Abstract
Trypanosoma cruzi is the protozoan parasite that causes Chagas disease or American trypanosomiasis. Kinetoplastid parasites could be considered as model organisms for studying factors involved in posttranscriptional regulation because they control gene expression almost exclusively at this level. The PUF (Pumilio/FBF1) protein family regulates mRNA stability and translation in eukaryotes, and several members have been identified in trypanosomatids. We used a ribonomic approach to identify the putative target mRNAs associated with TcPUF6, a member of the T. cruzi PUF family. TcPUF6 is expressed in discrete sites in the cytoplasm at various stages of the parasite life cycle and is not associated with the translation machinery. The overexpression of a tandem affinity purification-tagged TcPUF6 protein allowed the identification of associated mRNAs by affinity purification assays and microarray hybridization yielding nine putative target mRNAs. Whole expression analysis of transfected parasites showed that the mRNAs associated with TcPUF6 were down-regulated in populations overexpressing TcPUF6. The association of TcPUF6 with the TcDhh1 helicase in vivo and the cellular co-localization of these proteins in epimastigote forms suggest that TcPUF6 promotes degradation of its associated mRNAs through interaction with RNA degradation complexes. Analysis of the mRNA levels of the putative TcPUF6-regulated genes during the parasite life cycle showed that their transcripts were up-regulated in metacyclic trypomastigotes. In these infective forms no co-localization between TcPUF6 and TcDhh1 was observed. Our results suggest that TcPUF6 regulates the half-lives of its associated transcripts via differential association with mRNA degradation complexes throughout its life cycle.
The kinetoplastid parasite Trypanosoma cruzi is the causative agent of Chagas disease or American trypanosomiasis, which affects several million people in South and Central America. During its life cycle, T. cruzi invades two different hosts: a reduviid insect and a mammal. The parasite has at least four distinct and well defined developmental stages: replicative epimastigotes, the infective metacyclic and cellular trypomastigotes, and replicative intracellular amastigotes (1). The developmental forms differ in terms of the environment they inhabit, metabolic activity, and ability to invade different host cells. Consequently, tight regulation of gene expression is required to allow the rapid adaptation essential for parasite survival. Gene expression in trypanosomes differs from that of higher eukaryotes and involves unusual mechanisms, such as polycistronic transcription, the editing of mitochondrial transcripts, and the addition of a 5′ mini exon sequence by trans-splicing and of the 3′ poly(A) tail to mRNA in a coupled reaction. No canonical RNA polymerase II promoter has yet been identified, and there is no clear evidence of transcriptional regulation for protein coding genes (2). Because the individual genes present in a given polycistronic unit may show different temporal expression patterns, the regulation of gene expression in trypanosomes is thought to be posttranscriptional (2, 3).
In eukaryotes, posttranscriptional regulation mechanisms allow rapid and efficient responses to environmental changes and physiological conditions. Detailed characterization of the regulatory processes involved and of the various components of the posttranscriptional machinery is needed. However, such studies have been hampered by the need to separate the effects of posttranscriptional control from the mechanisms regulating transcription initiation. Trypanosomatids, which lack transcriptional regulation, provide an excellent noise-free model for studying the posttranscriptional control of gene expression, the gene regulatory networks controlled at this level, and the macromolecular machineries involved. Posttranscriptional regulation involves specific interactions between regulatory trans-acting factors and conserved cis-elements present in the 5′- and 3′-untranslated regions (UTR)2 of the transcripts (4). RNA-binding proteins bind to sequence-specific and/or structural elements in the UTR regions of functionally related mRNAs, modulating their expression (5, 6). RNA-binding proteins can be classified into different families according to their structural features. The PUF (Pumilio/FBF1) family of posttranscriptional regulators is widespread among eukaryotes and has been the focus of intense studies in recent years. PUF proteins are defined by the presence of a Pumilio domain consisting of eight imperfect repeats and carboxyl- and amino-terminally flanking regions. Each repeat folds into three α-helices, which face the internal side of a rainbow-shaped structure. RNA binds to the α-helices present on the concave surface of the protein, this RNA recognition system being highly modular (7). The recognition motif has been identified for Drosophila, yeast, Caenorhabditis elegans, mouse, and human PUF proteins. Most PUF family proteins bind sequences containing an UGUR core motif, with the flanking sequences controlling the target specificity of each PUF protein (7–9). PUF proteins regulate mRNA stability and translation by enhancing the deadenylation and subsequent degradation of mRNAs or repressing translation initiation (8, 10). Interactions between the yeast PUF protein Mpt5p (or PUF5) and deadenylating complexes have recently been described, and such interactions are conserved throughout evolution in eukaryotes (11). Mpt5p interacts directly with the Pop2p nuclease, which is part of the Ccr4p-Pop2p-Not deadenylase complex. This protein bridges the interaction with the Dhh1 helicase, which is also involved in mRNA storage and degradation (12). Affinity tag purification and microarray analysis have shown that each PUF protein binds and regulates a functionally and cytotopically related subpopulation of mRNAs in yeast (9).
Several conserved PUF proteins have been identified in the genomes of kinetoplastida. In T. cruzi, the PUF protein family has 10 members. Based on in silico analysis, they can be assigned to three groups according to putative binding specificity (13). We have previously described and characterized TcPUF6, a member of this family (14). TcPUF6 is an ortholog of the Trypanosoma brucei protein TbPUF1 (15). T. cruzi PUF6 protein is produced constitutively, throughout the life of the parasite. The protein is present in the cytoplasm of the replicative epimastigote forms in multiple discrete foci, a characteristic pattern also observed for members of the yeast PUF family (9) and other trypanosome PUF proteins (13, 15). We used a ribonomic approach (16, 17) to identify the putative target mRNAs associated with TcPUF6. The overexpression of a TAP-tagged TcPUF6 protein did not affect growth rate or morphology. Microarray analysis of affinity-purified transcripts and whole expression of the transfected parasites showed that mRNAs associated with TcPUF6 were down-regulated in parasites overexpressing TcPUF6. We assessed the in vivo association of TcPUF6 with the TcDhh1 helicase by immunoprecipitation, TAP-Tag assays, and immunofluorescence co-localization assays.
