Abstract
Ikaros encodes a zinc finger protein that is involved in heritable gene silencing. In hematopoietic cells, Ikaros localizes to pericentromeric heterochromatin (PC-HC) where it recruits its target genes, resulting in their activation or repression via chromatin remodeling. The function of Ikaros is controlled by post-translational modifications. CK2 kinase has been shown to phosphorylate Ikaros at its C terminus, affecting cell cycle progression. Using in vivo labeling of murine thymocytes followed by phosphopeptide mapping, we identified four novel Ikaros phosphorylation sites. Functional analysis of phosphomimetic mutants showed that the phosphorylation of individual amino acids determines the affinity of Ikaros toward probes derived from PC-HC. In vivo experiments demonstrated that targeting of Ikaros to PC-HC is regulated by phosphorylation. The ability of Ikaros to bind the upstream regulatory elements of its known target gene terminal deoxynucleotidyltransferase (TdT) was decreased by phosphorylation of two amino acids. In thymocytes, Ikaros acts as a repressor of the TdT gene. Induction of differentiation of thymocytes with phorbol 12-myristate 13-acetate plus ionomycin results in transcriptional repression of TdT expression. This process has been associated with increased binding of Ikaros to the upstream regulatory element of TdT. Phosphopeptide analysis of in vivo-labeled thymocytes revealed that Ikaros undergoes dephosphorylation during induction of thymocyte differentiation and that dephosphorylation is responsible for increased DNA binding affinity of Ikaros toward the TdT promoter. We propose a model whereby reversible phosphorylation of Ikaros at specific amino acids controls the subcellular localization of Ikaros as well as its ability to regulate TdT expression during thymocyte differentiation.
Ikaros encodes a C2H2 zinc finger protein with expression that is restricted to hematopoietic cells and the pituitary gland (1–3). Experiments in knock-out mice established Ikaros as a protein essential for immune response and for normal hematopoiesis of the lymphoid, myeloid, and erythroid lineages (4–7). Ikaros negatively regulates cell cycle progression and acts as a tumor suppressor (8). The mechanism through which Ikaros controls the expression of its target genes remains unclear. Ikaros contains an activation domain (9) and has been shown to act as a direct transcriptional activator, although abundant data support a role for Ikaros in heritable gene silencing.
The majority of Ikaros protein has been shown to produce a punctate pattern of staining in the nuclei of lymphoid cells. Co-staining with antibodies against the murine homologue of Drosophila melanogaster heterochromatin protein-1 has shown that the Ikaros protein is localized to pericentromeric heterochromatin (PC-HC)3 (10). Using a combined immunofluorescence in situ hybridization approach, Ikaros has been shown to co-localize with γ-satellite-labeled centromeric regions (10). Localization of Ikaros within PC-HC was further confirmed by immunogold electron microscopy (11) and by correlating the ability of Ikaros to bind probes derived from the PC-HC region with its localization to PC-HC (12). Thus, the punctate pattern of staining observed for Ikaros protein is due to its pericentromeric localization.
Ikaros associates with Mi-2β, a catalytic subunit of the histone deacetylase complex, NuRD, as well as with BRG1, a catalytic subunit of the SWI/SNF nucleosome remodeling complex that acts as an activator of gene expression (13–15). Ikaros can also associate with two co-repressors, Sin3 and the C-terminal-binding protein (CtBP), which supports the hypothesis that Ikaros has a role in transcriptional repression (13, 16). The current hypothesis is that Ikaros binds the upstream region of target genes and aids in their recruitment to PC-HC, resulting in repression or activation of the gene (10, 17).
Studying the mechanism of Ikaros action is further complicated by the paucity of credible known Ikaros target genes. Ikaros has been shown to repress expression of the λ5 gene (18), whereas it positively regulates expression of the CD8α gene (19). The regulation of terminal deoxynucleotidyltransferase (TdT) gene expression during thymocyte differentiation has been extensively studied (20–23). Ikaros has been shown to bind in vivo to the D′ upstream regulatory element of the TdT gene where it competes with the Elf-1 transcription factor to regulate expression of TdT during thymocyte differentiation (20).
The activity of the most abundant Ikaros isoform is regulated by association with other Ikaros isoforms (24, 25) as well as association with other members of the Ikaros family (26). The association with smaller Ikaros isoforms that lack the DNA binding domain results in impaired Ikaros function; thus, these isoforms act as dominant negative mutants (24).
The function of Ikaros is also controlled by post-translational modifications. Sumoylation of Ikaros was found to regulate its interaction with Sin3, Mi-2β, and CtBP corepressors of transcription (27). During mitosis Ikaros is inactivated in a cell cycle-specific manner by phosphorylation at its evolutionarily conserved linker sequences (28). Ikaros is constitutively phosphorylated at multiple sites. A previous study identified several phosphorylated amino acids located primarily within the C-terminal region of Ikaros. Phosphorylation of Ikaros at its C-terminal region by CK2 kinase was shown to regulate the ability of Ikaros to control G1/S cell cycle progression (29). Recently, additional phosphorylation sites have been identified, although their functional and biological significance were not reported (30).
Here we identify and provide functional analysis of four additional Ikaros phosphorylation sites. Results show that the phosphorylation of particular amino acids alters Ikaros subcellular localization as well as its DNA binding affinity toward probes derived from PC-HC and from the regulatory elements of its target genes. We also provide evidence for the physiological role of reversible phosphorylation of these amino acids in controlling expression of TdT, a known Ikaros target gene, during thymocyte differentiation. Our results also suggest that these sites are targets for CK2 kinase or another kinase in the CK2 pathway. These data provide new evidence for the mechanism by which Ikaros controls expression of the TdT gene during T cell development and for the role of CK2 kinase in regulating Ikaros function.
EXPERIMENTAL PROCEDURES
Cells—The murine VL3-3M2 thymocyte leukemia cell line has been described previously (31). The human 293T endothelial kidney cell line and the murine the NIH/3T3 (3T3) fibroblast cell line were obtained from American Type Culture Collection (ATCC), Manassas, VA. Murine thymocytes were isolated as described previously (32) under an Institutional Animal Care and Use Committee-approved protocol. Treatment of thymocytes with PMA plus ionomycin has been described previously (20, 32).
Antibodies—The antibodies used to detect the C terminus (IK-CTS) of Ikaros (comprising amino acids (320–515 of the murine IK-VI isoform) have been described previously (33).
In Vivo Labeling and Phosphopeptide Mapping—For in vivo labeling, cells were incubated with radioactive orthophosphate. Cells were cultured in RPMI 1640 (Invitrogen) with 10% fetal calf serum (FCS) (CD4+/CD8+ murine thymocytes, murine peripheral T cells, and VL3-3M2) or with Dulbecco's modified Eagle's medium with 10% FCS (HEK293T). Cells were washed twice with phosphate-free RPMI 1640 medium and incubated for 4 h with 0.5 mCi/ml [32P]orthophosphate (PerkinElmer Life Sciences) in phosphate-free medium. Cells were collected by centrifugation, lysed on ice for 20 min in solubilizing buffer (50 mm Tris-HCL pH 7.2, 1% v/v Nonidet P-40, 150 mm NaCl, 5 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, and 5 μm leupeptin), and centrifuged at 15,000 rpm at 4 °C for 20 min. Lysates were incubated with anti-IK-CTS antibodies for 1 h at 4 °C, and the resulting immunocomplexes were absorbed to protein G-Sepharose (GE Healthcare), washed 4 times with solubilizing buffer, separated by SDS-PAGE, transferred to a nylon membrane, and subjected to autoradiography.
