Abstract
Fatty acid translocase (FAT/CD36) plays an important role in facilitating long chain fatty acid transport. FAT/CD36 gene deletion protects mice from high fat diet-induced obesity. In this study we have investigated the regulatory mechanism of FAT/CD36 expression at the transcription level. FAT/CD36 expression was activated during 3T3-L1 adipocyte differentiation, and FAT/CD36 protein levels were positively correlated with CCAAT/enhancer-binding protein α (C/EBPα) and peroxisome proliferator-activated receptor γ. However, a negative correlation was detected between FAT/CD36 and C/EBPβ. Overexpression of C/EBPα or C/EBPβ increased FAT/CD36 mRNA and protein levels in several types of cells. Restoration of C/EBPα or C/EBPβ expression in C/EBPα- or C/EBPβ-deficient mouse embryonic fibroblasts increased FAT/CD36 expression. However, in mouse embryonic fibroblasts C/EBPα was a more potent activator of FAT/CD36 expression than was C/EBPβ. Expression of C/EBPα robustly increased FAT/CD36 proximal promoter-directed luciferase expression in human embryonic kidney 293 cells. A C/EBP-responsive element was identified in the FAT/CD36 promoter by using 5′ and specific site mutations. The binding of C/EBPα in the FAT/CD36 promoter was detected by chromatin immunoprecipitation in 3T3-L1 adipocytes. These results demonstrated that C/EBPα regulates FAT/CD36 gene expression at the transcriptional level.
Fatty acid translocase (FAT/CD36)4 is an 88-kDa single chain membrane glycoprotein expressed in many tissues and cell types, including adipocytes, heart, skeletal muscle, platelets, monocytes, and macrophages. Because of its broad ligand binding, FAT/CD36 has been defined as a class B scavenger receptor with remarkably diverse biological functions such as long chain fatty acid transport and signal transduction (1, 2). Studies using FAT/CD36-deficient mice have demonstrated that FAT/CD36 plays an important role in fatty acid (FA) uptake and oxidation in skeletal muscle and lipid accumulation in adipocytes (3–6). FAT/CD36–/– mice are lean and protected from high fat diet-induced obesity (7). In contrast, overexpression of FAT/CD36 in skeletal muscle increases FA oxidation and decreases blood triglycerides and FA (8).
Although many studies have illustrated the function and tissue expression of FAT/CD36, understanding of the regulation of FAT/CD36 expression is rather limited. The tissue pattern of FAT/CD36 expression indicates that transcriptional regulation might play an important role. It has been suggested that oxidized low density lipoprotein increases FAT/CD36 expression in macrophages through PPARγ-mediated FAT/CD36 transcriptional activation (9). However, a later study indicated there is no peroxisome proliferator-activated receptor γ (PPAR)-responsive element in the FAT/CD36 promoter and PPARγ or PPARα ligands increase FAT/CD36 transcription through an indirect mechanism (10).
CCAAT/enhancer-binding protein α (C/EBPα) is the prototypical member of the C/EBP transcription factor family (11). C/EBPα is highly expressed in hepatocytes, adipocytes, granulocytes, macrophages, and also in skeletal muscle (11, 12), which mirrors the FAT/CD36 expression pattern. C/EBPα is a master adipogenic transcription factor that modulates gene expression not only during adipocyte differentiation but also adipose-specific gene expression in mature adipocytes (13). Compelling evidence indicates that C/EBPα plays a critical role in maintaining lipid and glucose metabolism. Our recent studies demonstrated that C/EBPα transactivation activity is up-regulated in type 2 diabetes by p38-mediated serine phosphorylation (14). The results of the current study show that both C/EBPα and C/EBPβ increase FAT/CD36 expression in different types of cells. However, C/EBPα exhibits a significantly higher potency than C/EBPβ. A C/EBP-responsive element was identified in the FAT/CD36 promoter. These data demonstrated that C/EBPα plays a key role in regulating FAT/CD36 expression.
EXPERIMENTAL PROCEDURES
Materials—Insulin, 3-isobutyl-1-methylxanthine, and dexamethasone were purchased from Sigma. SYBR Green and luciferin were purchased from Invitrogen. Anti-C/EBPα, C/EBPβ, PPARγ, and α-tubulin antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). FAT/CD36 antibody was from R&D Systems, Inc. (Minneapolis, MN).
Experimental Animals—C/EBPβ-deficient mice were bred as previously described in 129/SV and C57BL/6 mixed genetic background (15). 8-week-old male C/EBPβ–/– mice and wild type littermates were used. Tissue samples were collected after overnight fasting. Male C57BL/6J mice were purchased from the Jackson Laboratory. All mice were housed in a pathogen-free animal facility with 12/12-h light/dark cycle with free access to food and water. The experiments using mouse models were carried out under the Association for Assessment and Accreditation of Laboratory Animal Care guidelines with approval of the University of Kentucky Animal Care and Use Committee. Purified adenoviruses were diluted in phosphate-buffered saline immediately prior to injection. Adenovirus was injected through the tail vein at a dose of 1 × 109 plaque-forming units/mouse in 100 μl. Liver tissues were collected 3 days after injection. Adenovirus encoding GFP was used for control.
Cell Culture—FAO, C2C12, HEK293, and 3T3-L1 fibroblasts were purchased from ATCC. RAW264.7 macrophages were generously provided by Dr. Alan Daugherty (University of Kentucky, Lexington, KY). 3T3-L1CARΔ1 cells stably express the coxsackie-adenovirus receptor, which improves adenoviral transduction efficiency (16). Mouse C/EBPα gene-deficient (C/EBPα–/–) fibroblasts were a gift from Dr. Gretchen J. Darlington (Baylor College of Medicine, Houston, TX). C/EBPβ–/– mouse embryo fibroblasts (MEFs) were harvested from mouse embryos (17). The cells were maintained at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA). Adipocyte differentiation was induced by a commonly used protocol (18). For adenovirus transduction studies, purified adenoviruses were added to medium at 100 plaque-forming units/cell for all cells except RAW264.7, which were transduced with 200 plaque-forming units/cell.
