Abstract
We describe the use of antisense morpholino oligonucleotides (AMOs) to restore normal splicing caused by intronic molecular defects identified in methylmalonic acidemia (MMA) and propionic acidemia (PA). The three new point mutations described in deep intronic regions increase the splicing scores of pseudoexons or generate consensus binding motifs for splicing factors, such as SRp40, which favor the intronic inclusions in MUT (r.1957ins76), PCCA (r.1284ins84), or PCCB (r.654ins72) messenger RNAs (mRNAs). Experimental confirmation that these changes are pathogenic and cause the activation of the pseudoexons was obtained by use of minigenes. AMOs were targeted to the 5′ or 3′ cryptic splice sites to block access of the splicing machinery to the pseudoexonic regions in the pre-mRNA. Using this antisense therapeutics, we have obtained correctly spliced mRNA that was effectively translated, and propionyl coenzyme A (CoA) carboxylase (PCC) or methylmalonylCoA mutase (MCM) activities were rescued in patients’ fibroblasts. The effect of AMOs was sequence and dose dependent. In the affected patient with MUT mutation, close to 100% of MCM activity, measured by incorporation of 14C-propionate, was obtained after 48 h, and correctly spliced MUT mRNA was still detected 15 d after treatment. In the PCCA-mutated and PCCB-mutated cell lines, 100% of PCC activity was measured after 72 h of AMO delivery, and the presence of biotinylated PCCA protein was detected by western blot in treated PCCA-deficient cells. Our results demonstrate that the aberrant inclusions of the intronic sequences are disease-causing mutations in these patients. These findings provide a new therapeutic strategy in these genetic disorders, potentially applicable to a large number of cases with deep intronic changes that, at the moment, remain undetected by standard mutation-detection techniques.
In the past few years, special attention has been given in the field of genetic diseases to research on mutations affecting splicing, which generally account for 10%–30% of the total mutant alleles1 and for which novel pharmacological and molecular therapies have begun to be tested.2 Most of the mutations affecting splicing disrupt conserved sequences at the exon-intron junctions—namely, the 5′ donor site, the 3′ acceptor site, the polypyrimidine tract and the branch-point sequence—with different consequences (exon skipping, activation of cryptic splice sites, etc.) depending on the local sequence context.3–5 Some mutations affect less well-conserved auxiliary splicing sequences—that is, exonic and intronic splicing enhancers or silencers—which are recognized by specific SR proteins.6 Other types of mutations, rather than disrupting conserved splice sites, create novel ones that are erroneously used by the splicing machinery, resulting in the generation of aberrant transcripts.4,5 These mutant-activated splice sequences generally occur deep in introns, causing the abnormal inclusion of intron sequences (pseudoexons) in the mRNA.7,8 The true prevalence of this type of mutations is probably underestimated because few laboratories analyze intron sequences far from coding regions, and, in cDNA, the corresponding transcripts (usually with a frameshift and a premature termination codon [PTC]) are degraded by the nonsense-mediated mRNA decay (NMD) mechanism. NMD is a well-conserved mechanism that occurs naturally in cells and that actively degrades PTC-bearing transcripts, thus preventing the generation of truncated proteins that are potentially toxic to cells.9
Alleles with intronic mutations activating cryptic splice sites are particularly amenable to therapeutic correction if use of the aberrant splice sites can be blocked, because the wild-type splice sites remain intact, thus retaining the potential for normal splicing. In this respect, antisense oligonucleotides have been used successfully to restore normal splicing in several disease models, such as β-thalassemia/HbE disorder,10 cystic fibrosis,11 ocular albinism type I,8 and ataxia telangiectasia.12 Antisense oligonucleotides modulate the splicing pattern by steric hindrance of the recognition and binding of the splicing apparatus to the selected cryptic sequences, thus forcing the machinery to use the natural sites. This strategy has also been used in Duchenne muscular dystrophy to force the skipping of a deleterious exon containing a premature stop codon.13
In this work, we report the identification of three novel deep intronic mutations that lead to the insertion of a pseudoexon or cryptic exon in the mRNA of patients with methylmalonic acidemia (MMA [MIM 251000]) or propionic acidemia (PA [MIM 606054]). These are the two most frequent organic acidemias affecting the propionate oxidation pathway in the catabolism of several amino acids, odd-chain fatty acids, and cholesterol.14 Both are life-threatening diseases that appear in the neonatal or infantile period and are caused by different gene defects inherited in autosomal recessive fashion and affecting the synthesis or function of two of the major enzymes of the pathway, propionyl coenzyme A (CoA) carboxylase (PCC [EC 6.4.1.3]) and methylmalonylCoA mutase (MCM [EC 5.4.99.2]), or of their coenzymes (biotin and adenosylcobalamin, respectively). Three consecutive enzymatic reactions are responsible for the conversion of proponylCoA to the succinylCoA that enters the Krebs cycle. The first reaction involves PCC that catalyzes the carboxylation of propionylCoA to d-methylmalonylCoA, which is then converted to the l form by a racemase, and, finally, the MCM enzyme catalyzes the isomerization of L-methylmalonylCoA to succinylCoA.14
Mutations in any of the two genes, PCCA or PCCB, which encode both subunits of the PCC enzyme, cause PA, whereas mutations in the MUT gene, which encodes the MCM enzyme, or in the genes MMAA and MMAB (responsible for the intramitochondrial synthesis of adenosylcobalamin) cause isolated MMA. The molecular bases of these disorders are well known, with >50 different mutations described for each of the PCCA, PCCB, and MUT genes. Missense mutations are the most frequent defects, followed by splicing mutations, which account for 15%–20% of the total alleles.15,16
In this work, we describe three genomic alterations—one in the MUT gene, one in the PCCA gene, and one in the PCCB gene—that are responsible for the aberrant insertion of intronic sequences in patients’ mRNA. The intronic pseudoexons aberrantly inserted in the mRNA were targeted with antisense morpholino oligonucleotides (AMOs) that prevent aberrant splicing, thus generating normal mRNA, which is translated into functional protein, achieving therapeutic correction of the defect.