The phosphopeptide mapping of Ikaros from labeled cells was performed by two-dimensional separation on thin layer cellulose (TLC) plates. We have used a protocol described by Boyle et al. (34). Briefly, Ikaros was isolated by immunoprecipitation with anti-IK-CTS antibodies, transferred to the nitrocellulose membrane, and detected by autoradiography. The part of the membrane that contained the Ikaros band was excised and soaked in 0.5% polyvinylpyrrolidone (PVP-360; Sigma) in 0.1 m acetic acid at 37 °C for 30 min to block nonspecific absorption of trypsin. After brief washing with water, the membrane was resuspended in 50 mm ammonium bicarbonate and simultaneously digested with 10 μg of trypsin and 10 μg of chymotrypsin. Tryptic peptides were released from the membrane during digestion. After digestion, ammonium bicarbonate was removed by repeated lyophilization and resuspended in sterile water. Thin layer electrophoresis was carried out on a TLC plate in pH 1.9 buffer in the Hunter thin layer electrophoresis unit (HTLE 7000; CBS Scientific, Inc., Del Mar, CA) at 1000 V for 20 min. After electrophoresis, thin layer ascending chromatography was done in the second dimension using organic solvent buffer to separate peptides based on their ability to partition between mobile (buffer) and stationary (cellulose) phases. The mobility of phosphopeptides in the electrophoresis dimension is determined by the electrical charge and the mass of the peptide, whereas the mobility of the peptide in the ascending chromatography is determined by the hydrophobicity of the peptide. After chromatography, plates were dried and exposed.
Expression and Purification of Recombinant Ikaros Protein—Murine full-length Ikaros isoform IK-VI cDNA was amplified and cloned into a pGEX-2T glutathione S-transferase-containing vector (GE Healthcare) that was cleaved with BamH1 and EcoR1 as described previously (35). IK-VI-glutathione S-transferase-containing pGEX-2T plasmid was transformed and expressed into the SCS-1 strain of Escherichia coli (Stratagene, La Jolla, CA). E. coli were induced with 1 mm isopropylthiogalactopyranoside for 2 h in the presence of 100 μm ZnCl2. The glutathione S-transferase-IK-VI fusion protein was purified from E. coli according to the standard procedures (GE Healthcare) except that all of the buffers contained 10 μm ZnCl2 as described previously (35). Fractions that contained purified glutathione S-transferase-IK-VI protein were pooled and stored at –80 °C.
In Vitro Kinase Assay—The in vitro kinase reaction was carried out with 10 ng of purified recombinant full-length Ikaros IK-VI protein incubated with 4 units of CK2 kinase (New England Biolabs) for 30 min at 30 °C in the presence of [γ-32P]ATP. Immunoprecipitated Ikaros protein was resolved by SDS-PAGE and transferred to nitrocellulose membrane as described previously (28). Proteolytic digestion of the Ikaros band was performed simultaneously with trypsin and chymotrypsin. Phosphopeptide mapping was performed as described above and previously (34).
Plasmids, Transfection, and Retroviral Transduction—Alanine or aspartate phosphomimetic substitution mutants for IK-VI were generated using the QuikChange method (Stratagene). For retrovirus generation, phosphomimetic IK-VI mutants were amplified by PCR and cloned between BglII and EcoRI sites of the murine stem cell virus internal ribosome entry site green fluorescent protein vector as described previously (12, 28). The full-length Ikaros isoform IK-VI was cloned into the mammalian expression vector pcDNA3 (Invitrogen), and its phosphomimetic mutants were transfected into 293T cells via the calcium phosphate method. 3T3 cells were transduced with ecotropic retrovirus.
Confocal Microscopy—3T3 cells were transduced with ecotropic retrovirus and analyzed by confocal microscopy as described previously (12). Images were acquired at room temperature by a Leica TCS-SP MP Confocal and Multiphoton Microscope with a Leica DM-LFS body (upright fixed-stage microscope) using a 100× Leica HX PLAPO (Planapo) oil immersion lens with numerical aperture of 1.4 (Heidelberg, Germany).
Biochemical Experiments—Nuclear extractions, Western blots, and gel-shift experiments were performed as described previously (12, 20), The γ satellite A, γ satellite B, and TdT D′ gel shift probes have been described previously (12).
Mass Spectrometry—After the in vitro kinase reaction with CK2 kinase, samples were digested with trypsin in solution. Peptides were loaded onto a self-packed column using Michrom (Auburn, CA) “magic C18” media. The peptides were then eluted using a reverse phase gradient and injected into the instrument via nanospray electrospray ionization. Spectra were collected using a linear trap quadrupole Fourier-transformed from Thermo Fisher Scientific (San Jose, CA). We screened for phosphopeptides using a neutral loss experiment during the acquisition. MS3 scanning events were triggered by detection of a neutral loss during the MS2 scan event of 98.0, 49.0, or 32.7 m/z. The data were searched against a human protein data base. Identified phosphopeptides from the search which were triggered by the neutral loss experiment were then manually inspected to verify the sites of phosphorylation.
RESULTS
Identification of New Ikaros Phosphorylation Sites by in Vivo Labeling and Phosphopeptide Mapping—Previous reports provided evidence that Ikaros is phosphorylated at multiple sites by CK2 kinase. Phosphorylation of several amino acids had been mapped in the region between amino acids 383 and 404 within exon 7 as well as amino acid 63 (29), but sites that are less abundantly phosphorylated remained unknown. To identify the additional phosphorylation sites, we performed in vivo phosphopeptide mapping of Ikaros in murine thymocytes and in the murine thymocyte leukemia cell line VL3-3M2 (Fig. 1A and data not shown). To identify less abundantly phosphorylated sites, we used 5 × 107 cells for in vivo labeling with [32P]orthophosphate. After immunoprecipitation and separation on SDS-PAGE, a double digestion of the phosphorylated protein was performed with both trypsin and chymotrypsin to enhance resolution of the phosphorylated peptides.
FIGURE 1.
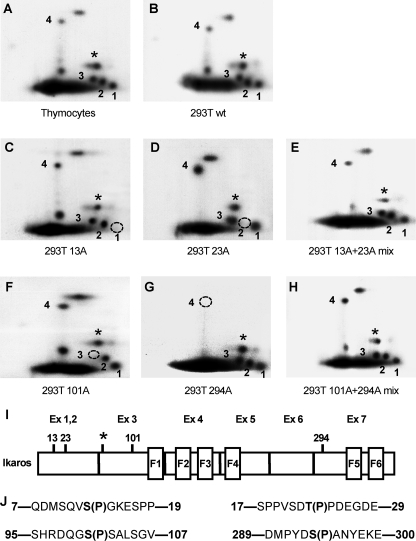
Identification of novel Ikaros phosphorylation sites. Two-dimensional phosphopeptide maps were generated with wild-type Ikaros from murine thymocytes (A) and with wild-type and mutant Ikaros proteins expressed in HEK293T cells (B–H). Phosphopeptides representing novel phosphorylation sites are numbered 1–4. A star indicates a phosphopeptide representing a previously identified phosphorylation site (amino acid 63). Phosphomimetic Ikaros mutants contain substitutions with alanine (A) or aspartate (D) at the indicated amino acids. Phosphopeptides that are absent in each mutant protein are indicated by a dashed circle. Simultaneous loading of 13A and 23A or 101A and 294A restores all phosphopeptides. I, positions of novel and previously identified (*) phosphorylation sites within the Ikaros protein. J, sequences surrounding novel Ikaros phosphorylation sites.
