Abstract
Cytochrome P450 170A1 (CYP170A1) is encoded by the sco5223 gene of the Gram-positive, soil-dwelling bacterium Streptomyces coelicolor A3(2) as part of a two-gene cluster with the sco5222 gene. The SCO5222 protein is a sesquiterpene synthase that catalyzes the cyclization of farnesyl diphosphate to the novel tricyclic hydrocarbon, epi-isozizaene (Lin, X., Hopson, R., and Cane, D. E. (2006) J. Am. Chem. Soc. 128, 6022–6023). The presence of CYP170A1 (sco5223) suggested that epiisozizaene might be further oxidized by the transcriptionally coupled P450. We have now established that purified CYP170A1 carries out two sequential allylic oxidations to convert epi-isozizaene to an epimeric mixture of albaflavenols and thence to the sesquiterpene antibiotic albaflavenone. Gas chromatography/mass spectrometry analysis of S. coelicolor culture extracts established the presence of albaflavenone in the wild-type strain, along with its precursors epi-isozizaene and the albaflavenols. Disruption of the CYP170A1 gene abolished biosynthesis of both albaflavenone and the albaflavenols, but not epi-isozizaene. The combined results establish for the first time the presence of albaflavenone in S. coelicolor and clearly demonstrate that the biosynthesis of this antibiotic involves the coupled action of epi-isozizaene synthase and CYP170A1.
Biosynthetic pathways for many antibiotics in bacteria and plants are often associated with the presence of cytochromes P450 (P450, CYP).3 The CYPs perform late-stage stereo- and regiospecific metabolic reactions, such as hydroxylation, epoxidation, dehydrogenation, and C–C bond coupling, to modify the parent structural skeletons to give biosynthetic intermediates and/or bioactive products (1–4). Members of the genus Streptomyces produce a wide variety of bioactive secondary metabolites used in human and veterinary therapeutics, including a majority of the medically useful antibiotics (5, 6). Although terpenoids are one of the largest classes of naturally occurring, biologically active substances, with more than 50,000 known compounds, including anti-cancer drugs, anti-parasitic, and antibacterial agents, only a relative handful of such terpene metabolites have been isolated from Streptomyces spp (7, 8).
P450 monooxygenases are a superfamily of heme proteins utilizing atmospheric dioxygen and electrons supplied by NAD(P)H almost always through redox partner proteins to generate, most commonly, oxidized organic products and a molecule of water (9). P450s play an important role in drug metabolism and in the biosynthesis of steroids, lipids, vitamins, antibiotics, and other natural secondary metabolites.
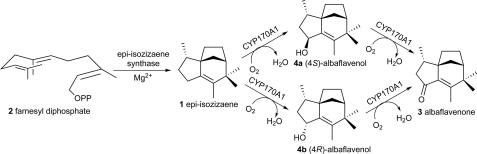
Streptomyces coelicolor A3(2) is genetically one of the most studied actinomycetes and has been a model for genetic analysis of antibiotic production. The complete genome sequence of S. coelicolor has revealed the presence of 18 CYP, 6 ferredoxin, and 4 ferredoxin reductase genes (10, 11). The endogenous biological function of CYP170A1, belonging to the 18-member P450 CYPome of S. coelicolor, has not previously been identified (12).4 The sco5223 gene encoding CYP170A1 in S. coelicolor is part of a two-gene cluster with a sesquiterpene cyclase gene (sco5222) with which it shares a four-nucleotide ATGA transcriptional overlap at its 5′-end. The sco5222 gene product has been shown to catalyze the synthesis of a novel sesquiterpene, epi-isozizaene (1, Fig. 1), by cyclization of the universal sesquiterpene synthase substrate farnesyl diphosphate (FPP, 2, Fig. 1) (13). We therefore hypothesized that epi-isozizaene might be the substrate for CYP170A1. Although epi-isozizaene had never previously been isolated from any natural source, the corresponding 5-keto derivative, albaflavenone (3, Fig. 1), which has been reported to have an earthy, camphor-like odor and to exhibit antibacterial activity, has previously been isolated from Streptomyces albidoflavus (14, 15). We now report that CYP170A1 catalyzes the two-step allylic oxidation of epiisozizaene to albaflavenone through the intermediacy of an epimeric mixture of albaflavenols (4a and 4b, Fig. 1) and that all four compounds can be detected in extracts of S. coelicolor.
FIGURE 1.
Albaflavenone biosynthetic pathway in S. coelicolor.
