Abstract
Proteins with a weak sequence similarity to tubulin and FtsZ are expressed from large plasmids of Bacillus anthracis and Bacillus thuringiensis and are probably involved in plasmid segregation. Previously designated RepX and TubZ, we designate them here as TubZ-Ba and TubZ-Bt. We have expressed and purified the proteins for in vitro studies. TubZ-Ba and TubZ-Bt share only 21% amino acid identity, but they have remarkably similar biochemical properties. They both assemble into two-stranded filaments and larger bundles above a critical concentration, and they hydrolyze GTP at a very high rate, ∼20 GTP min–1 TubZ–1. Assembly is also supported by GTPγS. A tiny amount of GTPγS stabilizes polymers assembled in GTP and inhibits the GTPase by a mechanism involving cooperativity. The nucleotide in the polymers is almost 100% GDP, which is similar to microtubules but very different from the 20–30% GDP in FtsZ polymers. This suggests that the TubZ polymers have a capping mechanism that may be related to the GTP cap that produces dynamic instability of microtubules.
FtsZ and tubulin have the same complex folding patterns and are therefore considered homologs, i.e. they are derived from a common ancestor. FtsZs share 40–50% sequence identity across most bacterial and archaeal species, and tubulins are even more highly conserved across eukaryotic species. However, the sequence identity between FtsZ and tubulin is so low, 10–15% as to be hardly recognizable. Erickson (1) has suggested that the reason for this extreme divergence is that FtsZ is performing the same function, cytokinesis, in all prokaryotic species, but tubulin has lost this function and acquired new ones.
Several Bacillus species have large, low-copy plasmids that code for toxins. These plasmids also code for a protein that is distantly related to FtsZ. Although originally termed “FtsZ-like” (2), further analysis showed a similar 15–20% sequence identity to either tubulin or FtsZ, and the proteins have some conserved regions that align with FtsZ, and others that align with tubulin (3). Remarkably, the FtsZ-like proteins produced by the plasmids of different Bacillus species are almost as divergent from each other as they are from FtsZ and tubulin. The ones that have been studied so far are from the pXO1 plasmid of Bacillus anthracis and the pBtoxis plasmid of Bacillus thuringiensis. These show only 21% amino acid identity to each other (4).
The FtsZ/tubulin-like protein from pXO1 was shown to be essential for maintaining the plasmid in B. anthracis (5). That study went on to show that this was the only plasmid-encoded protein that was necessary for plasmid stability. A minireplicon containing only a short DNA sequence from pXO1 (presumably a centromere-like segment) could be maintained provided the FtsZ/tubulin-like protein was produced, either from the same plasmid or in trans. Tinsley and Khan (5) suggested that this protein might be involved in replicating the plasmid DNA and named the protein RepX. An alternative is that RepX is involved in partitioning the low-copy plasmids to daughter cells, as originally suggested by Berry et al. (4). We now believe this to be the case, especially in light of the more detailed analyses of TubZ from pBtoxis.
The FtsZ/tubulin-like protein from pBtoxis, ORF156, was identified as one of two plasmid-encoded proteins needed for plasmid maintenance in B. thuringiensis (6). Similar to the study of pXO1, a minireplicon expressing these two proteins and containing a short DNA sequence with an iteron repeat (a centromere-like segment) was stably maintained. Tang et al. (6) suggested that the protein was functioning for plasmid partitioning. Larsen et al. (3) were interested in this protein as a distant relative of tubulin and FtsZ, and named it TubZ. They constructed a GFP-tagged version that they expressed in B. thuringiensis and in Escherichia coli. In both species the TubZ assembled into a single filament that spanned most of the length of the cell. The filaments were remarkably uniform in fluorescence, both along the length of the filament and between filaments in different cells. The filaments demonstrated a treadmilling behavior, growing at one end and shrinking at the other, at a rate of about 30 nm per s. Margolin (7) has reviewed the discoveries and mechanisms of these two proteins.
The likely function of TubZ from B. thuringiensis is plasmid partitioning (3, 4, 6). We believe this also applies to RepX. To avoid the confusion of different names for the FtsZ/tubulin-like proteins from different species, we propose a simple and uniform nomenclature. We will use the name TubZ, which was meant to designate the similarity to tubulin and FtsZ (3), and we will add a species identification. Thus, we will designate the proteins from B. anthracis and B. thuringiensis TubZ-Ba and TubZ-Bt. This nomenclature can be easily extended to the larger group of FtsZ/tubulin-like proteins on other plasmids and Archaea (3). An advantage of this nomenclature is that it implies nothing about function.
Assembly of TubZ-Bt has been characterized in the cytoplasm of B. thuringiensis and E. coli as discussed above (3). Assembly has not been studied in vitro. Assembly of TubZ-Ba has not been studied either in vitro or in vivo. Their extreme sequence divergence from each other suggests that their functions could be as different as those of FtsZ and tubulin. To explore these proteins further, we have expressed and purified both of them and characterized their assembly properties in vitro.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—We used PCR to clone the TubZ-Ba sequence from plasmid pDSW208-FtsZPXO1-GFP, a gift from Dr. Theresa Koehler, University of Texas Medical Center, which was generated from B. anthracis strain 7702. BamHI and EcoRI restriction sites were added at the ends and used to insert TubZ-Ba into the pGEX2T expression vector, which adds an N-terminal GST tag. The TubZ-Ba-pGEX2T vector was then transferred into E. coli strain BL21.
