Abstract
The Tor1,2 protein kinases globally influence many cellular processes including nitrogen-responsive gene expression that correlates with intracellular localization of GATA transcription activators Gln3 and Gat1/Nil1. Gln3-Myc13 and Gat1-Myc13 are restricted to the cytoplasm of cells provided with good nitrogen sources, e.g. glutamine. Following the addition of the Tor1,2 inhibitor, rapamycin, both transcription factors relocate to the nucleus. Gln3-Myc13 localization is highly dependent upon Ure2 and type 2A-related phosphatase, Sit4. Ure2 is required for Gln3 to be restricted to the cytoplasm of cells provided with good nitrogen sources, and Sit4 is required for its location to the nucleus following rapamycin treatment. The paucity of analogous information concerning Gat1 regulation prompted us to investigate the effects of deleting SIT4 and URE2 on Gat1-Myc13 localization, DNA binding, and NCR-sensitive transcription. Our data demonstrate that Tor pathway control of NCR-responsive transcription bifurcates at the regulation of Gln3 and Gat1. Gat1-Myc13 localization is not strongly influenced by deleting URE2, nor is its nuclear targeting following rapamycin treatment strongly dependent on Sit4. ChIP experiments demonstrated that Gat1-Myc13 can bind to the DAL5 promoter in the absence of Gln3. Gln3-Myc13, on the other hand, cannot bind to DAL5 in the absence of Gat1. We conclude that: (i) Tor pathway regulation of Gat1 differs markedly from that of Gln3, (ii) nuclear targeting of Gln3-Myc13 is alone insufficient for its recruitment to the DAL5 promoter, and (iii) the Tor pathway continues to play an important regulatory role in NCR-sensitive transcription even after Gln3-Myc13 is localized to the nucleus.
Increasing use of rapamycin analogues clinically and in Phase II and III clinical trials has greatly stimulated investigation of its cellular target mTor (1–4). This global regulator influences many cellular processes, and the model organism, Saccharomyces cerevisiae has been particularly useful in elucidating the biochemical mechanisms through which such regulation is achieved. In contrast with higher eukaryotes, S. cerevisiae contains two Tor serine/threonine protein kinases, Tor1 and Tor2 (5–7). Activities of the nitrogen catabolite repression (NCR)-sensitive4 GATA family transcription activators, Gln3 and Gat1, have been used as downstream reporters of Tor1,2-mediated gene regulation, and this has increased interest in their regulation as well (8–16). The utility of GATA factor localization as a Tor reporter derives from the correlation that Gln3 and Gat1 respond similarly to rapamycin inhibition of Tor1,2, to nitrogen starvation, or when cells are provided with a poor nitrogen source (proline); they localize to the nucleus, and NCR-sensitive transcription increases. On the other hand, with good nitrogen sources (e.g. glutamine and in some strains ammonia), transcription of NCR-sensitive genes encoding proteins required for the transport and utilization of poor nitrogen sources is minimal, which correlates with Gln3-Myc13 and Gat1-Myc13 being sequestered in the cytoplasm, a response that, in the case of Gln3, requires Ure2 (8–18). The findings that Gln3 interacts with Ure2 in vivo and can be isolated as a Gln3-Ure2 complex in vitro extended the above correlations and offered a possible mechanism of how cytoplasmic sequestration of the GATA factors might be achieved (8, 11, 19, 20).
These and other correlations led to the proposal that excess nitrogen activates Tor1,2 (8–14, 21–24), although the precise mechanism remains unknown. They in turn phosphorylate Tap42, which inhibits the protein phosphatase Sit4. Upon rapamycin treatment, Tor is inactivated, and Tap42 dissociates from Sit4, which dephosphorylates Gln3 and thereby dissociates the Gln3-Ure2 complex. Dephosphorylated Gln3 can then enter the nucleus and mediate NCR-sensitive transcription. Gat1 was reported to be similarly regulated (8). Subsequently, protein kinase Npr1 was posited to be a negative regulator of nuclear Gln3 localization (25).
This model has stimulated detailed studies of the steps outlined above. Although intracellular Gln3 phosphorylation and localization sometimes positively correlated, as predicted by the model of Tor pathway structure and operation, in other cases experimental observations were inconsistent with the predictions. Detailed investigations of expected correlations that failed to occur repeatedly led to alternative explanations of existing data and concomitantly revised and increased our understanding of Tor1,2 and GATA factor regulation. Among the most important findings have been the observations that: (i) in its active form, Sit4 is in a complex with Tap42 (21, 22, 27); (ii) although methionine sulfoximine, an inhibitor of glutamine biosynthesis, and rapamycin treatment both cause nuclear Gln3-Myc13 localization (28, 29), they produce opposite effects on Gln3-Myc13 phosphorylation, i.e. the former increases phosphorylation, whereas the latter decreases it (29); (iii) Sit4 remains active with respect to Gln3 dephosphorylation in the presence of both good and poor nitrogen sources, i.e. its activity is not demonstrably nitrogen source-responsive (30); (iv) nitrogen source-dependent changes in Gln3-Myc13 phosphorylation become demonstrable when SIT4 is deleted, suggesting that nitrogen-responsive protein kinase activity rather than Sit4 phosphatase activity is the primary determining link between nitrogen availability and Gln3-Myc13 phosphorylation (30); and (v) Npr1 protein kinase participates in Gln3 regulation indirectly by influencing the uptake of ammonia (31, 32).
The studies described above evaluated the influence of nutrients, Tor1,2 inhibitors, and type 2A-related phosphatase activities (Sit4, Pph3) on Gln3-Myc13 phosphorylation and localization. Missing from these analyses, however, are data that analyze and correlate the requirements of GATA factor localization with in vivo DNA binding and NCR-sensitive transcription. Also missing are data that address the regulation of Gat1. Although Gat1 was concluded to be regulated analogously to Gln3 (8), several predicted responses have eluded demonstration (8). Gat1-mediated transcription is NCR-sensitive, but it has not yet been possible to demonstrate a Gat1-Ure2 complex in vitro (7, 8, 33). Further, nitrogen source or rapamycin-dependent changes in Gat1-Myc13 phosphorylation have not been demonstrated, even though changes in Gat1-Myc13 phosphorylation in response to carbon starvation can be readily observed (33).
To provide the missing information cited above, we investigated the requirements of type 2A-related phosphatases (Sit4 and Pph3) for NCR-sensitive gene expression and compared them with those expected from previous studies of Gln3-Myc13 localization (30). This led us to investigate rapamycin-induced Gat1-Myc13 localization, DNA binding, and NCR-sensitive transcription in wild type and type 2A-related phosphatase mutant strains. These investigations demonstrate that Tor1,2 pathway regulation of NCR-sensitive gene transcription bifurcates at the level of the GATA factors Gln3 and Gat1.
MATERIALS AND METHODS
Strains and Culture Conditions: S. cerevisiae—Strains used in this work are listed in Table 1. Growth conditions were identical to those described in Tate et al. (30) and Scherens et al. (34). Rapamycin (Sigma and LC Laboratories) and methionine sulfoximine (Sigma) were prepared as described earlier (30) and used as indicated in the figure legends.
TABLE 1.