EXPERIMENTAL PROCEDURES
Parasites—T. cruzi clone Dm28c (18) was used throughout this work. Epimastigote forms were maintained at 28 °C in liver infusion tryptose medium supplemented with 10% heat-inactivated fetal bovine serum. Metacyclic trypomastigotes and extracellular amastigotes were prepared as described by Contreras et al. (19, 20).
Immunofluorescence and Imaging—Immunofluorescence assays were performed as previously described by Holetz et al. (21). Serum dilutions were: rabbit anti-TcPUF6 1:50 and mouse anti-TcDhh1 1:20. Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 546-conjugated anti-rabbit secondary antibodies (1:400) (Molecular Probes, Invitrogen) were used. The cells were stained with 4′,6′-diamino-2-phenylindole (1:2000; Sigma). Subcellular localization images were acquired using a Nikon Eclipse E600 microscope coupled to a Cool SANP-PRO color camera (Media Cybernetics). The merged images were obtained by superimposing image files in the Image-PRO PLUS software V6.2 (Media Cybernetics).
Sucrose Density Gradient Separation and Polysome Analysis—T. cruzi polysomes were purified and separated on sucrose gradients, using a modified version of the method described by Brecht and Parsons (22). Exponentially growing cultures of 5 × 108 epimastigotes were lysed in buffer A (300 mm KCl, 10 mm MgCl2, 10 mm Tris-HCl, pH 7.4, 10% Nonidet P-40, and 2 m sucrose) and centrifuged at 16,000 × g for 5 min at 4 °C. The supernatant (400 μl) was layered onto 15 to 55% sucrose density gradients prepared in buffer B plus inhibitors (300 mm KCl, 10 mm MgCl2, 10 mm Tris-HCl, pH 7.4, 100 μg/ml cycloheximide, 10 μm E-64, 1 mm phenylmethylsulfonyl fluoride, 1 mg/ml heparin) and centrifuged at 4 °C for 2 h at 39,000 rpm in a Beckman SW40 rotor. As a control, the cells were preincubated with 2 mm of Puromycin for 1 h at 28 °C before being lysed. Gradients were fractionated using an ISCO Foxy Jr. collection system with pump speed set to 1 ml/min, a fraction time of 30 s/fraction, and a chart speed of 60 cm/h. For Western blot analysis, 30-μl aliquots of the indicated fractions were boiled for 5 min with SDS-PAGE loading buffer.
RNA Preparations—Total RNA was extracted from 5 × 108 exponentially growing epimastigote forms, using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Linearly amplified RNA was generated from 1 μg of total RNA (single round), using the MessageAmp amplified RNA kit (Ambion) according to the manufacturer's manual. cDNA was synthesized from 1 μg of total or affinity-purified RNAs using an oligo(dT) primer (US Biochemical Corp.) and reverse transcriptase (IMPROM II, Promega) as recommended.
Microarray Hybridization and Analysis—Fluorescent cyanin (Cy) dyes Cy3 or Cy5, as appropriate were incorporated into the second strand of the cDNA, using 2 μg of amplified RNA as the starting material for each sample. The labeled cDNA was purified with a Microcom 30 device (Millipore). Microarray hybridizations and washes were carried out in a GeneTac automated hybridization station (Genomic Solutions). The Cy3- and Cy5-labeled cDNAs were mixed and added to 120 μl of hybridization solution and allowed to hybridize for 14–16 h at 42 °C. The microarray slides were then washed in buffers of increasing stringency (0.5× and 0.05× SSC) and dried by centrifugation at 280 × g for 5 min. The dried slides were scanned in a 428 Array Scanner (Affymetrix). The images were analyzed with Spot software. The resulting data were corrected for background and normalized, using the normexp and PrintTip-Loess methods, respectively, within the Limma package (24). Microarray results were confirmed by quantitative PCR, as previously described (25). Two-step real time reverse transcription-PCR assays were carried out with an ABI PRISM 7000 sequence detection system (Applied Biosystems).
Tandem Affinity Purification and Immunoprecipitation Assays—For TAP-Tag assays, the coding sequence of the TcPUF6 gene inserted into a pTEXTAPTAG vector3 encoding the amino acid sequences of the IgG-binding domain of Staphylococcus aureus protein A and the human calmodulin-binding peptide as a double tag (26). T. cruzi epimastigotes (5 × 107) were transfected with 50 μg of vector DNA by electroporation in 4-mm cuvettes, using the Bio-Rad GenePulser II electroporator, with two pulses of 450 V and 500 millifarads. We added 250 μg/ml geneticin 24 h after electroporation, for the selection of transfected parasites. After 72 h, the cultures were diluted 1:4, and the concentration of geneticin was increased to 500 μg/ml. Transfectants were cloned by serial dilution in 24-well plates and cultured in the presence of 500 μg/ml of geneticin. TAP-Tag assays of bound proteins and RNAs were performed as previously described (9, 17). The soluble cytosolic fraction was prepared from 1010 exponentially growing epimastigote forms at a final concentration of 109/ml of polysome lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm Hepes, pH 7.0, and 1% Nonidet P-40). The parasites were lysed for 1 h at 4 °C (17). Lysed parasites were centrifuged at 10,000 × g for 20 min at 4 °C, and the supernatants were immediately used in affinity purification assays. The tagged proteins were purified on IgG-Sepharose columns (Amersham Biosciences). They were washed three times and treated with 200 units/ml of TEV protease (Invitrogen), as previously described (26). Eluted RNAs were purified using the RNeasy mini kit (Qiagen) and amplified using the cRNA amplification kit (Ambion), following the manufacturer's instructions.
