing support for their potential role in higher cognition and language. Some FOXP2 targets, such as FGF8 and PAX3, have known roles in cerebral cortical patterning, immediately connecting FOXP2 transcriptional regulation to brain patterning during development.36,61
FOXP2 Targets and Transcriptional Activity
FOXP2 promoter binding demonstrated by ChIP-chip across multiple samples and multiple independent replicate experiments provides strong support for the targets identified. By comparing targets identified in different CNS tissues, we were able to identify a large set of core nervous-system targets with genes specific to the different regions. Further, we confirm a cross-section of targets, using ChIP-PCR in vitro and in vivo with use of a second antibody, an additional level of confirmation.
We also note that a subset of FOXP2 targets were identified in two brain regions and not in lung. Although we hypothesize that some of these may be CNS-specific FOXP2 targets, it remains possible that some could be targets in other nonneural tissues, a question that can be explored in subsequent studies. Additionally, this distinction of CNS specificity does not necessarily make these genes higher priorities for further study than genes identified in lung. Many FOXP2 targets were also identified in the lung, including several genes known to play important roles in nervous-system development or maturation, such as MEF2D, GDF5, POU4F3, SEMA3B, A2BP1, and PAX3, which is correlative to the key function of FOXP2 in both lung and brain development.
It is also supportive that we identified consensus sites described elsewhere on the basis of the binding structure of FOXP244 in a majority of the FOXP2 target genes. Statistical analysis supports the enrichment of putative FOXP2 binding sites within the target genes identified in BG. However, there was only a trend toward significance in frontal cortex and lung, and not all candidate genes identified have a consensus binding sequence, suggesting that other as-yet unidentified FOXP2 regulatory regions exist in these genes or that FOXP2 may act in a complex that permits binding to different sites. These results indicate the need for further investigation into the sequence-specific DNA binding parameters of FOXP2.
Here, we took an additional validation step by showing that >25% of genes whose promoters were bound by FOXP2 protein were indeed regulated by manipulation of FOXP2 levels in a neuronal cell line. Since the basic ChIP experiment was done in tissue, and it was necessary to perform functional confirmation in vitro, the cell line is distinct from the original in vivo tissue; yet, we were still able to confirm transactivation in a percentage of targets very similar to that in other published studies.39,40,42,43 We were able to show that, in general, overexpression of FOXP2 resulted in the expected decrease in expression of target genes. However, overexpression of FOXP2 increased expression of TAGLN, CALCRL, and CER1, suggesting transcriptional activation. This, along with the study by Vernes et al.,19(in this issue) provides the first evidence that FOXP2 can potentially act as a transcriptional activator in certain situations. ChIP-chip identifies promoter regions that are bound to FOXP2 but does not indicate more-complex transcriptional regulation that may have tissue- and cell-specific requirements for regulating transcription. Thus, the behavior of the subset of genes not altered after forced FOXP2 overexpression may be attributed to the lack of additional binding partners necessary for FOXP2 transcriptional regulation of certain genes. However, we do expect a percentage of false-positive results contained within our subset of genes.
