Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) mediates the transcriptional response of cells to oxidative stress and is translocated into the nucleus following, or concomitant with, its activation by electrophiles or reactive oxygen species. The mechanism of its translocation into the nucleus is not entirely elucidated. Here we have identified two novel nuclear localization signal (NLS) motifs in murine Nrf2, one located near the N-terminal region (amino acid residues 42–53) and the other (residues 587–593) located near the C-terminal region. Imaging of green fluorescent protein (GFP)-tagged Nrf2 revealed that mutation(s) in any of these sequences resulted in decreased nuclear fluorescence intensity compared with the wild-type Nrf2 when Nrf2 activation was induced with the electrophile tert-butylhydroquinone. The mutations also impaired Nrf2-induced transactivation of antioxidant response element-driven reporter gene expression to the same extent as the Nrf2 construct bearing mutation in a previously identified bipartite NLS that maps at residues 494–511. When linked to GFP or to GFP-PEPCK-C each of the novel NLS motifs was sufficient to drive nuclear translocation of the fusion proteins. Co-immunoprecipitation assays demonstrated that importins α5 and β1 associate with Nrf2, an interaction that was blocked by the nuclear import inhibitor SN50. SN50 also blocked tert-butylhydroquinone-induced nuclear fluorescence of GFP-Nrf2 in cells transfected with wild-type GFP-Nrf2. Overall these results reveal that multiple NLS motifs in Nrf2 function in its nuclear translocation in response to pro-oxidant stimuli and that the importin α-β heterodimer nuclear import receptor system plays a critical role in the import process.
Nuclear factor erythroid 2-related factor 2 (Nrf2),4 in association with the cytoskeleton-associated Kelch-like protein Keap1, functions as a sensor of oxidative and electrophilic stress in cells (1–4). In non-stressed cells, Nrf2 is transcriptionally inactive because of the repressive effect of Keap1 in the cytoplasm (4–6). Reactive oxygen species or electrophilic agents induce modifications of this complex that allow Nrf2 to translocate into the nucleus to mediate activation of a variety of genes (2, 5–12). The promoters of such genes contain antioxidant response element(s) (AREs), at which Nrf2, in association with small Maf proteins or other basic region-leucine zipper transcription factors (1, 13–18), interacts to regulate gene transcription. As determined by microarray analyses, such genes include those that code for proteins that function in DNA repair, enzymes that catalyze phase II reactions in drug metabolism, signal transduction proteins, and many others that function in protein trafficking, chaperone system/stress response, and apoptosis (19, 20).
Electrophile-induced Nrf2 release from the Keap1-Nrf2 complex appears to involve not only modification of specific cysteine residues in Keap1 (7–10) but also switching of Cullin 3-dependent ubiquitination from Nrf2 to Keap1, leading to the degradation of Keap1 and stabilization and activation of Nrf2 (11, 12). Karapetian et al. (21) have proposed a nuclear-cytoplasmic shuttling model for Nrf2 and Keap1 in which Nrf2 is disengaged from Keap1, within the nucleus, by the nuclear protein prothymosin α, thus liberating Nrf2 to interact with the ARE on target gene promoters. Their model, as well as that proposed by Velichkova and Hasson (22), implicates the nuclear export signal in Keap1 as playing a key role in this process. The authors showed that mutation within this nuclear export signal or interference (using leptomycin B) with the chromosome region maintenance exportin system resulted in accumulation of Nrf2 and Keap1 in the nucleus. In another model of the regulation of Nrf2 activity, Nrf2 is proposed to be constitutively expressed and to be directly translocated into the nucleus following its synthesis on ribosomes (23). According to that model, Keap1 is independently shuttled into the nucleus to target Nrf2 for degradation, although the mechanism of this degradation was not determined. In a recent report in which the subcellular localization of Keap1 was assessed with antibody that specifically detects endogenous Keap1 (6), it was clear that Keap1 is predominantly (∼81%) localized in the cytoplasm under basal conditions and that this localization is not changed when cells are treated with electrophiles to induce release of Nrf2.
Irrespective of the model thus far proposed for the activation of Nrf2, translocating it into the nucleus is integral to its gene expression-inducing effect. The mechanism by which this translocation occurs is not entirely elucidated. A number of reports have indicated that phosphorylation at Ser40 of Nrf2 (in the cytoplasm) by protein kinase C, which appears to be concomitant with its activation, is not necessary for its nuclear import (24, 25). Those results, however, do not rule out the involvement of phosphorylation at other sites, by other kinases, or phosphorylation of potential accessory protein(s) that might impact its nuclear translocation. For example, although Nrf2 is not a direct target of MAPK (i.e. ERK), Zipper and Mulcahy (26) showed that MAPK-directed phosphorylation is a requirement for nuclear localization of Nrf2 during pyrrolidine dithiocarbamate-induced expression of glutamate cysteine ligase. They suggest a model involving ERK-mediated phosphorylation of some type of accessory protein that might be required for Nrf2 nuclear translocation.
Recently we showed that the synthetic peptide dubbed SN50 blocks tert-butylhydroquinone (tBHQ)-induced nuclear accumulation of Nrf2 (27). Because SN50 had been demonstrated previously to block the nuclear translocation of NF-κB, AP-1, NFAT, and STAT1 (28, 29) by interfering with the action of the importin α-β heterodimer (29), we suggested (27) that importins might be involved in the nuclear translocation of Nrf2. Because importins recognize NLS-containing proteins, it seemed reasonable to surmise that Nrf2 would possess nuclear localization signal(s). Indeed Jain et al. (30) have identified a bipartite NLS in Nrf2 that they showed to be involved in its translocation into the nucleus. In their studies on the degradation of Nrf2, Yamamoto and co-workers (31, 32) demonstrated that the fusion protein Neh2-EGFP, in which the Neh2 domain of Nrf2 (which mediates Nrf2 interaction with Keap1) is linked to EGFP, localizes to the nucleus. Because the Neh2 domain (amino acid residues 1–99) does not include the aforementioned bipartite NLS, their results suggest the existence of other NLS motif(s) in Nrf2. By computer-based search, we have identified two such motifs, one of which is located near the N-terminal region (amino acid residues 42–53) and the other (residues 587–593) of which is located near the C-terminal region. Using HepG2 and K562 cells, we demonstrate in this report that these two sequences are functional nuclear localization signals. We also demonstrate that importin α5 and importin β1 are involved in the nuclear import of Nrf2.
EXPERIMENTAL PROCEDURES
DNA Constructs—Plasmid that expresses the fusion protein GFP-Nrf2 was constructed by subcloning the coding sequence of mouse Nrf2 to the C terminus of pEGFP-C2 (Clontech) via restriction enzyme sites of XhoI and SmaI. Plasmids pEGFP-NLS1 and pEGFP-NLS3 were constructed by fusing the EGFP coding sequence in pEGFP-C2 to the coding sequence for NLS1 (RQKDYELEKQKK) and NLS3 (PKSKKPD), respectively. Plasmid pEGFP-Pck1 was constructed via fusing the EGFP coding sequence in pEGFP-C2 to the coding sequence for rat liver cytoplasmic phosphoenolpyruvate carboxykinase (PEPCK-C). Plasmids pEGFP-Pck1-NLS1 and pEGFP-Pck1-NLS3 were constructed by linking pEGFP-Pck1 to the coding sequence for NLS1 (RQKDYELEKQKK) and NLS3 (PKSKKPD), respectively. The preparation of other plasmid constructs has been described previously (33).
Reporter Gene Assays—K562 cells, obtained from the American Type Culture Collection (Manassas, VA), were maintained in culture as described previously (34, 35). For co-transfection of expression plasmids, cells (1 × 105 cells in 1 ml of medium/well) seeded in 24-well plates for 24 h were co-transfected with 0.2 or 0.3 μg of luciferase reporter plasmid (human Gαi2 gene promoter or human NAD(P)H:quinone oxidoreductase1-ARE-luc reporter) and wild type or mutant forms of Nrf2 plasmid using FuGENE™ 6 transfection reagent (Roche Applied Science) at a 3:1 ratio of FuGENE 6 reagent (μl) to DNA (μg). The total amount of DNA was adjusted, if necessary, by adding the empty plasmid. When used, tBHQ was added 1 h after the transfections. The cells were harvested 20–24 h later, by centrifugation at 12,000 × g (45 s) in 1.5-ml microcentrifuge tubes, and processed for luciferase assay as described previously (27, 35).
