Abstract
Glucocorticoid-induced tumor necrosis factor receptor (GITR), found constitutively expressed on human primary natural killer (NK) cells at low levels was up-regulated upon stimulation by either Toll-like receptor ligand or NK cell growth factor, interleukin (IL)-15. cDNA microarray analysis showed that engagement of GITR primarily suppressed the activation of NF-KB pathway of NK cells and up-regulated anti-inflammatory genes heme oxygenase-1 and IL-10. Further analysis revealed that GITR activation suppressed NK cell proliferation in response to IL-15. GITR activation also suppressed proinflammatory cytokine secretion and increased NK cell apoptosis. GITR activation resulted in blocked phosphorylation of Stat5 and Akt, which may have contributed to the observed antiproliferative effect of GITR on NK cells. Increased apoptosis was independent of the Fas-FasL pathway, but Bcl-XL and phospho-Bad protein expressions were diminished, suggesting involvement of the mitochondrial apoptosis pathway. The results suggest that although GITR is an activation marker for NK cells similar to that for T cells, GITR serves as a negative regulator for NK cell activation. Our studies demonstrate a novel physiological role of GITR on NK cells.
Natural killer (NK)3 cells are an important component of the innate immune system. They use cytolytic granules and cytokine production to mediate their effector functions (1). By dynamic interactions with other immune cells, such as dendritic cells, macrophages, and T cells, NK cells can also mediate the transition from innate to adaptive immune response (2–4). NK cell activity is regulated delicately by a large number of putative activating and inhibitory receptors (5, 6). Under normal physiological conditions, mature NK cells are relatively quiescent. Following infection, NK cells undergo proliferative expansion that is either nonspecific (e.g. mediated by cytokines) or specific (e.g. induced by engagement of activating receptors), after which these cells return to relative quiescence.
Glucocorticoid-induced tumor necrosis factor receptor (GITR, or TNFRSF18) is a recently identified member of the tumor necrosis factor receptor superfamily. It was initially identified as a glucocorticoid-responsive gene in a murine hybridoma T cell line (7). Initial studies suggested that GITR was selectively expressed on CD4+CD25+ regulatory T cells, and receptor engagement abrogated the suppressive function of T regulatory cells (8). Subsequent studies showed GITR expression on CD4+CD25– and CD8+ T cells (9, 10). It is markedly induced in T cells after TCR activation in mice, and it is a general consensus that GITR provides a strong co-stimulatory signal for T cells when stimulated by its natural ligand or its agonistic antibody (8, 11–16).
GITR is not confined to T cells. It is expressed on macrophages, B cells (8, 13, 15–17), and NK cells (18, 19), whereas its cognate ligand (GITRL) is constitutively expressed on antigen-presenting cells, such as dendritic cells and B cells (20, 21). Manipulation of the GITR-GITRL interaction is suggested as a target for tumor immunotherapy, treatment of viral infection, and treatment of autoimmune diseases (22–30). This possibility has intensified the significance of studying GITR-GITRL interactions.
Although there is a consensus for the role of GITR in T cells, its role in NK cells remains controversial. Hanabuchi et al. (19) showed that GITRL expression on activated plasmacytoid dendritic cell precursors enhanced both NK cell cytotoxic activity and IFN-γ production by NK cells via GITR-GITRL interactions. A recent report by Baltz et al. (18) suggested that GITRL-expressing tumor cells diminish NK cell cytotoxicity and inhibit IFN-γ release. In light of the role of GITR in the regulation of T cells and its constitutive and increased expression upon activation on human NK cells, we investigated GITR signaling mechanisms and the potential role of GITR in regulating NK cell activation.
EXPERIMENTAL PROCEDURES
Normal Donors—A total of 20 healthy human donors were studied. Samples from normal donors were obtained from the National Institutes of Health blood bank after informed consent (protocol 99-C-0168).
Isolation of Human Peripheral Blood Mononuclear Cells (PBMCs) and NK Cells and Cell Culture—Human PBMCs from normal donors were isolated from their buffy coat using Ficoll gradient centrifugation as previously described (31). NK cells were obtained from isolated PBMCs by a magnetic sorting technique using a negative NK isolation kit II (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. For cell stimulation, PBMCs (2 × 106/ml) or purified NK cells (2 × 106/ml) in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum (Gemini Bioproducts, West Sacramento, CA) supplemented with 2 mm glutamine and 1× antibiotics were plated in 6-well plates (Nalge Nunc International, Rochester, NY) precoated with anti-GITR at 10 μg/ml in the presence or absence of either 10 ng/ml IL-15 (Peprotech Inc., Rocky Hill, NJ) or poly(I-C) (Sigma) at 25 μg/ml for stimulation. Cells were cultured at 37 °C in 5% CO2.
Flow Cytometry Analysis—To study GITR expression in resting and activated NK cells, PBMCs were cultured for 48 h with or without stimulation. Whole blood or PBMCs were analyzed using a three-color flow cytometry staining protocol. Specifically, 2 × 106/ml cells were stained with 5 μg/ml biotinylated anti-human GITR antibody (R & D Systems, Minneapolis, MN) in staining buffer (1× phosphate-buffered saline, 0.5% bovine serum albumin (Sigma)) at 4 °C for 30 min. After two washes with phosphate-buffered saline, cells were stained with 4 μg/ml allophycocyanin-labeled streptavidin (BD Biosciences) and counterstained with anti-human PE-CD56 and FITC-CD3 antibodies (BD Biosciences) at room temperature for 15 min. After two washes with phosphate-buffered saline and fixation with 300 μl of 1% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, PA), cells were acquired by a FACSCalibur flow cytometer (BD Biosciences) and analyzed by FlowJo software (TreeStar, San Jose, CA). Briefly, lymphocytes were gated based on cell optic characteristics (forward scatter versus side scatter). CD3+, CD56+, CD3–, and CD56– populations were gated based on antibody staining. A subpopulation was derived from its parent population. The gate for the positive staining of GITR was set based on the nonspecific staining of an isotype-matched biotinylated control antibody. The percentage of GITR-positive cells in a defined subpopulation was calculated as the number of GITR-positive cells in the subpopulation divided by the total number of cells in the subpopulation. For example, the percentage of GITR in the CD3–CD56+ population was calculated as follows: (number of GITR+CD3–CD56+ cells/number of CD3–CD56+ cells) × 100%.