EXPERIMENTAL PROCEDURES
Expression and Purification of CYP170A1—CYP170A1 was expressed and purified as reported earlier (12). To improve protein expression, CYP170A1 was coexpressed with molecular chaperones GroES/GroEL (16). Briefly, the cells cotransformed with the plasmids CYP170A1pET17b and pGro12 were cultured overnight in Luria Bertani broth containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. After inoculation (1:100) in 1.5 liters of Terrific Broth containing 100 μg/ml ampicillin and 50 μg/ml kanamycin, growth was carried out at 37 °C and 240 rpm until the optical density at 600 nm reached ∼1.0. After the addition of 1 mm δ-aminolevulinic acid for heme synthesis and induction with 1 mm isopropyl-β-d-thiogalactopyranoside, chaperone expression was induced by addition of arabinose to a final concentration of 4 mg/ml. Cell growth was continued for an additional 20 h at 27 °C and 190 rpm. The cells were harvested by centrifugation and resuspended in 100 ml of lysis buffer (100 mm potassium phosphate buffer, pH 7.7, 20% (v/v) glycerol, and 100 mm NaCl). Cells were broken by freeze-thawing and the cytosol containing CYP170A1 isolated following centrifugation at 100,000 × g. The soluble CYP170A1 was purified by metal (Ni2+) affinity chromatography (Qiagen) by washing with 20 mm imidazole followed by 50 mm imidazole and elution with 100 mm histidine.
UV-visible Spectra and Substrate Binding Assay—All absorbance spectra were recorded using a double beam Shimadzu UV-2401PC spectrophotometer. A reduced CO difference spectrum was carried out as described previously (12). The interaction of epi-isozizaene with CYP170A1 was examined by perturbation of the heme Soret spectrum. CYP170A1 (2.5 μm) in 20 mm potassium phosphate buffer (2.0 ml, pH 7.7) was divided between two tandem cuvettes. After thermal equilibration at 25 °C a baseline was established between 350 and 450 nm and sequential portions (1–5 μl) of concentrated epiisozizaene (1 mm in Me2SO) were added to the CYP170A1 chamber of the sample cuvette and the buffer chamber of the reference cuvette to give a final ligand concentration in the range of 0–30 μm. An equal volume of Me2SO was added to the CYP170A1 chamber of the reference cuvette and the difference spectrum recorded after each titration. Kd values were estimated by fitting plots of ΔA390–420 versus [epi-isozizaene].
Disruption of CYP170A1 Gene in S. coelicolor—The CYP170A1 knock-out strain was isolated from a library of 1 × 105 independent mutants generated by in vivo transposition in S. coelicolor, as described previously (12).
CYP170A1 Activity Assay in Vitro—CYP170A1 (1 nmol), Escherichia coli flavodoxin (10 nmol), and E. coli flavodoxin reductase (2 nmol) were reconstituted in 400 μl of 50 mm Tris-HCl buffer (pH 8.2) containing 20% (v/v) glycerol and epiisozizaene (20 nmol) plus 1% Me2SO. Following incubation of this mixture for 5 min on ice, the reconstituted enzyme solution was placed in a shaking bath at 35 °C. The reaction was started by the addition of NADPH to a final concentration of 1 mm and 60 μl of an NADPH-regenerating system (glucose-6-phosphate (60 mm), β-nicotinamide adenine dinucleotide phosphate (3 mm), and glucose-6-phosphate dehydrogenase (0.2 unit)). The reaction was carried out for 1.5 h in a 10-ml test tube, at which time it was quenched with 4 μl of concentrated HCl and extracted three times with 400 μl of pentane:methylene chloride (4:1). The extracts were concentrated under a stream of N2, and 2 μl of extract was analyzed by gas chromatography/mass spectrometry (GC/MS). The turnover rate for the epi-isozizaene oxidation was determined by carrying out a series of 30-min incubations with the concentration of 5–90 μm substrate. The mixtures were extracted and analyzed by GC/MS. The turnover number was calculated by a nonlinear regression fit to the Michaelis-Menten equation using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA).
Isolation and Identification of Terpenoids Produced by S. coelicolor—S. coelicolor A3(2) M145 and the CYP170A1 knockout strain were cultured side by side at 30 °C in yeast extract-malt extract medium (17) for 2–5 days at 280 rpm. 500-ml fermentation cultures were centrifuged for 10 min at 4,000 rpm (3500 × g), which was extracted three times with 10 ml of pentane :methylene chloride (4:1) at pH 5 for 2 h. The extracts were dried over Na2SO4 for 20 min on ice, concentrated under a stream of N2 to 200 μl, and 2 μl of extract was analyzed by GC/MS.
GC/MS Analysis of in Vitro and in Vivo Products—For GC/MS analysis, electron impact (EI) mass spectra were obtained using a Finnigan DSQ Quantum mass spectrometer in the EI positive ion full-scan mode with ion source at 250 °C. GC separations were achieved using a fused silica HP5 (25-m, 0.32-mm inside diameter) capillary column, with energy of ionization 70 electron volts and temperature program (70 °C hold for 4 min, to 300 °C at 10 °C/min, 300 °C hold for 2 min).