Localization of Nrf2 in HepG2 Cells by Fluorescence Microscopy—HepG2 cells, obtained from the American Type Culture Collection (ATCC), were grown in Opti-MEM + GlutaMAX medium supplemented with 1 mm sodium pyruvate, 10% fetal bovine serum, 100 units of penicillin/ml of medium, and 100 μg of streptomycin/ml of medium. About 2 × 105 cells/well were seeded onto coverslips in 6-well plates in 2 ml of medium and incubated overnight at 37 °C. Cells were then transfected with 2 μg of plasmid pEGFP-Nrf2 construct (wild type or mutants), pEGFP-NLS1, pEGFP-NLS3, pEGFP-Pck1-NLS1, pEGFP-Pck1-NLS3, or empty vector (pEGFP-C2) using FuGENE HD transfection reagent (Roche Applied Science) at a 3:1 ratio of FuGENE HD (μl) to DNA (μg). Twenty-four hours after transfection, the cells were incubated with or without 20 μm tBHQ (for 1 h) to induce activation and nuclear accumulation of Nrf2. In some experiments, the cells were incubated with SN50 or SN50M (36 μm) before the addition of tBHQ (20 μm) 1 h later. The cells were then harvested by removing the medium and rinsing once with 1× phosphate-buffered saline followed by fixation with 1 ml of ice-cold methanol for 2 min and rinsing with phosphate-buffered saline. After fixation, the cells were incubated with 100 μg/ml RNase A (Sigma) for 20 min at 37 °C and rinsed three times with 1× phosphate-buffered saline. To stain the nuclei, the cells were incubated for 2 min at room temperature in 3 μg/ml propidium iodide, rinsed with phosphate-buffered saline, and then rinsed once with H2O. Coverslips were then mounted onto slides using Aqua-Polymount (Polysciences, Inc., Warrington, PA), kept overnight at 4 °C, and visualized under a fluorescence microscope at excitation and emission wavelengths, respectively, of 536 and 617 nm for red fluorescence and 485 and 530 nm for green fluorescence. Using Adobe Photoshop, images of the propidium iodide and green fluorescent protein (GFP) fluorescence patterns were merged to visualize nuclear localization. The images were quantified by using Nikon Elements Advanced Research Software (Melville, NY); access to this software was provided by the Morphology Core Facility at Meharry Medical College.
Identification of Putative Nuclear Localization Signals in Murine Nrf2—The PSORT II program was used to identify sequences in murine Nrf2 (Entrez Protein accession number Q60795) containing highly charged, basic amino acid residues (indicated in boldface in Fig. 1A) that could potentially function as nuclear localization sequences.
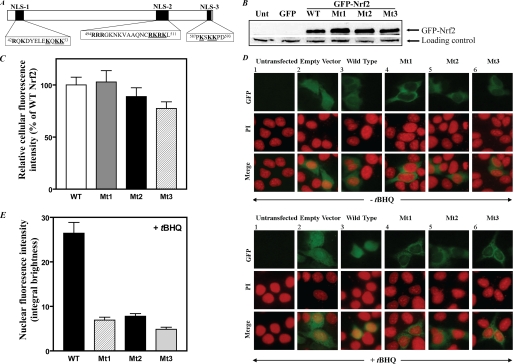
FIGURE 1.
Mutations of the three NLS motifs in Nrf2 impair nuclear translocation of Nrf2 as determined by fluorescence imaging analyses. A, schematic of putative nuclear localization sequences (NLS1, NLS2, and NLS3) in murine Nrf2. Charged, basic amino acid residues are indicated in boldface. In the experiments reported in this study, mutations were introduced at the three NLS loci by replacing the underlined basic amino acid residues with alanine residues. B, expression of Nrf2 in HepG2 cells transfected with wild-type pEGFP-Nrf2 or with pEGFP-Nrf2 bearing mutations at the NLS1, NLS2, or NLS3 loci. HepG2 cells (2 × 105 cells/2 ml/well in 6-well plates) grown for 24 h were transfected with 2 μg of pEGFP-C2 or pEGFP-Nrf2 (wild type or mutants) using FuGENE HD transfection reagent. After 40 h, whole-cell lysates were prepared (33) and used for Western blotting (50 μg of protein/lane) with anti-GFP antibody (sc-9996, Santa Cruz Biotechnology, Inc.) to measure the degree of expression of wild-type versus mutant GFP-Nrf2. A nonspecific band was used as loading control. The blots are representative of three experiments. C, quantification of fluorescence images indicating similar expression of wild-type versus mutant GFP-Nrf2. Green fluorescence under basal conditions (no tBHQ treatment) was quantified for at least 20 cells similar to and including those shown in the top horizontal row of the upper panel in D. The entire green region was delineated, and the average fluorescence intensity of the green channel was measured using Nikon Elements Advanced Research Software (Melville, NY). The experiments were repeated three times. Values plotted are means ± S.E. In D, HepG2 cells were grown and processed for analysis by fluorescence microscopy as described under “Experimental Procedures.” In the upper panel, cells were not treated with tBHQ; in the lower panel, cells were treated with tBHQ (20 μm) for 1 h to induce nuclear accumulation of Nrf2. Images of the propidium iodide (PI) and GFP fluorescence patterns were merged to visualize nuclear localization. E, the relative nuclear fluorescence of GFP in cells treated with tBHQ (20 μm). The region co-localizing with the propidium iodide (nuclear area) was delineated, and the integral brightness in this region (nuclear green fluorescence) was quantified by analyzing the merged images for at least 20 cells similar to and including those shown in the lower panel of D. The experiments were repeated three times. Values plotted are means ± S.E. Empty Vector, pEGFP-C2; WT, cells transfected with wild-type pEGFP-Nrf2; Mt1, Mt2, and Mt3, cells transfected with pEGFP-Nrf2 in which the underlined residues in NLS1, NLS2, and NLS3, respectively (see A), were mutated to alanine residues; Unt, untransfected.
Site-directed Mutagenesis—Nrf2 was mutated at the putative nuclear localization sequences by using the QuikChange™ site-directed mutagenesis kit from Stratagene (La Jolla, CA) to replace the underlined basic amino acids residues (Fig. 1A) with alanine residues. Mutants Mt1, Mt2, and Mt3 (corresponding to mutations at the NLS1, NLS2, and NLS3 loci, respectively) were created by using the following primers: 5′-agcgacagaaggactatgagctggaaGCacagGCCGCactcgaaaaggaaagacaagagcaactc-3′, 5′-gtcgccgcccagaactgtGCTGCTGCCGCCctTgagaacattgtcgagctggag-3′, and 5′-ggcaatgtgttccttgttcccGCTagcGCTGCTccagatacaaagaaaaactaggcg-3′ (capital letters are mutated bases). Mutations were confirmed by DNA sequencing at the Molecular Biology Core Facility at Meharry Medical College.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assays (EMSAs)—Nuclear extracts were prepared as described previously (27). For EMSA, the annealed 5′-overhang oligonucleotide sequence of the Gαi2 gene promoter, containing the ARE-binding motif in the promoter, was radiolabeled with [α-32P]dCTP by using the Klenow fill-in reaction and purified as described previously (33). After electrophoresis, the gel was dried and then exposed to Classic Blue Autoradiography Film BX (Molecular Technologies, St. Louis, MO) at –80 °C. The radiolabeled bands were detected by autoradiography.
Co-immunoprecipitation—Immunoprecipitation was performed with nuclear and cytoplasmic extracts from K562 cells that were incubated for various time periods with tBHQ. Briefly the extracts (20 μg of protein), prepared as described previously (27), were diluted with chromatin immunoprecipitation dilution buffer (Upstate Cell Signaling Solutions, Lake Placid, NY) and incubated with 2 μg each of either normal rabbit IgG or antibody to a specified karyopherin/importin overnight at 4 °C with gentle rotation. Immunocomplexes were collected by incubation at 4 °C for 2 h to overnight with 20 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) followed by centrifugation at 1,000 × g for 1 min. The agarose beads were washed four times with 1 ml of wash buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5 mm sodium vanadate, 0.2% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A) and boiled for 5 min with 30 μl of 2× SDS sample buffer. After centrifugation, the supernatant solution was subjected to Western blotting using antibody against Nrf2.
Western Blotting—Western blotting analyses, using various antibodies, were performed as described previously (27).