Two-dimensional phosphopeptide mapping of double-digested phosphoprotein revealed that Ikaros is phosphorylated constitutively at multiple sites with several phosphopeptides being easily detectable and resolved (Fig. 1A). The observed phosphorylation pattern was identical in VL3-3M2 cells, activated murine peripheral T cells, and the murine B cell line Bal17 (data not shown). A similar phosphopeptide mapping pattern for IK-VI was detected when this isoform was ectopically expressed in 293T (human embrionyl kidney carcinoma) cells (Fig. 1B), suggesting that this phosphorylation was not tissue-specific.
Phosphopeptide analysis of deletion mutants that span the entire Ikaros protein revealed that three different phosphopeptides are localized within the N-terminal end of Ikaros, whereas the fourth additional phosphopeptide is localized within exon 7 (data not shown). An examination of the potential phosphoacceptors within the N-terminal region of Ikaros suggested that amino acids 13, 23, and 101 might be phosphorylated. In addition, based on theoretical mobilities of the potential Ikaros phosphopeptides (34), we calculated that amino acid 294 might be phosphorylated as well. To test this hypothesis and to identify phosphorylated amino acids, potential phosphoacceptor sites were mutated into alanine, and phosphopeptide mapping of these mutants was performed. Results showed that mutation of serine 13 abolishes phosphopeptide 1 (Fig. 1C), whereas mutation of threonine 23 abolishes phosphopeptide 2 (Fig. 1D). Simultaneous loading of the 13A and 23A samples restored all of the phosphopeptides observed with the wild-type protein (Fig. 1E). Similarly, alanine mutation of serine 101 abolished the phosphopeptide 3 (Fig. 1F), and mutation of serine 294 abolished phosphopeptide 4 (Fig. 1G). When 101A and 294A samples were simultaneously loaded, the wild-type map was again restored (Fig. 1H). These experiments identified four novel phosphorylation sites in the Ikaros protein. The positions of these sites are shown in Fig. 1, panel I. We also identified one additional phosphopeptide (data not shown) that represented a previously described phosphorylation at amino acid 63 (labeled as a star in Fig. 1). All four novel phosphorylated amino acids along with their surrounding sequences (Fig. 1J) are evolutionarily conserved between mouse and human, which suggests their biological relevance.
Phosphorylation of Specific Amino Acids Determines the Ability of Ikaros to Bind DNA Probes Derived from Pericentromeric Heterochromatin—The above results show that Ikaros is phosphorylated at multiple N-terminal amino acids. Phosphorylation of the Ikaros protein has been postulated to control its DNA binding ability. To determine whether phosphorylation of amino acids 13, 23, 63, 101, or 294 affects the ability of Ikaros to bind DNA, gel shift experiments were performed with nuclear extracts from HEK293T cells that overexpressed wild-type Ikaros or its phosphomimetic mutants. In phosphomimetic mutants the mutation of phosphorylation sites to alanine mimics constitutive dephosphorylation, whereas mutation into aspartate mimics constitutive phosphorylation.
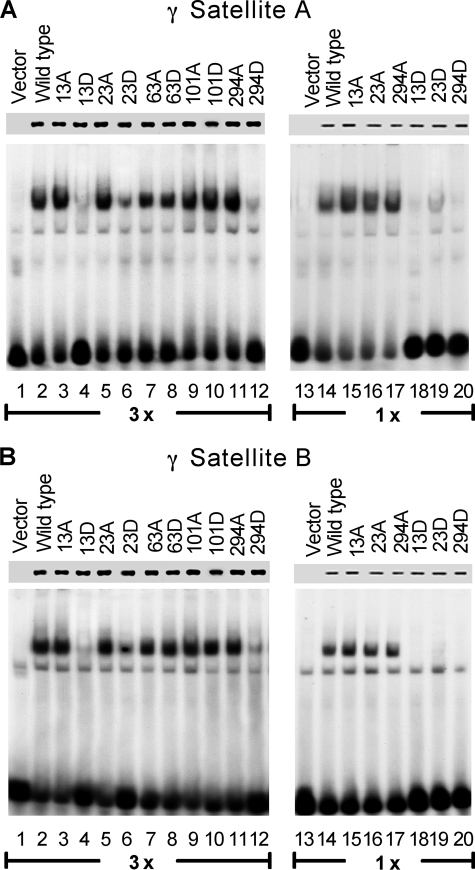
A major component of Ikaros function is localization to PC-HC. Ikaros binds to repetitive sequences within PC-HC that contain consensus Ikaros binding sites, and its localization to the PC-HC is directly related to its ability to bind these sequences (12). We used radiolabeled DNA probes that contain a high affinity binding site from γ satellite repeat sequence that has previously been shown to confer Ikaros with the ability to localize to PC-HC in mouse (12). The first probe, γ satellite A, contains two consensus Ikaros binding sites in close proximity to each other and is a target for high affinity binding of wild-type Ikaros. The second probe, γ satellite B, contains a single consensus Ikaros binding site that provides a target for low affinity binding of wild-type Ikaros protein. Potential differences in DNA binding affinity were quantified by phosphoimaging using the ImageQuant 5.1 program to measure the strength of shifted bands. Results showed that phosphomimetic substitutions to aspartate at amino acids 13 or 294 decrease the ability of Ikaros to bind the γ satellite A probe by 5-fold. Aspartate mutation of amino acid 23 reduces the ability of Ikaros to bind the same probe by 3-fold in both high extract (Fig. 2A, lanes 1–12) and low extract concentrations (Fig. 2A, lanes 13–20). Substitutions of the same amino acids to alanine did not change the DNA binding affinity of Ikaros for the γ satellite A probe (Fig. 2A). Similar results were obtained when γ satellite B probe was used (Fig. 2B). Phosphomimetic substitutions at positions 13, 23, or 294 reduced binding ability toward γ satellite B probe by 3–4-fold in high extract concentrations (Fig. 2B, lanes 1–12), whereas DNA binding was abolished in low extract concentrations (Fig. 2B, lanes 13–20). No changes in DNA binding affinity toward the γ satellite A or γ satellite B probes were observed with phosphomimetic substitutions at positions 63 or 101. These data demonstrate that phosphorylation of a single amino acid can determine DNA binding affinity of Ikaros toward γ satellite repeats.
FIGURE 2.
Phosphomimetic substitutions reduce the ability of Ikaros to bind a DNA probe derived from the pericentromeric heterochromatin region. Nuclear extracts were prepared from HEK293T cells expressing wild-type or mutant Ikaros proteins. Phosphomimetic Ikaros mutants contain substitutions with alanine or aspartate at the indicated amino acids. Equal amounts of Ikaros proteins in nuclear extracts were confirmed by Western blot (upper lane). Gel shifts were performed with two extracts concentrations (lanes 1–12, 3×, 6 μg per reaction; lanes 13–20, 1×, 2 μg per reaction). Gel shifts were performed using radiolabeled probes derived from the murine γ satellite A repeat (A) or the murine γ satellite B repeat (B).