Immunoprecipitation assays were performed as previously described (17). Antiserum against TcPUF6 or TcDhh1 (50 μl) was incubated with 100 μl of protein A-Sepharose or protein G-agarose beads (Sigma), respectively, overnight at 4 °C. The beads were blocked by incubation in 5% skim milk powder in phosphate-buffered saline. The soluble cytosolic fraction was prepared from 109 exponentially growing epimastigote forms. Conjugated beads were incubated for 1 h at room temperature in the presence of the soluble protein fraction. Bound proteins were washed three times with ten column volumes of polysome lysis buffer and eluted with one volume of 0.2 m glycine, pH 2.3. For RNA immunoprecipitation assays 50 μl of anti-TcPUF6 and preimmune serum were used as previously described. Eluted RNAs were purified using the RNeasy mini kit (Qiagen) concentrated on microcon devices and immediately converted to cDNA as described. Similar volumes of the resulting cDNAs were subjected to PCR amplification. PCR conditions were as follows: 95 °C for 1 min, followed by 26 cycles of 58 °C for 45 s, and 72 °C for 1 min for all the genes studied. Primer sets are listed in supplemental Table S2.
Western Blot Analysis—Protein extracts from epimastigotes in the exponential growth phase were prepared and separated by SDS-PAGE in 10% acrylamide gels, electrotransferred onto Hybond-C membranes (Amersham Biosciences), and incubated with the different polyclonal sera as previously described (14). Bound antibodies were detected using alkaline phosphatase-conjugated goat anti-rabbit IgG (H+L) (Sigma) diluted 1/7.500. For relative protein quantification, band densitometry was performed with One DScan E X 3.1 software (Scanalytic), using wild-type TcPUF6 protein levels as a reference. Before each microarray hybridization or affinity purification assay, Western blots were performed to confirm the overexpression of the tagged protein.
Quantitative PCR—Two-step real time reverse transcription-PCR assays were performed using the ABI PRISM 7000 sequence detection system (Applied Biosystems) as previously described (25). In brief, amplifications were carried out in triplicate with 10 ng of cDNA and the recommended concentration of SYBR green master mix (Applied Biosystems). Primer sets are listed in supplemental Table S2. PCR conditions were as follows: 50 °C for 2 min and 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s for all genes studied or 45 cycles of 95 °C for 15 s and 70 °C for 1 min for control genes. Thermal dissociation confirmed that reverse transcription-PCR generated a single amplicon. For relative quantification, the standard curve method was used, based on cycle threshold values. 1:5 dilutions of known concentrations of cDNA were used to generate curves extending from 80 pg to 50 ng of cDNA. A standard curve was generated for each of the genes studied and both control genes. Gene expression was normalized against TcL9 or TcH2B control genes (25). The results are expressed as fold changes of gene expression in metacyclic trypomastigotes using epimastigotes as the reference population.
RESULTS
Cellular Localization of TcPUF6 in the Different Stage Forms—We have previously shown that TcPUF6 is present in multiple discrete foci in the cytoplasm of the replicative epimastigote forms. However, given the differences between the developmental stages of T. cruzi, we conceived that TcPUF6 might be distributed differently in nonreplicative, infective metacyclic trypomastigotes and in replicative intracellular amastigote forms. We used a previously described polyclonal TcPUF6 antiserum (14) to determine the distribution of the protein in cells. In both cellular forms, TcPUF6 was localized in discrete cytoplasmic foci (supplemental Fig. S1).
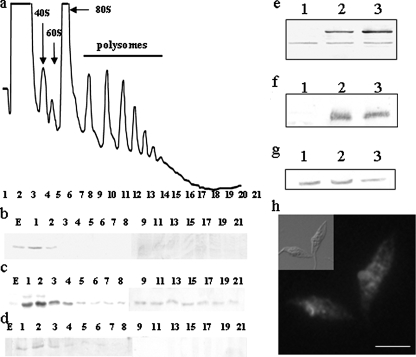
TcPUF6 Is Localized in Ribosome-free Regions of the Cytoplasm—It has been suggested that PUF proteins bind to functionally related mRNAs and localize them in specific cytoplasmic regions. We analyzed the possible association of TcPUF6 with polysomes. Exponentially growing epimastigotes were treated with cycloheximide to trap the mRNAs on the elongating ribosomes. Polysome profiles were obtained from sucrose gradients loaded with the soluble cytoplasmic fractions in three independent assays (Fig. 1a). As a control, the polysome-enriched fraction was treated with EDTA or puromycin prior centrifugation to dissociate the ribosomal subunits from the mRNAs (not shown). Western blot analysis of the different fractions showed that TcPUF6 co-sedimented with the ribosome-free fractions in all three experiments (collected fractions 1–3). No signal was detected with polysomes or monosomes (Fig. 1b). An antiserum against T. cruzi eIF3 translation initiation factor subunit Tif34 was used as a control of a polysome associated protein.4 A signal corresponding to the TcTif34 protein was found in fractions along the entire gradient in all of the experiments (Fig. 1c). As negative control, an anti-PEPCK serum was tested, and the corresponding signal was found up to fractions 6–9 depending on the assay analyzed (Fig. 1d). Furthermore, we used confocal microscopy to perform co-localization assays with the anti-TcPUF6 serum and a commercial antiserum against human ribosomal P-proteins (Immunovision). Our results showed that TcPUF6 was not present in the ribosome-rich regions of the cytoplasm (supplemental Fig. S2g). We also investigated possible associations between TcPUF6 and actively translating ribosomes. Co-localization assays were performed with two different antisera raised against two subunits, Tif34 and Ptr1, of T. cruzi eIF3. No clear co-localization was observed with either the Ptr1 subunit (supplemental Fig. S2h) or the Tif34 subunit (not shown). Thus, TcPUF6 is present in the cytoplasm and is not associated with ribosomes or polysomes.
FIGURE 1.