Protein was expressed by adding 0.5 mm isopropyl-1-thio-β-d-galactopyranoside to the culture when its absorbance at 600 nm was ∼1.0. After 3 h, cells were centrifuged and resuspended in 50 mm Tris-HCl, pH 7.9, 300 mm KCl. 1 mm phenylmethylsulfonyl fluoride and 0.2 mg/ml lysozyme were added, and cells were incubated for 30 min at 4 °C. Cells were lysed with a French pressure cell and centrifuged at 32,000 rpm for 20 min. The supernatant was then mixed with 5 ml of glutathione-agarose (Sigma, G4510) for 1 h at 4 °C. The agarose was loaded onto a column and washed with 50 mm Tris, pH 7.9, 300 mm KCl. GST-TubZ-Ba protein was eluted with 10 mm reduced glutathione, in the same buffer. The GST tag was removed by adding 2 units/ml of thrombin for 2 h at 4 °C. The protein was further purified by chromatography on a Source Q 10/10 column (GE Health Care, Piscataway, NJ), eluted with a linear gradient of 50 mm to 500 mm KCl in 50 mm Tris-HCl, pH 7.9, 1 mm EDTA, 10% glycerol. Peak fractions were identified by SDS-PAGE and stored at –80 °C. For most experimental measurements, the protein was dialyzed into assembly buffer, sometimes referred to as HMK100: 50 mm Hepes, pH 7.7, 100 mm KAc, 5 mm MgAc, 1 mm EGTA.
The TubZ-Bt gene was obtained from MosquitoDunks® (Summit Chemical Co.), a commercial sample of B. thuringiensis subsp Isrealiensis. Bacteria were propagated and genomic DNA, including the DNA of the plasmid pBtoxis, was isolated. The TubZ-Bt gene was obtained by PCR and transferred to pET15. The His tag protein was expressed and purified by affinity chromatography on a Talon column (Clontech Lab, Inc.). The His tag was removed with 2 units/ml of thrombin, and the protein further purified on a Source Q column.
GTPase Assay—To measure the GTPase activity, we used the continuous, regenerative coupled GTPase assay of Ingerman et al. (8). Our assay mixture included 0.4 mm phosphoenolpyruvate, 0.3 mm NADH, 20 units/ml each pyruvate kinase and lactate dehydrogenase (Sigma), and 0.1–0.5 mm GTP. Each GDP released from TubZ-Ba is regenerated to GTP with the loss of one molecule of NADH. The NADH concentration was monitored by its absorbance at 340 nm (extinction coefficient 6220 m–1 cm–1) using a Shimadzu UV-2401PC spectrophotometer. Following addition of GTP, the absorbance showed a linear decrease over time. We measured the slope of the straight line after 100 s, typically between 100 and 300 s, and used the extinction coefficient of NADH to determine the GTP hydrolysis rate. We then plotted the rate versus TubZ concentration, and the slope of this line (above the critical concentration) gave the overall rate of GTP hydrolysis in GTP per min per TubZ.
Light Scattering Measurements—90 degree light scattering was measured using a Shimadzu RF-5301 PC spectrofluorometer, with both excitation and emission set at 350 nm. Assembly reactions were initiated by adding 20–500 μm GTP, and/or the indicated concentrations of GTP analogue, into 3–10 μm TubZ. Light scattering measurements were begun about 2–3 s after nucleotide addition.
Electron Microscopy—TubZ filaments were visualized by negative stain EM. About 10 μl of the sample (5–10 μm) in the appropriate buffer was incubated with GTP for 1–2 min and applied to a carbon-coated copper grid. Samples were negatively stained with 2% uranyl acetate, and photographed using a Philips 301 electron microscope at ×50,000 magnification.
Measurement of Filament-bound Guanine Nucleotide—We used the method of Romberg and Mitchison (9) to determine the content of nucleotide bound to TubZ polymers. TubZ at 0, 5, 10, 15, and 20 μm was added to 10 μl of a GTP-regenerating system (2 mm phosphoenolpyruvate and 40 units/ml pyruvate kinase). 20 μm GTP and 0.8 μCi of [α-32P]GTP were added, and after 100 s, the samples were denatured with 30 μl of 1 m perchloric acid and 10 mm EDTA. The mixture was neutralized with 20 μl of 1 m Na2CO3, and the samples were centrifuged at 10,000 rpm for 5 min to remove the precipitate.
The nucleotides in the sample were analyzed on poly(ethylenimine)-cellulose thin-layer chromatography plates. Plates were pre-run in water and dried, and a 5-μl sample was spotted. The plates were run in 1 m LiCl. The dried plates were exposed to a Fujix bas 1000 phosphorimager plate, which was read in a Typhoon 9400 Variable Mode imager and analyzed by Image-Quant software. The amount of GDP as a fraction of total nucleotide was determined. Because all free GDP is immediately converted to GTP by the regenerating system, the measured GDP must come from nucleotide bound to filaments. We repeated the experiment with FtsZ from E. coli (EcFtsZ) as a control.