Microarray Analysis—Human NK cells were stimulated with poly(I-C) (25 μg/ml) and cultured in plates with or without precoated anti-GITR antibody (10 μg/ml). After 6 h of stimulation, NK cells were collected and lysed, and total RNA was purified using an RNAeasy isolation kit according to the manufacturer's instructions (Qiagen). For cDNA microarray analysis, an array of genes selectively involved in the NF-κB pathway (SuperArray Inc., Gaithersburg, MD) was used for analysis. The cDNA microarray analysis was performed following the manufacturer's instructions. Briefly, an equal amount of total RNA was biotinylated, and probes were then hybridized onto the membranes containing NF-κB pathway-specific genes. After extensive washing, specific hybridization onto the membrane was detected by chemiluminescence and recorded by a digital CCD camera (UVP, Upland, CA). Differential gene expression was then analyzed using GEArray Expression Analysis Suite software (SuperArray Inc.) designed by the manufacturer.
NK Cell Proliferation Assay—Purified NK cells were cultured in 96-well flat bottom plates (Nalge Nunc International) at 2 × 106/ml in 0.2 ml/well and stimulated with or without IL-15 in the presence of either 0.5 or 5 μg/ml anti-GITR agonist antibody. All experiments were done in triplicates. Cells were cultured at 37 °C in 5% CO2 for 3 or 5 days. Eight to ten hours before harvest, cells were pulsed with [3H]thymidine (2.5 μCi/ml) (Amersham Biosciences). Cells were then harvested, and [3H]thymidine uptake was measured with a β counter (PerkinElmer Life Sciences). The extent of proliferation was expressed as stimulation index (SI), which is represented by the ratio of radioactive [3H]thymidine (cpm) incorporated by cells treated with the agonist antibody to that by cells without treatment.
SDS-PAGE and Western Blotting—A total of 5 million NK cells were lysed in 100 μl of lysis buffer (50 mm Tris-Cl, 1% Triton X-100, 100 mm NaCl, 2 mm EDTA, 50 mm NaF, 50 mm glycerol-phosphate, 1 mm NaVO4, and 1× protease inhibitor mixture) (Roche Applied Science). Complete cell lysis was achieved by immediately vortexing the cells and then boiling in an equal amount of 2× SDS protein loading buffer at 95 °C for 5 min. Cell debris was removed by centrifugation at 12,000 rpm for 3 min. Twenty microliters of each sample was loaded into a 12% SDS-polyacrylamide gel containing a 4% stacking gel. Immunoblotting was carried out. Primary antibodies of anti-Bcl-XL, anti-cleaved caspase 3, anti-cleaved PARP, anti-phospho-Bad, anti-phospho-Akt, anti-Akt, and anti-phospho-PDK1 were purchased from Cell Signaling Technology (Beverly, MA). Anti-β-actin antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Apoptosis Assay—Apoptotic cells were detected by staining cells with the combination of annexin-V-FITC and Via-probe™ (7-amino-actinomycin D; 7-AAD) according to the manufacturer's instructions (BD Biosciences). Early apoptotic cells (FITC+7-AAD–) and late apoptotic cells (FITC+7-AAD+) were counted for total apoptosis (32). Briefly, 2.5 × 105 cells were incubated with saturating concentrations of annexin V-FITC and 7-AAD for 15 min at room temperature and immediately analyzed by flow cytometry as described above.
Fas Neutralization Assay—Purified NK cells were preincubated with a Fas neutralization antibody, clone ZB4 (10 μg/ml; Upstate Biotechnology, Inc., Lake Placid, NY) for 1 h at 37 °Cin 5% CO2 to allow antibody/cell interaction. Cells were then plated in 6-well plates, precoated with anti-GITR at 10 μg/ml, in the presence or absence of IL-15. After 48 h of incubation, cells were collected for apoptosis assays.
Statistical Analysis—Analysis of NK proliferation was done using independent Student's t test. Analysis of cytokine secretion and apoptosis were performed using Student's t test, paired two samples for means.
RESULTS
Gene Expression Profiling Revealed Predominantly Negative Effects of GITR on Activated Human NK Cells—To assess the impact of GITR on NK cell activation, we used an NF-κB-selective microarray assay to study overall effects of GITR on human NK cells. Purified human NK cells were activated by TLR3 ligand poly(I-C) in the presence or absence of GITR stimulation. Surprisingly, the overall effect of GITR engagement was to inhibit human NK cell activation. Of 113 genes analyzed, 21 genes qualified as differentially regulated by GITR when a typical 2-fold difference standard was applied as cut-off. Among those 21 genes differentially regulated by GITR, only 4 of 21 (19%) genes were up-regulated by GITR, whereas 17 of 21 (81%) genes were suppressed by GITR stimulation (Table 1). It was noteworthy that among the four genes that were up-regulated by GITR, heme oxygenase-1 and IL-10 are both well known protective genes against oxidative stress and inflammatory response (33, 34). Table 1 lists all 21 genes that were differentially regulated by GITR and their -fold changes.
TABLE 1.
Genes differentially regulated by GITR in human NK cells
Purified human NK cells were cultured in the presence or absence of poly(I-C) with or without GITR stimulation. Microarray analysis was performed as described under “Experimental Procedures.” Changes of gene expression were calculated as -fold changes between two groups after normalization on control group (medium alone). The poly(I-C) alone group was calculated as gene expression in the presence of poly(I-C) normalized on medium alone. The GITR + poly(I-C) group was calculated as gene expression of poly(I-C) + GITR group normalized on control group divided by the poly(I-C) alone group. All numbers in the poly(I-C) alone and GITR + poly(I-C) groups are -fold changes.