Enzymatic Synthesis of Epi-isozizaene and Synthesis of Albaflavenols/Albaflavenone—Farnesyl diphosphate (12.0 mg) was incubated overnight at 30 °C with epi-isozizaene synthase (0.6 μmol), isolated, and purified as previously described (13). A mixture of epi-isozizaene (2 mg) and selenium dioxide (20 mg) was stirred in 3 ml of methylene chloride overnight at room temperature. The resulting suspension was applied to a 1-cm silica gel column and eluted with methylene chloride, collecting fractions of 0.5 ml. Fractions containing pure albaflavenol (Rf 0.15, methylene chloride, SiO2 TLC) were combined and concentrated under vacuum. GC/MS analysis indicated the formation of a 1.4:1 mixture of epimeric alcohols, m/z 220, 4a (tR 11.4 min) and 4b (tR 11.6 min) high resolution mass spectrum (4a/4b): m/z 220.1830 observed (calculated for C15H24O: 220.1827). 1H NMR and 13C NMR spectra were recorded on the mixture of 4a and 4b and the individual signals assigned based on the 1.4:1 ratio of the two components, using a combination of one- and two-dimensional NMR, including 13C DEPT135, 1H1H COSY, HMBC, and HSQC spectra (Table 1).
TABLE 1.
1H and 13C NMR assignments of albaflavenols (4a and 4b) and albaflavenone (3)
a Positionnumbering for 3 is different in Ref. 14.
A mixture of 0.5 mg of albaflavenols (4a and 4b), 20 mg of celite, and 10.0 mg of CrO3 in 2 ml of ether:methylene chloride (3:1) was stirred for 4 h at room temperature. After removal of the solids by filtration, the solvent was evaporated and the resulting product 3 analyzed directly by GC/MS (m/z 218, tR 12.3 min) and NMR, including 1H and 13C NMR, 13C DEPT135, and HSQC NMR (Table 1).
RESULTS AND DISCUSSION
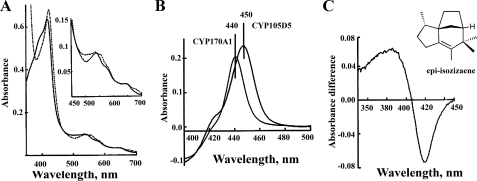
Unique Reduced CO Difference Spectra—To investigate the possible biochemical function of CYP170A1, we tested whether epi-isozizaene could be the substrate of CYP170A1 because the CYP170A1 gene (sco5223) is directly downstream of that encoding the sesquiterpene cyclase (sco5222) (13). We purified a histidine-tagged CYP170A1 protein (supplemental Fig. S1). The oxidized absolute spectrum of CYP170A1 shows Soret bands at 393 and 417 nm that indicate a mixture of high spin and low spin states (Fig. 2A), the ratios between the spin states varying somewhat between different preparations. It could be that some very hydrophobic ligand from E. coli bound into the heme pocket and leads to the partial high spin state of CYP170A1. The position of these Soret bands are typical for high and low spin P450s. Surprisingly, the purified CYP170A1 displays a unique reduced carbon monoxide difference spectrum, with a maximum ∼440 nm instead of the typical P450 difference absorption maximum at ∼450 nm (Fig. 2B). Lamb et al. (12) reported previously that this monooxygenase could have a reduced carbon monoxide difference spectral maximum at or below 447 nm in assays. Under our experimental conditions, we have examined more than 10 preparations of purified enzyme and the Soret maximum reproducibly seen at 440 nm. All other P450s reported to date show the Soret maximum of the reduced CO difference spectra at or about 450 nm, from which the name of these monooxygenases arises (18). The 10-nm hypsochromic shift of the absorbance maximum was the same whether the reduction was carried out chemically (Na2S2O4) or with biological redox proteins (E. coli flavodoxin and flavodoxin reductase). The molecular basis for the observed Soret maximum at 440 nm in the reduced CO difference spectrum of CYP170A1 remains unknown at present. It is also an open question whether there is any biological significance for this unusual shift to shorter wavelength.
FIGURE 2.
UV-visible absorption spectra of purified CYP170A1 in Tris-HCl buffer (50 mm, pH 8.2). A, oxidized absolute spectrum in solid line; reduced spectrum using Na2S2O4 in dashed line. B, reduced CO difference spectrum showing unique 440 nm compared with the normal reduced CO difference spectrum at 450 nm of CYP105D5. C, type I binding spectrum resulting from addition of epi-isozizaene (0–30 μm) to CYP170A1 (2.5 μm). The substrate epi-isozizaene structure is shown.