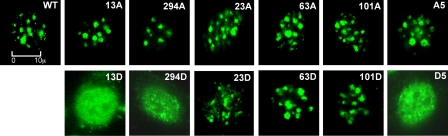
Phosphorylation Regulates the Targeting of Ikaros to Pericentromeric Heterochromatin—In hematopoietic cells, wild-type Ikaros localizes in pericentromeric foci and produces a punctate pattern when visualized by confocal microscopy (10). To determine whether the decreased DNA binding of Ikaros toward γ satellite probes affects its subcellular localization, we expressed wild-type and phosphomimetic Ikaros mutants in 3T3 cells by retroviral transduction. Retrovirus-introduced Ikaros has been shown to localize to regions of PC-HC in the nucleus of 3T3 cells in the same manner as in hematopoietic cells. Because 3T3 cells do not express endogenous, wild-type Ikaros, these cells have previously been used to study the effects of Ikaros mutations on the subcellular localization of Ikaros proteins (12). The introduction of wild-type Ikaros showed punctate staining, which is characteristic of its localization at regions of PC-HC in 3T3 cells, as demonstrated previously (12) (Fig. 3). Results showed that a single aspartate phosphomimetic mutation at amino acids 13 or 294 caused redistribution of Ikaros protein in the nucleus from pericentromeric localization to a diffuse nuclear pattern (Fig. 3, 13D and 294D). The aspartate phosphomimetic mutation at amino acid 23 preserved pericentromeric localization, although non-centromeric, diffuse nuclear localization was increased compared with the wild-type protein (Fig. 3, 23D). In contrast, alanine mutants did not display any changes in subcellular localization of Ikaros (Fig. 3, 13A, 23A, and 294A) compared with the wild-type. The aspartate phosphomimetic mutations at positions 63 and 101 did not affect pericentromeric localization of Ikaros (Fig. 3, 63D and 101D). The combined aspartate phosphomimetic mutation of all five amino acids caused re-distribution of Ikaros protein in the nucleus to a diffuse nuclear pattern (Fig. 3, D5), whereas simultaneous mutation of all five amino acids to alanine did not alter subcellular localization of Ikaros (Fig. 3, A5). These data suggest that targeting of Ikaros to PC-HC is regulated by phosphorylation at specific amino acids.
FIGURE 3.
Phosphomimetic substitutions abolish pericentromeric targeting of Ikaros. Confocal fluorescence was used to examine the subnuclear localization of wild-type (WT) and mutant Ikaros after retroviral transduction into 3T3 cells. Phosphomimetic Ikaros mutants contain substitutions with alanine or aspartate at indicated amino acids. A5 and D5 represent alanine or aspartate mutants of all 5 amino acids 13, 23, 63, 101, and 294. Single cell images are shown.
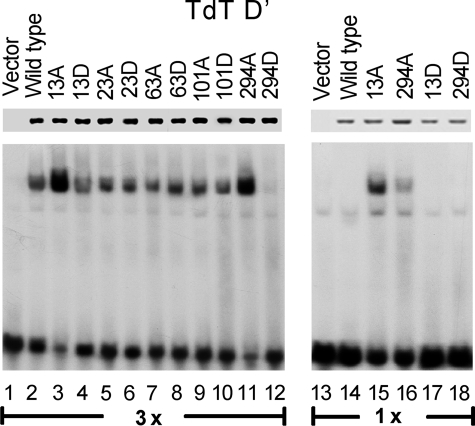
Phosphorylation Regulates the DNA Binding Affinity of Ikaros to Regulatory Elements of the TdT Gene—Ikaros regulates expression of several genes during lymphoid development. We sought to determine how phosphorylation of Ikaros affects its DNA binding affinity toward its known in vivo target genes. Previously Ikaros has been shown to bind to the upstream regulatory sequence D′ of the TdT gene and to negatively regulate TdT expression during T cell development (20). We tested the ability of phosphomimetic Ikaros mutants to bind the TdT D′ regulatory sequence by gel shift assay (Fig. 4). Results showed that alanine mutation of amino acids 13 or 294 increase DNA binding affinity toward the TdT D′ regulatory sequence, whereas aspartate mutations of amino acid 294 decreased its ability to bind DNA 3-fold compared with wild-type (Fig. 4, lanes 1–12). This difference was even stronger when the low extract concentration was used (Fig. 4, lanes 13–20). These data suggest that phosphorylation of Ikaros controls its ability to bind to the regulatory sequence of the TdT gene.
FIGURE 4.
Phosphomimetic substitutions alter the DNA binding affinity of Ikaros toward the upstream regulatory element of the TdT gene. Nuclear extracts were prepared from HEK293T cells expressing wild-type or mutant Ikaros proteins. Phosphomimetic Ikaros mutants contain substitutions with alanine or aspartate at the indicated amino acids. Equal amounts of Ikaros proteins in nuclear extracts were confirmed by Western blot (upper lane). Gel shifts were performed with two extracts concentrations (lanes 1–12, 3×, 6μg per reaction; lanes 13–20, 1×, 2μg per reaction) using a radiolabeled probe that contains the TdT D′ regulatory element.
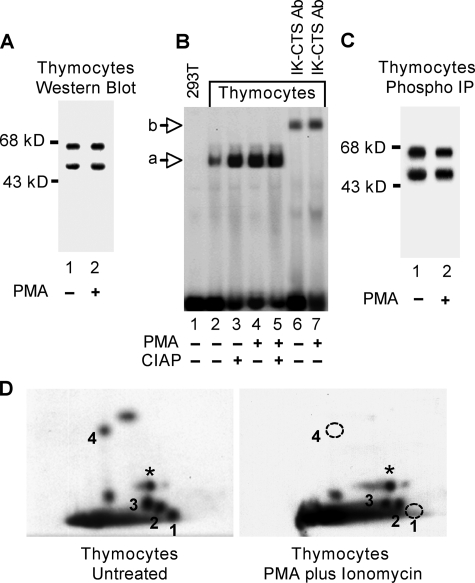
Dephosphorylation of Ikaros during Thymocyte Differentiation Increases Its DNA Binding Affinity toward the TdT Promoter—The regulation of TdT gene expression by Ikaros during T cell differentiation has been studied extensively. In immature CD4+/CD8+ lymphocytes and in VL3-3M2 cells, the TdT gene is well expressed, and its expression is positively regulated by the Ets family member Elf-1 (21). Both Ikaros and Elf-1 have been shown to bind the element in the TdT promoter that is required for transcription (21, 23, 33, 35). Treatment of thymocytes with PMA plus ionomycin leads to rapid down-regulation of TdT expression. The binding of Ikaros to the TdT D′ regulatory sequence after PMA plus ionomycin treatment has been shown to be directly responsible for the down-regulation of TdT expression (20). A competition model has been proposed in which lymphocyte stimulation is followed by a switch from Elf-1 to Ikaros occupancy of the TdT D′ regulatory sequence leading to repression of TdT gene expression (20). The molecular mechanism for this switch is unknown. We hypothesize that changes in phosphorylation of Ikaros during thymocyte differentiation contribute to its increased affinity to the TdT D′ regulatory element.