RESULTS
Putative Nuclear Localization Sequences in Nrf2—We used the PSORT II program, which predicts the probability of protein sorting signals, to identify sequences in murine Nrf2 containing highly charged, basic amino acid residues that could potentially function as nuclear localization sequences. Besides a previously studied bipartite NLS (30), we identified two novel putative nuclear localization signals in this protein, one located near the N-terminal region (amino acid residues 42–53 and occurring within the Neh2 domain) and the other (residues 587–593) located near the C-terminal region. In the schematic in Fig. 1A, we have designated these sequences NLS1 and NLS3, respectively. Both of these sequences are monopartite clusters and are present in rat, mouse, chicken, and human as can be seen in a cross-species comparison of amino acid sequence alignment of Nrf2 from these species (26). These highly conserved sequences are distinctly different from the previously identified (30) classical bipartite NLS that maps at residues 494–511, which we have designated as NLS2 in Fig. 1A. Jain et al. (30) have shown that NLS2 indeed mediates translocation of Nrf2 into the nucleus, but no information is currently available on whether the monopartite clusters can function as authentic nuclear localization signals.
Mutating the Putative NLS Motifs in Nrf2 Results in Failure to Localize Nrf2 to the Nucleus as Detected by Fluorescence Microscopy—To facilitate monitoring of the cellular localization of Nrf2, we prepared the plasmid pEGFP-Nrf2 and mutated the putative NLS motifs by changing the underlined basic amino acid residues shown in Fig. 1A to alanine residues. We then used fluorescence imaging to study the impact of these mutations on the nuclear translocation of Nrf2 in two cell types (K562 and HepG2 cell lines) in the presence or absence of tBHQ. Because nuclei of K562 cells are large relative to the total cell space, making it difficult to clearly discern the cytoplasmic from the nuclear compartment, the fluorescence imaging data shown here were generated by using only HepG2 cells because cytoplasmic and nuclear compartments are clearly visualized in these cells. Introducing mutations at the putative NLS motifs did not adversely affect expression of the GFP-Nrf2 as can be seen from (i) identical expression (assessed by Western blotting of whole-cell extracts) of wild-type GFP-Nrf2 and GFP-Nrf2 mutated at any of the three putative NLS motifs (Fig. 1B) and (ii) the similar intensity of green fluorescence in cells transfected with the wild-type plasmid versus cells transfected with the mutant plasmids (Fig. 1C).
Addition of tBHQ to cells transfected with wild-type EGFP-Nrf2 increased nuclear fluorescence compared with basal conditions (Fig. 1D, compare column 3 in the lower panel with column 3 in the upper panel). This is consistent with our previously published Western blotting data, which showed that tBHQ enhances nuclear accumulation of Nrf2 (27). Contrary to cells transfected with wild-type pEGFP-Nrf2, tBHQ-treated cells transfected with plasmids mutated at either the NLS1, the NLS2, or the NLS3 site exhibited much decreased fluorescence in their nuclei compared with cells treated with the wild-type plasmid that displayed intense green fluorescence in the nuclear compartment (Fig. 1D, lower panel, compare columns 4–6 with column 3). This difference is even more dramatic when one compares the merged images. The merged image in column 3 (Fig. 1D, lower panel) clearly shows that in cells transfected with the wild-type plasmid much of the green fluorescence of the EGFP-Nrf2 co-localizes with the propidium iodide stain (used as counterstain to localize the nucleus). In contrast, the nuclei of cells transfected with the mutant plasmids (Fig. 1D, lower panel, columns 4–6) retain the intense red color of the propidium iodide, whereas much of the green fluorescence remains outside the nuclear compartment. Quantification of the intensity of green fluorescence in the nuclear compartment indicated a substantially decreased content of GFP fusion protein (GFP-Nrf2) in the nuclei of cells transfected with all three mutants compared with cells transfected with the wild-type plasmid (Fig. 1E). Overall the data in Fig. 1 (D and E) show that mutation in the putative nuclear localization sequences substantially decreases nuclear localization of Nrf2.
Two additional approaches were used to provide further support for the idea that NLS1 and NLS3 function as authentic nuclear localization signals. In the first approach, the coding sequence of NLS1 as well as that of NLS3 was cloned into that of GFP or PEPCK-C to monitor NLS-driven nuclear translocation of the fusion proteins, the rationale being that the functionality of an NLS motif should be transferable to a heterologous protein (36–39). In the second approach, we used site-directed mutagenesis to show that mutating the NLS motifs in Nrf2 results in failure of Nrf2 to transactivate ARE-driven reporter gene constructs.
Fusion of NLS1 or NLS3 to GFP or PEPCK-C Enhances Nuclear Localization of the Fusion Protein—We created fusion proteins of NLS motifs linked to GFP or PEPCK-C (see schematics in Fig. 2A) as described under “Experimental Procedures” and assessed the localization of the NLS-containing fusion proteins compared with GFP alone or GFP-PEPCK-C alone. Although the molecular sizes of GFP, GFP-NLS1, and GFP-NLS3 are far less than the diffusion limit (∼50 kDa) of the nuclear pore complex (40), our prediction was that the NLS domain(s), if functionally competent, would enhance localization of the fusion protein(s) to the nucleus. HepG2 cells transfected with pEGFP alone exhibited dispersed localization of GFP, i.e. they contain GFP fluorescence in both the nuclear and cytoplasmic compartments (Fig. 2B). In contrast, cells transfected with pEGFP-NLS1 or pEGFP-NLS3 exhibited enhanced localization of the fusion proteins to the nucleus; this is quite apparent from the merged images depicting co-localization of GFP-NLS1 and GFP-NLS3 with propidium iodide (Fig. 2B).
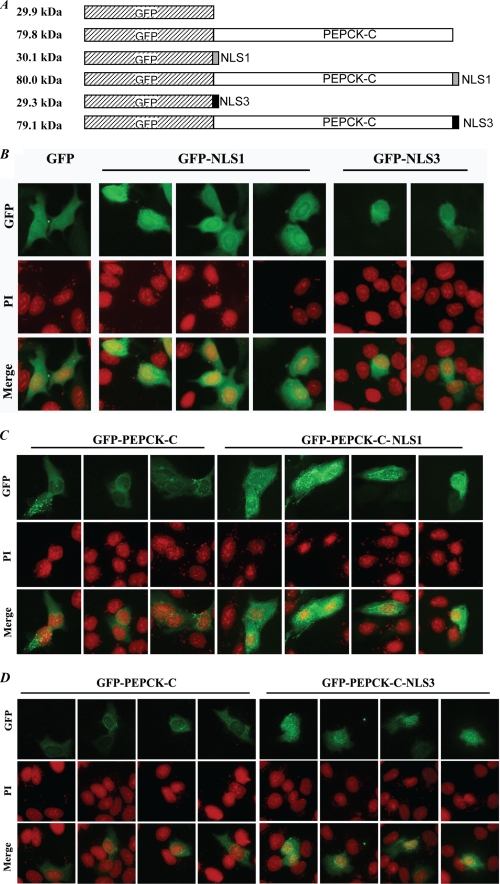
FIGURE 2.
NLS1 and NLS3 drive localization of NLS fusion proteins to the nucleus. A, schematics of GFP fusion proteins used. When the coding sequence of NLS1 or NLS3 was fused to that of GFP or GFP-Pck1, a portion of the multiple cloning site (polylinker) sequence was deleted; this accounts for the somewhat smaller size of GFP-NLS3 compared with GFP or GFP-PEPCK-C-NLS3 compared with GFP-PEPCK-C. In B through D, HepG2 cells transfected with plasmids bearing coding sequences for the GFP fusion proteins indicated in the schematics in A were processed for analysis by fluorescence microscopy as described under “Experimental Procedures.” Construction of the plasmids is described under “Experimental Procedures.” Images in B–D, respectively, are from cells transfected with plasmids harboring coding sequences for GFP (control), GFP-NLS1, or GFP-NLS3 (B); GFP-PEPCK-C (control) or GFP-PEPCK-C-NLS1 (C); and GFP-PEPCK-C (control) or GFP-PEPCK-C-NLS3 (D). The images shown are from different fields. Two experiments were performed for each panel. NLS1 and NLS3 are sequences designated NLS1 and NLS3, respectively, in Fig. 1A. PI, propidium iodide.
Because the sizes of the GFP-NLS1 and GFP-NLS3 are smaller than the diffusion limit (∼50 kDa) of the nuclear pore complex, we used additional fusion proteins, i.e. GFP-PEPCK-C, GFP-PEPCK-C-NLS1, and GFP-PEPCK-C-NLS3, which provided unique advantages over GFP-NLS fusion constructs. For example, besides the fact that PEPCK-C is a purely cytoplasmic protein, the size of the GFP-PEPCK-C fusion protein (used as control) is 79.8 kDa, which is much larger than the diffusion limit of the nuclear pore complex. Therefore one would not expect it to be localized in the nucleus or to be dispersed throughout the cell as would GFP alone. Thus, if NLS1 and NLS3 functioned as nuclear localization signals, one would expect them to drive localization of GFP-PEPCK-C-NLS1 or GFP-PEPCK-C-NLS3 to the nuclear compartment.
