Abstract
Factor H is a regulatory glycoprotein of the complement system. We expressed the three N-terminal complement control protein modules of human factor H (FH1-3) and confirmed FH1-3 to be the minimal unit with cofactor activity for C3b proteolysis by factor I. We reconstructed FH1-3 from NMR-derived structures of FH1-2 and FH2-3 revealing an ∼105-Å-long rod-like arrangement of the modules. In structural comparisons with other C3b-engaging proteins, factor H module 3 most closely resembles factor B module 3, consistent with factor H competing with factor B for binding C3b. Factor H modules 1, 2, and 3 each has a similar backbone structure to first, second, and third modules, respectively, of functional sites in decay accelerating factor and complement receptor type 1; the equivalent intermodular tilt and twist angles are also broadly similar. Resemblance between molecular surfaces is closest for first modules but absent in the case of second modules. Substitution of buried Val-62 with Ile (a factor H single nucleotide polymorphism potentially protective for age-related macular degeneration and dense deposit disease) causes rearrangements within the module 1 core and increases thermal stability but does not disturb the interface with module 2. Replacement of partially exposed (in module 1) Arg-53 by His (an atypical hemolytic uremic syndrome-linked mutation) did not impair structural integrity at 37 °C, but this FH1-2 mutant was less stable at higher temperatures; furthermore, chemical shift differences indicated potential for small structural changes at the module 1-2 interface.
Complement factor H (FH)3 is a soluble multiple-domain glycoprotein (155 kDa) that is abundant (500-800 μg/ml) in human plasma (1). It regulates the alternative pathway (AP) of activation of the complement system, a key molecular component of immune defense. Sequence variations in FH have been linked to three complement-mediated diseases; dense deposit disease (DDD, or membranoproliferative glomerulonephritis type II), atypical hemolytic uremic syndrome (aHUS), and age-related macular degeneration (AMD) (2-4). Factor H and five FH-related proteins form a subgroup within the family of homologous proteins encoded by the regulators of complement activation (RCA) gene cluster (5-7). The other subgroup includes C4b-binding protein α-chain (C4BPα), membrane cofactor protein (MCP, CD46), decay accelerating factor (DAF, CD55), and complement receptor type 1 (CR1, CD35).
Activation of the complement system via either the alternative or classical pathways involves a proteolytic cascade (8, 9). During this process complement components are enzymatically cleaved into active forms and surface-deposited. Deposition subsequently triggers destruction and immune clearance and is accompanied by release of pro-inflammatory anaphylatoxins. The AP of complement is permanently switched on; if left unchecked, a positive-feedback loop amplifies deposition of activated components onto any nearby surface. Factor H controls amplification via the AP both in fluid phase and selectively on self-surfaces (10, 11), thereby helping to direct complement toward its target, foreign or unwanted cells or particles. The remaining RCA family members are membrane proteins (CR1, DAF, and MCP) or act primarily on the classical pathway of complement (C4BP).
Like the other RCA proteins, FH intervenes at the level of enzymatically active bi- and trimolecular complexes called the C3 and C5 convertases (C3b,Bb and C3b2,Bb, respectively, in the AP). These assemble on surfaces during the activation process and drive the proteolytic cascade by cleaving C3 and C5 into active fragments. Via unknown mechanisms, FH competes with factor B (FB) for binding to C3b, accelerates the irreversible decay of the convertases into their components (C3b and Bb), and acts as a cofactor for the factor I (FI)-mediated cleavage of C3b.
The 20 homologous domains that make up the whole of FH, known as complement control protein modules (CCPs), are each ∼60 amino acid residues in length and stabilized by conserved disulfide bonds (1, 12). Neighboring CCPs are linked by sequences containing between three and eight residues (13). Low-resolution structural studies suggest a “beads-on-a-string” arrangement of CCPs within an elongated FH molecule that may bend back on itself (14). Within FH, C3b-binding sites have been mapped to CCPs 1-4 and CCPs 19-20 (15-18). A third C3b-binding site was inferred to occupy CCPs 12-14 (17, 18). The C-terminal C3b-binding site additionally binds polyanions and acts as an anchor for attachment of FH to cell-surface bound C3b (19-21). The N-terminal C3b-binding site on the other hand is considered critical for engaging and disrupting convertases given that a recombinant protein consisting of FH modules 1-4 displays the full fluid-phase activity of FH and retains a capability to accelerate decay of cell-surface bound convertases (22-24). At least one single nucleotide polymorphism mapped to this region of FH has been linked to disease; V62I has been suggested to be protective against AMD and DDD (4, 25). Moreover, a mutation of Arg-53 to His was discovered in a patient with aHUS (26).
Each RCA family member has a distinct functional profile (27), but the structural basis for complement regulation remains to be established. This is despite availability of experimentally determined three-dimensional structures of C4BPα CCPs 1-2 (28), MCP CCPs 1-2 (29), DAF CCPs 1-4 (30), CR1 CCPs 15-17 (31), and the vaccinia virus complement control protein (VCP) CCPs 1-4 (32), all of which correspond to complete or partial functional sites within their respective parent proteins (Fig. 1). The N terminus of the principal soluble regulator of the AP, FH, therefore provides an attractive structural target. We report the production of a functionally active recombinant fragment of FH composed of CCPs 1-3 (FH1-3) together with the three-dimensional structure of FH1-3. We also report on the structural consequences of two disease-linked sequence variations in this region.
FIGURE 1.
Sequence alignment for some CCPs of known three-dimensional structure. The three blocks of sequences represent first, second, and third modules of functional sites. FB CCP 1 and CR1 CCP 17 have insertions compared with other sequences as indicated. Conservation of residues in four or more neighboring sequences is indicated by shading. Residues that lie in β-strands are underlined, and corresponding regions are assigned numbers that reflect the occurrence of up to eight β-strands (as identified by STRIDE (77)) in some examples of this module family. Bold font indicates a residue considered to be functionally critical on the basis of mutagenesis; black arrows indicate such residues in DAF that are conserved in FH; white arrows indicate residues critical for C3b interaction in FB CCP 3 conserved in FH CCP 3. Filled squares indicate exposed side chains, and half-filled squares correspond to partly exposed side chains in FH (as identified by GETAREA (57)). Arg-53 and Val-62 are indicated in italics, and hv loop indicates the position of the hypervariable loop.
EXPERIMENTAL PROCEDURES
Expression of Protein—The DNA fragments encoding human FH residues 20-142 for FH1-2, 84-206 for FH2-3, and 20-206 for FH1-3 (native sequence numbering, i.e. before cleavage of signal peptide) were cloned into Pichia pastoris expression vector pPICZαΒ. These clones have a valine at position 62 and, for the purposes of this study, will be referred to as wild type. The coding region of FH1-2 in pPICZαΒ was mutated to the variants FH1-2(H53) and FH1-2(I62) using the QuikChange™ site-directed mutagenesis kit (Stratagene). The expressed proteins were directed to the secretory pathway by placing the Saccharomyces cerevisiae α-factor secretion sequence upstream from the coding sequence. FH like-1 (a splice variant containing modules 1-7) was cloned into an insect cell expression system and expressed and purified as described previously (33).
After transformation into P. pastoris strain KM71H, protein was expressed and isotopically labeled in batches of 0.6 liters (initial volume) of cell culture using a fermentor. Samples of FH1-2 and FH2-3 were enriched with >98% 15N or with 13C and 15N by providing (15NH4)2SO4 as the sole nitrogen source and [13C]glucose, [13C]glycerol, and [13C]methanol as carbon sources as previously described (34). Additionally, a 15N sample of FH1-3 was enriched with >75% 2H by using 95% D2O and 1% methanol-d4 in the growth medium. After an induction period of 4-6 days, cells were pelleted, supernatant was harvested and diluted 5-fold in the presence of 5 mm EDTA and 1 mm phenylmethylsulfonyl fluoride, and the pH was adjusted to 4.0. Purification of all three constructs was achieved by cation-exchange chromatography (SP Sepharose Fast Flow™ Sigma-Aldrich) followed by reverse phase high performance liquid chromatography (Supelco Discovery® BIO Wide Pore C5 column, Supelco Inc., PA). The resulting peak fractions were lyophilized. N-terminal sequencing confirmed the identity of the protein constructs as well as the presence of an N-terminal cloning artifact, EAEAAG, left over from incomplete processing of the α-factor secretion signal by the aminodipeptidase Ste13. Mass spectrometry confirmed the predicted sequence and the expected oxidation state of all cysteines (as contributors to disulfide bonds). Yields of purified protein were in the region of 1 mg of protein per g of wet cells.