To test this hypothesis we examined the relationship between the differentiation status of CD4+/CD8+ thymocytes and the ability of Ikaros to bind the TdT D′ regulatory element. Nuclear extracts from untreated CD4+/CD8+ thymocytes and from thymocytes 24 h after induction of differentiation with PMA plus ionomycin were normalized to contain the same amount of Ikaros isoforms V and VI as evidenced by Western blot (Fig. 5A). Electromobility shift assay was performed under conditions that prevent binding of Elf-1 (20). Results revealed that the affinity of Ikaros toward the TdT D′ regulatory region is reduced by 3-fold in untreated thymocytes as compared with PMA plus ionomycin-treated cells (Fig. 5B, lanes 2 and 4). Phosphatase treatment of nuclear extracts from untreated thymocytes increases the ability of Ikaros to bind the TdT D′ regulatory region to the same level observed in stimulated cells, whereas the same treatment does not induce significant changes in PMA plus ionomycin-treated cells (Fig. 5B, lanes 2–5). These results suggest that decreased affinity of Ikaros toward the TdT D′ regulatory element in unstimulated thymocytes is due to direct phosphorylation. The complete super-shifting of the DNA-protein complex by incubation with anti IK-CTS antibodies confirmed that the complex observed in the gel shift assay contains Ikaros proteins and not Elf-1 (Fig. 5B, lanes 6 and 7).
FIGURE 5.
Dephosphorylation of Ikaros in thymocytes after PMA plus ionomycin treatment. Nuclear extracts of unstimulated and PMA plus ionomycin-treated thymocytes were obtained and normalized for Ikaros concentrations by Western blot (A). B, DNA binding activities of Ikaros toward the TdT D′ regulatory elements were compared by gel shift in the absence lanes (2 and 4) or presence (lanes 3 and 5) of calf-intestine alkaline phosphatase (CIAP, 10 units). Ab, antibody. Unstimulated and PMA plus ionomycin-treated murine thymocytes were grown in the presence of [32P]orthophosphate. Ikaros proteins were immunoprecipitated (IP) from nuclear extract using anti-Ikaros antibodies (C) and used to generate two-dimensional phosphopeptide maps (D). Phosphopeptides representing novel phosphorylation sites are numbered 1–4. A star indicates a phosphopeptide representing a previously identified phosphorylation site (amino acid 63). The two phosphopeptides that were not detected in PMA plus ionomycin-treated thymocytes are indicated with a dashed circle.
Two-dimensional phosphopeptide mapping was used to determine whether the phosphorylation of Ikaros changes after the induction of differentiation. Unstimulated and PMA plus ionomycin treated thymocytes were in vivo labeled with [32P]orthophosphate. Immunoprecipitation of Ikaros from nuclear extracts followed by SDS-PAGE separation and exposure to film showed that Ikaros isoforms were phosphorylated in both samples (Fig. 5C). Two-dimensional phosphopeptide mapping of the largest murine Ikaros isoform, IK-VI, revealed that Ikaros undergoes dephosphorylation of amino acids 13 and 294 after PMA plus ionomycin treatment of CD4+/CD8+ thymocytes (Fig. 5).
These data show that the DNA binding affinity of Ikaros toward the TdT D′ regulatory element during thymocyte differentiation is controlled by phosphorylation at specific amino acids. Results imply that the reversible phosphorylation of Ikaros at amino acids 13 and 294 is physiologically significant in regulating the expression of the TdT gene during thymocyte differentiation. Our studies suggest that dephosphorylation of Ikaros is one of the major factors regulating the switch from Elf-1 to Ikaros occupancy of the TdT D′ regulatory element and the subsequent repression of TdT transcription.
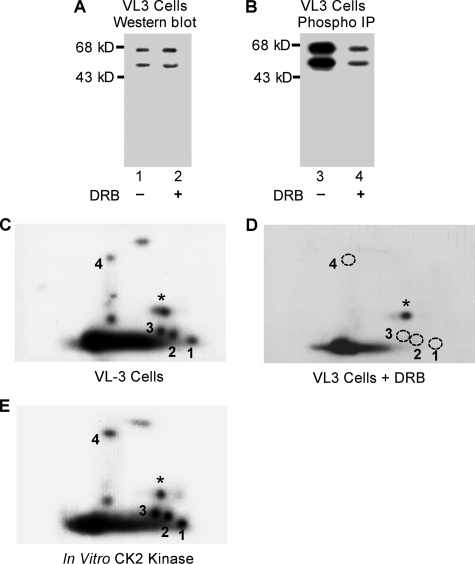
Phosphorylation of the Four Novel Sites Is Dependent on CK2 Kinase Activity—Ikaros is phosphorylated by CK2 kinase at multiple sites. It has been suggested that CK2 kinase is responsible for the majority of Ikaros phosphorylation (29). We studied the phosphorylation of Ikaros in the VL3-3M2 murine thymocyte cell line (31). This cell line exhibits properties of CD4+/CD8+ thymocytes, and PMA plus ionomycin treatment leads to repression of TdT expression (20, 31). We performed in vivo [32P]orthophosphate labeling of VL3-3M2 cells that were either untreated or treated for 1 h with DRB, a specific inhibitor of CK2 kinase. Immunoprecipitation of equal amounts of Ikaros (evidenced by Western blot, Fig. 6A) from nuclear extracts of untreated and DRB-treated cells showed that phosphorylation of Ikaros is reduced by 6-fold after treatment with CK2 inhibitors (Fig. 6B). These data are in agreement with previously published results and suggest that CK2 kinase is the major kinase responsible for phosphorylation of Ikaros, although the role of other kinases cannot be excluded.
FIGURE 6.
Phosphorylation of the four novel sites is dependent on CK2 kinase activity. A, VL3-3M2 cells were treated with 50 μm DRB to inhibit CK2 activity, and Ikaros protein concentrations in treated and untreated cells were determined by Western blot. B, Ikaros proteins were immunoprecipitated (IP), resolved on SDS-PAGE gel, transferred to nitrocellulose membrane, and exposed to measure 32P incorporation. Two-dimensional phosphopeptide maps of Ikaros from untreated (C) or DRB-treated (D) VL3-3M2 cells were generated. Phosphopeptides representing novel phosphorylation sites are numbered 1–4. A star indicates a phosphopeptide representing a previously identified phosphorylation site (amino acid 63). The phosphopeptides that were not detected in VL3-3M2 cells after DRB treatment are indicated with a dashed circle. E, phosphopeptide mapping of an in vitro kinase assay with recombinant Ikaros and CK2 kinase.
We compared phosphopeptide maps from in vivo labeled endogenous Ikaros in untreated VL3-3M2 cells (Fig. 6C) and Ikaros from VL3-3M2 cells after treatment with CK2 inhibitors (Fig. 6D). Phosphopeptide mapping of the Ikaros from VL3-3M2 cells treated with CK2 inhibitors produced a pattern in which phosphorylation was undetectable at amino acids 13, 23, 101, and 294. At the same time phosphorylation at amino acid 63 was easily detectable along with phosphorylation at several other amino acids, which were difficult to resolve (Fig. 6D). These data suggest that the physiological activity of CK2 kinase is necessary for in vivo phosphorylation of amino acids 13, 23, 101, and 294. It appears that phosphorylation of these four amino acids requires high levels of CK2 kinase activity, whereas low CK2 activity is sufficient for phosphorylation of amino acid 63.
