Abstract
DNA methylation is a heritable modification of genomic DNA central to development, imprinting, transcriptional regulation, chromatin structure, and overall genomic stability. Aberrant DNA methylation of individual genes is a hallmark of cancer and has been shown to play an important role in neurological disorders such as Rett syndrome. Here, we asked whether normal DNA methylation might distinguish individual brain regions. We determined the quantitative DNA methylation levels of 1,505 CpG sites representing 807 genes with diverse functions, including proliferation and differentiation, previously shown to be implicated in human cancer. We initially analyzed 76 brain samples representing cerebral cortex (n=35), cerebellum (n=34), and pons (n=7), along with liver samples (n=3) from 43 individuals. Unsupervised hierarchical analysis showed clustering of 33 of 35 cerebra distinct from the clustering of 33 of 34 cerebella, 7 of 7 pons, and all 3 livers. By use of comparative marker selection and permutation testing, 156 loci representing 118 genes showed statistically significant differences—a ⩾17% absolute change in DNA methylation (P<.004)—among brain regions. These results were validated for all six genes tested in a replicate set of 57 samples. Our data suggest that DNA methylation signatures distinguish brain regions and may help account for region-specific functional specialization.
Epigenetics—the study of information heritable during cell division, other than the DNA sequence itself—might help to explain the mechanism by which one tissue is distinguished from another developmentally.1 Although cells of varying lineage in a single individual share the same DNA sequence, they remember their tissue of origin when they divide. DNA methylation is an epigenetic mark involving a covalent modification of the nucleotide cytosine that occurs in vertebrates at CpG dinucleotides and is generally associated with gene silencing, with notable exceptions, such as some regulatory elements controlling genomic imprinting.2 Disruption of DNA methylation is a hallmark of cancer, with genomewide and gene-specific hypomethylation and hypermethylation of genes and genomic regions.3 DNA methylation is also likely to be important in normal brain development. Rett syndrome (MIM 312750), which involves loss of the normal MeCP2 methylcytosine-recognition protein, causes loss of neurodevelopmental milestones, severe mental retardation, and motor dysfunction.4 Most studies of DNA methylation in normal and disease-affected tissues have been focused on individual genes. However, recent technology allows high-throughput methylation analysis of larger numbers of CpG sites (from hundreds to thousands) across the genome.5
The human brain is a complex organ, and, although a great deal is now known about variations in gene expression that distinguish brain regions, an epigenetic connection to brain anatomy has not been explored. Several studies have examined brain region–specific large-scale gene-expression variation in the mouse. Sandberg et al. found that gene expression in the cerebellum was most distinct when its profile was compared with that of the cerebral cortex, the midbrain, and the hippocampus, with 23 genes expressed uniquely in cerebellum and 28 genes absent there, although expressed in other regions.6 A second study found 1,489–3,220 genes (depending on the threshold used) differentially expressed across five brain regions.7 Finally, a third large-scale study of mouse brain gene expression, The Allen Brain Atlas project, identified region-specific gene expression across 12 brain regions.8 Similarly, microarray studies in human postmortem brain samples have revealed substantial gene-expression differences among the caudate nucleus, cerebellum, and cerebral cortex, as well as differences of smaller magnitude among cerebral cortex regions, including Broca’s area, prefrontal cortex, premotor cortex, primary visual cortex, and anterior cingulate cortex.9 More recently, Roth and colleagues10 profiled 65 distinct human tissues, including 20 CNS tissues. They demonstrated a robust distinction between CNS tissues as a whole and various non-CNS tissues. Furthermore, within the CNS tissues, they discovered region-specific transcriptional expression and a strikingly distinct cerebellar profile, as compared with all other CNS tissues.10 Although much research has elucidated brain region–specific gene-expression differences, no comprehensive survey of methylation in rodent or human brain has yet been attempted.
The recent application of high-throughput technology to the field of epigenetics enables researchers to perform large-scale studies of DNA methylation at thousands of CpG dinucleotides across hundreds of genes. Here, we measured methylation levels at 1,505 CpG sites representing 807 genes, using the Illumina GoldenGate Methylation Cancer Panel I platform,11 to determine whether methylation profiles vary in brain tissues.
Snap-frozen tissues were acquired from the Harvard Brain Tissue Resource Center at McLean Hospital, Belmont, MA, and from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore. A summary of characteristics for the 76 brain samples representing 43 individuals analyzed in our study can be found in table 1. Care was taken to dissect tissue away from white matter and blood vessels. Genomic DNA was isolated for all samples included in the study by use of the MasterPure DNA purification kit (Epicentre Biotechnologies) in accordance with the manufacturer’s specifications. Bisulfite conversion of 500 ng of genomic DNA was achieved through use of the EZ DNA methylation gold kit (Zymo Research). Bisulfite treatment of genomic DNA results in unmethylated cytosine nucleotides being changed to thymidine while methylated cytosines remain unchanged. This difference is then detected as a C/T nucleotide polymorphism at each CpG site. A total of 83 samples (76 brain samples with matched specimens) were run on the Illumina platform, and a β value of 0–1.0 was reported for each CpG site, signifying the percentage of methylation. β values were calculated by subtracting background with use of negative controls on the array and taking the ratio of the methylated signal intensity to the sum of both methylated and unmethylated signals.11 The Illumina GoldenGate methylation assay is reported to be accurate for β value differences ⩾0.17, which was thus chosen as our threshold for methylation differences. Sample quality was assessed by computing mean methylation levels across all samples (0.33), excluding two outliers (four samples with means of 0.22 and 0.40). Array quality was also assessed through linear regression of the correlation of two pairs of replicate samples showing r2 values of 0.991 and 0.982. Computational analysis was performed using the GenePattern12 comparative marker–selection module, to define specific loci with the greatest difference in methylation levels between tissues. In defining our list of significantly different loci, we applied two constraints: (1) P<.004, as determined by comparative marker–selection analysis (with use of a two-sided t test statistic), and (2) mean methylation-level difference across brain regions ⩾0.17.
Table 1. .
Characteristics of 43 Individuals from Whom 76 Brain Samples Were Analyzed
| Characteristic | CBLa (n=34) | CMb (n=35) | PNc (n=7) |
| Aged (years) | 20.6±13.6 | 28.6±22.1 | 64.4±24.3 |
| Sex: | |||
| Female | 6 | 7 | 1 |
| Male | 27 | 27 | 6 |
| Indeterminate | 1 | 1 | 0 |
| PMId,e (h) | 19.2±12.4 | 18.7±10.6 | 20.5±7.4 |
| Race: | |||
| White | 11 | 13 | 0 |
| African American | 12 | 12 | 0 |
| Unknown | 11 | 10 | 7 |
| Cause of death: | |||
| Accident | 8 | 5 | 0 |
| Asphyxia | 1 | 1 | 0 |
| Cardiopulmonary | 6 | 7 | 1 |
| Drowning | 6 | 7 | 0 |
| Epiglottitis | 1 | 1 | 0 |
| Pancreatitis | 1 | 1 | 0 |
| Pneumonia | 0 | 1 | 1 |
| Thermal burns | 0 | 1 | 0 |
| Seizures | 1 | 1 | 0 |
| Suicide | 1 | 1 | 0 |
| Unknown | 9 | 9 | 5 |
| Diagnosis: | |||
| Normal | 18 | 16 | 3 |
| Autism | 16 | 15 | 0 |
| Bipolar | 0 | 4 | 4 |
CBL = cerebellum.
