Abstract
Alterations in signaling pathway activity have been implicated in the pathogenesis of Duchenne muscular dystrophy, a degenerative muscle disease caused by a deficiency in the costameric protein dystrophin. Accordingly, the notion of the dystrophin-glycoprotein complex, and by extension the costamere, as harboring signaling components has received increased attention in recent years. The localization of most, if not all, signaling enzymes to this subcellular region relies on interactions with scaffolding proteins directly or indirectly associated with the dystrophin-glycoprotein complex. One of these scaffolds is myospryn, a large, muscle-specific protein kinase A (PKA) anchoring protein or AKAP. Previous studies have demonstrated a dysregulation of myospryn expression in human Duchenne muscular dystrophy, suggesting a connection to the pathophysiology of the disorder. Here we report that dystrophic muscle exhibits reduced PKA activity resulting, in part, from severely mislocalized myospryn and the type II regulatory subunit (RIIα) of PKA. Furthermore, we show that myospryn and dystrophin coimmunoprecipitate in native muscle extracts and directly interact in vitro. Our findings reveal for the first time abnormalities in the PKA signal transduction pathway and myospryn regulation in dystrophin deficiency.
The major macromolecular protein complex within the muscle costamere is the dystrophin-glycoprotein complex (DGC).2 The DGC harbors the large, actin-binding protein dystrophin, the absence of which results in Duchenne muscular dystrophy (DMD), a lethal, X-linked degenerative muscle disease (1–4). It is undisputed that defects within the DGC are one of the major triggers of muscular dystrophy, yet the mechanisms by which a defective DGC leads to muscle degeneration remain unclear. Hence, intense efforts have been put forth to molecularly dissect the function of dystrophin as well as the DGC (5, 6) and to identify novel regulators of this complex in striated muscle.
Dystrophin serves as a scaffold for numerous protein-protein interactions with components of the DGC as well as non-DGC proteins at the level of the costamere (7, 8). The prevailing model suggests a structural and mechanical role for dystrophin (9), but an additional function in signaling has been proposed (10, 11). Although elucidating the role of dystrophin in signaling has been a challenging task, there is evidence of altered signaling activity in muscular dystrophies. Alterations in calcium levels and the neuronal nitric oxide synthase pathway in DMD have been known for some time (12–14). Perturbations in the activity of additional signaling molecules such as calcineurin, Akt, JNK1, and IκB kinase/NF-κB have been linked to muscular dystrophies (15–19).
Recently, we showed that the costameric protein myospryn functions as a muscle-specific protein kinase A (PKA) anchoring protein or AKAP (20). This was the first demonstration of myospryn coordinating the PKA signaling pathway at the level of the costamere. The AKAP family of scaffolding proteins is found in every cell type, and since PKA is a ubiquitous, diffusible enzyme, substrate specificity is achieved by localizing the activity of the enzyme within discrete subcellular compartments through the anchoring function of AKAPs (21). Thus, the ability of myospryn to anchor PKA at the costamere may have profound regulatory implications given that dystrophin has been shown to be phosphorylated by PKA (22, 23). A potential consequence of these associations in muscle disease is implied by the observation that myospryn transcripts are reduced in human DMD (24).
Regarding PKA signaling, it now appears that myospryn is one of a growing number of muscle proteins within the costamere that have the ability to anchor PKA. Two recent reports have described an association between PKA and dystrobrevin and synemin, respectively (25, 26). Thus, the identification of PKA anchoring at the level of the costamere and the DGC points to a potential novel role for this conserved signaling pathway in regulating costameric function. Using the mdx mouse model of DMD, we demonstrate for the first time that PKA activity is significantly reduced in dystrophic muscle. We also report that myospryn and the RIIα regulatory subunit of PKA are mislocalized in mdx muscle from their normal location at the costamere, providing some insight into a possible mechanism of altered PKA activity. Furthermore, we demonstrate that dystrophin and myospryn coimmunoprecipitate from native muscle extracts in vivo and directly interact in vitro. Therefore, our results suggest that misregulated myospryn expression along with impaired PKA activity and localization in dystrophin-deficient muscle contribute to the pathogenesis of DMD.
EXPERIMENTAL PROCEDURES
Plasmids—Dystrophin/Dp71 (amino acids 3028–3679) (27) was cloned in pcDNA3.1-FLAG vector. Myc-myospryn constructs were described previously (20). The TRIM domain of myospryn (Spe) (28) was cloned into pGEX-2T-KG.
