Abstract
Plants, unlike other higher eukaryotes, possess all the necessary enzymatic equipment for de novo synthesis of methionine, an amino acid that supports additional roles than simply serving as a building block for protein synthesis. This is because methionine is the immediate precursor of S-adenosylmethionine (AdoMet), which plays numerous roles of being the major methyl-group donor in transmethylation reactions and an intermediate in the biosynthesis of polyamines and of the phytohormone ethylene. In addition, AdoMet has regulatory function in plants behaving as an allosteric activator of threonine synthase. Among the AdoMet-dependent reactions occurring in plants, methylation of cytosine residues in DNA has raised recent interest because impediment of this function alters plant morphology and induces homeotic alterations in flower organs. Also, AdoMet metabolism seems somehow implicated in plant growth via an as yet fully understood link with plant-growth hormones such as cytokinins and auxin and in plant pathogen interactions. Because of this central role in cellular metabolism, a precise knowledge of the biosynthetic pathways that are responsible for homeostatic regulation of methionine and AdoMet in plants has practical implications, particularly in herbicide design.
Keywords: methionine, S-adenosyl methionine, plant development
Methionine is the only sulfur-containing amino acid that is essential for mammals and must therefore be derived entirely from the diet. In contrast, methionine is synthesized de novo by plants and most microorganisms after the initial steps of inorganic sulfate assimilation and cysteine or homocysteine (Hcy) syntheses (1–4). Because of its central importance in cellular metabolism, the metabolic sequence ensuring the conversion of cysteine into methionine has been extensively studied in enteric bacteria. The pioneer work of John Giovanelli (5) established that the enzymatic reactions leading to methionine were similar in the plant kingdom (1). In plants, as in bacteria, methionine belongs to the aspartate family of amino acids, which also comprises lysine, threonine, and isoleucine (1–4). These biosynthetic pathways deserve considerable attention for several reasons. First, the study of their biochemical and molecular control would provide new insights into the mechanisms involved in the homeostatic regulation of amino acids in plants. Second, these metabolic pathways give rise to essential amino acids that limit the nutritional quality of crop plants as diet for human beings and monogastric animals because seeds of cereals and legumes are deficient in lysine and methionine, respectively (6). Third, the inhibition of acetolactate synthase, an enzyme involved in the biosynthesis of isoleucine, valine, and leucine, by sulfonylureas or imidazolinones is lethal for plants (7). Thus, it is anticipated that key regulatory enzymes of the aspartate-derived amino acid branches also would be suitable targets for efficient herbicides.
The synthesis of aspartate-derived amino acids as well as the assimilation of sulfate and the synthesis of sulfur amino acids in plants have been covered recently in several reviews (7–10). The present article is intended to serve three purposes. The first is to provide a general background on the physiology of methionine synthesis in higher plants. The second is to highlight some recent findings linked to the metabolism of S-adenosylmethionine (AdoMet) in plants due to its regulatory influence on the aspartate pathway and its implication in plant growth and plant–pathogen interactions. The third is to present and discuss an integrative view of our present understanding of the functioning of the methionine and AdoMet biosynthetic/recycling pathways, notably in relation with the unique compartmentation of metabolism in higher plants. The trends that emerge here open new research directions for the study of these essential metabolites in plants.
De Novo Synthesis of Methionine.
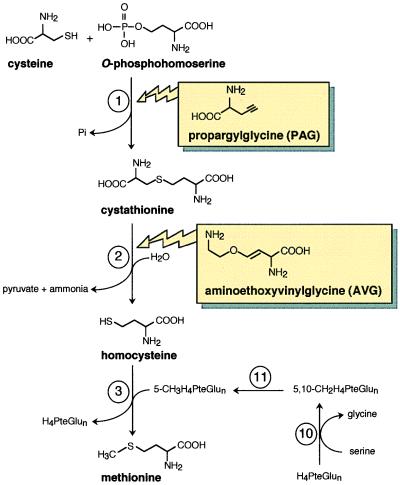
The methionine molecule originates from three convergent pathways: the carbon backbone deriving from aspartate, the sulfur atom from cysteine, and the methyl group from the β-carbon of serine (Fig. 1). In higher plants, starting from O-phosphohomoserine (OPH), the three consecutive reactions catalyzed by cystathionine γ-synthase, cystathionine β-lyase, and methionine synthase are unique to methionine synthesis (11). The first two are catalyzed by pyridoxal 5′-phosphate (PLP)-dependent enzymes and constitute the transsulfuration pathway that consists in the transfer of the sulfur atom of cysteine (C3 skeleton) to Hcy (C4 skeleton) with the thioether cystathionine as intermediate (Fig. 1). First, cystathionine γ-synthase (EC 4.2.99.9) catalyzes the synthesis of cystathionine from cysteine and OPH in a γ-replacement reaction. This is the only known enzyme with a physiological function to catalyze a replacement reaction at the γ-carbon of an amino acid, i.e., the substitution of an electronegative group by another one on the fourth carbon of an amino acid. Cystathionine β-lyase (EC 4.4.1.8) subsequently catalyzes an α, β-elimination of cystathionine to produce Hcy, pyruvate, and ammonia (Fig. 1). The terminal step in methionine synthesis involves the transfer of the methyl group from N5-methyl-tetrahydrofolate (5-CH3H4PteGlun) to Hcy (Fig. 1). In plants, this reaction is catalyzed by a cobalamin-independent methionine synthase (EC 2.1.1.14) (12).
Figure 1.
Methionine biosynthesis and structure of enzyme inhibitors. Enzymes: 1, cystathionine γ-synthase; 2, cystathionine β-lyase; 3, methionine synthase; 10, serine hydroxymethyltransferase; 11, 5,10-CH2H4PteGlun reductase.