| Gene symbol | Description | RefSeq number | Poly(I-C) alone | GITR + poly(I-C) |
|---|---|---|---|---|
| Up-regulated by GITR | ||||
| HMOX1 | Heme oxygenase (decycling) 1 | NM_002133 | 1.1 | 11.6 |
| IL-10 | Interleukin 10 | NM_000572 | 0.4 | 6.4 |
| PPP5C | Protein phosphatase 5, catalytic subunit | NM_006247 | 0.5 | 2.0 |
| IL-8 | Interleukin 8 | NM_000584 | 3.1 | 2.0 |
| Down-regulated by GITR | ||||
| GPR89 | G protein-coupled receptor 89 | NM_016334 | 0.5 | 0.4 |
| RHOC | Ras homolog gene family, member C | NM_175744 | 1.6 | 0.4 |
| RHOA | Ras homolog gene family, member A | NM_001664 | 3.5 | 0.4 |
| MYD88 | Myeloid differentiation primary response gene 88 | NM_002468 | 1.5 | 0.4 |
| MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | NM_003954 | 1.3 | 0.3 |
| STAT1 | Signal transducer and activator of transcription 1, 91 kDa | NM_007315 | 1.6 | 0.3 |
| BIRC2 | Baculoviral IAP repeat-containing 2 | NM_001166 | 1.5 | 0.3 |
| TLR8 | Toll-like receptor 8 | NM_016610 | 2.1 | 0.3 |
| TNFAIP3 | Tumor necrosis factor, α-induced protein 3 | NM_006290 | 2.2 | 0.3 |
| RELA | V-rel reticuloendotheliosis viral oncogene homolog A | NM_021975 | 1.6 | 0.3 |
| TLR2 | Toll-like receptor 2 | NM_003264 | 2.0 | 0.2 |
| TRADD | TNFRSF1A-associated via death domain | NM_003789 | 1.6 | 0.2 |
| TNFRSF10B | Tumor necrosis factor receptor superfamily, member 10b | NM_003842 | 1.9 | 0.2 |
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A | NM_001065 | 1.8 | 0.2 |
| IL-1B | Interleukin 1, β | NM_000576 | 4.4 | 0.2 |
| C3 | Complement component 3 | NM_000064 | 2.7 | 0.1 |
| CCL2 | Chemokine (C-C motif) ligand 2 | NM_002982 | 3.5 | 0.1 |
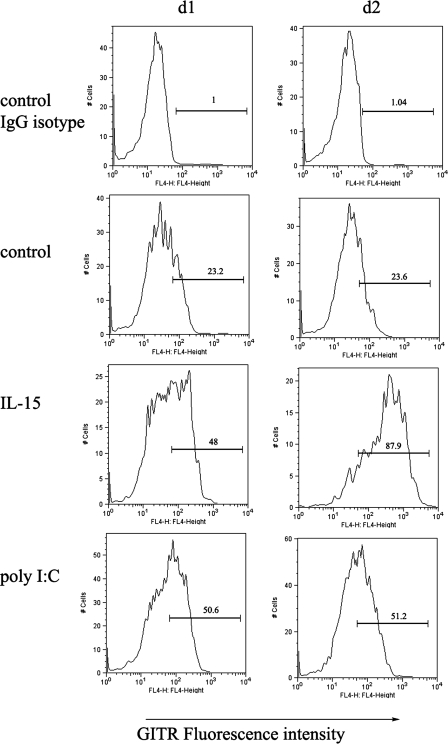
GITR Expression Is Up-regulated by either TLR Ligand Poly(I-C) or NK Cell Growth Factor IL-15—We stimulated NK cells with either TLR3 ligand poly(I-C) or IL-15 to test their ability and effectiveness at regulating GITR expression in activated NK cells. Human PBMCs were treated with IL-15 or poly(I-C) as described, and GITR expression in CD3–CD56+ NK cells was monitored by flow cytometry. As shown in Fig. 1, there was constitutively low GITR expression (23%) in resting human primary NK cells (Fig. 1, control) compared with nonspecific staining of an isotype-matched biotinylated control antibody (Fig. 1, control IgG isotype), which is consistent with a recent report (19). In addition, both poly(I-C) and IL-15 up-regulated GITR expression in NK cells by more than 2-fold over control (medium versus stimuli). Interestingly, the up-regulation of GITR by poly(I-C) seemed more rapid than that seen with IL-15, since GITR expression plateaued after 24 h of stimulation by poly(I-C), whereas it took 48 h to reach a plateau following IL-15 stimulation. Same staining has been repeated in three donors, and similar results have been obtained.
FIGURE 1.
GITR expression was up-regulated in activated NK cells. Purified PBMCs were cultured in the presence or absence of either 10 ng/ml IL-15 or 25 μg/ml poly(I-C) for 1 or 2 days. Cells were collected; stained with anti-human FITC-CD3, PE-CD56, and anti-GITR antibody; and analyzed by flow cytometry. The x axis depicts relative fluorescence intensity (expression level of GITR), whereas the y axis represents relative cell numbers. IgG isotype control indicates that the cells without treatment were stained with nonspecific isotypematched biotinylated control antibody. Control indicates that the cells without treatment were stained with 5 μg/ml biotinylated anti-human GITR antibody to show constitutive expression of GITR.
GITR Inhibited NK Cell Proliferation and Production of Proinflammatory Cytokines upon Activation—We next examined NK cell proliferation in response to IL-15, with or without GITR engagement (Fig. 2). An agonist monoclonal antibody (clone 110416) against GITR was used to stimulate NK cells. IL-15, along with 0.5 μg/ml or 5 μg/ml anti-GITR antibody, was included in the culture medium for 3 or 5 days. As shown in Fig. 2A, after 3 days, the mean stimulation index was significantly less in the anti-GITR-treated groups (24.2 ± 1.9 for 0.5 μg/ml anti-GITR (p < 0.01) and 18.7 ± 4.0 for 5 μg/ml anti-GITR (p < 0.05)) when compared with cultures receiving IL-15 and an irrelevant mouse IgG negative control (34.6 ± 3.3), suggesting that GITR activation inhibited IL-15-induced NK proliferation. Similarly, in the day 5 treatment group (Fig. 2B), the stimulation index for the irrelevant IgG control (41.1 ± 0.8) and anti-GITR antibody treatment of 0.5 μg/ml (32.6 ± 0.9) and 5 μg/ml (26.2 ± 2.4) showed similar patterns.
FIGURE 2.
GITR inhibited NK proliferation. Stimulation index of NK cells was used to evaluate NK proliferation responses to IL-15 and GITR stimulation. An agonist monoclonal antibody of GITR was used to stimulate GITR. 10 ng/ml IL-15 or 0.5 and 5 μg/ml soluble GITR antibody were added into medium when cells were plated. After 3 days (A) and 5 days (B) of incubation, [3H]thymidine assay was performed, and the stimulation index was calculated and represented as mean ± S.E. (*, p < 0.05; **, p < 0.01).