Determination of Complement-regulating Activity—Cofactor activity was initially determined using an end point fluid-phase cofactor assay. Molar equivalent amounts (1 μm) of constructs FH1-2, FH2-3, and FH1-3 were added to 5 μg (1.8 μm) of purified complement C3b mixed with 2 μg (1.4 μm) of FI (both from Comptech) in a total volume of 15 μl. A negative control where no FH constructs were present was also included. After incubation for 1 h at 37 °C, SDS-containing sample buffer including the reducing agent dithiothreitol was added, and the reactions were then heat-inactivated. The samples were subjected to SDS-PAGE to detect the extent of proteolytic cleavage of C3b α-chain by FI. To obtain a semiquantitative estimation of FH1-3 activity, a concentration titration cofactor assay was performed using the above assay conditions on full-length FH and FH1-3 at a series of molar-equivalent concentrations in a total reaction volume of 15 μl and incubated for 10 min at 37 °C. The decay acceleration assay was performed on FH1-3, FH-like-1, and FH as previously described (20).
Data Collection and Processing—The addition of free l-Arg and l-Glu was previously reported to increase long-term stability and solubility of concentrated protein NMR samples (35). The present study utilized deuterated versions (l-Arg-d7 and l-Glu-d5) to prevent their signals dominating 13C,1H-resolved experiments.
Standard suites of NMR spectra (36) were acquired on Bruker AVANCE 600 and 800 MHz spectrometers using 5-mm triple-resonance probes. Data for resonance assignments were acquired at 37 °C on 1 mm 13C,15N FH1-2, 2 mm 13C,15N FH2-3, and 0.6 mm 2H,15N FH1-3 in 20 mm potassium phosphate, 0.05% (w/v) NaN3, 10% (v/v) D2O, 50 mm arginine-d7, 50 mm glutamate-d5 at pH 6.2. 15N-Edited nuclear Overhauser effect spectroscopy-HSQC (NOESY-HSQC) (37) experiments were acquired on all constructs at 800 MHz with mixing times of 100 ms. 13C-Edited NOESY-HSQC (38) experiments were acquired at 800 MHz for FH1-2 and FH2-3 with mixing times of 100 ms. Steady-state (1H,15N) nuclear Overhauser effects (NOEs) (39) were measured at 600 and 800 MHz and were calculated from the ratio of the intensities of the cross-peaks in the reference spectra to those recorded with saturation of the 1H signal. Additionally, a 1H,15N-states time-proportional phase incrementation transverse relaxation-optimized spectroscopy spectrum (40) with watergate was acquired for FH1-3 at 800 MHz.
Subsequently, the FH1-2 and FH2-3 samples were aligned in 12 mg/ml Pseudomonas filamentous phage (Profos, Regensburg, Germany) for measurements of residual dipolar couplings (RDCs); buffer conditions were kept identical to those used for NOE measurement. The degree of alignment was monitored from the splitting of the 2H signal, 6.2 Hz for FH1-2 at 600 MHz and 7.1 Hz for FH2-3 measured at 800 MHz. 1DNH (41), 1DCαHα (42), and 1DCαC′ (43) RDC couplings were measured for aligned and unaligned samples of FH1-2 and FH2-3. The RDCs from residues deemed flexible based on relaxation data criteria were not used in the structure calculations (44).
NMR data were processed using the Azara suite of programs provided by Wayne Boucher and the Department of Biochemistry, University of Cambridge, UK. Maximum entropy reconstruction was used for the F1 and F2 dimensions of the three-dimensional experiments. The RDC data were analyzed using an in-house macro developed for the Collaborative Computing Project for the NMR community Analysis software (45). Relaxation data were analyzed using the rates analysis function provided in the Analysis software (45). Analysis of NMR relaxation data and scrutiny of FH1-2 and FH2-3 NOESY spectra provided no evidence of non-transient self-association.
Resonance Assignment and Structure Calculation—Processed spectra were viewed, and nuclei were assigned using Analysis (45). For example, ∼96 and ∼98% of observable protons were assigned within FH1-2 and FH2-3, respectively.
Resolved peaks in 15N NOESY and 13C NOESY spectra were picked, and where possible, the root resonances were assigned unambiguously. All proline residues were defined as trans based on chemical shift differences between Cβ and Cγ resonances (46) and observation of appropriate strong NOE cross-peaks. A list of chemical shifts and a set of largely unassigned NOESY spectra were supplied as input for CYANA 2.1, an automated structure-calculation software (47). The disulfide linkages were inferred from the proximity and geometry of Cys side chains in an initial set of structure calculations using CYANA 2.1 that did not incorporate disulfide bonds. Hydrogen bonds were inferred on the basis of a series of 15N HSQC spectra recorded on lyophilized samples resuspended in 99.96% D2O (Sigma-Aldrich) over a time-course of 2 h with 15-min intervals. No significant difference in the protected amides signals was observed after 1 h. Hydrogen-bond derived distance constraints were then created for protected protons possessing a feasible proton-accepting partner, as judged from first-round structure calculations in CYANA 2.1. A seven-cycle routine of target function minimization was subsequently performed using the combined automated NOE assignment and structure determination module within CYANA 2.1. The upper-limit distance constraints, 2722 for FH1-2 and 2733 for FH2-3, generated from CYANA 2.1 were converted to the Crystallography and NMR System (CNS) format (48) using FormatConverter (45). These constraints were incorporated into a final round of structure calculations, performed using CNS, during which the RDC constraints, 91 for FH1-2 and 169 for FH2-3, were incorporated into the final stages of refinement by including a TENSO energy term (49) with a harmonic potential. A lack of NOE or RDC violations and the low root mean square deviation (r.m.s.d.) for backbone atoms (Table 1) confirmed the three-dimensional structure of each protein is well defined by experimental data. Overall quality as assessed by WHAT IF (50) and favorable Ramachandran statistics (51) was checked for consistency with a high resolution structure. The FH1-2(H53) model was created on the basis on the closest-to-mean FH1-2(R53) NMR structure employing the side-chain replacement program, SCWRL Version 3 (52, 53).
TABLE 1.
Structural statistics for the lowest energy structures
| FH1-2 | FH2-3 | |
|---|---|---|
| Number of lowest energy structuresa,b | 30 | 29 |
| Upper limit distance constraints | ||
| Intraresidue | 563 | 562 |
| Sequential | 837 | 789 |
| Intermediate range (2 ≤ |i - j| ≤ 4) | 323 | 337 |
| Long range (|i - j| > 4) | 999 | 1045 |
| Total | 2722 | 2733 |
| Intermodular | 20 | 8 |
| Module 1 to linker | 58 | |
| Module 2 to linker | 48 | 57 |
| Module 3 to linker | 35 | |
| Hydrogen bonds | 20 | 16 |
| Residual dipolar couplings | ||
| 1DNH | 55 | 75 |
| 1DCαC′ | 36 | 59 |
| 1DCαHα | N/Ac | 35 |
| Root mean square deviations (residues from CysI to CysIV of each module) | ||
| All heavy atoms | 1.25 | 1.28 |
| Backbone atoms Cα, N, CO | ||
| Module 1 | 0.52 | |
| Module 2 | 0.69 | 0.32 |
| Module 3 | 0.96d | |
| All modules | 0.92 | 0.90 |
| Skew, twist, and tilt angles (°) | ||
| Skew | ||
| Minimum | 236 | 46 |
| Maximum | 280 | 145 |
| Mean | 259 | 91 |
| S.D. | 12 | 19 |
| Twist | ||
| Minimum | 163 | 131 |
| Maximum | 185 | 150 |
| Mean | 173 | 141 |
| S.D. | 6 | 5 |
| Tilt | ||
| Minimum | 15 | 6 |
| Maximum | 35 | 24 |
| Mean | 23 | 15 |
| S.D. | 5 | 4 |
| Ramachandran assessment (%) | ||
| Most favored | 76.4 | 79.3 |
| Additionally allowed | 20.9 | 19.6 |
| Generously allowed | 1.2 | 0.8 |
| Disallowed | 1.5 | 0.3 |
| Coarse packing WHAT IF score | −1.574 | −1.254 |
| Buried surface area of intermodular junction (Å2) | 549 | 517 |
| Buried surface area of intermodular junction in FH1-3 (Å2) | 534 | 509 |
Less than two NOE violations above 0.5 Å for each structure.