TcPUF6 is present in the ribosome-free fractions of sucrose gradients. a, the positions of the 40 and 60 S subunits, the 80 S ribosome monomer, and polysomes are indicated in the sucrose density gradient. b–d, Western blots of the collected fractions (30 μl) probed with anti-TcPUF6 (1/500) (b), anti-TcTif34 (1/500) (c), and anti-PEPCK (1/250) sera (d). Transfected parasites overexpress a TcPUF6-tagged protein. e, Western blot analysis of protein extracts from epimastigotes (lane 1), epimastigotes transfected with the pTEX-TAPTAG-transfected clones (lane 2), and the PUF6 A- and (4) B-transfected clones (lane 3), with antiserum against TcPUF6 protein (1/500 dilution). f, protein extracts were also tested with an anti-CBP serum (1/1000) (Upstate). g, the same blot was re-probed with an anti-PEPCK serum (1/500). h, immunolocalization of T. cruzi TcPUF6 in TcPUF6-TAPTAG-transfected epimastigote forms with anti-CBP antiserum (1/100). Bars, 5 μm.
Overexpression of a TAP-tagged TcPUF6 Protein Does Not Affect Differentiation Phenotype—A TAP-tagged TcPUF6 protein was overexpressed in epimastigote forms to identify the mRNAs associated with TcPUF6 and to obtain additional information about protein function. We used the pTEX vector, which is maintained as a low copy number episomal DNA with discrete overexpression (27), to facilitate the purification of mRNP complexes without affecting the cellular distribution of the protein. Two clonal populations derived from independent transfection assays were selected. These populations, PUF6A and PUF6B, overexpressed a protein of about 90 kDa in size resulting from the fusion of TcPUF6 with the 30-kDa tag. The level of overexpression was analyzed by Western blotting, using different concentrations of total protein extract, with in silico quantification by peptide densitometry, using the wild-type protein as a reference. The fusion protein was overexpressed by factors of three and five in the PUF6 A and B populations, respectively (Fig. 1e). Immunofluorescence microscopy was performed with both the anti-TcPUF6 and an anti-tag serum and showed that the characteristic cellular distribution of TcPUF6 was maintained in the transfected parasites (Fig. 1h). Parasites overexpressing the TcPUF6-tagged protein showed no significant change in cell growth rate or morphology; parasites transfected with the pTEX-TAPTAG vector were used as the reference population. The ability of the TcPUF6-overexpressing parasites to differentiate was also analyzed in vitro, and no significant difference in differentiation rates following metacyclogenesis and amastigogenesis in vitro were observed (not shown).
Microarray Analysis of TcPUF6-overexpressing Parasites—We investigated the phenotypic changes caused by TcPUF6 at the molecular level, using DNA microarray hybridization to study the steady-state mRNA levels of parasites transfected with vector alone and epimastigotes overexpressing TcPUF6. A T. cruzi cDNA microarray containing ∼6,200 independent probes was analyzed by competitive hybridization, using an amplified cRNA derived from the total RNA of both populations (supplemental Figs. S3 and S4). TcPUF6 overexpression had a considerable effect on global gene expression. We simplified the analysis by considering only genes showing changes in mRNA levels by at least a factor of three. Overall, 269 genes fulfilled this condition, corresponding to about 4% of the sequences present in the microarray. From these genes, 232 were up-regulated in the TcPUF6 transfectants, whereas 37 were clearly down-regulated (supplemental Table S1). The TcPUF6 transcript itself increased in abundance by a factor of 4.5, consistent with Western blotting data (Fig. 1e). The genes selected were identified as putative RNA-binding proteins (9), proteases (11), protein kinases (18), transporters (13), and 48 enzymes involved in central metabolism. However, about 50% of the sequences analyzed were classified as hypothetical proteins (122 sequences) (supplemental Table S1). An analysis of the biological process potentially affected by changes in TcPUF6 levels showed that the up-regulated probes included genes encoding proteins involved in DNA metabolism, stress responses, cytoskeleton and mitochondrial proteins, and several surface antigens. An analysis of the down-regulated genes showed that they included one encoding a putative cyclin and six genes encoding proteins involved in RNA processing, including the TcPUF7 protein. However, most of the down-regulated genes were identified as hypothetical proteins. These results were corroborated by quantitative PCR.
Identification of Putative TcPUF6 Target mRNAs—Ribonucleoprotein complexes formed by mRNAs, and the tagged TcPUF6 protein were purified from soluble protein lysates prepared from transfected exponentially growing epimastigote forms by affinity chromatography on IgG-Sepharose columns, as previously described (9). RNA was purified from the eluted fractions and amplified; the resulting cRNA was labeled and hybridized to the T. cruzi cDNA microarray. Parasites transfected with vector alone were used as a control in affinity purification experiments; however, the levels of RNA obtained were almost undetectable, even after amplification (not shown). Total epimastigote RNA was used as a control in microarray competitive hybridization analyses to eliminate nonspecific hybridization signals with the most abundant mRNAs. Eight independent affinity purifications were performed, only three of which generated enough RNA for microarray hybridization, including dye swap controls.
Nine probes were selected based on increased signals obtained in at least two of three experiments (Table 1). Five corresponded to genes encoding hypothetical proteins. The others corresponded to a serine-threonine phosphorilase, the XRND1 exonuclease, a putative amino acid transporter (PAT6), and the 69-kDa PFR2 (paraflagellar rod protein 2). A detailed analysis of the hypothetical proteins showed that two had predicted transmembrane domains and one had a signal peptide. One of the proteins with transmembrane domains presented some similarity to the cytochrome B5 protein.
TABLE 1.