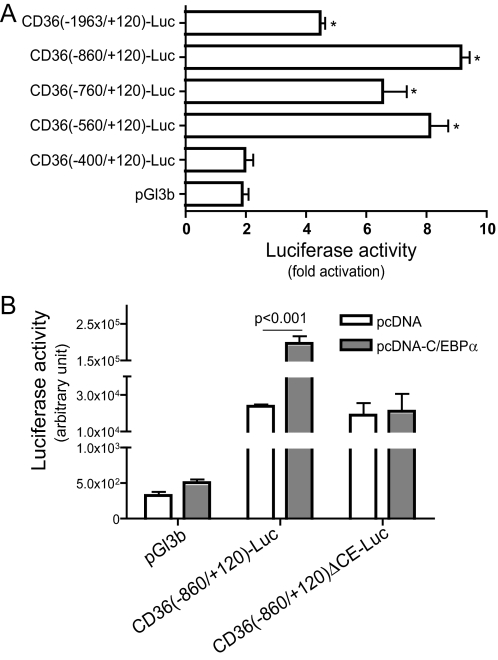
Plasmid Construction and Adenovirus Vector Preparation—The plasmids and adenoviruses encoding C/EBPα, C/EBPβ, or C/EBPα siRNA were previously described (19, 20). The FAT/CD36 promoter was cloned by PCR using mouse genomic DNA as template. The FAT/CD36 proximal promoter-directed luciferase gene reporter constructs were created by inserting the PCR product of FAT/CD36 5′-flanking regions into pGl3 basic vector (Promega). The structures of these gene reporter constructs are illustrated in Fig. 4A. Site-directed mutations of C/EBPα element (CE), (5′-ACATGTCGTAAGGA-3′ to 5′-ACATGTCcgAcGGA-3′) in the mouse FAT/CD36 promoter were constructed using a QuikChange mutagenesis kit following the instruction provided by the supplier (Stratagene). The plasmids were verified by sequencing.
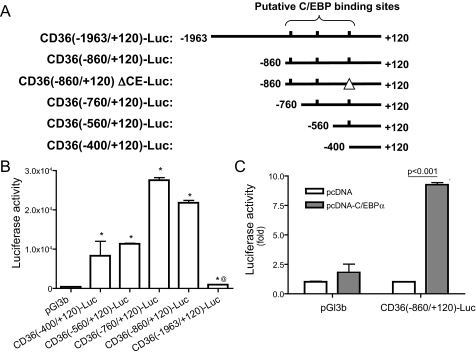
FIGURE 4.
C/EBPα stimulates FAT/CD36 promoter activity. A, luciferase gene reporter constructs were created by inserting the mouse FAT/CD36 proximal promoter with indicated 5′-deletions or specific CE mutation (CD36(–860/+120)ΔCE-Luc). Putative C/EBP binding sites are indicated as short vertical lines. The perfect palindromic consensus site is 5′-ATTGCGCAAT-3′. B, luciferase gene reporter constructs were transfected into HEK293 cells. C, C/EBPα-encoding plasmid and CD36(–860)-Luc or control vector were co-transfected into HEK293 cells. 24 h after transfection, luciferase activities were measured. Transfection efficiency was normalized by cytomegalovirus promoter-directed β-galactosidase. n = 6, Error bars indicate ± S.E. *, p < 0.05 versus pGl3b-transfected cells, @, p < 0.001 versus CD(–860/+120)-Luc-transfected cells.
Quantitative RT-PCR Analysis—Total RNA was prepared from cells or tissue samples using TRIzol reagent following the manufacturer's protocol (Invitrogen). RNA samples were treated with DNase to remove any contaminating DNA. cDNA was synthesized using SuperScript III Reverse Transcriptase and oligo(dT)12–18 primers (Invitrogen). Real-time PCR was performed using the Mx3000P Real-Time PCR system (Stratagene) using SYBR Green dye (Molecular Probes). The sequences for the primers are: FAT/CD36, 5′-TGGCCTTACTTGGGATTGG-3′ and 5′-CCAGTGTATATGTAGGCTCATCCA-3′, and for 18 S rRNA, 5′-CGAAAGCATTTGCCAAGAAT-3′ and 5′-AGTCGGCATCGTTTATGGTC-3′.
Luciferase Assay—Reporter constructs and expression plasmids were transfected into cells using FuGENE 6. A pCMV-β-galactosidase plasmid (Clontech Laboratories, Inc., Palo Alto, CA) was co-transfected for transfection efficiency control. 24 h after transfection, cells were lysed and luciferase activity was measured (21). Luciferase activities were normalized to β-galactosidase luminescence for transfection efficiency and expressed as relative luciferase activity.
Chromatin Immunoprecipitation Assays—Chromatin immunoprecipitation was previously described in detail (18). Briefly, 3T3-L1 preadipocytes and adipocytes were cross-linked with 1% formaldehyde (Sigma) in serum-free Dulbecco's modified Eagle's medium at room temperature for 5–10 min. Chromatin was sheared by sonicating on ice, yielding chromatin fragments of 100–600 bp. Chromatin complexes were immunoprecipitated for 12–18 h at 4 °C while rotating with 5 μg of anti-C/EBPα antibody or with normal rabbit serum to provide controls. DNA was isolated and purified using Quick PCR Purification kit (Qiagen, Valencia, CA). The binding of C/EBPα to the FAT/CD36 promoter was detected by PCR. The sequences for primers over –470 to –321 nt of the mouse FAT/CD36 promoter were: forward, 5′-GGTTCTGTTTGGGTGGAGAA-3′, reverse, 5′-TGCACATTAATCCCTTCGTG-3′. PCR produced a 149-bp product.