PDB accession codes 2RLP and 2RLQ respectively.
Not available due to poor spectral quality.
High value due to the flexible and ill-defined hypervariable loop.
Deriving the Structure of the Triple Module Construct FH1-3—The structure of CCP 2 was found to be very similar in both FH1-2 and FH2-3. The largest context-dependent structural differences occur in loops located close to the C terminus of CCP 2, adjacent to CCP 3. On the other hand, the structures of the loops of CCP 2 adjacent to its N terminus and the CCP 1-2 interface appear context-independent. This observation was corroborated by a comparison of chemical shifts between the two module pairs (not shown). A 15N HSQC spectrum and 15N HSQC transverse relaxation-optimized spectroscopy spectrum of the FH1-3 construct were recorded on a 0.6 mm sample of 2H,15N FH1-3 in 20 mm potassium phosphate, 50 mm arginine-d7, 50 mm glutamate-d5 at pH 6.2. The spectra were compared with those of the two pairs, FH1-2 and FH2-3, to assess the degree of conservation in chemical shifts (supplemental Fig. 1). This subsequently provided a basis for rational selection of CCP 2 residues from each module-pair structure to use in creating the template for the modeling procedure (Fig. 2, C and D). In this regard, care was taken to preserve the structure of the loops involved in the intermodular junctions, as identified from the module-pair structures using the Protein Interaction Calculator (54) server. The closest-to-mean structures on CCP 2 for the pairs were then superimposed on their mutual CCP 2, and appropriate complementary segments of CCP 2 were deleted to create the template. Modeler 9v1 (55, 56) was used to reconstruct 20 models of FH1-3, and the one with the lowest objective function score was selected as the representative model.
FIGURE 2.
NMR-derived structures of FH1-2 and FH2-3 and reconstructed FH1-3. A, backbone representations showing separate ensembles of FH1-2 (cyan) and FH2-3 (magenta) structures, each superimposed on the Cα atoms of their respective CCP 2s. B, backbone representations showing both the ensembles superposed on their mutual CCP 2s (Cα r.m.s.d. 0.78 Å). C, schematic representation of the structure of FH1-3 derived from structures of overlapping pairs and showing the boundaries of the FH1-2 and FH2-3 structures used as templates in Modeler 9v1 (55, 56). D, orthogonal view of the schematic representation in C.
The solvent-accessible surface area for exposed side chains in FH1-3 and buried surface areas at bimodular interfaces were calculated using GETAREA version 1.1 (57). Intermodular tilt, twist, and skew angles for FH1-2 and FH2-3 were calculated using the same protocol as previously described (13, 58). Visualization of structures and generation of figures was performed using PyMOL (59).
RESULTS
FH1-3, the Minimal Unit Capable of Cofactor Activity—To determine the minimal unit of FH capable of serving as a cofactor for FI-catalyzed proteolysis of C3b, the constructs FH1-2, FH2-3, and FH1-3, expressed and purified from P. pastoris, were tested for activity in a SDS-PAGE based cofactor assay (Fig. 3A). This experiment demonstrated that FH1-3 displayed measurable cofactor activity, whereas the pairs FH1-2 and FH2-3, at molar equivalent concentrations, did not. An assay (Fig. 3B) in which activity was measured as a function of the amount of cofactor indicated that about 10-fold more FH1-3 than full-length FH is required to achieve a similar extent of C3b cleavage, reflecting a contribution from the other modules of FH. Thus, FH1-3 represents the minimal unit capable of cofactor activity and is a worthwhile structural target. We also assayed FH1-3 for decay acceleration activity (data not shown) and compared it to full-length FH and to FH-like 1. We could not detect any activity of FH1-3 at a concentration >100-fold higher that the concentration of FH or ∼6-fold higher than the concentration of FH like-1, required to release 50% of Bb from cell-surface-attached convertase.
FIGURE 3.
Assays of cofactor activity for factor I-catalyzed cleavage of C3b. Inferred identities of Coomassie-stained protein bands on SDS-PAGE gels are indicated. MWM, molecular weight markers. A, results of an end point C3b cofactor assay (excess C3b and FI in the absence (lane 2) and presence of 1 μm FH1-2 (lane 4), 1 μm FH2-3 (lane 5), and 1 μm FH1-3 (lane 6). Molecular weight markers are in lane 1. B, C3b cofactor assay with excess FI and C3b in the presence of FH at 0, 0.84, 2.53, 7.6 nm (lanes 1-4) or FH1-3 at 0.84, 2.53, 7.6, 22.8, 68.4, and 205 nm (lanes 5-10).
Structure Determination of FH1-3—We elected to reconstruct the three-dimensional structure of FH1-3 computationally from the structures of FH1-2 and FH2-3 rather than determine directly the structure of FH1-3. This decision was dictated by issues of yield, solubility, and spectral quality. Similar problems were encountered in a study of a triple-CCP module segment of CR1 (31).
Using 13C,15N-labeled samples of recombinant FH1-2 and FH2-3, conventional heteronuclear NMR experiments led to good-quality structure determinations (Table 1). The ensembles of lowest energy FH1-2 and FH2-3 structures displayed in Fig. 2 are consistent with 2722 and 2733 upper-limit distance constraints, respectively. Sets of 20 and 8 NOE-derived inter-modular distance constraints along with 91 and 169 RDC constraints help define the 1-2 and 2-3 module-module interfaces, respectively. The angles (13, 58) that describe the intermodular orientations of each member of the ensemble are summarized in Table 1 and graphed in Fig. 4.
FIGURE 4.
Structural comparison of FH1-3 with DAF-(1-4) and CR1-(15-17). Schematic representations are drawn in PyMol: DAF-(1-4) (green), CR1-(15-17) (blue), and FH1-3 (CCP 1, red; CCP 2, magenta; CCP 3, orange). Overlays, performed on the cysteine Cα atoms, are as follows: A, DAF CCP 2 and FH1-3 CCP 1; B, DAF CCP 3 and FH1-3 CCP 2; C, DAF CCP 4 and FH1-3 CCP 3; D, CR1 CCP 15 and FH1-3 CCP 1; E, CR1 CCP 16 and FH1-3 CCP 2; F, CR1 CCP 17 and FH1-3 CCP 3. Plots to show comparison of intermodular angles (mean ± 1 S.D. for the ensemble in each case) between first and second modules (G) and second and third modules (H) of the functional sites within FH, DAF, and CR1 (site 2).
Fig. 2 shows the lowest energy structures of FH1-2 and FH2-3 overlaid on the backbone atoms of CCP 2, with a low r.m.s.d. of 0.78 Å. A comparison, taking advantage of a partial backbone assignment of FH1-3, revealed no evidence to suggest that the presence of CCP 1 affects the chemical shifts of residues at the CCP 2-3 interface nor that the presence of CCP 3 perturbs chemical shifts in the CCP 1-2 interface (supplemental Fig. 1). It was, therefore, legitimate to derive the structure of FH1-3 by computationally concatenating the two module-pair structures ensuring that the appropriate module-module interfaces and orientations are retained.
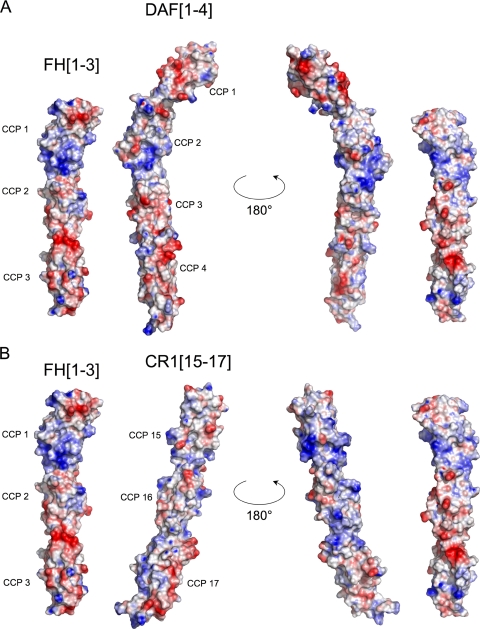
The Three Modules of FH1-3 Are Organized in a Rod-like Arrangement—The three compactly folded, oblate CCPs that comprise FH1-3 are arranged in an end-to-end rod-like conformation spanning ∼105 Å from tip to tip (Fig. 2). The variation in intermodular angles among the ensemble of calculated structures likely reflects some limited flexibility between modules. There are several exposed apolar residues in each module (see Fig. 1), but inspection of the overall surface of FH1-3 revealed no extensive hydrophobic patches. A representation of the electrostatic surface of FH1-3 demonstrates that CCP 1 exposes more extensive and prominent electro-positive regions (Fig. 5) than the other modules, and this is particularly evident toward its interface with CCP 2.