RESULTS
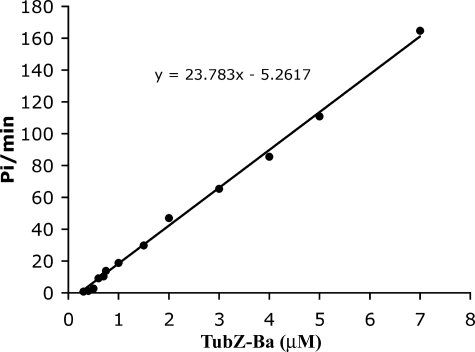
TubZ-Ba Has a Very High GTPase Activity and Assembles into Double Helical Filaments—Fig. 1 shows the GTP hydrolysis rate for increasing concentrations of TubZ-Ba. The GTPase was linearly proportional to the TubZ-Ba concentration above a critical concentration of 0.2–0.4 μm (depending on the buffer), giving 24 GTP TubZ-Ba–1 min–1 (Fig. 1). The rate did not vary significantly with pH, being 21∼25 GTP TubZ-Ba–1 min–1 at room temperature in most buffers (pH 6.5, 7.0, and 7.7 with 100 mm KAc and 5 mm MgAc). We have reported that the GTPase activity of E. coli FtsZ (EcFtsZ)2 is around 4–7 GTP FtsZ–1 min–1 at room temperature (10). Thus the GTPase of TubZ-Ba is 3–4-fold higher than that of EcFtsZ.
FIGURE 1.
GTP hydrolysis at increasing concentrations of TubZ-Ba. This experiment was in HMK100 buffer at pH 7.7, and gave a hydrolysis rate of 24 GTP TubZ-Ba–1 min–1.
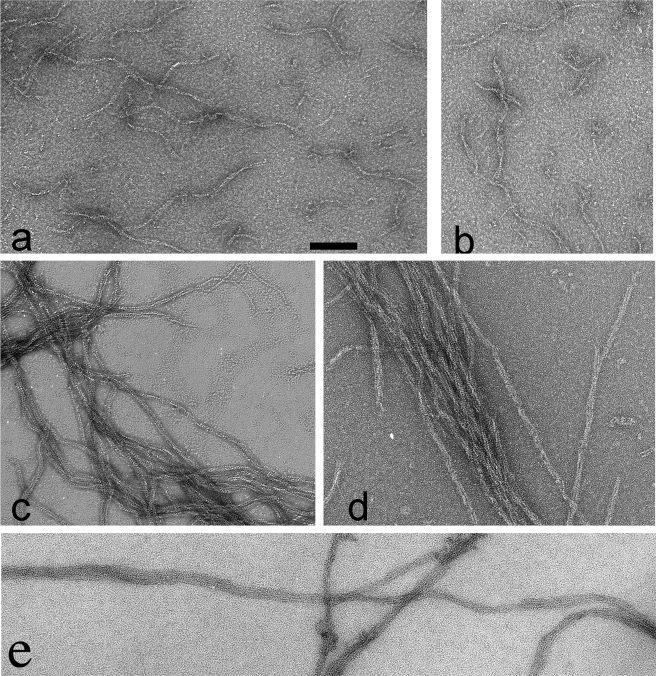
Negative stain EM showed that TubZ-Ba assembled into short, twisted two-stranded filaments and some larger bundles (Fig. 2). The filaments alternate between wider and straight segments that show the two strands and narrow edge views that show a gentle curvature. The flattened structure in the negative stain image suggests a three-dimensional structure in which the two-stranded ribbon forms a helix, with a diameter of ∼7 nm and pitch of 75 nm. Occasionally the ribbon showed a third strand. The assembly required GTP or a GTP analogue.
FIGURE 2.
TubZ-Ba filament structure observed by negative stain EM. a and b show TubZ-Ba filaments assembled in 100 μm GTP. The filaments mostly appear as wavy two-stranded helices, but occasional polymers have three strands (bottom 2b). c, shows TubZ-Ba filaments assembled in 20 μm GTPγS. Filaments are longer and tend to associate into bundles. In the upper right, a two-stranded filament is seen separating into single strands, d shows TubZ-Ba filaments assembled in 100 μm GTP plus 1 μm GTPγS. e shows TubZ-Ba assembled in GTP in the presence of 10% polyvinyl alcohol, a crowding agent. The TubZ-Ba concentration was 5 μm in all measurements. The scale bar is 100 nm.
We then tested TubZ-Ba in crowding solution conditions that should mimic the conditions of the bacterial cytoplasm (11–13). When assembled in buffer containing 10% polyvinyl alcohol, TubZ-Ba retained its very high GTPase activity, about 18 GTP TubZ-Ba–1 min–1. Assembly, however, was substantially changed. Instead of the short, twisted two-stranded filaments, in polyvinyl alcohol TubZ-Ba assembled very long bundles of ∼2–10 protofilaments (Fig. 2e).