CM = cerebrum.
PN = pons.
Values are given as mean±SD.
PMI = postmortem interval (time from death to tissue freezing).
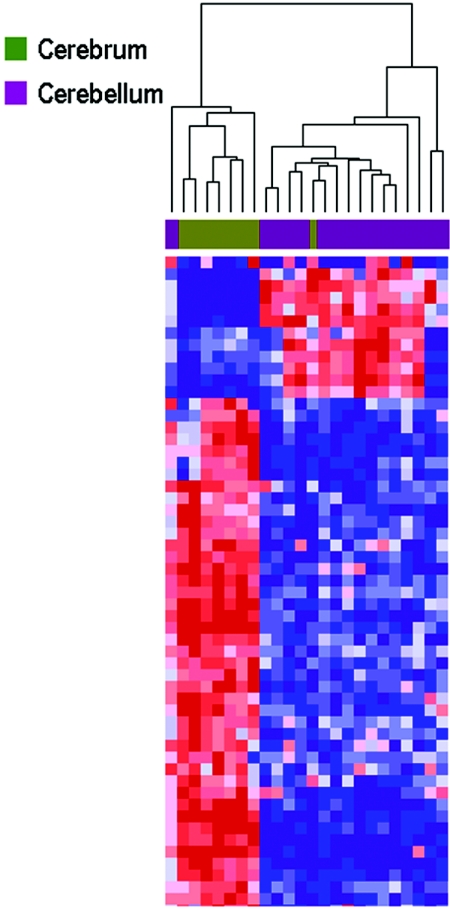
In the first experiment, we examined 8 cerebral cortex samples and 16 cerebellar samples from 24 individuals, analyzing 1,532 loci representing 473 genes from two sources. The first was a group of 380 CpG sites, including those normally found in methylated CpG islands and in GC-rich sequences and those with methylation changes in response to trichostasin A (TSA) or 5-aza-deoxycytidine treatment that we had identified earlier through a systematic genomewide screen.13,14 The second was a group of 1,152 loci from the Illumina Golden Gate Methylation Cancer Panel I.11 The Illumina panel was employed because it has already been validated on human tissue samples, including colon, lung, ovary, breast, and prostate, and the set of genes included in the panel are growth- and development-related and thus might also influence brain development.11 Hierarchical clustering analysis revealed a striking separation of gene methylation between specimens from the two brain regions, with clustering of 7 of 8 cerebral cortex samples and 15 of 16 cerebellar samples (fig. 1). The greatest number of methylation differences was related to brain region rather than to age, sex, postmortem interval, race, diagnosis, or cause of death (fig. 2). Please note that, in the subsequent experiments described below, comparisons were made in the same individual, thereby negating differences due to these other factors. The top 20 differentially methylated probes, with P<.004 and a minimum mean methylation change of 17% across the two tissues, are provided in table 2. Genes showing significant relative hypomethylation in the cerebral cortex compared with the cerebellum included engrailed 2 (EN2 [MIM 131310]) (table 2), which influences cerebellar development15 and may play a role in autism (MIM 209850),16 and HDAC7A (MIM 606542), which encodes part of a family of enzymes that regulate chromatin remodeling in the brain. Among those hypomethylated in cerebellum was HTR2A (MIM 182135), which is epigenetically regulated17 and encodes a serotonin receptor implicated in many neuropsychiatric phenotypes.18 One limitation of this experiment is that the samples were not paired from the same individual and thus could represent interindividual variation.
Figure 1. .
Hierarchical clustering of methylation data from cerebral cortex and cerebellum samples analyzed in experiment 1. Methylation profiles of 1,532 CpG sites from 24 brain samples (16 cerebella and 8 cerebra) from 24 individuals were clustered using uncentered correlation and pairwise average linkage. Columns represent samples; rows correspond to CpG sites. Two major branches are defined by our methylation data and correlate with brain region, one containing 7 of 8 cerebra and one containing 15 of 16 cerebella. A heat map showing relative methylation differences (red indicates more methylated; blue indicates less methylated) from a handful of analyzed loci is represented in the clustering dendrogram.
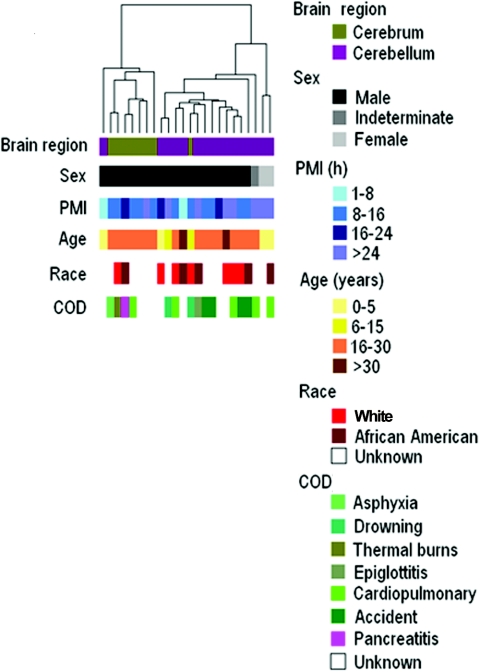
Figure 2. .
Hierarchical clustering of methylation data from cerebral cortex and cerebellum samples analyzed in experiment 1. Methylation profiles of 1,532 CpG sites from 24 brain samples (16 cerebella and 8 cerebra) from different individuals were clustered using uncentered correlation and pairwise average linkage. Columns represent samples; rows are color bars that correspond to sample characteristics. As shown by the color bars, the two major dendrogram branches defined by our methylation data correlate most strongly with brain region, as opposed to age, sex, postmortem interval (PMI), cause of death (COD), or race.
Table 2. .
Loci Demonstrating Significant Differential Methylation (P<.004; β>0.17) between Cerebellum and Cerebral Cortex from Unmatched Individuals
| Cerebellum |
Cerebral Cortex |
||||||
| Feature IDa | Meanb | SD | Meanb | SD | Differenceb | P | False-Discovery Rate |
| SFTPA1_E340_R | .16 | .15 | .73 | .36 | .57 | .002 | .02 |
| RBP1_P426_R | .80 | .08 | .31 | .29 | .48 | .002 | .02 |
| TIMP2_S1512_Fc | .58 | .12 | .18 | .21 | .41 | .002 | .02 |
| EN2_B_S1503_Fc | .52 | .15 | .16 | .24 | .35 | .002 | .02 |
| PRSS8_E134_R | .37 | .09 | .72 | .22 | .35 | .002 | .02 |
| HTR2A_P1387_R | .27 | .07 | .60 | .20 | .33 | .002 | .02 |
| HDAC7A_P344_F | .75 | .03 | .43 | .24 | .33 | .002 | .02 |
| IL16_P93_R | .79 | .07 | .46 | .23 | .32 | .002 | .02 |
| CDH3_P779_Fc | .15 | .05 | .46 | .25 | .31 | .002 | .02 |
| PDGFRB_P273_F | .50 | .22 | .20 | .20 | .30 | .004 | .03 |
| ZP3_P220_F | .24 | .09 | .53 | .21 | .30 | .002 | .02 |
| SLC5A5_E60_F | .78 | .15 | .50 | .23 | .28 | .002 | .02 |
| ASB4_P391_F | .40 | .05 | .68 | .18 | .28 | .002 | .02 |
| GSTM2_P453_R | .76 | .13 | .48 | .22 | .27 | .002 | .02 |
| KCNK4_P257_Fc | .22 | .08 | .49 | .19 | .27 | .002 | .02 |
| BLK_E202_Fc | .08 | .05 | .35 | .22 | .27 | .002 | .02 |
| PTPN6_P126_Rc | .04 | .02 | .31 | .18 | .26 | .002 | .02 |
| APOA1_P261_F | .84 | .04 | .58 | .21 | .25 | .002 | .02 |
| HLA_DPB1_S746_Fc | .17 | .13 | .42 | .17 | .25 | .004 | .03 |
| SERPINA5_P156_F | .53 | .08 | .28 | .14 | .25 | .002 | .02 |
Gene symbols are contained within the Feature ID before the first underscore.