Material and Methods
Genetic Analysis of Fibroblast Cell Lines
The study included fibroblast cell lines from one Spanish patient with MMA described elsewhere17 and from two patients from Turkey with PA, one PCCA deficient and the other PCCB deficient. Genetic analysis was performed using fibroblast cell lines as the source of mRNA and genomic DNA (gDNA). Total mRNA was isolated by Tripure Isolation reagent (Roche), and subsequent RT-PCR was done using primers and conditions described elsewhere.17,18 The PCR products were sequenced with the same primers used for amplification, with BigDye Terminator v.3.1 mix and subsequent analysis by capillary electrophoresis on an ABI Prism 3700 Genetic Analyzer (Applied Biosystems). BLAST analysis was used to localize the inserted sequence. Intronic gDNA was amplified using primers located in intron 14 (5′-GTAACCCGTTTACTAGTTGCC-3′ and 5′-CACTATAACATACCTGAAGGG-3′) for the PCCA gene insertion, primers located in intron 5 (5′-TATCTTTCCACAGATAATGCCTC-3′) and intron 6 (5′-AAGCAAGGTTTGAGATGAATGG-3′) for the PCCB gene insertion, and primers located in intron 11 (5′-GGCTTCCAGCTTCATCCATG-3′ and 5′-TGGCACGTGCCTGTAGTACC-3′) for the MUT gene insertion. The insertions and gDNA mutations were described as recommended by the Human Genome Variation Society (HGVS). The DNA mutations are numbered on the basis of cDNA sequence and intronic positions described in Ensembl. The genomic changes were studied in 300 control alleles by restriction analysis with use of NlaIII, BsaAI, and BbsI to detect the PCCA, PCCB, and MUT intronic mutations, respectively.
Splice scores of the natural and cryptic donor and acceptor sites were determined using the analysis tools from the Berkeley Drosophila Genome Project (BDGP), and prediction of the presence of exonic splice enhancer or silencer sequences was performed using ESEfinder,6 Rescue-ESE19 (RESCUE-ESE Web Server), and PESX20 (PESXs Server).
Oligonucleotide Treatment and Analysis
The 25-mer AMOs were designed, synthesized, and purified by Gene Tools and were targeted to donor or acceptor cryptic splice sites in the pre-mRNA for each of the intronic inserted sequences in accordance with the manufacturer’s criteria.21 The sequence of the AMOs used is shown in figure 1. Endo-Porter (Gene Tools) was used as the delivery mechanism. For AMO treatment, 4–5×105 fibroblast cells were grown in 6-well plates, and, after overnight culture, different concentrations of AMO with 6-8 μl/ml of Endo Porter were added to the culture medium. Cells were harvested at different times, and mRNA was isolated as described above.
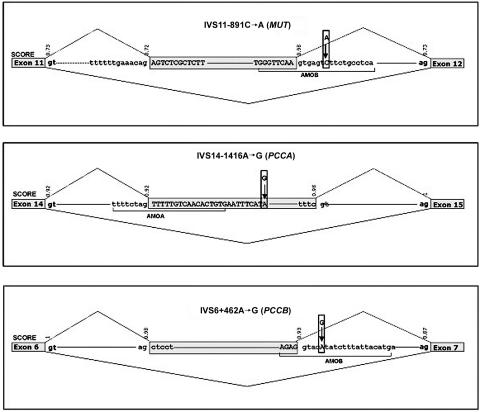
Figure 1. .
Schematic representation of MUT, PCCA, and PCCB regions around the pseudoexons. Exons and pseudoexons are boxed. The inserted intronic sequence is shown in uppercase letters, and the surrounding intronic sequence is in lowercase letters. The sequence of the AMO used is underlined, and splice scores calculated with the BDGP software are denoted above the corresponding 5′ and 3′ splice sites. The mutations are denoted by arrows.