Compared with the control fusion protein (GFP-PEPCK-C), which exhibited green fluorescence predominantly in the cytoplasm, cells transfected with either pEGFP-Pck1-NLS1 or pEGFP-Pck1-NLS3 exhibited enhanced localization of the fusion proteins to the nucleus as evident from the co-localization of GFP-PEPCK-C-NLS1 (Fig. 2C) and GFP-PEPCK-C-NLS3 (Fig. 2D) with propidium iodide (merged images). These data strengthen our conclusion that NLS1 and NLS3 are functional nuclear localization signals, a conclusion that is consistent with our findings (Fig. 1D) that mutating NLS1 or NLS3 impairs nuclear localization of Nrf2.
Mutations in the Putative Nuclear Localization Sequences in Nrf2 Result in Failure of Nrf2 to Transactivate ARE-driven Reporter Gene Constructs—The functionality of the NLS motifs was further studied by monitoring the impact of mutating the identified NLS motifs on the transcriptional activity of Nrf2. For these experiments, we transfected K562 cells with expression plasmids harboring the gene for wild-type Nrf2 or Nrf2 mutated at the three NLS motifs and measured Nrf2-induced gene transcription using two reporter gene constructs, hNQO1-luc (41, 42) and Gαi2-luc (27, 43), that contain the ARE, the response element that is established as the transcriptional binding site for Nrf2 in partnership with other basic region-leucine zipper transcription factors such as small Maf proteins (1, 12–17). As expected, promoter activity of these reporter gene constructs was much enhanced in cells treated with tBHQ (Fig. 3). However, mutation at any of the NLS motifs, as indicated in Fig. 1A, failed to stimulate promoter activity not only under basal conditions but also in cells treated with tBHQ. These results complement the imaging data shown in Fig. 1D and support the conclusion that all three nuclear localization sequences in Nrf2 are relevant for the nuclear import and function of this transcription factor.
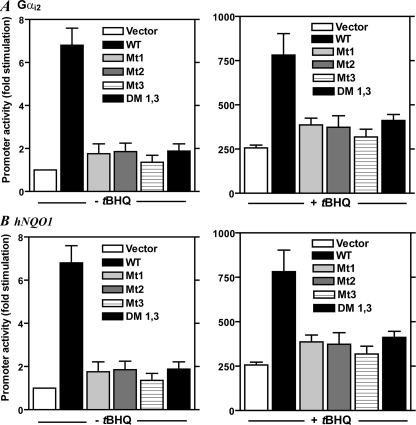
FIGURE 3.
Mutations of the three NLS motifs in Nrf2 impair Nrf2-induced reporter gene promoter activity. In A, K562 cells were co-transfected with 0.2 μg of Gαi2 gene promoter construct (pGαi2(–1214/+115)-luc) and wild-type or mutated Nrf2 (0.3 μg) or the empty vector (pCI-Neo) followed by the addition of tBHQ (20 μm) 1 h later. In B, K562 cells were co-transfected with hNQO1-ARE-luc reporter gene construct (0.3 μg) and wild-type or mutated Nrf2 (0.2 μg) or the empty vector (pCI-Neo) followed by the addition of tBHQ (20 μm) 1 h later. For both panels, the cells were harvested for luciferase assay 20 h later as described under “Experimental Procedures.” The plotted values are means ± S.E. of duplicate assays for five experiments. Vector, empty vector (pCI-Neo); WT, plasmid containing cDNA for Nrf2 (pCI-Nrf2); Mt1, Mt2, and Mt3, pCI-Nrf2 mutated at NLS1, NLS2, and NLS3, respectively, as indicated in legend to Fig. 1A; DM 1,3, double mutant containing mutations at NLS1 and NLS3 as indicated in Fig. 1A.
Nrf2 Mutated at the NLS1 or NLS3 Does Not Inhibit Promoter-inducing Activity of Wild-type Nrf2 or Stimulus-induced Nuclear Translocation of Wild-type Nrf2—Because mutation in any of the NLS motifs in Nrf2 resulted in its failure to localize to the nucleus as well as in its failure to transactivate ARE-driven reporter gene constructs, we considered the possibility that such mutations may confer a dominant negative property on the mutant species. If this were the case, the NLS mutants should override the action of the wild-type Nrf2 either in the fluorescence imaging assay or in the reporter gene assay. To test this idea, we assessed the impact of co-transfecting NLS1 or NLS3 mutants with the wild-type Nrf2. As shown in Fig. 4A, none of these mutants diminished wild-type Nrf2-induced reporter gene activity either in the absence or presence of tBHQ. Also NLS1 and NLS3 mutants had no effect on tBHQ-induced nuclear translocation of the wild-type GFP-Nrf2 as measured by fluorescence imaging (Fig. 4B). Taken together, these data indicate that mutations at the NLS1 or NLS3 motif in Nrf2 do not confer dominant negative properties to these mutants.
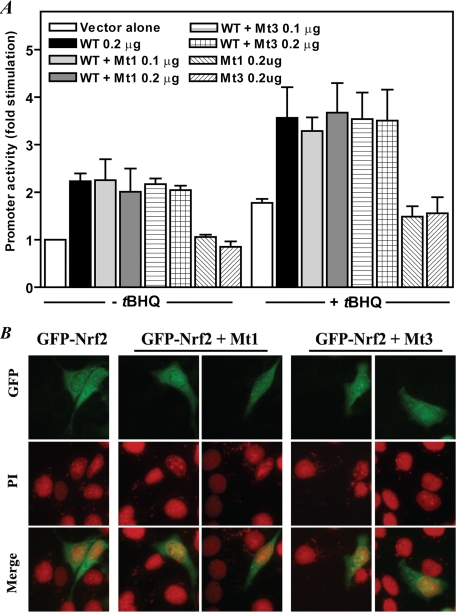
FIGURE 4.
Nrf2 mutated at the NLS1 or NLS3 motif does not inhibit transactivating activity or stimulus-induced nuclear translocation of wild-type Nrf2. A, reporter gene assay. K562 cells were co-transfected with 0.1 μg of Gαi2 gene promoter construct and wild-type Nrf2 (0.2 μg) in the absence or presence of mutant Nrf2 (0.1 and 0.2 μg) followed by the addition of tBHQ (20 μm) 1 h later. The cells were harvested for luciferase assay 24 h later as described under “Experimental Procedures.” The values are means ± S.E. of duplicate assays for three to four experiments. Vector, pCI-Neo; WT, pCI-Nrf2; Mt1 and Mt3, pCI-Nrf2 in which the underlined residues in NLS1 and NLS3, respectively (see Fig. 1A), were mutated to alanine residues. B, GFP-Nrf2 mutated at the NLS1 or NLS3 motif does not inhibit tBHQ-induced nuclear translocation of wild-type GFP-Nrf2. HepG2 cells grown as described in Fig. 1D were transfected with wild-type pEGFP-Nrf2 (WT) alone or co-transfected with wild-type pEGFP-Nrf2 and pEGFP-Nrf2 in which the underlined residues in NLS1 (Mt1) and NLS3 (Mt3), respectively (see Fig. 1A), were mutated to alanine residues followed 24 h later by addition of tBHQ (20 μm) to induce nuclear translocation of Nrf2. The cells were processed for fluorescence microscopy analysis as in Fig. 1D. The experiments were repeated two times. PI, propidium iodide.
Nuclear Translocation of Nrf2 Involves Importins/Karyopherins—The gene for Nrf2 predicts a 66-kDa protein (44), but the protein migrates on SDS-polyacrylamide gels as an 85–96-kDa protein (41, 44, 45). Because it is much larger than the diffusion limit (∼50 kDa) of the nuclear pore complex (40), one would expect its translocation into the nucleus to be mediated by a carrier-dependent import mechanism. Cargo proteins that contain nuclear localization signal(s) are recognized, in the cytoplasm, through their nuclear localization signal(s) by the soluble adaptor proteins termed importins/karyopherins (α and/or β), which upon binding the cargo proteins result in a complex that is then ferried through the nuclear pore complex in the nuclear membrane into the nucleoplasm (40, 46–52). SN50 is a well characterized cell-penetrating synthetic peptide that blocks nuclear translocation of stress-responsive transcription factors, specifically NF-κB, NFAT, AP-1, and STAT1 (28, 29, 53, 54), by interfering with the action of the importin α (Rch1)-β heterodimer (28). Torgerson et al. (29) showed that in EMSAs SN50 decreased transcription factor binding to cognate DNA response elements in the probes used. In a previous study (27) we showed that SN50 blocks tBHQ-induced nuclear accumulation of Nrf2. Therefore, we used SN50 as a reagent to explore the involvement of importins in the nuclear translocation of Nrf2. First, fluorescence imaging was used to assess the impact of SN50 on tBHQ-induced nuclear localization of Nrf2. Second, we used EMSA to monitor the impact of SN50 on tBHQ-induced Nrf2 binding to ARE-containing DNA probe. Third, we assessed Nrf2-dependent gene transcription in cells treated with SN50.