Western Blot Analysis—Total and nuclear proteins were extracted as described previously (19). Relative protein levels of C/EBPα, C/EBPβ, PPARγ, and FAT/CD36 were measured by Western blot using specific antibodies. The bands from Western blots were quantified using the ChemiDoc XRS gel documentation system and Quantity One software (Bio-Rad).
Data Analysis—The data are expressed as the means ± S.E. Statistical analyses were performed using Student's t test or analysis of variance, followed by a contrast test with Tukey or Dunnett error protection. The differences were considered significant at p < 0.05.
RESULTS
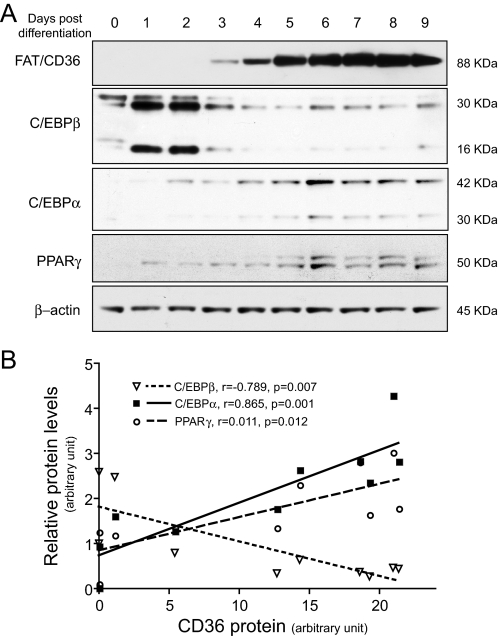
Parallel Expression of C/EBPα and FAT/CD36 during 3T3-L1 Adipocyte Differentiation—The role of FAT/CD36 in FA transport and lipid accumulation in adipocytes has been well studied. FAT/CD36 is highly expressed in adipocytes and plays an important role in FA uptake and lipid accumulation in adipocytes. Because transcriptional regulation of adipocyte differentiation has been extensively studied, we used 3T3-L1 cells as a cellular model to study the expression of FAT/CD36 and adipogenic transcription factors during adipocyte differentiation. Consistent with previous reports, no FAT/CD36 protein was detected in 3T3-L1 preadipocytes (Fig. 1A). A clear band of FAT/CD36 protein was detected from cells 3 days post differentiation. FAT/CD36 protein levels reached maximal levels at days 7 and 8 (Fig. 1A). The expression of C/EBPα, C/EBPβ, and PPARγ was also measured (Fig. 1A). The protein levels of FAT/CD36 were positively correlated with the total protein levels of C/EBPα (p = 0.0012, r = 0.865) and PPARγ (p = 0.0119, r = 0.753) (Fig. 1B). Interestingly, C/EBPβ protein levels were negatively correlated with FAT/CD36 protein levels during 3T3-L1 adipocyte differentiation (p = 0.0067, r =–0.7886) (Fig. 1B). These results led us to hypothesize that C/EBPα modulates FAT/CD36 gene transcription during adipocyte differentiation. Because the regulatory effect of PPARγ on FAT/CD36 expression has been reported (9), we focused our study on C/EBPα and C/EBPβ.
FIGURE 1.
FAT/CD36 protein levels positively correlate with C/EBPα or PPARγ during 3T3-L1 adipocyte differentiation. Adipocyte differentiation was induced by treatment with a hormone mixture. Total or nuclear protein was extracted at indicated time points. Relative protein levels were determined by Western blotting (A). The correlation of FAT/CD36 protein levels with C/EBPα, C/EBPβ, or PPARγ was analyzed by Pearson correlation (B). The image and graph represent four independent studies.
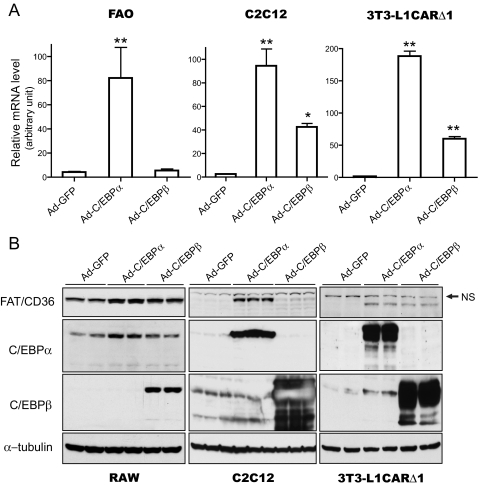
C/EBPα Increases FAT/CD36 Gene Expression—To determine the effects of C/EBPα and C/EBPβ on FAT/CD36 expression, C/EBPα or C/EBPβ were overexpressed using adenovirus vectors. Several types of cells were used, including C2C12 myotubes, FAO hepatocytes, RAW264.7 macrophages, and 3T3-L1CARΔ1 preadipocytes and adipocytes. Both mRNA and protein levels of FAT/CD36 were measured 24 h after adenovirus transduction. C/EBPα or C/EBPβ protein levels were significantly increased by Ad-C/EBPα and Ad-C/EBPβ transduction, as monitored by Western blot (Fig. 2B or data not shown). As shown in Fig. 2A, overexpression of C/EBPα robustly increased FAT/CD36 mRNA in all three types of cells. Similarly, C/EBPβ overexpression increased FAT/CD36 mRNA levels in FAO, C2C12 myotubes, and 3T3-L1CARΔ1 adipocytes. However, the magnitude of FAT/CD36 mRNA increase in Ad-C/EBPβ-transduced cells was much less than in Ad-C/EBPα-transduced cells (Fig. 2A). Significant increases of FAT/CD36 protein levels also occurred in Ad-C/EBPα- or Ad-C/EBPβ-transduced C2C12 myotubes, RAW264.7 macrophages, and 3T3-L1CARΔ1 preadipocytes (Fig. 2B). These results indicate that both C/EBPα and C/EBPβ increase FAT/CD36 gene expression.