As we have described elsewhere,17 the affected patient with an MUT mutation is compound heterozygous for the intronic insertion and a splicing mutation in the last nucleotide of exon 10 (c.1808G→A), which produces two aberrant transcripts as a result of the use of cryptic splice sites. For RT-PCR in this patient, we used a forward primer placed at the junction of exons 10 and 11 (5′-GCTATCAAGAGGGTTCATAAATT-3′) and a reverse primer located in exon 13 (5′-CTTAGAAGAAGAGATTTT-3′) to amplify only the allele corresponding to the pseudoexon insertion between exons 11 and 12. In some cases, a forward primer placed at the junction of exon 11 and inserted intronic sequence (5′-TCTTTTCCAGAGTCTCGCTCTTT-3′) was used to selectively amplify the cDNA containing the intronic insertion. For RT-PCR analysis of PCCA- and PCCB-deficient cell lines, we used primers described elsewhere.18
PCC activity was assayed as described elsewhere,22 and 14C propionate incorporation into acid-precipitable material was determined in intact cells grown in basal medium as propionate metabolism via MCM.23 For the detection of biotin-bound proteins, fibroblasts were harvested by trypsinization and were freeze thawed, and protein concentration in cell extracts was determined by the Bradford assay. Equal amounts of total protein (30–50 μg) from each sample were loaded onto a denaturing 6% polyacrylamide gel. After electrophoresis, proteins were transferred to PVDF membranes (Immobilon-P [Millipore]), and the biotin-containing proteins were detected with an avidin alkaline phosphatase conjugate as described elsewhere.24
Minigene Construction and in Vitro Splicing Analysis
For evaluation of in vitro splicing, the pSPL3 vector (Life Technologies [Gibco BRL], kindly provided by Dr. B. Andresen) was used. Gene fragments corresponding to each pseudoexon and flanking regions were amplified from patients and from control DNA and were cloned into the TOPO vector (Invitrogen). For the PCCB minigene, the amplified fragment included exon 6. The insert was excised with EcoRI and subsequently was cloned into pSPL3. Clones containing the desired normal and mutant inserts in the correct orientation were identified by restriction-enzyme analysis and automated DNA sequencing. Samples of 2 μg of the wild-type or mutant minigenes were transfected into Hep3B cells by use of Jetpei (Polyplus transfections), in accordance with the manufacturer’s recommendations. At 24–48 h after transfection, cells were harvested, RNA was purified, and RT-PCR analysis was performed using the pSPL3-specific primers SD6 and SA2 (Exon Trapping System [Gibco BRL]). Amplified products were separated by agarose gel electrophoresis and were further analyzed by excising the bands from the gel by means of the Qiaex Gel extraction kit (Qiagen) and subsequent direct sequencing.
Results
Genetic Analysis of Patients
The three patients with MMA and PA exhibited aberrantly spliced mRNA with an amplified band that was larger than normal after RT-PCR analysis. Direct sequencing of the products obtained by RT-PCR and subsequent BLAST search revealed that the insertions corresponded to intronic sequences flanked by cryptic 5′ and 3′ splice sites, thus resembling a pseudoexon (fig. 1). In all three patients the presence of intronic mutations presumably activates the pseudoexon (table 1), whereas the naturally used adjacent splice sites of the surrounding exons remain functional (fig. 1). These intronic variants were not present in the National Center for Biotechnology (NCBI) dbSNP, and none were found in 300 control alleles analyzed.
Table 1. .
Inserted Intronic Sequences, Genomic Intronic Variations, and in Silico Analysis of Genomic Change
| Gene (IDa) | Origin of Inserted Sequence | mRNA Changeb | gDNA Changeb | In Silico Effect of gDNA Changec |
| MUT (4594) | Intron 11 | r.1957ins76 | C→A in position +7 of 5′ donor site of pseudoexon (IVS11-891C→A) | Increase in splicing score (from .98 to .99) |
| PCCA (5095) | Intron 14 | r.1284ins84 | A→G in the middle of pseudoexon (IVS14-1416A→G) | Creates SRp40 binding site and eliminates SRp55 binding site |
| PCCB (5096) | Intron 6 | r.654ins72 | A→G in position +5 of 5′ donor site of pseudoexon (IVS6+462A→G) | Increase in splicing score (from .93 to 1) in 5′ donor site of pseudoexon |
Gene identification (ID) numbers correspond to the NCBI Entrez Gene database.
In the MUT-deficient affected patient, we had previously detected a 76-bp insertion between exons 11 and 12 (r.1957ins76) in the heterozygous state and corresponding to an exon-like region in intron 11.17 In controls and patients with other mutations, the insertion transcript could be detected using a specific primer.17 The patient’s DNA was found to have a new C→A change in intron 11 at position +7 relative to the inserted sequence (IVS11-891C→A) (fig. 1). The scores of the 5′ and 3′ cryptic splice sites were 0.72 and 0.98, respectively, and the C→A mutation increased the 5′ cryptic splice site to 0.99. Additional in silico analysis revealed no significant changes in splicing regulatory sequences caused by the mutation (no exonic splicing enhancer [ESE] or exonic splicing suppressor [ESS] predicted by Rescue-ESE and PESX programs and just a slight decrease in the scores for SC35, SRp40, and SRp55 by ESEfinder analysis).
In the PCCA-deficient affected patient, we have identified an 84-bp insertion between exons 14 and 15 of the PCCA gene (r.1284ins84) in a homozygous state. This 84-bp insertion corresponds to a pseudoexon in intron 14 and is readily detected in patients with mRNA-destabilizing mutations and even at low levels in control cell lines25 but never in a homozygous state. No other sequence changes were detected in the amplified cDNA from the patient. The pseudoexon was amplified from gDNA of the patient and was sequenced, revealing an A→G substitution (IVS14-1416A→G) in the middle of the inserted sequence. In silico analysis with ESEfinder prediction software showed that the change created an SRp40 binding site and eliminated an SRp55 binding site. Rescue-ESE predicted loss of an ESE sequence and creation of two novel ones. No change was predicted by the PESX program.