Putative TcPUF6 targets identified by affinity purification assays are down-regulated in the transfected parasites and up-regulated in the metacyclic forms
| GeneID TSKa | Gene annotation | Immunoprecipitation assaysb | Fold change in transfectantsc | Fold change in metacyclicsd |
|---|---|---|---|---|
| Tc00.1047053508039.70 | Hypothetical protein | +,+,- | -4.9 | 0.34 ± 0.02 |
| Tc00.1047053506753.110 | Hypothetical protein (cytochrome b5-like) | -,-,- | -3.5 | NDe |
| Tc00.1047053504013.110 | Serine/threonine-protein phosphatase 2A | +,+,+ | -3.6 | 8.67 ± 1.02 |
| Tc00.1047053511523.20 | Hypothetical protein | +,+,- | -3.6 | 6.57 ± 0.53 |
| Tc00.1047053507641.80 | XRND1 exonuclease | +,+,- | -4.0 | 4.23 ± 0.35 |
| Tc00.1047053509179.50 | Hypothetical protein | +,+,- | -3.4 | 70.3 ± 6.48 |
| AAS47054 | PAT6 amino acid transporter | +,+,+ | -3.0 | 1.54 ± 0.14 |
| Tc00.1047053511215.119 | 69 kDa paraflagellar rod protein putative | +,+,+ | -8.4 | 7.81 ± 0.68 |
| EH035829 | Hypothetical protein | ND | -4.7 | ND |
Gene code from Trypanosoma cruzi Sequencing Consortium (when this is not available, the GenBank™ sequence ID is provided instead).
Presence (+) or absence (-) of the affinity purified transcripts in the three independent immunoprecipitation assays.
Fold change value of decreased expression determined by microarray analysis.
Fold change value of transcript expression levels in metacyclic trypomastigotes compared with epimastigote forms by quantitative PCR assays.
ND, not determined.
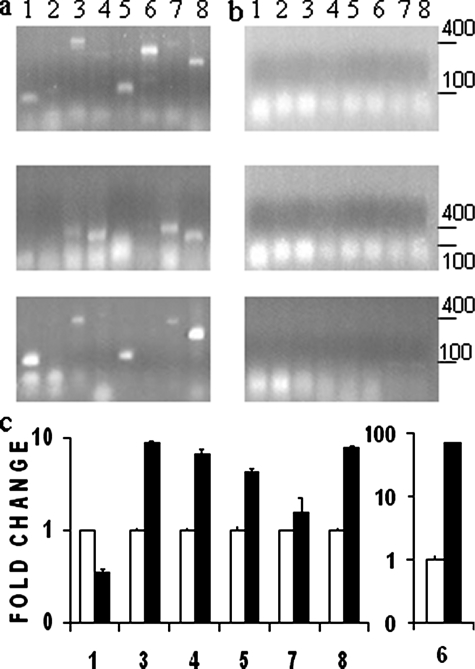
Alternatively, to confirm the TAP-Tag results, the ribonucleoprotein complexes were purified by immunoprecipitation assays using soluble lysates prepared from wild-type epimastigote forms. The anti-TcPUF6 serum was used in the immunoprecipitation experiments (Fig. 2a) using preimmune serum as a negative control (Fig. 2b). The presence of the previously identified mRNAs in the eluted complexes was detected by reverse transcription-PCR. We obtained positive results for seven of the eight sequences tested in at least two of three independent assays confirming the TAP-Tag results. Only the mRNA corresponding to the cytochrome B5-like gene was not detected in any of the experiments.
FIGURE 2.
Tag affinity-purified transcripts are also present in anti-TcPUF6 immunoprecipitation elutes. Reverse transcription-PCR analysis of eluted fractions from three independent Immunoprecipitation assays using anti-TcPUF6 sera (a) or preimmune serum as a control (b). PCR products were resolved in 1.2% agarose gels. c, TcPUF6 associated mRNAs are preferentially expressed in metacyclic trypomastigotes. Total RNA from epimastigotes (white bars) and metacyclic trypomastigotes (black bars) was analyzed. Metacyclic trypomastigotes gene expression results are expressed in terms of fold change in relation to epimastigotes gene expression. The results were normalized against those for the L9 ribosomal protein. Standard deviations for triplicate experiments are shown. The numbers correspond to: Tc00.1047053508039.70 (bar 1), cytochrome b5-like (bar 2), Ser/Thr phosphatase 2A (bar 3), Tc00.1047053511523.20 (bar 4), XRND1 (bar 5), Tc00.1047053509179.50 (bar 6), PAT6 (bar 7), and 69-kDa PFR2 (bar 8).
PUF proteins bind conserved elements present in the 3′-UTR of mRNAs. In most proteins characterized to date, the consensus sequence bears a core motif with the UGUR sequence. As a first approach to characterize the TcPUF6 target sequence, the MEME algorithm (28) was used to search for the presence of the UGUR motif in the 3′-UTR of seven selected genes annotated in the T. cruzi GeneDB. Because we have no information about the 3′-UTR of the mRNAs studied, we analyzed 200 nucleotides downstream from the stop codon as the putative UTR of these genes. At least one UGUR sequence was found, though no clear motif could be determined in silico (not shown).
Transcripts Associated with TcPUF6 Are Down-regulated in Transfected Parasites—It can be inferred that in the case of transcripts bound and regulated by TcPUF6, its overexpression should affect their expression levels in a coordinated manner. Analysis of the microarray data showed significantly lower mRNA levels in the transfected parasites than in control parasites for all of the selected genes (Table 1). Microarray results were confirmed by quantitative PCR using total RNA prepared from control and TcPUF6-transfected parasites. The quantitative PCR assays confirmed the microarray data for all the selected genes, although differences in expression levels were smaller (not shown). Thus, overexpression of TcPUF6 results in down-regulation of the transcript levels of its putative mRNA targets.
