Abstract
Lumbar disc herniation (LDH), degeneration and herniation of the nucleus pulposus of the intervertebral disc (IVD) of the lumbar spine, is one of the most common musculoskeletal diseases. Its etiology and pathogenesis, however, remain unclear. Type XI collagen is important for cartilage collagen formation and for organization of the extracellular matrix. We identified an association between one of the type XI collagen genes, COL11A1, and LDH in Japanese populations. COL11A1, which encodes the α1 chain of type XI collagen, was highly expressed in IVD, but its expression was decreased in the IVD of patients with LDH. The expression level was inversely correlated with the severity of disc degeneration. A single-nucleotide polymorphism (c.4603C→T [rs1676486]) had the most significant association with LDH (P=3.3×10-6), and the transcript containing the disease-associated allele was decreased because of its decreased stability. These observations indicate that type XI collagen is critical for IVD metabolism and that its decrease is related to LDH.
Lumbar disc herniation (LDH), degeneration and herniation of the nucleus pulposus of intervertebral disc (IVD) of the lumbar spine, is one of the most common musculoskeletal diseases.1–3 Its etiology and pathogenesis, however, remain unclear. Genetic factors have been implicated in the etiology of lumbar disc degeneration.4,5 Genetic abnormalities of the extracellular matrix (ECM) are implicated in disc degeneration and LDH. Phenotypes of transgenic mice and human mutations underscore the candidacy of ECM genes as susceptibility genes for LDH.6,7 Several researchers have reported the association of ECM protein genes, including genes for type IX collagen8,9 and aggrecan,10 with lumbar disc disease (LDD). We reported elsewhere that cartilage intermediate layer protein and asporin—ECM proteins highly expressed in IVD, as well as articular cartilage—are implicated in LDD.11,12
Type XI collagen is a cartilage-specific ECM protein important for cartilage collagen fibril formation and for ECM organization.13–16 Type XI collagen is composed of three α-chains, α1(XI), α2(XI), and α3(II), which are encoded by COL11A1, COL11A2, and COL2A1, respectively. The three chains fold into triple-helical heterotrimers to form procollagen, which is secreted into the ECM, where it participates in fibril formation with other cartilage-specific collagens, type II and IX collagens.13 Type XI collagen regulates the diameter of cartilage collagen fibrils. Its N-terminal noncollagenous region limits the appositional lateral growth of the fibril by blocking further accretion of type II collagen.14,15 Chondrodysplasia in mouse (cho) is an autosomal recessive disorder due to a frame-shift mutation of COL11A1.16 The collagen fibrils of cho mice are much thicker than normal.16,17 Thus, type XI collagen has a critical role in the organization of the supramolecular architecture of cartilage collagen.
Type XI collagen is present in IVD, both in the annulus fibrosus and nucleus pulposus,18 but its significance in LDH is not known. Type XI collagen is a quantitatively minor component of cartilage collagen fibrils, but it is essential for the interaction between proteoglycan (PG) aggregates and collagens. It binds with high affinity to PG, which is important in vivo for anchoring cartilage PG to the collagen fibrillar network.19 Mutations in type XI collagen cause various types of chondrodysplasias in human, including Stickler syndrome type II (MIM #604841), Marshall syndrome (MIM #154780), and oto-spondylo-mega-epiphyseal dysplasia (MIM #215150). These disorders are collectively termed “type XI collagenopathies,”20 and all are complicated by abnormalities of the spine, including narrowing of the IVD. In particular, patients with Stickler syndrome have spondylar abnormalities and Schmorl’s node (disc herniation into the vertebral body).21 These human mutations are in vivo evidence that type XI collagen is critical for IVD integrity; thus, the type XI collagen genes are good candidates for the gene that causes LDH.
Here, we present evidence that COL11A1, one of the type XI collagen genes, contributes to the genetic risk of LDH in Japanese. We have observed significant association between LDH and a functional SNP in COL11A1 in independent Japanese populations. COL11A1 was highly expressed in IVD, but its expression was decreased in the IVD of patients with LDH. COL11A1 expression level was inversely correlated with the severity of disc degeneration in patients with LDH, and the transcript containing the disease-associated allele of the SNP was decreased.