In the PCCB-deficient affected patient, we have identified a new 72-bp insertion between exons 6 and 7 in the PCCB gene (r.654ins72) in a homozygous state, corresponding to an intron 6 region resembling an exon with 3′ and 5′ splice sites with high scores (fig. 1). Direct sequencing of the genomic region identified an A→G substitution at position +5 relative to the inserted sequence (IVS6+462A→G), increasing the cryptic 5′ donor splicing score from 0.93 to 1. Additional in silico analysis predicted creation of an SRp55 site (ESEfinder) and elimination of a putative silencer (PESX). Rescue-ESE predicted no changes.
Functional Analysis of the Intronic Changes
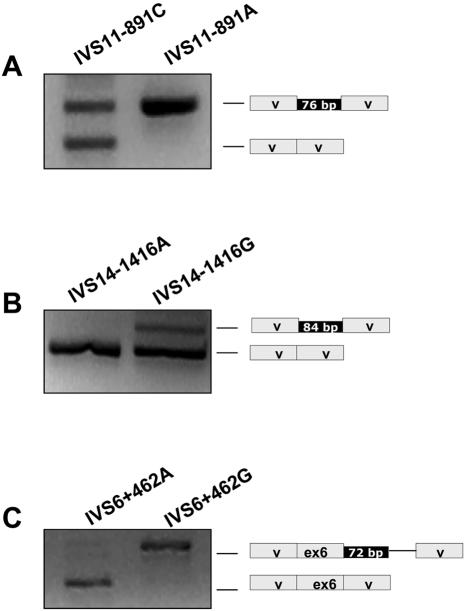
To provide evidence that the observed intronic changes are the cause of the pseudoexon inclusion in the patients’ mRNA, the splicing pattern associated with these changes was further evaluated using minigenes. Minigene constructs with wild-type and mutant pseudoexons and flanking sequences were generated in the pSPL3 vector. The results of splicing analysis after transfection in Hep3B cells are shown in figure 2. The wild-type PCCA and PCCB minigene constructs showed practically total absence of pseudoexon inclusion. The MUT wild-type minigene produced two bands corresponding to the pseudoexon insertion and to a splicing event between the vector splice sites. The mutant constructs resulted in the pseudoexon inclusion in all cases, representing the major transcript for the MUT and PCCB minigenes and part of the transcripts for the PCCA minigene. Sequence analysis confirmed the identity of the transcripts in all cases. The major transcript obtained from the PCCB mutant minigene corresponds to the pseudoexon inclusion along with the insertion of a vector sequence caused by the cloning-derived creation of a 3′ splice acceptor site, which was chosen along with a cryptic 5′ splice site present in the vector (Exon Trapping System [Life Technologies]).
Figure 2. .
Splicing assay for the wild-type and mutant minigenes corresponding to the MUT (A), PCCA (B), and PCCB (C) intronic changes. The results of the RT-PCR analysis using vector-specific primers are shown along with the schematic representation of the transcripts obtained, which were characterized by sequence analysis. V = vector exonic sequences. The solid line in panel C corresponds to a cryptic exon generated during the cloning process.
Restoration of Correct Splicing in MUT-, PCCA-, and PCCB-Mutated Fibroblast Cell Lines
To demonstrate that these changes are disease-causing mutations in the patients and to try to rescue PCCA, PCCB, and MUT expression in the patients’ fibroblasts, we have investigated the possibility of redirecting transcript processing by modified AMOs. The AMOs used were complementary to the 5′ or 3′ cryptic splice sites of the intronic sequences inserted (fig. 1), to block access of the splicing machinery to the pre-mRNA. In all three cases, RT-PCR analysis demonstrated that the AMO used abolished the alternatively spliced transcript induced by the mutations (fig. 3).
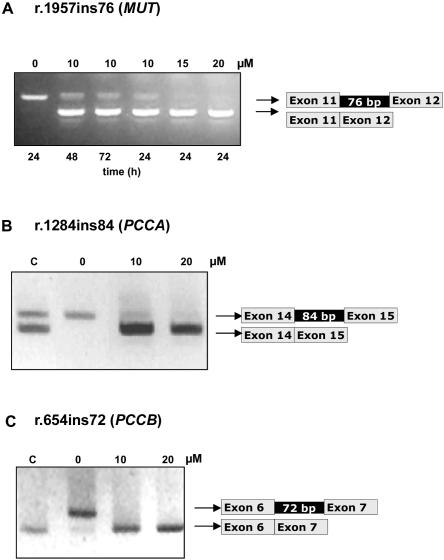
Figure 3. .