FIGURE 5.
Comparison of electrostatic surfaces of FH1-3 with DAF-(1-4) (A) and CR1-(15-17) (B). The surfaces were generated using the Adaptive Poisson-Boltzmann Solver (78) plug-in within PyMol (using the PARSE (79) parameter set with the nonlinear form of the Poisson-Boltzmann equation, a protein dielectric constant of 20, solvent dielectric constant of 80, solvent ion radius of 1.4 Å, temperature 310 K, and assumed ion concentration of 100 mm). Red is negative charge, and blue is positive charge within a range of -5/+5kT(k = Boltzmann's constant, T = absolute temperature). The orientation of FH1-3 is the same as in Fig. 2D. The other structures were orientated by superposing DAF CCP 2 (A) or CR1 CCP 15 (B) on FH CCP 1 and then transposing.
Five predominantly extended stretches of residues connected by turns or loops run back and forth in approximate alignment with the long axis of each of the three CCP modules. These form β-strands and small β-sheets varying between the modules in number and extent (Fig. 1) and held together by a compact hydrophobic core plus two disulfide bonds. For the purposes of cross-reference and further discussion, regions within the FH modules were assigned numbers that reflect the occurrence of up to eight β-strands in other examples of this module family (see Fig. 1). The N-terminal CCP of FH is dissimilar from the other two (Cα r.m.s.d. of 3.1 and 3.0 Å versus CCP 2 and 3, respectively), whereas CCPs 2 and 3 overlay with aCα r.m.s.d. of 1.8 Å even though the highest level of sequence identity is between CCPs 1 and 2 (Table 2). A stretch of residues before strand 3 (see Fig. 1) has previously been called the hypervariable loop (27); in CCP 3 this has 1H,15N NOE intensities lower than 0.5 and, thus, is less well structured than the hypervariable loops of CCP 1 and CCP 2. Otherwise, heteronuclear NOEs for residues in all three modules and in the linking sequences between them are predominantly uniform (data not shown).
TABLE 2.
Structure and sequence pair-wise similarities between FH and other RCA-CCPs of known structure
Domain boundaries considered from one residue before Cysl to three residues after CysIV (where available). Values on the left-hand side specify Cα r.m.s.d. values in Angstroms (Å) Values in parentheses represent the structural alignment lengths in terms of the numbers of Cα atoms for which the r.m.s.d. values were calculated. Values in italics on the right-hand side represent pair-wise percentage (%) sequence identities calculated using the Percentage Identity Matrix option under ClustalX (80).
| Factor H modules | |||
|---|---|---|---|
| RCA-CCP (PDB code (81)) | FH1 | FH2 | FH3 |
| FH 1 (2RLP) | |||
| FH 2 (2RLQ) | 3.12 (59); 27 | ||
| FH 3 (2RLQ) | 3.00 (54); 22 | 1.80 (55); 20 | |
| FB 1 (2OK5) | 4.13 (64); 20 | 2.34 (55); 19 | 2.04 (51); 32 |
| FB 2 (2OK5) | 3.35 (60); 20 | 2.20 (57); 24 | 1.80 (58); 25 |
| FB 3 (2OK5) | 3.38 (58); 23 | 1.91 (57); 23 | 1.30 (56); 26 |
| C4BPα 1 (2A55) | 2.23 (60); 27 | 3.48 (69); 24 | 3.24 (61); 19 |
| C4BPα 2 (2A55) | 3.22 (58); 22 | 1.81 (57); 31 | 1.70 (57); 24 |
| CR1 15 (1GKN) | 1.98 (58); 26 | 3.03 (59); 20 | 2.76 (55); 18 |
| CR1 16 (1GKN) | 3.38 (58); 24 | 1.57 (58); 28 | 1.67 (57); 24 |
| CR1 17 (1GKG) | 2.74 (58); 26 | 2.31 (58); 36 | 2.12 (57); 32 |
| DAF 1 (1OK3) | 2.48 (60); 22 | 2.54 (59); 15 | 2.59 (59); 17 |
| DAF 2 (1OK3) | 2.22 (61); 31 | 2.92 (60); 21 | 2.80 (59); 20 |
| DAF 3 (1H03) | 3.31 (58); 22 | 1.59 (58); 29 | 1.54 (57); 34 |
| DAF 4 (1H03) | 2.68 (58); 28 | 1.91 (57); 25 | 1.63 (58); 31 |
| MCP 1 (2CKL) | 2.56 (60); 23 | 2.73 (58); 20 | 2.73 (58); 22 |
| MCP 2 (2CKL) | 3.32 (53); 27 | 1.77 (59); 36 | 1.63 (56); 34 |
| VCP 1 (1G40) | 3.41 (61); 27 | 3.09 (59); 24 | 2.82 (60); 21 |
| VCP 2 (1G40) | 3.33 (58); 22 | 2.44 (57); 39 | 2.04 (56); 28 |
| VCP 3 (1G40) | 2.79 (58); 30 | 2.57 (58); 30 | 2.47 (56); 35 |
| VCP 4 (1G40) | 3.06 (58); 25 | 2.21 (55); 21 | 2.29 (58); 38 |
The elongated nature of FH1-3 reflects small intermodular tilt angles (see Fig. 4, G and H). Twist angles on the other hand are large so that the hypervariable loops, labeled in Fig. 2, of modules 1 and 3 project from the same face of the FH1-3 structure (module 2 does not possess a prominent hypervariable loop due to a deletion in this region; see Fig. 1). Each of the two intermodular interfaces is stabilized by H-bonds between the third linker residue and the extremities of long strands 4 within the adjacent modules; i.e. between the amide of linker residue Arg-83 or Val-144 and the carbonyl of Gly-55 or Gly-117 (both in 4-5 loops), respectively, and between the carbonyl of Arg-83 or Val-144 and the amide of Tyr-106 or Phe-170 (both in 3-4 loops), respectively. The CCP 1-CCP 2 and CCP 2-CCP 3 interfaces bury 549 and 517 Å2 of surface area, respectively. In each case the start of strand 5 of the preceding module contributes hydrophobic portions of side chains (e.g. Tyr-56 from CCP 1) to a hydrophobic cluster along with the alkyl segments of linker side-chains (e.g. Lys-82 and Arg-83 in the CCP 1-CCP 2 linker) and side chains from the 3-4 loop and the 6-7 loop (e.g. Thr-131 in CCP 2) of the next module.
Structural Consequences of Disease-linked Sequence Variations V62I and R53H—We inspected the structure of FH1-3 to ascertain the likely structural consequences of disease-linked sequence variations. The side-chain of Val-62 is completely buried toward the C-terminal end of the hydrophobic core of CCP 1. Replacement with isoleucine occurs as a result of a common single nucleotide polymorphism that appears to be protective against AMD and DDD (4, 25). This insertion of an additional methylene group into the core of a small globular domain could have profound effects on its folding and stability (60). To address this issue, we expressed a 15N-labeled NMR sample of the V62I variant of FH1-2 (termed FH1-2(I62) to distinguish it from the original sample of FH1-2(V62)) in P. pastoris and collected a 15N HSQC spectrum (supplemental Fig. 2A). The overall similarity of this spectrum to the 15N HSQC spectrum of FH1-2(V62) proves that both CCP modules in FH1-2(I62) fold correctly. Further analysis investigated more subtle structural differences between the two variants. On the basis of comparison with the FH1-2(V62) 15N HSQC spectrum, combined 15N and 1H chemical shift perturbation values (61) were calculated for FH1-2(I62) and plotted against residue number; amides exhibiting the largest changes in chemical shifts were then highlighted on the FH1-2 structure (Fig. 6). Notably, differences are exclusive to CCP 1, and there are negligible changes in CCP 2 chemical shifts. Differences are wide-spread throughout CCP 1 but centered on residue 62, as expected. The cross-peak corresponding to the amide of Tyr-50, H-bonded to the carbonyl of residue 62, exhibits one of the biggest changes in amide chemical shift. Other differences in chemical shift are attributable to small rearrangements in the hydrophobic core to accommodate the extra methylene group of Ile-62. The exchange rates with solvents of amide protons reflects the extent of hydrogen-bonded secondary structure and conformational flexibility. Observation of slowly exchanging amides, i.e. those still observable after 1 h, indicates that FH1-2(I62) is at least as rigid in this respect as FH1-2(V62) (supplemental Fig. 3). A series of 15N HSQC spectra collected at increasing temperatures demonstrates that FH1-2(I62) is slightly more thermally stable than FH1-2(V62) (supplemental Figs. 2, A-C, and 4).