To determine whether CK2 kinase can directly phosphorylate the novel sites that we identified, an in vitro kinase reaction was performed with CK2 kinase and recombinant Ikaros. Phosphopeptide mapping of Ikaros phosphorylated in vitro by CK2 kinase (Fig. 6E) produced a similar phosphopeptide map to the one obtained from VL3-3M2 cells phosphorylated in vivo (Fig. 6C). This suggests that most of the in vivo phosphopeptides observed in our experiments are the result of phosphorylation by CK2 kinase. These phosphopeptides co-migrated with those from VL3-3M2 cells when VL3-3M2 samples were loaded together with in vitro CK2 kinase samples (data not shown). These results suggest that CK2 kinase can phosphorylate amino acids 13, 23, 63, 101, and 294 in vitro.
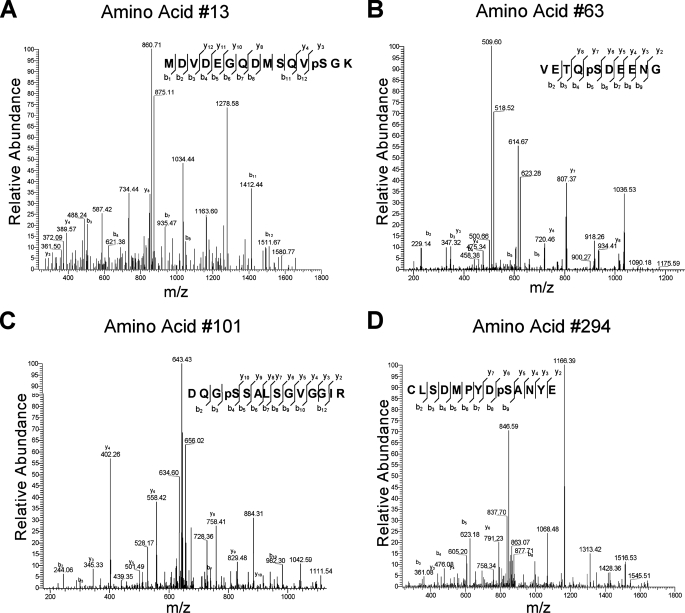
To provide additional confirmation that CK2 kinase can phosphorylate Ikaros at the novel sites, a mass spectrometry analysis was performed on recombinant Ikaros after an in vitro kinase assay with CK2. Phosphorylated Ikaros was digested with trypsin. Peptides were analyzed by nano-liquid chromatography/electrospray ionization/tandem mass spectroscopy. Phosphopeptides were screened using a neutral loss experiment during acquisition. Identified peptides containing phosphorylation sites and triggered by the neutral loss experiment were then manually inspected to verify the sites of phosphorylation. Results identified phosphopeptides that contain three of the four novel phosphorylation sites as well as amino acid 63 (Fig. 7). These data confirm that CK2 kinase can phosphorylate three of the four novel phosphorylation sites.
FIGURE 7.
Mass spectrometry analysis of recombinant Ikaros after in vitro phosphorylation by CK2 kinase. Tandem mass spectrum of four phosphopeptides (A–D). A sufficient number of complementary Bn anions (N terminus-derived fragment ions) and Yn ions (C terminus-derived fragment ions) were detected to assign the phosphorylation sites to particular amino acids.
Results obtained by in vitro kinase assay with mass spectrometry and phosphopeptide mapping as well as phosphopeptide mapping of VL3-3M2 cells that are treated with a CK2 inhibitor suggest that the activity of CK2 kinase is essential for phosphorylation of amino acids 13, 23, 101, and 294. In addition, the phosphorylation of these sites appears to be more sensitive to the effects of CK2 kinase inhibitors as compared with the other phosphoacceptor sites.
DISCUSSION
To detect novel phosphorylation sites of Ikaros, we enhanced the sensitivity of assays and the resolution of phosphopeptides. To achieve this we used large numbers of cells for in vivo labeling followed by double trypsin/chymotrypsin digestion of phosphorylated Ikaros. This approach allowed us to identify four novel phosphorylation sites that we confirmed by phosphopeptide mapping of their corresponding alanine mutants.
Protein phosphorylation is a reversible, dynamic process. Below we present evidence for two possible physiological functions of Ikaros phosphorylation at novel sites.
Regulation of Subcellular Localization—The majority of Ikaros protein is localized within PC-HC, with a smaller amount detectable in euchromatin (11). Targeting of Ikaros to PC-HC is achieved by its direct binding to the γ satellite repeats located within PC-HC (12). The function of Ikaros is to bind to the upstream regions of target genes and to aid in recruiting them to PC-HC, leading to their repression (10). This model assumes the existence of a mechanism that reversibly regulates the dissociation of Ikaros protein from pericentromeric repeats and from the upstream regulatory regions of its target genes. We propose that phosphorylation of specific amino acids represents such a mechanism and that the balance between the amounts of dephosphorylated and phosphorylated Ikaros proteins regulates the physiological function of Ikaros. Because the majority of the Ikaros protein is localized in PC-HC in hematopoietic cells (10, 12, 36), candidate regulatory amino acids are expected to be less frequently phosphorylated than other residues. An increase in the proportion of Ikaros with phosphorylation at these amino acids would cause redistribution of the Ikaros complex to other parts of the nucleus. In 3T3 cells where endogenous wild-type Ikaros is not present, the introduction of Ikaros-containing single phosphomimetic mutations at amino acids 13 or 294 produces a diffuse nuclear distribution. This is most likely due to the Ikaros decreased DNA binding affinity toward repetitive sequences located within the γ satellite region of PC-HC since it has been demonstrated that a 3-fold decrease in the DNA binding affinity of Ikaros toward γ satellite repetitive sequence is associated with the loss of PC-HC localization (28).
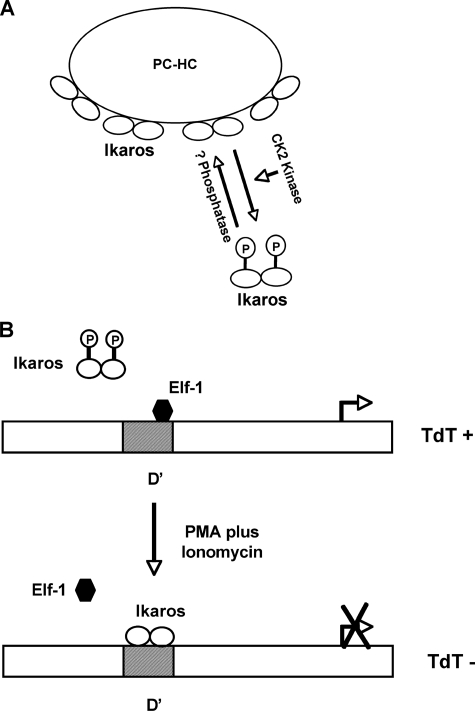
We propose that a transient increase in the amount of phosphorylated proteins (due to increased activity of a particular kinase or decreased activity of phosphatases) would lead to dissociation of Ikaros from PC-HC and its targeting to other parts of the nucleus (Fig. 8A). Continuous alteration of the phosphorylation status of Ikaros would, thus, severely impact its activity.
FIGURE 8.