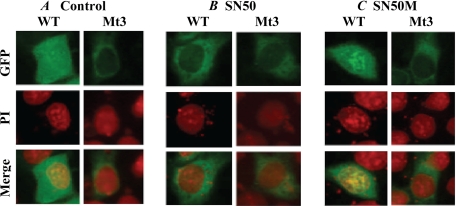
To assess the impact of SN50 on tBHQ-induced nuclear localization of Nrf2, HepG2 cells were transfected with wild-type pEGFP-Nrf2 or pEGFP-Nrf2 mutated at the NLS3 locus. The cells were then incubated with SN50 (36 μm) or SN50M followed 1 h later with tBHQ (20 μm). Fluorescence imaging was then performed as described for Fig. 1D. The results (Fig. 5) show that SN50 impaired nuclear localization of Nrf2 in cells transfected with the wild-type pEGFP-Nrf2; it had no effect in cells transfected with the NLS3 mutant. As would be predicted, the scrambled peptide SN50M, which does not block nuclear translocation (28, 29), had no effect when either the wild-type or the mutant pEGFP-Nrf2 was used. Because SN50 had been demonstrated to interact with importin α-β heterodimer that recognizes NLS-containing cargo proteins (29) these results suggest that SN50 competed effectively against such importins for binding to wild-type Nrf2 that had intact NLS motifs, whereas in the absence of such motifs no such effects were observed. Extrapolated to the NLS1 motif, these results complement the data in Fig. 1D and are consistent with the interpretation that NLS1 and NLS3 are authentic nuclear localization sequences.
FIGURE 5.
SN50 decreases nuclear localization of Nrf2 as measured by fluorescence imaging. HepG2 cells were transfected with wild-type (WT) pEGFP-Nrf2 or pEGFP-Nrf2 mutated at the NLS3 locus. The cells were then incubated with SN50 (36 μm) or SN50M (36 μm) followed 1 h later with tBHQ (20 μm). Fluorescence imaging and visualization were then performed as described for Fig. 1D. PI, propidium iodide.
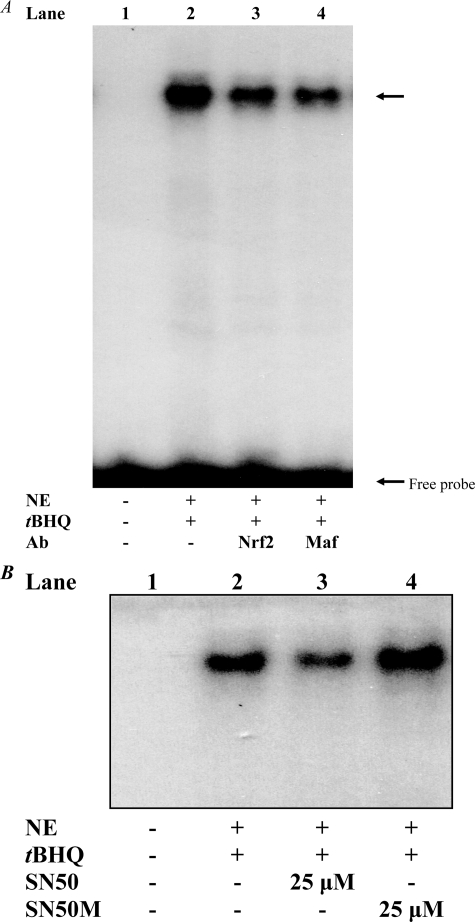
In the second approach (Fig. 6) in which we used EMSA to monitor the impact of SN50 on tBHQ-induced Nrf2 binding to ARE-containing DNA probe, the DNA probe used was a labeled double-stranded DNA probe, 5′-GCCCGCCCCGGCCCAGTCACAGGCTTGGTTC-3′, which contains the ARE (underlined) motif that maps at –84/–76 in the Gαi2 gene promoter. We have shown previously (by gel shift assays) that this probe forms a complex with Nrf2 and the small Maf proteins (27, 43), its prototypic binding heterodimer partners at the ARE (1, 12–17). Specificity of the binding was demonstrated by showing that an unrelated oligonucleotide (Sp1 consensus oligonucleotide) had no effect on the binding (27, 43). In the presence of antibody to either Nrf2 or small Maf proteins, the intensity of the protein-DNA complex decreases (27, 43), indicating that binding of these antibodies prevents association of Nrf2 or small Mafs with the labeled probe and therefore that these transcription factors are in the complex. This fact is recapitulated in Fig. 6A. Using this probe as a test probe to study the effect of SN50 on Nrf2 binding to its DNA cognate element, we show that the Nrf2-DNA binding activity was decreased in nuclear extracts from K562 cells treated with SN50 (Fig. 6B, compare lane 3 with lane 2) but not with the control peptide (SN50M) (Fig. 6B, compare lane 4 with lane 3), a cell-penetrating peptide that has a mutated NLS (28, 29, 53).
FIGURE 6.
Nuclear import inhibitor SN50 decreases Nrf2-DNA binding activity of nuclear extracts. K562 cells were incubated for 1 h with or without 25 μm SN50 (Biomol, Plymouth Meeting, PA) or the control peptide SN50M followed by addition of tBHQ (20 μm). Nuclear extracts were prepared 1 h later and used for EMSA. The EMSA was performed as described previously (27, 43) using 32P-labeled double-stranded DNA probe 5′-GCCCGCCCCGGCCCAGTCACAGGCTTGGTTC-3′, which contains the ARE (underlined) motif that maps at –84/–76 in the Gαi2 gene promoter. The reactions were carried out with 2 μg of nuclear extract protein for each lane. When antibodies were used (A), the nuclear extract was incubated with the labeled probe for 30 min at 25 °C prior to addition of each antibody and then incubated for an additional 30 min followed by electrophoresis (27). Antibodies against Nrf2 and small Maf (Santa Cruz Biotechnology, Inc.) were used at 2 μg for each lane. Nrf2-DNA binding complex is indicated by the upper arrow. B, SN50, but not SN50M, decreases the Nrf2-DNA binding activity of nuclear extracts. Ab, antibody; NE, nuclear extract.
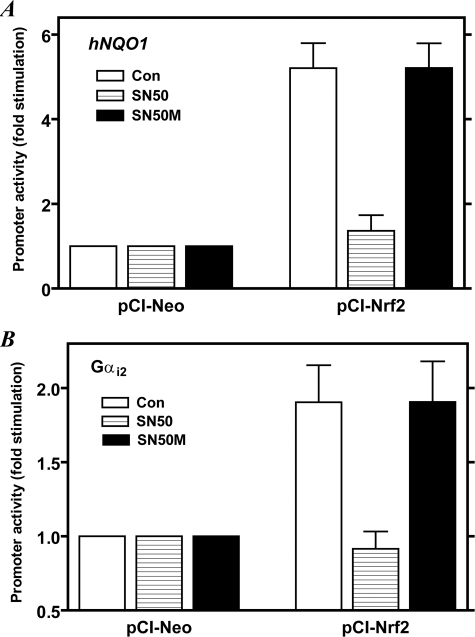
Next we assessed the impact of SN50 on Nrf2-dependent gene transcription by assaying reporter gene activity of two ARE-containing reporter gene constructs, hNQO1-luc (41, 42) and Gαi2-luc (27, 43). As shown in Fig. 7, SN50 (at 5 μm) decreased Nrf2-induced promoter activities of both of these reporter gene constructs to base-line levels, whereas the control peptide (SN50M) had no effect. The concentration of SN50 used here is 3–4 times lower than that originally demonstrated by Torgerson et al. (29) to block nuclear import of NF-κB, NFAT, or STAT1 in Jurkat cells and is much lower than that (13.5 μm) used byÁlvarez-Maqueda et al. (54) to demonstrate the involvement of NF-κB in the induction of heme oxygenase-1 in human lymphocytes by 15-deoxy-Δ12,14-prostaglandin J2. Taken together with the data in Figs. 5 and 6, we conclude that the dampened transactivation effect of Nrf2 in SN50-treated cells resulted from impaired nuclear translocation of Nrf2, consequently resulting in decreased Nrf2-DNA binding activity and attenuation of Nrf2-induced promoter activity. Because SN50 interferes with the action of the importin α-β heterodimer (29), these results can be interpreted to implicate importins/karyopherins in the nuclear translocation of Nrf2.