Cell Culture and coIP—COS cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine. COS cells were transfected with the appropriate plasmids using TransIT-LT1 (Mirus), and coimmunoprecipitation (coIP) assays were performed as described elsewhere (20).
Immunoblots—Proteins were fractionated, transferred onto Immun-Blot polyvinylidene difluoride membrane (Bio-Rad), and immunoblotted with primary antibodies anti-FLAG (Sigma) or c-Myc (9E10) (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated secondary antibodies and reacted with Western Lightning chemiluminescent reagent (PerkinElmer Life Sciences). For myospryn analysis in wild type and mdx muscle, extracts were separated by SDS-PAGE electrophoresis, electroblotted, and probed with anti-myospryn antibodies at 1:2,000 (28).
For endogenous immunoprecipitations, adult mouse hind limb muscle was homogenized in radioimmune precipitation assay (RIPA) buffer. Extracts were incubated with protein G-Sepharose 4 Fast Flow beads (GE Healthcare) and either anti-GAL4 or anti-myospryn antibodies for 2 h at 4 °C. Immunoprecipitated protein extracts were fractionated and immunoblotted with anti-dystrophin antibody (Sigma).
GST Pulldown Assays—GST and GST-Spe proteins expressed in Escherichia coli (DH5α) were preincubated with glutathione-Sepharose 4B beads (GE Healthcare) followed by the addition of FLAG-Dp71 for an additional 2 h at 4 °C. Washed beads were resuspended, fractionated, and immunoblotted with anti-FLAG to detect Dp71.
Immunohistochemistry—Hind limb muscle sections from adult wild type and mdx (The Jackson Laboratory, Bar Harbor) mice were prepared as before (20). PKA-RII immunodetection was carried out as described previously (20). All other primary antibodies were diluted 1:100 in blocking solution (3% normal horse serum in 1× phosphate-buffered saline) incubated for 1 h, washed in 1× phosphate-buffered saline followed by secondary antibodies diluted at 1:200: chicken anti-goat FITC (Santa Cruz Biotechnology) for anti-PKA RII and horse antimouse Texas Red (Vector Laboratories) for vinculin. Primary rabbit polyclonal anti-myospryn antibodies, mouse monoclonal anti-dystrophin antibodies (Sigma), and mouse monoclonal anti-vinculin antibodies (Sigma) were diluted 1:100 and incubated as above. Slides were washed, and secondary antibodies were applied for co-localization: goat anti-rabbit Texas Red (Vector Laboratories) and horse anti-mouse-FITC (Vector Laboratories) for myospryn/dystrophin or goat anti-rabbit FITC (Vector Laboratories) and horse anti-mouse Texas red for myospryn/vinculin.
Real-time PCR—Pooled equimolar cDNA samples were prepared from 3–6-month-old wild type and mdx hind limb muscle. PCR was performed using the SYBR® Green PCR master mix and the ABI Prism 7900 sequence detection system (Applied Biosystems). Primers used were as follows: Gravin, 5′GCTGAGTCCCAAGCTAATGAC-3′, 5-GGACGGTATCTGACTTTTCGTT-3′ (PBID 13626040a2); Akap-Lbc, 5′-GAGTCTGTCTGCCGATTCAAA-3′,5′-GCTGATTACAATGGAGTGCCTC-3′ (PBID 14192799a3); Akap15, 5′-CACCAACAAAAAGGCCCAAA, 5′-TGAGCTGTCATTGGATCCCTG3′; makap, 5′-TTGCGTCTCACAAAGCAGGA-3′,5′-TCTGAGAATCCAGCTGTGTTGC-3′; Akap79/150, 5′-AGAACCCAGTAATGGCATCCG-3′,5′-TCTTTCAGATCTTCTGCCTGGC-3′; myospryn, 5′-GACAAGGGGAGACCTCATGGT-3′, 5′-GGTTGGTATAGTCCAGCAGAATG-3′; gapdh, 5′-TGGCAAAGTGGAGATTGTTGCC-3′,5′-AAGATGGTGATGGGCTTCCCG-3′. Primers with PBID designation were obtained from the Primer Bank web site (29).
PKA Activity Assay—Three-month-old adult wild type and mdx hind limb muscle were homogenized in Tris buffer. Protein extracts were subjected to a commercially available PKA activity assay (Promega).