Mean β value (fractional methylation from 0 to 1).
Features not present on the Illumina 1505 Cancer Methylation Panel I array.
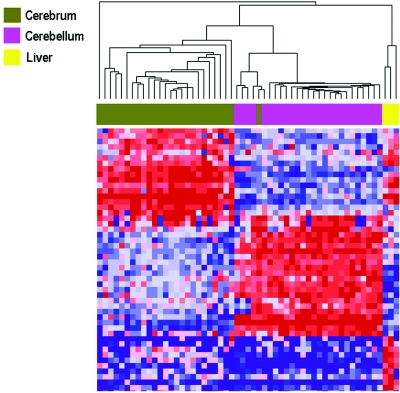
In the second experiment, we tested the potential confounding effect of interindividual variation by examining tissues from 26 individuals, including additional brain regions of 14 individuals from experiment 1, thus providing perfectly matched cerebral cortex and cerebellum, as well as matched liver from three individuals. In this experiment, we examined 1,505 CpGs from 807 genes on the Illumina Golden Gate Methylation Cancer Panel I.11 This set of genes included 621 loci also represented on the array in experiment 1. In the second experiment, there was just as dramatic a separation of gene methylation as that seen in the first experiment, with clustering of 25 of 26 cerebral cortex samples, 26 of 26 cerebellar samples, and all 3 liver samples (fig. 3). Furthermore, analysis of variation (ANOVA) showed that the methylation pattern correlated much more strongly within a brain region across individuals (728 of 1,505 CpG with correlation at P<.05) than within an individual across brain regions (151 loci of 1,505 CpG with correlation at P<.05). Table 3 shows the 20 most differentially methylated probes, whereas table 4 contains a complete list of the 131 that differed significantly. Of the 46 brain-region methylation markers discovered in experiment 1, 32 were present on the second array, and 26 of these were rediscovered as significantly different in experiment 2. Among the genes hypomethylated in cerebral cortex was SLC22A3 (MIM 604842), which encodes an extraneuronal monoamine transporter that inactivates catecholamine neurotransmitters and is thus a candidate gene for neuropsychiatric disease.19 It has been shown to be imprinted in a tissue-specific and temporally restricted fashion.20
Figure 3. .
Hierarchical clustering of methylation data from cerebral cortex, cerebellum, and liver samples analyzed in experiment 2. Methylation profiles of 1,505 CpG sites from 55 samples (26 cerebra, 26 cerebella, and 3 livers) from the same individuals were clustered using uncentered correlation and pairwise average linkage. Columns represent samples; rows correspond to CpG sites. Clustering of 26 of 26 cerebella, 25 of 26 cerebra, and all 3 livers is shown by the dendrogram. A heat map showing relative methylation differences (red indicates more methylated; blue indicates less methylated) from a handful of loci analyzed is represented below the clustering dendrogram. The heat map shows genes with greatest difference between the groups (complete list in table 4).
Table 3. .
Loci Demonstrating Greatest Differential Methylation (P<.004; β>0.17) between Matched Cerebral Cortex and Cerebellum Samples from the Same Individual
| Feature IDa | Cerebellum Meanb | Cerebral Cortex Meanb | Differenceb |
| SFTPA1_E340_R | .18 | .96 | .78 |
| RBP1_P426_R | .98 | .30 | .68 |
| HPN_P823_F | .12 | .79 | .67 |
| MAPK9_P1175_F | .31 | .94 | .63 |
| JAK3_P1075_R | .15 | .77 | .63 |
| BCR_P422_F | .14 | .76 | .62 |
| MYCL2_P19_F | .83 | .22 | .61 |
| ACVR1_P983_F | .12 | .71 | .59 |
| MAPK4_E273_R | .13 | .72 | .59 |
| IL1RN_P93_R | .13 | .67 | .55 |
| IL8_E118_R | .08 | .62 | .54 |
| SLC22A3_P528_F | .90 | .37 | .53 |
| PIK3R1_P307_F | .18 | .70 | .52 |
| FGF3_P171_R | .56 | .04 | .52 |
| PLA2G2A_P528_F | .39 | .90 | .50 |
| CD9_P585_R | .11 | .59 | .48 |
| SERPINE1_P519_F | .07 | .55 | .48 |
| TRIP6_P1090_F | .10 | .57 | .47 |
| LIMK1_P709_R | .11 | .58 | .47 |
| BLK_P14_F | .10 | .56 | .46 |
Gene symbols are contained within the Feature ID before the first underscore.
Mean β value (fractional methylation from 0 to 1).
Table 4. .