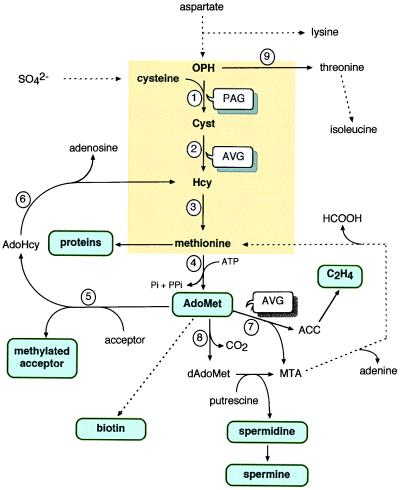
Methionine occupies a central position in cellular metabolism in which the processes of protein synthesis, methyl-group transfer through AdoMet, polyamines, and ethylene syntheses are interlocked (13–15). Among these pathways, the synthesis of proteins is the only one pathway consuming the entire methionine molecule. The synthesis of AdoMet is, however, the major route for methionine metabolism; 80% of this amino acid is being engaged in this reaction (Fig. 2) (16). Two pathways using the methyl or the 4-carbon moieties of methionine and involving AdoMet as a first intermediate have been described in plants. Both routes result in a recycling of the sulfur atom of methionine. First, >90% of AdoMet is used for transmethylation reactions in which the methyl group of methionine is transferred to acceptors, the major end products being choline and its derivatives, including phosphatidylcholine (the major polar lipid) (16). These reactions are accompanied by a recycling of the homocysteinyl moiety to regenerate methionine. In breif, S-adenosyl-l-Hcy (AdoHcy) produced during methylation reactions is converted into Hcy via a reaction catalyzed by AdoHcy hydrolase (EC 3.3.1.1). Methionine is then regenerated through methylation of Hcy (Fig. 2). Thus, methionine synthase not only catalyzes the last reaction in de novo methionine synthesis but also serves to regenerate the methyl group of AdoMet. Second, the use of the 4-carbon moiety of AdoMet for the synthesis of polyamines and, in some plant tissues, ethylene also is accompanied by recycling of the methylthio moiety and regeneration of methionine (Fig. 2). Here, 5-methylthioadenosine is a by-product that is converted into methionine by a series of reactions involving the intermediates 5-methylthioribose, 5-methylthioribose-1-phosphate, and 2-keto-4-methylthiobutyrate, in which the methylthio moiety and 4 about the 5 ribose carbons are converted to the methylthio and 4-carbon moieties of methionine, respectively (Fig. 2) (17). This route is far less to be negligible because it represents approximately one-third of the amount of methionine accumulating in protein (16).
Figure 2.
Relationships among methionine, threonine, AdoMet, polyamine, biotin, and ethylene biosynthetic pathways in higher plants. Enzymes: 1, cystathionine γ-synthase; 2, cystathionine β-lyase; 3, methionine synthase; 4, AdoMet synthetase; 5, AdoMet-dependent methylase; 6, AdoHcy hydrolase; 7, 1-aminocyclopropane-1-carboxylic acid synthase; 8, AdoMet decarboxylase; 9, threonine synthase. Note that AVG inhibits both cystathionine γ-synthase and 1-aminocyclopropane-1-carboxylic acid synthase. For more details about reactions 1, 2, and 3, see Fig. 1.
Cystathionine Synthesis.
In plants, two pathways converge to provide the substrates for the reaction catalyzed by cystathionine γ-synthase: the reduction of inorganic sulfate followed by the incorporation of sulfide into cysteine and the synthesis of OPH from aspartate (Figs. 1 and 2). All of the reactions ensuring OPH synthesis and reductive sulfate assimilation to sulfide occur in the chloroplast (7–9, 18). Sulfide is incorporated into O-acetylserine via O-acetylserine (thiol) lyase (EC 4.2.99.8) and thus forming cysteine. O-Acetylserine synthesis is catalyzed by serine acetyltransferase (EC 2.3.1.30) from serine and acetyl-CoA. Both enzymes have been demonstrated in chloroplasts (18–20).
The characterization of a genomic fragment of Arabidopsis thaliana containing the 3′-end of the cystathionine γ-synthase gene was presented in the first molecular report concerning a plant methionine-synthesizing enzyme (21). The corresponding cDNA was then isolated by functional complementation of an Escherichia coli mutant strain lacking cystathionine γ-synthase and the entire gene structure was determined (22). As compared with the bacterial metB gene product, the Arabidopsis enzyme possesses an amino-terminal extension sharing many features with signal sequences for plastid import (22). This result confirmed previous biochemical characterization in plants showing that cystathionine γ-synthase activity was associated exclusively with the stromal space of chloroplasts (23, 24). The enzyme was obtained in pure form from spinach chloroplasts (25). From this purification, its calculated representation was only ≈0.004% of the cell soluble protein content. Heterologous production of the Arabidopsis cystathionine γ-synthase as a functional mature enzyme was achieved subsequently in recombinant bacteria (25). The plant enzyme is a tetramer of 200–215 kDa consisting in identically sized subunits of 53 kDa. The absorption spectrum of the purified enzyme displays a maximum at 425–427 nm, which is characteristic of PLP-dependent proteins. The cofactor is covalently attached to the enzyme via the formation of an aldimine between the carboxyl moiety of PLP and the ɛ-NH2 moiety of Lys379 (24, 25).
In contrast with cystathionine γ-synthase from bacterial sources that uses either the succinyl or the acetyl derivatives of homoserine as substrates (2, 26), the corresponding plant enzyme uses OPH as the physiological α-aminobutyryl donor for cystathionine synthesis (27). Because threonine synthase (EC 4.2.99.2) also is present in the chloroplast and uses OPH as substrate, OPH is therefore a branch point intermediate for methionine and threonine syntheses in plants (Fig. 2) (28, 29). Importantly, plant threonine synthase is strongly activated by AdoMet in a cooperative manner (28, 30), suggesting that the intraplastidial AdoMet concentration could determine the relative flux of OPH toward methionine and threonine syntheses.
The physiological replacement reaction catalyzed by cystathionine γ-synthase follows a ping-pong mechanism in which OPH binds first to the enzyme, and cystathionine is the last product released (25, 31). Despite a high catalytic constant (30 s−1), the high Km values for OPH and cysteine (2.5 and 0.46 mM, respectively) exhibited by the Arabidopsis enzyme raise the question of cystathionine γ-synthase efficiency in vivo (25). This is because the OPH and cysteine concentrations are below 100 μM in the chloroplast compartment (32, 33). It is worth noting that, contrarily to cystathionine γ-synthase, the Km value of threonine synthase for OPH in the presence of its activator AdoMet is extremely low, being of the order of 5–10 μM (28–30). Thus, a decrease of the intraplastidial concentration of AdoMet could entail an increase of the OPH concentration in this compartment because of a depressed threonine synthase activity. So, the net rate of cystathionine synthesis could increase because, as stated above, the Km of this enzyme for OPH is considerably higher than the intraplastidial OPH concentration. Thus, the kinetic properties of threonine synthase and cystathionine γ-synthase seem well-suited to ensure a rapid adaptation of the partitioning of OPH between the two diverging metabolic pathways in response to fluctuations in the concentrations of OPH and AdoMet. This interpretation does not exclude other types of regulation of this partitioning (see below).