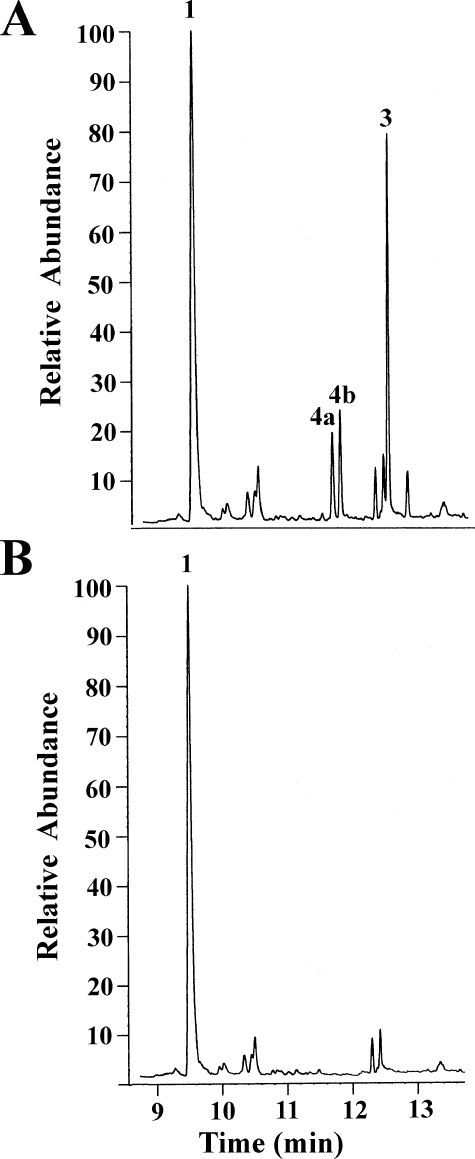
In Vitro Activity of CYP170A1—Titration of CYP170A1 with epi-isozizaene produced a typical type I P450 binding spectrum in which the low spin heme Soret peak at 420 nm is shifted to a high spin peak at 390 nm (Fig. 2C). The dissociation constant for epiisozizaene (Kd) was calculated to be 1.3 ± 0.2μm, strongly suggesting that epi-isozizaene can specifically bind to the active site of CYP170A1 and supporting the proposed role of epi-isozizaene as a substrate. Analysis of the CYP170A1-catalyzed reaction by GC/MS revealed the time-dependent formation of two isomeric products with m/z 220 (tR 11.4 min; tR 11.6 min) in a ratio of ∼1:1.2 and a third product with m/z 218 (tR 12.3 min) (Fig. 3). Negative control incubations lacking CYP170A1, flavodoxins, or NADPH did not generate any products.
FIGURE 3.
In vitro catalytic activity of CYP170A1 supported by flavodoxin and flavodoxin reductase. Oxidation reactions were carried out as described under “Experimental Procedures.” A, product profile obtained following the flavodoxin- and flavodoxin reductase-supported reaction using CYP170A1 (1 nmol) and epi-isozizaene (40 nmol) plus 1% Me2SO at 35 °C for 1.5 h. Three products are noted, 4a and 4b (albaflavenol epimers) and 3 (albaflavenone), respectively; substrate epi-isozizaene as 1. B, negative control incubation, as in A but flavodoxin/flavodoxin reductase or NADPH was omitted.
Synthesis of Albaflavenols and Albaflavenone—To identify these metabolites produced by CYP170A1, we synthesized reference standards of the epimeric albaflavenols and albaflavenone. Allylic oxidation of epi-isozizaene with SeO2 gave a 1.4:1 mixture of two albaflavenol epimers for which the structures were readily assigned based on the appearance of the characteristic 1H NMR signals atδ 4.56 (doublet, J = 5.40 Hz) and 4.61 (triplet, J = 7.82 Hz) for the H-5, allylic CHOH protons in the major and minor isomers, respectively, as well as the observation of the corresponding C-5 carbinyl 13C signals at 70.80 and 71.85 ppm. Comparison with the spectra of epi-isozizaene confirmed the disappearance of the H-5 allylic methylene proton signals at δ 2.2 (doublet of doublet of quartets) and 2.06 (doublet of doublet of doublet of quartets) and the C-5 methylene at 27.3 ppm. Full assignments, based on a combination of standard one- and two-dimensional NMR experiments are given in Table 1. Although we did not attempt to separate the individual epimers, we have provisionally assigned the configuration of the two alcohols, using Hyperchem 7.5 and MM+ to predict the coupling constants for the H-5 carbinyl protons, based on the calculated ground state conformation of each epimer. In this manner 4a was assigned the (5S)-configuration (doublet, J = 5.40 Hz; predicted: doublet, J = 6.2 Hz) and epimer 4b the (5R)-configuration (triplet, J = 7.82 Hz; predicted: doublet of doublets, J = 5.5, 7.2 Hz). Oxidation of the mixture of 4a and 4b with CrO3 on celite gave a single product, albaflavenone, whose 1H and 13C NMR spectra as well as electron impact mass spectrum were in complete agreement with those reported for the antibiotic albaflavenone previously isolated from S. albidoflavus (14, 15) (Table 1).