RESULTS
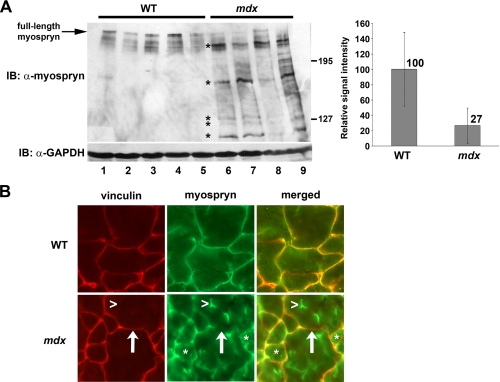
Degradation and Mislocalization of Myospryn in mdx Muscle—Since myospryn is localized to the costamere, a structure destabilized in dystrophin deficiency, we asked whether myospryn expression and localization are affected in dystrophic muscle. Immunoblot analysis revealed a significant 73% decrease (p < 0.03) in full-length myospryn protein in mdx hind limb muscle (Fig. 1A, arrows and right panel). This decrease was associated with an accumulation of smaller fragments in mdx but not in wild type extracts (Fig. 1A, asterisks). These smaller myosprynimmunoreactive bands suggest that myospryn is a specific target of increased protease activity, a cellular mechanism previously linked to muscular dystrophy (30).
FIGURE 1.
Impaired myospryn protein expression and localization in mdx mice. A, left panel, Western blot analysis of myospryn protein in wild type (WT) and mdx hind limb muscle. WT (lanes 1–5) and mdx (lanes 6–9) samples were immunoblotted (IB) with anti-myospryn. (*, degradation products.) Right panel, quantification of full-length myospryn. Signal intensity was quantified by ImageQuant (GE Healthcare), and the background was subtracted for each sample. Signal intensities for each group were averaged and normalized to WT set at 100 ± S.D. WT = 100 ± 48%, mdx = 27 ± 23%. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, double label immunohistochemistry on wild type and mdx mouse hind limb muscle (transverse sections). Left panels, anti-vinculin antibody delineates the myofiber periphery. Note that the costameric protein vinculin is largely unaffected in mdx hind limb muscle. Middle panels, anti-myospryn antibody reveals severe mislocalization of myospryn in mdx tissue. Right panels, merged images for vinculin and myospryn. (*, myofiber cytoplasm; ↑, myofiber periphery/sarcolemma; >, punctuate foci of myospryn staining in mdx muscle.)
Next, we sought to determine whether myospryn is mislocalized in this dystrophin-deficient model. As depicted in Fig. 1B, immunostaining of adult wild type hind limb muscle detected myospryn along the myofiber periphery (see WT-myospryn, upper middle panel) as previously published by our group and others (28, 31, 32). In striking contrast, mdx muscle exhibited a discontinuous localization of myospryn at the myofiber periphery with extensive regions devoid of myospryn (Fig. 1B; compare WT-myospryn with mdx-myospryn and note the arrows in mdx-vinculin and mdx-myospryn). This highly irregular pattern was associated with an increase in myospryn in the cytoplasm (Fig. 1B, asterisks), particularly in discrete foci within myofibers (see arrowheads; compare vinculin with myospryn in mdx). Vinculin, a representative costameric protein, was localized normally at the myofiber periphery in both wild type and mdx muscle (Fig. 1B). These results clearly demonstrate a specific and pronounced mislocalization of myospryn in dystrophin-deficient skeletal muscle.
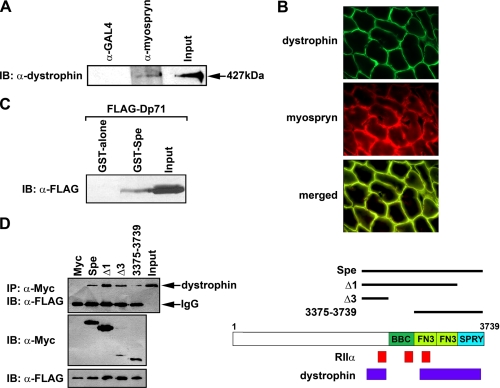
Direct Interaction between Dystrophin and Myospryn—Given the above results and the previously reported biochemical fractionation assays demonstrating the presence of dystrophin and myospryn in microsomal fractions (31), we were prompted to test whether a dystrophin-myospryn complex exists in vivo. Adult mouse hind limb muscle extracts were subjected to an immunoprecipitation assay using anti-myospryn antibodies followed by SDS-PAGE fractionation and immunoblotting. As shown in Fig. 2A, Western blot analysis revealed the presence of dystrophin in the anti-myospryn precipitated complex (arrowhead) but not in the anti-GAL4 complex, demonstrating the specificity of this association. Double label immunohistochemistry was subsequently performed on adult hind limb muscle, which revealed a co-localization of both proteins at the myofiber periphery (Fig. 2B) consistent with the immunoprecipitation results.