In the absence of cysteine, cystathionine γ-synthase from enteric bacteria efficiently catalyzes a γ-elimination reaction of O-succinylhomoserine yielding α-ketobutyrate, succinate and ammonia (34). For the plant enzyme using OPH as substrate, this reaction is ≈2,700-fold slower than the physiological γ-replacement reaction (i.e., in the presence of cysteine) and 1,500-fold slower than the elimination reaction catalyzed by the E. coli enzyme (25). This difference results from the inability of the plant cystathionine γ-synthase to accumulate a long wavelength-absorbing species that is characteristic for the efficient elimination reaction catalyzed by the enterobacterial enzyme (25, 35). Presumably, despite the similarities suggested by the amino acid sequence comparisons, the PLP environments of the E. coli and Arabidopsis enzymes are different. It is worth noting that the Km values for cysteine in the replacement reaction markedly differ with the bacterial and plant enzymes. Although the former is on the order of 50 μM (34), the latter is one order of magnitude higher (25, 31). Hence, a low rate of the elimination reaction prevents consumption of OPH through this futile process, especially in view of the low intracellular cysteine concentration in plants (32).
Plant cystathionine γ-synthase exhibits a broad specificity for its sulfur-containing substrate. In particular, the enzyme can catalyze the synthesis of Hcy when cysteine is replaced by sodium sulfide (24, 25, 31, 36). There is strong experimental evidence to suggest, however, that this reaction has no physiological significance in plants. Indeed, 35S inorganic sulfate-labeling studies showed that the direct sulfhydration pathway could contribute no more than 3% of the total Hcy synthesis (37, 38). Also, a Nicotiana plumbaginifolia mutant impaired in cystathionine β-lyase is unable to grow without a methionine or Hcy supply (39).
DL-Propargylgylcine (PAG) is a suicide, active site-directed, inhibitor of cystathionine γ-synthase (40) (Fig. 1). Fig. 3 shows the strong impediment of Arabidospsis growth caused by this compound and its reversal upon administration of methionine. Inactivation by PAG of the enzyme from spinach, wheat, or Arabidopsis was consistent with the existence of an intermediate reversible-enzyme inhibitor complex (Kiapp = 45–140 μM) preceding the formation of the final enzyme-inhibitor complex (24, 25, 31). The partial restoration of the activity by exogenous PLP suggested that PAG interacts with the PLP prosthetic group of the enzyme. Kinetic and equilibrium studies showed that PAG binding to PLP was considerably enhanced in the enzyme-binding pocket as compared with that with PLP free in solution (24). Enzyme inactivation by PAG was counteracted by OPH but not by cysteine, consistent with the finding that in the γ-replacement reaction cysteine cannot bind to the enzyme active site without prior action of OPH. Interestingly, in the presence of 100 μM PAG, the pseudo-first-order rate constant for the inactivation process was reduced by approximately 50% at 1 μM OPH, a concentration that is 2,500-fold lower than the apparent Km value for OPH. Thus, the plant enzyme has a much higher affinity for the OPH substrate than inferred from examination of its Km value, meaning that the kcat for the γ-replacement reaction is considerably higher than the dissociation rate constant of the enzyme-OPH complex (25).
Figure 3.
Impediment of Arabidopsis growth by PAG and its reversal by methionine. Left, control plants; Center, plants sprayed daily (from 16 days after sowing) with a PAG solution (2 mM); Right, plants sprayed daily (from 16 days after sowing) with a PAG (2 mM) plus methionine (2 mM) solution. Plants shown are 23-days-old.
Hcy Synthesis.
In spinach, two types of enzymes that can catalyze Hcy formation were found in the cytosol and in the chloroplast (41). Yet, these enzymes exhibited variant kinetic properties (10). On the one hand, the cytosolic enzyme catalyzed the cleavage of cystathionine at a very low rate and with a high Km value (>10 mM). As it proved to react very efficiently with cystine to form cysteine-persulfide, pyruvate, and ammonia, this enzyme is now referred to as a cystine β-lyase. The physiological significance of this cytosolic enzyme remains obscure. On the other hand, the chloroplastic enzyme was shown to be the only cystathionine β-lyase involved in methionine synthesis because it supported cleavage of cystathionine at a high rate (10). This was confirmed in Echinochloa colonum and in Arabidopsis in which only one form of cystathionine β-lyase could be detected, exhibiting no reactivity or only a weak reactivity toward cystine, and being exclusively located in plastids (42, 43).
A unique Arabidopsis cDNA encoding cystathionine β-lyase was cloned by functional complementation of an E. coli mutant lacking endogenous enzyme activity (43). Southern blot experiments suggested that a single-copy gene codes for the enzyme in Arabidopsis. As deduced from the nucleotide sequence, the enzyme contains a N-terminal extension of 70 amino acids that is not found in its E. coli counterpart. This extension contains several hydroxylated and small hydrophobic residues and has a net positive charge, features that are characteristic of plastid transit peptides. Western blot analyses revealed a single polypeptide of 46 kDa in a crude Arabidopsis protein extract, thus showing that the enzyme is synthesized as a precursor of ≈50 kDa and further imported within the chloroplast to yield a 46-kDa mature subunit (44). The amino acid sequence predicted for the Arabidopsis mature enzyme showed 25% identity with its E. coli counterpart and, surprisingly, higher homology (between 28 and 36% identity) with cystathionine γ-synthase from plant and bacterial sources and cystathionine γ-lyase from Saccharomyces cerevisiae (43). All of these enzymes are involved in the cysteine/methionine biosynthetic pathways and belong to the same class of PLP-dependent enzymes that catalyze γ-replacement reactions of C4 amino acids and β- or γ-cleavage of cystathionine (45). This suggested that plant cystathionine γ-synthase and cystathionine β-lyase derived from a common ancestor, as proposed earlier for the corresponding bacterial metB and metC genes (46).
Cystathionine β-lyase was purified from spinach chloroplasts (41) and from an E. colonum cell suspension (42). As for cystathionine γ-synthase, this enzyme was present in trace amounts in the stromal space of plastids (41). Cystathionine β-lyase also was isolated in pure form and at much higher yield from an E. coli strain overexpressing the Arabidopsis cDNA coding the mature form of the enzyme (44). The native enzyme is a homotetramer of 160–170 kDa with a subunit molecular mass of 44–46 kDa (41, 42, 44). It covalently binds PLP with a stoichiometry of 1 mol of cofactor per mol of subunit, via Schiff-base formation with Lys278 of the protein (44). The Arabidopsis enzyme showed a broad substrate specificity, being most active with cystathionine and the dithioacetal djenkolate, an amino acid isolated from the djenkol bean (44). This substrate selectivity does not appear to derive from substrate binding but rather from chemical steps. The strength of the cleavage seems to be controlled by the following points: (i) all the active substrates contain a C3 L-α-amino acid moiety, (ii) the nature of the sulfur-containing bond (thioether, thioacetal or disulfide) influences the rate of the elimination process, and (iii) the length of the second moiety and therefore the spatial separation of the two carboxyl groups of the substrate has a critical role in the efficiency of the cleavage (1, 44).