Loci Demonstrating Significant Differential Methylation (P<.004; β>0.17) between Cerebellum and Cerebral Cortex from the Same Individual
| Cerebellum |
Cerebral Cortex |
||||||
| Featurea | Meanb | SD | Meanb | SD | Differenceb | P | False-Discovery Rate |
| SFTPA1_E340_R | .18 | .12 | .96 | .07 | .78 | .002 | .007 |
| RBP1_P426_R | .98 | .01 | .30 | .20 | .68 | .002 | .007 |
| HPN_P823_F | .12 | .06 | .79 | .24 | .67 | .002 | .007 |
| MAPK9_P1175_F | .31 | .10 | .94 | .12 | .63 | .002 | .007 |
| JAK3_P1075_R | .15 | .05 | .77 | .16 | .63 | .002 | .007 |
| BCR_P422_F | .14 | .10 | .76 | .17 | .62 | .002 | .007 |
| MYCL2_P19_F | .83 | .07 | .22 | .21 | .61 | .002 | .007 |
| ACVR1_P983_F | .12 | .07 | .71 | .14 | .59 | .002 | .007 |
| MAPK4_E273_R | .13 | .04 | .72 | .15 | .59 | .002 | .007 |
| IL1RN_P93_R | .13 | .03 | .67 | .16 | .55 | .002 | .007 |
| IL8_E118_R | .08 | .04 | .62 | .17 | .54 | .002 | .007 |
| SLC22A3_P528_F | .90 | .05 | .37 | .24 | .53 | .002 | .007 |
| PIK3R1_P307_F | .18 | .05 | .70 | .15 | .52 | .002 | .007 |
| FGF3_P171_R | .56 | .25 | .04 | .12 | .52 | .002 | .007 |
| PLA2G2A_P528_F | .39 | .10 | .90 | .13 | .50 | .002 | .007 |
| CD9_P585_R | .11 | .05 | .59 | .21 | .48 | .002 | .007 |
| SERPINE1_P519_F | .07 | .04 | .55 | .26 | .48 | .002 | .007 |
| TRIP6_P1090_F | .10 | .05 | .57 | .17 | .47 | .002 | .007 |
| LIMK1_P709_R | .11 | .04 | .58 | .17 | .47 | .002 | .007 |
| BLK_P14_F | .10 | .04 | .56 | .21 | .46 | .002 | .007 |
| PDGFRB_P273_F | .69 | .17 | .23 | .16 | .46 | .002 | .007 |
| SPARC_P195_F | .74 | .10 | .30 | .13 | .44 | .002 | .007 |
| STK23_E182_R | .25 | .30 | .68 | .18 | .44 | .002 | .007 |
| SERPINA5_P156_F | .86 | .07 | .42 | .17 | .43 | .002 | .007 |
| TRPM5_P721_F | .28 | .07 | .71 | .18 | .43 | .002 | .007 |
| IL16_P93_R | .91 | .03 | .48 | .15 | .43 | .002 | .007 |
| ACVR1_E328_R | .85 | .10 | .42 | .20 | .43 | .002 | .007 |
| LCN2_P86_R | .43 | .12 | .85 | .19 | .42 | .002 | .007 |
| MPO_P883_R | .16 | .08 | .58 | .17 | .41 | .002 | .007 |
| GSTM2_P453_R | .91 | .09 | .50 | .16 | .41 | .002 | .007 |
| FGF1_E5_F | .12 | .03 | .51 | .16 | .39 | .002 | .007 |
| PDGFRB_E195_R | .61 | .21 | .22 | .15 | .39 | .002 | .007 |
| ZP3_P220_F | .29 | .06 | .68 | .14 | .39 | .002 | .007 |
| PRSS8_E134_R | .52 | .07 | .91 | .08 | .39 | .002 | .007 |
| IL1RN_E42_F | .56 | .10 | .93 | .09 | .38 | .002 | .007 |
| MPL_P657_F | .83 | .15 | .45 | .20 | .38 | .002 | .007 |
| ALOX12_P223_R | .16 | .07 | .54 | .17 | .38 | .002 | .007 |
| FGF1_P357_R | .05 | .02 | .43 | .15 | .38 | .002 | .007 |
| TNF_P158_F | .83 | .09 | .46 | .30 | .37 | .002 | .007 |
| IL6_P213_R | .79 | .06 | .42 | .17 | .36 | .002 | .007 |
| GSTM2_P109_R | .59 | .17 | .23 | .15 | .36 | .002 | .007 |
| PTPN6_P282_R | .56 | .14 | .92 | .10 | .36 | .002 | .007 |
| HDAC7A_P344_F | .96 | .02 | .61 | .20 | .36 | .002 | .007 |
| MYCL2_E44_R | .46 | .09 | .11 | .16 | .35 | .002 | .007 |
| POMC_P400_R | .06 | .05 | .41 | .20 | .35 | .002 | .007 |
| FAS_P322_R | .55 | .13 | .21 | .10 | .35 | .002 | .007 |
| TRIP6_P1274_R | .06 | .04 | .41 | .16 | .34 | .002 | .007 |
| WRN_P969_F | .05 | .03 | .39 | .14 | .34 | .002 | .007 |
| GP1BB_P278_R | .87 | .05 | .53 | .16 | .34 | .002 | .007 |
| SEMA3C_P642_F | .87 | .04 | .54 | .22 | .33 | .002 | .007 |
| JAK3_P156_R | .77 | .07 | .44 | .15 | .33 | .002 | .007 |
| TEK_P526_F | .26 | .06 | .59 | .11 | .33 | .002 | .007 |
| PTPN6_E171_R | .10 | .06 | .43 | .18 | .33 | .002 | .007 |
| STAT5A_E42_F | .96 | .03 | .63 | .12 | .32 | .002 | .007 |
| TEK_P479_R | .29 | .05 | .60 | .16 | .32 | .002 | .007 |
| TGFB3_E58_R | .39 | .09 | .70 | .12 | .31 | .002 | .007 |
| TNK1_P221_F | .08 | .02 | .39 | .15 | .31 | .002 | .007 |
| FGFR3_P1152_R | .75 | .20 | .44 | .15 | .31 | .002 | .007 |
| KRT5_P308_F | .37 | .07 | .68 | .07 | .30 | .002 | .007 |
| ASB4_P391_F | .64 | .05 | .94 | .07 | .30 | .002 | .007 |
| EGF_E339_F | .68 | .10 | .97 | .05 | .29 | .002 | .007 |
| MC2R_P1025_F | .11 | .03 | .40 | .20 | .29 | .002 | .007 |
| PYCARD_P393_F | .06 | .03 | .35 | .11 | .29 | .002 | .007 |
| MMP10_E136_R | .12 | .05 | .41 | .13 | .29 | .002 | .007 |
| MATK_P190_R | .05 | .03 | .34 | .19 | .29 | .002 | .007 |
| SLC5A5_E60_F | .89 | .08 | .61 | .13 | .29 | .002 | .007 |
| GP1BB_E23_F | .85 | .04 | .56 | .18 | .29 | .002 | .007 |
| EVI2A_P94_R | .97 | .02 | .69 | .19 | .28 | .002 | .007 |
| IL16_P226_F | .72 | .17 | .44 | .17 | .28 | .002 | .007 |
| CD40_P372_R | .66 | .12 | .38 | .17 | .28 | .002 | .007 |
| SLC22A2_P109_F | .48 | .10 | .76 | .15 | .28 | .002 | .007 |
| SLC14A1_P369_R | .35 | .12 | .62 | .15 | .27 | .002 | .007 |
| MMP2_P303_R | .06 | .03 | .33 | .17 | .27 | .002 | .007 |
| SNCG_E119_F | .57 | .09 | .30 | .16 | .26 | .002 | .007 |
| HDAC1_P414_R | .92 | .06 | .66 | .11 | .26 | .002 | .007 |