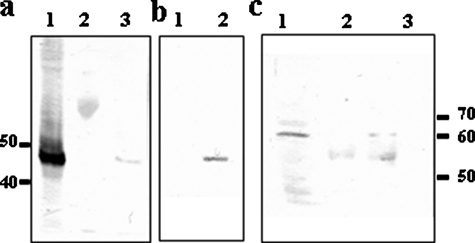
TcPUF6 and TcDhh1 Are Present in the Same Ribonucleoprotein Complexes—Our results strongly suggest that TcPUF6 promotes the degradation of target mRNAs, consistent with an interaction between this protein and RNA-degrading complexes. We have recently developed a mouse polyclonal antiserum against a recombinant T. cruzi Dhh1 helicase ortholog (21). This antiserum was used to investigate the interaction of TcPUF6 and TcDhh1 by immunoprecipitation assays followed by Western blotting with rabbit anti-TcPUF6 and mouse anti-TcDhh1 sera. The anti-TcPUF6 serum bound to protein A-Sepharose was used in immunoprecipitation assays with soluble protein extracts of epimastigote forms. Western blots of the eluted proteins with the mouse anti-TcDhh1 serum showed the presence of a 50-kDa band corresponding to TcDhh1 in three independent experiments (Fig. 3a). The preimmune serum bound to protein A-Sepharose was used as a negative control. These results were confirmed by carrying out TAP-Tag affinity purification with soluble protein extracts from the transfected epimastigote forms and from parasites transfected with vector alone as a control. Tandem affinity-purified fractions gave no signal corresponding to TcDhh1 in Western blots. No protein band was detected on the silver-stained SDS-PAGE gel of the eluted fractions (not shown). We attempted only one purification protocol, consisting of the use of IgG-Sepharose columns followed by TEV protease treatment. In this case, a clear band corresponding to the 50-kDa protein TcDhh1 was observed in Western blots of the eluted fractions (Fig. 3b).
FIGURE 3.
TcPUF6 is part of a complex bearing the TcDhh1 helicase. a, Western blot using anti-TcDhh1 antibody (1/250) of total protein input (lane 1) and eluates immunoprecipitated with preimmune (lane 2) and anti-TcPUF6 antisera (lane 3). b, Western blots with anti-TcDhh1 antibody of TAG affinity-purified proteins from vector-transfected parasites (lane 1) and TcPUF6-tagged expressing parasites (lane 2). c, Western blot using anti-TcPuf6 antibody (1/500) of total protein input (lane 1), and eluates immunoprecipitated using preimmune (lane 2) and anti-TcDhh1 antibody (lane 3).
Immunoprecipitation assays were also performed using the mouse anti-TcDhh1 serum bound to protein G-agarose beads. Western blotting for TcPUF6 revealed the presence of a faint 65-kDa band corresponding to the protein in the eluted fractions (Fig. 3c). The various purification assays were carried out in at least three independent assays, and similar results were obtained in each case.
TcPUF6-associated mRNAs Are Up-regulated in the Infective Metacyclic Trypomastigotes—Microarray analysis of gene expression during the parasite life cycle showed that some of the transcripts associated with TcPUF6 were preferentially expressed in metacyclic forms.5 Because no clear functional relationship could be established for the previously identified transcripts, we considered the possibility that TcPUF6 could be regulating their temporal expression. Quantitative PCR analysis was performed to determine the levels of mRNA expression of these genes in epimastigotes and metacyclic trypomastigote forms. Total RNA was used in the assays, and the results showed that six of the seven genes have their transcript levels up-regulated in the infective metacyclic forms when compared with epimastigote forms. The results were normalized with respect to expression of the ribosomal L9 gene (Fig. 2c and Table 1) and the histone H2B gene (not shown) rendering similar results.
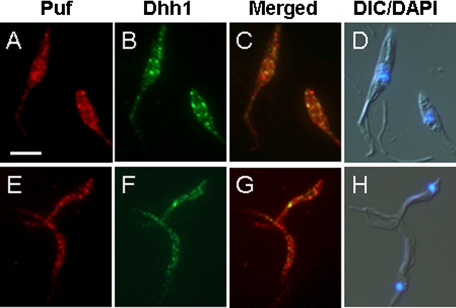
TcPUF6 and TcDhh1 Do Not Co-localize in Metacyclic Trypomastigotes—We have previously showed that mRNAs associated with TcPUF6 are negatively regulated in the transfected parasites. Interaction with TcDhh1 suggests that TcPUF6 enhances degradation of its molecular targets through association with mRNA degradation complexes. However, in vivo these mRNAs are up-regulated in the metacyclic forms where TcPUF6 is also expressed. This could be the result of differential localization of the regulated mRNAs in the infective forms. We performed co-localization experiments between TcPUF6 and TcDhh1 both in epimastigotes and metacyclic trypomastigotes. In epimastigotes all of the signal corresponding to TcDhh1 co-localizes with TcPUF6 (Fig. 4c), whereas part of the foci where TcPUF6 is detected remain as an independent signal. When metacyclic trypomastigotes were analyzed, TcDhh1 signal was overall clearly decreased in comparison with epimastigotes, and no co-localization between the two proteins was observed (Fig. 4g).
FIGURE 4.
TcPUF6 and TcDhh1 show different co-localization patterns during the parasite life cycle. Localization of TcPUF6 (A and E) and TcDhh1 (B and F) in T. cruzi epimastigotes (A–D) and metacyclic trypomastigotes (E–H) is shown. C and G show superimposed TcPUF6 and TcDhh1 images. D and H show 4′,6′-diamino-2-phenylindole images merged with differential interference contrast images. Scale bar, 5 μm.
DISCUSSION
In this work, we used a ribonomic approach to characterize the biological role of TcPUF6 and its molecular targets. It is now accepted that discrete subpopulations of mRNAs are bound to RNA-binding proteins and have their expression co-regulated by these proteins through their function as posttranscriptional inducers or repressors of functionally related genes. Functionally related mRNAs are regulated in ribonucleoprotein complexes and degraded or stored for subsequent translation in defined cytoplasmic regions, defining what has been termed “posttranscriptional operons” (16).