The cystathionine analogs l-aminoethoxyvinylglycine (AVG; an amino acid first isolated from Streptomyces sp.) (Fig. 1) and rhizobitoxine (a β,γ-unsaturated amino acid isolated from Rhizobium japonicum), which are antibacterial and phytotoxic amino acids, behave as suicide substrates for plant and bacterial cystathionine β-lyase (41, 44, 47). For the plant enzyme, the inactivation process by AVG followed a two-step mechanism in which the irreversible step of inactivation of the enzyme is preceded by the formation of a reversible enzyme-inhibitor complex (Kiapp= 110–160 μM) (41, 44). Also, AVG can react with the polypeptide-bound PLP (Kd = 235 μM) and with PLP free in solution (Kd = 350 μM) (41, 44). Thus, the prosthetic group of the plant enzyme is freely accessible to the inhibitor. The interaction between the E. coli cystathionine β-lyase and AVG has been investigated recently by a combination of kinetic methods and x-ray crystallography (47). Upon AVG treatment, time-dependent slow-binding inhibition was observed. Kinetic analyses revealed a one-step inhibition of the bacterial enzyme by AVG with a dissociation constant of the order of 1 μM, that is 150-fold smaller than that for the plant enzyme. For the E. coli enzyme, the inhibitor is bound strongly by several ionic, hydrogen bonding, and van der Waals interactions involving eight amino acids (47). Among these residues, five are conserved in the Arabidopsis enzyme and the other three, Tyr238, Tyr338, and Trp340 are replaced by Asn307, Val404, and Phe406, respectively, in the plant cystathionine β-lyase. Presumably, these changes account for the marked difference in reactivity between the plant and bacterial cystathionine β-lyases with AVG. The results highlight the fact that, although the plant and bacterial enzymes appear to be quite similar, their kinetic behavior can vary considerably due to punctual changes in amino acids participating to the respective active sites. In other words, conclusions drawn from bacterial enzymes as a model system can be misleading when trying to interpret the reactivity and regulatory behavior of the corresponding enzymes in plants.
Methionine Synthesis.
Contrarily to the transsulfuration enzymes that are present only in organisms catalyzing de novo methionine synthesis, methionine synthase is required in all organisms to ensure regeneration of the methyl group of AdoMet (Fig. 2). Two types of enzymes can fulfill this function: a cobalamin-dependent methionine synthase that contains a vitamin B12 (cobalamin) cofactor, requires catalytic amounts of AdoMet for activity and uses either mono- or polyglutamate forms of 5-CH3H4PteGlun (n = 1 or n ≥ 3) as methyl donors, and a cobalamin-independent methionine synthase that can use only the polyglutamate forms of 5-CH3H4PteGlun (n ≥ 3) as substrates and does not require a vitamin prosthetic group to ensure methionine synthesis. It is generally admitted that organisms that can synthesize or take up vitamin B12 possess only the cobalamin-dependent enzyme whereas those that do not contain vitamin B12 use the cobalamin-independent route for methionine synthesis. To date, the cobalamin-dependent type of enzyme has been demonstrated in mammals, Euglena gracilis and some enteric bacteria such as E. coli or Salmonella typhimurium, whereas the cobalamin-independent type is found in plants, yeast, E. gracilis and some bacteria (2, 4, 12, 48–50). Organisms such as E. coli or E. gracilis are unusual in that they contain both types of methionine synthase. It is not clear why both methylating systems should occur in these organisms and what is the relative contribution of each form of the enzyme in methylation of Hcy (2, 49). Perhaps a form is involved in the net synthesis of methionine whereas the other plays a role in the recycling of the homocysteinyl moiety of AdoMet to regenerate methionine.
Recently, plant cDNAs encoding methionine synthase were obtained from C. roseus (12), Coleus blumei (50), and from the green algae Chlamydomonas reinhardtii (51). The C. roseus cDNA was further used to clone the complementary and genomic DNA encoding this protein in Arabidopsis (GenBank accession no. U97200). The plant cDNAs encode slightly acidic proteins with predicted Mr of ≈85,000. None of these polypeptides exhibit a transit peptide-like sequence thus indicating that the proteins are localized in the cytosol. This result was confirmed in Arabidopsis by Western blot experiments using polyclonal antibodies raised against the Arabidopsis recombinant methionine synthase (B.G., S.R., D.J., and R.D., unpublished results). Amino acid sequences deduced from the plant cDNAs are highly conserved among themselves (>80% identity) and show near 50% identity with the cobalamin-independent methionine synthase from E. coli.
The methyl group for methylation of Hcy comes from 5-CH3H4PteGlun (Figs. 1 and 2), a folate compound that has no other known metabolic fate (52). The recombinant enzyme from C. roseus expressed in E. coli exhibited activity with the triglutamate form of 5-CH3H4PteGlun (Km value of 28 μM) but not with the monoglutamate form. Neither AdoMet nor cobalamin had any effect on the enzyme activity and did not lead to the use of the monoglutamate form as a methyl donor for the reaction (12). These facts strongly suggest that in higher plants methionine synthase is of the cobalamin-independent type.
In most organisms, the single carbons involved in folate-dependent processes are derived from the β-carbon of serine (52). First, serine hydroxymethyltransferase (EC 2.1.2.1) catalyzes the transfer of a methylene group from serine to tetrahydrofolate (H4PteGlun). In plants, the enzyme has been characterized from mitochondria, plastids, and cytosol (53, 54). Second, the reduction of 5,10-CH2H4PteGlun to 5-CH3H4PteGlun is catalyzed by 5,10-CH2H4PteGlun reductase. This enzyme is a flavoprotein that has been well-studied in bacterial and mammalian cells. The bacterial (EC 1.7.99.5) and mammalian (EC 1.5.1.20) enzymes exhibit variant structural properties as well as differences in substrate preference for the donor of reducing equivalents (FADH2 vs. NADPH, respectively) (55, 56). Interestingly, the N-terminal part of the mammalian enzyme contains a binding site for AdoMet, a potent allosteric inhibitor of the reaction (57). 5,10-CH2H4PteGlun reductase activity also was detected in plants such as carrot (58) and Lemna minor (59). However, the catalytic properties of the enzyme were not studied, and it is not clear whether the plant enzyme is of the bacterial or mammalian type. Also, its subcellular localization is not yet determined in plants.
AdoMet Synthesis and Its Metabolism.