FIGURE 6.
Differences in chemical shifts arising from substitutions of amino acid residues in FH1-2. Combined (1H,15N) chemical shift difference (Δδ = √((ΔδNH)2 + (Δδ15N/5)2)) from amino acid residue substitutions, FH1-2(H53) (A) and FH1-2(I62) (C). Mapping of the largest chemical shift perturbation of amides (Δδ > 0.04 ppm) for FH1-2(H53) (B) and for FH1-2 (I62) (D) onto the closest to mean FH1-2 structure shown as schematics (PyMol) with the side chain of the relevant residue highlighted. The sphere radius of the HN atom is proportional to the magnitude of chemical shift perturbation.
Mutation of Arg-53 to histidine has been found in FH of an aHUS patient (26). According to a sequence alignment (Fig. 1), this residue is conserved in several modules that represent the N-terminal CCPs of functional sites in RCAs. From the structure of FH1-3 it is apparent that the guanidyl group of this side chain is exposed on the side of CCP 1, but most of its long alkyl chain is buried. In the structure of the wild type the side chain of Arg-53 lies alongside that of Tyr-56 (Fig. 7). The latter is a strictly conserved hydrophobic residue of CCP 1 that is also proximal to several residues of CCP 2, including Thr-131 and (the well conserved) Tyr-106. The question thus arises when these contacts are disrupted by replacement of Arg-53 with a histidine residue, Does CCP 1 still adopt a stable folded structure? As with FH1-2(I62), the aHUS-associated mutant FH1-2(H53) was expressed and labeled with 15N. The 15N HSQC spectrum of FH1-2(H53) at 37 °C (supplemental Fig. 5A) is clearly that of a fully folded protein, very similar in structure to FH1-2 and consistent with the outcome of a computational model of the mutant protein (Fig. 7). Further examination revealed that cross-peaks in NMR spectra for the FH1-2(H53) amides of residues His-53 and Gly-55 were untraceable (residue 54 is a proline), whereas cross-peaks for the amides of residues Lys-51, Cys-52, and Tyr-56 had substantially altered chemical shifts. All of these residues are in the 4-5 loop of CCP 1 that extends toward CCP 2 and makes contact with the CCP 1-CCP 2 linker (Fig. 4). Cross-peaks corresponding to residues in the adjacent region of CCP 1 (Asn-29—Ile-32), whose side chains partly bury the alkyl side-chain of Arg-53 in the wild-type, undergo large changes in chemical shift. These two loops account for most of the chemical shift differences between mutant and wild type within CCP 1. Interestingly, the amide chemical shifts of Gln-81 in the intermodular linker of FH1-2(H53) are also perturbed with respect to the wild-type module pair as are the amides of two CCP 2 residues, Tyr-106 and Thr-131; both of these lie close to Tyr-56 within the interface between CCPs 1 and 2. Slowly exchanging amides in FH1-2(H53) were found to be fewer in number than in FH1-2(V62) or FH1-2(I62); this observation is consistent with a greater degree of conformational flexibility in the mutant (supplemental Fig. 3). A series of 15N HSQCs recorded at increasing temperatures suggested that FH1-2(H53) reversibly loses its compactly folded nature at a lower temperature than either the Val-62 or Ile-62 variants (supplemental Fig. 5, A-C). Module 1 appears to be more strongly affected as a result of the R53H mutation, but both modules lose thermal stability (supplemental Fig. 4). Unlike FH1-2(V62) or FH1-2(I62), FH1-2(H53) shows some loss of spectral intensity over the 37-45 °C range.
FIGURE 7.
Residues whose amide shifts are most affected by the R53H mutation. A, schematic-type representation focusing on the CCP 1-CCP 2 interface of FH1-2 (closest-to-mean structure), oriented as in Fig. 2C. Residues exhibiting combined amide chemical shift differences >0.1 ppm (in FH1-2(H53) versus wild type) are shown in space-fill representation and colored blue-green. Residues in CCP 2 exhibiting combined amide chemical shift differences >0.04 ppm are shown in space-fill representation and colored light blue. The side chain of the substituted residue, Arg-53, is drawn as sticks, and its atomic surface is represented as dots. B, the same as A but rotated by 45° about the y axis. C and D, the same views as in A and B but showing a model of FH1-2(H53). The color scheme is as before except His53 is colored pink.
DISCUSSION
Extended and Twisted Three-CCP Segments Are Minimal Functional Units of RCAs—Pure properly folded protein consisting of the three CCPs at the FH N terminus (i.e. FH1-3) has cofactor activity for FI-catalyzed cleavage of C3b, although 10-fold more FH1-3 compared with full-length FH is required to achieve the same level of activity. This observation supports previous studies of material secreted from Chinese hamster ovary cells (22) but disagrees with a negative result for a version of FH1-3 expressed in a baculovirus system (23). Triple-CCP module segments of CR1 (i.e. CCPs 1-3, 8-10, and 15-17) are also functionally active, although, as in FH, neighboring modules boost these activities (62). Three consecutive modules of C4BPα (CCPs 1-3) (63), MCP (CCPs 2-4) (64), and VCP (CCPs 1-3 and 2-4) (65) likewise constitute minimal functional sites. Thus, three CCP modules appear to be generally required to bind C3b/C4b in the process of preparing these molecules for cleavage by FI. We could not detect decay-accelerating activity for FH1-3 in a cell-surface assay but note that in the case of DAF, three modules (CCPs 2-4) are necessary and sufficient to engage with C3b,Bb (66) and accelerate its decay. The simplest interpretation of the fact that FH has >100 times more decay accelerating activity (and FH-like 1 (effectively CCPs 1-7 of FH) is at least 6 times more active) is that each of the GAG binding CCP modules of factor H, including CCPs 7 and 20, contribute strongly to the ability of FH to bind to the cell surface in the vicinity of the convertase, and CCPs 19 and 20 provide additional affinity for cell surface-bound C3b. It remains possible that FH1-3 plays a principal role in the dismantling of the convertase once FH is anchored to the surface, but based on previous reports of baculovirus-expressed FH[1-4] having measurable decay accelerating activity (24), it seems likely that CCP 4 of FH also contributes to direct engagement with the convertase. It is notable that the proenzyme FB also contains three CCP modules. These are stowed away in a compact fashion but loosely associated with the remainder of FB before the encounter with C3b (67), whereupon they presumably swing out and engage with C3b; subsequently the FB CCPs are cleaved and dissociate, leaving active C3 convertase (C3b,Bb). In summary, three-CCP module segments of complement proteins represent functional units whose structures can be usefully compared and contrasted.
The overall structure of FH1-3 is highly extended (∼105Åin length) like the structures of CR1-(15-17) (∼118 Å) and CCP modules 2-4 (∼112 Å) of the DAF-(1-4) crystal structure (Fig. 4, A-F) but unlike the compact arrangement of the three FB CCPs. Agreement between the NMR-derived FH1-3 structure and the same modules within a best-fit model of FH modules 1-5 built on the basis of small-angle x-ray scattering data and homology (68) is poor (12.6 Å for Cα). Indeed, in the most favored scattering-based model, modules 1-3 form a bent rather than a straight structure. Note, however, that better agreement in terms of the extended nature of modules 1-3 is obtained when comparing the NMR-derived FH1-3 structure with “model 11” from the deposited ensemble of modeled structures that reportedly fit with the scattering data. This implies that CCPs 4 and 5 must be highly tilted relative to one another and to CCP 3 in an otherwise straight structure of FH modules 1-5 to be consistent with both NMR spectra and current interpretation of the small-angle x-ray scattering data.