Strains used in the work
| Strain | Background | Parent | Genotype | Primer |
|---|---|---|---|---|
| TB50 | TB | (12) | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa | None |
| TB123 | TB | (12) | MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX] | None |
| TB136-2a | TB | (12) | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], sit4::kanMX | None |
| TB138-1a | TB | (12) | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, ure2::URA3, GLN3-MYC13[KanMX] | None |
| FV003 | TB | TB123 | MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], pph3::natMX | pph3: 5′, -400 to -379 and -22 to -1 3′ 927 to 950 and 1206 to 1228 |
| FV004 | TB | TB136-2a | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], sit4::kanMX, pph3::natMX | pph3: 5′, -400 to -379 and -22 to -1 3′ 927 to 950 and 1206 to 1228 |
| FV005 | TB | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gln3::kanMX | gln3: 5′, -438 to -421 and -15 to -1 3′ 2194 to 2211 and 2597 to 2614 |
| FV006 | TB | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gat1::natMX | gat1: 5′, -422 to -405 and -15 to -1 3′ 1534 to 1555 and 1879 to 1896 |
| FV008 | TB | TB136-2a | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], sit4::kanMX, gat1::natMX | gat1: 5′, -422 to -405 and -15 to -1 3′ 1534 to 1555 and 1879 to 1896 |
| FV018 | TB | TB123 | MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], gat1::natMX | gat1: 5′, -422 to -405 and -15 to -1 3′ 1534 to 1555 and 1879 to 1896 |
| FV029 | TB | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, sit4::natMX | sit4: 5′, -450 to -429 and -23 to -1 3′ 937 to 955 and 1380 to 1400 |
| FV030 | TB | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, sit4::natMX, gln3::kanMX | sit4: 5′, -450 to -429 and -23 to -1 3′ 937 to 955 and 1380 to 1400 |
| gln3: 5′, -438 to -421 and -15 to -1 3′ 2194 to 2211 and 2597 to 2614 | ||||
| FV063 | TB | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3] | 5′ GAT1-F2, 3′ GAT1-R1 |
| FV064 | TB | FV005 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gln3::kanMX, GAT1-MYC13[HIS3] | 5′ GAT1-F2, 3′ GAT1-R1 |
| FV065 | TB | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3], pph3::natMX | pph3: 5′, -400 to -379 and -22 to -1 3′ 927 to 950 and 1206 to 1228 |
| FV066 | TB | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3], sit4::kanMX | sit4: 5′, -450 to -429 and -23 to -1 3′ 937 to 955 and 1380 to 1400 |
| FV067 | TB | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3], pph3::natMX, sit4::kanMX | pph3: 5′, -400 to -379 and -22 to -1 3′ 927 to 950 and 1206 to 1228 |
| sit4: 5′, -450 to -429 and -23 to -1 3′ 937 to 955 and 1380 to 1400 | ||||
| FV071 | TB | TB136-2a | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, ure2::natMX, GLN3-MYC13[KanMX], sit4::kanMX | ure2: 5′, -300 to -279 and -21 to -1 3′ 1066 to 1084 and 1325 to 1345 |
| FV072 | TB | TB138-1a | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, ure2::URA3, GLN3-MYC13[KanMX], sit4::natMX | sit4: 5′, -450 to -429 and -23 to -1 3′ 937 to 955 and 1380 to 1400 |
| FV088 | TB | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3], ure2::natMX | ure2: 5′, -300 to -279 and -21 to -1 3′ 1066 to 1084 and 1325 to 1345 |
| FV089 | TB | FV066 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3], sit4::kanMX, ure2::natMX | ure2: 5′, -300 to -279 and -21 to -1 3′ 1066 to 1084 and 1325 to 1345 |
Strain Construction—Deletion strains involving insertion of kanMX or natMX cassettes were constructed using the long flanking homology strategy of Wach (35), as described in Tate et al. (30). Chromosomal GLN3 or GAT1 were tagged at their C termini with 13 copies of the Myc epitope (Myc13) as described by Longtine et al. (36), using primers 5′-agcaattgctgacgaattggattggttaaaatttggtataCGGATCCCCGGGTTAATTAA-3′ (GLN3-F2) and 5′-TTATTAACATAATAAGAATAATGATAATGATAATACGCGGgaattcgagctcgtttaaac-3′ (GLN3-R1) for GLN3 and 5′-AAATGGCAATCTGAGCCTGGATTGGTTGAATCTGAATTTACGGATCCCCGGGTTAATTAA-3′ (GAT1-F2) and 5′-CATGGAAAGAAGCGAGTACTTTTTTTTTTTTGGGGGATCTAGAATTCGAGCTCGTTTAAAC-3′ (GAT1-R1) for GAT1.
Northern Blot Analysis—Total RNA was extracted as described earlier (37). Northern blot analysis was performed as described by Foury and Talibi (38). Digoxigenin DNA probes (about 500 bp) were synthesized by PCR, using primers 5′-AGTGTTGTCACACCTTGC-3′ and 5′-ACCCATTAATAGGGTTTC-3′ for DAL5 and 5′-AAACAGCAAGAAAGTCCACTGG-3′ and 5′-ACCTCTTAATCTTCTAGCCAAC-3′ for HHT1 and labeled using a PCR digoxigenin probe synthesis kit (Roche Applied Science). Treatment of the Hybond-N+ nylon membranes was as described earlier (31).
Chromatin Immunoprecipitation—The cells (100-ml cultures grown to an absorbance (A660 nm = 0.6) corresponding to 6 × 106 cells/ml) were treated with 1% formaldehyde for 30 min at 25 °C and mixed by orbital shaking. Glycine was then added to a final concentration of 500 mm and incubation continued for 5 min. The cells were collected, washed once with cold 10 mm Tris-HCl, pH 8, washed once with cold FA-SDS buffer (50 mm HEPES-KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride), and resuspended in 1 ml of cold FA-SDS buffer. An equal volume of glass beads (0.5 mm in diameter) was added, and the cells were disrupted by vortexing for 30 min in a cold room. The lysate was diluted into 4 ml of FA-SDS buffer, and the glass beads were discarded. The cross-linked chromatin was then pelleted by centrifugation (17,000 × g for 35 min), washed for 60 min with FA-SDS buffer, resuspended in 1.6 ml of FA-SDS buffer for 15 min at 4 °C, and sonicated three times for 30 s. each (Branson Sonifier 250, Pulse 60%, Power 2) to yield an average DNA fragment size of 700 base pairs. Finally, the sample was clarified by centrifugation at 14,000 × g for 30 min and diluted 4-fold in FA-SDS buffer, and aliquots of the resultant chromatin containing solution were stored at –80 °C.
Myc13-tagged proteins were immunoprecipitated by incubating 100 μl of the chromatin containing solution for 180 min at 4 °C with 2 μl of mouse anti-Myc antibodies (Santa Cruz) prebound to 10 μl of Dynabeads Pan Mouse IgG (Dynal) according to the manufacturer's instructions. Immune complexes were washed six times in FA-SDS buffer and recovered by treating with 50 μl of Pronase Buffer (25 mm Tris, pH 7.5, 5 mm EDTA, 0.5% SDS) at 65 °C with agitation. Input (IN) and immunoprecipitated (IP) fractions were then subjected to Pronase treatment (0.5 mg/ml; Roche Applied Science) for 60 min at 37 °C, and formaldehyde cross-links were reversed by incubating the eluates overnight at 65 °C. Finally, the samples were treated with RNase (50 μg/ml) for 60 min at 37 °C. DNA from the IP fractions was purified using the High Pure PCR Product Purification Kit (Roche Applied Science) and eluted in 50 μl of 20 mm Tris buffer, pH 8. IN fractions were boiled 10 min and diluted 500-fold with no further purification prior to quantitative PCR analysis.
Concentrations of specific DNA targets in IN and IP samples were measured by real time PCR using a LightCycler 1.5 instrument and the FastStart DNA Master Plus SYBR Green I kit according to the manufacturer's (Roche Applied Science) protocol. Primers amplified a 161-bp region in the promoter of DAL5 (DAL5P1, 5′-CGAGGAGCTATCATTTGCTG-3′; DAL5P2, 5′-ATCTTTTGCCCCGATAATCC-3′) or a 150-bp region 2.5 kb upstream of the DAL5 AUG as the unbound control (DAL5U1, 5′-GTTCATTAGTCGCCTACAGC-3′; DAL5U2, 5′-CAGAGCCCCGCATATTTTGA-3′). A standard curve was generated for each primer pair with five successive 10-fold dilutions of an IN sample. This standard curve was used to assess PCR efficiency and determine the relative concentration of target DNA in all other samples. Specificity of the PCR products was assessed by melting curve analysis. Primer pairs generating different products, identified by more than one melting temperature peak, were discarded.