FIGURE 7.
SN50, but not SN50M, inhibits Nrf2-induced transcription from ARE-driven gene promoters. In A, K562 cells were co-transfected with hNQO1-ARE-luc reporter gene construct (0.3 μg) without or with plasmid (0.2 μg) containing cDNA for Nrf2 (pCI-Nrf2) or the empty vector (pCI-Neo) followed by the addition of 5 μm SN50 or SN50M 1 h later. In B, the cells were co-transfected with pGαi2(–1214/+115)-luc reporter gene construct (0.2 μg) without or with plasmid (0.3 μg) containing cDNA for Nrf2 (PCI-Nrf2) or the empty vector (PCI-Neo) followed by the addition of 5 μm SN50 or SN50M 1 h later. For both panels, the cells were harvested after 24 h and processed for luciferase assay as described under “Experimental Procedures.” The hNQO1-ARE-luc reporter construct contains a single copy of ARE, derived from the human NQO1 promoter, placed upstream of a minimal promoter containing a TATA box fused to the luciferase gene (41, 42). The Gαi2-luc reporter gene construct contains only one ARE sequence (5′-TGACTGGGC-3′) that maps at –84/–76 in the promoter (27, 43). Values shown are means ± S.E. for duplicate assays from four different experiments. Values obtained for cells that were transfected with the empty vector (pCI-Neo) were set as 1.0. Con, control (no peptide added).
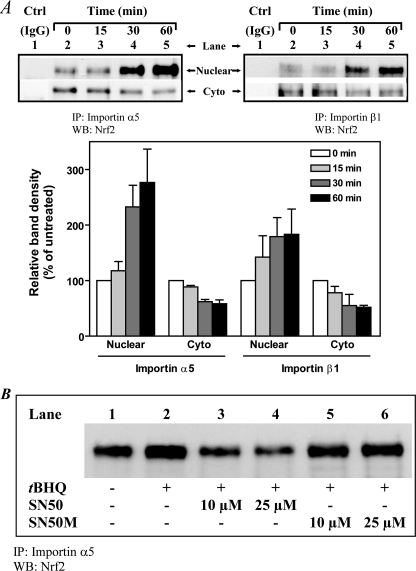
Co-immunoprecipitation Assay Reveals Association of Karyopherins/Importins and Nrf2—At least six isoforms of the importin α are present in human cells (55, 56). If Nrf2 is recognized by one or more of these importins, the association of the two protein species, in the context of the intact cell, should be detectable by a co-immunoprecipitation assay. Furthermore such association should be disrupted by SN50 as a result of competition between SN50 and Nrf2 for the importin α molecule(s). To test these predictions, we immunoprecipitated nuclear and cytoplasmic extracts with antibody against karyopherin α1 (alternative name, importin α5) or karyopherin β1 (alternative name, importin β1) at various time points after addition of tBHQ; after extensive washing, the immunoprecipitates were processed for analysis for the presence of Nrf2 by Western blotting. As shown in Fig. 8A (upper panel), Nrf2 was detected in anti-importin α5 precipitates as well as in anti-importin β1 precipitates but not in extracts immunoprecipitated with normal IgG (control). In the reverse experiments in which extracts were first immunoprecipitated with anti-Nrf2 antibody followed by detection of importin(s) by Western blotting, the importins were detected in such blots (data not shown). Association between importin (α5 or β1) and Nrf2 was detectable in both the cytoplasmic and nuclear fractions within 15–30 min after addition of tBHQ. Quantitation of the data indicated that the band densities obtained with the nuclear fractions increased within the same time frame as the band densities obtained with the cytoplasmic fractions decreased (Fig. 8A, lower panel). Notably the time frame of these reciprocal changes coincides with our earliest detection of tBHQ-induced nuclear accumulation of Nrf2 (27). These reciprocal dynamics are consistent with an interpretation of rapid nuclear uptake of the cargo protein, i.e. Nrf2, through a carrier-mediated process.
FIGURE 8.
Association of karyopherins/importins and Nrf2 as revealed by their co-immunoprecipitation. K562 cells (4 × 106/8 ml of medium in T25 flask) were cultured for 24 h followed by the addition of tBHQ (20 μm). The cells were harvested at the indicated time points (up to 60 min) after the addition of tBHQ. Nuclear and cytoplasmic extracts were prepared as described previously (27, 43). A, representative blots of immunoprecipitates generated with antibody against karyopherin α1 (importin α5) or karyopherin β1 (importin β1) followed by Western blotting for Nrf2. Immunoprecipitation was performed as described under “Experimental Procedures” using nuclear extract or cytoplasmic fraction (20 μg of protein in each case) and 2 μg each of anti-karyopherin α1 (importin α5) antibody (sc-6918, Santa Cruz Biotechnology, Inc.), anti-karyopherin β1 (importin β1) antibody (sc-11367, Santa Cruz Biotechnology, Inc.), or normal rabbit IgG. Immunoprecipitated material (corresponding to 5–10 μg of protein from the nuclear extract or cytoplasmic fraction) was analyzed by Western blotting using anti-Nrf2 antibody (sc-13032, Santa Cruz Biotechnology, Inc.). Upper panel, lane 1, normal IgG (control); lanes 2–5, immunoprecipitates with antibody against karyopherin α1 (importin α5) or karyopherin β1 (importin β1) at 0, 15, 30, and 60 min after addition of tBHQ. Lower panel, graphic representation of the data in the upper panel. Quantitation of the density of bands representing co-immunoprecipitated Nrf2 was assessed by densitometric scanning and expressed relative to the band at zero time (no tBHQ), which was set as 100%. Data are means ± S.E. for three different experiments. B, effect of SN50 on the association of Nrf2 with importin α. K562 cells were pretreated for 1 h with or without 10 or 25 μm SN50 or the control peptide SN50M followed by incubation with tBHQ (20 μm) for 1 h. Nuclear extracts were then prepared and used for co-immunoprecipitation analysis as in A. IP, immunoprecipitate; WB, Western blot; Cyto, cytoplasmic extract; Ctrl, control; nuclear, nuclear extract.
Fig. 8B shows that the association between importin α and Nrf2 was weak in cells treated with SN50 (compare lanes 3 and 4 with lane 2), whereas the control peptide (SN50M) had no effect (compare lanes 5 and 6 with lane 2). These data confirm our prediction that SN50 would disrupt the association of Nrf2 with importin(s) and are consistent with the idea that SN50 competes with an NLS-containing cargo protein for binding to importin α (29). Overall the results of these co-immunoprecipitation experiments (Fig. 8) indicate that during the nuclear transfer process Nrf2 associates with importin α5 and importin β1. This indicates that the importin α-β heterodimer nuclear import system plays a critical role in the import process.
DISCUSSION
In this work, we have identified two previously uncharacterized monopartite nuclear localization sequences that map at amino acid residues 42–53 and 587–593 in murine Nrf2. Both sequences are also present in Nrf2 from other species, notably chicken and human (26). We have designated these sequences NLS1 and NLS3, respectively, to distinguish them from a previously identified bipartite sequence (designated NLS2 in Fig. 1A), which had been implicated in the nuclear translocation of this transcription factor (30, 57). Interestingly NLS1 occurs within the Neh2 domain of Nrf2, the domain that mediates interaction of Nrf2 with Keap1 (31, 32). Given the central importance of Nrf2 in cell biology (1, 2, 58), the presence of multiple nuclear localization signals may confer advantages that allow this protein to respond to different types of signals or to target different regulatory pathways that impinge on nuclear endpoints.