GTPγS Can Support TubZ-Ba Filament Assembly, and a Tiny Amount Can Stabilize Assembly with GTP—GTPγSisa GTP analogue that can substitute for GTP in many GTP-binding proteins and is either slowly hydrolyzed or not hydrolyzed at all. When GTPγS was added to TubZ-Ba, it assembled into filaments similar to those assembled in GTP. However, the GTPγS filaments were much longer and tended to cluster into somewhat larger irregular bundles (Fig. 2c). We note that in rare cases the two-stranded filaments unwound and left lengths of single filaments that were apparently stable without the lateral contacts. Polymers assembled with 100 μm GTP plus 1 μm GTPγS appeared similar to those assembled with 20 μm GTPγS (Fig. 2d).
A light scattering assay showed that in 100 μm GTP, polymers rapidly assembled and immediately began to disassemble (Fig. 3a). The 5 μm TubZ-Ba would have consumed 50% of the 100 μm GTP in 25 s. The disassembly began even before this, so it is probably induced by the accumulation of GDP. Light scattering was back to the baseline after 80 s. When assembly was initiated with 1.5 μm GTPγS, the light scattering rose somewhat more slowly and remained at the plateau. With 1.5 μm GTPγS, we expect only 1.5 μm TubZ-Ba to assemble. When 100 μm GTP was added to this assembly at 400 s, the light scattering increased further and then decreased back to the original plateau. We believe that the initial assembly of TubZ-Ba in GTPγS formed filaments that were very stable. When GTP was added later, the remaining TubZ subunits assembled into filaments that were either separate from or attached to the end of the GTPγS filaments. These new filaments rapidly hydrolyzed the GTP and disassembled, leaving the stable GTPγS filaments. The GTPγS subunits, once assembled into their original polymers, apparently did not mix with the GTP subunits.
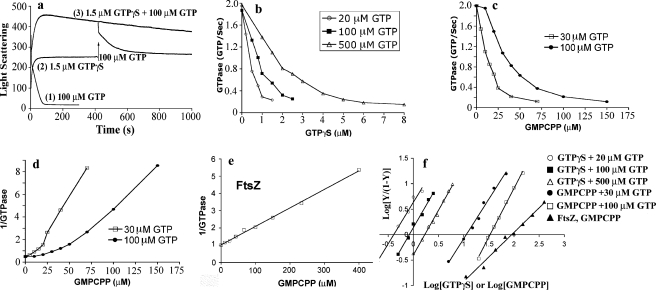
FIGURE 3.
Light scattering and GTPase activity of TubZ-Ba measured in the presence of non-hydrolyzable GTP analogues, GTPγS or GMPCPP. a, assembly measured by light scattering of 5 μm TubZ-Ba in 100 μm GTP; in 1.5 μm GTPγS, with 100 μm GTP added at 400 s; and in 1.5 μm GTPγS plus 100 μm GTP added together at time 0. b, inhibition of GTPase activity of TubZ-Ba by GTPγS. Data are shown for 5 μm TubZ-Ba and increasing GTPγS, plus 20, 100, and 500 μm GTP. c, inhibition of GTPase activity by GMPCPP. d, plot of 1/GTPase activity versus GMPCPP concentration, showing that the relationship is not linear. The curvature indicates that the inhibition is cooperative. e, plot of 1/GTPase activity versus GMPCPP concentration for 5 μm FtsZ and 100 μm GTP. In contrast to TubZ, this plot is linear. f, Hill plot of the GTPase inhibition by GTPγS and GMPCPP. The five lines on the left are for TubZ-Ba, with Hill coefficients of 1.7–1.9, and the line on the right is for FtsZ, with a Hill coefficient of 1.
We then tested assembly initiated by a mixture of 1.5 μm GTPγS plus 100 μm GTP. The light scattering rose to a value twice that with 1.5 μm GTPγS alone, and when it reached the plateau it decreased only very slowly. EM showed that the filaments assembled in 1.0 μm GTPγS plus 100 μm GTP were very similar to those assembled in 20 μm GTPγS (Fig. 2d).
To investigate this further, we assayed the rate of GTP hydrolysis for 5 μm TubZ-Ba, with 20, 100, and 500 μm GTP, and variable amounts of GTPγS. For this experiment we assayed the steady state GTP hydrolysis rate as the slope of the absorbance curve as soon as it became linear, typically from 100 to 300 s following initiation of assembly. As shown in Fig. 3b, the GTPase was completely inhibited at 1, 2, and 5 μm GTPγS for the three concentrations of GTP.
GMPCPP is a slowly hydrolyzable GTP analog that supports assembly of tubulin and FtsZ. We found that it induced assembly of TubZ-Ba into bundles that were similar to those assembled in GTPγS. GMPCPP also inhibited the GTPase activity of TubZ-Ba, but only at much higher concentrations. GMPCPP needed to be approximately equimolar to GTP to completely inhibit the GTPase of 5 μm TubZ-Ba (Fig. 3c). These results suggest that GMPCPP binds TubZ-Ba with an affinity similar to that of GTP, while GTPγS binds with much higher affinity.