The data were analyzed with LightCycler software, version 5.32. The IP/IN ratio corresponds to the concentration of target DNA in the IP sample relative to that in the corresponding IN sample, multiplied by 10. IP/IN values obtained for the unbound control (DAL5U) were substracted from IP/IN values obtained for the DAL5 promoter (DAL5P). To counterbalance variation generated by the immunoprecipitation step, we treated all of our data as follows. The wild type-induced value was set as 1, and the IP/IN value of every simultaneously immunoprecipitated sample was normalized accordingly. For every independent culture, the mean of the IP/IN ratios for two to four replicate immunoprecipitations was calculated. The values in Figs. 4 and 7 correspond to the mean IP/IN value of at least two independent cultures. The mean normalized IP/IN values of both DAL5U and DAL5P are displayed in the supplemental material.
FIGURE 4.
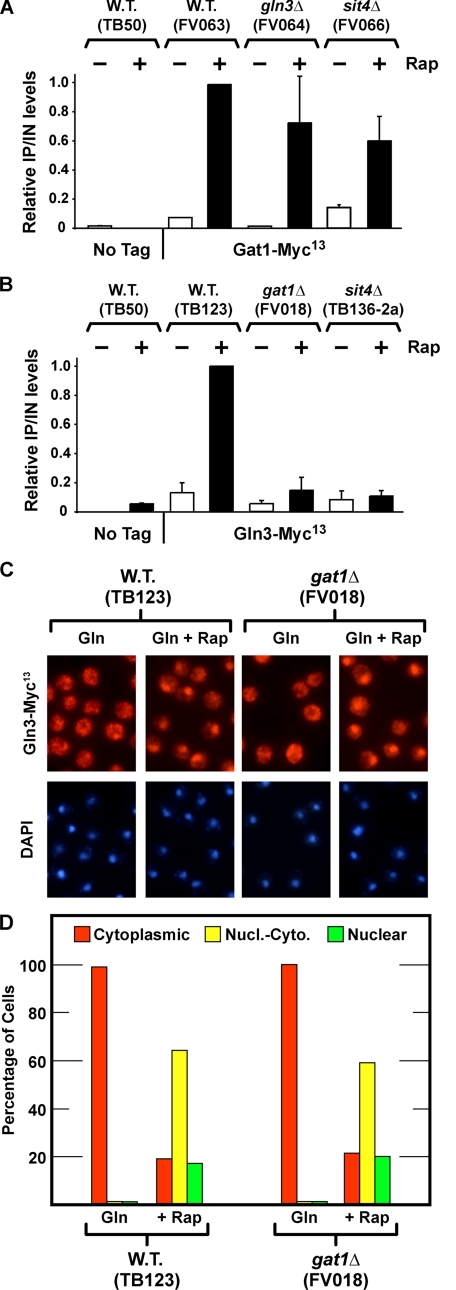
A and B, effects of sit4Δ, gln3Δ, and gat1Δ on rapamycin-induced binding of Gat1-Myc13 and Gln3-Myc13 to the DAL5 promoter. C and D, effect of deleting GAT1 on Gln3-Myc13 localization. Wild type (W.T.) untagged (TB50), wild type GAT1-MYC13 (FV063), gln3Δ GAT1-MYC13 (FV064), and sit4Δ GAT1-MYC13 (FV066) (A), and wild type untagged (TB50), wild type GLN3-MYC13 (TB123), gat1Δ GLN3-MYC13 (FV018), sit4Δ GLN3-MYC13 (TB136-2a) (B) strains were grown in YNB-glutamine medium with or without the addition of rapamycin (0.2 μg/ml) for 30 min. ChIP was performed using antibodies against c-Myc as described under “Materials and Methods.” Quantitative PCR of IP and IN fractions was performed with primers for DAL5 promoter (DAL5P) and for a region 2.5 kb upstream of DAL5 open reading frame as a control (DAL5U). For each immunoprecipitation, IP/IN values were calculated as follows: [DAL5P]IP/[DAL5P]IN – [DAL5U]IP/[DAL5U]IN, normalized to the value obtained with wild type-induced cells. Histograms represent the average of at least two experiments from independent cultures. The error bars indicate standard errors. C and D, wild type and gat1Δ strains were grown in YNB-glutamine medium. Split cultures were left untreated (Gln) or treated with rapamycin (0.2 μg/ml) for 20 min (+Rap), sampled, and processed for immunofluorescence microscopy as described under “Materials and Methods” and in the legend to Fig. 3.
FIGURE 7.
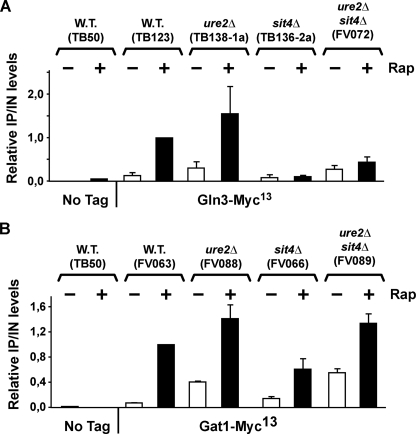
ChIP analysis of rapamycin-induced recruitment of Gln3-Myc13 and Gat1-Myc13 to the DAL5 promoters in wild type, sit4Δ, ure2Δ, and ure2Δsit4Δ strains. Wild type untagged (TB50), wild type GLN3-MYC13 (TB123), ure2Δ GLN3-MYC13 (TB138-1a), sit4Δ GLN3-MYC13 (TB136-2a), ure2Δsit4Δ GLN3-MYC13 (FV072), wild type GAT1-MYC13 (FV063), ure2Δ GAT1-MYC13 (FV088), sit4Δ GAT1-MYC13 (FV066), and ure2Δsit4Δ GAT1-MYC13 (FV089) strains were grown in YNB-glutamine medium with or without addition of rapamycin (0.2 μg/ml) for 30 min. ChIP and subsequent quantitative PCR were performed as in Fig. 4. W.T., wild type.
Quantitative RT-PCR—cDNA was generated from 100–500 ng of total RNA using a Transcriptor First Strand cDNA synthesis kit (Roche Applied Science) with oligo(dT) as primer following the manufacturer's recommended protocol. cDNAs were quantified by real time PCR as described above. Primers amplified a 154-bp region of DAL5 (DAL5O1, 5′-TTCGAATGCTTCCCTAGACG-3′; DAL5O2, 5′-CTTCATGGCCTCATCAACCT-3′) or a 125-bp region of TBP1 (TBP1O1, 5′-TATAACCCCAAGCGTTTTGC-3′; TBP1O2, 5′-GCCAGCTTTGAGTCATCCTC-3′). Expression values correspond to the ratio of DAL5- over TBP1-specific mRNAs determined in each sample.
Indirect Immunofluorescence Microscopy—Cell collection and fixation for indirect immunofluorescence was performed using the method of Cox et al. (39) as modified by Tate et al. (30, 32). Gln3-Myc13 localization was visualized using primary monoclonal antibody 9E10 (c-Myc; Covance MMS-150P) at a dilution of 1:1000 and 1:5000 for Gat1-Myc13 visualization. Alexa Fluor 594 goat anti-mouse IgG antibody (Molecular Probes, at a dilution of 1:200) was used as secondary antibody in both cases. DNA was visualized using 4′,6′-diamino-2-phenylindole (DAPI) contained in the mounting medium (Sigma) (39). Some strains, those containing the sit4Δ and especially mutant strains containing GAT1-MYC13, required greater amounts of zymolyase and/or times of digestion to achieve high quality results.
The cells were imaged using a Zeiss Axioplan 2 imaging microscope with a 100× Plan-Apochromat 1.40 oil objective at room temperature. The images were acquired using a Zeiss Axio camera and AxioVision 3.0 and 4.6.3 (Zeiss; 4, 2007) software, processed with Adobe Photoshop and Illustrator programs. Gamma settings were altered where necessary to decrease background fluorescence in areas that did not contain cells and to avoid any change or loss in cellular detail. Changes were applied uniformly to the image presented.