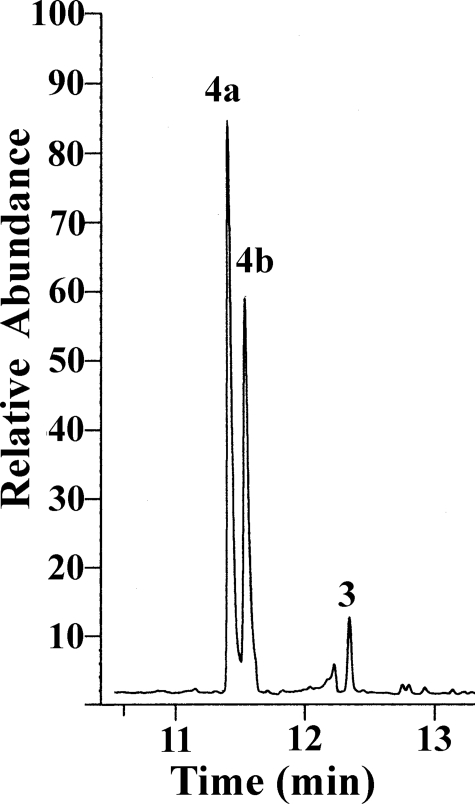
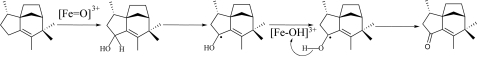
The m/z 220 components of the CYP170A1-catalyzed oxidation of epi-isozizaene were identified as albaflavenol epimers 4a and 4b by direct comparisons of their retention time and their mass spectral fragmentation patterns with the authentic albaflavenols (supplemental Fig. S2), whereas the m/z 218 product was identical to synthetic albaflavenone in GC retention time and mass spectrum (supplemental Fig. S3). These results clearly demonstrate that CYP170A1 catalyzes the two-step oxidation of epi-isozizaene to the antibiotic albaflavenone. CYP170A1 first carries out a non-stereo-specific oxidation of epi-isozizaene to give a mixture of the albaflavenol epimers 4a and 4b, each of which can serve as substrate for the second oxidation to yield albaflavenone 3 (Fig. 1). This is quite different from most other P450s which catalyze regio- and stereospecific oxidation. For example, Hyoscyamus premnaspirodiene oxygenase (CYP71D55) has very high stereospecificity to produce the single alcohol solavetivol intermediate for solavetivone (19). In addition, incubation of the mixture of epimeric albaflavenols 4a and 4b with CYP170A1 in the presence of flavodoxin/flavodoxin reductase and the NADPH regenerating system led to formation of albaflavenone, confirming that both epimers can serve as substrates for this P450 (Fig. 4). It is interesting to note, when comparing the amount of albaflavenone produced by epiisozizaene or albaflavenols as substrate, that CYP170A1 can generate ∼3-fold more albaflavenone from epi-isozizaene than from the mixture of albaflavenols as substrate even when the alcohol concentration is 10-fold higher than that of epi-isozizaene. This could indicate that release of the alcohols into the medium followed by rebinding to CYP170A1 is inefficient and is not important in the catalytic activity. This could also indicate that the mechanism of the oxidation of the allylic alcohols to albaflavenone by CYP170A1 involves abstraction of two hydrogen atoms (Fig. 5). It would seem less likely that the alcohol intermediates could flip in the active site without dissociation to form the geminal 5,5-diol followed by loss of a water to produce albaflavenone. However, it is very possible that the alcohol intermediates may re-enter the active site via different orientations to form the unstable gem-diol intermediate because this enzyme generates the epimeric albaflavenols nonstereoselectively. The two mechanisms might be distinguished by analyzing albaflavenone obtained from CYP170A1-catalyzed oxidation of 18O-labeled albaflavenols (20, 21). At this stage we cannot exclude any of the possible mechanisms discussed here or that this enzyme may proceed through a combination of gem-diol and double hydrogen abstraction mechanism. None of the proposed CYP170A1 reaction mechanisms is without precedent. Nevertheless, only a small number of CYPs have been shown to catalyze two sequential oxidation steps (3, 19). It will be interesting to examine in detail the CYP170A1 active site that can bind epi-isozizaene in two orientations, producing two alcohol epimers, and then convert both to the same ketone. X-ray crystallography studies are underway to permit this analysis. Using the flavodoxin/flavodoxin reductase couple, the turnover number is ∼0.32 min–1 for conversion of epiisozizaene to albaflavenone. The natural ferredoxin-ferredoxin reductase electron transport couple for CYP170A1 from among the 24 possible combinations in S. coelicolor is unknown, and therefore the rate of conversion of epi-isozizaene will probably be increased with the endogenous reductase system. However, as noted above, the binding constant of epi-isozizaene to CYP170A1 clearly predicts it to be an endogenous substrate.