Material and Methods
Study Population
All subjects were Japanese who were living in the middle part of the Honshyu island in Japan (table 1). They visited the participating hospitals and received medical examinations. For the initial screening, we recruited 188 case patients with LDD and 179 control subjects. The mean ages of the case and control groups were 26.5 and 58.7 years, respectively. The case group included 58 patients who had no herniation (disc degeneration only) and 130 patients with LDH. The mean age of the LDH case patients was 25.5 years. For the second screening (replication study), we recruited 359 patients with LDH and 286 control subjects. The mean ages of the case and control groups were 41.4 and 69.6 years, respectively. For the third screening, we recruited 334 patients with LDH and 376 control subjects. The mean ages of the case and control groups were 41.8 and 53.9 years, respectively. Subjects for the initial, second, and third screenings were recruited at the participating hospitals in the Toyama, Tokyo, and Kyoto areas, respectively. All LDH case patients had unilateral pain radiating from the back along the femoral or sciatic nerve to the corresponding dermatome of the nerve root with duration of >3 mo. Radiographic examination, including functional four-direction images and magnetic resonance imaging (MRI) (sagittal and axial images obtained with a 1.5-T imaging system), revealed positive findings indicating disc herniation. The degree of disc degeneration was evaluated by MRI and was scored according to Schneiderman’s classification.22 Of the affected individuals, 787 case patients underwent surgical treatment, and the other individuals with LDH were treated conservatively. All were followed up for >1 year. We excluded from the study individuals with spinal canal stenosis, spondylolisthesis, spondylosis, synovial cysts, spinal tumor, and trauma. We also excluded those who had occupational and/or habitual risk factors, such as heavy manual laborers, occupational drivers, and heavy smokers. We obtained informed consent from each subject, as approved by the ethical committees at the SNP Research Center of RIKEN and the participating hospitals.
Table 1. .
Clinical Characteristics of Subjects
| Age(years) |
|||||
| Screening and Group |
No. of Subjects | Mean ± SD | Range | Male (%) |
BMIa |
| 1st: | |||||
| Case: | |||||
| LDDb | 188 | 26.5 ± 10.4 | 13–74 | 40.0 | 21.0 |
| LDH only | 130 | 25.5 ± 6.9 | 13–66 | 54.0 | 21.1 |
| Control | 179 | 58.7 ± 11.7 | 23–81 | 6.0 | 23.0 |
| 2ndc: | |||||
| Case | 359 | 41.4 ± 14.6 | 15–77 | 62.4 | 23.1 |
| Control | 286 | 69.6 ± 9.2 | 38–87 | 58.4 | 24.3 |
| 3rdc: | |||||
| Case | 334 | 41.8 ± 15.1 | 11–83 | 61.3 | 23.4 |
| Control | 376 | 53.9 ± 9.7 | 13–86 | 47.6 | 22.2 |
BMI calculated as body weight in kilograms divided by the square of height in meters.
Includes disc degeneration only and LDH.
Case group in the 2nd and 3rd screenings has LDH only.
Genotyping
We selected sequence variations of the type XI collagen genes (COL11A1, COL11A2, and COL2A1) for the first screening from the International HapMap Project database and JSNP Database. The SNPs covered >90% of the alleles with an r2 value ⩾0.8. We identified additional sequence variations in COL11A1 by direct sequencing of a 230-kb region of DNA from 24 case patients. We extracted genomic DNA for genotyping from peripheral blood leukocytes of the subjects and genotyped SNPs as described elsewhere.11,12
Haplotype Structure and Statistical Analyses
We estimated haplotype frequencies, using the expectation-maximization algorithm and pairwise linkage-disequilibrium index (D′ and Δ in 465 control individuals, as described elsewhere).23 χ2 tests were used to compare cases with controls for allelic and genotypic frequencies; the odds ratio (OR) and its 95% CI were calculated. We used a permutation test to adjust significance in the analysis of association between the COL11A1 SNPs and LDH.24 We performed 107 permutations of the cases and the controls. Bonferroni correction was applied when significance was adjusted for the number of SNPs genotyped. MRI data, real-time PCR data, and mRNA stability data were tested using Student’s t test.
Analysis of COL11A1 Expression
We extracted and purified total RNAs and synthesized randomly primed cDNAs, using Multiscribe reverse transcriptase (PE Applied Biosystems). We performed quantitative real-time PCR using the ABI PRISM 7700 (Applied Biosystems) and QuantiTect SYBR Green PCR (QIAGEN) according to the manufacturer’s instructions.