Using a similar experimental paradigm, Vernes et al.19(in this issue) identified FOXP2 targets in vitro in SH-SY5Y cells by using a commercial antibody that recognizes a different region of the FOXP2 protein and found 303 FOXP2 targets. Of our core set of 34 targets that we found in vivo in both IFC and BG, 47% (16) overlap the targets identified in SH-SY5Y cells by Vernes et al., and there is a 22% overlap between all 285 genes we identified in either IF or BG and the 303 genes that Vernes et al. identified in SY5Y (62 of 285). This highly significant overlap provides another level of independent confirmation of our results, and our data speak to the relevance of at least a subset of the in vitro findings for normal human brain development in vivo. Vernes et al. also found up-regulation of certain targets’ expression after FOXP2 overexpression, consistent with the notion that some targets may not be repressed. Although we each confirmed that FOXP2 binding regulates the targets identified, and both studies note down-regulation of HSP7 and PM5 with FOXP2 expression in SH-SY5Y cells, in some cases, such as for CALCRL, we observed different directions of regulation in vitro after FOXP2 overexpression than did Vernes et al. Testing the possibility that this difference could be the result of identification of different isoforms, each group tested both CALCRL primer pairs on our respective cDNA. The results were consistent across primer pairs in each cell line and therefore are not due to testing distinct isoforms (data not shown). Many factors could explain these differences between the studies. For example, variations in the levels of FOXP2 expression in each of the cell lines used could explain these discrepancies. SH-SY5Y cells also contain abundant amounts of FOXP1 protein, and it is possible that FOXP2 and FOXP1 share a subset of common transcriptional targets. FOXP2 has been shown to heterodimerize with FOXP1,13 and heterodimers of FOXP2 with FOXP1 may result in different transcriptional outcomes than homodimers of FOXP2 or FOXP1. Thus, one could imagine a scenario in which low levels of FOXP2 repress transcription through heterodimerization with FOXP1, but, with increasing amounts of FOXP2, there is competition of FOXP2 homodimers with endogenous FOXP1, leading to transcriptional activation. Thus, as has been the case with other transcription factors, it will be important to define a subset of genes coregulated by FOXP2 and FOXP1, as well as the repertoire of FOXP2 partners and their regulation, to understand the context in which FOXP2 acts as a transcriptional repressor or activator.
Function of Targets
The target genes identified provide a foundation for exploring the transcriptional network downstream of FOXP2 in the developing brain. Functions such as growth regulation, embryonic development, and signal transduction identified as enriched GO categories for the candidate targets of FOXP2 are also common functions for targets of other forkhead-containing genes.62–64 GO analysis of the potential targets specific to IFC indicated that many of these genes are involved in growth and morphogenesis, whereas other classes, such as development, were enriched for in targets identified in BG, indicating different downstream functional differences in the different regions of the brain. Furthermore, FOXP2 continues to be expressed in the adult and is modulated by vocal learning in adult song birds.55 Thus, the identification of FOXP2 target genes involved in neurite outgrowth, calcium signaling, and learning, by Ingenuity pathway analysis, may provide a potential molecular link to ongoing behavioral plasticity throughout development and into adulthood.
Previous work has suggested that FOXP2 may be under positive selection in humans, so we reasoned that some FOXP2 targets may be components of neurodevelopmental pathways also under positive selection.17,18 Large-scale gene coexpression network analysis of human and chimpanzee brain confirmed the previously reported action of positive selection on the pathway of the electron transport chain.57,65–67 Further, this work showed that entire functional modules of coexpressed genes appear to be under positive selection in humans.65 Other genomewide analyses investigating alleles with accelerated increase of frequency between three different human populations have demonstrated that multiple genes in the phosphatidylinositol pathway, in addition to the electron transport chain pathway, have undergone positive selection.68 These precedents support the notion that positive selection of genes regulated by FOXP2 may direct us to molecular pathways of particular interest in human evolution and human cognitive specialization, since language is a complex trait that likely can be modulated by many genes.11 Some of the FOXP2 target genes that we identified are genes known to be involved in human brain development and patterning, such as EFNB3, HESX1, and CER1. This provides further support for the notion that targets of FOXP2 showing these significant differences between humans and our closest relative, the chimpanzee, are particularly important in development of human cognitive specializations.
Similarly, 15 of the potential FOXP2 targets identified here (APOD,69 CCK,70,71 CCK-AR,72–75 CCND2,76 CD5,77,78 DISC1,79–81 DRD2,82 GABBR1,83 MT2A,84 NOS1,85–87 PMX2B,88 TDO2,89 TIMELESS,90 WNT1,91 and ZNF7492) have shown some evidence of association with schizophrenia (SCZD [MIM 181500]), a disease that has been suggested to involve primary language dysfunction. Although most of these associations are preliminary or marginal, DISC1 has been clearly replicated. The association of SNP rs2396753, located within an intron of FOXP2, with schizophrenia with auditory hallucinations,93 further suggests that targets of FOXP2 may also be candidates for involvement in schizophrenia. The concept that complex genetic disorders might involve multiple genes within common pathways implies that the FOXP2 targets identified here are realistic candidates for a variety of neurodevelopmental disorders involving higher cognitive functions.