FIGURE 4.
Incubation of albaflavenols (4a and 4b) with CYP170A1 supported by flavodoxin and flavodoxin reductase.
FIGURE 5.
Proposed mechanism of biosynthesis of albaflavenone catalyzed by CYP170A1.
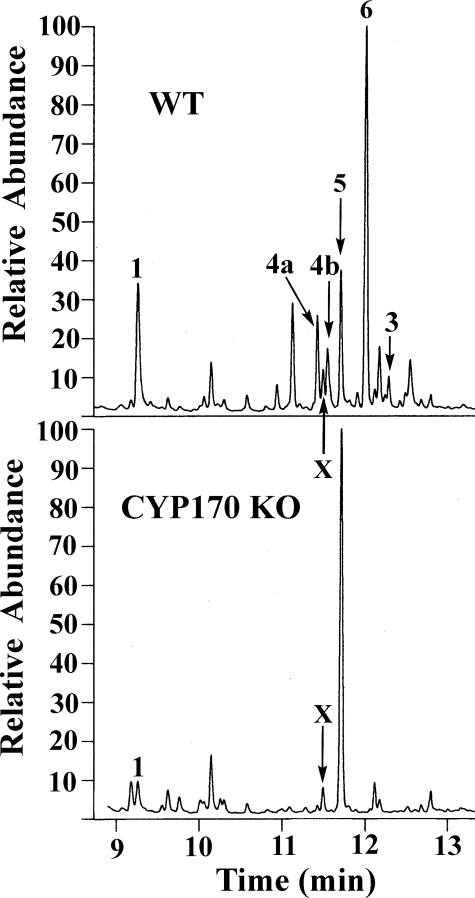
Involvement of CYP170A1 in Albaflavenone Biosynthesis—The oxidation of epi-isozizaene to albaflavenols (4a and 4b) and albaflavenone by CYP170A1 led us to focus on the hydrophobic secondary metabolites produced by the parent S. coelicolor strain. Culture extracts from both wild type and the CYP170A1 knock-out strain were analyzed by GC/MS. Both epimers of albaflavenol as well as albaflavenone were produced by the wild-type strain, whereas none of these three metabolites could be detected in the extracts of the CYP170A1 knock-out stain (Fig. 6). Epi-isozizaene was detected in the extracts of the wild-type culture, with the titer gradually decreasing during 5 days of fermentation. 1 was found in the CYP170A1 knock-out culture as well as at levels 7-fold lower than in wild type (Fig. 6). A possible explanation is that the transcript of epi-isozizaene synthase gene (sco5222) may be unstable or at low levels when the transposon is present in the CYP170A1 gene (sco5223), which is translationally coupled to sco5222 by a 4-nucleotide 3′-5′-ATGA overlap. In addition, we found germacradienol (5, a precursor of geosmin, Fig. 6) had accumulated in the CYP170A1 knock-out extract. Because there are only two sesquiterpene cyclases in S. coelicolor, epi-isozizaene synthase and geosmin synthase, the germacradienol accumulation would be expected when epi-isozizaene synthase was affected due to the competition of the common substrate, farnesyl diphosphate. Another major component (6, tR 12 min, m/z 218, Fig. 6) that is present in the extracts of the wild type, but absent in the CYP170A1 knock-out, has not been characterized yet. At present it is not clear whether this unknown compound is related to the biological function of CYP170A1 or to some other biological process, and this requires further investigation. Nevertheless, the in vivo observation is that epi-isozizaene is still produced, whereas all the derived albaflavenol and albaflavenone oxidation products are absent in the mutant, fully consistent with the results of the in vitro experiments and thereby confirming the deduced biochemical role of CYP170A1 in the two-step oxidative conversion of epi-isozizaene to the sesquiterpene antibiotic albaflavenone.
FIGURE 6.