Besides its well-known role as a methyl donor in a myriad of transmethylation reactions (60), in plants AdoMet is involved in ethylene biosynthesis through the activity of 1-aminocyclopropane-1-carboxylate synthase (EC 4.4.1.14) (17), spermidine and spermine biosyntheses through the activity of AdoMet decarboxylase (EC 4.1.1.50) (61), and biotin biosynthesis through the activity of 7,8-diaminopelargonic acid aminotransferase (EC 2.6.1.62), an unusual enzyme that uses AdoMet as an amino group donor (62, 63) (Fig. 2). All of these metabolic reactions are considered important to plant growth and development (17, 61, 64).
AdoMet Synthesis.
The formation of AdoMet from methionine and ATP is catalyzed by AdoMet synthetase (EC 2.5.1.6) (Fig. 2). Inorganic tripolyphosphate is an enzyme-bound intermediate that is cleaved by a tripolyphosphatase activity of the enzyme, where the α- and β-phosphate groups of ATP give rise to PPi, whereas the terminal γ-phosphate is released as inorganic phosphate (65). The activity of the yeast and bacterial AdoMet synthetases is modulated by AdoMet in a complex manner. Thus, high concentrations of AdoMet inhibit the enzyme, whereas low concentrations of AdoMet allosterically activate the enzyme by stimulation of the tripolyphosphate cleavage reaction (2, 11). The partially purified enzyme from pea seedlings is strongly inhibited by tripolyphosphate suggesting that the reaction catalyzed by the plant enzyme follows the same mechanism as that examplified for the yeast and bacterial AdoMet synthetase (66).
Genes encoding plant AdoMet synthetase have been recently cloned from various species. Characterization of these genes revealed that plant AdoMet synthetase is highly conserved in all organisms and encoded by a gene family. The absence of a detectable signal sequence at the N-terminal part of AdoMet synthetase isoforms predicted from the rice (67) and C. roseus (68) cDNA sequences indicated that these isoenzymes are located in the cytosol. The specific localization of AdoMet synthetases in the cytosolic compartment raises the problem of AdoMet transport across the limiting border of mitochondria and chloroplasts to sustain internal transmethylation reactions.
The properties of three C. roseus isoenzymes were compared after heterologous expression of the individual enzymes in recombinant bacteria (68). In particular, inhibition by the tripolyphosphate intermediate and the AdoMet reaction product was observed. It is not known from that study whether plant AdoMet synthetases are activated by low concentrations of AdoMet, as demonstrated for the bacterial and yeast enzymes. The close similarities in physicochemical properties of the C. roseus isoenzymes suggested that the existence of several isoforms of AdoMet synthetase in plants reflects specificities in the association with enzymes that use AdoMet (68).
AdoMet Recycling During Transmethylation Reactions.
Because AdoHcy, which is produced during transmethylation reactions, is a strong competitive inhibitor of the AdoMet-dependent methylases, it must be removed for proper functioning of these enzymes. This removal is ensured by AdoHcy hydrolase that catalyzes the hydrolysis of AdoHcy to produce Hcy and adenosine (Fig. 2). Moreover, because the equilibrium of the enzyme-catalyzed reaction is strongly in favor of AdoHcy synthesis (60), hydrolysis is driven in vivo by the irreversible conversion of Hcy into methionine via methionine synthase and the removal of adenosine (the enzymes involved in this adenosine metabolism are not yet precisely identified; however, in yeast a mutant impaired in adenosine kinase accumulates AdoHcy; see ref. 4).
Given its importance in the regulation of biological methylation reactions through a control of the intracellular AdoHcy:AdoMet ratio (69), AdoHcy hydrolase has been the focus of recent investigations in plants. To date, the enzyme has been characterized in tobacco (70) and parsley (71), and its gene is cloned. The predicted amino acid sequence is highly homologous to that of AdoHcy hydrolase from various organisms. Although the subcellular localization of the plant enzyme has not been established yet, its N-terminal sequence lacks the typical features of transit peptide signals, thereby suggesting that the plant enzyme is localized to the cytosol. Again, whether the enzyme is specifically localized in this compartment, we are forced to conclude that AdoHcy produced in the various cell organelles must be transported toward the cytosolic compartment where it can be hydrolyzed.
Interestingly, the plant enzyme may play a role in signal transduction mediated by cytokinins (plant growth hormones that are implicated in the regulation of cell division and differentiation) because it is a cytokinin-binding protein (72). Moreover, the AdoHcy hydrolase gene is induced by cytokinins and auxin (also a chief member of plant growth-promoting hormones) in suspension-cultured tobacco cells (70). Clearly, the biochemical and molecular bases of these intriguing properties need further investigation if one considers the key role of this enzyme in C1 metabolism. Transgenic tobacco plants constitutively expressing an antisense AdoHcy hydrolase tobacco gene showed resistance to infection by various viruses (73). It is not yet known whether this increased resistance was caused by undermethylation at the 5′ terminus of viral mRNA or to a cytokinin effect. Also, there exists a close metabolic link between plant defense against fungal pathogens and increased turnover of activated methyl groups because a fungal elicitor derived from the fungus Phytophtora megasperma f. sp. glycinea rapidly activated transcription of AdoMet synthetase and AdoHcy hydrolase genes in cultured parsley cells as well as leaves of this plant (71).
Regulatory Considerations.
In yeast, biosynthesis of the sulfur amino acids is specifically controlled via AdoMet-mediated negative transcriptional regulation. This repression involves a complex set of interacting trans-acting factors, one of which is the protein Cbf1p that also is implicated in chromosome segregation (for an excellent review, see ref. 4). In enteric bacteria, methionine synthesis also is negatively regulated by AdoMet. Here, two types of control mechanisms are superimposed: metabolic control of enzyme activities and control of enzyme amounts (2, 26). On the one hand, homoserine succinyltransferase, which catalyzes the first reaction specific for the pathway, is feedback inhibited by the synergetic action of methionine and AdoMet. On the other hand, AdoMet acts as a corepressor and on binding to the metJ gene product (aporepressor) forms the holorepressor that negatively regulates transcription of several genes involved in the methionine pathway.