Regulation of Ikaros activity by phosphorylation at the novel sites. A, the balance between hyperphosphorylation of Ikaros (by CK2 kinase or other unidentified kinases) and its dephosphorylation by unknown phosphatases cause Ikaros to reversibly localize at PC-HC. B, a working model of the regulation of TdT expression during the early stages of thymocyte differentiation (modified from Trinh et al. (20)). Dephosphorylation of Ikaros during thymocyte differentiation (mimicked by induction of differentiation with PMA plus ionomycin) increases the DNA binding affinity of Ikaros toward the D′ regulatory region, resulting in the displacement of Elf-1 and the repression of the TdT gene.
Regulation of the Expression of the TdT Gene during Lymphoid Development—Extensive studies of the regulation of TdT expression have identified Ikaros and Elf-1 as regulatory proteins that compete for overlapping sequences of the regulatory element D′ within the TdT promoter (21, 23, 33, 35). Our results suggest that dephosphorylation of amino acids 13 and 294 increase the DNA binding affinity of Ikaros toward the D′ element (Fig. 4). This suggests that the presence of small amounts of hyperphosphorylated Ikaros decreases the ability of the Ikaros complex to bind the TdT D′ regulatory element. Further experiments showed that the DNA binding affinity of Ikaros toward the D′ element is increased in stimulated VL3-3M2 cells and that phosphorylation of Ikaros is responsible for its decreased DNA binding affinity in unstimulated cells (Fig. 5B). Phosphopeptide mapping revealed that Ikaros is dephosphorylated after PMA plus ionomycin treatment at amino acids 13 and 294 (Fig. 5C). We propose that dephosphorylation of Ikaros at amino acids 13 and 294 after PMA plus ionomycin treatment leads to increased DNA binding affinity toward the D′ regulatory element, displacement of Elf-1, and repression of TdT expression (Fig. 8B). This event also leads to changes in chromatin remodeling and to repositioning of the TdT gene to PC-HC and changes described above. Subsequently, changes in histone methylation along with CpG methylation lead to irreversible gene silencing (22, 32). This model does not rule out the possibility that other events, changes in expression of Elf-1, or its post-translational modification, or changes in expression of other Ikaros family members (e.g. Helios), might also contribute to TdT silencing, but our data suggest that dephosphorylation of Ikaros is a major mechanism in the initiation of TdT repression during thymocyte differentiation.
Ikaros is a physiological substrate for CK2 kinase (29). Inhibition of CK2 kinase activity in vivo leads to severely decreased Ikaros phosphorylation. Treatment of VL3-3M2 cells with CK2 inhibitors decreased the phosphorylation of other known constitutive phosphorylation sites but abolished phosphorylation of the novel four phosphorylation sites (Fig. 6D). This suggests that phosphorylation of these four amino acids is dependent on CK2 activity and that their phosphorylation is part of the CK2 signaling pathway. Phosphopeptide mapping of Ikaros after in vitro phosphorylation by CK2 kinase closely resembles the phosphopeptide map obtained by in vivo labeling (Fig. 6, C and E), suggesting that CK2 kinase can phosphorylate all four newly identified sites. Mass spectrometry confirmed that CK2 kinase can directly phosphorylate three of four in vivo phosphorylated amino acids (Fig. 7). However, all of the four phosphorylation sites are surrounded by sequences that are not optimal for CK2 kinase activity, which makes them weak substrates for CK2 kinase (37, 38). It should be noted that at least three physiological substrates of CK2, p53 (39), HMG14 (40), and clathrin β light chain (41), lack the typical consensus motif. It has been suggested that tertiary structure might also affect the ability of CK2 kinase to act on a particular substrate (42). At this moment it is unclear whether the novel phosphorylation sites are directly phosphorylated by CK2 kinase or whether they are substrates for additional kinase(s) that act along the CK2 pathway. More studies will need to be done to further address that question.
Increased expression of the catalytic subunit of CK2 kinase causes T cell leukemia/lymphoma in transgenic animals via various pathways (43–46). Overexpression of the CK2 catalytic subunit has also been associated with lymphoproliferative and autoimmune disease (45). Interestingly, decreased activity of Ikaros has been associated with T cell lymphoma and hyperimmune response (5, 8, 47). Phosphorylation of amino acids 13 and 294 by CK2 kinase as well as phosphorylation of additional amino acids that have been previously defined can provide a mechanistic explanation of this effect of increased CK2 kinase. According to our hypothesis, the pro-oncogenic activity of overexpressed CK2 kinase would be mediated partially through altered phosphorylation of Ikaros and subsequent changes in Ikaros activity.
In summary, we report four novel phosphorylation sites of the Ikaros protein. Phosphorylation of two amino acids regulates targeting of Ikaros to PC-HC as well as its affinity toward the promoter of the TdT gene. During thymocyte differentiation, an alteration in phosphorylation at these sites contributes to the down-regulation of TdT expression, thus providing evidence of the physiological significance of this process. We suggest that the activity of CK2 kinase directly or indirectly controls phosphorylation of these amino acids. Future studies that test the models shown in Fig. 8 will aid in understanding the regulatory mechanisms of lymphocyte development.
This work was supported by National Institutes of Health Grants K22 CA 111392 (to S. D.) and 5K01 DK066163 (to K. J. P.), a Midwest Athletes Against Childhood Cancer grant award, and a Cure Kids Cancer Coalition grant (to S. D.). This work was also supported by the University of Wisconsin Medical Education and Research Committee New Investigator Program (to S. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PC-HC, pericentromeric heterochromatin; TdT, terminal deoxynucleotidyltransferase; IK-CTS, C terminus of Ikaros; PMA, phorbol 12-myristate 13-acetate; DRB, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole.