GC/MS analysis of in vivo products from 4-day culture. Upper panel, GC trace of compounds produced by wild-type S. coelicolor; bottom panel, GC trace of compounds produced by CYP170A1 knock-out culture extract. The epi-isozizaene, two isomers of albaflavenol, and albaflavenone are noted as 1, 4a, 4b, and 3, an unknown compound as X, and germacradienol as 5. An uncharacterized peak was noted as 6, which was absent in CYP170A1 knock-out.
S. avermitilis, the industrially important producer of the potent anthelminthic agent avermectin, harbors a two-gene cluster homologous to the sco5222-sco5223 pair of S. coelicolor (23). We have recently established that the sav3032 gene of S. avermitilis encodes a terpene synthase of 363 amino acids with 76% identity to S. coelicolor epi-isozizaene synthase that catalyzes the cyclization of farnesyl diphosphate to epi-isozizaene.5 Notably, sav3032 is transcriptionally coupled with a four-nucleotide ATGA overlap to the downstream sav3031 gene encoding an apparent P450 (UniProt Q82IV2) of 456 amino acids with 76% identity to CYP170A1 of S. coelicolor (sco5223p) (24). It is therefore very likely that Streptomyces avermitilis will also produce albaflavenone and that the biochemical pathway is identical to that uncovered here in S. coelicolor.
CONCLUSIONS
We have determined the endogenous biological function of the sco5222–sco5223 operon and the gene product (sco5223) CYP170A1 from S. coelicolor. Both in vitro and in vivo results firmly establish that CYP170A1 plays an important role in the biosynthesis of the sesquiterpene antibiotic albaflavenone. Although there are multiple examples of three-step oxidations of methyl groups to carboxylic acids that are catalyzed by a single P450 (25–27), the two-step oxidation of an allylic methylene to a conjugated ketone by a single P450 is less common. Notably, in the biosynthesis of oxidized monoterpenes, P450-catalyzed allylic oxidation is normally followed by a conventional NAD(P)+-dependent dehydrogenation, as for example in the conversion of (–)-limonene to (–)-isopiperitenone by way of (–)-trans-isopiperitenol in peppermint catalyzed by successive P450 and dehydrogenase enzymes (22, 28).
Supplementary Material
Acknowledgments
We thank K. Wilkins for a copy of the mass spectrum of albaflavenone isolated from S. albidoflavus 246.
This work was supported by National Institutes of Health Grants GM69970 (to M. R. W.), ES00267 (to M. R. W.), and GM30301 (to D. E. C.) and by a Leverhulme Trust research fellowship (to D. C. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: P450 or CYP, cytochrome P450 monooxygenase; GC/MS, gas chromatography/mass spectrometry.
Lamb et al. reported that this monoxygenase has sterol 14α-demethylase activity (12). Although CYP170A1 has sequence similarity to CYP51 and can give Type I binding with lanosterol, the previous exogenous activity in lanosterol demethylation was not reproduced in this work, and as noted previously S. coelicolor does not produce sterols.
X. Lin, H. Ikeda, and D. Cane, unpublished observations.
References
- 1.Donadio, S., Staver, M. J., McAlpine, J. B., Swanson, S. J., and Katz, L. (1991) Science 252 675–679 [DOI] [PubMed] [Google Scholar]
- 2.Zhao, B., Guengerich, F. P., Bellamine, A., Lamb, D. C., Izumikawa, M., Lei, L., Podust, L. M., Sundaramoorthy, M., Reddy, L. M., Kelly, S. L., Kalaitzis, J. A., Stec, D., Voehler, M., Falck, J. R., Moore, B. S., Shimada, T., and Waterman, M. R. (2005) J. Biol. Chem. 280 11599–11607 [DOI] [PubMed] [Google Scholar]
- 3.Quaderer, R., Omura, S., Ikeda, H., and Cane, D. E. (2006) J. Am. Chem. Soc. 128 13036–13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solenberg, P. J., Matsushima, P., Stack, D. R., Wilkie, S. C., Thompson, R. C., and Baltz, R. H. (1997) Chem. Biol. 4 195–202 [DOI] [PubMed] [Google Scholar]
- 5.Bérdy, J. (2005) J. Antibiot. (Tokyo) 58 1–26 [DOI] [PubMed] [Google Scholar]