FIGURE 2.
Association between dystrophin and myospryn. A, dystrophinmyospryn complex in native hind limb muscle. Protein extracts were immunoprecipitated with anti-myospryn or anti-GAL4 antibodies and immunoblotted (IB) using anti-dystrophin antibodies (427 kDa, endogenous dystrophin). B, myospryn and dystrophin colocalize at the muscle costamere. Double immunohistochemistry of transverse sections from mouse hind limb muscle is shown. Upper panel, dystrophin localizes to myofiber periphery. Middle panel, myospryn localizes to peripheral myofiber as described previously (28). Lower panel, merged images indicating co-localization. C, direct interaction between myospryn and dystrophin. Western blot analysis of FLAG-dystrophin (Dp71) protein extracts subjected to a GST pulldown assay with myospryn (GST-Spe) or GST alone is shown. D, mapping the dystrophin interaction regions in myospryn. Left panel, Western analysis of coIP assays with several Myc-myospryn constructs co-transfected with FLAG-dystrophin (Dp71). Right panel, schematic of myospryn depicting dystrophin interaction regions.
To confirm the above observations, we tested whether myospryn and dystrophin can directly interact by GST pulldown assay. We generated a recombinant GST fusion protein consisting of the carboxyl-terminal TRIM region of myospryn (Spe). In addition, we constructed a carboxyl-terminal dystrophin expression construct known as Dp71, which harbors docking sites for dystrobrevin, syntrophin, and dystroglycan (27). As shown in Fig. 2C, myospryn (GST-Spe) effectively precipitated dystrophin (Dp71), whereas GST alone failed to do so. To begin to delineate the dystrophin interaction sites on myospryn, coIP assays were initiated. Co-transfection of Dp71 along with several myospryn deletion constructs in COS cells revealed that dystrophin interacts within at least two independent regions in the TRIM domain (Fig. 2D, left and right panels). Notably, these dystrophin interaction regions overlap with our previously reported RIIα anchoring sites (20). The specificity of the interaction between Dp71 and the myospryn TRIM region was confirmed by performing immunoprecipitations in the reverse direction using anti-FLAG antibodies (supplemental Fig. S1). Taken together, the above results demonstrate a novel, direct association between dystrophin and myospryn in striated muscle.
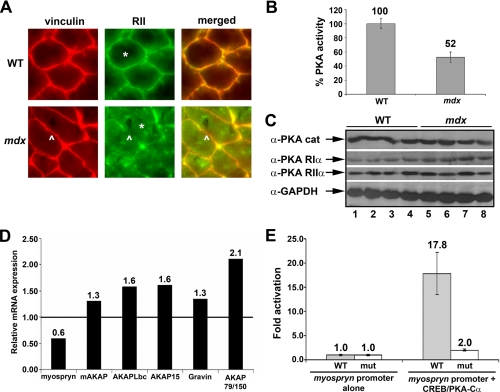
Altered PKA Activity and Anchoring in mdx Hind Limb Muscle—Having previously established that myospryn functions as a muscle-specific AKAP and now finding that it is mislocalized and degraded in dystrophin deficiency, we evaluated the PKA signaling pathway in mdx muscle. Because myospryn specifically binds to the RIIα regulatory subunit of PKA, we performed immunohistochemistry on mdx hind limb muscle with anti-RII antibodies. We demonstrate that RIIα, which is normally present and co-localized with myospryn at the costamere (20), is mislocalized in mdx muscle with considerably reduced staining at the sarcolemma (Fig. 3A, arrowheads). Moreover, a preponderance of cytoplasmic staining is evident in mdx but not wild type muscle (compare asterisks, WT-RIIα versus mdx-RIIα). To our knowledge, this is the first demonstration of deregulated PKA localization in muscular dystrophy and is consistent with the ability of myospryn to anchor the PKA holoenzyme.