FIGURE 2.
C/EBPα and C/EBPβ increase FAT/CD36 expression. C2C12 myotubes and 3T3-L1CARΔ1 adipocytes were differentiated as outlined under “Experimental Procedures.” FAO and RAW264.7 were cultured to near confluency before adenovirus transduction. 24 h after adenovirus transduction, mRNA and protein were prepared. The expression levels of FAT/CD36 mRNA were measured by real-time PCR (A) and protein by Western blotting (B). 18 S rRNA was used as internal control for real-time PCR. FAT/CD36 mRNA levels were normalized by 18 S rRNA. The same membranes were stripped and probed with indicated antibodies. Error bars indicate ± S.E. *, p < 0.05, **, p < 0.001 versus Ad-GFP-transduced cells. NS, nonspecific binding.
As shown in Figs. 1 and 2B, no FAT/CD36 protein was detected in preadipocytes. However, overexpression of C/EBPα or C/EBPβ activated FAT/CD36 expression in these cells (Fig. 2B). These results suggest that C/EBPα and C/EBPβ play a critical role in FAT/CD36 gene expression.
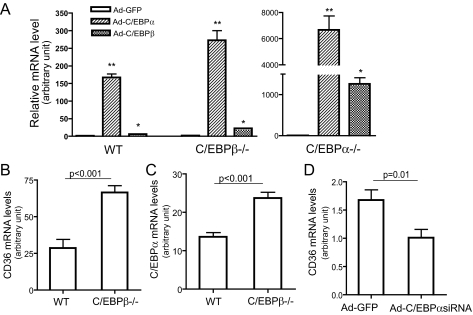
C/EBPα Is the More Potent Inducer of FAT/CD36 Gene Expression—The negative correlation of C/EBPβ and FAT/CD36 protein levels during adipocyte differentiation appears contradictory to the stimulating effect of C/EBPβ on FAT/CD36 expression observed in the above overexpression studies. It also raises the question of whether C/EBPα or C/EBPβ is predominant in controlling FAT/CD36 expression. To address these questions, the following studies were carried out. First, we compared FAT/CD36 expression levels between C/EBPα–/– and C/EBPβ–/– MEFs. Interestingly, unlike 3T3-L1 preadipocytes, FAT/CD36 mRNA and protein were detectable in wild type, C/EBPα–/–, and C/EBPβ–/– MEFs. However, basal FAT/CD36 protein levels were too low to be quantitatively analyzed by Western blot. Therefore, FAT/CD36 mRNA levels were analyzed using real-time PCR. The results showed that FAT/CD36 mRNA in C/EBPβ–/– MEFs was ∼9-fold higher than that in C/EBPα–/– MEF (data not shown). Consistent with the above studies, overexpression or restoration of C/EBPα or C/EBPβ in C/EBPα–/– or C/EBPβ–/– MEF, respectively, robustly increased FAT/CD36 mRNA levels (Fig. 3A). FAT/CD36 mRNA levels were increased more than 6000-fold by C/EBPα restoration in C/EBPα–/– MEFs, whereas FAT/CD36 mRNA levels were only increased ∼21-fold in Ad-C/EBPβ-transduced C/EBPβ–/– MEFs. Overexpression of C/EBPα induced much more dramatic increase of FAT/CD36 mRNA in all three MEFs compared with Ad-C/EBPβ-transduced cells (Fig. 3A). For an unknown reason, the same dosage of C/EBPβ adenoviruses led to significantly higher amounts of C/EBPβ protein compared with C/EBPα (Fig. 2B). However, FAT/CD36 protein levels were higher in Ad-C/EBPα-transduced cells than Ad-C/EBPβ-transduced cells (Fig. 2B). Together, these results indicate that C/EBPα is more potent than C/EBPβ in inducing FAT/CD36 expression.
FIGURE 3.
C/EBPα is more potent than C/EBPβ in inducing FAT/CD36 expression. A, effects of C/EBPα or C/EBPβ on FAT/CD36 expression in MEFs. Immortalized wild type, C/EBPα–/–, and C/EBPβ–/– MEFs were transduced with adenovirus vectors encoding C/EBPα, C/EBPβ, or GFP. mRNA levels of FAT/CD36 were detected by real-time PCR and normalized by 18 S rRNA, n = 4. B and C, increased FAT/CD36 and C/EBPα expression in the livers of C/EBPβ–/– mice. Total RNA was isolated from livers of C/EBPβ–/– mice after overnight fasting, n = 5. FAT/CD36 and C/EBPα mRNA levels were expressed relative to 18 S rRNA. D, knocking down C/EBPα reduces FAT/CD36 expression. Adenovirus vectors encoding C/EBPα siRNA or GFP were injected via the tail vein. Liver tissues were collected 3 days later. FAT/CD36 mRNA (normalized by 18 S rRNA) was measured using real-time PCR, n = 6. Error bars indicate ± S.E. *, p < 0.05, **, p < 0.001 versus Ad-GFP-treated cells.