RNA Stability Assay
We amplified by PCR ∼1,700-bp of COL11A1 cDNA that contained the entire ORF. We cloned the COL11A1 cDNA containing the associated SNP c.4603C→T into pCR-Blunt II-TOPO (Invitrogen) and confirmed the sequence of the inserts. Vectors were digested using HindIII, and COL11A1 mRNAs were transcribed using RiboMax Large Scale RNA Production System-T7 (Promega) and were purified by SV Total RNA Isolation System (Promega). The whole-cell extract was prepared by washing OUMS-27 cells in PBS and resuspending them in an extraction buffer. After incubation on ice for 30 min and microcentrifugation at 4°C, we transferred supernatants to new tubes and stored them at −80°C until use. We mixed and incubated each 5 μg of synthesized RNA and the diluted (1:1,000) whole-cell extract at room temperature for the tested time (5 or 10 min). We stopped the reaction with addition of a formamide dye. The samples were then heated at 95°C for 5 min and were placed on ice immediately. We detected COL11A1 mRNAs of the samples by northern-blot analysis and quantified their signal intensities, using the Esper-Scanner (Epson) and Adobe Photoshop 6.0.
Immunohistochemistry for Type XI Collagen
We processed and embedded tissue samples in paraffin by the AMeX method. We predigested the tissue sections with 500 U/ml of testicular hyaluronidase (Sigma) for 30 min at 37°C. For immunofluorescent visualization, we blocked nonspecific labeling with blocking reagent (DakoCytomation) for 10 min at room temperature and then incubated the sections with the rabbit polyclonal antibody against bovine type XI collagen (1:500) at 4°C overnight. For the staining of the negative control, we applied nonimmune rabbit IgG (DakoCytomation) to the section instead of primary antibody. After washing them with Tris-buffered saline, we incubated the sections with secondary antibody conjugated to horseradish peroxidase–labeled polymer (Envision+ [DakoCytomation]) for 30 min at room temperature. We visualized the immunoreactive products using a diaminobenzidine reagent and counterstained them with hematoxylin.
Results
We first examined the association of the type XI collagen genes (COL11A1, COL11A2, and COL2A1) with LDD, which included patients with and without LDH. We tested tag SNPs that were selected from the JSNP Database and the International HapMap Project database. A comparison of 188 LDD cases and 179 controls revealed no association with any of the SNPs; however, there was a significant association with COL11A1 when cases were stratified on the basis of the presence or absence of LDH (table 2). In a comparison of 130 patients with LDH with 179 controls, one SNP (c.4603C→T [rs1676486]) had a significant association. To confirm the association, we examined another 359 LDH cases and 286 controls for the COL11A1 SNP. Again, we identified the significant association between the SNP and LDH (table 2). Adjusted P=.00030 was obtained by 107 permutations
Table 2. .
Association between LDH and c.4603C→T (rs1676486) in COL11A1
| No. of Cases with Genotype |
No. of Controls with Genotype |
T Allele Frequency |
||||||||||
| Screening and Case Group |
CC | CT | TT | Total No. of Cases | CC | CT | TT | Total No. of Controls | Case | Control | P | OR (95% CI) |
| 1st: | ||||||||||||
| LDD a | 85 | 86 | 17 | 188 | 99 | 67 | 13 | 179 | .31 | .26 | .076 | 1.34 (.97–1.84) |
| LDH only | 55 | 60 | 15 | 130 | 99 | 67 | 13 | 179 | .34 | .26 | .020 | 1.51 (1.07–2.14) |
| 2nd: | ||||||||||||
| LDH only | 149 | 163 | 47 | 359 | 154 | 108 | 21 | 283 | .35 | .26 | .00038 | 1.55 (1.21–1.97) |
Includes disc degeneration only and LDH.
To identify the disease-causing sequence variation, we examined sequence variations in COL11A1 exons and their flanking regions from a public database and by resequencing 24 patients with LDH. A total of 23 sequence variations were identified and were tested for association. SNP c.4603C→T had the most significant association (table 3), which remained significant after Bonferroni correction for multiple testing. We examined whether confounding effects, such as age and sex, affect the associations with LDH and found no relationship between the genotype and these factors (table 4). The association was positive in both sexes (table 5).
Table 3. .