FIGURE 3.
Deregulated PKA signaling and regulation of myospryn gene expression by PKA-CREB. A, double label immunohistochemistry of wild type and mdx hind limb muscle (transverse sections). Left panels, anti-vinculin immunoreactivity along myofiber periphery. Middle panels, anti-RII reveals mislocalization of this PKA regulatory subunit in mdx muscle. Right panels, merged images for vinculin and RII immunostaining. (*; myofiber cytoplasm. ^; sarcolemma. >; punctuate foci in mdx muscle). B, reduced PKA activity in mdx hind limb muscle. Protein extracts from wild type (n = 3) and mdx (n = 3) muscle were subjected to a PKA activity assay. The difference in group means was significant (p < 0.003). C, PKA subunit expression in wild type and mdx muscle. Immunoblots of protein extracts from wild type (n = 4) and mdx (n = 4) hind limb muscle demonstrating the predominant catalytic (cat) and regulatory PKA subunits are shown. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. D, quantitative RT-PCR of AKAP expression. cDNA reactions from pooled mdx hind limb muscle mRNA (n = 3) relative to wild type were normalized to 1.0. All samples were performed in triplicate, and Ct values for each gene were averaged and normalized to glyceraldehyde-3-phosphate dehydrogenase. E, myospryn is transcriptionally activated by PKA-CREB signaling. Transient transfection reporter assays in COS cells of the myospryn reporter containing the wild type CRE (TAACGTTA) and a myospryn reporter with a mutant (mut) CRE (TCCCGGGA) were performed. Luciferase data were performed in triplicate, and the difference in promoter activity was significant (p < 0.01).
Because myospryn functions as an AKAP and because of our observation that the RIIα regulatory subunit is mislocalized, we asked whether PKA activity was altered in dystrophic muscle. It is known that PKA anchoring plays an important role in regulating PKA enzymatic activity (19). Therefore, protein extracts from adult wild type (n = 3) and mdx (n = 3) hind limb muscle were subjected to a PKA activity assay. As shown in Fig. 3B, PKA activity is reduced by 50% (p < 0.003) in mdx muscle pointing to a significant alteration of the PKA signaling pathway in dystrophic muscle. Western analyses did not detect differences in expression of the catalytic or regulatory subunits of PKA in mdx muscle (Fig. 3C). Furthermore, cAMP assays failed to reveal differences in cAMP levels in mdx hind limb muscle (data not shown). These results indicate that the decrease in activity is the result of aberrant PKA anchoring and not due to reduced expression of the holoenzyme subunits or upstream alterations in the second messenger cAMP.
Myospryn Is a Uniquely Down-regulated AKAP in mdx Muscle—Previously published data revealed down-regulated myospryn gene expression in human DMD muscle biopsies (24). To determine whether myospryn gene expression is also reduced in mdx muscle, we performed quantitative RT-PCR between pooled wild type (n = 3) and mdx (n = 3) hind limb muscle cDNAs. Our results revealed a 40% decrease in myospryn transcripts in mdx muscle (Fig. 3D) consistent with the finding in DMD. We then asked whether this effect on myospryn transcription reflects a more global phenomenon of reduced AKAP gene expression in dystrophic muscle. By performing quantitative RT-PCR on five well characterized AKAPs, mAKAP, AKAPLbc, AKAP15, gravin, and AKAP79/150, we found that myospryn is the only AKAP down-regulated in mdx hind limb muscle (Fig. 3D). These results reinforce the notion that the down-regulation of myospryn is highly specific and not the result of a global reduction in AKAP gene expression in dystrophic muscle.
Myospryn Is Regulated by the PKA-CREB Signaling Pathway—Given the specific reduction of myospryn gene expression in both DMD and mdx muscle and our new finding of reduced PKA activity, we asked whether this down-regulation may be the consequence of altered PKA signaling. Thus, we scanned the myospryn promoter for potential cAMP-response element (CRE)-binding protein (CREB) binding sites. CREB is one of the major effectors of PKA signaling (33). We found a CRE, TAACGTTA, located at –220 bp upstream of the myospryn transcription start site. The importance of this CRE site was tested by co-transfecting COS cells with a luciferase reporter construct containing 0.8 kb of the myospryn promoter along with CREB plus constitutively active PKA. As depicted in Fig. 3E, luciferase assays revealed a 17.8 ± 4.4-fold activation (p < 0.01) over myospryn promoter alone. The transcriptional activation occurred specifically through the CRE in the myospryn promoter since a mutation within the CRE site (TCCCGGGA) resulted in a 90% reduction (2.0 ± 0.26-fold activation; p < 0.01) of PKA-CREB dependent activation. These results demonstrate that myospryn is a novel target gene downstream of the PKA-CREB signal transduction pathway. Thus, reduced myospryn gene expression in dystrophin-deficient muscle may also contribute to reduced PKA activity by having less available anchoring protein at the costamere.