Tilt angles within the functional sites of FH, CR1, and DAF are uniformly small, whereas twist angles between first and second modules are close to 180°, and twist angles between second and third modules are between 90 and 135° (Fig. 4, G and H). The relatively small surface areas buried between neighboring CCP modules (∼500 Å2) have similar values in FH1-3, DAF, and CR1-(15-17) (13). These observations suggest that a twisted, end-to-end arrangement of CCPs capable of spanning non-proximal subsites within the convertase is crucial for function. Conformational mobility among the multiple domains of C3b is probable (69-72), and it is conceivable that FH shares with CR1 (and MCP) the ability to bind and stabilize a domain rearrangement that renders the first cleavage site in C3b accessible by FI; interactions of these RCAs with FI are also likely. The CCPs of the viral mimic of RCAs, VCP, are more tilted than their mammalian equivalents, bury more surface area between them (800-900 Å2 in two of the interfaces (13)), and produce less-extended functional sites (CCPs 1-3 measure ∼95 Å and CCPs 2-4 measure ∼91 Å in length).
FH CCPs 1-3 Structurally Resemble Equivalently Positioned CCPs in DAF and CR1—The program Combinatorial Extension (73) was used to structurally align and calculate Cα r.m.s.d. values (Table 2) for each of CCPs 1, 2, and 3 of FH versus other CCPs of known structure that interact with C3/C4 or C3b/C4b.
There is a striking overlay of the backbones of FH CCP 3 and FB CCP 3 (1.3 Å over 56 Cα atoms). Moreover FB CCP 3 residues known to be critical for interaction with C3b are conserved in FH CCP 3 (Fig. 1). The other CCPs of FB and FH are less similar, suggesting that competition by FH for FB binding to C3b is likely a consequence of a shared binding site on C3b for their respective third modules. It is probable that module 4 extends the footprint of FH on C3b, enhancing affinity and helping to explain the relatively poor complement regulatory activity of the three-module construct.
Comparison of backbone structures shows that the N-terminal CCP of FH is more similar to the first modules of other RCA functional sites than it is to their second and third modules. Similarity is highest in the cases of the first CCP in site 2 of CR1, i.e. CCP 15, and the first CCP of the DAF functional site, i.e. CCP 2 (Table 2). Not only does the FH CCP 1 backbone superimpose well on the backbones of these modules, but there is also conservation of residues that contributes to the interface with the next module. In addition, six of seven DAF CCP 2 residues critical for function are conserved in FH CCP 1, some of which (Arg-53, Lys-82, and Arg-83) correspond also to interface residues. Indeed, there is resemblance between electrostatic surface representations of FH CCP 1 and DAF CCP 2 (Fig. 5). A smaller set of functionally critical residues is conserved, and a lesser degree of electrostatic resemblance is evident when FH CCP 1 is compared with CR1 CCP 15. In contrast, there is no conservation of functionally critical residues and poor backbone overlay when comparing FH CCP 1 with CCP 1 or CCP 2 of MCP.
There is a close resemblance at the backbone level between the second module of FH and the second modules of functional sites in the other regulators, CR1 CCP 16, DAF CCP 3, and C4BPα CCP 2 (Table 2). Despite their conserved structural framework, inspection of the electrostatic surface representations of these DAF and FH modules does not reveal obvious similarities (Fig. 5). Furthermore, residues identified as functionally critical in DAF CCP 3, e.g. Phe-169 and Leu-171 (74), are not conserved in FH CCP 2.
In addition to its resemblance to FB CCP 3, the third FH module also shares a highly similar (Cα r.m.s.d. = 1.6 Å) backbone structure with the third CCP of the DAF functional site (i.e. DAF CCP 4). The resemblance is less marked, however, when comparing FH CCP 3 to CR1 CCP 17 (Cα r.m.s.d. = 2.1 Å). Two of three DAF CCP 4 residues that were identified as important in mutagenesis studies are conserved in FH CCP 3 (Fig. 1). Surprisingly, there was relatively little obvious structural similarity between CCPs 1, 2, or 3 of FH and modules 1, 2, or 3 of VCP, respectively, despite high percentage sequence identity.
Implications of Structural Similarities among CCPs for Functional Properties—That the N-terminal modules of FH and CR1 and the second module of DAF (all of which form parts of sites with decay accelerating activity), but not the equivalent module of MCP (that does not have decay accelerating activity), share backbone and surface similarities suggests they recognize the same subsite within the C3b,Bb complex. This is likely to be the von Will-ebrand factor type A domain of Bb (75). The subsequent two modules within the decay accelerators are arranged in a similar way with respect to the first modules of their respective triple-module functional units (Fig. 4, A-F). Assuming these intermodular arrangements are preserved in the complex, it follows that FH CCP 2, CR1 CCP 16, and DAF CCP 3 would be positioned in similar orientations and locations relative to the convertase surface. But the fact that these modules, although highly similar in their backbone structures, differ in their surface properties implies they are unlikely to make equivalent interactions with the convertase. Indeed, the functionally critical surface hydrophobic patch present on DAF CCP 3 (74) does not occur anywhere on the surface of FH CCP 2. This observation is consistent with the suggestion of Harris et al. (75) that the bulk of DAF CCP 3 acts primarily as a spacer. Thus, the second modules of FH and CR1 site 2 could play a similar role with respect to their decay acceleration properties but might have the additional job of recruiting FI to the complex. The fourth DAF module was proposed to interact weakly with C3b (74) once DAF CCP 2 has docked onto Bb; FH CCP 3 and, to a lesser extent, CR1 CCP 17 are likely to be functionally equivalent to DAF CCP 4 since they all share a similar structure and two of three functionally critical residues in DAF CCP 4 are conserved in FH CCP 3 and CR1 CCP 17. Thus, structural comparisons support a common mechanism of decay acceleration among these three RCAs in which separate binding sites in the first and third modules, for C3b and Bb, are so-positioned by the middle module that they bind to and stabilize an intermediate on the pathway to convertase dissociation. We are in a weaker position to speculate about cofactor activity due to the more limited nature of the structural comparison we can make with MCP. The different architecture of VCP indicates that it has evolved an alternative mechanism for decay acceleration.
Disease-linked Sequence Variations—Thus, the first modules within decay acceleration sites on RCAs are inferred to perform the common role of binding to factor Bb of the C3 convertase. Mutations and sequence variations in FH CCP 1, therefore, have the capacity to modulate its decay accelerating activity. The mutation R53H in FH CCP 1 corresponds to a conserved DAF CCP 2 residue (Arg-96) that is important for decay accelerating activity according to DAF mutagenesis (74, 76) and is also conserved in CR1 CCP 15 as Arg-933. NMR studies and molecular modeling (Fig. 7) show His-53 can replace Arg-53 in FH without loss of CCP 1 structural integrity. The R53H mutation, however, results in chemical shift perturbations among nearby residues, implying a possible effect on local structure, and this is consistent with the fact that the alkyl part of the Arg-53 side chain is largely buried. There are also chemical shift changes in CCP 2 relative to the wild type, presumably due to the close association of the Arg-53, or His-53, side chain with interface-critical residue Tyr-56 that makes hydrophobic contacts with Tyr-106 in CCP 2 (Fig. 7); thus, we cannot rule out a small mutation-induced rearrangement of the modules. This mutant is thermally less stable than wild type, and indeed there is some spectral evidence for a slight deterioration of the structure of the mutant at temperatures just above 37 °C. Taken together with the probable loss of charge resulting from replacement of positively charged arginine with histidine (at physiological pH) and, hence, the disturbance of an electrostatic surface that is common to C3b-engaging DAF and FH modules, these results are consistent with a detrimental effect of the Arg-53 to His mutation on the decay accelerating capabilities of FH. Thus, the presence of a mutation in a patient with aHUS is probably not coincidental even though most other mutations linked to this disease occur in FH CCPs 19 and 20 and disrupt other aspects of FH function. On the other hand, the single nucleotide polymorphism in FH corresponding to V62I seems unlikely to have any consequences for the ability of FH to regulate complement. The Ile-62 allotype of FH1-2 appears slightly more thermally stable and rigid than the Val-62 version, so it is conceivable that its potentially protective roles in AMD and DDD arise from a more robust nature of the full-length protein within a physiological setting, but this would require further investigation.