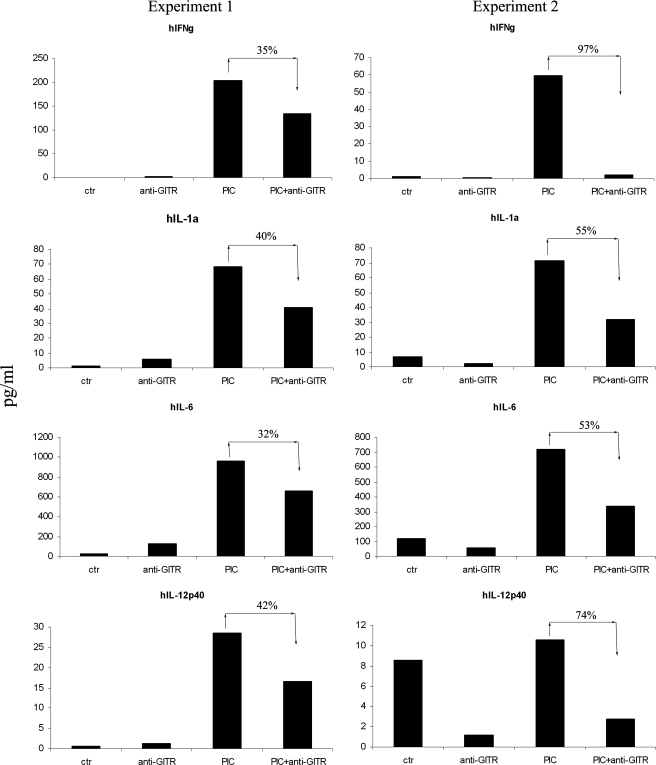
Secretion of proinflammatory cytokines is one of the hallmarks for NK cell activation and its functions. To further understand the functional impact of GITR on NK cell activation, culture supernatants of NK cells, stimulated in the presence or absence of GITR, were analyzed. Fig. 3 showed two sets of data side by side; GITR engagement significantly reduced poly(I-C)-induced production of proinflammatory cytokines (e.g. IFN-γ and IL-6 (p = 0.03 in both groups)). Productions of IL-12 and IL-1α were also slightly reduced (p = 0.06 and p = 0.05 separately). There was no significant difference of cytokine production between GITR engagement compared with control group (IFN-γ, p = 0.5; IL-1a, p = 0.49; IL-6, p = 0.43; IL-12p40, p = 0.28).
FIGURE 3.
GITR inhibited the production of proinflammatory cytokines upon activation. Purified NK cells were stimulated by poly(I-C) in the presence or absence of GITR stimulation. Two sets of data are presented side by side. In each experiment, each treatment was done in triplicates. After 48 h of culture, the triplicates were pooled, and culture supernatants were collected for cytokine analysis. The y axis represents cytokine concentration (pg/ml). Suppression percentage in each treatment was also shown.
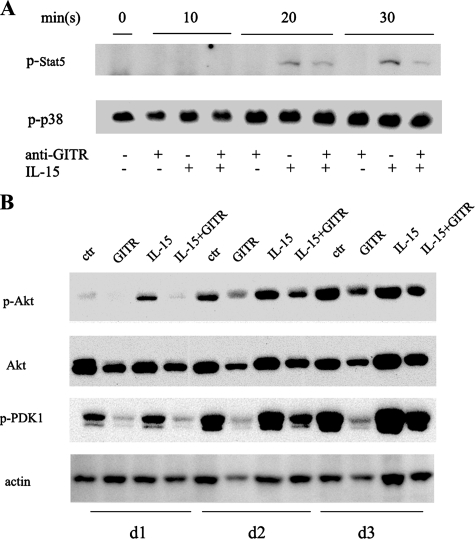
GITR Engagement Blocked Phosphorylation of Stat5 and Akt but Did Not Affect the Mitogen-activated Protein Kinase Pathway—To investigate possible molecular mechanisms for the antiproliferative effect of GITR on activated NK cells, we next examined the signaling pathways of IL-15 on NK cells that may have been affected by the engagement of GITR, such as the Jak3/Stat5 and the phosphatidylinositol 3-kinase/Akt pathways. As shown in Fig. 4A, GITR activation abrogated IL-15-induced Stat5 phosphorylation at time points of 20 and 30 min following treatments. As shown in Fig. 4B, IL-15 induced Akt phosphorylation at 1 and 2 days after the initiation of treatments. GITR activation abrogated IL-15-induced phosphorylation. Consistent with the Akt results, another molecule in the phosphatidylinositol 3-kinase pathway, the phosphorylation of PDK1 induced by IL-15, was also abrogated by GITR activation. However, there was little change in phosphorylated p38 (Fig. 4A) or ERK1/2 (data not shown).
FIGURE 4.
A, GITR stimulation decreased phosphorylation of Stat5 in IL-15-treated NK cells. Purified NK cells were cultured in the presence/absence of 10 ng/ml IL-15 in plates coated with the control IgG or anti-GITR antibody for 10, 20, and 30 min. Cells were collected and processed for Western blot analysis using anti-p-Stat5 and anti-p-p38 antibodies. B, GITR stimulation decreased phosphorylation of Akt and PDK1 in IL-15-treated NK cells. Purified NK cells were cultured in the presence/absence of 10 ng/ml IL-15 in plates coated with control IgG or anti-GITR antibody for 1, 2, or 3 days. Cells were collected and processed for Western blot analysis using anti-p-Akt, anti-Akt, or anti-p-PDK1 antibodies.
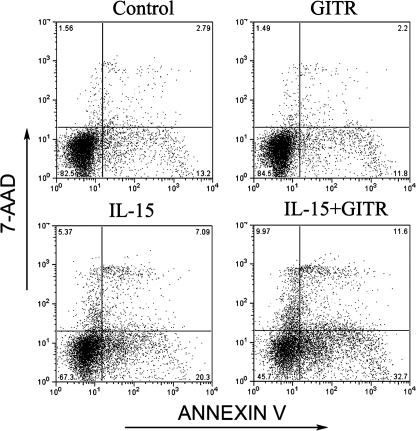
GITR Activation Induced NK Cell Apoptosis—Earlier studies suggested that GITR prevents T cells from undergoing T cell receptor-induced apoptosis (14). Therefore, we wanted to determine if GITR engagement affects apoptosis of activated NK cells. NK cells stimulated with either poly(I-C) or IL-15 were stained with FITC-annexin V in conjunction with 7-AAD and subsequently analyzed by flow cytometry. This method allows discrimination between living (FITC–7-AAD–), early apoptotic (FITC+7-AAD–), and late apoptotic cells (FITC+7-AAD+) (32). Fourteen normal donors were used for IL-15 treatment, and six normal donors were used for poly(I-C) treatment. As shown in Tables 2 and 3, we summarized the apoptosis information of 20 donors that have been analyzed. Briefly, after 48 h, it showed a significant increase of apoptosis compared with IL-15/poly(I-C) plus GITR engagement with IL-15/poly(I-C) treatment only (p = 0.0016 < 0.01 in the IL-15 group and p = 0.04 < 0.05 in the poly(I-C) group). GITR activation in 10 of 14 donors (71.5%) in the IL-15 treatment group and four of six (66.7%) in the poly(I-C) treatment group showed more apoptosis if we used 5% more apoptosis as a cut-off line. Fig. 5 shows a typical flow cytometry analysis for NK cell apoptosis. Early apoptotic cells (FITC+7-AAD–) and late apoptotic cells (FITC+7-AAD+) were counted for total apoptosis (32).