To gain a better understanding of the potential broader involvement of the PKA-CREB signaling pathway in muscular dystrophy, we performed a comparative analysis between genes dysregulated in mdx muscle (Refs. 35 and 36 and supplemental Refs. 38–41) and known CREB targets (Refs. 33 and 37 and supplemental Ref. 42). As seen in supplemental Table S1, there are nearly two dozen genes dysregulated in mdx muscle that function downstream of CREB. All of these genes are cAMPregulated, reinforcing the notion that the PKA signal transduction pathway is defective in dystrophin-deficient muscle.
DISCUSSION
The results of our study uncover a novel defect in the PKA signal transduction pathway in the mdx mouse model of dystrophin deficiency. The perturbation in this signaling cascade is consistent with reduced expression of the AKAP myospryn and the mislocalization of its gene product and the regulatory subunit of PKA.
The intricate costameric network is believed to be responsible for maintaining the integrity and stability of the muscle cell membrane. The absence of dystrophin, members of the DGC, and possibly other costameric proteins triggers a series of cellular and molecular events resulting in muscle degeneration, thereby establishing this region as a critical support system in proper muscle function. Because of the multitude of scaffolding proteins within the costamere, signaling components are likely to be key participants in the pathogenesis of muscle disease. As such, the role of signaling pathways in muscular dystrophy has received increased attention in recent years (10–12).
We demonstrate that the muscle-specific AKAP myospryn is degraded and mislocalized from the costamere in mdx hind limb muscle consistent with our finding that myospryn and dystrophin physically interact. This is a significant finding given that the predominant costameric components affected in dystrophin deficiency are those proteins directly associated with the DGC (1–4). To our knowledge, myospryn represents one of a limited number of non-DGC costameric proteins impaired in dystrophin deficiency.
It is worth noting that the cumulative intensities of the degradation products of myospryn in mdx muscle appear equal to or greater than the intensity of full-length myospryn in wild type tissue. This contrasts with reduced myospryn transcripts in mdx muscle. One interpretation for this apparent discrepancy is that myospryn protein is indeed reduced and that the observed increase in signal intensity is the result of enhanced immunoreactivity of these smaller polypeptides toward the antibody. Alternatively, these collective signal intensities may reflect actual protein levels. In this case, the lack of correlation between transcripts and protein could be explained by the stability and low turnover of myospryn fragments. Regardless of the mechanism, full-length myospryn is clearly diminished in mdx muscle.
Consistent with the ability of myospryn to bind PKA, we find that the RIIα regulatory subunit is mislocalized from the costamere. Accordingly, we see a substantial reduction in PKA activity in mdx muscle. This is the first, direct demonstration of deregulated PKA activity in dystrophic muscle. It is worth noting that a previous study revealed altered activity of the l-type Ca2+ channel, whose function is regulated by PKA, in mdx muscle (34). Therefore, our findings provide a potential molecular explanation for these observations.
This global reduction of PKA activity in dystrophic muscle is also likely to affect numerous downstream PKA-dependent pathways such as its ability to regulate gene expression. Intriguingly, we observed a reduction in myospryn gene expression in mdx muscle, which prompted us to examine the potential regulation of myospryn expression through one of the major PKA substrates, the CREB transcription factor. Indeed, we detected a robust and specific activation of myospryn through PKACREB signaling. Taken together, our findings suggest that perturbation of myospryn expression and localization may, in part, be responsible for the altered PKA activity in dystrophic muscle. These studies highlight the importance of tightly regulating myospryn expression in striated muscle.