In plants, neither cystathionine γ-synthase nor cystathionine β-lyase is significantly inhibited by methionine pathway intermediates or end-products such as AdoMet. Also, these enzymes are not sensitive to feedback inhibition by any of the aspartate-derived amino acids (25, 31, 36, 74, 75). There are strong indications, however, for a control of methionine biosynthesis in plants at the level of gene expression and that the major site for such a regulation is probably cystathionine γ-synthase. Thus, it was observed that, (i) when Lemna paucicostata was grown in the presence of 2 μM methionine, the intracellular pool of this amino acid increased up to 200-fold (a proton-coupled methionine symport in the plant plasma membrane mediates methionine transport into the plant cells and its accumulation) and the level of extractable cystathionine γ-synthase activity was reduced by 85% (76), (ii) administration of 2 μM methionine to growing Lemna had essentially no effect on the accumulation of sulfate into cysteine but specifically depressed accumulation into cystathionine and its products to as low as 15–20% that of the control plants (36, 76), (iii) culture conditions causing methionine starvation, e.g., in the presence of AVG or in the presence of lysine plus threonine mixtures that inhibit an early step in OPH synthesis catalyzed by aspartate kinase (EC 2.7.2.4) (7), were associated with a substantial increase of extractable cystathionine γ-synthase activity (36, 77), and (iv) the constitutive expression of the AdoMet-insensitive E. coli threonine synthase in tobacco cells resulted in a 3.5-fold increase in cystathionine γ-synthase activity (78). In this context, Fig. 4A illustrates the inhibitory effect of a combination of lysine and threonine on the growth of Arabidopsis and the restoration of plant growth by methionine.
Figure 4.
Methionine is essential for Arabidopsis growth. (A) Sensitivity of Arabidopsis to exogeneously added lysine plus threonine (Center) and restoration of growth by exogeneously added methionine (Right). Plants were sprayed every 2 days (from 8 days after sowing) with 10 mM lysine and 10 mM threonine (Center) or with 10 mM lysine, 10 mM threonine, or 2 mM methionine (Right). Control plants, Left. Plants shown are 15-days-old. (B) Growth of wild-type Arabidopsis (Left) and cystathionine γ-synthase antisense plants (Right). Plants shown are 33-days-old.
Studies of Lemna growing in the presence of PAG established that 10% of control cystathionine γ-synthase activity was essential for viability. Yet, the use of sublethal concentrations of PAG indicated that only 16% of control activity is necessary and sufficient to maintain normal rates of growth. Despite this 6-fold reduction in enzyme activity the net rate of methionine biosynthesis remained unaffected (40). Thus, repressing cystathionine γ-synthase activity to approximately the same extent by methionine or PAG (e.g., to ≈15% of control activity) was associated with markedly different effects on the flux from sulfur into methionine, meaning that cystathionine γ-synthase is by itself not sufficient to regulate methionine synthesis. Because the methionine-treated Lemna showed a substantial elevation in AdoMet concentration, Thompson et al. (36, 40) suggested that an additional factor could be the activation of threonine synthase by AdoMet and thus directing OPH toward threonine synthesis at the expense of the methionine branch. This ought not to be the case because activities of threonine synthase measured in Lemna were inversely related to the methionine nutrition of the plants, presumably to limit the overproduction of threonine that could result from allosteric stimulation of the enzyme by AdoMet (28). Therefore, there are no conclusive results on the mechanisms by which methionine regulates its own synthesis in plants and the specific regulatory molecular mechanisms remain poorly understood.
It must be stressed that these previous studies only addressed the question of the regulation of methionine biosynthesis by measurements of extractable enzyme activities. Therefore, it is difficult to decipher whether modulation of such enzyme activities reflects transcriptional and/or post-transcriptional control. Also, disadvantages in the use of methionine biosynthetic inhibitors are that they are not strictly specific to the target enzymes and may pose problems with respect to permeability and stability in cells. For example, AVG that has been used largely as a cystathionine β-lyase inhibitor is also a well-known inhibitor of 1-aminocyclopropane-1-carboxylic acid synthase (Fig. 2). The recent cloning of plant cDNAs for the methionine and AdoMet biosynthetic enzymes as well as for enzymes involved in AdoMet metabolism provided unique opportunities to manipulate more accurately the expression of the endogenous genes by using reverse genetics approaches. In this way, Boerjan et al. (79) observed that cosuppression (silencing) of the AdoMet synthetase genes is associated with a marked elevation in free methionine levels in transformed tobacco leaves (up to 20–50 mM in the cytosol, assuming that this compartment occupies 10% of the total cell volume). It is clear, therefore, that methionine itself does not regulate the synthesis of any of the methionine-synthesizing enzymes. Also, when a cDNA coding for AdoMet decarboxylase was introduced in an antisense orientation in plants to suppress endogenous enzyme expression, there was a significant reduction (3-fold) in spermidine and spermine contents and a marked elevation (50-fold) in the rate of ethylene biosynthesis (61). Taken together, these results point out a role of AdoMet, and not of methionine, in the regulation of expression of cystathionine γ-synthase in plants, as demonstrated in bacteria (2, 26).
To investigate the consequence of repressing cystathionine γ-synthase in plants, we have transformed Arabidopsis with a T-DNA construct containing the cDNA coding the Arabidopsis enzyme in an antisense orientation. The most salient results of that study are presented here (B.G., S.R., D.J., and R.D., unpublished results). Transgenic plants exhibited two types of altered phenotypes, both of which being reversed by administration of exogenous methionine. The one, corresponding to plants homozygous for the transgene construct, was lethal, i.e., plants turned yellow and died soon after germination. The other, corresponding to hemizygous plants, was less severe, being essentially characterized by a strong reduction in growth (Fig. 4B). Presumably, the expression level of antisense mRNA was higher in the homozygous than in the hemizygous plants. These observations were in agreement with the PAG-inhibition data of Thompson et al. (40) showing that a basal level of cystathionine γ-synthase activity is essential for viability. As expected, the accumulation of cystathionine γ-synthase was specifically depressed in the transformed hemizygous plants (to ≈20% the level of wild type). Yet, the transgenic plants exhibited approximately a 3-fold increased level of cystathionine β-lyase, which could therefore compensate for the depressed cystathionine γ-synthase activity, and approximately a 2-fold increase of threonine synthase, presumably to compensate for a decrease in the availability of the AdoMet activator. Clearly, other steps than the first committed reaction in methionine synthesis are important to account for the homeostatic regulation of this amino acid in plants. In addition to a control of some enzyme activities by internal metabolites (e.g., allosteric regulation of threonine synthase by AdoMet), these results suggest a model in which the regulation of at least three genes, i.e., those coding for cystathionine γ-synthase, cystathionine β-lyase, and threonine synthase, is mediated by trans-acting factors working in association with methionine or a derivative of it, most presumably AdoMet. The observed phenotypic abnormalities (see Fig. 4B) raise the interesting possibility that alterations in the methionine/AdoMet intracellular levels somehow affect the activity of meristemic cells that are, within the apices, responsible for primary plant growth because they are capable of undergoing repeated division. We do not know yet whether this arose from an alteration of AdoMet-dependent enzyme reactions—e.g., DNA methylation, which may have a general regulatory influence on gene expression—or from a modification in plant growth hormone levels (see above for a possible link between cytokinin-mediated transduction pathways and AdoMet metabolism).