| SEPT5_P464_R | .04 | .02 | .31 | .12 | .26 | .002 | .007 |
| CSF3_P309_R | .93 | .03 | .67 | .12 | .26 | .002 | .007 |
| PADI4_P1158_R | .82 | .06 | .56 | .13 | .26 | .002 | .007 |
| TJP2_P518_F | .02 | .03 | .28 | .17 | .25 | .002 | .007 |
| SOD3_P460_R | .94 | .02 | .69 | .15 | .25 | .002 | .007 |
| PDGFRB_P343_F | .31 | .14 | .06 | .07 | .25 | .002 | .007 |
| PRSS1_E45_R | .51 | .16 | .76 | .08 | .25 | .002 | .007 |
| NPR2_P1093_F | .13 | .05 | .37 | .17 | .25 | .002 | .007 |
| RAN_P581_R | .13 | .05 | .37 | .10 | .25 | .002 | .007 |
| MCF2_E195_F | .22 | .28 | .46 | .20 | .25 | .004 | .012 |
| SEPT9_P374_F | .99 | .01 | .74 | .15 | .25 | .002 | .007 |
| MT1A_P600_F | .37 | .10 | .61 | .14 | .24 | .002 | .007 |
| PTHLH_E251_F | .94 | .02 | .70 | .12 | .24 | .002 | .007 |
| CCR5_P630_R | .28 | .10 | .52 | .16 | .24 | .002 | .007 |
| BMP4_P123_R | .10 | .04 | .33 | .14 | .24 | .002 | .007 |
| BAX_E281_R | .10 | .05 | .34 | .11 | .24 | .002 | .007 |
| APOA1_P261_F | .98 | .01 | .74 | .19 | .23 | .002 | .007 |
| CASP10_P186_F | .87 | .03 | .64 | .12 | .23 | .002 | .007 |
| CD40_E58_R | .56 | .17 | .33 | .17 | .23 | .002 | .007 |
| NOS2A_P288_R | .60 | .17 | .83 | .10 | .23 | .002 | .007 |
| NOS3_P38_F | .66 | .08 | .89 | .09 | .23 | .002 | .007 |
| TRIM29_E189_F | .63 | .07 | .86 | .07 | .23 | .002 | .007 |
| DNAJC15_P65_F | .17 | .12 | .40 | .16 | .23 | .002 | .007 |
| ALOX12_E85_R | .03 | .04 | .25 | .12 | .23 | .002 | .007 |
| RUNX1T1_E145_R | .04 | .03 | .27 | .10 | .22 | .002 | .007 |
| GRB7_P160_R | .03 | .03 | .26 | .14 | .22 | .002 | .007 |
| MST1R_P392_F | .94 | .03 | .73 | .13 | .22 | .002 | .007 |
| DDR2_E331_F | .99 | .00 | .77 | .15 | .21 | .002 | .007 |
| DNMT2_P199_F | .66 | .05 | .45 | .09 | .21 | .002 | .007 |
| PRKCDBP_E206_F | .03 | .02 | .24 | .16 | .21 | .002 | .007 |
| GSTM2_E153_F | .35 | .18 | .14 | .11 | .21 | .002 | .007 |
| RAB32_P493_R | .50 | .09 | .30 | .15 | .21 | .002 | .007 |
| NBL1_E205_R | .52 | .07 | .72 | .13 | .20 | .002 | .007 |
| CD9_P504_F | .03 | .02 | .24 | .11 | .20 | .002 | .007 |
| TEK_E75_F | .85 | .05 | .65 | .12 | .20 | .002 | .007 |
| RARRES1_P426_R | .03 | .03 | .23 | .15 | .20 | .002 | .007 |
| EPHA2_P203_F | .06 | .03 | .26 | .10 | .20 | .002 | .007 |
| VAV1_P317_F | .58 | .07 | .78 | .12 | .20 | .002 | .007 |
| SEPT9_P58_R | .98 | .01 | .79 | .11 | .19 | .002 | .007 |
| CPA4_E20_F | .78 | .04 | .58 | .09 | .19 | .002 | .007 |
| PPARG_P693_F | .85 | .07 | .66 | .16 | .19 | .002 | .007 |
| HRASLS_P353_R | .19 | .08 | .38 | .17 | .19 | .002 | .007 |
| BCR_P346_F | .09 | .06 | .28 | .14 | .18 | .002 | .007 |
| FASTK_P598_R | .27 | .05 | .45 | .11 | .18 | .002 | .007 |
| CDK2_P330_R | .01 | .01 | .19 | .13 | .18 | .002 | .007 |
| TNFRSF10D_E27_F | .03 | .05 | .21 | .14 | .18 | .002 | .007 |
| IGF1_E394_F | .60 | .06 | .42 | .15 | .18 | .002 | .007 |
| MFAP4_P197_F | .02 | .01 | .20 | .14 | .18 | .002 | .007 |
| HBII-13_E48_F | .53 | .10 | .71 | .19 | .18 | .002 | .007 |
| MAPK10_E26_F | .78 | .09 | .60 | .15 | .17 | .002 | .007 |
| CSF1_P339_F | .01 | .01 | .18 | .10 | .17 | .002 | .007 |
| CCKAR_E79_F | .25 | .06 | .43 | .17 | .17 | .002 | .007 |
| EPHA8_P456_R | .60 | .06 | .78 | .11 | .17 | .002 | .007 |
| MAS1_P469_R | .81 | .05 | .98 | .02 | .17 | .002 | .007 |
| CD34_P780_R | .86 | .04 | .69 | .13 | .17 | .002 | .007 |
| CRK_P721_F | .77 | .09 | .60 | .12 | .17 | .002 | .007 |
Gene symbols are contained within the Feature ID before the first underscore.
Mean β value (fractional methylation from 0 to 1).
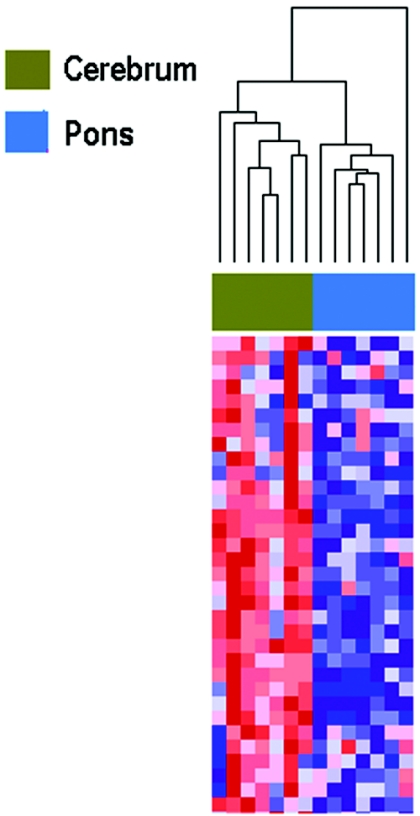
In the third experiment, we again paired brain regions from the same individuals, using seven from whom we had matched cerebral cortex and pons regions, on the same array as that in experiment 2. Again, there was striking separation of gene methylation, with clustering of seven of seven cerebral cortex samples and seven of seven pons (fig. 4). Similarly, ANOVA showed that the methylation pattern correlated much more strongly within a brain region across individuals (292 of 1,505 CpG with correlation at P<.05) than within an individual across brain regions (116 of 1,505 CpG with correlation at P<.05). Thus, methylation consistently distinguished brain regions in a given individual. Table 5 shows the most differentially methylated probes between cerebral cortex and pons. Among the genes hypomethylated in cerebral cortex in this experiment was insulin-like growth factor 1 (IGF1 [MIM 147440]), previously shown to have distinct developmental patterns of expression in differing brain regions.21 Hypomethylated genes in pons included FGF1 (MIM 131220) and FGFR2 (MIM 176943), fibroblast growth-factor system genes that are part of a signaling pathway that plays a role in brain development and differentiation.22
Figure 4. .