TABLE 2.
Apoptosis occurred in IL-15 and IL-15 plus GITR treatment groups of human NK cells
Control, NK cells in culture without treatment; GITR, NK cells treated with GITR engagement for 2 days; IL-15, NK cells treated with 10 ng/ml IL-15 for 2 days; IL-15 + GITR, NK cells treated with IL-15 and GITR engagement for 2 days.
| Donor number | Control | GITR | IL-15 | IL-15 + GITRa |
|---|---|---|---|---|
| 1 | 28.9 | 55.04 | 57 | 63.45 |
| 2 | 16.02 | 14.33 | 28.12 | 46.3 |
| 3 | 27.85 | 32.47 | 33.6 | 48.6 |
| 4 | 42.5 | 46 | 50.1 | 63.1 |
| 5 | 18.87 | 8.56 | 9.53 | 13.85 |
| 6 | 65.7 | 67.1 | 71.9 | 78.4 |
| 7 | 54.49 | 77.09 | 54.4 | 81.69 |
| 8 | 34.61 | 72.19 | 44.2 | 88.64 |
| 9 | 50 | 52.58 | 66.3 | 75.63 |
| 10 | 36.7 | 35.3 | 39.2 | 41.5 |
| 11 | 25.66 | 23.72 | 33.7 | 40.6 |
| 12 | 41 | 45.5 | 43.7 | 51.7 |
| 13 | 62 | 55.28 | 69.9 | 74.7 |
| 14 | 56.3 | 49.2 | 37.3 | 40.9 |
Significant increase occurred in IL-15 plus anti-GITR treatment group compared with IL-15 treatment only (p < 0.01).
TABLE 3.
Apoptosis occurred in poly(I-C) and poly(I-C) plus GITR treatment groups of human NK cells
Control, NK cells in culture without treatment; GITR, NK cells treated with GITR engagement for 2 days; IL-15, NK cells treated with 25 μg/ml poly(I-C) for 2 days; poly(I-C) + GITR, NK cells treated with poly(I-C) and GITR engagement for 2 days.
| Donor number | Control | GITR | Poly(I-C) | Poly(I-C) + GITRa |
|---|---|---|---|---|
| 1 | 42.2 | 44.4 | 39.9 | 48.7 |
| 2 | 57.9 | 59.9 | 43.9 | 63.47 |
| 3 | 84.45 | 82.69 | 72.65 | 79.63 |
| 4 | 47.3 | 63.08 | 42.21 | 61.93 |
| 5 | 46.6 | 95.98 | 48 | 91.34 |
| 6 | 59.33 | 63.16 | 59.39 | 61.36 |
Significant increase occurred in poly(I-C) plus anti-GITR treatment group compared with poly(I-C) treatment only (p < 0.05).
FIGURE 5.
GITR engagement enhanced cytokine-induced NK cell apoptosis. A typical flow cytometry analysis for apoptosis is shown. Purified NK cells were treated with or without 10 ng/ml IL-15/or 25 μg/ml poly(I-C) in the presence or absence of GITR stimulation for 48 h. Samples were stained for FITC-annexin V in conjunction with 7-AAD. Staining for FITC–7-AAD–, FITC+7-AAD–, or FITC+7-AAD+ represents living, early apoptotic, and late apoptotic cells, respectively.
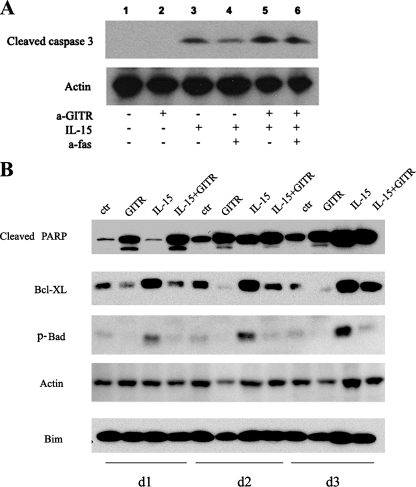
GITR-enhanced Apoptosis Is Independent of the Fas-FasL Pathway but Mediated by a Mitochondria-dependent Pathway—Fas-FasL interaction is responsible for cytokine-induced T cell death (35, 36). To test whether the Fas-FasL pathway causes GITR-induced NK cell apoptosis, purified NK cells were preincubated with a Fas-neutralizing antibody before IL-15 treatment and GITR engagement. As shown in Fig. 6A, consistent with our finding that GITR enhances cytokine-induced apoptosis, the level of activated/cleaved caspase 3 for IL-15 plus GITR-treated NK cells was greatly increased when compared with the IL-15 treatment group (Fig. 6A, lane 5 versus lane 3). After Fas was blocked, the level of cleaved caspase 3 was dramatically decreased in the IL-15 treatment group (Fig. 6A, lane 4 versus lane 3), indicating that Fas-FasL interaction contributed to IL-15-induced cell death of NK cells. However, Fas blockade did not further block caspase 3 cleavage induced by GITR engagement (Fig. 6A, lane 5 versus lane 6), indicating that Fas-FasL interaction is not involved in the enhanced apoptosis seen with GITR engagement. FITC-annexin V, in conjunction with 7-AAD staining, showed similar results (data not shown).
FIGURE 6.
GITR-induced apoptosis is independent of the Fas-FasL pathway but involves the mitochondrial pathway. A, Fas neutralization antibody did not block GITR enhancement of apoptosis. Purified NK cells were preincubated with Fas neutralization antibody, 10 μg/ml ZB4, for 1 h and plated in 6-well plates precoated with anti-GITR in the presence or absence of 10 ng/ml IL-15. After 48 h, cells were collected for Western blot analysis using anti-cleaved caspase 3 and anti-actin antibodies. B, GITR engagement decreased Bcl-XL and p-Bad protein level but increased cleaved PARP in IL-15-treated NK cells. Purified NK cells were cultured in the presence/absence of 10 ng/ml IL-15 in plates coated with control IgG and anti-GITR antibody. After 2 days, cells were collected and processed for Western blot analysis using the indicated antibodies.