An attempt to deplete the pool of AdoMet in plants was effected by constitutive expression in tomato of AdoMet hydrolase (EC 3.3.1.2), an enzyme from bacteriophage T3 that catalyzes the conversion of AdoMet to 5-methylthioadenosine and homoserine (80). Unexpectedly, constitutive AdoMet hydrolase expression was not lethal to plants, suggesting in agreement with our results on antisense expression of the cystathionine γ-synthase gene in Arabidopsis (see above) that the transgenic plants may be conditioned to overcome the decrease in the AdoMet pool, that is, the expression levels of enzymes involved in the methionine synthesizing and/or recycling pathways could be induced to maintain an AdoMet pool sufficient for growth.
Finally, the role of AdoHcy hydrolase in plants was addressed recently by expressing antisense RNA of the tobacco enzyme in tobacco (81). The transgenic plants displayed distinct morphological changes (e.g., stunting, loss of apical dominance, delayed senescence) as well as abnormalities in flowers including a floral homeotic change. Also, in these plants, a repetitive DNA sequence appeared less methylated than controls, suggesting that AdoHcy hydrolase regulates DNA methylation in planta and therefore plays a general role in regulation of gene expression, particularly in primary growth and flower morphogenesis. Consistent with this finding Arabidopsis plants transformed with an antisense construct of an Arabidopsis DNA methyltransferase cDNA (METI) had reduced cytosine methylation in GC dinucleotides resulting in hypomethylation of centromeric and ribosomal DNA as well as aberrant expression of floral homeotic genes, and exhibited phenotypic and developmental abnormalities similar to those described above for transgenic tobacco plants expressing an antisense construct of the AdoHcy hydrolase gene (82).
In general, these studies aimed at impairing AdoMet-dependent processes in plants also unraveled an unexpected finding in that plants do have an astonishing capability to maintain methionine, AdoMet, and threonine fluxes in the face of adverse conditions, thereby suggesting a high plasticity in regulatory mechanisms. An understanding of the regulatory enzymes in such complex metabolic networks has obvious implications at both the academic and industrial levels, particularly in herbicide design.
Integration of the Methionine and AdoMet Synthetic/Recycling Pathways Within the Plant Cell.
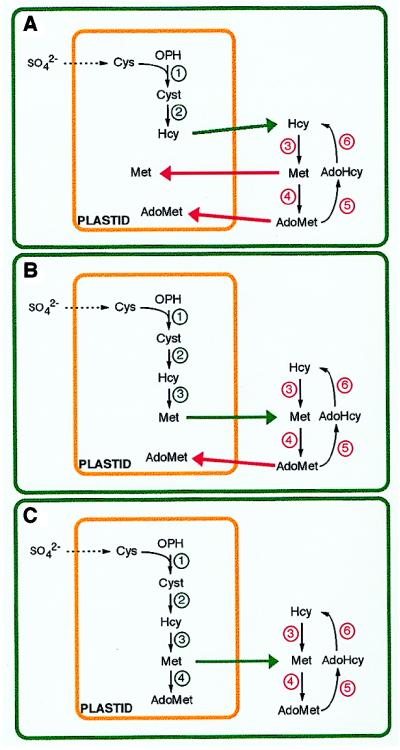
The available experimental evidence compiles the view that all biochemical steps for conversion of aspartate to Hcy are localized exclusively to the plastidial compartment. In contrast, the available data would indicate a cytosolic localization for methionine synthase, AdoMet synthetase, and AdoHcy hydrolase activities. Based on these observations, Fig. 5 illustrates three possibilities to account for the integration of the methionine and AdoMet biosynthetic/recycling pathways within the plant cell.
Figure 5.
Three models for the compartmentation of methionine- and AdoMet-synthesizing/recycling enzymes in plant cells. (A) All enzymes up to Hcy synthesis are present in plastids. The last steps of methionine and AdoMet syntheses occur in the cytosol. (B) The plastid is autonomous for methionine synthesis and the last step of AdoMet synthesis occurs in the cytosol. (C) The plastid is autonomous for methionine and AdoMet biosyntheses. Recycling of methionine from AdoMet metabolism occurs in the cytosol. Nomenclature for enzymes as in Figs. 1 and 2.
In the first (Fig. 5A), plastids catalyze the conversion of aspartate to Hcy, whereas further metabolism, i.e., methionine and AdoMet syntheses occur in the cytosol. This model is supported by a number of molecular and biochemical data (see above) and by biochemical localization studies of enzymes involved in methionine and AdoMet biosyntheses in pea and barley leaves (83). Obviously, methionine can cross the plastid envelope to meet the chloroplast requirements for protein synthesis (for a review, see ref. 10). This is less certain for AdoMet and Hcy. For example, in animals, whether AdoMet can cross biological membranes is still controversial (69). This scheme therefore implies a transport of Hcy from the plastid to the cytosol, as well as a transport of AdoMet to the plastid. AdoMet should at least be present in this compartment to account for the well-demonstrated regulation of threonine synthase activity. Unfortunately, there is at present no information regarding the cellular traffic of Hcy and AdoMet in plants. Nevertheless, the possible existence of an AdoMet transporter in plants is supported by the observation that, unlike enteric bacteria (2), S. cerevisiae can actively transport AdoMet (4).