Hierarchical clustering of methylation data from cerebral cortex and pons samples analyzed in experiment 3. Methylation profiles of 1,505 CpG sites from 14 brain samples (7 cerebra and 7 pons) from the same individual were clustered using uncentered correlation and pairwise average linkage. Columns represent samples; rows correspond to CpG sites. Two major branches are defined by our methylation data and correlate with brain region: one containing seven of seven cerebra and one containing seven of seven pons. A heat map showing relative methylation differences (red indicates more methylated; blue indicates less methylated) from a handful of loci analyzed is represented below the clustering dendrogram.
Table 5. .
Loci Demonstrating Significant Differential Methylation (P<.004; β>0.17) between Cerebral Cortex and Pons from the Same Individual
| Cerebral Cortex |
Pons |
||||||
| Feature IDa | Meanb | SD | Meanb | SD | Differenceb | P | False-Discovery Rate |
| HTR2A_E10_R | .62 | .06 | .84 | .04 | .22 | .002 | .05 |
| MATK_P190_R | .35 | .08 | .55 | .09 | .20 | .002 | .05 |
| WRN_P969_F | .31 | .05 | .50 | .10 | .19 | .002 | .05 |
| BCR_P346_F | .20 | .05 | .38 | .09 | .19 | .002 | .05 |
| IGF1_E394_F | .42 | .09 | .60 | .04 | .18 | .002 | .05 |
| DKFZP564O0823_P386_F | .39 | .04 | .57 | .07 | .18 | .002 | .05 |
| TNFSF10_E53_F | .54 | .13 | .20 | .05 | .34 | .004 | .05 |
| SERPINE1_P519_F | .62 | .13 | .32 | .05 | .30 | .004 | .05 |
| FGFR2_P460_R | .52 | .12 | .23 | .06 | .30 | .004 | .05 |
| AXIN1_P995_R | .53 | .16 | .82 | .06 | .29 | .004 | .05 |
| CD40_E58_R | .48 | .06 | .20 | .07 | .28 | .004 | .05 |
| SPARC_P195_F | .39 | .09 | .13 | .05 | .26 | .004 | .05 |
| PRKCDBP_P352_R | .72 | .07 | .48 | .07 | .24 | .004 | .05 |
| TJP2_P518_F | .29 | .11 | .07 | .03 | .22 | .004 | .05 |
| ZMYND10_P329_F | .19 | .08 | .39 | .11 | .21 | .004 | .05 |
| CASP10_P186_F | .72 | .06 | .52 | .06 | .20 | .004 | .05 |
| TNF_P158_F | .65 | .07 | .45 | .20 | .20 | .004 | .05 |
| CDK2_P330_R | .20 | .08 | .01 | .01 | .20 | .004 | .05 |
| FGF1_E5_F | .52 | .11 | .32 | .05 | .20 | .004 | .05 |
| CASP10_P334_F | .71 | .09 | .52 | .05 | .19 | .004 | .05 |
| MPO_P883_R | .70 | .05 | .51 | .08 | .18 | .004 | .05 |
| RAB32_P493_R | .53 | .08 | .35 | .06 | .18 | .004 | .05 |
| IL8_E118_R | .60 | .06 | .42 | .07 | .18 | .004 | .05 |
| STAT5A_E42_F | .66 | .06 | .48 | .04 | .18 | .004 | .05 |
| MMP9_P189_F | .88 | .05 | .71 | .04 | .17 | .004 | .05 |
Gene symbols are contained within the Feature ID before the first underscore.
Mean β value (fractional methylation from 0 to 1).
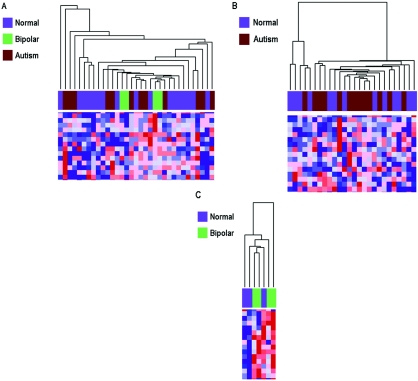
We performed analyses of reliability and of potential confounding, as well as validation experiments, to assess the robustness of our findings. We tested the reproducibility of methylation measurements of the arrays, examining seven samples at 1,505 CpG sites. Linear-regression analysis was performed, and correlation coefficients ranged from 0.94 to 0.99. To control for any effects that might be attributable to disease state, we assessed for differences in DNA methylation between normal and disease-affected samples. We found no correlation, using all 1,505 CpG loci, comparing unaffected individuals (n=16) with those with bipolar disorder (n=4) and autism (n=13) in cerebral cortex (fig. 5A), cerebellum (fig. 5B), or pons (fig. 5C).
Figure 5. .
Hierarchical clustering of methylation data from cerebral cortex, cerebellum, and pons samples analyzed in experiments 2 and 3. Columns represent samples, rows correspond to CpG sites, and heat maps showing relative methylation differences (red indicates more methylated; blue indicates less methylated) from a handful of loci analyzed are represented below the clustering dendrograms. All clustering analyses were performed using uncentered correlation and pairwise average linkage. A, Methylation profiles of 1,505 CpG sites from 33 cerebra samples (16 from normal individuals, 13 from individuals given diagnoses of autism, and 4 from individuals with bipolar disorder) were clustered. Clustering does not reveal disease-specific branches. B, Methylation profiles of 1,505 CpG sites from 26 cerebella samples (13 from normal individuals and 13 from individuals given diagnoses of autism) were clustered. Clustering does not reveal an autism-specific branch. C, Methylation profiles of 1,505 CpG sites from seven pons samples (three from normal individuals and four from individuals with bipolar disorder) were clustered. Clustering does not reveal a bipolar-specific branch.
Finally, we sought to validate the observed methylation data by an independent method, bisulfite pyrosequencing, which measures methylation variation at >90% precision.23 Bisulfite-pyrosequencing validation was performed by bisulfite treatment of genomic DNA (Qiagen Epitect kit), PCR amplification, and pyrosequencing of CpG sites. We obtained sequences for all Illumina probes and designed flanking primers (methylation-unbiased nested PCR and sequencing primers) to the CpG site for which a β value was reported by Illumina (available from the authors on request). Pyrosequencing was performed on a Biotage PSQ HS96 Pyrosequencer. Bisulfite conversion controls and quantitative levels of methylation for each CpG dinucleotide were evaluated with Pyro Q-CpG software.