In summary, these data are the first identification of human FOXP2 targets in the developing brain. This is also the first time, to our knowledge, that ChIP-chip has been used to assess transcription factor targets in the human fetal brain. Many of the identified FOXP2 targets have been previously characterized as having critical roles in several important neuronal features, such as neurite outgrowth and axon pathfinding. In addition, we have uncovered targets of FOXP2 that may have vital functions in the evolution of the mammalian brain. Since FOXP2 has a direct link to speech in humans, together these findings provide insight into signaling pathways that may be important both in the development and evolution of language.
Acknowledgments
This work is submitted in grateful memory of Zheng Luo, who performed the pilot ChIP-chip experiments in the Geschwind laboratory. His technical skills were essential to establishing this technique, and his presence is very much missed. We thank the NICHD Brain and Tissue Bank for Developmental Disorders, under contracts N01-HD-4-3368 and N01-HD-4-3383, for providing human fetal brain tissue. We also thank Eric Wexler, M.D., Ph.D., and other members of the Geschwind laboratory for helpful discussions, as well as an anonymous reviewer whose insightful comments brought several interesting targets to our attention and highlighted some of the key evolutionary context. This research was supported by grants R21MH075028 (to D.H.G.), T32GM008243 (to E.S.), and T32HD007032 (to G.K.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for blastp and megablast)
- DAVID Bioinformatics Resources, http://david.abcc.ncifcrf.gov/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for FOXP2 isoforms (accession numbers NP_055306 and NP_683697 or NP_683698) and accession numbers in tables 1–3)
- NCBI GEO, http://www.ncbi.nlm.nih.gov/geo/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SPCH1, Coffin-Lowry syndrome, and SZCD)
References
- 1.Marcus GF, Fisher SE (2003) FOXP2 in focus: what can genes tell us about speech and language? Trends Cogn Sci 7:257–262 10.1016/S1364-6613(03)00104-9 [DOI] [PubMed] [Google Scholar]
- 2.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- 3.MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, et al (2005) Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet 76:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE (2006) Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet 15:3154–3167 10.1093/hmg/ddl392 [DOI] [PubMed] [Google Scholar]
- 5.Watkins KE, Dronkers NF, Vargha-Khadem F (2002) Behavioural analysis of an inherited speech and language disorder: comparison with acquired aphasia. Brain 125:452–464 10.1093/brain/awf058 [DOI] [PubMed] [Google Scholar]
- 6.Belton E, Salmond CH, Watkins KE, Vargha-Khadem F, Gadian DG (2003) Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp 18:194–200 10.1002/hbm.10093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watkins KE, Gadian DG, Vargha-Khadem F (1999) Functional and structural brain abnormalities associated with a genetic disorder of speech and language. Am J Hum Genet 65:1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA (2004) Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci 24:3152–3163 10.1523/JNEUROSCI.5589-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ (2003) FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain 126:2455–2462 10.1093/brain/awg247 [DOI] [PubMed] [Google Scholar]
- 10.Corballis MC (2004) FOXP2 and the mirror system. Trends Cogn Sci 8:95–96 10.1016/j.tics.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 11.Crespi BJ (2007) Sly FOXP2: genomic conflict in the evolution of language. Trends Ecol Evol 22:174–175 10.1016/j.tree.2007.01.007 [DOI] [PubMed] [Google Scholar]
- 12.Shu W, Yang H, Zhang L, Lu MM, Morrisey EE (2001) Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem 276:27488–27497 10.1074/jbc.M100636200 [DOI] [PubMed] [Google Scholar]