Correction of aberrant splicing of MUT, PCCA, and PCCB genes by AMO targeted to the pseudoexon 5′ or 3′ splice sites. A, RT-PCR analysis of the MUT-mutated cell line of total RNA extracted from untreated cells (0 μM) and treated for different times with the specified amounts of AMO targeted to the 5′ cryptic splice site. The sense primer used is located at the junction of exons 10 and 11, and the reverse primer is located in exon 13. B, RT-PCR analysis of the PCCA-deficient cell line untreated or treated for 72 h with 10 or 20 μM of the corresponding AMO targeted to the 3′ splice site. C, RT-PCR analysis of the PCCB-deficient cell line untreated or treated for 72 h with 10 or 20 μM of AMO targeted to the 5′ splice site. Lane C, Control cell line.
The optimal conditions for AMO treatments were determined in each fibroblast cell line. The exclusion of intronic sequences in all cases was oligonucleotide sequence–specific, since AMO targeted to another gene had no effect on pseudoexon inclusion (data not shown). In untreated cell lines, practically no correctly spliced mRNA was detected, as shown by the lack of band comigrating with that of the control one (fig. 3). The correctly spliced fragment was generated 24 h after treatment of the cell lines with AMO complexed with peptide carrier. The products of aberrant splicing were either absent or present at much lower levels (fig. 3). The identity of the PCR products was always confirmed by sequencing.
In the MUT-mutated fibroblast cell line, AMO targeted to the 5′ splice site prevented the inclusion of the intronic sequence 24 h after treatment. The experiments showed dose-dependent correction of splicing (fig. 3A). The larger band containing the intronic insertion was detected with 10 μM AMO but not with 15 and 20 μM AMO. Using a primer located in the junction of exon 11 and the inserted pseudoexon to rescue the larger band, we observed essentially the same results (data not shown).
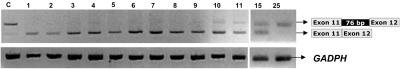
To test the stability of the restored, correctly spliced MUT mRNA, the fibroblast cell line was treated with AMO and was harvested at 1–25 d. In this experiment, the correctly spliced mRNA was still at high levels at 10 d, and trace levels of aberrantly spliced mRNA were obtained. At 15 d, similar amounts of normal and larger fragment were detected, and no normal transcript was obtained at 25 d (fig. 4).
Figure 4. .
Time course of correctly spliced MUT mRNA stability in fibroblasts treated with AMO. Shown is RT-PCR analysis using primers to amplify MUT and GAPDH genes before treatment and after treatment for up to 25 d with 10 μM AMO targeted to the 5′ splice site.
In PCCA- and PCCB-mutated fibroblasts, we used one AMO targeted to the 3′ splice site and the 5′ splice site of the pseudoexon, respectively, and analysis was performed 72 h after AMO delivery. In PCCA RT-PCR analysis, low levels of the larger band with the 84-bp insertion were detected in the control sample, as has been described elsewhere.25 In the PCCA-deficient patient, the larger band was completely absent when fibroblasts were exposed to 20 μM of AMO (fig. 3B). In the PCCB-deficient patient, the larger band also completely disappeared at both concentrations of AMO (fig. 3C).
Restoration of Enzymatic Activity in MUT-, PCCA-, and PCCB-Mutated Fibroblast Cell Lines
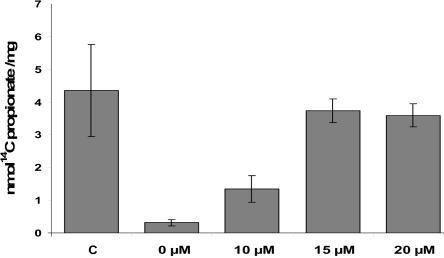
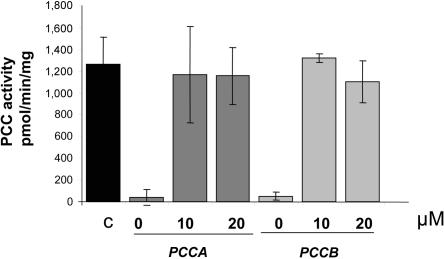
To determine whether the RT-PCR studies correlated with restoration of activities, we measured incorporation of 14C-propionate to acid-precipitable material (as determination of MCM activity) and PCC activity in MUT-mutated and PCC-deficient cell lines, respectively. PCC and MCM activities were rescued in the corresponding patients’ fibroblasts 48–72 h after treatment with AMO. In the MUT-mutated heterozygous patient, dose-dependent activity was rescued at 48 h (fig. 5). The 14C-propionate incorporation levels were close to 40% compared with control levels by use of 10 μM AMO and were close to 100% by use of 15 μM and 20 μM of AMO. In PCC-deficient cell lines, maximum activities were obtained 72 h after AMO delivery, reaching control levels of PCC activity (fig. 6). No effect on MCM or PCC activities was obtained after AMO treatment in cell lines bearing different mutations and exhibiting some levels of intronic MUT17 or PCCA gene insertions.
Figure 5. .
Functional correction of MCM activity after AMO treatment. MCM activity was measured by incorporation of [14C] into trichloroacetic acid–precipitable material in the control cell line (C) and the MUT-mutated cell line untreated or treated with 10, 15, or 20 μM of AMO targeted to the 5′ cryptic splice site. The cells were harvested 48 h after transfection. The data show the mean±SD from at least four independent experiments, and control data were obtained from four independent cell lines.
Figure 6. .