Polymorphisms in COL11A1 and Their Association with LDH[Note]
| No. in the Three Genotype Groupsa |
Allelic Frequency |
Pb |
|||||||
| Location in COL11A1 and Nucleotide Sequence Change |
Amino Acid Change | dbSNP | Case | Control | Case | Control | Allele | Genotyped | OR (95% CI)c |
| IVS1: | |||||||||
| 9284T→C | … | rs1415359 | 423/63/1 | 422/42/1 | .07 | .05 | .068 | .16 | .69 (.47–1.03) |
| IVS6: | |||||||||
| 82274A→C | … | … | 437/49/1 | 424/38/1 | .05 | .04 | .35 | .61 | .82 (.53–1.25) |
| IVS10: | |||||||||
| 90221G→A | … | rs945748 | 426/62/1 | 414/48/1 | .07 | .05 | .29 | .54 | .82 (.56–1.19) |
| IVS11: | |||||||||
| 90406A→G | … | rs3767272 | 396/76/3 | 401/55/0 | .09 | .06 | .032 | .049 | 1.47 (1.03–2.10) |
| IVS20: | |||||||||
| 104122A→T | … | rs2622877 | 438/47/2 | 400/46/0 | .05 | .05 | .94 | .38 | .98 (.65–1.48) |
| IVS26: | |||||||||
| 111262T→C | … | rs2786125 | 428/49/1 | 429/33/1 | .05 | .04 | .11 | .24 | .70 (.45–1.08) |
| IVS41: | |||||||||
| 146354T→C | … | rs1012282 | 425/62/1 | 415/47/1 | .07 | .05 | .24 | .47 | 1.26 (.86–1.84) |
| IVS42: | |||||||||
| 165864A→C | … | rs1841838 | 381/104/3 | 374/84/6 | .11 | .1 | .52 | .27 | 1.10 (.82–1.47) |
| IVS44: | |||||||||
| 169351A→G | … | rs2126643 | 378/100/3 | 373/79/6 | .11 | .1 | .44 | .23 | 1.12 (.84–1.51) |
| 172702C→G | … | rs3767273 | 382/103/3 | 372/84/4 | .11 | .1 | .41 | .5 | .88 (.66–1.19) |
| IVS50: | |||||||||
| 192606G→A | … | rs4908273 | 231/211/43 | 271/167/23 | .31 | .23 | .00023 | .001 | 1.47 (1.20–1.80) |
| Exon 52: | |||||||||
| 193817(c.3968)T→C | L1323P | rs3753841 | 193/230/65 | 238/187/38 | .37 | .28 | .000081 | .00041 | 1.47 (1.21–1.79) |
| IVS52: | |||||||||
| 194187T→C | … | … | 218/214/48 | 258/178/26 | .32 | .25 | .00038 | .0016 | .69 (.57–0.85) |
| IVS54: | |||||||||
| 200918A→G | … | rs3767274 | 399/73/4 | 367/86/5 | .09 | .1 | .15 | .34 | .79 (.58–1.08) |
| 206255G→T | … | rs3767275 | 457/30/0 | 442/15/1 | .03 | .02 | .088 | .068 | .60 (.33–1.09) |
| 208970T→A | … | rs1676500 | 443/45/1 | 425/33/1 | .05 | .04 | .29 | .53 | 1.27 (.81–1.99) |
| IVS58: | |||||||||
| 218282C→G | … | … | 431/46/1 | 430/32/1 | .05 | .04 | .15 | .32 | .72 (.46–1.13) |
| Exon 62: | |||||||||
| 219597(c.4603)C→T | P1535S | rs1676486 | 204/223/62 | 252/177/33 | .35 | .26 | .000015 | .000099 | 1.54 (1.27–1.88) |
| Exon 63: | |||||||||
| 221284(c.4770)C→T | I1590I | rs2229783 | 169/236/83 | 214/201/47 | .41 | .32 | .000028 | .00017 | 1.49 (1.24–1.80) |
| IVS63: | |||||||||
| 221659G→A | … | rs1463048 | 169/235/83 | 212/199/50 | .41 | .32 | .000081 | .00047 | 1.46 (1.21–1.76) |
| IVS65: | |||||||||
| 225275T→A | … | rs3753844 | 207/223/55 | 239/186/33 | .34 | .28 | .0014 | .0056 | 1.38 (1.13–1.68) |
| Exon 67 (3′ UTR): | |||||||||
| 230265C→T | … | rs1031820 | 443/45/1 | 430/33/1 | .05 | .04 | .27 | .5 | .78 (.50–1.21) |
| 230461A→G | … | … | 439/45/1 | 429/33/0 | .05 | .04 | .17 | .3 | .73 (.46–1.15) |
Note.— The cDNA (accession number NM001854.2) and genomic DNA (accession numbers AC093150.4, AL627203.7, and AC099567.2) sequences of COL11A1 are based on data from GenBank. The A of the ATG translation initiation codon in the reference sequence corresponds to position +1.
Homozygote of the major allele/heterozygote/homozygote of the minor allele.
By the χ2 test.
Calculated for the alleles.
Calculated for the homozygotes of the major allele versus the heterozygotes and the homozygotes of the minor allele.
Table 4. .