Of note, the major classes of genes regulated by the PKACREB pathway are those involved in cellular metabolism, cell survival, and proliferation (33). Incidentally, the gene expression profiles in dystrophin-deficient muscle underscore the molecular hallmarks of dystrophy such as muscle cell death, the inability to regenerate muscle effectively, and altered metabolic activity (35, 36, 38–41). Therefore, it is formally possible that these defects arise from the deregulation of PKA signaling observed in mdx muscle. It is also worth noting that a recent report described a muscular dystrophy phenotype in transgenic mice overexpressing a dominant negative CREB in skeletal muscle (37). Studies are in progress to examine the activity of CREB in mdx muscle and to further dissect the PKA-CREB signaling pathway in dystrophin deficiency.
Supplementary Material
Acknowledgments
We thank Marc Montminy (Salk Institute for Biological Sciences, La Jolla, CA) for the CREB expression vector and CREB-dependent reporter and G. Stanley McKnight (University of Washington, Seattle, WA) for the constitutively active PKA-C subunit. We also thank Geof Cooper and Ulla Hansen (Boston University) for critical reading of the manuscript.
This work was supported by grants from the National Institute of Health and the Muscular Dystrophy Association (to F. J. N.) and an Undergraduate Research Opportunities Program (UROP-Boston University) award (to J. A. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure, a supplemental table, and supplemental references.
Footnotes
The abbreviations used are: DGC, dystrophin-glycoprotein complex; DMD, Duchenne muscular dystrophy; PKA, protein kinase A; AKAP, PKA anchoring protein; GST, glutathione S-transferase; FITC, fluorescein isothiocyanate; WT, wild type; RT-PCR, reverse transcription-PCR; PBID, PrimerBank identification; RII, type II regulatory subunit; TRIM, tripartite motif; CRE, cAMP-response element; CREB, cAMP-response element-binding protein; coIP, coimmunoprecipitation.
References
- 1.Dalkilic, I., and Kunkel, L. M. (2003) Curr. Opin. Genet. Dev. 13 231–238 [DOI] [PubMed] [Google Scholar]
- 2.Davies, K. E., and Nowak, K. J. (2006) Nat. Rev. Mol. Cell Biol. 7 762–773 [DOI] [PubMed] [Google Scholar]
- 3.Durbeej, M., and Campbell, K. P. (2002) Curr. Opin. Genet. Dev. 12 349–361 [DOI] [PubMed] [Google Scholar]
- 4.Heydemann, A., and McNally, E. M. (2007) Trends Cardiovasc. Med. 17 55–59 [DOI] [PubMed] [Google Scholar]
- 5.Sweeney, H. L., and Barton, E, R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13464–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake, D. J., Weir, A., Newey, S. E., and Davies, K. E. (2002) Physiol. Rev. 82 291–329 [DOI] [PubMed] [Google Scholar]
- 7.Ervasti, J. M. (2007) Biochim. Biophys. Acta. 1772 108–117 [DOI] [PubMed] [Google Scholar]
- 8.Ervasti, J. M. (2003) J. Biol. Chem. 278 13591–13594 [DOI] [PubMed] [Google Scholar]
- 9.Judge, L. M., Haraguchiln, M., and Chamberlain, J. S. (2006) J. Cell Sci. 119 1537–1546 [DOI] [PubMed] [Google Scholar]
- 10.Lapidos, K. A., Kakkar, R., and McNally, E. M. (2004) Circ. Res. 94 1023–1031 [DOI] [PubMed] [Google Scholar]
- 11.Rando, T. A. (2001) Muscle Nerve 24 1575–1594 [DOI] [PubMed] [Google Scholar]
- 12.Batchelor, C. L., and Winder, S. J. (2006) Trends Cell Biol. 16 198–205 [DOI] [PubMed] [Google Scholar]
- 13.Brenman, J. E., Chao, D. S., Xia, H., Aldape, K., and Bredt, D. S. (1995) Cell 82 743–752 [DOI] [PubMed] [Google Scholar]
- 14.Chang, W. J., Iannaccone, S. T., Lau, K. S., Masters, B. S., McCabe, T. J., McMillan, K., Padre, R. C., Spencer, M. J., Tidball, J. G., and Stull, J. T. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9142–9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakkalakal, J. V., Harrison, M. A., Carbonetto, S., Chin, E., Michel, R. N., and Jasmin, B. J. (2004) Hum. Mol. Genet. 13 379–388 [DOI] [PubMed] [Google Scholar]