To determine the regulatory effects of C/EBP on FAT/CD36 expression in vivo, wild type C57BL/6J or C/EBPβ gene-deficient liver cells were transduced with Ad vectors encoding C/EBPα or C/EBPβ protein. Consistent with the results from in vitro studies, FAT/CD36 mRNA and protein levels were significantly increased in the livers of Ad-C/EBPα- or Ad-C/EBPβ-treated mice (p < 0.05 versus Ad-GFP treated mice, data not shown). We then studied FAT/CD36 expression in liver and white fat tissues of C/EBPβ–/– mice and livers of Ad-C/EBPα siRNA-treated mice. To our surprise, FAT/CD36 expression levels were significantly increased in both liver and epididymal adipose tissues of C/EBPβ–/– mice compared with their control (Fig. 3B). These results are in line with results from a recent study that reported that the FAT/CD36 protein was significantly increased in livers of C/EBPβ–/– C57BL/6 mice (22). C/EBPα and C/EBPβ usually share DNA binding consensus sequences and may functionally compensate for each other in regulating certain target genes (23). Our previous study showed that reducing C/EBPα in mouse liver induced a compensatory increase in C/EBPβ levels (24). Therefore, we measured the C/EBPα expression levels in C/EBPβ-deficient mouse livers. As expected, C/EBPα mRNA levels were significantly elevated in C/EBPβ–/– livers (Fig. 3C), which implies that increased FAT/CD36 expression in C/EBPβ–/– mice may be caused by elevated C/EBPα. However, other mechanisms cannot be ruled out.
As we previously reported, Ad-C/EBPα siRNA tail vein injection almost exclusively transduced liver tissues (24). Three days after virus administration, endogenous liver C/EBPα protein levels were reduced ∼70% whereas C/EBPβ protein levels were significantly increased in Ad-C/EBPα siRNA-transduced mice (data not shown) (24). In contrast to C/EBPβ-deficient mice, FAT/CD36 mRNA levels were remarkably decreased in livers of C/EBPα siRNA adenovirus-treated mice (Fig. 3D). Together, these results indicate that C/EBPα plays an important role in FAT/CD36 gene expression.
C/EBPα Activates the FAT/CD36 Promoter—Because C/EBPα is a transcription factor and the above studies revealed that overexpression of C/EBPα increases FAT/CD36 mRNA levels, we conducted the following studies to determine whether C/EBPα regulates FAT/CD36 at the transcriptional level. Initially, we characterized the proximal promoter of FAT/CD36 at basal conditions. A series of luciferase gene reporter constructs was created by inserting deletion mutants of the mouse FAT/CD36 promoter into pGL3-basic vector (Fig. 4A). These gene reporter constructs were transfected into HEK293 cells. Luciferase activities directed by the 5′-flanking sequences from –1963 to +120 nt were only 2-fold higher than that in pGL3-basic control vector-transfected cells (Fig. 4B). However, significantly higher levels of luciferase activities (50–80-fold versus pGl3b) were detected in cells transfected with reporter constructs containing the FAT/CD36 promoter from –860 to +120 or from –400 to +120 nt (Fig. 4B). These results are consistent with results from a previous study that reported a similar FAT/CD36 promoter activation pattern in Mono Mac 6 cells (25). Our results also indicate that there is (are) repressive regulatory element(s) between –1963 and –860 nt of the 5′-flanking region of the mouse FAT/CD36 gene. Because there is no endogenous FAT/CD36 expression in HEK293 cells (26), we speculate that the repressive region or/and other repressive elements in our tested FAT/CD36 promoter sequences may be responsible for the silencing of FAT/CD36 gene expression in HEK293 cells. The lack of endogenous C/EBP expression in HEK293 cells (19) may result from silencing. However, more studies are required to verify this hypothesis.
To determine whether C/EBPα regulates FAT/CD36 transcription, pcDNA-C/EBPα and CD36(–860/+120)-Luc were co-transfected into HEK293 cells. As shown in Fig. 4C, ectopic expression of C/EBPα increased luciferase activities ∼9-fold, suggesting that C/EBPα enhances FAT/CD36 promoter activation. As expected, ectopic expression of the active C/EBPβ isoform liver active protein (LAP) also increased FAT/CD36 promoter activity (data not shown). These results indicate that C/EBP regulates FAT/CD36 expression at the transcriptional level, although the involvement of other mechanisms such as translation cannot be ruled out from the present studies.
C/EBPα-responding Element in the FAT/CD36 Promoter—We scanned the 1.0-kb 5′-flanking region of the mouse FAT/CD36 gene to identify putative C/EBP consensus binding sequences. Three putative C/EBP binding sites were identified (Fig. 4A, top line). As shown in Fig. 5A, overexpression of C/EBPα remarkably increased luciferase activities in the cells cotransfected with reporter constructs with different 5′-deletions of the FAT/CD36 promoter from –1963 to –560 nt. However, luciferase activities in CD36(–400/+120)-Luctransfected cells were not further increased by C/EBPα overexpression (Fig. 5A), which suggests that this construct does not respond to C/EBPα. The difference in C/EBPα-induced luciferase expression between construct CD36(–560/+120)-Luc and CD36(–400/+120)-Luc and the lack of response of CD36(–400/+120)-Luc to C/EBPα demonstrate that there is a C/EBPα-responsive element(s) within –560 to –400 nt. Based on sequence scanning, we hypothesized that –415 to –400 nt contains the CE in the FAT/CD36 promoter. We used a luciferase gene reporter construct containing the FAT/CD36 promoter (–860 to +120 nt) with mutation of the putative CE to further test this idea. As shown in Fig. 5B, mutation of the CE of the FAT/CD36 promoter diminished C/EBPα-stimulated luciferase gene expression. Together, these results demonstrate that there is one C/EBPα-responding element in the FAT/CD36 promoter.
FIGURE 5.