One possible mechanism for the inhibition of GTPase would be simple competitive binding to block GTP binding. In this case, a plot of 1/GTPase versus GTPγS or GMPCPP concentration should be linear. A linear plot was obtained for the case of FtsZ and GMPCPP (Fig. 3e), but both analogs gave non-linear curves for TubZ-Ba (Fig. 3d and data not shown). The non-linearity was suggestive of cooperativity, so we re-plotted the data in the form of a Hill plot. We assumed the equation log[Y/(1 – Y)] = nlog[M] + k, where Y is the fractional inhibition of the GTPase activity and M is the concentration of GTPγSor GMPCPP. As shown in Fig. 3f, both GTPγS and GMPCPP gave linear Hill plots for TubZ-Ba. For GTPγS, the Hill coefficient was about 1.7 for the three GTP concentrations, and for GMPCPP it was 1.6 and 1.9 for 30 and 100 μm GTP. These values are all close to 2, so we take this as the measure of the cooperativity for the analogs inhibiting the GTPase. For a comparison we tested the inhibition of the GTPase of FtsZ by GMPCPP. The plot of 1/GTPase versus GMPCPP was linear (Fig. 3e), and the Hill coefficient was 1 (Fig. 3f).
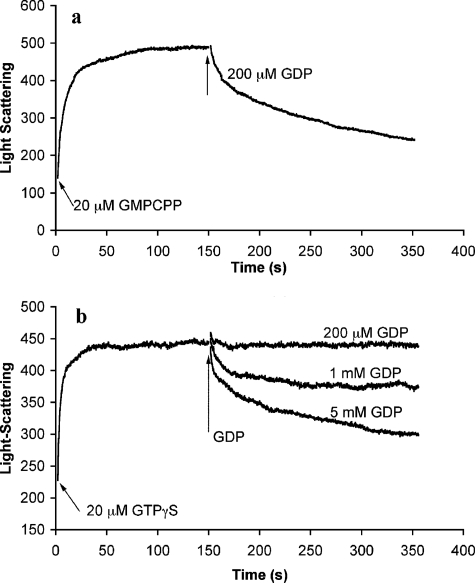
To explore the differences between GMPCPP and GTPγS, we tested how excess GDP would affect polymers of TubZ-Ba assembled with the GTP analogs. We had already observed that polymers assembled in GTP rapidly disassembled as the GTP was depleted and GDP accumulated (Fig. 3a). Polymers assembled in 20 μm GMPCPP showed a complex disassembly when a 10-fold excess of GDP was added. An initial rapid phase of disassembly was followed by a slow phase (Fig. 4a). However, for TubZ-Ba assembled in 20 μm GTPγS, 200 μm GDP had almost no effect. Significant filament disassembly was only obtained with 1–5 mm GDP (Fig. 4b). The disassembly induced by 5 mm GDP for the GTPγS assembly was similar to that produced by 0.2 mm GDP for GMPCPP assembly. This is consistent with the suggestion that TubZ-Ba protein has a much higher binding affinity for GTPγS than for GMPCPP, GTP, and GDP.
FIGURE 4.
Excess GDP can depolymerize TubZ-Ba assembled in GMPCPP (a) or GTPγS (b).
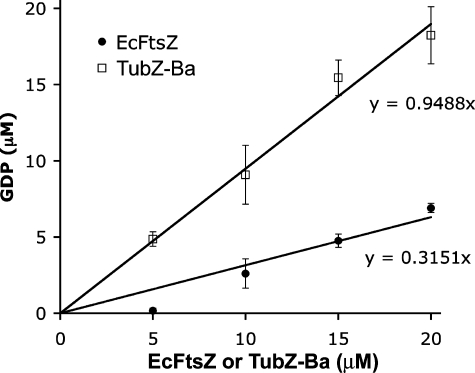
The Nucleotide in TubZ-Ba Filaments Is Almost Exclusively GDP—Romberg and Mitchison (9) confirmed that microtubules contained almost 100% GDP, but the nucleotide content of FtsZ filaments was only 20% GDP (9). We used their assay to determine the nucleotide content of TubZ-Ba filaments. TubZ-Ba was incubated with 20 μm GTP and 0.8 μCi of [α-32P]GTP in the presence of a GTP-regenerating system (see details under “Experimental Procedures”). The GTP-regenerating system rapidly converts all free GDP to GTP, so the only GDP that exists in the assembly mixture is that which is protected from regeneration by being bound to polymers. Fig. 5 shows the GDP in increasing concentrations of TubZ-Ba protein, with a comparable assay of FtsZ as a control. For FtsZ about 30% of the nucleotide was GDP, similar to the 20% value determined previously by Romberg and Mitchison (9). (The previous measure was at pH 6.5, 50 mm potassium, and ours was at pH 7.7, 100 mm potassium.) In contrast, in the TubZ-Ba polymers 95% of the nucleotide was GDP. In this respect TubZ-Ba polymers are similar to microtubules.
FIGURE 5.
The amount of GDP in polymer was plotted as a function of total protein for TubZ-Ba and E. coli FtsZ. 95% of the nucleotide in TubZ-Ba was GDP, while only 30% was GDP in FtsZ.