Determination of Intracellular Gln3-Myc13 and Gat1-Myc13 Distribution—To provide more representative and complete descriptions of Gln3-Myc13 and Gat1-Myc13 behavior than obtainable from an isolated image, we manually scored Gln3-Myc13 or Gat1-Myc13 localization in 200 or more cells in multiple, randomly chosen microscopic fields from which each image presented was taken. Scoring was performed exclusively using unaltered primary image files viewed with Zeiss Axio-Vision 3.0 and 4.6.3 software. The cells were classified into one of three categories: cytoplasmic (cytoplasmic fluorescence only), nuclear-cytoplasmic (fluorescence appeared in the cytoplasm as well as co-localizing with DAPI-positive material), and nuclear (co-localizing only with DAPI-positive material). Although scoring limitations and reproducibility were described in Tate et al. (30, 32), we emphasize again, as we did earlier (40), that the nuclear-cytoplasmic category is, of necessity, arbitrary. Placing cells in that category is based on subjective visual evaluation by the individual scoring the cells; it is not an objective instrument-based measurement. When the fluorescent signal is not restricted to a single cellular compartment, scoring depends upon repeated decisions of whether it is nuclear-cytoplasmic or a category flanking it. They will undoubtedly differ in detail from those of another observer, who sets their category dividing lines differently. Although our intracellular distributions were scored as consistently as possible, conclusions are most prudently made when primarily based on straightforwardly detected changes in overall distribution patterns that are apparent in the microscopic images. Similar experiments were generally repeated two or more times with similar results. Experiment to experiment variation can be ascertained by comparing similar experimental conditions in this work and previous work (30, 32, 40). During this work, we noticed that unless the fluorescent signal was exclusively localized to a single cellular compartment, there was greater variability in scoring Gat1-Myc13 than Gln3-Myc13.
RESULTS
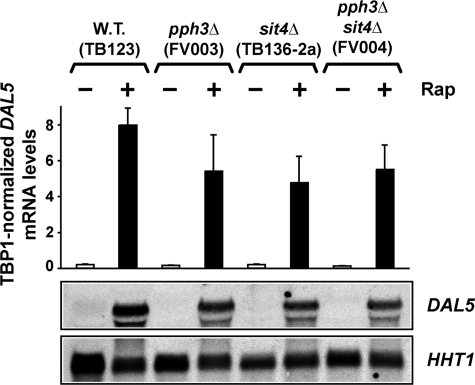
Type 2A-related Phosphatase (Sit4 and Pph3) Requirements for Rapamycin-induced DAL5 Transcription—Our studies began by evaluating the type 2A-related Sit4 and Pph3 protein phosphatase requirements of DAL5 gene expression. This permitted comparisons with previously obtained information about Gln3-Myc13 localization and the ability to test predictions generated by it. Wild type (TB123) and isogenic pph3Δ (FV003), sit4Δ (TB136-2a), and pph3Δ sit4Δ (FV004) strains were grown in YNB-glutamine medium to mid-log phase (A660 nm = 0.6). Following a 30-min treatment with 0.2 μg/ml rapamycin, expression of DAL5, a representative NCR-sensitive gene, was analyzed by quantitative RT-PCR and Northern blot assays (Fig. 1). As expected, DAL5 expression in untreated cells was minimal in all four strains, i.e. NCR-sensitive transcription was repressed because of growth with a good nitrogen source (Fig. 1). Quite surprisingly, however, rapamycin-induced DAL5 expression was only slightly reduced in all three mutants, demonstrating that Sit4 and Pph3 were dispensable (Fig. 1). This lack of a Sit4 requirement differed sharply from the absolute Sit4 requirement previously shown for rapamycin-induced nuclear Gln3-Myc13 localization under identical conditions (see Figs. 4 and 5 of Ref. 30). The presence of rapamycin-induced DAL5 transcription in sit4Δ cells, where Gln3-Myc13 was excluded from the nucleus, suggested that our current view of Tor1,2 regulation of NCR-sensitive (DAL5) transcription required revision.
FIGURE 1.
Effect of deleting type 2A-related phosphatase genes SIT4 and PPH3 on rapamycin-induced DAL5 expression. Total RNA was isolated from wild type (TB123), pph3Δ (FV003), sit4Δ (TB136-2a), and pph3Δ sit4Δ (FV004) cells expressing GLN3-MYC13 that replaced the native GLN3 gene. Cells were grown in YNB-glutamine medium and treated with rapamycin (Rap) (0.2 μg/ml) for 30 min. Control cells were similarly grown but untreated. DAL5 mRNA levels were quantified by quantitative RT-PCR, as described under “Materials and Methods.” DAL5 values were normalized with TBP1. The values represent the averages of at least three experiments from independent cultures, and the error bars indicate standard errors. 30 μg of total RNA from each sample were subjected to Northern blot analysis. HHT1 was used as the loading and transfer efficiency control.
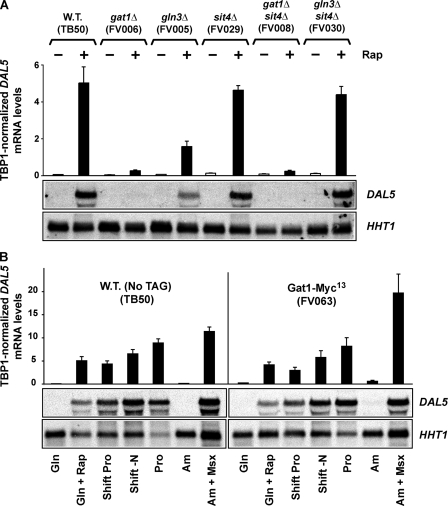
The most plausible explanation of rapamycin-responsive DAL5 transcription in the sit4Δ was that it derived from a transcription factor other than Gln3. Gat1 was the most likely candidate to support this transcription because it had been previously shown to mediate NCR-sensitive gene expression, to be regulated by Tor1,2, and hence to be responsive to rapamycin treatment (5, 7, 17, 18, 41–44). To test this hypothesis, we measured the individual contributions of Gln3 and Gat1 to DAL5 transcription. Deleting GLN3 (gln3Δ, FV005) reduced rapamycin-induced DAL5 expression to about a third of the wild type (TB50) level (Fig. 2A). This, however, was a much weaker effect than the essentially background levels observed when GAT1 was deleted (Fig. 2A, gat1Δ, FV006). Positively correlating with these observations, rapamycin-induced DAL5 transcription was also absent in a sit4Δgat1Δ (FV008), but unaffected in a sit4Δgln3Δ (FV030). In fact, additionally deleting SIT4 in a gln3Δ inexplicably suppressed the effect of the gln3Δ. Thus, Gat1 activated DAL5 expression following rapamycin treatment and did not require Sit4 activity to do so.
FIGURE 2.
A, relative contributions of Gat1 and Gln3 to rapamycin-induced DAL5 expression. Total RNA was isolated from wild type (TB50), gat1Δ (FV006), gln3Δ (FV005), sit4Δ (FV029), gat1Δsit4Δ (FV008), and gln3Δsit4Δ (FV030) cultures grown in YNB-glutamine medium. The cells were treated and analyses performed as in Fig. 1. B, functionality and normal regulation of the integrated GAT1-MYC13 construct. Total RNA was isolated from wild type (TB50) and wild type GAT1-MYC13 (FV063) cells grown in glutamine (Gln) medium in the presence or absence of 0.2 μg/ml rapamycin (Rap) for 30 min, 60 min after transfer from glutamine to proline (shift Pro), or nitrogen-free medium (shift -N), proline (Pro), or ammonium (Am) medium in the presence or absence of 2 mm methionine sulfoximine (Msx) for 20 min. The cells were treated and analyses performed as in Fig. 1. W.T., wild type.
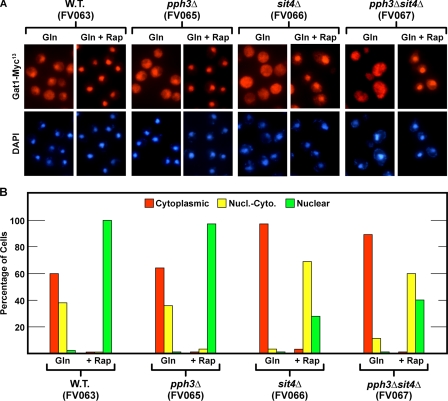
Rapamycin-induced Nuclear Gat1-Myc13 Localization Is Largely Sit4-independent—The above observations suggested that rapamycin-induced nuclear Gat1-Myc13 localization might possess different protein phosphatase (Sit4, Pph3) requirements than Gln3-Myc13 and that the Tor signal transduction pathway bifurcated at the control of GATA factor localization. To test these suggestions directly, we replaced chromosomal GAT1 with GAT1-MYC13. Before further analyses, however, we used quantitative RT-PCR and Northern blot assays to validate the functionality and native regulation of the construct (Fig. 2B). Steady state DAL5 mRNA levels were comparable in wild type untagged (TB50) and Gat1-Myc13-tagged (FV063) cells cultured under conditions previously used to analyze Gln3-Myc13 localization (30). Thus, by these criteria, our Myc13-tagged Gat1 protein was normally regulated.