- 6.Watve, M. G., Tickoo, R., Jog, M. M., and Bhole, B. D. (2001) Arch. Microbiol. 176 386–390 [DOI] [PubMed] [Google Scholar]
- 7.Dewick, P. M. (2002) Nat. Prod. Rep. 19 181–222 [DOI] [PubMed] [Google Scholar]
- 8.Dubey, V. S., Bhalla, R., and Luthra, R. (2003) J. Biosci. (Bangalore) 28 637–646 [DOI] [PubMed] [Google Scholar]
- 9.Groves, J. T. (2005) in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano, P. R., ed) 3rd Ed., pp. 1–80, Kluwer Academic/Plenum Publishers, New York
- 10.Bentey, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., Bateman, A., Brown, S., Chandra, G., Chen, C. W., Collins, M., Cronin, A., Fraser, A., Goble, A., Hidalgo, J., Hornsby, T., Howarth, S., Huang, C. H., Kieser, T., Larke, L., Murphy, L., Oliver, K., O' Neil, S., Rabbiowitsch, E., Rajandream, M. A., Rutherford, K., Rutter, S., Seeger, K., Saunders, D., Sharp, S., Squares, R., Squares, S., Taylor, K., Warren, T., Wietzorrek, A., Woodward, J., Barrell, B. G., Parkhill, J., and Hopwood, D. A. (2002) Nature 417 141–147 [DOI] [PubMed] [Google Scholar]
- 11.Lamb, D. C., Skaug, T., Song, H.-L., Jackson, C. J., Podust, L. M., Waterman, M. R., Kelly, D. B., Kelly, D. E., and Kelly, S. L. (2002) J. Biol. Chem. 277 24000–24005 [DOI] [PubMed] [Google Scholar]
- 12.Lamb, D. C., Fowler, K., Kieser, T., Manning, N., Podust, L. M., Waterman, M. R., Kelly, D. E., and Kelly, S. L. (2002) Biochem. J. 364 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, X., Hopson, R., and Cane, D. E. (2006) J. Am. Chem. Soc. 128 6022–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurtler, H., Pedersen, R., Anthoni, U., Christophersen, C., Nielsen, P. H., Wellington, E. M., Pedersen, C., and Bock, K. (1994) J. Antibiot. (Tokyo) 47 434–437 [DOI] [PubMed] [Google Scholar]
- 15.Scholler, C. E., Gurtler, H., Pedersen, R., Molin, S., and Wilkins, K. (2002) J. Agric. Food Chem. 50 2615–2621 [DOI] [PubMed] [Google Scholar]
- 16.Nishihara, K., Kanemori, M., Kitagawa, M., Yanagi, H., and Yura, T. (1998) Appl. Environ. Microbiol. 64 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000) in Practical Streptomyces Genetics, pp. 405–418, The John Innes Foundation, Norwich, United Kingdom
- 18.Omura, T., and Sato, R. (1964) J. Biol. Chem. 239 2370–2378 [PubMed] [Google Scholar]
- 19.Takahashi, S., Yeo, Y. S., Zhao, Y., O'Maille, P. E., Greenhagen, B. T., Noel, J. P., Coates, R. M., and Chappell, J. (2007) J. Biol. Chem. 282 31744–31754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood, A. W., Swinney, D. C., Thomas, P. E., Ryan, D. E., Hall, P. F., Levin, W., and Garland, W. A. (1988) J. Biol. Chem. 263 17322–17332 [PubMed] [Google Scholar]
- 21.Bellucci, G., Chiaape, C. Pucci, L., and Gervasi, P. G. (1996) Chem. Res. Toxicol. 9 871–874 [DOI] [PubMed] [Google Scholar]
- 22.Karp, F., Mihaliak, C. A., Harris, J. L., and Croteau, R. (1990) Arch. Biochem. Biophys. 276 219–226 [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H., Shiba, T., Sakaki, Y., Hattori, M., and Omura, S. (2003) Nat. Biotechnol. 21 526–531 [DOI] [PubMed] [Google Scholar]
- 24.Lamb, D. C., Ikeda, H., Nelson, D. R., Ishikawa, J., Skaug, T., Jackson, C., Omura, S., Waterman, M. R., and Kelly, S. L. (2003) Biochem. Biophys. Res. Commun. 307 610–619 [DOI] [PubMed] [Google Scholar]
- 25.Helliwell, C. A., Chandler, P. M., Poole, A., Dennis, E. S., and Peacock, W. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pikuleva, I. A., Babiker, A., Waterman, M. R., and Bjorkhem, I. (1998) J. Biol. Chem. 273 18153–18160 [DOI] [PubMed] [Google Scholar]
- 27.Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., Ho, K. A., Eachus, R. A., Ham, T. S., Kirby, J., Chang, M. C., Withers, S. T., Shiba, Y., Sarpong, R., and Keasling, J. D. (2006) Nature 440 940–943 [DOI] [PubMed] [Google Scholar]
- 28.Wise, M. L., and Croteau, R. (1999) in Comprehensive Natural Products Chemistry. Isoprenoids Including Carotenoids and Steroids (Cane, D. E., ed) Vol. 2, pp. 97–153, Elsevier, Oxford [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.