Correlation between Age and Genotype at c.4603C→T (rs1676486) in COL11A1
| Mean ± SD Age (in years) for Genotype |
||||
| Population | CC | CT | TT | Pa |
| Case | 36.8 ± 15.0 | 36.9 ± 14.5 | 36.8 ± 14.5 | .58 |
| Control | 64.8 ± 12.1 | 63.9 ± 11.1 | 63.1 ± 13.1 | .54 |
P value was calculated using the Kruskal-Wallis test.
Table 5. .
Genotype at c.4603C→T (rs1676486) in COL11A1, Stratified by Sex
| Male |
Female |
|||||
| Measure | Case | Control | Total | Case | Control | Total |
| No. of subjects: | ||||||
| All | 298 | 177 | 475 | 191 | 285 | 476 |
| CC | 116 | 98 | 214 | 88 | 154 | 242 |
| CT | 144 | 65 | 209 | 79 | 112 | 191 |
| TT | 38 | 14 | 52 | 24 | 19 | 43 |
| T allele frequency (%) | .37 | .26 | .33 | .33 | .26 | .29 |
| P valuea | … | … | .00074 | … | … | .021 |
P value for allelic difference between the patients with LDH and the control groups for each sex, by the χ2 test.
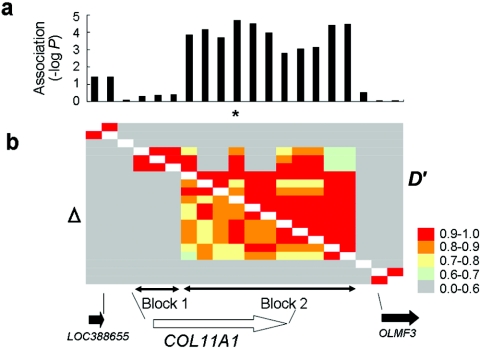
Using the 20 SNPs in and around COL11A1 that had a minor-allele frequency >10%, we analyzed the linkage-disequilibrium structure of the region and found highly structured linkage-disequilibrium blocks (fig. 1). COL11A1 was covered by two blocks, and the SNP with a significant association (c.4603C→T) was contained in block 2. We further analyzed the haplotype structure of block 2 and identified seven haplotypes with frequencies >0.01 that covered >97% of both the case and control groups (table 6). The association was weaker than that of c.4603C→T alone, suggesting the absence of a hidden causal SNP. We further examined the association of the SNP, using an additional 334 patients with LDH and 376 controls. Our findings of the association between this SNP and LDH were replicated (P=.044; OR 1.27 [95% CI 1.01–1.59]. Therefore, this SNP is strongly associated with LDH (combined P=3.3×10-6 in allelic frequency) (table 7).
Figure 1. .
Case-control association study and linkage-disequilibrium mapping. a, Association of COL11A1 with LDH. The −log10 transformation of the corrected P value (allele 1 vs. allele 2) was plotted on the Y-axis. The asterisk (*) indicates c.4603C→T. b, Pairwise linkage disequilibrium between SNPs in and around COL11A1 measured by D′ and Δ in 465 controls. The COL11A1 region is divided into two linkage-disequilibrium blocks.
Table 6. .
Haplotype Association Analysis of COL11A1 with LDH[Note]
| Frequency |
|||
| Haplotype | Case | Control | Pa |
| H1 | .527 | .616 | .000154 |
| H2 | .302 | .222 | .000150 |
| H3 | .038 | .039 | .90 |
| H4 | .041 | .037 | .63 |
| H5 | .045 | .034 | .27 |
| H6 | .014 | .014 | .91 |
| H7 | .011 | .008 | .50 |
Note.— Results are for the haplotypes of block 2 that contained the susceptibility SNP, c.4603C→T.
By the χ2 test.
Table 7. .
Association between Genotype at c.4603C→T (rs1676486) in COL11A1 and LDH in the Japanese Population
| No. with Genotype |
||||||
| Group | CC | CT | TT | Allelic Frequency | P | OR (95% CI) |
| Case | 360 | 367 | 96 | .34 | .0000033 | 1.42 (1.23–1.65) |
| Control | 453 | 325 | 60 | .265 | ||
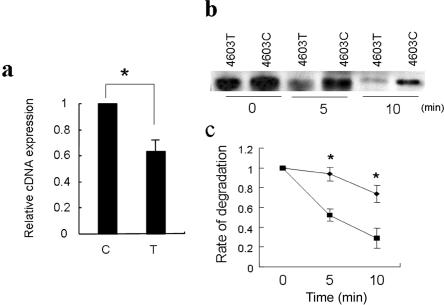
To clarify the functional impact of c.4603C→T, we quantified the allelic difference of the mRNA expression by real-time RT-PCR. The expression level of the susceptibility allele c.4603T was significantly lower than that of the c.4603C allele (fig. 2a). We hypothesized that this SNP affects COL11A1 transcription by altering mRNA stability and examined the stability of COL11A1 mRNA containing the SNP. We mixed mRNAs produced by in vitro transcription with cell lysate and assessed mRNA degradation by endogenous components of the cells, using northern-blot analysis. The transcript containing the susceptible allele degraded faster (fig. 2b and 2c).