References
- 1.Georgopoulos, K., Moore, D. D., and Derfler, B. (1992) Science 258 808–812 [DOI] [PubMed] [Google Scholar]
- 2.Yu, S., Asa, S. L., and Ezzat, S. (2002) Mol. Endocrinol. 16 1069–1078 [DOI] [PubMed] [Google Scholar]
- 3.Yoshida, T., Ng, S. Y., Zuniga-Pflucker, J. C., and Georgopoulos, K. (2006) Nat. Immunol. 7 382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu, L., Nichogiannopoulou, A., Shortman, K., and Georgopoulos, K. (1997) Immunity 7 483–492 [DOI] [PubMed] [Google Scholar]
- 5.Avitahl, N., Winandy, S., Friedrich, C., Jones, B., Ge, Y., and Georgopoulos, K. (1999) Immunity 10 333–343 [DOI] [PubMed] [Google Scholar]
- 6.Winandy, S., Wu, L., Wang, J. H., and Georgopoulos, K. (1999) J. Exp. Med. 190 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumortier, A., Kirstetter, P., Kastner, P., and Chan, S. (2003) Blood 101 2219–2226 [DOI] [PubMed] [Google Scholar]
- 8.Winandy, S., Wu, P., and Georgopoulos, K. (1995) Cell 83 289–299 [DOI] [PubMed] [Google Scholar]
- 9.Molnar, A., and Georgopoulos, K. (1994) Mol. Cell. Biol. 14 8292–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, K. E., Guest, S. S., Smale, S. T., Hahm, K., Merkenschlager, M., and Fisher, A. G. (1997) Cell 91 845–854 [DOI] [PubMed] [Google Scholar]
- 11.Klug, C. A., Morrison, S. J., Masek, M., Hahm, K., Smale, S. T., and Weissman, I. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobb, B. S., Morales-Alcelay, S., Kleiger, G., Brown, K. E., Fisher, A. G., and Smale, S. T. (2000) Genes Dev. 14 2146–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koipally, J., Renold, A., Kim, J., and Georgopoulos, K. (1999) EMBO J. 18 3090–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J., Sif, S., Jones, B., Jackson, A., Koipally, J., Heller, E., Winandy, S., Viel, A., Sawyer, A., Ikeda, T., Kingston, R., and Georgopoulos, K. (1999) Immunity 10 345–355 [DOI] [PubMed] [Google Scholar]
- 15.O'Neill, D. W., Schoetz, S. S., Lopez, R. A., Castle, M., Rabinowitz, L., Shor, E., Krawchuk, D., Goll, M. G., Renz, M., Seelig, H. P., Han, S., Seong, R. H., Park, S. D., Agalioti, T., Munshi, N., Thanos, D., Erdjument-Bromage, H., Tempst, P., and Bank, A. (2000) Mol. Cell. Biol. 20 7572–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koipally, J., and Georgopoulos, K. (2000) J. Biol. Chem. 275 19594–19602 [DOI] [PubMed] [Google Scholar]
- 17.Liberg, D., Smale, S. T., and Merkenschlager, M. (2003) Trends Immunol. 24 567–570 [DOI] [PubMed] [Google Scholar]
- 18.Thompson, E. C., Cobb, B. S., Sabbattini, P., Meixlsperger, S., Parelho, V., Liberg, D., Taylor, B., Dillon, N., Georgopoulos, K., Jumaa, H., Smale, S. T., Fisher, A. G., and Merkenschlager, M. (2007) Immunity 26 335–344 [DOI] [PubMed] [Google Scholar]
- 19.Harker, N., Naito, T., Cortes, M., Hostert, A., Hirschberg, S., Tolaini, M., Roderick, K., Georgopoulos, K., and Kioussis, D. (2002) Mol. Cell 10 1403–1415 [DOI] [PubMed] [Google Scholar]
- 20.Trinh, L. A., Ferrini, R., Cobb, B. S., Weinmann, A. S., Hahm, K., Ernst, P., Garraway, I. P., Merkenschlager, M., and Smale, S. T. (2001) Genes Dev. 15 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst, P., Hahm, K., Trinh, L., Davis, J. N., Roussel, M. F., Turck, C. W., and Smale, S. T. (1996) Mol. Cell. Biol. 16 6121–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su, R. C., Sridharan, R., and Smale, S. T. (2005) Semin. Immunol. 17 129–140 [DOI] [PubMed] [Google Scholar]
- 23.Ernst, P., Hahm, K., and Smale, S. T. (1993) Mol. Cell. Biol. 13 2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun, L., Liu, A., and Georgopoulos, K. (1996) EMBO J. 15 5358–5369 [PMC free article] [PubMed] [Google Scholar]
- 25.Ronni, T., Payne, K. J., Ho, S., Bradley, M. N., Dorsam, G., and Dovat, S. (2007) J. Biol. Chem. 282 2538–2547 [DOI] [PubMed] [Google Scholar]
- 26.Dovat, S., Montecino-Rodriguez, E., Schuman, V., Teitell, M. A., Dorshkind, K., and Smale, S. T. (2005) J. Immunol. 175 3508–3515 [DOI] [PubMed] [Google Scholar]
- 27.Gomez-del Arco, P., Koipally, J., and Georgopoulos, K. (2005) Mol. Cell. Biol. 25 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dovat, S., Ronni, T., Russell, D., Ferrini, R., Cobb, B. S., and Smale, S. T. (2002) Genes Dev. 16 2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-del Arco, P., Maki, K., and Georgopoulos, K. (2004) Mol. Cell. Biol. 24 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sridharan, R., and Smale, S. T. (2007) J. Biol. Chem. 282 30227–30238 [DOI] [PubMed] [Google Scholar]
- 31.Groves, T., Katis, P., Madden, Z., Manickam, K., Ramsden, D., Wu, G., and Guidos, C. J. (1995) J. Immunol. 154 5011–5022 [PubMed] [Google Scholar]
- 32.Su, R. C., Brown, K. E., Saaber, S., Fisher, A. G., Merkenschlager, M., and Smale, S. T. (2004) Nat. Genet. 36 502–506 [DOI] [PubMed] [Google Scholar]
- 33.Hahm, K., Cobb, B. S., McCarty, A. S., Brown, K. E., Klug, C. A., Lee, R., Akashi, K., Weissman, I. L., Fisher, A. G., and Smale, S. T. (1998) Genes Dev. 12 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle, W. J., van der Geer, P., and Hunter, T. (1991) Methods Enzymol. 201 110–149 [DOI] [PubMed] [Google Scholar]
- 35.Hahm, K., Ernst, P., Lo, K., Kim, G. S., Turck, C., and Smale, S. T. (1994) Mol. Cell. Biol. 14 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown, K. E., Baxter, J., Graf, D., Merkenschlager, M., and Fisher, A. G. (1999) Mol. Cell 3 207–217 [DOI] [PubMed] [Google Scholar]
- 37.Meggio, F., and Pinna, L. A. (2003) FASEB J. 17 349–368 [DOI] [PubMed] [Google Scholar]
- 38.Marin, O., Meggio, F., Draetta, G., and Pinna, L. A. (1992) FEBS Lett. 301 111–114 [DOI] [PubMed] [Google Scholar]
- 39.Meek, D. W., Simon, S., Kikkawa, U., and Eckhart, W. (1990) EMBO J. 9 3253–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walton, G. M., Spiess, J., and Gill, G. N. (1985) J. Biol. Chem. 260 4745–4750 [PubMed] [Google Scholar]
- 41.Hill, B. L., Drickamer, K., Brodsky, F. M., and Parham, P. (1988) J. Biol. Chem. 263 5499–5501 [PubMed] [Google Scholar]
- 42.Carroll, D., Santoro, N., and Marshak, D. R. (1988) Cold Spring Harbor Symp. Quant. Biol. 53 91–95 [DOI] [PubMed] [Google Scholar]
- 43.Seldin, D. C., and Leder, P. (1995) Science 267 894–897 [DOI] [PubMed] [Google Scholar]
- 44.Kelliher, M. A., Seldin, D. C., and Leder, P. (1996) EMBO J. 15 5160–5166 [PMC free article] [PubMed] [Google Scholar]
- 45.Rifkin, I. R., Channavajhala, P. L., Kiefer, H. L., Carmack, A. J., Landesman-Bollag, E., Beaudette, B. C., Jersky, B., Salant, D. J., Ju, S. T., Marshak-Rothstein, A., and Seldin, D. C. (1998) J. Immunol. 161 5164–5170 [PubMed] [Google Scholar]
- 46.Channavajhala, P., and Seldin, D. C. (2002) Oncogene 21 5280–5288 [DOI] [PubMed] [Google Scholar]
- 47.Wojcik, H., Griffiths, E., Staggs, S., Hagman, J., and Winandy, S. (2007) Eur. J. Immunol. 37 1022–1032 [DOI] [PubMed] [Google Scholar]