We then used the GAT1-MYC13 constructs to evaluate intracellular Gat1-Myc13 localization in glutamine-grown, rapamycin-treated wild type (FV063), sit4Δ (FV066), pph3Δ (FV065), and sit4Δpph3Δ (FV067) cells. Rapamycin induced nuclear localization of Gat1-Myc13 in glutamine-grown wild type cells (Fig. 3, A and B, W.T.). However, unlike the situation with Gln3-Myc13 (Figs. 4 and 5 of Ref. 30), deleting SIT4 only modestly reduced nuclear Gat1-Myc13 localization following rapamycin treatment. This concomitantly increased the number of cells in which Gat1-Myc13 was nuclear-cytoplasmic (Fig. 3, A and B, sit4Δ) but not those in which Gat1-Myc13 was exclusively localized to the cytoplasm. This localization profile closely paralleled the limited decrease in DAL5 expression observed in the sit4Δ (Fig. 1) and led us to conclude that nuclear Gat1-Myc13 localization possessed at most only a limited Sit4 requirement rather than the absolute requirement observed for Gln3-Myc13. Deletion of PPH3, either alone or in conjunction with SIT4, did not yield a phenotype that significantly differed from a wild type in the former case or a sit4Δ in the latter (Fig. 3, A and B, pph3Δ and pph3Δsit4Δ). This argued that, as occurred with Gln3-Myc13, Pph3 did not play a demonstrable role in nuclear Gat1-Myc13 localization under these conditions.
FIGURE 3.
Effects of rapamycin on the intracellular localization of Gat1-Myc13 in wild type, sit4Δ, and pph3Δ strains. Wild type (W.T.) and mutant strains were grown in YNB-glutamine medium. Split cultures were left untreated (Gln) or treated with rapamycin (0.2 μg/ml) for 20 min (+Rap), sampled, and processed for immunofluorescence microscopy as described under “Materials and Methods.” Strain numbers appear below the pertinent genotype. The images are presented in pairs with Gat1-Myc13-dependent fluorescence above and DAPI-stained cells below. The images and corresponding histograms below them were taken from the same slides. Intracellular distributions of Gat1-Myc13 (using criteria described under “Materials and Methods”) are indicated by the bar color in the histograms: red, cytoplasmic; yellow, nuclear-cytoplasmic; green, nuclear.
Additional smaller differences appeared when Gln3-Myc13 and Gat1-Myc13 localization data were compared (compare Figs. 4 and 5 in Ref. 30 with Fig. 3 of this work). Fluorescence signals emanating from Gat1-Myc13 were stronger than those from Gln3-Myc13, and Gat1-Myc13 was somewhat more nuclear under most conditions. For example, Gln3-Myc13 could not be detected in the nuclei of glutamine-grown wild type cells (Figs. 4 and 5 of Ref. 30), whereas Gat1-Myc13 was frequently observed to be nuclear-cytoplasmic (Fig. 3, A and B, W.T. Gln). This correlated with earlier observations that Gat1-dependent transcription was less NCR-sensitive than that mediated by Gln3 (41–44). Additionally, Gat1-Myc13 was nuclear in nearly all rapamycin-treated, glutamine-grown wild type cells, whereas Gln3-Myc13 was more nuclear-cytoplasmic (Figs. 4 and 5 of Ref. 30). Overall, rapamycin generated a stronger response with Gat1-Myc13 than Gln3-Myc13. We also observed greater variability in data scoring Gat1-Myc13 localization in glutamine-grown but not rapamycin-treated wild type cells than we had previously encountered with Gln3-Myc13.
Gat1-Myc13 and Gln3-Myc13 Possess Different Requirements for Rapamycin-induced Binding to the DAL5 Promoter—The previous view that Gln3 and Gat1 were similarly regulated was challenged by the strikingly different Sit4 requirements they possessed for rapamycin-induced nuclear localization. Therefore, we used chromatin immunoprecipitation (ChIP) assays to determine whether these differences extended to GATA factor binding to the NCR-sensitive DAL5 promoter. Gat1-Myc13 was effectively recruited to the promoter of rapamycin-treated, glutamine-grown wild type (FV063), gln3Δ (FV064), and sit4Δ (FV066) cells (Fig. 4A and supplemental Fig. S1). In other words, rapamycin-induced Gat1-Myc13 binding upstream of DAL5 was almost completely independent of Sit4, which was consistent with the predominantly nuclear and nuclear-cytoplasmic Gat1-Myc13 localization in rapamycin-treated wild type and sit4Δ cells (Fig. 3). Further, efficient Gat1-Myc13 binding did not require the presence of a second GATA factor, Gln3, because it occurred in the gln3Δ (Fig. 4A and supplemental Fig. S1). In contrast, Gln3-Myc13 binding to the DAL5 promoter required not only Sit4, because of the necessity of the phosphatase for nuclear Gln3-Myc13 localization, but also the presence of Gat1, i.e. very little Gln3-Myc13 bound upstream of DAL5 in a gat1Δ (Fig. 4B and supplemental Fig. S1).
To determine whether the loss of Gln3-Myc13 binding to DAL5 in a gat1Δ derived from Gat1 being required for nuclear Gln3-Myc13 localization, we evaluated intracellular Gln3-Myc13 distribution. Deleting GAT1 had little if any affect on nuclear Gln3-Myc13 localization following rapamycin treatment (Fig. 4, C and D).
DAL5 Transcription Remains Highly Rapamycin-inducible in Strains Lacking Ure2—The differences in Sit4 requirements for rapamycin-induced Gln3-Myc13 and Gat1-Myc13 localization and DNA binding prompted us to inquire whether Gln3 and Gat1 were also regulated differently downstream of Sit4, i.e. by Ure2.
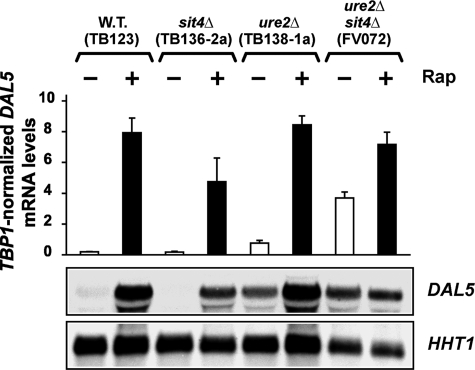
Although recently questioned (15, 16, 22, 27, 29, 30–32, 45, 46), rapamycin was originally posited to abrogate excess nitrogen-dependent negative regulation of Sit4 by Tor1,2, thereby enabling Sit4 phosphatase to dephosphorylate Gln3 (8). This, in turn, brought about dissociation of the Gln3-Ure2 complex that permitted Gln3 to enter the nucleus. Importantly, Gat1 was reported to be similarly regulated (8). This model predicted that rapamycin-induced DAL5 expression in a wild type strain would be the same as in an untreated or rapamycin-treated ure2Δ, where the Gln3-Ure2 and presumably Gat1-Ure2 interactions are lost. In other words, expression should be high and constitutive in the ure2Δ regardless of the conditions tested.
We tested this prediction using quantitative RT-PCR and Northern blot assays of DAL5 transcription. As expected since the initial discovery of the ure2 locus (47, 48), deleting URE2 increased DAL5 expression in untreated, glutamine-grown cells (Fig. 5). Surprisingly, however, rapamycin treatment dramatically increased DAL5 expression far beyond the level observed in an untreated ure2Δ and did so whether or not Sit4 was active (Fig. 5). DAL5 transcription levels were nearly identical in rapamycin-treated wild type, ure2Δ, and sit4Δure2Δ cells. Beyond these remarkable observations, which we will address further below, we found that deleting both SIT4 and URE2 yielded a marked synergistic positive effect on DAL5 transcription in untreated, glutamine-grown cells (Fig. 5). These results and their magnitude supported the idea that Tor1,2 regulation of DAL5 transcription might be more complicated than simple Ure2-mediated control of intracellular GATA factor localization.