We next examined the activation status of essential apoptosis signaling molecules. Bcl-2 family members play a central role in mitochondria-mediated apoptosis (37). The proapoptotic protein, Bad, which in its dephosphorylated state is able to interact with and inactivate the antiapoptotic proteins, Bcl-2 and Bcl-XL, resulted in the release of mitochondrial cytochrome c (38, 39). Phosphorylation of Bad at serine residue 112 prevents its interaction with Bcl-2 and Bcl-XL, preventing mitochondrial damage and the release of cytochome c, subsequently reducing cell apoptosis (40). In this study, we detected the expression of mitochondria-related apoptosis molecules, such as cleaved PARP, Bcl-XL, and phospho-Bad, following GITR stimulation. We did Western blots to detect apoptotic signals in four donors from 10 donors showing apoptosis of the IL-15 treatment group. All of these four donors showed similar expression patterns of apoptotic signals. Fig. 6, A and B, showed two independent Western blots for apoptosis analysis from two different donors. Compared with the untreated control, cleaved PARP expressions in GITR and GITR plus IL-15 groups were significantly increased with concomitant decreases in the levels of Bcl-XL and phospho-Bad (Fig. 6B). The IL-15 treatment group had obvious expression of both Bcl-XL and phospho-Bad. However, following GITR engagement, the expression of Bcl-XL and phospho-Bad was abrogated in IL-15-treated cultures.
DISCUSSION
Recently, the role of GITR in the development and pathophysiology of T cell immune responses has been actively explored. Most of the evidence comes from the functional studies of murine GITR. GITR activation, either by anti-GITR antibody or by its natural ligand GITRL, increases TCR-induced T cell proliferation and cytokine production and rescues T cells from anti-CD3-induced apoptosis (7). GITR engagement can also inhibit T regulatory cells and/or render effector T cells more resistant to T regulatory cell-mediated suppression (16, 41). Therefore, GITR-GITRL interactions are thought to potentiate T cell activation and function (14, 15, 41–43). Consistent with the co-stimulatory role of GITR for T cells and T cell immune responses, a series of studies has shown that manipulation of the GITR-GITRL interaction can be an effective strategy for immunotherapies for tumors, viral infections, and autoimmune diseases (22–30). Recent studies have suggested that GITR is also expressed in the innate immune system and can modulate the functions of innate immune response (13, 17). However, the role of GITR on NK cells remains controversial. Earlier reports suggested that the expression of GITRL on plasmacytoid dendritic cell precursors enhanced NK cell function by interacting with GITR expressed on NK cells (19). Recently, Baltz et al. (18) reported that engagement of GITR on NK cells with GITRL expressed on tumor cells inhibited both cytotoxic responses and the secretion of proinflammatory cytokine, IFN-γ, by NK cells.
We investigated the role of GITR in regulation of human NK cell function. Several lines of evidence indicate that GITR primarily signals through the NF-KB pathway (7, 18, 42, 43), which prompted us to use an NF-κB-selective cDNA microarray to globally assess the potential roles of GITR on human NK cells. Surprisingly, 17 of 21 (81%) genes, differentially regulated by GITR stimulation, were down-regulated, whereas only 4 of 21 (19%) of genes were up-regulated (Table 1). More intriguingly, among the four up-regulated genes, both heme oxygenase 1 and IL-10 are well established for their protective roles against oxidative stress and inflammation (33, 34). Our findings that GITR has a globally suppressive effect on the NF-κB pathway and that it can up-regulate heme oxygenase-1 and IL-10 genes not only assert our proposal, along with the recent studies by Balz et al. (18) that GITR negatively regulates NK activation, but also may point to new directions to further dissect molecular mechanisms of the biological roles of GITR. We are currently investigating the mechanisms and impact of GITR in up-regulation of heme oxygenase-1 and IL-10.
NK cells initiate the first line of defense against infection by sensing conserved microbial structures through TLRs. Although IL-15 is a well established NK cell activation and growth factor, TLR stimulation has also been shown to potently activate NK cells (44, 45). We have shown that GITR is constitutively expressed at low levels on human primary NK cells but up-regulated upon activation by both TLR ligand and NK cell growth factor, IL-15 (Fig. 1). This observation strongly suggests that GITR is an activation marker for human NK cells. However, in agreement with cDNA microarray data, GITR activation led to inhibition of NK cell proliferation in response to IL-15 (Fig. 2) and increased apoptosis (Tables 2 and 3). This apparently causes suppression of NK cell activation. Collectively, our results indicate that GITR on activated NK cells primarily serves as negative regulator of NK cell activation.
The underlying mechanism for the antiproliferative effect of GITR activation seems multifaceted. We observed decreased phosphorylation of Stat5 protein (Fig. 4A), which is consistent with the notion that GITR directly blocks IL-15 signaling. However, we also observed decreased phosphorylation of Akt (Fig. 4B), which is not known to directly associate with the Stat5 signaling pathway but is an essential component for the phosphatidylinositol 3-kinase and cell survival (46). Our observation of the antagonistic effect of GITR to both Stat5 and Akt revealed the potential molecular mechanisms of antiproliferative effects of GITR. Furthermore, consistent with what has been reported by Balz et al. (18), that GITR-GITRL engagement suppressed NK cell cytotoxicity and interferon γ secretion, GITR activation decreased the production of poly(I-C)-induced proinflammatory cytokines (e.g. IFN-γ, IL-1α, IL-6, and IL-12). Our results further showed that GITR negatively regulated NK cell activation.
Our data also suggested that GITR suppresses NK cell activation and function by enhancing NK cell apoptosis. Earlier studies suggested that constitutive expression of GITR decreased T cell activation induced cell death (7), whereas a later study showed that GITR stimulation differentially regulates T cell survival (47). It enhances alloreactive CD8+CD25– T cell proliferation, whereas it decreases allo-reactive CD4+CD25– T cell proliferation. Allo-stimulated CD4+CD25– cells show increased apoptosis upon GITR stimulation that is dependent on the Fas-FasL pathway (47). However, we showed that Fas-FasL interaction was responsible for cytokine (IL-15)-induced activation-induced cell death of NK cells (Fig. 6A) but not for GITR-induced enhancement of cell death (Fig. 6A), suggesting that cytokine-induced activation-induced cell death and GITR-induced enhancement of apoptosis were mediated through different mechanisms. Bcl-2 family members play a central role in the regulation of mitochondria-mediated apoptosis. The ratio of proapoptotic Bcl-2 family members (e.g. Bax and Bad) to antiapoptotic family members (e.g. Bcl-2 and Bcl-XL) determines mitochondrial membrane permeabilization and apoptosis occurrence (48). We observed that cytokines (IL-15) increased Bcl-XL and phospho-Bad protein levels, which supports the report of IL-15 promoting NK cell survival by maintaining Bcl-2 and Bcl-XL protein expression (49), whereas GITR activation abrogated Bcl-XL protein expression (Fig. 6B), suggesting that the mitochondrial pathway is involved in GITR-induced apoptosis enhancement.