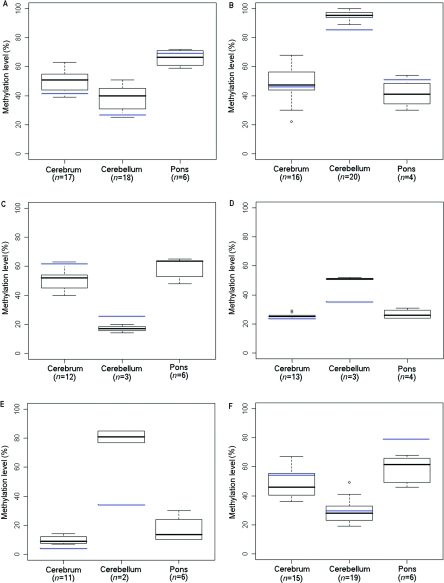
We chose genes for validation that showed a range of variation between tissues on the arrays, including 12%, 24%, and 36% differences. Six genes in 2–20 samples were analyzed for quantitative methylation by bisulfite pyrosequencing. All the loci tested showed substantial differences in DNA methylation across brain regions in the same direction and magnitude as we had found using Illumina arrays (fig. 6). For example, the Illumina data had revealed hypermethylation of HDAC7A in cerebellum compared with cerebrum by a magnitude of 0.33 in experiment 1 and 0.36 in experiment 2; in pyrosequencing, we also saw hypermethylation of cerebellum relative to cerebrum by a magnitude of 0.46. Linear regression was performed, comparing the percentage of methylation reported by pyrosequencing and Illumina β values, and correlation coefficients equaled 0.99 (EN2), 0.89 (HTR2A), 0.96 (GABRB3), 0.72 (MT1A), 0.74 (RASSF1), and 0.76 (HDAC7A).
Figure 6. .
Box plots of methylation data from bisulfite-pyrosequencing analysis. A, RASSF1. B, HDAC7A. C, HTR2A. D, GABRB3. E, EN2. F, MT1A. Mean methylation levels across all Illumina experiments are denoted by blue lines. n is the number of samples analyzed by pyrosequencing.
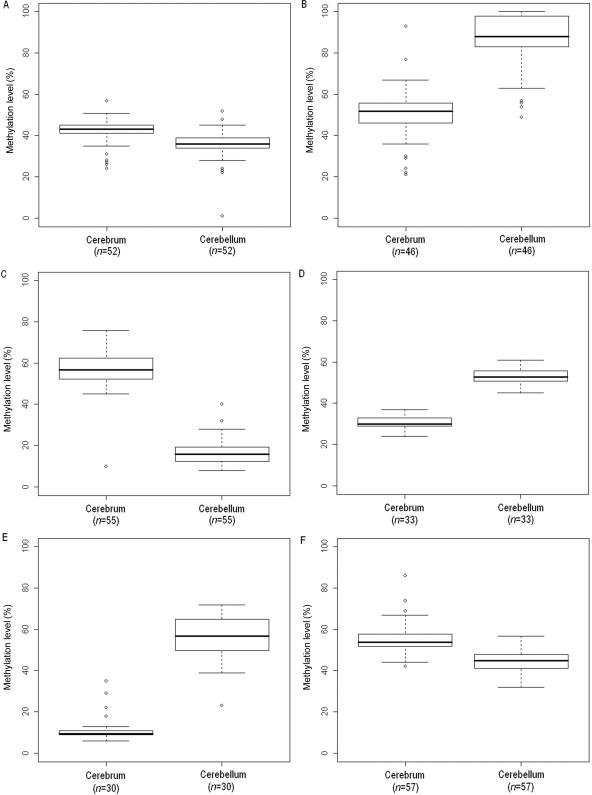
To assess the reproducibility of our methylation results in an independent set of individuals, we obtained snap-frozen brain tissue (donated by The Stanley Medical Research Institute’s brain collection, courtesy of Drs. Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken) and performed bisulfite pyrosequencing of six genes (RASSF1, HDAC7A, HTR2A, GABRB3, EN2, and MT1A) in 52, 46, 55, 33, 30, and 57 paired cerebral cortex and cerebellum samples, respectively. All six of the loci confirmed our initial brain-region methylation findings in this independent set of individuals (fig. 7).
Figure 7. .
Box plots of methylation data from bisulfite-pyrosequencing analysis. A, RASSF1. B, HDAC7A. C, HTR2A. D, GABRB3. E, EN2. F, MT1A. n is the number of samples analyzed by pyrosequencing.
Finally, we examined the pattern of gene expression of five genes, RASSF1, HDAC7A, GABRB3, EN2, and HTR2A. We chose these genes because they represent a diverse set of cellular functions, including regulation of cell proliferation, chromatin structure modification, development, and neurotransmission, and they represent three genes (RASSF1, HDAC7A, and GABRB3) in which the analyzed methylation sites were within the promoter and two genes (EN2 and HTR2A) in which the sites were >1 kb from the promoter. Two of these genes, one in which the CpG locus analyzed was within the promoter (HDAC7A) and one in which it was distal to the promoter (EN2), were not in the commercial Illumina GoldenGate Methylation Cancer Panel I but were added to a custom array on the basis of our identification of genes with altered expression in response to TSA, a chromatin-modifying drug, or 5-aza-deoxycytidine, a drug known to decrease DNA methylation.14 In all three cases in which the methylated sites were within the promoter, the difference in expression was as expected—that is, the more-methylated tissue showed the lower mean expression, although, in one of these cases, the results did not achieve statistical significance (table 6). Interestingly, in both cases in which the methylated sites lay outside the promoter, the more-methylated tissue also showed the greater expression. Similar to our findings that EN2 expression is decreased in brain tissue with less methylation, Gius et al.14 previously discovered that expression of EN2 is down-regulated 1.7-fold in response to TSA, a chromatin-modifying drug that normally results in increased gene expression.
Table 6. .
Quantitative Real-Time PCR Results of Five Genes Assayed in Paired Cerebellum and Cerebral Cortex Samples from 15 Individuals[Note]
| Measure | RASSF1 | HDAC7A | GABRB3 | EN2 | HTR2A |
| Methylationa | CM > CBL | CBL > CM | CBL > CM | CBL > CM | CM > CBL |
| Distanceb | 200 | 200 | 440 | 6500 | 1400 |
| Fold changec | −2.0 | 1.5 | 1.4 | −100 | 40.4 |
| P | .011 | .007 | .210 | <.001 | <.001 |
| Expressiond | CBL > CM | CM > CBL | CM > CBL | CBL > CM | CM > CBL |
Note.— CBL = cerebellum; CM = cerebral cortex.
Methylation level from greatest to least, based on β value.
Distance (in bp) to transcriptional start site from the locus showing differential methylation across brain regions.
Fold change = 2−ΔΔCT; ΔΔCT = (CT cerebrum target gene−CT phosphglycerate kinase 1)−(CT cerebellum target gene−CT phosphglycerate kinase 1).
Quantitative real-time PCR result. For each gene, brain-region expression is ranked from greatest to least.