Figure 2. .
Difference in transcription and stability of COL11A1 mRNA containing the LDH-associated SNP. a, Relative cDNA expression of c.4603C→T evaluated by real-time PCR. Data represent the ratios of cDNA to genomic DNA, and expression of the C allele is converted to 1 (an asterisk [*] indicates P<.05, by Student's t test). Data represent the mean ± SD in triplicate assays. b, Sequential change of COL11A1 mRNA analyzed by northern blotting. “4603C” and “4603T” indicate COL11A1 mRNA produced by in vitro transcription with c.4603C and c.4603T, respectively. c, Rate of degradation of the transcripts. Diamonds indicate the transcript with c.4603C; squares indicate the transcript with c.4603T. The difference of the rate of degradation was significant at both 5 min and 10 min after the reaction (an asterisk (*) indicates P<.05, by Student's t test). Data represent the mean ± SD in triplicate assays.
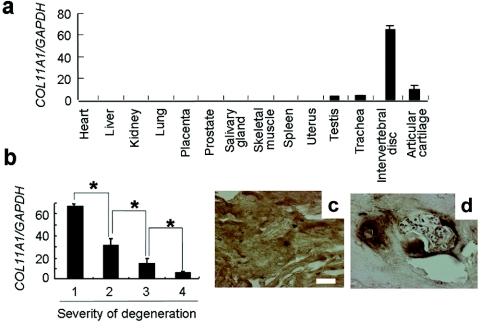
To gain insight into the role of type XI collagen in LDH, we examined COL11A1 expression in tissues and cells by quantitative real-time PCR. COL11A1 mRNA was predominantly expressed in IVD (fig. 3a). We investigated the correlation between the COL11A1 mRNA expression level and a variety of LDH phenotypes and found that severity of disc degeneration evaluated by MRI was inversely correlated with COL11A1 expression in IVDs of patients with LDH (fig. 3b). We further analyzed the expression and localization of type XI collagen in IVD by immunohistochemistry. Normal discs had a highly uniform ECM structure, with intense immunostaining of type XI collagen in the nucleus pulposus cells and ECM (fig. 3c). In degenerative discs, however, we observed weak immunostaining of type XI collagen around the nucleus pulposus cells (fig. 3d). These findings implicate a decrease of type XI collagen in the pathogenesis of LDH.
Figure 3. .
Type XI collagen expression in human. a, COL11A1 expression in different tissues. COL11A1 mRNA was predominantly expressed in IVD. b, Inverse correlation between COL11A1 expression and severity of degeneration of IVD in patients with LDH (an asterisk [*] indicates P<.05, by Student's t test). The degree of disc degeneration is evaluated by MRI and is scored according to the classification of Schneiderman. c and d, Immunostaining of type XI collagen in IVDs from an unaffected individual (c) and a patient with LDH (Schneiderman’s grade 3) (d). Ubiquitous and intense staining was found in the normal disc. In contrast, the staining was found only in and around the territorial matrices of clustered cells in the degenerative disc. The white scale bar indicates 50 nm.
Discussion
Through a case-control association study focusing on type XI collagen, we identified COL11A1 as a susceptibility gene for LDH. COL11A1 mRNA was substantially expressed in IVD, and the expression in patients with LDH was decreased according to the severity of degeneration. Our findings further indicate that the susceptibility SNP produces unstable COL11A1 transcripts. A few cis-elements have been implicated in mRNA stabilization.25 The 4856–4865 nucleotides (caaaaaatct) in COL11A1 mRNA closely match the consensus for a mRNA stability motif, “g/tanaaaag/tcc/t.”26 The sequence variation might affect the mRNA stability motif and disrupt the cis-element critical for mRNA stability, although they are >200 bp apart. Alternatively, the sequence variation might induce a conformational change in the mRNA that would decrease mRNA stability or increase the sensitivity to RNase. The decrease of the COL11A1 transcript would lead to a decrease in type XI collagen in the ECM of IVD.