- 13.Li S, Weidenfeld J, Morrisey EE (2004) Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24:809–822 10.1128/MCB.24.2.809-822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perrone L, Pasca di Magliano M, Zannini M, Di Lauro R (2000) The thyroid transcription factor 2 (TTF-2) is a promoter-specific DNA-binding independent transcriptional repressor. Biochem Biophys Res Commun 275:203–208 10.1006/bbrc.2000.3232 [DOI] [PubMed] [Google Scholar]
- 15.Yang Q, Kong Y, Rothermel B, Garry DJ, Bassel-Duby R, Williams RS (2000) The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem J 345:335–343 10.1042/0264-6021:3450335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan SA, Akers L, Moody SA (2001) FoxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transcriptional repression that is dependent on the C-terminal domain. Dev Biol 232:439–457 10.1006/dbio.2001.0191 [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Webb DM, Podlaha O (2002) Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics 162:1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S (2002) Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418:869–872 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- 19.Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE (2007) High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet 81:1232–1250 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA (2003) Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol 460:266–279 10.1002/cne.10654 [DOI] [PubMed] [Google Scholar]
- 21.Ren B, Dynlacht BD (2004) Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol 376:304–315 [DOI] [PubMed] [Google Scholar]
- 22.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al (2000) Genome-wide location and function of DNA binding proteins. Science 290:2306–2309 10.1126/science.290.5500.2306 [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 25.Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA (2003) Identifying biological themes within lists of genes with EASE. Genome Biol 4:R70 10.1186/gb-2003-4-10-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C (2003) Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA 100:13030–13035 10.1073/pnas.2135499100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al (2002) Intra- and interspecific variation in primate gene expression patterns. Science 296:340–343 10.1126/science.1068996 [DOI] [PubMed] [Google Scholar]
- 28.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al (2004) Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14:1462–1473 10.1101/gr.2538704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S (2005) Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309:1850–1854 10.1126/science.1108296 [DOI] [PubMed] [Google Scholar]
- 30.Galaburda AM, LeMay M, Kemper TL, Geschwind N (1978) Right-left asymmetrics in the brain. Science 199:852–856 10.1126/science.341314 [DOI] [PubMed] [Google Scholar]
- 31.Geschwind DH, Miller BL, DeCarli C, Carmelli D (2002) Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci USA 99:3176–3181 10.1073/pnas.052494999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins KE, Vargha-Khadem F, Ashburner J, Passingham RE, Connelly A, Friston KJ, Frackowiak RS, Mishkin M, Gadian DG (2002) MRI analysis of an inherited speech and language disorder: structural brain abnormalities. Brain 125:465–478 10.1093/brain/awf057 [DOI] [PubMed] [Google Scholar]
- 33.Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA (2005) Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308:1794–1798 10.1126/science.1110324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TH, Xiong H, Zhang Z, Ren B (2005) β-Catenin activates the growth factor endothelin-1 in colon cancer cells. Oncogene 24:597–604 10.1038/sj.onc.1208237 [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B (2003) A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA 100:8164–8169 10.1073/pnas.1332764100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuchi-Shimogori T, Grove EA (2001) Neocortex patterning by the secreted signaling molecule FGF8. Science 294:1071–1074 10.1126/science.1064252 [DOI] [PubMed] [Google Scholar]