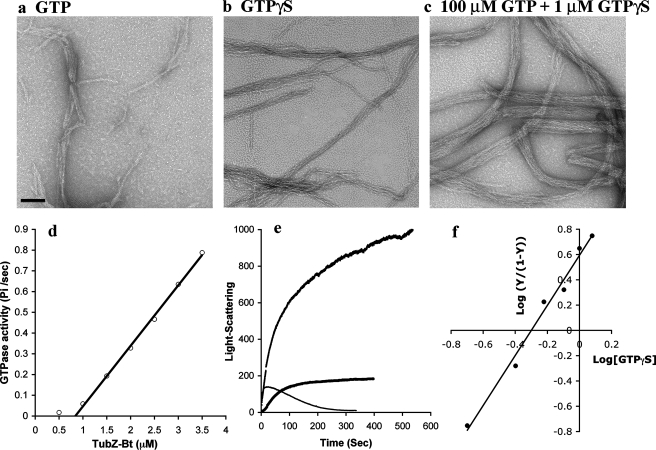
Assembly and GTPase of TubZ-Bt—We did a similar analysis of TubZ-Bt, the tubulin-like protein from the plasmid pBtoxis of B. thuringiensis (3, 6, 14). Negative stain EM showed that assembly in 100 μm GTP produced filaments similar to those of TubZ-Ba, although the two-stranded structure was less obvious (Fig. 6a). Assembly in 1 μm GTPγS gave filaments that were much longer and somewhat more bundled, and showed more clearly the two-stranded structure (Fig. 6b). Assembly in 100 μm GTP plus 1 μm GTPγS gave much larger bundles (Fig. 6c). In crowding condition (10% polyvinyl alcohol), TubZ-Bt assembled into long bundles similar to TubZ-Ba (not shown).
FIGURE 6.
Biochemical properties of TubZ-Bt. a–c, images of negatively stained polymers assembled in 1 mm GTP (a), 1 μm GTPγS(b), and a mixture of 100 μm GTP and 1 μm GTPγS(c). The bar is 100 nm. d, GTPase activity of TubZ-Bt showed a critical concentration of 0.8 μm, and above this a linear activity of 18 GTP TubZ-Bt–1 min–1. e, assembly of 3 μm TubZ-Bt with different combinations of nucleotides. In 0.1 mm GTP, rapid assembly was followed by disassembly. In 1 μm GTPγS assembly occurred more slowly and after 100 s remained at the plateau. In 100 μm GTP plus 1 μm GTPγS assembly rose much higher, and then continued a slower rise after 100 s. This second phase is probably due to bundling seen in c. f, Hill plot for inhibition of GTPase by GTPγS gives a Hill coefficient of 1.9.
The biochemical properties of TubZ-Bt were remarkably similar to those of TubZ-Ba. TubZ-Bt had a very high GTPase activity, 16∼18 GTP TubZ-Bt–1 min–1, and a critical concentration of 0.8 μm (Fig. 6d). The nucleotide in TubZ-Bt filaments was almost exclusively GDP, similar to TubZ-Ba and tubulin. Light scattering showed a very rapid assembly in 100 μm GTP, followed by disassembly as the GTP was consumed (Fig. 6e). In GTPγS the assembly occurred more slowly, but when the plateau was reached at ∼80 s, it was stable. In the mixture of 100 μm GTP plus 1 μm GTPγS assembly occurred in a first phase at an intermediate rate to 40 s, followed by a steady increase in scattering. The second phase is probably due to the increased bundling seen by EM (Fig. 6c). Finally, we found that the GTPase was strongly inhibited by very small amounts of GTPγS. In 100 μm GTP, the GTPase of 3 μm TubZ-Bt was reduced by 80% by 1 μm GTPγS (not shown). The plot of 1/GTPase versus GTPγS was non-linear (not shown). The Hill plot was linear with a Hill coefficient of 1.9 (Fig. 6f).
DISCUSSION
Assembly of TubZ-Ba and TubZ-Bt both showed two aspects of cooperativity, which may not be related to each other. The first aspect of cooperativity is the characteristic critical concentration (Cc) at steady state. Below Cc there is no polymer, and above Cc all excess protein assembles into polymer. Actin and microtubules are among the best characterized cooperative assemblies, and they show a classic Cc. FtsZ also shows a Cc, although the basis for cooperativity is still obscure for this one-subunit thick filament (15).
The second aspect of cooperativity is the Hill coefficient of ∼2 found for the inhibition of GTP hydrolysis by GTPγS and GMPCPP. This is an unusual application of the Hill equation but we think it is informative. The normal application of the Hill equation is to describe binding of a ligand to multiple sites on an enzyme or other substrate. In our application we used it to describe the inhibition of GTPase by GTPγS, in particular how the inhibition depends on the concentration of GTPγS. Steady state GTP hydrolysis probably involves several steps. By analogy with tubulin, and probably also FtsZ, exchange of nucleotide probably occurs only on free subunits. Following a hydrolysis event the subunit must disassemble and exchange its GDP for GTP. The subunit can then reassemble and undergo another round of hydrolysis. We suggest that the inhibition of steady state GTP hydrolysis by GTPγS may involve a block in the disassembly stage. The Hill coefficient of 2 then suggests that to block disassembly there needs to be two adjacent subunits with GTPγS. This may in turn be related to the two-stranded structure of the filaments, which is especially obvious for TubZ-Ba. Note that for FtsZ, which assembles primarily one-stranded protofilaments, the Hill coefficient for GMPCPP inhibition was 1.