IVD has a highly structured ECM to resist mechanical forces. The highly oriented network of the fibrillar collagens offers tensile strength,27,28 and highly hydrated aggregating PG resists comprehensive forces. They form a mesh suited to holding water molecules, which further increases their ability to withstand mechanical forces. Therefore, the structural integrity of ECM and the physiologic balance of its components are critical to IVD function. Perturbation of ECM metabolism would increase the mechanical load of the IVD, leading to its degeneration. The reduction in type XI collagen, the critical organizer of ECM, ultimately causes disintegration of ECM and hence IVD degeneration, although it could occur as a secondary event of LDH. The present study underscores the importance of ECM proteins in the pathogenesis of common bone and joint diseases, including LDH. Our results should lead to a better understanding of the pathogenic mechanisms of LDH and suggest promising targets for a novel treatment strategy for LDH.
Acknowledgments
We thank Drs. S. Seki, T. Koyanagi, Y. Fukui, T. Kono, H. Kono, H. Hirabayashi, K. Kono, M. Ishikawa, M. Tamura, K. Nojiri, H. Morisue, and N. Hosogane, for help in collecting samples and performing the experimental study, and Y. Takanashi, for technical assistance. This work was supported by grants-in-aid from Ministry of Education, Culture, Sports and Science of Japan (grant 19209049 [to S.I.]).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Applied Biosystems, http://www.appliedbiosystems.com/index.cfm
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for COL11A1 sequences [accession numbers NM001854.2, AC093150.4, AL627203.7, and AC099567.2])
- International HapMap Project, http://hapmap.org/
- JSNP Database, http://snp.ims.u-tokyo.ac.jp/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Stickler syndrome type II, Marshall syndrome, and oto-spondylo-mega-epiphyseal dysplasia)
References
- 1.Andersson GB (1999) Epidemiological features of chronic low-back pain. Lancet 354:581–585 10.1016/S0140-6736(99)01312-4 [DOI] [PubMed] [Google Scholar]
- 2.Skovron ML (1992) Epidemiology of low back pain. Baillieres Clin Rheumatol 6:559–573 10.1016/S0950-3579(05)80127-X [DOI] [PubMed] [Google Scholar]
- 3.Videman T, Battie MC (1996) A critical review of epidemiology of idiopathic low back pain. In: Weinstein JN, Gordon SL (eds) Low back pain: a scientific and clinical overview. American Academy of Orthopaedic Surgeons, Rosemont, IL, pp 317–332 [Google Scholar]
- 4.Matsui H, Kanamori M, Ishihara H, Yudoh K, Naruse Y, Tsuji H (1998) Familial predisposition for lumbar degenerative disc disease: a case-control study. Spine 23:1029–1034 10.1097/00007632-199805010-00013 [DOI] [PubMed] [Google Scholar]
- 5.Sambrook PN, MacGregor AJ, Spector TD (1999) Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis Rheum 42:366–372 [DOI] [PubMed] [Google Scholar]
- 6.Kimura T, Nakata K, Tsumaki N, Miyamoto S, Matsui Y, Ebara S, Ochi T (1996) Progressive degeneration of articular cartilage and intervertebral discs: an experimental study in transgenic mice bearing a type IX collagen mutation. Int Orthop 20:177–181 10.1007/s002640050058 [DOI] [PubMed] [Google Scholar]
- 7.Watanabe H, Nakata K, Kimata K, Nakanishi I, Yamada Y (1997) Dwarfism and age-associated spinal degeneration of heterozygote cmd mice defective in aggrecan. Proc Natl Acad Sci USA 94:6943–6947 10.1073/pnas.94.13.6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annunen S, Paassilta P, Lohiniva J, Perala M, Pihlajamaa T, Karppinen J, Tervonen O, Kroger H, Lahde S, Vanharanta H, et al (1999) An allele of COL9A2 associated with intervertebral disc disease. Science 285:409–412 10.1126/science.285.5426.409 [DOI] [PubMed] [Google Scholar]
- 9.Paassilta P, Lohiniva J, Goring HH, Perala M, Raina SS, Karppinen J, Hakala M, Palm T, Kroger H, Kaitila I, et al (2001) Identification of a novel common genetic risk factor for lumbar disk disease. JAMA 285:1843–1849 10.1001/jama.285.14.1843 [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi Y, Osada R, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T (1999) Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine 24:2456–2460 10.1097/00007632-199912010-00006 [DOI] [PubMed] [Google Scholar]