C/EBP-responding element in the FAT/CD36 promoter. A, luciferase gene reporter constructs containing the FAT/CD36 proximal promoter with 5′-deletions were co-transfected with pcDNA-C/EBPα into HEK293 cells. B, C/EBPα-encoding plasmid and luciferase gene reporter constructs with intact FAT/CD36 promoter or FAT/CD36 promoter with CE deletion or mutation were co-transfected into HEK293 cells. 24 h after transfection, cells were lysed and luciferase activities were measured with normalization for transfection efficiency. Luciferase activities were presented as C/EBPα-induced luciferase fold activation (A) compared with pcDNA empty vector-treated cells or assay readout (B). Error bars indicate ± S.E. *, p < 0.05 versus pGl3b and pcDNA-C/EBPα-treated cells.
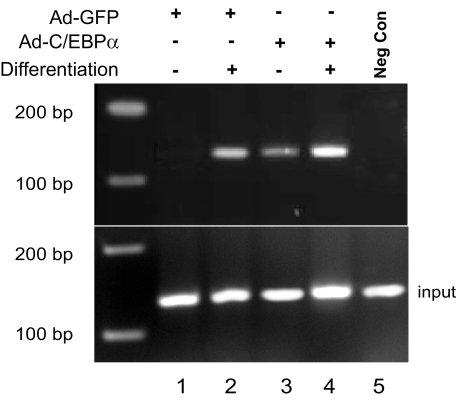
Binding of C/EBPα at the FAT/CD36 Promoter—3T3-L1CARΔ1 cells and chromatin immunoprecipitation assays were employed to investigate the direct association of C/EBPα with the FAT/CD36 promoter. Both 3T3-L1CARΔ1 preadipocytes and fully differentiated 3T3-L1CARΔ1 adipocytes were lysed for chromatin immunopreparation as described previously (21). A strong band was detected in the 3T3-L1CARΔ1 adipocytes, and C/EBPα overexpression further increased the band densities (Fig. 6, lanes 2 and 4). The binding of C/EBPα to the FAT/CD36 promoter was barely detectable in 3T3-L1CARΔ1 preadipocytes (lane 1), whereas a band was detected in Ad-C/EBPα-transduced preadipocytes (lane 3). Taken together with the results from the above promoter activation studies, the data indicate that C/EBPα binds to the mouse FAT/CD36 proximal promoter.
FIGURE 6.
Association of C/EBPα with the FAT/CD36 promoter. Confluent 3T3-L1CARΔ1 preadipocytes (lanes 1 and 3) and differentiated adipocytes (lanes 2 and 4) were transduced with adenovirus encoding C/EBPα or GFP. 24 h later, chromatin was extracted and binding of C/EBPα to the FAT/CD36 promoter was analyzed by chromatin immunoprecipitation using a pair of primers specifically targeting the mouse FAT/CD36 proximal promoter region. The negative control was prepared by immunoprecipitating chromatin from Ad-C/EBPα-treated adipocytes with normal rabbit serum instead of anti-C/EBPα antibody. The image is representative of four independent experiments. Lanes 1 and 3 are from undifferentiated 3T3-L1CARΔ1 cells, and 2, 4, and 5 are from mature 3T3-L1CARΔ1 adipocytes.
DISCUSSION
Because of the diverse ligand binding characteristics of FAT/CD36, multiple biological functions of FAT/CD36 have been identified. Although the mechanism of FA transport is still under debate, FAT/CD36 appears to play an important role in FA transport and metabolism. FAT/CD36 facilitates FA transport in skeletal muscle (28). Deletion of the FAT/CD36 gene leads to reduced FA transport and oxidation in skeletal muscle under both basal and insulin stimulation conditions (29). Compelling evidence has demonstrated the importance of FAT/CD36 in adipocyte differentiation and lipid accumulation (5, 30). Importantly, most studies have shown that FAT/CD36 expression is elevated in a variety of tissues in insulin-resistant and type 2 diabetic subjects or animals (31–35). High level FAT/CD36 might result in lipid accumulation, which is supported by studies using FAT/CD36 null mice (3). Lipid accumulation in adipose and skeletal muscle has been directly linked to the development of obesity and insulin resistance. In addition, lipid accumulation in macrophages plays an important role in atherogenesis. Insulin resistance is the common feature of obesity and type 2 diabetes. However, it is not clear whether increased FAT/CD36 induces insulin resistance or insulin resistance leads to increased FAT/CD36 expression. Defective insulin signaling has been reported as the underlying mechanism for increased FAT/CD36 expression in macrophages (33). In contrast, some studies have indicated that insulin up-regulates FAT/CD36 expression and translocation from an endosomal pool to the plasma membrane (28, 36). Apparently, more studies are required to address how FAT/CD36 expression is elevated in obesity and type 2 diabetes. Our current study provides evidence indicating C/EBPα up-regulates FAT/CD36 expression at the transcription level. Our previous studies have demonstrated that, although the C/EBPα protein level is not significantly altered under diabetic or hyperglycemia conditions, C/EBPα transactivation is increased by p38-mediated serine phosphorylation (14, 24). Therefore, we speculate that increased C/EBPα transcriptional activity may be another mechanism for the elevated expression of FAT/CD36 in diabetes.
Previous studies have suggested that increased FAT/CD36 protein in insulin resistance or diabetes results from elevation of FAT/CD36 protein synthesis or reduction of FAT/CD36 protein degradation (31, 33). However, several studies indicate that FAT/CD36 mRNA levels usually parallel protein levels (27, 30, 37). Work from our laboratory has shown that both FAT/CD36 mRNA and protein are increased in liver and skeletal muscle tissues of high fat diet-induced obese mice.5 Although mRNA levels can be determined by many factors, transcription is the first and most important step. Therefore, we propose that elevated FAT/CD36 gene transcription contributes to increased FAT/CD36 expression in obesity and diabetes.