A Possible Capping Mechanism—The TubZ-Ba and TubZ-Bt polymers share several features with microtubules that may indicate a capping mechanism related to dynamic instability. Most important, nearly 100% of the nucleotide in the polymer is GDP for both microtubules and TubZ. Because the GDP polymer is unstable and disassembles, a mechanism is needed to stabilize the polymers and promote growth. This has been explored in detail for microtubules, where a small GTP cap at each end is thought to stabilize the GDP tubulin in the core (16, 17). A similar capping mechanism for TubZ is suggested by the fact that the filaments are maintained in solution, despite being nearly 100% TubZ-GDP.
Capping is also suggested by the substoichiometric stabilization of polymers by GTPγS. The GTPase of 5 μm FtsZ in 100 μm GTP was inhibited 50 and 100% by 1 and 2 μm GTPγS (Fig. 3b), this correlated with the almost complete stabilization of polymers at 1.5 μm GTPγS (Fig. 3a). Assuming that GTPγS binds much more tightly than GTP, the polymers from this mix would contain 1.5 μm GTPγS and 3.5 μm GTP (a more accurate analysis would require knowledge of the KD for binding each nucleotide). That the 3.5 μm GTP subunits are mostly stabilized by the minority GTPγS subunits implies a cooperative mechanism for filament stabilization, or capping. The Hill coefficient of ∼2 further suggests that the cap might consist of two GTP or GTPγS subunits at the end of the two-stranded filament. This is at present a preliminary speculation, but further study may shed light on mechanisms common to TubZ and tubulin.
GTPγS does not support assembly of FtsZ by itself. However, when protofilament bundles were assembled with GTP in the presence of Ca, GTPγS could stabilize these polymers (18). A capping mechanism was suggested to explain this stabilization (18). This mechanism appears to be quite different from what we observe for TubZ, where GTPγS can generate assembly by itself, and is incorporated into the polymers.
A capping mechanism generates dynamic instability in microtubules, but TubZ-Bt behaves very differently from microtubules in vivo. Microtubules show dynamic instability at both ends, elongating for several μm, and then shortening by several μm. TubZ-Bt showed a very different treadmilling behavior when assembled in bacteria (3). The filament grew continually at one end and shrank at the other. It is possible that the long filament of TubZ-Bt seen in bacteria has a GTP cap at the growing end, and GDP on all other subunits, including the shrinking minus end. Continuous growth at the GTP end and disassembly at the GDP end would generate treadmilling. An alternative possibility is that there is a GTP cap at each end, at one end the growth phase greatly exceeds shrinking, and at the other end the shrinking phase exceeds growth. This kind of differential dynamic instability can produce treadmilling in microtubules (17). In this case, GTPγS would stabilize the polymers by placing permanent caps at both ends.
Divergent Sequences but Identical Biochemistry—The weak sequence similarity of TubZ-Bt to FtsZ suggested a possible role in plasmid partitioning (3, 4, 6, 14). Although TubZ-Ba was originally suggested to function in DNA replication (hence the name RepX), we believe that it functions like TubZ-Bt in plasmid partitioning. However, TubZ-Ba showed only 21% sequence identity to TubZ-Bt, suggesting that the two proteins may have functions as different as those of FtsZ and tubulin (1).
Our in vitro analyses show, however, that TubZ-Ba and TubZ-Bt have remarkably similar biochemical properties. How can one reconcile the extremely divergent sequences with the apparently identical biochemistry? One possibility would be that they had separate origins as divergent genes, and acquired the virtually identical biochemistry by convergent evolution. A second is that they had a common origin and somehow diverged rapidly in the different Bacillus species.
A comparison of the two plasmids favors the second possibility, a common origin followed by divergence of sequence. 29 of 125 predicted pBtoxis proteins showed sequence similarity to predicted proteins from pXO1(4). Several groups of these genes map to the two plasmids with similar spacing, suggesting a common ancestry. In particular, the TubZ-Ba and TubZ-Bt genes are both located just before the likely replication origin (4). Why then does FtsZ have 40–50% sequence identity across almost all bacterial and archaeal species, yet TubZ-Ba and TubZ-Bt share only 21% sequence identity? We suggest that the function of TubZ in plasmid partition may be much less stringent than the function of FtsZ in cytokinesis, and permit the greater divergence.
Apart from the TubZ proteins of Bacillus plasmids, a number of very divergent FtsZ/tubulin-like sequences have been identified in the genomes of various archaea (2, 3). These do not appear to be involved in cell division, because each species has one or more conventional FtsZs. It may be that these divergent FtsZ's function primarily to assemble long cytoskeletal filaments, as a part of some still unknown mechanism.