- 11.Seki S, Kawaguchi Y, Chiba K, Mikami Y, Kizawa H, Oya T, Mio F, Mori M, Miyamoto Y, Masuda I, et al (2005) A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptility to intervertebral disc disease of the lumbar spine. Nat Genet 37:607–612 10.1038/ng1557 [DOI] [PubMed] [Google Scholar]
- 12.Kizawa H, Kou I, Iida A, Sudo A, Miyamoto Y, Fukuda A, Mabuchi A, Kotani A, Kawakami A, Yamamoto S, et al (2005) An asparatic acid repeat polymorphism in asporin negatively affects chondrogenesis and increase susceptibility to osteoarthritis. Nat Genet 37:138–144 10.1038/ng1496 [DOI] [PubMed] [Google Scholar]
- 13.Keene DR, Oxford JT, Morris NP (1995) Ultrastructural localization of collagen types II, IX, and XI in the growth plate of human rib and fetal bovine epiphyseal cartilage: type XI collagen is restricted to thin fibrils. J Histochem Cytochem 43:967–979 [DOI] [PubMed] [Google Scholar]
- 14.Blaschke UK, Eikenberry EF, Hulmes DJ, Galla HJ, Bruckner P (2000) Collagen XI nucleates self-assembly and limits lateral growth of cartilage fibrils. J Biol Chem 275:10370–10378 10.1074/jbc.275.14.10370 [DOI] [PubMed] [Google Scholar]
- 15.Gregory KE, Oxford JT, Chen Y, Gambee JE, Gygi SP, Aebersold R, Neame PJ, Mechling DE, Bachinger HP, Morris NP (2000) Structural organization of distinct domains within the non-collagenous N-terminal region of collagen type XI. J Biol Chem 275:11498–11506 10.1074/jbc.275.15.11498 [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Lacerda DA, Warman ML, Beier DR, Yoshioka H, Ninomiya Y, Oxford JT, Morris NP, Andrikopoulos K, Ramirez F, et al (1995) A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell 80:423–430 10.1016/0092-8674(95)90492-1 [DOI] [PubMed] [Google Scholar]
- 17.Seegmiller RE, Myers RA, Dorfman A, Horwitz AL (1981) Structural and associative properties of cartilage matrix constituents in mice with hereditary chondrodysplasia. Connect Tissue Res 9:69–77 [DOI] [PubMed] [Google Scholar]
- 18.Oegema TR Jr (1993) Biochemistry of the intervertebral disc. Clin Sports Med 12:419–439 [PubMed] [Google Scholar]
- 19.Smith GN Jr, Williams JM, Brandt KD (1985) Interaction of proteoglycans with the pericellular (1α, 2α, 3α) collagens of cartilage. J Biol Chem 260:10761–10767 [PubMed] [Google Scholar]
- 20.Spranger J (1998) The type XI collagenopathies. Pediatr Radiol 28:745–750 10.1007/s002470050459 [DOI] [PubMed] [Google Scholar]
- 21.Rose PS (2001) Thoracolumbar spinal abnormalities in Stickler syndrome. Spine 26:403–409 10.1097/00007632-200102150-00017 [DOI] [PubMed] [Google Scholar]
- 22.Schneiderman G, Flannigan B, Kingston S, Thomas J, Dillin WH, Watkins RG (1987) Magnetic resonance imaging in the diagnosis of disc degeneration: correlation with discography. Spine 12:276–281 10.1097/00007632-198704000-00016 [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, Ikari K, Furushima K, Okada A, Tanaka H, Furukawa K, Yoshida K, Ikeda T, Ikegawa S, Hunt SC, et al (2003) Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet 73:812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885 10.1038/nature05616 [DOI] [PubMed] [Google Scholar]
- 25.Mullner EW, Kuhn LC (1988) A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell 53:815–825 10.1016/0092-8674(88)90098-0 [DOI] [PubMed] [Google Scholar]
- 26.Capon F, Allen MH, Ameen M, Burden AD, Tillman D, Barker JN, Trembath RC (2004) A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet 13:2361–2368 10.1093/hmg/ddh273 [DOI] [PubMed] [Google Scholar]
- 27.Kempson GE, Muir H, Pollard C, Tuke M (1973) The tensile properties of cartilage of human femoral condyles related to the content of collagen and glycosaminoglycans. Biochem Biophys Acta 297:456–472 [DOI] [PubMed] [Google Scholar]
- 28.Schmidt MD, Mow VC, Chun LE, Eyre DR (1990) Effects of proteoglycan extraction on the tensile behavior of articular cartilage. J Orthop Res 8:353–363 10.1002/jor.1100080307 [DOI] [PubMed] [Google Scholar]