FIGURE 5.
Effects of sit4Δ, ure2Δ, and sit4Δure2Δ mutations on DAL5 expression. Total RNA was isolated from TB wild type (TB123), sit4Δ (TB136-2a) ure2Δ (TB138-1a), and ure2Δsit4Δ (FV072) cells grown in YNB-glutamine medium that were untreated or treated with 0.2 μg/ml rapamycin for 30 min. The cells were treated, and analyses were performed as for Fig. 1. W.T., wild type.
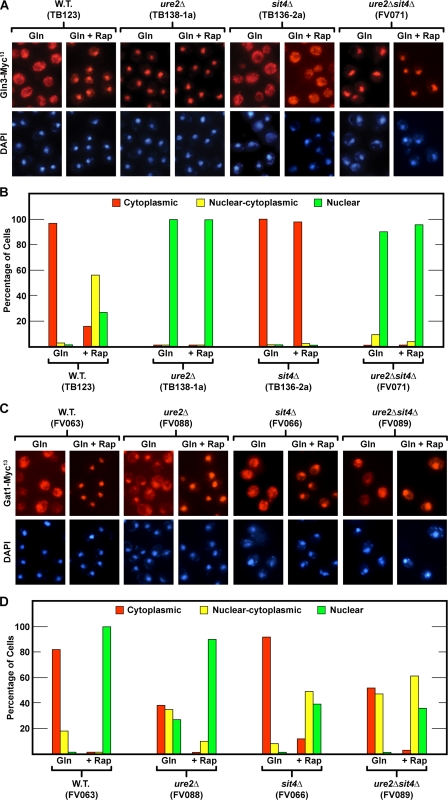
Gat1-Myc13 and Gln3-Myc13 Localization Responds Differently to Deletion of Ure2—To investigate the unexpected ability of rapamycin to greatly increase DAL5 transcription in ure2Δ mutants, we focused on processes occurring between the action of Sit4 and DAL5 transcription, i.e. intracellular GATA factor localization and binding to the DAL5 promoter. To that end, we determined the effects of deleting URE2 on Gln3-Myc13 and Gat1-Myc13 localization and then queried whether or not the absolute and limited Sit4 requirements observed for nuclear Gln3-Myc13 and Gat1-Myc13 localization, respectively, were abrogated in ure2Δ.
Gln3-Myc13 localization in glutamine-grown cells responded to deleting URE2 as predicted, i.e. Gln3-Myc13 uniformly localized to the nuclei of essentially all ure2Δ cells whether or not they were treated with rapamycin, and in both cases Gln3-Myc13 was more nuclear than in rapamycin-treated wild type cells (Fig. 6, A and B, TB123 versus TB138-1a).
FIGURE 6.
Effects of rapamycin treatment on the intracellular localization of Gln3-Myc13 and Gat1-Myc13 in ure2Δ, sit4Δ, and ure2Δsit4Δ mutant strains. The formats for the experiments and presentation of the data were the same as in Fig. 3. A and B, Gln3-Myc13 was visualized. C and D, Gat1-Myc13 was visualized. Note that FV071 and FV072 have the same genotypes and were constructed in the same genetic background (see Table 1). In a similar experiment, FV072 gave results similar to those depicted here for FV071. W.T., wild type.
Parallel experiments demonstrated that Gat1-Myc13 localization was regulated quite differently. Although Gat1-Myc13 became a bit more nuclear in an untreated, glutamine-grown ure2Δ compared with wild type, i.e. the fraction of ure2Δ cells with nuclear-cytoplasmic or nuclear Gat1-Myc13 localization increased somewhat, it remained exclusively cytoplasmic in roughly 40% of the cells. This represented about a 2-fold decrease relative to wild type (Fig. 6, C and D, FV063 versus FV088). In other words, Gat1-Myc13 localization was not negatively regulated by Ure2 to the same degree as Gln3-Myc13, where nuclear localization was observed in nearly all untreated ure2Δ cells (Fig. 6, compare A and B with C and D). Also in contrast with Gln3-Myc13, the addition of rapamycin to the ure2Δ increased nuclear Gat1-Myc13 localization to the point where it was now nuclear in most cells just as in the wild type (Fig. 6, C and D, wild type +Rap versus ure2Δ+Rap). From these data we concluded that a protein other than or in addition to Ure2 was potentially responsible for maintaining Gat1-Myc13 in the cytoplasm of glutamine-grown cells and its ability to function depended upon Tor1,2, i.e. it too responded to rapamycin treatment.
Experiments addressing the epistasis of ure2Δ and sit4Δ mutations for Gln3-Myc13 localization showed that a ure2Δ was clearly epistatic to a sit4Δ in untreated and rapamycin-treated glutamine-grown cells in that Gln3-Myc13 was cytoplasmic in the sit4Δ but nuclear in the sit4Δure2Δ double mutant. The parallel epistasis analysis of Gat1-Myc13 localization yielded results that were much less straightforward than with Gln3-Myc13, again pointing to differences in their regulation. The untreated sit4Δure2Δ mutant phenotype was overall perhaps more like that of the ure2Δ than the sit4Δ, whereas the rapamycin-treated sit4Δure2Δ behaved more like a sit4Δ. In neither case were the phenotypes sufficiently strong or different to confidently draw firm conclusions about epistasis. What could be confidently concluded was that Sit4 and Ure2 exerted much less control over Gat1-Myc13 than Gln3-Myc13 localization.
More Than Gln3-Myc13 Nuclear Localization Is Required for It to Bind to the DAL5 Promoter—We finally investigated Sit4- and Ure2-mediated regulation of Gln3-Myc13 and Gat1-Myc13 interactions with the DAL5 promoter (Fig. 7 and supplemental Fig. S2). Using ChIP assays, we performed epistasis experiments parallel to those described in Fig. 6. The most striking and unexpected observation was that nuclear Gln3-Myc13 localization and binding to the DAL5 promoter did not correlate with one another. Gln3-Myc13 was uniformly nuclear in both untreated and rapamycin-treated ure2Δ cells (Fig. 6, A and B). In contrast, its binding to the DAL5 promoter remained rapamycin-inducible (Fig. 7A).
In another example, Gln3-Myc13 binding to the DAL5 promoter was 3-fold less in an untreated ure2Δ compared with the rapamycin-treated wild type (Fig. 7A and supplemental Fig. S2), whereas nuclear Gln3-Myc13 localization was greater in the former instance than in the latter (Fig. 6, A and B). In yet a third example, the addition of a sit4Δ to a ure2Δ strain substantially diminished Gln3-Myc13 binding to the DAL5 promoter in rapamycin-treated cells (Fig. 7A and supplemental Fig. S2), despite its exclusively nuclear localization (Fig. 6, A and B). These failed correlations indicated that more than just nuclear Gln3-Myc13 localization dictated its binding to DAL5 DNA especially in the presence of rapamycin. Moreover, this binding was somehow influenced by Sit4.
There was, however, one positive correlation we could see in the ure2Δ strains. Increased Gln3-Myc13 binding to the DAL5 promoter following the addition of rapamycin to a ure2Δ correlated with rapamycin-induced nuclear Gat1-Myc13 localization under the same conditions, which is in agreement with the observation that Gln3-Myc13 binding to the DAL5 promoter requires Gat1.
In contrast with Gln3-Myc13, more positive correlations were observed with Gat1-Myc13. Gat1-Myc13 binding to the DAL5 promoter in untreated and rapamycin-induced ure2Δ cells (Fig. 7B and supplemental Fig. S2) roughly paralleled its nuclear localization and Gat1-supported transcription (Figs. 5 and 6, C and D). Nevertheless, rapamycin-induced Gat1-Myc13 binding to the DAL5 promoter in a ure2Δsit4Δ was comparable with that in a ure2Δ, even though somewhat less Gat1-Myc13 was present in the nucleus in the former situation.