- 37.Bel-Vialar S, Itasaki N, Krumlauf R (2002) Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129:5103–5115 [DOI] [PubMed] [Google Scholar]
- 38.Prince VE, Price AL, Ho RK (1998) Hox gene expression reveals regionalization along the anteroposterior axis of the zebrafish notochord. Dev Genes Evol 208:517–522 10.1007/s004270050210 [DOI] [PubMed] [Google Scholar]
- 39.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA (2007) Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445:931–935 10.1038/nature05478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY (2007) Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445:936–940 10.1038/nature05563 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 102:4459–4464 10.1073/pnas.0501076102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124:207–219 10.1016/j.cell.2005.10.043 [DOI] [PubMed] [Google Scholar]
- 43.Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS (2006) T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem 281:11992–12000 10.1074/jbc.M513613200 [DOI] [PubMed] [Google Scholar]
- 44.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L (2006) Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14:159–166 10.1016/j.str.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 45.Wang B, Lin D, Li C, Tucker P (2003) Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem 278:24259–24268 10.1074/jbc.M207174200 [DOI] [PubMed] [Google Scholar]
- 46.McMahon AP, Bradley A (1990) The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62:1073–1085 10.1016/0092-8674(90)90385-R [DOI] [PubMed] [Google Scholar]
- 47.Thomas KR, Capecchi MR (1990) Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346:847–850 10.1038/346847a0 [DOI] [PubMed] [Google Scholar]
- 48.Zeniou M, Ding T, Trivier E, Hanauer A (2002) Expression analysis of RSK gene family members: the RSK2 gene, mutated in Coffin-Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum Mol Genet 11:2929–2940 10.1093/hmg/11.23.2929 [DOI] [PubMed] [Google Scholar]
- 49.Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel JL, Sassone-Corsi P, Hanauer A (1996) Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384:567–570 10.1038/384567a0 [DOI] [PubMed] [Google Scholar]
- 50.Ooashi N, Futatsugi A, Yoshihara F, Mikoshiba K, Kamiguchi H (2005) Cell adhesion molecules regulate Ca2+-mediated steering of growth cones via cyclic AMP and ryanodine receptor type 3. J Cell Biol 170:1159–1167 10.1083/jcb.200503157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith MD, Dawson SJ, Latchman DS (1997) Inhibition of neuronal process outgrowth and neuronal specific gene activation by the Brn-3b transcription factor. J Biol Chem 272:1382–1388 10.1074/jbc.272.2.1382 [DOI] [PubMed] [Google Scholar]
- 52.Hobson SA, Holmes FE, Kerr NC, Pope RJ, Wynick D (2006) Mice deficient for galanin receptor 2 have decreased neurite outgrowth from adult sensory neurons and impaired pain-like behaviour. J Neurochem 99:1000–1010 10.1111/j.1471-4159.2006.04143.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivins JK, Yurchenco PD, Lander AD (2000) Regulation of neurite outgrowth by integrin activation. J Neurosci 20:6551–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oinuma I, Ishikawa Y, Katoh H, Negishi M (2004) The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science 305:862–865 10.1126/science.1097545 [DOI] [PubMed] [Google Scholar]
- 55.Teramitsu I, White SA (2006) FoxP2 regulation during undirected singing in adult songbirds. J Neurosci 26:7390–7394 10.1523/JNEUROSCI.1662-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT (2004) Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119:1027–1040 10.1016/j.cell.2004.11.040 [DOI] [PubMed] [Google Scholar]
- 57.Goodman M, Grossman LI, Wildman DE (2005) Moving primate genomics beyond the chimpanzee genome. Trends Genet 21:511–517 10.1016/j.tig.2005.06.012 [DOI] [PubMed] [Google Scholar]