A variety of proteins are now known to contain multiple nuclear localization signals (36, 59–68), including some (e.g. 5-lipoxygenase (36) and Fli-1 (65)) that contain atypical nuclear localization signals that do not conform to either the classical bipartite or the monopartite motifs. There is bourgeoning interest in deciphering whether individual nuclear localization signals in multiple NLS-containing proteins might exhibit different properties. For example, the glucocorticoid receptor possesses two nuclear localization signals, one of which (NL1) appears to mediate the nuclear translocation of the unliganded receptor and requires binding to importin α, whereas the other (NL2) appears to mediate slower translocation of the receptor, a phenomenon that is agonist-specific and independent of binding to importin α (62). Mammalian high mobility group box transcription factors also possess two nuclear localization signals, one that is a classical RanGTP-dependent signal and a second one that binds calmodulin (68). In our work, mutations in any of the nuclear localization signals in Nrf2 resulted in its failure to localize to the nucleus as well as in its failure to transactivate ARE-driven reporter gene constructs, indicating that all three nuclear localization sequences in Nrf2 are critical for the nuclear import and function of this transcription factor. We considered the possibility that such mutations may confer a dominant negative property on the mutant species. However, this did not appear to be the case as none of the mutants diminished wild-type Nrf2-induced transactivation of the Gαi2 gene promoter (Fig. 4A) or translocation of wild-type GFP-Nrf2 into the nucleus as seen from fluorescence imaging (Fig. 4B). Perhaps the NLS domains might be in close proximity to one another in the native conformation of the protein such that mutation in any of them affects the interaction of Nrf2 with the importin(s) that recognizes such domains. A test of this idea would require availability of crystal structure of the Nrf2 protein.
Our study demonstrates, for the first time, that the importin α-β complex (specifically importin α5 and importin β1) is involved in the nuclear translocation of Nrf2. Given that there are up to six isoforms of importin α in mammalian cells (55, 56), further studies are warranted to decipher whether other importin molecules participate in binding to, or display selectivity in binding to Nrf2, during its nuclear translocation.
In addition to nuclear localization signals, nucleocytoplasmic shuttling proteins also contain sequences that function as nuclear export signals, which interact with exportins (51, 69, 70). Two functional nuclear export signals have been identified in Nrf2 (71, 72), and Fyn kinase-mediated phosphorylation of Tyr568 in Nrf2 was recently reported to regulate nuclear export and degradation of Nrf2 (73). Thus, it seems reasonable to conclude that the transcriptional action of Nrf2 must depend on regulation of its nucleocytoplasmic shuttling, an area that poses an important challenge for further study.
Acknowledgments
Access to the Nikon Elements Advanced Research Software used to quantify green fluorescence signals was provided by the Meharry Medical College Morphology Core Laboratory, which is supported in part by National Institutes of Health Grants U54NS041071-06, G12RR03032-19, U54CA91408, and U54RR019192-04. We thank Dr. Marilyn E. Thompson (Meharry Medical College) for providing us instructions and free access to the use of the fluorescence microscope used by her research group, Dr. Richard W. Hanson (Case Western Reserve University School of Medicine, Cleveland, OH) for allowing us the use of resources in his laboratory to clone EGFP fusion proteins, Dr. Jeffrey Johnson (University of Wisconsin, Madison, WI) for providing us with hNQO1-ARE-luc reporter gene construct, and Dr. Jacek Hawiger (Vanderbilt University School of Medicine, Nashville, TN) for comments on the manuscript.
This work was supported in part by National Institutes of Health Grants SO6-GM08037 and HL-66109. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Nrf2, nuclear factor erythroid 2-related factor 2; ARE, antioxidant response element; GFP, green fluorescent protein; EGFP, enhanced GFP (also known as red-shifted variant of wild-type GFP); Keap1, Kelch-like ECH-associated protein 1; NLS, nuclear localization signal; Pck1, coding sequence of rat liver cytoplasmic phosphoenolpyruvate carboxykinase (PEPCK-C); tBHQ, tert-butylhydroquinone; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; NFAT, nuclear factor of activated T-cells; EMSA, electrophoretic mobility shift assay.
References
- 1.Nguyen, T., Sherratt, P. J., and Pickett, C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43 233–260 [DOI] [PubMed] [Google Scholar]
- 2.Motohashi, H., and Yamamoto, M. (2004) Trends Mol. Med. 10 549–557 [DOI] [PubMed] [Google Scholar]
- 3.Wakabayashi, N., Dinkova-Kostova, A. T., Holtzclaw, W. D., Kang, M.-I., Kobayashi, A., Yamamoto, M., Kensler, T. W., and Talalay, P. (2004) Proc. Natl. Acad. Sci., U. S. A. 101 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang, M.-I., Kobayashi, A., Wakabayashi, N., Kim, S.-G., and Yamamoto, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., Igarashi, K., Engel, J. D., and Yamamoto, M. (1999) Genes Dev. 13 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watai, Y., Kobayashi, A., Nagase, H., Mizukami, M., McEvoy, J., Singer, J. D., Itoh, K., and Yamamoto, M. (2007) Genes Cells 12 1163–1167 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, D. D., Lo, S.-C., Cross, J. V., Templeton, D. J., and Hannink, M. (2004) Mol. Cell. Biol. 24 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W., and Diehl, J. A. (2004) Mol. Cell. Biol. 24 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, D. D., Lo, S.-C., Sun, Z., Habib, G. M., Lieberman, M. W., and Hannink, M. (2005) J. Biol. Chem. 280 30091–30099 [DOI] [PubMed] [Google Scholar]
- 10.Furukawa, M., and Xiong, Y. (2005) Mol. Cell. Biol. 25 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, F., Sekhar, K. R., Freeman, M. L., and Liebler, D. C. (2005) J. Biol. Chem. 280 31768–31775 [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, A., Kang, M.-I., Watai, Y., Tong, K. I., Shibata, T., Uchida, K., and Yamamoto, M. (2006) Mol. Cell. Biol. 26 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kataoka, K., Noda, M., and Nishizawa, M. (1994) Mol. Cell. Biol. 14 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh, K., Igarashi, K., Hayashi, N., Nishizawa, M., and Yamamoto, M. (1995) Mol. Cell. Biol. 15 4184–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., Yamamoto, M., and Nabeshima, Y. (1997) Biochem. Biophys. Res. Commun. 236 313–322 [DOI] [PubMed] [Google Scholar]
- 16.Motohashi, H., Shavit, J. A., Igarashi, K., Yamamoto, M., and Engel, J. D. (1997) Nucleic Acids Res. 25 2953–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venugopal, R., and Jaiswal, A. K. (1998) Oncogene 17 3145–3156 [DOI] [PubMed] [Google Scholar]
- 18.Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D., and Yamamoto, M. (2002) Gene (Amst.) 294 1–12 [DOI] [PubMed] [Google Scholar]
- 19.Li, J., Lee, J.-M., and Johnson, J. A. (2002) J. Biol. Chem. 277 388–394 [DOI] [PubMed] [Google Scholar]
- 20.Kwak, M.-K., Wakabayashi, N., Itoh, K., Motohashi, H., Yamamoto, M., and Kensler, T. W. (2003) J. Biol. Chem. 278 8135–8145 [DOI] [PubMed] [Google Scholar]