DISCUSSION
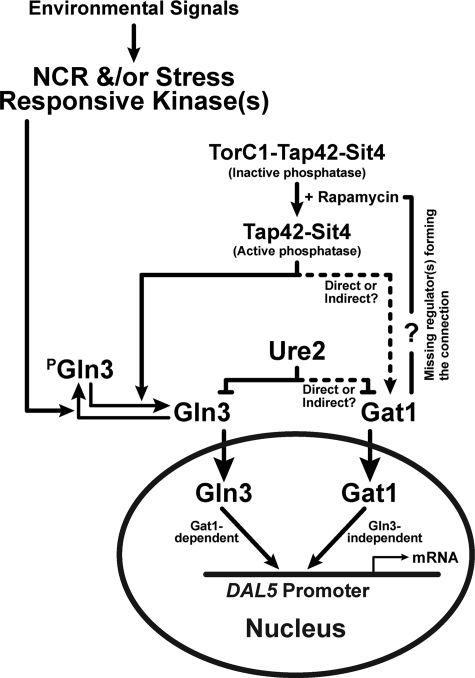
The most important mechanistic outcome of the above experiments is their demonstration that the Tor1,2 signal transduction pathway bifurcates at the level of GATA factor regulation in S. cerevisiae (Fig. 8). This conclusion is supported by three main lines of evidence: (i) rapamycin-induced nuclear Gln3-Myc13 localization in glutamine-grown cells possesses an absolute requirement for the type 2A-related phosphatase, Sit4, whereas nuclear Gat1-Myc13 localization exhibits only a limited Sit4 requirement at best; (ii) intracellular Gat1-Myc13 localization is largely immune to regulation by Ure2, the highly effective negative regulator absolutely required to sequester Gln3-Myc13 in the cytoplasm of cells provided with a good nitrogen source; and (iii) Gln3-Myc13 binding to the DAL5 promoter requires the presence of Gat1, but Gat1-Myc13 binds to this DNA independently of Gln3.
FIGURE 8.
Diagrammatic summary of data, showing bifurcation of Tor pathway at the level of GATA factor regulation. The arrows and bars indicate positive and negative regulation, respectively. The absence of arrows or bars indicates insufficient data are available to make such a characterization. This diagrammatic summary does not address the molecular mechanism of Gln3 regulation by Ure2 (two models have been suggested (8, 11)) or the transfer of environmental signals to Tor1,2 and other protein kinases.
Previous reports forecast that differences in Gln3 and Gat1 regulation might exist and encouraged us to look for them. Although a Ure2-Gln3-Myc13 complex was straightforwardly identified by in vitro co-immunoprecipitation (8, 11, 19), repeated attempts to identify a similar Ure2-Gat1 complex were unsuccessful (8, 11). Additionally, in vivo rapamycin-induced and in vitro alkaline phosphatase-dependent dephosphorylation of Gln3-Myc13 was easily demonstrated (8, 11, 33, 46). In contrast, similar attempts to demonstrate Gat1-Myc13 phosphorylation and rapamycin-induced dephosphorylation were unsuccessful, even though Gat1-Myc13 phosphorylation per se, i.e. Snf1-dependent Gat1-Myc13 phosphorylation, could be readily detected (33).
The above evidence demonstrating regulatory bifurcation of GATA factor regulation additionally raises significant new possibilities and questions. If the overall mechanisms of Gln3 and Gat1 regulation in response to rapamycin treatment are analogous, at least two components participating in Tor1,2 regulation of GATA factor localization likely remain to be identified: (i) molecule(s) responsible for Gat1-Myc13 sequestration in the cytoplasm of cells provided with good nitrogen sources, the functional counterpart of Ure2, and (ii) molecule(s) forming the regulatory connection between the site of rapamycin action, presumably the TorC1-Tap42-phosphatase complex, and the molecule(s) responsible for sequestering Gat1 in the cytoplasm during growth with excess nitrogen. The working diagram in Fig. 8 portrays this connection, but the lack of pertinent data does not justify defining it further.
A related question is prompted by the modest effects of sit4Δ and ure2Δ mutations on Gat1-Myc13 regulation. Do the phenotypes generated by these mutations derive from direct control of Gat1-Myc13 by Sit4 and Ure2 or alternatively represent indirect secondary effects? Stated in another way, do Sit4, Ure2, plus unknown proteins with analogous and somewhat redundant functions jointly regulate Gat1-Myc13? Alternatively, do these unknown regulatory proteins alone regulate Gat1-Myc13 and observed influences of the sit4Δ and ure2Δ derive as indirect consequences of regulatory cross-talk between branches of the bifurcated pathway?
Two earlier observations will likely have an impact on the answers to these questions: (i) Gat1-mediated expression of multiple genes associated with the transport and metabolism of nitrogenous compounds remains highly NCR-sensitive in a gln3Δure2Δ mutant (41–44) and (ii) overexpression of Ure2 can restrict EGFP-Gat1 to the cytoplasm under conditions in which it would otherwise be nuclear (49).
Next, two sets of correlations prompt us to speculate that Gat1-Myc13 and Gln3-Myc13 may positively influence one another's binding to the DAL5 promoter, conceivably through protein-protein interactions: (i) Following entry of the GATA factors into the nucleus, rapamycin induces Gat1-Myc13 binding to the DAL5 promoter independently of Gln3, whereas Gln3-Myc13 binding requires Gat1. In other words, gaining entry to the nucleus in response to rapamycin treatment is alone insufficient to bring about Gln3-Myc13 binding to the DAL5 promoter. Further, even though Gln3-Myc13 is fully nuclear in a ure2Δ, its binding to the DAL5 promoter was much less than observed when rapamycin was present, i.e. when Tor1,2 regulation was abrogated. This increased rapamycin-induced Gln3-Myc13 binding correlates with parallel increases in nuclear Gat1-Myc13 localization and binding to the DAL5 promoter in response to rapamycin addition. (ii) Conversely, increased nuclear Gln3-Myc13 localization that occurs in the sit4Δure2Δ relative to a sit4Δ and the roughly 2-fold greater Gln3-Myc13 binding to DAL5 in a ure2Δsit4Δ relative to that in a sit4Δ correlates with the roughly 2-fold increased Gat1-Myc13 binding to the DAL5 promoter in the double mutant.
If the possibility that Gat1 and Gln3 do interact with one another and thereby reciprocally promote each other's binding to the promoter is valid, it would contribute significantly toward explaining these correlations. That said, data supporting a positive effect of Gat1-Myc13 on Gln3-Myc13 binding to the DAL5 promoter are certainly stronger than those arguing in favor of the converse situation.
As we speculate about such models of GATA factor control, however, we keep two important caveats firmly in mind: (i) because of the unexpectedly high occurrence of strain-specific variations in nitrogen-responsive regulation, general characteristics of NCR-sensitive, GATA factor control must be distinguished from strain-specific traits (45), and (ii) models describing intra-nuclear GATA factor regulation must take the structures of the particular promoters being studied into account. For example, DAL5 is among the simplest of the NCR-sensitive promoters, and for that reason it is often used as a NCR-sensitive reporter. A small fragment of the DAL5 promoter, containing two functional GATAA sequences, is necessary and sufficient to support NCR-sensitive transcription in a heterologous expression vector assay (50). With more complex promoters, Gln3 binding to DNA may require one or more non-GATA factor DNA-binding proteins. Examples of this phenomenon have been observed with DAL7, PUT1, and GLN1 promoter fragments (26, 51).
Finally, multiple observations made in this work lead us to suspect that there may be yet undiscovered rapamycin-induced and/or Sit4-controlled events that regulate rapamycin-induced DAL5 transcription beyond the point of GATA factor entry into the nucleus. The most indicative observations of this possibility are the multiple effects of rapamycin addition and SIT4 deletion in ure2Δ strains. Such putative events could potentially occur at the level of GATA factor binding to the regulated gene promoter or thereafter at the level of transcriptional activation. Our observations also demonstrate that gross transcription levels of nitrogen-responsive genes are alone unlikely to be unambiguous reporters of Tor1,2 pathway regulation.