Functional correction of PCC activity after AMO treatment. PCC activity was measured 72 h after transfection with 0, 10, or 20 μM of the corresponding AMO in PCCA- and PCCB-deficient cell lines. The black bar corresponds to control PCC activity measured in four independent cell lines. Dark-gray bars show PCC activity in PCCA-deficient fibroblasts treated with AMO targeted to the 3′ cryptic splice site. Light-gray bars show PCC activity in PCCB-deficient fibroblasts treated with AMO targeted to the 5′ cryptic splice site. The data show the mean±SD from at least three independent experiments.
Restoration of Biotinylated Protein in PCCA-Mutated Fibroblast Cell Line
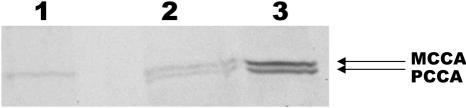
In the PCCA-deficient cell line, detection of biotinylated proteins was performed after treatment with the corresponding AMO. As shown in figure 7, PCCA protein, which is absent before treatment, is detected at close to normal levels 72 h after transfection with 20 μM AMO. The protein is already present 48 h after transfection (data not shown).
Figure 7. .
Recovery of biotinylated PCCA protein after AMO treatment. Biotinylated proteins were detected in total cellular extract by avidin-alkaline phospahatase assay in the PCCA-deficient cell line untreated (lane 1) or 72 h after treatment with the corresponding AMO (lane 2). Lane 3, Hepatoma cellular extract. Shown are the biotin-containing α-subunits of methylcrotonylCoA carboxylase (MCCA) and PCCA.
Discussion
In this work, we report, for the first time, the applicability of antisense therapeutics to the correction of aberrant splicing in two different organic acidemias (three gene defects), with recovery of functional protein and activity within the therapeutic range. The targeted sequences correspond to intronic sequences resembling an exon that are inserted in the mature mRNA, resulting in a PTC-bearing transcript predictably encoding a nonfunctional protein. The fact that prevention of the aberrant inclusion of the pseudoexons by use of AMO results in the recovery of enzymatic activity confirms that the insertions are the disease-causing mutations in the patients.
The in silico analysis of the pseudoexon 3′ and 5′ splice sites predict high scores in each case (fig. 1), thus suggesting that such sequences may be included in the mature mRNA. This prediction holds true for the PCCA gene insertion, which is detected with standard RT-PCR conditions at very low levels in control cell lines and at higher levels in samples from patients with frameshift or nonsense mutations that result in PTC-bearing transcripts degraded by NMD.25 The MUT gene insertion can also be rescued in control cell lines by use of a specific primer, suggesting that it may be a normally rare transcript part of the “background” noise of the splicing process.17 Potential 3′ and 5′ splice sites are highly abundant in intronic sequences, although they alone are insufficient to dictate exon recognition.26 The fact that pseudoexon inclusion is not a frequent event during normal splicing has been attributed to defects in splicing regions, despite their apparently good consensus values, and to the enrichment in splicing silencers.4 However, in the pseudoexons described here, a single point mutation activates the pseudoexon, thus pointing to their high resemblance to true exons. This raises the possibility that these pseudoexons might be part of a splice-controlling mechanism in which alternative splicing events can regulate gene expression by inducing the inclusion of the apparent pseudoexon and leading to NMD, as described for the tropomyosin gene.27
In the MUT-mutated and PCCB-deficient patients included in this study, a point mutation raises slightly the splicing score of the 5′ splice site of the corresponding pseudoexon, resulting in its aberrant inclusion in the mRNA. In the PCCA-deficient patient, the only change identified in the pseudoexonic region corresponds to an A→G change in the middle of the pseudoexon. Analysis with ESE prediction softwares (ESEfinder and Rescue-ESE) identified loss of an ESE and creation of novel ESE sequences, specifically a novel binding site for SRp40 (ESEfinder). This, in conjunction with the high-score cryptic 3′ and 5′ sites, most likely favors the pathogenic intronic inclusion. SRp40 knock-down experiments will also help to clarify the underlying mechanism.
In all three cases, evidence that the pseudoexon insertion is caused by the identified intronic mutations is provided by the results obtained with minigenes. The mutant constructs resulted in pseudoexon inclusion. In the MUT wild-type minigene, some amount of transcript with the insertion is detected, as would be expected given that it is also detectable in control cell lines. However, for PCCA and PCCB wild-type minigenes, practically no transcript with the pseudoexon insertion is detected. In the PCCA-mutant minigene, only part of the resulting transcripts corresponds to the insertion. This does not exactly mimic the results obtained in patients’ fibroblasts, probably because of the lack of a wide-enough genomic context of the cloned pseudoexon, as has been described for exon 37 in the NF1 gene.4,28 A cloning-created cryptic exon derived from the vector, as occurs in the PCCB minigene, is a common occurrence with pSPL3,29 but it is clear that the PCCB pseudoexon is included with the mutant construction but not with the wild-type one, confirming that the change leads to pseudoexon insertion.