- 58.Preuss TM, Caceres M, Oldham MC, Geschwind DH (2004) Human brain evolution: insights from microarrays. Nat Rev Genet 5:850–860 10.1038/nrg1469 [DOI] [PubMed] [Google Scholar]
- 59.Oldham MC, Geschwind DH (2006) Deconstructing language by comparative gene expression: from neurobiology to microarray. Genes Brain Behav Suppl 1 5:54–63 10.1111/j.1601-183X.2006.00195.x [DOI] [PubMed] [Google Scholar]
- 60.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917–925 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- 61.Terzic J, Saraga-Babic M (1999) Expression pattern of PAX3 and PAX6 genes during human embryogenesis. Int J Dev Biol 43:501–508 [PubMed] [Google Scholar]
- 62.Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L (1998) Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2:109–120 10.1016/S1097-2765(00)80119-7 [DOI] [PubMed] [Google Scholar]
- 63.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM (1999) Direct control of the forkhead transcription factor AFX by protein kinase B. Nature 398:630–634 10.1038/19328 [DOI] [PubMed] [Google Scholar]
- 64.Seoane J, Le HV, Shen L, Anderson SA, Massague J (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211–223 10.1016/S0092-8674(04)00298-3 [DOI] [PubMed] [Google Scholar]
- 65.Oldham MC, Horvath S, Geschwind DH (2006) Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA 103:17973–17978 10.1073/pnas.0605938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt TR, Goodman M, Grossman LI (2002) Amino acid replacement is rapid in primates for the mature polypeptides of COX subunits, but not for their targeting presequences. Gene 286:13–19 10.1016/S0378-1119(01)00800-9 [DOI] [PubMed] [Google Scholar]
- 67.Grossman LI, Wildman DE, Schmidt TR, Goodman M (2004) Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet 20:578–585 10.1016/j.tig.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 68.Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas EA, Dean B, Pavey G, Sutcliffe JG (2001) Increased CNS levels of apolipoprotein D in schizophrenic and bipolar subjects: implications for the pathophysiology of psychiatric disorders. Proc Natl Acad Sci USA 98:4066–4071 10.1073/pnas.071056198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA (2007) Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry (http://www.nature.com/mp/journal/vaop/ncurrent/abs/4002011a.html) (electronically published May 1, 2007; accessed July 15, 2007) [DOI] [PMC free article] [PubMed]
- 71.Wang Z, Wassink T, Andreasen NC, Crowe RR (2002) Possible association of a cholecystokinin promoter variant to schizophrenia. Am J Med Genet 114:479–482 10.1002/ajmg.10408 [DOI] [PubMed] [Google Scholar]
- 72.Sanjuan J, Toirac I, Gonzalez JC, Leal C, Molto MD, Najera C, De Frutos R (2004) A possible association between the CCK-AR gene and persistent auditory hallucinations in schizophrenia. Eur Psychiatry 19:349–353 [DOI] [PubMed] [Google Scholar]
- 73.Minato T, Tochigi M, Kato N, Sasaki T (2007) Association study between the cholecystokinin A receptor gene and schizophrenia in the Japanese population. Psychiatr Genet 17:117–119 10.1097/YPG.0b013e328011c02e [DOI] [PubMed] [Google Scholar]
- 74.Toirac I, Sanjuan J, Aguilar EJ, Gonzalez JC, Artigas F, Rivero O, Najera C, Molto MD, de Frutos R (2007) Association between CCK-AR gene and schizophrenia with auditory hallucinations. Psychiatr Genet 17:47–53 10.1097/YPG.0b013e3280298292 [DOI] [PubMed] [Google Scholar]
- 75.Tachikawa H, Harada S, Kawanishi Y, Okubo T, Shiraishi H (2000) Novel polymorphisms of the human cholecystokinin A receptor gene: an association analysis with schizophrenia. Am J Med Genet 96:141–145 [DOI] [PubMed] [Google Scholar]
- 76.Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M (2007) Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA 104:10164–10169 10.1073/pnas.0703806104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Printz DJ, Strauss DH, Goetz R, Sadiq S, Malaspina D, Krolewski J, Gorman JM (1999) Elevation of CD5+ B lymphocytes in schizophrenia. Biol Psychiatry 46:110–118 10.1016/S0006-3223(98)00307-2 [DOI] [PubMed] [Google Scholar]