- 21.Karapetian, R. N., Evstafieva, A. G., Abaeva, I. S., Chichkova, N. V., Filonov, G. S., Rubtsov, Y. P., Sukhacheva, E. A., Melnikov, S. V., Schneider, U., Wanker, E. E., and Vartapetian, A. B. (2005) Mol. Cell. Biol. 25 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velichkova, M., and Hasson, T. (2005) Mol. Cell. Biol. 25 4501–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen, T., Sherratt, P. J., Nioi, P., Yang, C. S., and Pickett, C. B. (2005) J. Biol. Chem. 280 32485–32492 [DOI] [PubMed] [Google Scholar]
- 24.Huang, H.-C., Nguyen, T., and Pickett, C. B. (2002) J. Biol. Chem. 277 42769–42774 [DOI] [PubMed] [Google Scholar]
- 25.Bloom, D. A., and Jaiswal, A. K. (2003) J. Biol. Chem. 278 44675–44682 [DOI] [PubMed] [Google Scholar]
- 26.Zipper, L. M., and Mulcahy, R. T. (2003) Toxicol. Sci. 73 124–134 [DOI] [PubMed] [Google Scholar]
- 27.Arinze, I. J., and Kawai, Y. (2005) J. Biol. Chem. 280 9786–9795 [DOI] [PubMed] [Google Scholar]
- 28.Lin, Y.-Z., Yao, S., Veach, R. A., Torgerson, T. R., and Hawiger, J. (1995) J. Biol. Chem. 270 14255–14258 [DOI] [PubMed] [Google Scholar]
- 29.Torgerson, T. R., Colosia, A. D., Donahue, J. P., Lin, Y.-Z., and Hawiger, J. (1998) J. Immunol. 161 6084–6092 [PubMed] [Google Scholar]
- 30.Jain, A. K., Bloom, D. A., and Jaiswal, A. K. (2005) J. Biol. Chem. 280 29158–29168 [DOI] [PubMed] [Google Scholar]
- 31.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., O'Connor, T., and Yamamoto, M. (2003) Genes Cells 8 379–391 [DOI] [PubMed] [Google Scholar]
- 32.Katoh, Y., Iida, K., Kang, M.-I., Kobayashi, A., Mizukami, M., Tong, K. I., McMahon, M., Hayes, J. D., Itoh, K., and Yamamoto, M. (2005) Arch. Biochem. Biophys. 433 342–350 [DOI] [PubMed] [Google Scholar]
- 33.Yang, J., Kawai, Y., Hanson, R. W., and Arinze, I. J. (2001) J. Biol. Chem. 276 25742–25752 [DOI] [PubMed] [Google Scholar]
- 34.Davis, M. G., Kawai, Y., and Arinze, I. J. (2000) Biochem. J. 346 455–461 [PMC free article] [PubMed] [Google Scholar]
- 35.Arinze, I. J., and Kawai, Y. (2003) J. Biol. Chem. 278 17785–17791 [DOI] [PubMed] [Google Scholar]
- 36.Jones, S. M., Luo, M., Peters-Golden, M., and Brock, T. G. (2003) J. Biol. Chem. 278 10257–10263 [DOI] [PubMed] [Google Scholar]
- 37.Maertens, G., Cherepanov, P., Debyser, Z., Engelborghs, Y, and Engelman, A. (2004) J. Biol. Chem. 279 33421–33429 [DOI] [PubMed] [Google Scholar]
- 38.Herrmann, F., Lee, J., Bedford, M. T., and Fackelmayer, F. O. (2005) J. Biol. Chem. 280 38005–38010 [DOI] [PubMed] [Google Scholar]
- 39.Malnou, C. E., Salem, T., Brockly, F., Wodrich, H., Piechaczyk, M., and Jariel-Encontre, I. (2007) J. Biol. Chem. 282 31046–31059 [DOI] [PubMed] [Google Scholar]
- 40.Macara, I. G. (2001) Microbiol. Mol. Biol. Rev. 65 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaiahgari, S., Kleeberger, S. R., Cho, H.-Y., Kalvakolanu, D. V., and Reddy, S. P. (2004) J. Biol. Chem. 279 42302–42312 [DOI] [PubMed] [Google Scholar]
- 42.Moehlenkamp, J. D., and Johnson, J. A. (1999) Arch. Biochem. Biophys. 363 98–106 [DOI] [PubMed] [Google Scholar]
- 43.Kawai, Y., and Arinze, I. J. (2006) Cancer Res. 66 6563–6569 [DOI] [PubMed] [Google Scholar]
- 44.Moi, P., Chan, K., Asunis, I., Cao, A., and Kan, Y. W. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart, D., Killeen, E., Naquin, R., Alam, S., and Alam, J. (2003) J. Biol. Chem. 278 2396–2402 [DOI] [PubMed] [Google Scholar]
- 46.Görlich, D., and Kutay, U. (1999) Annu. Rev. Cell Dev. Biol. 15 607–660 [DOI] [PubMed] [Google Scholar]
- 47.Chook, Y. M., and Blobel, G. (2001) Curr. Opin. Struct. Biol. 11 703–715 [DOI] [PubMed] [Google Scholar]
- 48.Stewart, M., Baker, R. P., Bayliss, R., Clayton, L., Grant, R. P., Littlewood, T., and Matsuura, Y. (2001) FEBS Lett. 498 145–149 [DOI] [PubMed] [Google Scholar]
- 49.Goldfarb, D. S., Corbett, A. H., Mason, D. A., Harreman, M. T., and Adam, S. A. (2004) Trends Cell Biol. 14 505–514 [DOI] [PubMed] [Google Scholar]
- 50.Mosammaparast, N., and Pemberton, L. F. (2004) Trends Cell Biol. 14 547–556 [DOI] [PubMed] [Google Scholar]
- 51.Pemberton, L. F., and Paschal, B. M. (2005) Traffic 6 187–198 [DOI] [PubMed] [Google Scholar]
- 52.Lange, A., Mills, R. E., Lange, C. J., Stewart, M., Devine, S. E., and Corbett, A. H. (2007) J. Biol. Chem. 282 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, X. Y., Robinson, D., Veach, R. A., Liu, D., Timmons, S., Collins, R. D., and Hawiger, J. (2000) J. Biol. Chem. 275 16774–16778 [DOI] [PubMed] [Google Scholar]
- 54.Álvarez-Maqueda, M., Bekay, R. E., Alba, G., Monteseirín, J., Chacón, P., Vega, A., Martín-Nieto, J., Bedoya, F. J., Pintado, E., and Sobrino, F. (2004) J. Biol. Chem. 279 21929–21937 [DOI] [PubMed] [Google Scholar]
- 55.Köhler, M., Ansieau, S., Prehn, S., Leutz, A., Haller, H., and Hartmann, E. (1997) FEBS Lett. 417 104–108 [DOI] [PubMed] [Google Scholar]
- 56.Köhler, M., Speck, C., Christiansen, M., Bischoff, F. R., Prehn, S., Haller, H., Görlich, D., and Hartmann, E. (1999) Mol. Cell. Biol. 19 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, D. D. (2006) Drug Metab. Rev. 38 769–789 [DOI] [PubMed] [Google Scholar]
- 58.Lee, J.-M., Li, J., Johnson, D. A., Stein, T. D., Kraft, A. D., Calkins, M. J., Jakel, R. J., and Johnson, J. A. (2005) FASEB J. 19 1061–1066 [DOI] [PubMed] [Google Scholar]
- 59.Vandromme, M., Cavadore, J., Bonnieu, A., Froeschle, A., Lamb, N., and Fernandez, A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4646–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen, S. T., Jackson, P. K., and Van Etten, R. A. (1996) EMBO J. 15 1583–1595 [PMC free article] [PubMed] [Google Scholar]
- 61.Russo, G., Ricciardelli, G., and Pietropaolo, C. (1997) J. Biol. Chem. 272 5229–5235 [DOI] [PubMed] [Google Scholar]
- 62.Savory, J. G. A., Hsu, B., Laquian, I. R., Giffin, W., Reich, T., Haché, R. J. G., and Lefebvre, Y. A. (1999) Mol. Cell. Biol. 19 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui, P., Qin, B., Liu, N., Pan, G., and Pei, D. (2004) Exp. Cell Res. 293 154–163 [DOI] [PubMed] [Google Scholar]
- 64.Krauer, K., Buck, M., Flanagan, J., Belzer, D., and Sculley, T. (2004) J. Gen. Virol. 85 165–172 [DOI] [PubMed] [Google Scholar]
- 65.Hu, W., Philips, A. S., Kwok, J. C., Eisbacher, M., and Chong, B. H. (2005) Mol. Cell. Biol. 25 3087–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Do, H.-J., Song, H., Yang, H.-M., Kim, D.-K., Kim, N.-H., Kim, J.-H., Cha, K.-Y., Chung, H.-M., and Kim, J.-H. (2006) FEBS Lett. 580 1865–1871 [DOI] [PubMed] [Google Scholar]
- 67.Bedard, J. E. J., Purnell, J. D., and Ware, S. M. (2007) Hum. Mol. Genet. 16 187–198 [DOI] [PubMed] [Google Scholar]
- 68.Hanover, J. A., Love, D. C., DeAngelis, N., O'Kane, M. E., Lima-Miranda, R., Schulz, T., Yen, Y.-M., Johnson, R. C., and Prinz, W. A. (2007) J. Biol. Chem. 282 33743–33751 [DOI] [PubMed] [Google Scholar]
- 69.Ossareh-Nazari, B., Gwizdek, C., and Dargemont, C. (2001) Traffic 2 684–689 [DOI] [PubMed] [Google Scholar]
- 70.Riddick, G., and Macara, I. G. (2005) J. Cell Biol. 168 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li, W., Jain, M. R., Chen, C., Yue, X., Hebbar, V., Zhou, R., and Kong, A.-N. T. (2005) J. Biol. Chem. 280 28430–28438 [DOI] [PubMed] [Google Scholar]
- 72.Li, W., Yu, S.-W., and Kong, A.-N. T. (2006) J. Biol. Chem. 281 27251–27263 [DOI] [PubMed] [Google Scholar]
- 73.Jain, A. K., and Jaiswal, A. K. (2007) J. Biol. Chem. 282 16502–16510 [DOI] [PubMed] [Google Scholar]