We characterized the putative mRNA targets of TcPUF6, using a strategy combining the analysis of overall gene expression in the transfected parasites and TAP-Tag affinity purification of the TcPUF6-associated mRNAs. Microarray analyses of total RNA from the transfected parasites indicated that the mRNA levels of several genes are affected by TcPUF6 overexpression. Some of the affected genes encode RNA-binding proteins, various kinases, and proteases, pointing to the existence of complex and interconnected posttranscriptional and posttranslational regulatory networks in T. cruzi. Changes in TcPUF6 protein level might therefore disturb different gene regulation cascades, although this does not necessarily imply that these mRNAs are targets of the protein.
TAP-Tag affinity purification assays and microarray hybridization were carried out to identify the molecular targets of TcPUF6. Despite the use of a genome-wide analysis, we were able to identify only a few target mRNAs reliably. Recent work on T. brucei did not lead to identification of the putative targets of the TcPUF6 ortholog, TbPUF1 (29). This may be due to the technical limitations of affinity purification assays. Consistent with this hypothesis, only small amounts of RNA were recovered in the affinity purification assays, making it necessary to carry out RNA amplification to obtain enough RNA for microarray experiments. The affinity-purified transcripts were down-regulated in TcPUF6-overexpressing parasites. This strongly suggests that TcPUF6 enhances the degradation of its associated mRNAs, as described for other PUF proteins in eukaryotes (30, 31). In this work, we overexpressed a tagged TcPUF6 protein to facilitate purification of the associated transcripts. However, this procedure may have affected purification efficiency, because overexpression of the tagged TcPUF6 protein may have reduced levels of the target mRNAs.
Caro et al. (13) classified the T. cruzi PUF family as a function of putative sequence targets. TcPUF6 was grouped with proteins capable of binding the conserved UGUR motif. We identified the UGUR motif in the putative 3′-UTR sequences of most of the purified transcripts. However, detailed binding assays are required to determine the specific target sequence in the UTRs of all the associated mRNAs. One of the identified transcripts corresponds to the PFR2 protein. In Leishmania mexicana, Mishra et al. (32) identified a 10-nucleotide negative regulator element present in the long 3′-UTR of the PFR2 mRNA. The paraflagellar rod protein regulator element bears an UGUR motif and acts by destabilizing the PFR2 mRNA in amastigotes.
It has been suggested that PUF proteins regulate the half-lives of associated mRNAs by interacting with proteins involved in mRNA degradation via a mechanism that is conserved throughout eukaryote evolution (11). We observed the interaction and co-localization between TcPUF6 and the decapping activator TcDhh1, supporting the notion that TcPUF6 might target mRNAs for degradation. TcDhh1-containing P body-like structures have been described in T. cruzi, and the number of such complexes per cell has been determined (21). The number of foci in which TcPUF6 is detected exceeds the estimated number of TcDhh1 foci/cell, suggesting that TcPUF6 is part of RNP complexes other than P-body-like structures. In yeast, PUF proteins do not localize with P-bodies, and it has been suggested that its association with these structures may be transient (11). The mammalian Pumilio2 protein has been localized in neurons and described as a novel member of dendrite stress granules, another macromolecular structure involved in mRNA degradation, being excluded from P-bodies under all conditions tested (33). The interaction of TcPUF6 with TcDhh1 may reflect differences in structure and function of T. cruzi P-body-like structures compared with higher eukaryotes. Recently, the formation of mRNA granules as a response to nutritional stress has been reported in T. cruzi. These RNA granules seem to be involved in the stabilization and storage of mRNAs, and they only partially co-localize with TcDhh1 (34). Our results suggest that the localization of TcPUF6 to specific loci in the cytoplasm and the interaction of this protein with TcDhh1 may be related to its putative involvement in RNA degradation mechanisms more than with mRNA storage mechanisms.
Conditional knock-out and interference assays have shown that the T. brucei ortholog, TbPUF1, is not essential for parasite survival (15, 29). It therefore seems that, rather than regulating in an “all-or-nothing” manner, TcPUF6 may control the fate of the associated transcripts so as to maintain the required levels of target mRNA expression. TcPUF6 is expressed throughout the parasite life cycle, and our results suggest that it is involved in mRNA degradation; however, analysis of the expression patterns of its associated mRNAs showed that they are up-regulated in the metacyclic forms. PUF proteins usually exert their control functions by interacting with other protein partners in combinatorial regulatory mechanisms (23, 35). Immunofluorescence assays showed that TcPUF6 and TcDhh1 do not co-localize in the infective metacyclic forms, hence suggesting the absence of interaction in these forms. Our results suggest that TcPUF6 might also play a role in the regulation of metacyclic trypomastigote transcripts. We propose a model in which TcPUF6 regulates the half-lives of its associated transcripts by its stage-dependent association with TcDhh1 containing complexes involved in mRNA degradation. The characterization of mRNA subsets and proteins interacting with TcPUF6 in parasites of different developmental stages might shed light on the mechanisms involved in posttranscriptional regulation in trypanosomes.
Supplementary Material
Acknowledgments
We thank Nilson Fidêncio for skilful technical assistance and Paulo Arauco for DNA sequencing. We also thank Dr. Esteban Serra for providing the pTEX-TAPTAG vector.
This work was supported by Fundação Oswaldo Cruz, Programa de Núcleos de Excelência (Fundação Araucária), Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (Conselho Nacional de Desenvolvimento Cientifico e Technologico (CNPq), PROSUL), and National Institutes of Health Grant 5R01AI050196-02. This work was also supported by CNPq research fellowships (to B. D., A. C., C. M. P., M. P. M., S. G., and M. A. K.) and a Programa de Estudio y Desarrollo de Ciencias Básicas fellowship and an AMSUD/Pasteur training fellowship (to P. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: UTR, untranslated region; TAP, tandem affinity purification; PEPCK, phosphoenolpyruvate carboxykinase.