- 16.Parsons, S. A., Millay, D. P., Sargent, M. A., Naya, F. J., McNally, E. M., Sweeney, H. L., and Molkentin, J. D. (2007) J. Biol. Chem. 282 10068–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langenbach, K. J., and Rando, T. A. (2002) Muscle Nerve 26 644–653 [DOI] [PubMed] [Google Scholar]
- 18.Kolodziejczyk, S. M., Walsh, G. S., Balazsi, K., Seale, P., Sandoz, J., Hierlihy, A. M., Rudnicki, M. A., Chamberlain, J. S., Miller, F. D., and Megeney, L. A. (2001) Curr. Biol. 11 1278–1282 [DOI] [PubMed] [Google Scholar]
- 19.Acharyya, S., Villalta, S. A., Bakkar, N., Bupha-Intr, T., Janssen, P. M., Carathers, M., Li, Z. W., Beg, A. A., Ghosh, S., Sahenk, Z., Weinstein, M., Gardner, K. L., Rafael-Fortney, J. A., Karin, M., Tidball, J. G., Baldwin, A. S., and Guttridge, D. C. (2007) J. Clin. Investig. 117 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds, J. G., McCalmon, S. A., Tomczyk, T., and Naya, F. J. (2007) Biochim. Biophys. Acta. 1773 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong, W., and Scott, J. D. (2004) Nat. Rev. Mol. Cell Biol. 5 959–970 [DOI] [PubMed] [Google Scholar]
- 22.Senter, L., Ceoldo, S., Petrusa, M. M., and Salviati, G. (1995) Biochem. Biophys. Res. Commun. 206 57–63 [DOI] [PubMed] [Google Scholar]
- 23.Michalak, M., Fu, S. Y., Milner, R. E., Busaan, J. L., and Hance, J. E. (1996) Biochem. Cell Biol. 74 431–437 [DOI] [PubMed] [Google Scholar]
- 24.Tkatchenko, A. V., Pietu, G., Cros, N., Gannoun-Zaki, L., Auffray, C., Leger, J. J., and Dechesne, C. A. (2001) Neuromuscul. Disord. 11 269–277 [DOI] [PubMed] [Google Scholar]
- 25.Ceccarini, M., Grasso, M., Veroni, C., Gambara, G., Artegiani, B., Macchia, G., Ramoni, C., Torreri, P., Mallozzi, C., Petrucci, T. C., and Macioce, P. (2007) J. Mol. Biol. 371 1174–1187 [DOI] [PubMed] [Google Scholar]
- 26.Russell, M. A., Lund, L. M., Haber, R., McKeegan, K., Cianciola, N., and Bond, M. (2006) Arch. Biochem. Biophys. 456 204–215 [DOI] [PubMed] [Google Scholar]
- 27.Crawford, G. E., Faulkner, J. A., Crosbie, R. H., Campbell, K. P., Froehner, S. C., and Chamberlain, J. S. (2000) J. Cell Biol. 150 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durham, J. T., Brand, O. M., Arnold, M., Reynolds, J. G., Muthukumar, L., Weiler, H., Richardson, J. A., and Naya, F. J. (2006) J. Biol. Chem. 281 6841–6849 [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., and Seed, B. (2003) Nucleic Acids Res. 31 e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramerova, I., Beckmann, J. S., and Spencer, M. J., (2007) Biochim. Biophys. Acta. 1772 128–144 [DOI] [PubMed] [Google Scholar]
- 31.Benson, M. A., Tinsley, C. L., and Blake, D. J. (2004) J. Biol. Chem. 279 10450–10458 [DOI] [PubMed] [Google Scholar]
- 32.Kouloumenta, A., Mavroidis, M., and Capetanaki, Y. (2007) J. Biol. Chem. 282 35211–35221 [DOI] [PubMed] [Google Scholar]
- 33.Mayr, B., and Montminy, M. (2001) Nat. Rev. Mol. Cell Biol. 2 599–609 [DOI] [PubMed] [Google Scholar]
- 34.Johnson, B. D., Scheuer, T., and Catterall, W. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 4191–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Y. W., Zhao, P., Borup, R., and Hoffman, E. P. (2000) J. Cell Biol. 151 1321–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haslett, J. N., Sanoudou, D., Kho, A. T., Bennett, R. R, Greenberg, S. A., Kohane, I. S., Beggs, A. H., and Kunkel, L. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15000–15005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berdeaux, R., Goebel, N., Banaszynski, L., Takemori, H., Wandless, T., Shelton, G. D., and Montminy, M. (2007) Nat. Med. 13 597–603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.