The unique FAT/CD36 expression pattern during adipocyte differentiation provides a very useful cellular model to study the transcriptional regulation of FAT/CD36 expression. C/EBPα and PPARγ are well known as master adipogenic transcription factors. Our study showed that the expression of FAT/CD36 is positively correlated with both C/EBPα and PPARγ. Despite uncertainty regarding the location of a PPARγ-responsive element in the FAT/CD36 gene, PPARγ has been identified as a transcription factor for the FAT/CD36 gene (9, 10). Interestingly, our study demonstrated that FAT/CD36 expression is induced in PPARγ-overexpressing and troglitazone-treated preadipocytes, but not in preadipocytes overexpressing PPARγ or treated with troglitazone alone (data not shown). These results indicate that PPARγ-directed FAT/CD36 expression is ligand-dependent, which fits with the nuclear receptor nature of PPARγ. In contrast, overexpression of C/EBPα or C/EBPβ alone can activate FAT/CD36 gene expression in preadipocytes. However, C/EBPα or C/EBPβ overexpression did not increase endogenous PPARγ expression in preadipocytes (data not shown). Furthermore, our current study has identified a C/EBP element in the FAT/CD36 proximal promoter. Therefore, we conclude that C/EBPα plays an important role in modulating FAT/CD36 gene transcription. Although our study cannot rule out the possibility of an increase of endogenous PPARγ ligand(s) and the involvement of PPARγ in C/EBP-induced FAT/CD36 gene transcription, the results from promoter activation studies demonstrated that C/EBPα most likely directly up-regulates FAT/CD36 transcription.
C/EBPα and C/EBPβ are two major members of the C/EBP family. Although C/EBPα and C/EBPβ usually share the same DNA binding consensus, different potencies in regulation of specific target genes have been well documented. Our previous study demonstrated a cluster of C/EBP elements in the intronic enhancer of the human adiponectin gene (19). However, C/EBPβ gene deletion did not alter adiponectin gene expression in adipose tissue of mice (19). In this study, we employed C/EBPα or C/EBPβ null MEFs to investigate the difference of these two C/EBP transcription factors in FAT/CD36 transcription. Our results indicate that C/EBPα is the more potent regulator of FAT/CD36 gene transcription and also may be responsible for the elevated FAT/CD36 expression in C/EBPβ-deficient mice. The underlying mechanism for the differential activity of C/EBPα and C/EBPβ in controlling FAT/CD36 gene transcription and the basis for cell-specific differences in their effects requires additional experimental investigation.
In summary, our study shows that both C/EBPα and C/EBPβ can directly up-regulate FAT/CD36 gene transcription through a C/EBP-responding element at the proximal promoter. However, C/EBPα is more potent than C/EBPβ in FAT/CD36 promoter activation and gene expression.
This work was supported in part by National Institutes of Health Grant RDK077643A (to J. S.), American Diabetes Association Grant 7-07-CD-23 (to J. S.), and American Heart Association Grant 0665289B (to J. S.) and by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (to P. F. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FAT/CD36, fatty acid translocase; FA, fatty acid; C/EBP, CCAAT/enhancer-binding protein; PPARγ, peroxisome proliferator-activated receptor γ; GFP, green fluorescent protein; HEK, human embryonic kidney; MEF, mouse embryonic fibroblast; siRNA, small interfering RNA; nt, nucleotide; Ad, adenovirus; CE, C/EBPα element.
L. Qiao and J. Shao, unpublished observation.
References
- 1.Febbraio, M., Hajjar, D. P., and Silverstein, R. L. (2001) J. Clin. Investig. 108 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaffer, J. E. (2002) Am. J. Physiol. 282 E239–E246 [DOI] [PubMed] [Google Scholar]
- 3.Febbraio, M., Abumrad, N. A., Hajjar, D. P., Sharma, K., Cheng, W., Pearce, S. F. A., and Silverstein, R. L. (1999) J. Biol. Chem. 274 19055–19062 [DOI] [PubMed] [Google Scholar]
- 4.Coburn, C. T., Knapp, F. F., Jr., Febbraio, M., Beets, A. L., Silverstein, R. L., and Abumrad, N. A. (2000) J. Biol. Chem. 275 32523–32529 [DOI] [PubMed] [Google Scholar]
- 5.Ibrahimi, A., Sfeir, Z., Magharaie, H., Amri, E.-Z., Grimaldi, P., and Abumrad, N. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohl, J., Ring, A., Korkmaz, U., Ehehalt, R., and Stremmel, W. (2005) Mol. Biol. Cell 16 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajri, T., Hall, A. M., Jensen, D. R., Pietka, T. A., Drover, V. A., Tao, H., Eckel, R., and Abumrad, N. A. (2007) Diabetes 56 1872–1880 [DOI] [PubMed] [Google Scholar]