The splicing correction by AMO was sequence specific, since control oligomers against another gene had no effect in each case. No obvious cytotoxicity was observed in the three cases. The persistence of correctly spliced MUT mRNA 15 d after termination of AMO delivery suggests that AMO are quite stable in the cell. In all cases, close to 100% of correctly spliced mRNA was detected after standard RT-PCR analysis, although, with a specific primer, the presence of aberrant transcript could be confirmed. This could be because the aberrant transcript is unstable as a result of the presence of PTC, and the normally spliced mRNA is amplified preferentially. In addition, the correctly spliced mRNA can be translated into significant amounts of active protein, reaching control levels, as judged by the detection of biotinylated PCCA protein in one case and enzymatic assays of all three cell lines.
All these results suggest that the antisense approach may be clinically promising for organic acidemias. Quantitative analysis of the restoration of enzymatic activity in a patient to 30%–40% of the normal level (the minimum value we obtained for the heterozygous MUT-mutated patient) is therapeutically significant since heterozygotes are asymptomatic. Interestingly, in both the heterozygous and homozygous patients, control activity levels can be reached with AMO treatment. This suggests that, in the treated-cell population, the amount of functional protein synthesized from the normally spliced mRNA is sufficient to correct the enzymatic defect. Studies in model animals, not feasible to date, would be highly important for discussions of the applicability of these therapies to organic acidemias. To date, treatment of PA and MMA relies on a protein-restricted diet and administration of carnitine and antibiotics, although management is sometimes poor, and long-term complications are common.30 Our results offer a novel mutation-specific therapeutic approach for these diseases, which may be applicable to a greater number of cases than is apparent at the moment, because aberrant intronic insertions may remain undetected with standard mutation-detection techniques. In this sense, we have detected only one mutation in several patients with PA and MMA (laboratory data) who could harbor this type of mutation that results in PTC transcripts that are degraded. Treatment of cell cultures with puromycin to avoid NMD before RNA analysis may reveal additional intronic insertions.
Morpholino analogs of oligonucleotides have several characteristics that render them suitable for therapeutic applications, such as high binding specificity and stability and resistance to nucleases and to degradation by RNAseH when forming hybrids with RNA. The major issues facing clinical applications concern safe delivery and optimal dose determination for each tissue involved. Efficient and nontoxic delivery of AMO to the liver, which would be the target tissue in these diseases, is one major challenge to be overcome before the practical use of AMO in patients with organic acidemia can be envisaged. In Duchenne muscular dystrophy, antisense oligonucleotides have been administered intravenously to patients, achieving splicing modulation to restore the coding frame for dystrophin.31 The efficacy of antisense therapeutics for splicing correction must be determined in each disease model and for each deleterious splicing event, although the results reported to date predict a broad applicability.7,11,12
Acknowledgments
We thank B. Merinero and C. Perez-Cerdá, for their excellent collaboration, and Dr. Gulden Gokcay, for sending the fibroblast samples. This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (SAF2004-06298) and Universidad Autónoma de Madrid–Comunidad Autónoma de Madrid (CCG06-UAM/BIO-0293). The institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa is gratefully acknowledged.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BDGP, http://www.fruitfly.org/seq_tools/splice.html
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Ensembl, http://www.ensembl.org/Homo_sapiens/index.html
- Entrez Gene, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene
- ESEfinder, http://rulai.cshl.edu/tools/ESE2/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for MUT [accession number NT_007592], PCCA [accession number NM_000282.2], and PCCB [accession number NM_000532.3])
- HGVS, http://www.hgvs.org/mutnomen/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PA and MMA) [PubMed]
- PESXs Server, http://cubweb.biology.columbia.edu/pesx/
- RESCUE-ESE Web Server, http://genes.mit.edu/burgelab/rescue-ese/
References
- 1.Krawczak M, Ball EV, Fenton I, Stenson PD, Abeysinghe S, Thomas N, Cooper DN (2000) Human gene mutation database—a biomedical information and research resource. Hum Mutat 15:45–51 [DOI] [PubMed] [Google Scholar]
- 2.Tazi J, Durand S, Jeanteur P (2005) The spliceosome: a novel multi-faceted target for therapy. Trends Biochem Sci 30:469–478 10.1016/j.tibs.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 3.Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54 10.1007/BF00210743 [DOI] [PubMed] [Google Scholar]
- 4.Buratti E, Baralle M, Baralle FE (2006) Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res 34:3494–3510 10.1093/nar/gkl498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pagani F, Baralle FE (2004) Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5:389–396 10.1038/nrg1327 [DOI] [PubMed] [Google Scholar]