Supplementary Material
Acknowledgments
We thank Juraj Bella and Dr. Graeme Ball for help with data acquisition and collection.
The atomic coordinates and structure factors (codes 2RLP and 2RLQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Medical Research Council and Wellcome Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-5.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: FH, factor H; aHUS, atypical hemolytic uremic syndrome; AMD, age-related macular degeneration; AP, alternative pathway; C4BP, C4b-binding protein; CCP, complement control protein module; CR1, complement receptor type 1; DAF, decay accelerating factor; DDD, dense deposit disease; FB, factor B; FI, factor I; HSQC, heteronuclear single quantum coherence; MCP, membrane cofactor protein; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; RCA, regulator of complement activation; RDC, residual dipolar coupling; VCP, vaccinia virus complement control protein; r.m.s.d., root mean square deviation.
References
- 1.Ripoche, J., Day, A. J., Harris, T. J., and Sim, R. B. (1988) Biochem. J. 249 593-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrera-Abeleda, M. A., Nishimura, C., Smith, J. L., Sethi, S., McRae, J. L., Murphy, B. F., Silvestri, G., Skerka, C., Jozsi, M., Zipfel, P. F., Hageman, G. S., and Smith, R. J. (2006) J. Med. Genet. 43 582-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kavanagh, D., Goodship, T. H., and Richards, A. (2006) Br. Med. Bull. 77-78 5-22 [DOI] [PubMed] [Google Scholar]
- 4.Hageman, G. S., Anderson, D. H., Johnson, L. V., Hancox, L. S., Taiber, A. J., Hardisty, L. I., Hageman, J. L., Stockman, H. A., Borchardt, J. D., Gehrs, K. M., Smith, R. J., Silvestri, G., Russell, S. R., Klaver, C. C., Barbazetto, I., Chang, S., Yannuzzi, L. A., Barile, G. R., Merriam, J. C., Smith, R. T., Olsh, A. K., Bergeron, J., Zernant, J., Merriam, J. E., Gold, B., Dean, M., and Allikmets, R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7227-7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez de Cordoba, S., Diaz-Guillen, M. A., and Heine-Suner, D. (1999) Mol. Immunol. 36 803-808 [DOI] [PubMed] [Google Scholar]
- 6.McRae, J. L., Murphy, B. E., Eyre, H. J., Sutherland, G. R., Crawford, J., and Cowan, P. J. (2002) Genetica 114 157-161 [DOI] [PubMed] [Google Scholar]
- 7.Soares, D. C., Gerloff, D. L., Syme, N. R., Coulson, A. F., Parkinson, J., and Barlow, P. N. (2005) Protein Eng. Des. Sel. 18 379-388 [DOI] [PubMed] [Google Scholar]
- 8.Walport, M. J. (2001) N. Engl. J. Med. 344 1058-1066 [DOI] [PubMed] [Google Scholar]
- 9.Walport, M. J. (2001) N. Engl. J. Med. 344 1140-1144 [DOI] [PubMed] [Google Scholar]
- 10.Meri, S., and Pangburn, M. K. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3982-3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pangburn, M. K., Pangburn, K. L., Koistinen, V., Meri, S., and Sharma, A. K. (2000) J. Immunol. 164 4742-4751 [DOI] [PubMed] [Google Scholar]
- 12.Kristensen, T., and Tack, B. F. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 3963-3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soares, D. C., and Barlow, P. N. (2005) in Structural Biology of the Complement System (Morikis, D., and Lambris, J. D., eds) pp. 19-62, CRC Press, Taylor & Francis Group, Boca Raton. FL
- 14.Perkins, S. J., Gilbert, H. E., Aslam, M., Hannan, J., Holers, V. M., and Goodship, T. H. (2002) Biochem. Soc. Trans. 30 996-1001 [DOI] [PubMed] [Google Scholar]
- 15.Hellwage, J., Jokiranta, T. S., Friese, M. A., Wolk, T. U., Kampen, E., Zipfel, P. F., and Meri, S. (2002) J. Immunol. 169 6935-6944 [DOI] [PubMed] [Google Scholar]
- 16.Jokiranta, T. S., Hellwage, J., Koistinen, V., Zipfel, P. F., and Meri, S. (2000) J. Biol. Chem. 275 27657-27662 [DOI] [PubMed] [Google Scholar]
- 17.Sharma, A. K., and Pangburn, M. K. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10996-11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jokiranta, T. S., Zipfel, P. F., Hakulinen, J., Kuhn, S., Pangburn, M. K., Tamerius, J. D., and Meri, S. (1996) FEBS Lett. 393 297-302 [DOI] [PubMed] [Google Scholar]
- 19.Blackmore, T. K., Hellwage, J., Sadlon, T. A., Higgs, N., Zipfel, P. F., Ward, H. M., and Gordon, D. L. (1998) J. Immunol. 160 3342-3348 [PubMed] [Google Scholar]
- 20.Ferreira, V. P., Herbert, A. P., Hocking, H. G., Barlow, P. N., and Pangburn, M. K. (2006) J. Immunol. 177 6308-6316 [DOI] [PubMed] [Google Scholar]
- 21.Jokiranta, T. S., Cheng, Z.-Z., Seeberger, H., Jozsi, M., Heinen, S., Noris, M., Remuzzi, G., Ormsby, R., Gordon, D. L., Meri, S., Hellwage, J., and Zipfel, P. F. (2005) Am. J. Pathol. 167 1173-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon, D. L., Kaufman, R. M., Blackmore, T. K., Kwong, J., and Lublin, D. M. (1995) J. Immunol. 155 348-356 [PubMed] [Google Scholar]
- 23.Kuhn, S., Skerka, C., and Zipfel, P. F. (1995) J. Immunol. 155 5663-5670 [PubMed] [Google Scholar]
- 24.Kuhn, S., and Zipfel, P. F. (1996) Eur. J. Immunol. 26 2383-2387 [DOI] [PubMed] [Google Scholar]
- 25.Biro, A., Prohaszka, Z., Fust, G., and Blasko, B. (2006) Mol. Diagn. Ther. 10 303-310 [DOI] [PubMed] [Google Scholar]
- 26.Saunders, R. E., Abarrategui-Garrido, C., Fremeaux-Bacchi, V., Goicoechea de Jorge, E., Goodship, T. H., Lopez Trascasa, M., Noris, M., Ponce Castro, I. M., Remuzzi, G., Rodriguez de Cordoba, S., Sanchez-Corral, P., Skerka, C., Zipfel, P. F., and Perkins, S. J. (2007) Hum. Mutat. 28 222-234 [DOI] [PubMed] [Google Scholar]
- 27.Kirkitadze, M. D., and Barlow, P. N. (2001) Immunol. Rev. 180 146-161 [DOI] [PubMed] [Google Scholar]
- 28.Jenkins, H. T., Mark, L., Ball, G., Persson, J., Lindahl, G., Uhrin, D., Blom, A. M., and Barlow, P. N. (2006) J. Biol. Chem. 281 3690-3697 [DOI] [PubMed] [Google Scholar]
- 29.Casasnovas, J. M., Larvie, M., and Stehle, T. (1999) EMBO J. 18 2911-2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukacik, P., Roversi, P., White, J., Esser, D., Smith, G. P., Billington, J., Williams, P. A., Rudd, P. M., Wormald, M. R., Harvey, D. J., Crispin, M. D., Radcliffe, C. M., Dwek, R. A., Evans, D. J., Morgan, B. P., Smith, R. A., and Lea, S. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1279-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, B. O., Mallin, R. L., Krych-Goldberg, M., Wang, X., Hauhart, R. E., Bromek, K., Uhrin, D., Atkinson, J. P., and Barlow, P. N. (2002) 108 769-780 [DOI] [PubMed]
- 32.Murthy, K. H., Smith, S. A., Ganesh, V. K., Judge, K. W., Mullin, N., Barlow, P. N., Ogata, C. M., and Kotwal, G. J. (2001) Cell 104 301-311 [DOI] [PubMed] [Google Scholar]