- 8.Ibrahimi, A., Bonen, A., Blinn, W. D., Hajri, T., Li, X., Zhong, K., Cameron, R., and Abumrad, N. A. (1999) J. Biol. Chem. 274 26761–26766 [DOI] [PubMed] [Google Scholar]
- 9.Nagy, L., Tontonoz, P., Alvarez, J. G., Chen, H., and Evans, R. M. (1998) Cell 93 229–240 [DOI] [PubMed] [Google Scholar]
- 10.Sato, O., Kuriki, C., Fukui, Y., and Motojima, K. (2002) J. Biol. Chem. 277 15703–15711 [DOI] [PubMed] [Google Scholar]
- 11.McKnight, S. L. (2001) Cell 107 259–261 [DOI] [PubMed] [Google Scholar]
- 12.Wagatsuma, A. Mol. Cell. Biochem. 304 25–33 [DOI] [PubMed]
- 13.Darlington, G. J., Ross, S. E., and MacDougald, O. A. (1998) J. Biol. Chem. 273 30057–30060 [DOI] [PubMed] [Google Scholar]
- 14.Qiao, L., MacDougald, O. A., and Shao, J. (2006) J. Biol. Chem. 281 24390–24397 [DOI] [PubMed] [Google Scholar]
- 15.Sterneck, E., Tessarollo, L., and Johnson, P. F. (1997) Genes Dev. 11 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlicky, D. J., DeGregori, J., and Schaack, J. (2001) J. Lipid Res. 42 910–915 [PubMed] [Google Scholar]
- 17.Sebastian, T., Malik, R., Thomas, S., Sage, J., and Johnson, P. F. (2005) EMBO J. 24 3301–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao, L., Schaack, J., and Shao, J. (2006) Endocrinology 147 865–874 [DOI] [PubMed] [Google Scholar]
- 19.Qiao, L., MacLean, P. S., Schaack, J., Orlicky, D. J., Darimont, C., Pagliassotti, M., Friedman, J. E., and Shao, J. (2005) Diabetes 54 1744–1754 [DOI] [PubMed] [Google Scholar]
- 20.Shao, J., Qiao, L., Janssen, R. C., Pagliassotti, M., and Friedman, J. E. (2005) Diabetes 54 976–984 [DOI] [PubMed] [Google Scholar]
- 21.Qiao, L., and Shao, J. (2006) J. Biol. Chem. 281 39915–39924 [DOI] [PubMed] [Google Scholar]
- 22.Schroeder-Gloeckler, J. M., Rahman, S. M., Janssen, R. C., Qiao, L., Shao, J., Roper, M., Fischer, S. J., Lowe, E., Orlicky, D. J., McManaman, J. L., Palmer, C., Gitomer, W. L., Huang, W., O'Doherty, R. M., Becker, T. C., Klemm, D. J., Jensen, D. R., Pulawa, L. K., Eckel, R. H., and Friedman, J. E. (2007) J. Biol. Chem. 282 15717–15729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, S. S., Chen, J. F., Johnson, P. F., Muppala, V., and Lee, Y. H. (2000) Mol. Cell. Biol. 20 7292–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao, L., MacLean, P. S., You, H., Schaack, J., and Shao, J. (2006) Endocrinology 147 3060–3069 [DOI] [PubMed] [Google Scholar]
- 25.Armesilla, A. L., Calvo, D., and Vega, M. A. (1996) J. Biol. Chem. 271 7781–7787 [DOI] [PubMed] [Google Scholar]
- 26.Tao, N., Wagner, S. J., and Lublin, D. M. (1996) J. Biol. Chem. 271 22315–22320 [DOI] [PubMed] [Google Scholar]
- 27.Chabowski, A., Chatham, J. C., Tandon, N. N., Calles-Escandon, J., Glatz, J. F. C., Luiken, J. J. F. P., and Bonen, A. (2006) Am. J. Physiol. 291 E675–E682 [DOI] [PubMed] [Google Scholar]
- 28.Chabowski, A., Coort, S. L. M., Calles-Escandon, J., Tandon, N. N., Glatz, J. F. C., Luiken, J. J. F. P., and Bonen, A. (2004) Am. J. Physiol. 287 E781–E789 [DOI] [PubMed] [Google Scholar]
- 29.Bonen, A., Han, X.-X., Habets, D. D. J., Febbraio, M., Glatz, J. F. C., and Luiken, J. J. F. P. (2007) Am. J. Physiol. 292 E1740–E1749 [DOI] [PubMed] [Google Scholar]
- 30.Sfeir, Z., Ibrahimi, A., Amri, E., Grimaldi, P., and Abumrad, N. (1999) Mol. Cell. Biochem. 192 3–8 [PubMed] [Google Scholar]
- 31.Griffin, E., Re, A., Hamel, N., Fu, C., Bush, H., McCaffrey, T., and Asch, A. S. (2001) Nat. Med. 7 840–846 [DOI] [PubMed] [Google Scholar]
- 32.Sampson, M. J., Davies, I. R., Braschi, S., Ivory, K., and Hughes, D. A. (2003) Atherosclerosis 167 129–134 [DOI] [PubMed] [Google Scholar]
- 33.Liang, C.-P., Han, S., Okamoto, H., Carnemolla, R., Tabas, I., Accili, D., and Tall, A. R. (2004) J. Clin. Investig. 113 764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coort, S. L., Hasselbaink, D. M., Koonen, D. P., Willems, J., Coumans, W. A., Chabowski, A., van der Vusse, G. J., Bonen, A., Glatz, J. F., and Luiken, J. J. (2004) Diabetes 53 1655–1663 [DOI] [PubMed] [Google Scholar]
- 35.Bonen, A., Parolin, M. L., Steinberg, G. R., Calles-Escandon, J., Tandon, N. N., Glatz, J. F., Luiken, J. J., Heigenhauser, G. J., and Dyck, D. J. (2004) FASEB J. 18 1144–1146 [DOI] [PubMed] [Google Scholar]
- 36.Chabowski, A., Coort, S. L. M., Calles-Escandon, J., Tandon, N. N., Glatz, J. F. C., Luiken, J. J. F. P., and Bonen, A. (2006) FEBS Lett. 579 2428–2432 [DOI] [PubMed] [Google Scholar]
- 37.Koonen, D. P. Y., Jacobs, R. L., Febbraio, M., Young, M. E., Soltys, C.-L. M., Ong, H., Vance, D. E., and Dyck, J. R. B. (2007) Diabetes 56 2863–2871 [DOI] [PubMed] [Google Scholar]