- 6.Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- 7.Lacerra G, Sierakowska H, Carestia C, Fucharoen S, Summerton J, Weller D, Kole R (2000) Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc Natl Acad Sci USA 97:9591–9596 10.1073/pnas.97.17.9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetrini F, Tammaro R, Bondanza S, Surace EM, Auricchio A, De Luca M, Ballabio A, Marigo V (2006) Aberrant splicing in the ocular albinism type 1 gene (OA1/GPR143) is corrected in vitro by morpholino antisense oligonucleotides. Hum Mutat 27:420–426 10.1002/humu.20303 [DOI] [PubMed] [Google Scholar]
- 9.Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5:89–99 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- 10.Suwanmanee T, Sierakowska H, Fucharoen S, Kole R (2002) Repair of a splicing defect in erythroid cells from patients with beta-thalassemia/HbE disorder. Mol Ther 6:718–726 10.1006/mthe.2002.0805 [DOI] [PubMed] [Google Scholar]
- 11.Friedman KJ, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R (1999) Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J Biol Chem 274:36193–36199 10.1074/jbc.274.51.36193 [DOI] [PubMed] [Google Scholar]
- 12.Du L, Pollard JM, Gatti RA (2007) Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc Natl Acad Sci USA 104:6007–6012 10.1073/pnas.0608616104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, van Ommen GJ, van Deutekom JC (2003) Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet 12:907–914 10.1093/hmg/ddg100 [DOI] [PubMed] [Google Scholar]
- 14.Fenton WA, Gravel RA, Rosenberg LE (2001) Disorders of propionate and methylmalonate metabolism. In: Scriver CR, Beaudet AL, Sly W, Valle D (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 2165–2190 [Google Scholar]
- 15.Desviat LR, Perez B, Perez-Cerda C, Rodriguez-Pombo P, Clavero S, Ugarte M (2004) Propionic acidemia: mutation update and functional and structural effects of the variant alleles. Mol Genet Metab 83:28–37 10.1016/j.ymgme.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 16.Worgan LC, Niles K, Tirone JC, Hofmann A, Verner A, Sammak A, Kucic T, Lepage P, Rosenblatt DS (2006) Spectrum of mutations in mut methylmalonic acidemia and identification of a common Hispanic mutation and haplotype. Hum Mutat 27:31–43 10.1002/humu.20258 [DOI] [PubMed] [Google Scholar]
- 17.Martinez MA, Rincon A, Desviat LR, Merinero B, Ugarte M, Perez B (2005) Genetic analysis of three genes causing isolated methylmalonic acidemia: identification of 21 novel allelic variants. Mol Genet Metab 84:317–325 10.1016/j.ymgme.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 18.Perez B, Desviat LR, Rodriguez-Pombo P, Clavero S, Navarrete R, Perez-Cerda C, Ugarte M (2003) Propionic acidemia: identification of twenty-four novel mutations in Europe and North America. Mol Genet Metab 78:59–67 10.1016/S1096-7192(02)00197-X [DOI] [PubMed] [Google Scholar]
- 19.Fairbrother WG, Yeh RF, Sharp PA, Burge CB (2002) Predictive identification of exonic splicing enhancers in human genes. Science 297:1007–1013 10.1126/science.1073774 [DOI] [PubMed] [Google Scholar]
- 20.Zhang XH, Chasin LA (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18:1241–1250 10.1101/gad.1195304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morcos PA (2007) Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem Biophys Res Commun 358:521–527 10.1016/j.bbrc.2007.04.172 [DOI] [PubMed] [Google Scholar]
- 22.Suormala T, Wick H, Bonjour JP, Baumgartner ER (1985) Rapid differential diagnosis of carboxylase deficiencies and evaluation for biotin-responsiveness in a single blood sample. Clin Chim Acta 145:151–162 10.1016/0009-8981(85)90282-7 [DOI] [PubMed] [Google Scholar]
- 23.Perez-Cerda C, Merinero B, Sanz P, Jimenez A, Garcia MJ, Urbon A, Diaz Recasens J, Ramos C, Ayuso C, Ugarte M (1989) Successful first trimester diagnosis in a pregnancy at risk for propionic acidaemia. J Inherit Metab Dis Suppl 2 12:274–276 [DOI] [PubMed] [Google Scholar]
- 24.Clavero S, Martinez MA, Perez B, Perez-Cerda C, Ugarte M, Desviat LR (2002) Functional characterization of PCCA mutations causing propionic acidemia. Biochim Biophys Acta 1588:119–125 [DOI] [PubMed] [Google Scholar]
- 25.Campeau E, Dupuis L, Leclerc D, Gravel RA (1999) Detection of a normally rare transcript in propionic acidemia patients with mRNA destabilizing mutations in the PCCA gene. Hum Mol Genet 8:107–113 10.1093/hmg/8.1.107 [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Chasin LA (2000) Multiple splicing defects in an intronic false exon. Mol Cell Biol 20:6414–6425 10.1128/MCB.20.17.6414-6425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grellscheid SN, Smith CW (2006) An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol Cell Biol 26:2237–2246 10.1128/MCB.26.6.2237-2246.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baralle M, Skoko N, Knezevich A, De Conti L, Motti D, Bhuvanagiri M, Baralle D, Buratti E, Baralle FE (2006) NF1 mRNA biogenesis: effect of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett 580:4449–4456 10.1016/j.febslet.2006.07.018 [DOI] [PubMed] [Google Scholar]
- 29.Vockley J, Rogan PK, Anderson BD, Willard J, Seelan RS, Smith DI, Liu W (2000) Exon skipping in IVD RNA processing in isovaleric acidemia caused by point mutations in the coding region of the IVD gene. Am J Hum Genet 66:356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard JV (1995) The management and outcome of propionic and methylmalonic acidaemia. J Inherit Metab Dis 18:430–434 10.1007/BF00710054 [DOI] [PubMed] [Google Scholar]
- 31.Takeshima Y, Yagi M, Wada H, Ishibashi K, Nishiyama A, Kakumoto M, Sakaeda T, Saura R, Okumura K, Matsuo M (2006) Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr Res 59:690–694 10.1203/01.pdr.0000215047.51278.7c [DOI] [PubMed] [Google Scholar]