Plasmid Partitioning—Plasmid partitioning can be achieved not only with very divergent tubulin homologs, but with at least two other protein systems unrelated to tubulin. The ParA/SopA system partitions a wide range of plasmids, and also bacterial chromosomes (19–21). Many of the ParA proteins from different plasmids and bacterial species show only 25–30% sequence identity to ParA of plasmid P1. Another well-studied partitioning system is based on the actin homolog ParM (22, 23). A second actin-homolog partitioning system is the newly discovered AlfA (24). AlfA forms long, dynamic filaments in the cytoplasm, and appears to function like ParM. Yet the amino acid sequence identity between AlfA and ParM is only 15%, less even than the 21% identity between TubZ-Ba and TubZ-Bt.
It is remarkable that a plasmid partitioning machine can apparently be constructed from three completely unrelated proteins. The one thing the proteins have in common is the ability to polymerize into long, thin filaments, which can apparently also bind a plasmid at each end. It may be that plasmid partitioning requires only a protein that can assemble a long filament, and some mechanism for attaching the plasmid to it. This apparently permits several different proteins to serve this cytoskeletal function, and for any given cytoskeletal protein a wide range of sequence divergence is apparently possible.
This work was supported by National Institutes of Health Grant GM66014. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EcFtsZ, FtsZ from E. coli; EM, electron microscopy; GTPγS, guanosine 5′-O-[γ-thio]triphosphate; GMPCPP, guanosine 5′-(a,b-methylene)triphosphate; GFP, green fluorescent protein; GST, glutathione S-transferase.
References
- 1.Erickson, H. P. (2007) Bioessays 29 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaughan, S., Wickstead, B., Gull, K., and Addinall, S. G. (2004) J. Mol. Evol. 58 19–29 [DOI] [PubMed] [Google Scholar]
- 3.Larsen, R. A., Cusumano, C., Fujioka, A., Lim-Fong, G., Patterson, P., and Pogliano, J. (2007) Genes Dev. 21 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, C., O'Neil, S., Ben-Dov, E., Jones, A. F., Murphy, L., Quail, M. A., Holden, M. T., Harris, D., Zaritsky, A., and Parkhill, J. (2002) Appl. Environ. Microbiol. 68 5082–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tinsley, E., and Khan, S. A. (2006) J. Bacteriol. 188 2829–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang, M., Bideshi, D. K., Park, H. W., and Federici, B. A. (2006) Appl. Environ. Microbiol. 72 6948–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolin, W. (2007) Curr. Biol. 17 R633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingerman, E., and Nunnari, J. (2005) Methods Enzymol. 404 611–619 [DOI] [PubMed] [Google Scholar]
- 9.Romberg, L., and Mitchison, T. J. (2004) Biochemistry 43 282–288 [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y., and Erickson, H. P. (2005) J. Biol. Chem. 280 22549–22554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minton, A. P. (2005) J. Pharm. Sci. 94 1668–1675 [DOI] [PubMed] [Google Scholar]
- 12.Minton, A. P. (2000) Curr. Opin. Struct. Biol. 10 34–39 [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, J. M., Jimenez, M., Velez, M., Mingorance, J., Andreu, J. M., Vicente, M., and Rivas, G. (2003) J. Biol. Chem. 278 37664–37671 [DOI] [PubMed] [Google Scholar]
- 14.Tang, M., Bideshi, D. K., Park, H. W., and Federici, B. A. (2007) J. Bacteriol. 189 8053–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Y., Bjornson, K., Redick, S. D., and Erickson, H. P. (2005) Biophys. J. 88 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai, A., and Mitchison, T. J. (1997) Annu. Rev. Cell Dev. Biol. 13 83–117 [DOI] [PubMed] [Google Scholar]
- 17.Erickson, H. P., and O'Brien, E. T. (1992) Annu. Rev. Biophys. Struct. Biol. 21 145–166 [DOI] [PubMed] [Google Scholar]
- 18.Scheffers, D. J., den Blaauwen, T., and Driessen, A. J. (2000) Mol. Microbiol. 35 1211–1219 [DOI] [PubMed] [Google Scholar]
- 19.Gerdes, K., Moller-Jensen, J., and Bugge Jensen, R. (2000) Mol. Microbiol. 37 455–466 [DOI] [PubMed] [Google Scholar]
- 20.Ebersbach, G., Ringgaard, S., Moller-Jensen, J., Wang, Q., Sherratt, D. J., and Gerdes, K. (2006) Mol. Microbiol. 61 1428–1442 [DOI] [PubMed] [Google Scholar]
- 21.Bouet, J. Y., Ah-Seng, Y., Benmeradi, N., and Lane, D. (2007) Mol. Microbiol. 63 468–481 [DOI] [PubMed] [Google Scholar]
- 22.Moller-Jensen, J., Borch, J., Dam, M., Jensen, R. B., Roepstorff, P., and Gerdes, K. (2003) Mol. Cell 12 1477–1487 [DOI] [PubMed] [Google Scholar]
- 23.Garner, E. C., Campbell, C. S., Weibel, D. B., and Mullins, R. D. (2007) Science 315 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker, E., Herrera, N. C., Gunderson, F. Q., Derman, A. I., Dance, A. L., Sims, J., Larsen, R. A., and Pogliano, J. (2006) EMBO J. 25 5919–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]