E. Serra, unpublished observations.
S. P. Fragoso, personal communication.
M. A. Krieger, manuscript in preparation.
References
- 1.De Souza, W. (2002) Kinetoplastid Biol. Dis. 31 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clayton, C. E. (2002) EMBO J. 21 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teixeira, S. M. (1998) Braz. J. Med. Biol. Res. 31 1503–1516 [DOI] [PubMed] [Google Scholar]
- 4.Mignone, F., Gissi, C., Liuini, S., and Pesole, G. (2002). Genome Biol. 3 0004.1–0004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auweter, S. D., Oberstrass, F. C., and Allain, F. H. T. (2006) Nucleic Acids Res. 34 4943–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keene, J. D., and Tenenbaum, S. A. (2002) Mol. Cell 9 1161–1167 [DOI] [PubMed] [Google Scholar]
- 7.Wang, X., McLachlan, J., Zamore, P. D., and Tanaka Hall, T. M. (2002). Cell 110 501–512 [DOI] [PubMed] [Google Scholar]
- 8.Wickens, M., Bernstein, D. S., Kimble, J., and Parker, R. (2002) Trends Genet. 18 150–157 [DOI] [PubMed] [Google Scholar]
- 9.Gerber, A. P., Herschlag, D., and Brown, P. O. (2004) PLoS Biol. 2 342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wharton, R. P., and Aggarwal, A. K. (2006). Sci. STKE 2006, pe37. [DOI] [PubMed]
- 11.Goldstrohm, A. C., Hook, B. A., Seay, D. J., and Wickens, M. (2006) Nat. Struct. Mol. Biol. 13 533–539 [DOI] [PubMed] [Google Scholar]
- 12.Coller, J. M., Tucker, M., Sheth, U., Valencia-Sanchez, M. A., and Parker, R. (2001) RNA 7 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caro, F., Bercovich, N., Atorrasagasti, C., Levin, M. J., and Vazquez, M. P. (2006) Exp. Parasitol. 113 112–124 [DOI] [PubMed] [Google Scholar]
- 14.Dallagiovanna, B., Perez, L., Sotelo-Silveira, J., Smircich, P., Duhagon, M. A., and Garat, B. (2005) Exp. Parasitol. 109 260–264 [DOI] [PubMed] [Google Scholar]
- 15.Hoek, M., Zanders, T., and Cross, G. A. (2001) Mol. Biochem. Parasitol. 57 706–716 [DOI] [PubMed] [Google Scholar]
- 16.Keene, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7018–7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenenbaum, S. A., Lager, P. J., Carson, C. C., and Keene, J. D. (2002) Methods 26 191–198 [DOI] [PubMed] [Google Scholar]
- 18.Contreras, V. T., Araujo-Jorge, T. C., Bonaldo, M. C., Thomaz, N., Barbosa, H. S., Meirelles, M., de, N., and Goldenberg, S. (1988). Mem. Inst. Oswaldo Cruz 83 123–133 [DOI] [PubMed] [Google Scholar]
- 19.Contreras, V. T., Salles, J. M., Thomas, N., Morel, C. M., and Goldenberg, S. (1985). Mol. Biochem. Parasitol. 16 315–327 [DOI] [PubMed] [Google Scholar]
- 20.Contreras, V. T., Navarro, M. C., DeLima, A. R., Arteaga, R., Duran, F., Askue, J., and Franco, Y. (2002). Mem. Inst. Oswaldo Cruz. 97 1213–1220 [DOI] [PubMed] [Google Scholar]
- 21.Holetz, F., Correa, A., Avila, A. R., Nakamura, C. V., Krieger, M. A., and Goldenberg, S. (2007) Biochem. Biophys. Res. Commun. 356 1062–1067 [DOI] [PubMed] [Google Scholar]
- 22.Brecht, M., and Parsons, M. (1998) Mol. Biochem. Parasitol. 97 189–198 [DOI] [PubMed] [Google Scholar]
- 23.Sonoda, J., and Wharton, R. P. (1999) Genes Dev. 13 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth, G. K. (2004). Stat. App. Genet. Mol. Biol. 3 article 3 [DOI] [PubMed]
- 25.Nardelli, S. C., Avila, A. R., Freund, A., Motta, M. C., Manhaes, L., de Jesus, T. C., Schenkman, S., Fragoso, S. P., Krieger, M. A., Goldenberg, S., and Dallagiovanna, B. (2007) Eukaryot. Cell 6 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. (2001). Methods 24 218–229 [DOI] [PubMed] [Google Scholar]
- 27.Kelly, J. M., Ward, H. M., Miles, M. A., and Kendall, G. (1992) Nucleic Acids Res. 20 3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey, T. L., and Elkan, C. (1994). Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press, Menlo Park, CA [PubMed]
- 29.Luu, V. D., Brems, S., Hoheisel, J. D., Burchmore, R., Guilbride, D. L., and Clayton, C. (2006) Mol. Biochem. Parasitol. 150 340–349 [DOI] [PubMed] [Google Scholar]
- 30.Olivas, W., and Parker, R. (2000) EMBO J. 19 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wreden, C., Verrotti, A. C., Schisa, J. A., Lieberfarb, M. E., and Strickland, S. (1997). Development 124 3015–3023 [DOI] [PubMed] [Google Scholar]
- 32.Mishra, K. K., Holzer, T. R., Moore, L. L., and LeBowitz, J. H. (2003) Eukaryot. Cell 2 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vessey, J. P., Vaccani, A., Xie, Y., Dahm, R., Karra, D., Kiebler, M. A., and Macchi, P. (2006) J. Neurosci. 26 6496–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassola, A., De Gaudenzi, J. G., and Frasch, A. C. (2007) Mol. Microbiol. 65 655–670 [DOI] [PubMed] [Google Scholar]
- 35.Jaruzelska, J., Kotecki, M., Kusz, K., Spik, A., Firpo, M., and Reijo-Pera, R. A. (2003) Dev. Genes Evol. 213 120–126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.