We studied the apoptotic effect of GITR on human NK cells. Our results are based on consecutive studies on 20 human donors, and we showed consistently a predominantly suppressive effect of GITR engagement on the activation of human NK cells despite donor-to-donor variation (Tables 2 and 3). This finding highlights the human heterogeneity and may partially explain some of the discrepancies among studies using either mice or human tissues (18, 19). On the other hand, GITR and GITRL have been reported to be expressed on a variety of cell types. GITRL is constitutively expressed on dendritic cells, B cells, and macrophages. Although GITR is considered as a co-stimulator in T cell immune responses, a more recent study demonstrated that reverse signaling through GITRL after engagement by soluble GITR in mouse plasmacytoid dendritic cells played a role in the anti-inflammatory action by the induction of indoleamine 2,3-dioxygenase (50). This provides strong evidence that GITR-GITRL interaction may play a much broader role than originally recognized. NK cells are important components of innate immune response. The interactions between NK cells with GITRL-expressing cells and the presence of reverse signaling probably contribute to the function and homeostasis of NK cells during inflammation. Our data, by directly stimulating GITR in NK cells, along with the results of Baltz et al. (18) through co-culture of NK cells with GITRL-expressing tumor cells, suggested that GITR negatively regulated NK function.
In summary, our work demonstrated that although GITR is an activation marker of NK cells, it functions as a negative regulator of NK cell activation. This study revealed a novel function of GITR in regulating NK cell function and may facilitate a better understanding of the role of GITR in regulating innate and adaptive immune responses.
Acknowledgments
We thank Drs. Igal Gery and Bobbie Austin for helpful comments during the preparation of the manuscript.
This research was supported by the Intramural Research Program of NEI, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NK, natural killer; GITR, glucocorticoid-induced tumor necrosis factor receptor; GITRL, GITR ligand; IFN, interferon; PBMC, peripheral blood mononuclear cell; IL, interleukin; FITC, fluorescein isothiocyanate; 7-AAD, 7-amino-actinomycin D; TLR, Toll-like receptor.
References
- 1.Chan, S. H., Perussia, B., Gupta, J. W., Kobayashi, M., Pospisil, M., Young, H. A., Wolf, S. F., Young, D., Clark, S. C., and Trinchieri, G. (1991) J. Exp. Med. 173 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bancroft, G. J., and Kelly, J. P. (1994) Immunobiology 191 424–431 [DOI] [PubMed] [Google Scholar]
- 3.Marcenaro, E., Ferranti, B., and Moretta, A. (2005) Autoimmun. Rev. 4 520–525 [DOI] [PubMed] [Google Scholar]
- 4.Shi, F. D., and Van Kaer, L. (2006) Nat. Rev. Immunol. 6 751–760 [DOI] [PubMed] [Google Scholar]
- 5.Karre, K., Ljunggren, H. G., Piontek, G., and Kiessling, R. (1986) Nature 319 675–678 [DOI] [PubMed] [Google Scholar]
- 6.Lanier, L. L. (2003) Curr. Opin. Immunol. 15 308–314 [DOI] [PubMed] [Google Scholar]
- 7.Nocentini, G., Giunchi, L., Ronchetti, S., Krausz, L. T., Bartoli, A., Moraca, R., Migliorati, G., and Riccardi, C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6216–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji, H. B., Liao, G., Faubion, W. A., Abadia-Molina, A. C., Cozzo, C., Laroux, F. S., Caton, A., and Terhorst, C. (2004) J. Immunol. 172 5823–5827 [DOI] [PubMed] [Google Scholar]
- 9.Nocentini, G., and Riccardi, C. (2005) Eur. J. Immunol. 35 1016–1022 [DOI] [PubMed] [Google Scholar]
- 10.Watts, T. H. (2005) Annu. Rev. Immunol. 23 23–68 [DOI] [PubMed] [Google Scholar]
- 11.Gurney, A. L., Marsters, S. A., Huang, R. M., Pitti, R. M., Mark, D. T., Baldwin, D. T., Gray, A. M., Dowd, A. D., Brush, A. D., Heldens, A. D., Schow, A. D., Goddard, A. D., Wood, W. I., Baker, K. P., Godowski, P. J., and Ashkenazi, A. (1999) Curr. Biol. 9 215–218 [DOI] [PubMed] [Google Scholar]
- 12.Kwon, B., Yu, K. Y., Ni, J., Yu, G. L., Jang, I. K., Kim, Y. J., Xing, L., Liu, D., Wang, S. X., and Kwon, B. S. (1999) J. Biol. Chem. 274 6056–6061 [DOI] [PubMed] [Google Scholar]
- 13.Li, Z., Mahesh, S. P., Kim, B. J., Buggage, R. R., and Nussenblatt, R. B. (2003) J. Autoimmun. 21 83–92 [DOI] [PubMed] [Google Scholar]
- 14.Ronchetti, S., Nocentini, G., Riccardi, C., and Pandolfi, P. P. (2002) Blood 100 350–352 [DOI] [PubMed] [Google Scholar]
- 15.Ronchetti, S., Zollo, O., Bruscoli, S., Agostini, M., Bianchini, R., Nocentini, G., Ayroldi, E., and Riccardi, C. (2004) Eur. J. Immunol. 34 613–622 [DOI] [PubMed] [Google Scholar]
- 16.Shimizu, J., Yamazaki, S., Takahashi, T., Ishida, Y., and Sakaguchi, S. (2002) Nat. Immunol. 3 135–142 [DOI] [PubMed] [Google Scholar]
- 17.Shin, H. H., Lee, M. H., Kim, S. G., Lee, Y. H., Kwon, B. S., and Choi, H. S. (2002) FEBS Lett. 514 275–280 [DOI] [PubMed] [Google Scholar]