- 33.Alam, M. N., Haque, A., Sreedhar, M., and Pangburn, M. K. (2004) J. Immunol. Methods 293 107-113 [DOI] [PubMed] [Google Scholar]
- 34.Herbert, A. P., Uhrin, D., Lyon, M., Pangburn, M. K., and Barlow, P. N. (2006) J. Biol. Chem. 281 16512-16520 [DOI] [PubMed] [Google Scholar]
- 35.Golovanov, A. P., Hautbergue, G. M., Wilson, S. A., and Lian, L. Y. (2004) J. Am. Chem. Soc. 126 8933-8939 [DOI] [PubMed] [Google Scholar]
- 36.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., III, and Skelton, N. J. (2006) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., Academic Press Inc., San Diego
- 37.Sklenar, V., Piotto, M., Leppik, R., and Saudek, V. (1993) J. Magn. Reson. 102 241-245 [Google Scholar]
- 38.Pascal, S. M., Muhandiram, D. R., Yamazaki, J. D., Forman-Kay, J. D., and Kay, L. E. (1994) J. Magn. Reson. 103 197-201 [Google Scholar]
- 39.Grzesiek, S., and Bax, A. (1993) J. Am. Chem. Soc. 115 12593-12594 [Google Scholar]
- 40.Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12366-12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ottiger, M., Delaglio, F., and Bax, A. (1998) J. Magn. Reson 131 373-378 [DOI] [PubMed] [Google Scholar]
- 42.Ball, G., Meenan, N., Bromek, K., Smith, B. O., Bella, J., and Uhrin, D. (2006) J. Magn. Reson. 180 127-136 [DOI] [PubMed] [Google Scholar]
- 43.Permi, P., Sorsa, T., Kilpelainen, I. I., and Annila, A. (1999) J. Magn. Reson. 141 44-51 [DOI] [PubMed] [Google Scholar]
- 44.Tjandra, N., Feller, S. E., Pastor, R. W., and Bax, A. (1995) J. Am. Chem. Soc. 117 12562-12566 [Google Scholar]
- 45.Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, M., Ulrich, E. L., Markley, J. L., Ionides, J., and Laue, E. D. (2005) Proteins 59 687-696 [DOI] [PubMed] [Google Scholar]
- 46.Schubert, M., Labudde, D., Oschkinat, H., and Schmieder, P. (2002) J. Biomol. NMR 24 149-154 [DOI] [PubMed] [Google Scholar]
- 47.Herrmann, T., Guntert, P., and Wuthrich, K. (2002) J. Mol. Biol. 319 209-227 [DOI] [PubMed] [Google Scholar]
- 48.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta. Crystallogr. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 49.Sass, H. J., Musco, G., Stahl, S. J., Wingfield, P. T., and Grzesiek, S. (2001) J. Biomol. NMR 21 275-280 [DOI] [PubMed] [Google Scholar]
- 50.Vriend, G. (1990) J. Mol. Graph. 8 52-56 [DOI] [PubMed] [Google Scholar]
- 51.Morris, A. L., MacArthur, M. W., Hutchinson, E. G., and Thornton, J. M. (1992) Proteins 12 345-364 [DOI] [PubMed] [Google Scholar]
- 52.Bower, M. J., Cohen, F. E., and Dunbrack, R. L., Jr. (1997) J. Mol. Biol. 267 1268-1282 [DOI] [PubMed] [Google Scholar]
- 53.Canutescu, A. A., Shelenkov, A. A., and Dunbrack, R. L., Jr. (2003) Protein Sci. 12 2001-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tina, K. G., Bhadra, R., and Srinivasan, N. (2007) Nucleic Acids Res. 35 473-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiser, A., and Sali, A. (2003) Methods Enzymol. 374 461-491 [DOI] [PubMed] [Google Scholar]
- 56.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
- 57.Fraczkiewicz, R., and Braun, W. (1998) J. Comput. Chem. 19 319-333 [Google Scholar]
- 58.Barlow, P. N., Steinkasserer, A., Norman, D. G., Kieffer, B., Wiles, A. P., Sim, R. B., and Campbell, I. D. (1993) J. Mol. Biol. 232 268-284 [DOI] [PubMed] [Google Scholar]
- 59.DeLano, W. L. (2002) The PyMOL Molecular Graphics System 0.99, DeLano Scientific, LLC, Palo Alto, CA
- 60.Jackson, S. E., Moracci, M., elMasry, N., Johnson, C. M., and Fersht, A. R. (1993) Biochemistry 32 11259-11269 [DOI] [PubMed] [Google Scholar]
- 61.Zuiderweg, E. R. (2002) Biochemistry 41 1-7 [DOI] [PubMed] [Google Scholar]
- 62.Krych-Goldberg, M., and Atkinson, J. P. (2001) Immunol. Rev. 180 112-122 [DOI] [PubMed] [Google Scholar]
- 63.Blom, A. M., Kask, L., and Dahlback, B. (2001) J. Biol. Chem. 276 27136-27144 [DOI] [PubMed] [Google Scholar]
- 64.Liszewski, M. K., Leung, M., Cui, W., Subramanian, V. B., Parkinson, J., Barlow, P. N., Manchester, M., and Atkinson, J. P. (2000) J. Biol. Chem. 275 37692-37701 [DOI] [PubMed] [Google Scholar]
- 65.Mullick, J., Bernet, J., Panse, Y., Hallihosur, S., Singh, A. K., and Sahu, A. (2005) J. Virol. 79 12382-12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brodbeck, W. G., Liu, D., Sperry, J., Mold, C., and Medof, M. E. (1996) J. Immunol. 156 2528-2533 [PubMed] [Google Scholar]
- 67.Milder, F. J., Gomes, L., Schouten, A., Janssen, B. J., Huizinga, E. G., Romijn, R. A., Hemrika, W., Roos, A., Daha, M. R., and Gros, P. (2007) Nat. Struct. Mol. Biol. 14 224-228 [DOI] [PubMed] [Google Scholar]
- 68.Perkins, S. J., Okemefuna, A. I., Fernando, A. N., Bonner, A., Gilbert, H. E., and Furtado, P. B. (2008) Methods Cell Biol. 84 375-423 [DOI] [PubMed] [Google Scholar]
- 69.Nishida, N., Walz, T., and Springer, T. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19737-19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janssen, B. J., Huizinga, E. G., Raaijmakers, H. C., Roos, A., Daha, M. R., Nilsson-Ekdahl, K., Nilsson, B., and Gros, P. (2005) Nature 437 505-511 [DOI] [PubMed] [Google Scholar]
- 71.Janssen, B. J. C., Christodoulidou, A., McCarthy, A., Lambris, J. D., and Gros, P. (2006) Nature 444 213-216 [DOI] [PubMed] [Google Scholar]
- 72.Wiesmann, C., Katschke, K. J., Yin, J., Helmy, K. Y., Steffek, M., Fairbrother, W. J., McCallum, S. A., Embuscado, L., DeForge, L., Hass, P. E., and van Lookeren Campagne, M. (2006) Nature 444 217-220 [DOI] [PubMed] [Google Scholar]
- 73.Shindyalov, I. N., and Bourne, P. E. (1998) Protein Eng. 11 739-747 [DOI] [PubMed] [Google Scholar]
- 74.Kuttner-Kondo, L., Hourcade, D. E., Anderson, V. E., Muqim, N., Mitchell, L., Soares, D. C., Barlow, P. N., and Medof, M. E. (2007) J. Biol. Chem. 282 18552-18562 [DOI] [PubMed] [Google Scholar]
- 75.Harris, C. L., Pettigrew, D. M., Lea, S. M., and Morgan, B. P. (2007) J. Immunol. 178 352-359 [DOI] [PubMed] [Google Scholar]
- 76.Kuttner-Kondo, L. A., Mitchell, L., Hourcade, D. E., and Medof, M. E. (2001) J. Immunol. 167 2164-2171 [DOI] [PubMed] [Google Scholar]
- 77.Frishman, D., and Argos, P. (1995) Proteins 23 566-579 [DOI] [PubMed] [Google Scholar]
- 78.Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10037-10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sitkoff, D., Ben-Tal, N., and Honig, B. (1996) J. Phys. Chem. 100 2744-2752 [Google Scholar]
- 80.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 25 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000) Nucleic Acids Res. 28 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.