- 78.McAllister CG, Rapaport MH, Pickar D, Podruchny TA, Christison G, Alphs LD, Paul SM (1989) Increased numbers of CD5+ B lymphocytes in schizophrenic patients. Arch Gen Psychiatry 46:890–894 [DOI] [PubMed] [Google Scholar]
- 79.Qu M, Tang F, Yue W, Ruan Y, Lu T, Liu Z, Zhang H, Han Y, Zhang D, Wang F, et al (2007) Positive association of the disrupted-in-schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am J Med Genet B Neuropsychiatr Genet 144:266–270 [DOI] [PubMed] [Google Scholar]
- 80.Porteous DJ, Millar JK (2006) Disrupted in schizophrenia 1: building brains and memories. Trends Mol Med 12:255–261 10.1016/j.molmed.2006.04.009 [DOI] [PubMed] [Google Scholar]
- 81.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, et al (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9:1415–1423 10.1093/hmg/9.9.1415 [DOI] [PubMed] [Google Scholar]
- 82.Noble EP (2003) D2 dopamine receptor gene in psychiatric and neurologic disorders and its phenotypes. Am J Med Genet B Neuropsychiatr Genet 116:103–125 10.1002/ajmg.b.10005 [DOI] [PubMed] [Google Scholar]
- 83.Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, Edenberg HJ, Kuczenski R, Geyer MA, Nurnberger JI Jr, et al (2007) Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet 144:129–158 [DOI] [PubMed] [Google Scholar]
- 84.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K (2007) Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry 62:711–721 10.1016/j.biopsych.2006.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reif A, Herterich S, Strobel A, Ehlis AC, Saur D, Jacob CP, Wienker T, Topner T, Fritzen S, Walter U, et al (2006) A neuronal nitric oxide synthase (NOS-I) haplotype associated with schizophrenia modifies prefrontal cortex function. Mol Psychiatry 11:286–300 10.1038/sj.mp.4001779 [DOI] [PubMed] [Google Scholar]
- 86.Shinkai T, Ohmori O, Hori H, Nakamura J (2002) Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Mol Psychiatry 7:560–563 10.1038/sj.mp.4001041 [DOI] [PubMed] [Google Scholar]
- 87.Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang KY, Huganir RL, et al (2005) Bipolar I disorder and schizophrenia: a 440–single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 77:918–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toyota T, Yoshitsugu K, Ebihara M, Yamada K, Ohba H, Fukasawa M, Minabe Y, Nakamura K, Sekine Y, Takei N, et al (2004) Association between schizophrenia with ocular misalignment and polyalanine length variation in PMX2B. Hum Mol Genet 13:551–561 10.1093/hmg/ddh047 [DOI] [PubMed] [Google Scholar]
- 89.Miller CL, Llenos IC, Dulay JR, Barillo MM, Yolken RH, Weis S (2004) Expression of the kynurenine pathway enzyme tryptophan 2,3-dioxygenase is increased in the frontal cortex of individuals with schizophrenia. Neurobiol Dis 15:618–629 10.1016/j.nbd.2003.12.015 [DOI] [PubMed] [Google Scholar]
- 90.Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, Kupfer DJ, Monk TH, Devlin B, Nimgaonkar VL (2006) Association study of eight circadian genes with bipolar I disorder schizoaffective disorder and schizophrenia. Genes Brain Behav 5:150–157 10.1111/j.1601-183X.2005.00147.x [DOI] [PubMed] [Google Scholar]
- 91.Miyaoka T, Seno H, Ishino H (1999) Increased expression of Wnt-1 in schizophrenic brains. Schizophr Res 38:1–6 10.1016/S0920-9964(98)00179-0 [DOI] [PubMed] [Google Scholar]
- 92.Takase K, Ohtsuki T, Migita O, Toru M, Inada T, Yamakawa-Kobayashi K, Arinami T (2001) Association of ZNF74 gene genotypes with age-at-onset of schizophrenia. Schizophr Res 52:161–165 10.1016/S0920-9964(00)00191-2 [DOI] [PubMed] [Google Scholar]
- 93.Sanjuan J, Tolosa A, Gonzalez JC, Aguilar EJ, Perez-Tur J, Najera C, Molto MD, de Frutos R (2006) Association between FOXP2 polymorphisms and schizophrenia with auditory hallucinations. Psychiatr Genet 16:67–72 10.1097/01.ypg.0000185029.35558.bb [DOI] [PubMed] [Google Scholar]