- 18.Baltz, K. M., Krusch, M., Bringmann, A., Brossart, P., Mayer, F., Kloss, M., Baessler, T., Kumbier, I., Peterfi, A., Kupka, S., Kroeber, S., Menzel, D., Radsak, M. P., Rammensee, H. G., and Salih, H. R. (2007) FASEB J. 21 2442–2454 [DOI] [PubMed] [Google Scholar]
- 19.Hanabuchi, S., Watanabe, N., Wang, Y. H., Wang, Y. H., Ito, T., Shaw, J., Cao, W., Qin, F. X., and Liu, Y. J. (2006) Blood 107 3617–3623 [DOI] [PubMed] [Google Scholar]
- 20.Mackay, F., and Kalled, S. L. (2002) Curr. Opin. Immunol. 14 783–790 [DOI] [PubMed] [Google Scholar]
- 21.Tuyaerts, S., Van Meirvenne, S., Bonehill, A., Heirman, C., Corthals, J., Waldmann, H., Breckpot, K., Thielemans, K., and Aerts, J. L. (2007) J. Leukocyte Biol. 82 93–105 [DOI] [PubMed] [Google Scholar]
- 22.Bushell, A., and Wood, K. (2007) Am. J. Transplant. 7 759–768 [DOI] [PubMed] [Google Scholar]
- 23.Cohen, A. D., Diab, A., Perales, M. A., Wolchok, J. D., Rizzuto, G., Merghoub, T., Huggins, D., Liu, C., Turk, M. J., Restifo, N. P., Sakaguchi, S., and Houghton, A. N. (2006) Cancer Res. 66 4904–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J., Choi, W. S., Kim, H. J., and Kwon, B. (2006) Exp. Mol. Med. 38 94–99 [DOI] [PubMed] [Google Scholar]
- 25.Ko, H. J., Kim, Y. J., Kim, Y. S., Chang, W. S., Ko, S. Y., Chang, S. Y., Sakaguchi, S., and Kang, C. Y. (2007) Cancer Res. 67 7477–7486 [DOI] [PubMed] [Google Scholar]
- 26.Kohm, A. P., Williams, J. S., and Miller, S. D. (2004) J. Immunol. 172 4686–4690 [DOI] [PubMed] [Google Scholar]
- 27.La, S., Kim, E., and Kwon, B. (2005) Exp. Mol. Med. 37 193–198 [DOI] [PubMed] [Google Scholar]
- 28.Patel, M., Xu, D., Kewin, P., Choo-Kang, B., McSharry, C., Thomson, N. C., and Liew, F. Y. (2005) Eur. J. Immunol. 35 3581–3590 [DOI] [PubMed] [Google Scholar]
- 29.Ramirez-Montagut, T., Chow, A., Hirschhorn-Cymerman, D., Terwey, T. H., Kochman, A. A., Lu, S., Miles, R. C., Sakaguchi, S., Houghton, A. N., and van den Brink, M. R. (2006) J. Immunol. 176 6434–6442 [DOI] [PubMed] [Google Scholar]
- 30.Suvas, S., Kim, B., Sarangi, P. P., Tone, M., Waldmann, H., and Rouse, B. T. (2005) J. Virol. 79 11935–11942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Z., Lim, W. K., Mahesh, S. P., Liu, B., and Nussenblatt, R. B. (2005) J. Immunol. 174 5187–5191 [DOI] [PubMed] [Google Scholar]
- 32.Lecoeur, H., Ledru, E., Prevost, M. C., and Gougeon, M. L. (1997) J. Immunol. Methods 209 111–123 [DOI] [PubMed] [Google Scholar]
- 33.Moore, K. W., de Waal Malefyt, R., Coffman, R. L., and O'Garra, A. (2001) Annu. Rev. Immunol. 19 683–765 [DOI] [PubMed] [Google Scholar]
- 34.Otterbein, L. E., Soares, M. P., Yamashita, K., and Bach, F. H. (2003) Trends Immunol. 24 449–455 [DOI] [PubMed] [Google Scholar]
- 35.Green, D. R., Droin, N., and Pinkoski, M. (2003) Immunol. Rev. 193 70–81 [DOI] [PubMed] [Google Scholar]
- 36.Lenardo, M. J. (1996) J. Exp. Med. 183 721–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz, M., Andrade-Navarro, M. A., and Gross, A. (2007) Apoptosis 12 869–876 [DOI] [PubMed] [Google Scholar]
- 38.Goodsell, D. S. (2004) Oncologist 9 226–227 [DOI] [PubMed] [Google Scholar]
- 39.Yang, E., Zha, J., Jockel, J., Boise, L. H., Thompson, C. B., and Korsmeyer, S. J. (1995) Cell 80 285–291 [DOI] [PubMed] [Google Scholar]
- 40.Chan, P. H. (2004) Neurochem. Res. 29 1943–1949 [DOI] [PubMed] [Google Scholar]
- 41.Kanamaru, F., Youngnak, P., Hashiguchi, M., Nishioka, T., Takahashi, T., Sakaguchi, S., Ishikawa, I., and Azuma, M. (2004) J. Immunol. 172 7306–7314 [DOI] [PubMed] [Google Scholar]
- 42.Esparza, E. M., and Arch, R. H. (2004) Cell Mol. Life Sci. 61 3087–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esparza, E. M., and Arch, R. H. (2005) J. Immunol. 174 7869–7874 [DOI] [PubMed] [Google Scholar]
- 44.Chalifour, A., Jeannin, P., Gauchat, J. F., Blaecke, A., Malissard, M., N′Guyen, T., Thieblemont, N., and Delneste, Y. (2004) Blood 104 1778–1783 [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, K. N., Leung, B., Kwong, M., Zarember, K. A., Satyal, S., Navas, T. A., Wang, F., and Godowski, P. J. (2004) J. Immunol. 172 138–143 [DOI] [PubMed] [Google Scholar]
- 46.Parry, R. V., Riley, J. L., and Ward, S. G. (2007) Trends Immunol. 28 161–168 [DOI] [PubMed] [Google Scholar]
- 47.Muriglan, S. J., Ramirez-Montagut, T., Alpdogan, O., Van Huystee, T. W., Eng, J. M., Hubbard, V. M., Kochman, A. A., Tjoe, K. H., Riccardi, C., Pandolfi, P. P., Sakaguchi, S., Houghton, A. N., and Van Den Brink, M. R. (2004) J. Exp. Med. 200 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hail, N., Jr. (2005) Apoptosis 10 687–705 [DOI] [PubMed] [Google Scholar]
- 49.Carson, W. E., Fehniger, T. A., Haldar, S., Eckhert, K., Lindemann, M. J., Lai, C. F., Croce, C. M., Baumann, H., and Caligiuri, M. A. (1997) J. Clin. Invest. 99 937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grohmann, U., Volpi, C., Fallarino, F., Bozza, S., Bianchi, R., Vacca, C., Orabona, C., Belladonna, M. L., Ayroldi, E., Nocentini, G., Boon, L., Bistoni, F., Fioretti, M. C., Romani, L., Riccardi, C., and Puccetti, P. (2007) Nat. Med. 13 579–586 [DOI] [PubMed] [Google Scholar]