In summary, we have found a DNA methylation signature that distinguishes three human brain regions. These brain methylation differences correlated much more strongly within a brain region across individuals than within an individual across brain regions. The result is surprising, since the genes analyzed were not preselected for known brain function. They came from a panel of genes previously known to show altered DNA methylation or a functional role in tumor development or progression. Of course, these same genes are themselves implicated generally in normal development and differentiation, and 80% of all genes are expressed in the normal brain.8 A substantial body of evidence shows brain region–specific differences in gene expression,9,10 and the region-specific patterns in DNA methylation shown here may help to explain these functional differences. Furthermore, because methylation variation in tissues is acquired developmentally, the differences in brain methylation may help to determine or stabilize normal brain differentiation. This idea is consistent with the fact that a failure to recognize DNA methylation marks caused by absence of the MECP2 protein in Rett syndrome causes progressive loss of neurodevelopmental milestones, as well as poorly regulated gene expression in affected brain tissue.24
These data highlight several areas for further study. First, the degree of complexity of the brain is greater than that of any other organ. In addition to the gross brain-region distinctions, such as cerebral cortex and cerebellum, the cerebral cortex itself is broadly divided into four lobes and is more finely divided into 47 Brodmann areas. Furthermore, in the classic studies by Mountcastle, the cortex is functionally organized into countless vertical columns ∼300–600 μm wide.25 In addition, cellular complexity involves both differing compositions of neurons and glia, as well as potential differences among subtypes of those classes, such as oligodendrocytes and astrocytes. This cellular complexity may also be a major contributor to differences among brain regions. Thus, it will be important to analyze DNA methylation across a great many brain subregions and cell types. However, such studies will likely require whole genome–based approaches, to discover the genes and potentially intergenic regions that epigenetically distinguish individual brain regions. Such technologies are emerging and should be generally available within the next few years. A second issue for further study is the degree to which brain epigenetic signatures might be altered in disease. Although we found no evidence of variation in bipolar disorder (also known as MAFD1 [MIM 125480]) or autism in these brain regions, our study was not designed to detect such variation. Both our sample size and our gene set were small and were not targeted to functional candidates. A third issue is the developmental role of methylation variation in the brain. Differential methylation may affect an early event in brain development, having an impact even in the absence of adult brain expression of the relevant genes. Sorting this out will require animal models involving both genetics and epigenetics. For example, one might knock out a brain region–specific methylation mark to determine its functional effect on the normal development of that region. A fourth issue is the relationship of brain methylation to expression. Although, in three instances, increased methylation in our samples correlated with gene silencing, in two others that were outside promoters, methylation sites likely represent regulatory regions in which methylation is associated with gene expression. Similarly, Gius et al.14 found that half of genes showing altered expression after demethylation become silenced rather than activated.
Finally, the identification of brain region–specific methylation differences shows that there are stable marks heritable during cell division that distinguish one brain region from another and are consistent in these differences from one individual to another. These differences in methylation appear to be widespread at the gene level, perhaps reflecting global regulation rather than discrete effects at a small number of genes. Although these results are in a relatively early stage, they do raise the intriguing possibility that epigenetic signatures in part determine the functional programs that have been traditionally associated with neuroanatomical distinctions.
Acknowledgments
Author contributions: C.L.-A. performed most of the experiments and data analysis; J.P., S.S., and R.Y. provided samples, expertise, and technical assistance; T.D. and P.A.C. assisted with experiments; J.-B.F. performed the initial Illumina hybridization; and J.B.P. was the clinical and A.P.F. the molecular senior investigator. We thank Rafael Irizarry for advice regarding statistical analysis. This work was supported by National Institutes of Health (NIH) grant HG003233. Some tissues were provided by the Harvard Brain Tissue Resource Center, which is supported in part by NIH grant MH68855, and by the University of Maryland Brain Bank, which is supported in part by NICHD contract NO1-HD-8-3283.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Rett syndrome, EN2, autism, HDAC7A, HTR2A, SLC22A3, IGF1, FGF1, FGFR2, and MAFD1)
References
- 1.Feinberg AP (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440 10.1038/nature05919 [DOI] [PubMed] [Google Scholar]
- 2.Razin A, Riggs AD (1980) DNA methylation and gene function. Science 210:604–610 10.1126/science.6254144 [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4:143–153 10.1038/nrc1279 [DOI] [PubMed] [Google Scholar]
- 4.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- 5.Callinan PA, Feinberg AP (2006) The emerging science of epigenomics. Hum Mol Genet 15:R95–R101 10.1093/hmg/ddl095 [DOI] [PubMed] [Google Scholar]
- 6.Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C (2000) Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci USA 97:11038–11043 10.1073/pnas.97.20.11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letwin NE, Kafkafi N, Benjamini Y, Mayo C, Frank BC, Luu T, Lee NH, Elmer GI (2006) Combined application of behavior genetics and microarray analysis to identify regional expression themes and gene-behavior associations. J Neurosci 26:5277–5287 10.1523/JNEUROSCI.4602-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- 9.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al (2004) Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14:1462–1473 10.1101/gr.2538704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A (2006) Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics 7:67–80 10.1007/s10048-006-0032-6 [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al (2006) High-throughput DNA methylation profiling using universal bead arrays. Genome Res 16:383–393 10.1101/gr.4410706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP (2006) GenePattern 2.0. Nat Genet 38:500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- 13.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP (2002) A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res 12:543–554 10.1101/gr.224102. Article published online before print in March 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, Young L, Brandenburg SA, Hu Y, Bisht KS, et al (2004) Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 6:361–371 10.1016/j.ccr.2004.08.029 [DOI] [PubMed] [Google Scholar]
- 15.Millen KJ, Wurst W, Herrup K, Joyner AL (1994) Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development 120:695–706 [DOI] [PubMed] [Google Scholar]
- 16.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfield S, Smith BJ, Zimmerman RA, et al (2005) Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet 77:851–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato MV, Shimizu T, Nagayoshi M, Kaneko A, Sasaki MS, Ikawa Y (1996) Genomic imprinting of the human serotonin-receptor (HTR2) gene involved in development of retinoblastoma. Am J Hum Genet 59:1084–1090 [PMC free article] [PubMed] [Google Scholar]
- 18.Norton N, Owen MJ (2005) HTR2A: association and expression studies in neuropsychiatric genetics. Ann Med 37:121–129 10.1080/07853890510037347 [DOI] [PubMed] [Google Scholar]
- 19.Lazar A, Walitza S, Jetter A, Gerlach M, Warnke A, Herpertz-Dahlmann B, Grundemann D, Grimberg G, Schulz E, Remschmidt H, et al (2007) Novel mutations of the extraneuronal monoamine transporter gene in children and adolescents with obsessive-compulsive disorder. Int J Neuropsychopharmacol (http://journals.cambridge.org/action/displayAbstract?fromPage=online&aid=1009648) (electronically published May 4, 2007; accessed October 31, 2007) [DOI] [PubMed]
- 20.Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP (2001) Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev 15:2361–2366 10.1101/gad.206201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bach MA, Shen-Orr Z, Lowe WL Jr, Roberts CT Jr, LeRoith D (1991) Insulin-like growth factor I mRNA levels are developmentally regulated in specific regions of the rat brain. Brain Res Mol Brain Res 10:43–48 10.1016/0169-328X(91)90054-2 [DOI] [PubMed] [Google Scholar]
- 22.Vaccarino FM, Schwartz ML, Raballo R, Rhee J, Lyn-Cook R (1999) Fibroblast growth factor signaling regulates growth and morphogenesis at multiple steps during brain development. Curr Top Dev Biol 46:179–200 [DOI] [PubMed] [Google Scholar]
- 23.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R (2003) Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 35:146–150 [DOI] [PubMed] [Google Scholar]
- 24.Tudor M, Akbarian S, Chen RZ, Jaenisch R (2002) Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA 99:15536–15541 10.1073/pnas.242566899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountcastle VB (1957) Modality and topographic properties of single neurons of cat’s somatic sensory cortex. J Neurophysiol 20:408–434 [DOI] [PubMed] [Google Scholar]