Supplementary Material
Acknowledgments
We thank Dr. Michael Hall for strains, Tim Higgins and André Feller for preparing the artwork, Fabienne Vierendeels for excellent technical assistance, Maxime Wéry for helpful advice on ChIP experiments, and the University of Tennessee Yeast Group for suggestions to improve the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is dedicated to Prof. Ronald Butow of the University of Texas Southwestern Medical Center (1936–2007), who contributed so much to the field of mitochondrial retrograde regulation and its interface with nitrogen and GATA factor regulation in S. cerevisiae.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: NCR, nitrogen catabolite repression; IN, input; IP, immunoprecipitated; RT, reverse transcription; DAPI, 4′,6′-diamino-2-phenylindole; ChIP, chromatin immunoprecipitation.
References
- 1.Schluter, M., and Schofer, J. (2005) Am. Heart Hosp. J. 3 182–186 [DOI] [PubMed] [Google Scholar]
- 2.Boulay, A., Rudloff, J., Ye, J., Zumstein-Mecker, S., O'Reilly, T., Evans, D. B., Chen, S., and Lane, H. A. (2005) Clin. Cancer Res. 11 5319–5328 [DOI] [PubMed] [Google Scholar]
- 3.Morgensztern, D., and McLeod, H. L. (2005) Anticancer Drugs. 16 797–803 [DOI] [PubMed] [Google Scholar]
- 4.Lorber, M. I., Mulgaonkar, S., Butt, M., Elkhammas, E., Mendez, R., Rajagopalan, P. R., Kahan, B., Sollinger, H., Li, Y., Cretin, N., and Tedesco, H. (2005) Transplantation 80 244–252 [DOI] [PubMed] [Google Scholar]
- 5.Thomas, G., Sabatini, D., and Hall, M. N. (eds) (2004) Current Topics in Microbiology and Immunology: Target of Rapamycin, Springer, New York
- 6.Inoki, K., Ouyang, H., Li, Y., and Guan, K. L. (2005) Microbiol. Mol. Biol. Rev. 69 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, T. G. (2004) in Nutrient-Induced Responses in Eukaryotic Cells Current Genetics (Winderickx, J., and Taylor, P. M., eds) Vol. 7, Chapter 9, pp. 225–257, Springer-Verlag Berlin [Google Scholar]
- 8.Beck, T., and Hall, M. N. (1999) Nature 402 689–692 [DOI] [PubMed] [Google Scholar]
- 9.Cardenas, M. E., Cutler, N. S., Lorenz, M. C., Di Como, C. J., and Heitman, J. (1999) Genes Dev. 13 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F., and Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram, P. G., Choi, J. H., Carvalho, J., Ai, W., Zeng, C., Chan, T. F., and Zheng, X. F. (2000) J. Biol. Chem. 275 35727–35733 [DOI] [PubMed] [Google Scholar]
- 12.Cox, K. H., Rai, R., Distler, M., Daugherty, J. R., Coffman, J. A., and Cooper, T. G. (2000) J. Biol. Chem. 275 17611–17618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuruvilla, F. G., Shamji, A. F., and Schreiber, S. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7283–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohde, J. R., Campbell, S., Zurita-Martinez, S. A., Cutler, N. S., Ashe, M., and Cardenas, M. E. (2004) Mol. Cell Biol. 24 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tate, J. J., and Cooper, T. G. (2003) J. Biol. Chem. 278 36924–36933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannattasio, S., Liu, Z., Thornton, J., and Butow, R. A. (2005) J. Biol. Chem. 280 42528–42535 [DOI] [PubMed] [Google Scholar]
- 17.Hofman-Bang, J. (1999) Mol. Biotechnol. 12 35–73 [DOI] [PubMed] [Google Scholar]
- 18.Magasanik, B., and Kaiser, C. A. (2002) Gene (Amst.) 290 1–18 [DOI] [PubMed] [Google Scholar]
- 19.Blinder, D., Coschigano, P. W., and Magasanik, B. (1996) J. Bacteriol. 178 4734–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni, A. A., Abul-Hamd, A. T., Rai, R., El Berry, H., and Cooper, T. G. (2001) J. Biol. Chem. 276 32136–32144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Como, C. J., and Arndt, K. T. (1996) Genes Dev. 10 1904–1916 [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Y., and Broach, J. R. (1999) EMBO J. 18 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacinto, E., Guo, B., Arndt, K. T., Schmelzle, T., and Hall, M. N. (2001) Mol. Cell 8 1017–1026 [DOI] [PubMed] [Google Scholar]
- 24.Carvalho, J., and Zheng, X. F. (2003) J. Biol. Chem. 278 16878–16886 [DOI] [PubMed] [Google Scholar]
- 25.Crespo, J. L., Helliwell, S. B., Wiederkehr, C., Demougin, P., Fowler, B., Primig, M., and Hall, M. N. (2004) J. Biol. Chem. 279 37512–37517 [DOI] [PubMed] [Google Scholar]
- 26.Rai, R., Daugherty, J. R., and Cooper, T. G. (1995) Yeast 11 247–260 [DOI] [PubMed] [Google Scholar]
- 27.Wang, H., Wang, X., and Jiang, Y. (2003) Mol. Biol. Cell 14 4342–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crespo, J. L., Powers, T., Fowler, B., and Hall, M. N. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6784–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tate, J. J., Rai, R., and Cooper, T. G. (2005) J. Biol. Chem. 280 27195–27204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tate, J. J., Feller, A., Dubois, E., and Cooper, T. G. (2006) J. Biol. Chem. 281 37980–37992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feller, A., Boeckstaens, M., Marini, A. M., and Dubois, E. (2006) J. Biol. Chem. 281 28546–28554 [DOI] [PubMed] [Google Scholar]
- 32.Tate, J. J., Rai, R., and Cooper, T. G. (2006) J. Biol. Chem. 281 28460–28469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulkarni, A., Buford, T. D., Rai, R., and Cooper, T. G. (2006) FEMS Yeast Res. 6 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherens, B., Feller, A., Vierendeels, F., Messenguy, F., and Dubois, E. (2006) FEMS Yeast Res. 6 777–791 [DOI] [PubMed] [Google Scholar]
- 35.Wach, A. (1996) Yeast 12 259–265 [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., McKenzie, A. 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998) Yeast 14 953–961 [DOI] [PubMed] [Google Scholar]
- 37.Schmitt, M. E., Brown, T. A., and Trumpower, B. L. (1990) Nucleic Acids Res. 18 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foury, F., and Talibi, D. (2001) J. Biol. Chem. 276 7762–7768 [DOI] [PubMed] [Google Scholar]
- 39.Cox, K. H., Tate, J. J., and Cooper, T. G. (2002) J. Biol. Chem. 277 37559–37566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tate, J. J., and Cooper, T. G. (2007) J. Biol. Chem. 282 18467–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffman, J. A., Rai, R., Cunningham, T., Svetlov, V., and Cooper, T. G. (1996) Mol. Cell Biol. 16 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coffman, J. A., el Berry, H. M., and Cooper, T. G. (1994) J. Bacteriol. 176 7476–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coffman, J. A., Rai, R., and Cooper, T. G. (1995) J. Bacteriol. 177 6910–6918; Correction (1996) J. Bacteriol. 178, 2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffman, J. A., Rai, R., Loprete, D. M., Cunningham, T., Svetlov, V., and Cooper, T. G. (1997) J. Bacteriol. 179 3416–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tate, J. J., Cox, K. H., Rai, R., and Cooper, T. G. (2002) J. Biol. Chem. 277 20477–20482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox, K. H., Kulkarni, A., Tate, J. J., and Cooper, T. G. (2004) J. Biol. Chem. 279 10270–10278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drillien, R., and Lacroute, F. (1972) J. Bacteriol. 109 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drillien, R., Aigle, M., and Lacroute, F. (1973) Biochem. Biophys. Res. Commun. 53 367–372 [DOI] [PubMed] [Google Scholar]
- 49.Cunningham, T. S., Andhare, R., and Cooper, T. G. (2000) J. Biol. Chem. 275 14408–14414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper, T. G., Rai, R., and Yoo, H. S. (1989) Mol. Cell Biol. 9 5440–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rai, R., Daugherty, J. R., Cunningham, T. S., and Cooper, T. G. (1999) J. Biol. Chem. 274 28026–28034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.