In the second (Fig. 5B), plastids could catalyze the final step of methionine synthesis. Two types of observations support this contention: (i) methylation of Hcy by using 5-CH3H4PteGlu as a substrate has been reported in pea leaf chloroplasts and mitochondria (84, 85), and (ii) the photosynthetic protozoan E. gracilis Z. contains, besides a cytosolic cobalamin-independent methionine synthase, three isoforms of cobalamin-dependent enzymes that are located in the chloroplast, the mitochondria, and the cytosol (49). A similar subcellular distribution for the last enzyme involved in methionine synthesis might, therefore, be expected in higher plant cells. The existence of this additional methionine synthase is attractive because it would render plastids able to synthesize methionine de novo, as is the case for other aspartate-derived amino acids (7). Accordingly, the role of the cytosolic compartment would be to recycle AdoMet during the transmethylation reactions, notably to control the level of AdoHcy, a powerful inhibitor of the AdoMet-dependent methyltransferases. In the frame of this hypothetical scheme, there are three related questions that concern (i) the existence of cobalamin in plants, a feature that has not yet been documented, (ii) the subcellular compartmentation of folate pools, particularly that of 5-CH3H4PteGlun, and (iii) the role of the polyglutamate chain in folate derivatives, notably in relation with the functioning of methionine synthase. Work on animal systems has shown that monoglutamyl folates are readily transported across cellular membranes, but permeability to polyglutamyl folates is considerably less (86). Indeed, in mammalian cells, folylpolyglutamate synthetase (EC 6.3.2.17) that catalyzes the elongation of the polyglutamate chain in folate compounds is present both in the cytosol and the mitochondria (87, 88). Yet, in higher plants, this enzyme was detected only in the mitochondria (89). In the absence of evidence for a transport system of polyglutamyl derivatives of folate from the mitochondria to the plastids, the de novo synthesis of methionine in plastids would therefore require the existence of a cobalamin-dependent form of methionine synthase because this enzyme can use 5-CH3H4PteGlu as a substrate. Indeed, this folate compound has been reported to account for 25% of total pteroylglutamates recovered from pea leaf chloroplast extracts whereas no evidence was obtained for the presence of 5-CH3H4PteGlu3 (85). Clearly the mechanisms involved in folate transport and folate traffic within the plant cell remain to be determined, particularly with subcellular fractions of demonstrated integrity. Also, although the existence of serine hydroxymethyltransferase in the chloroplast compartment is well-established (54) the subcellular localization of 5,10-CH2H4PteGlun reductase in the plant cell remains to be clarified.
In the third possibility (Fig. 5C), plastids could catalyze the final steps of both methionine and AdoMet syntheses. The existence of an isoform of AdoMet synthetase in the plastidial compartment could solve the problem of AdoMet import from the cytosol to the stromal space. In view of the available evidence, this isoform shall, however, differ substantially from those presently described in plants and that appear very likely to be localized to the cytosol.
CONCLUSION
Recent work has allowed an important progress in our knowledge concerning the enzymes involved in the synthesis and recycling of methionine and AdoMet in plants. The development of molecular probes and antibodies to these enzymes, along with their detailed biochemical characterization, now render possible a thorough investigation of the regulation of these pathways. Beyond its fundamental aspect, the understanding of the regulatory patterns of methionine biosynthesis in plants is crucial for improvement of animal nutrition and also may lead to the discovery of new targets for novel herbicides. Elucidation of the cellular and developmental regulation of the pathway will benefit from the cloning of the genes coding for methionine-synthesizing and recycling enzymes, particularly concerning the functioning of the promoters of these genes and the identification of the key transcription factors. Finally, an understanding of the intracellular compartmentation of the enzymes involved in the synthesis and recycling of methionine and AdoMet also requires a better comprehension of how the cell regulates its folate requirements between the cytosol, the mitochondria, and the chloroplasts. As we have discussed in this article, AdoMet is involved directly in a number of housekeeping functions in plants. The fact that it also might intervene in seemingly unrelated biological processes such as plant-growth-hormone-mediated processes and plant pathogen interactions highlights the paramount importance of this molecule that will be the prospect of future exciting research.
Acknowledgments
We thank Gilles Curien and Michel Droux for helpful discussions and Régis Pépin and Claudette Job for photography of the Arabidopsis plants.
ABBREVIATIONS
- AdoHcy
S-adenosylhomocysteine
- AdoMet
S-adenosylmethionine
- AVG
l-aminoethoxyvinylglycine
- Hcy
homocysteine
- H4PteGlun
tetrahydrofolate
- MTA
5-methylthioadenosine
- 5-CH3H4PteGlun
N5-methyltetrahydrofolate
- 5
10-CH2H4PteGlun, N5,N10-methylenetetrahydrofolate
- OPH
O-phosphohomoserine
- PAG
DL-propargylgylcine
- PLP
pyridoxal 5′-phosphate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U97200).
References
- 1.Giovanelli J. Methods Enzymol. 1987;143:419–426. [Google Scholar]
- 2.Saint-Girons I, Parsot C, Zakin M M, Barzu O, Cohen G N. CRC Crit Rev Biochem. 1988;23:S1–S42. doi: 10.3109/10409238809083374. [DOI] [PubMed] [Google Scholar]
- 3.Marzluf G A. Adv Genet. 1994;31:187–206. doi: 10.1016/s0065-2660(08)60398-3. [DOI] [PubMed] [Google Scholar]
- 4.Thomas D, Surdin-Kerjan Y. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giovanelli J, Mudd S H. Biochem Biophys Res Commun. 1967;27:150–156. doi: 10.1016/s0006-291x(67)80054-8. [DOI] [PubMed] [Google Scholar]
- 6.Habben J E, Larkins B A. In: Seed Development and Germination. Kigel J, Galili G, editors. New York: Dekker; 1995. pp. 791–810. [Google Scholar]
- 7.Azevedo R A, Arruda P, Turner W L, Lea P J. Phytochemistry. 1997;46:395–419. doi: 10.1016/s0031-9422(97)00319-1. [DOI] [PubMed] [Google Scholar]
- 8.Leustek T. Physiol Plant. 1996;97:411–419. [Google Scholar]
- 9.Hell R. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- 10.Ravanel S. C R Acad Sci Paris. 1997;320:497–504. [Google Scholar]
- 11.Giovanelli J, Mudd S H, Datko A H. In: The Biochemistry of Plants. Miflin B J, editor. Vol. 5. New York: Academic; 1980. pp. 468–487. [Google Scholar]
- 12.Eichel J, Gonzalez J C, Hotze M, Matthews R G, Schröder J. Eur J Biochem. 1995;230:1053–1058. doi: 10.1111/j.1432-1033.1995.tb20655.x. [DOI] [PubMed] [Google Scholar]
- 13.Zarembinski T I, Theologis A. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- 14.Walden R, Cordeiro A, Tiburcio A F. Plant Physiol. 1997;113:1009–1013. doi: 10.1104/pp.113.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cossins E A, Chen L. Phytochemistry. 1997;45:437–452. doi: 10.1016/s0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- 16.Giovanelli J, Mudd S H, Datko A H. Plant Physiol. 1985;78:555–560. doi: 10.1104/pp.78.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKeon T A, Yang S-F. In: Plant Hormones and Their Role in Plant Growth and Development. Davies P J, editor. Dordrecht: Kluwer; 1990. pp. 94–112. [Google Scholar]
- 18.Lunn J E, Droux M, Martin J, Douce R. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruffet M-L, Droux M, Douce R. Plant Physiol. 1994;104:597–604. doi: 10.1104/pp.104.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolland N, Droux M, Lebrun M, Douce R. Arch Biochem Biophys. 1993;300:213–222. doi: 10.1006/abbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 21.Le Guen L, Thomas M, Kreis M. Mol Gen Genet. 1994;245:390–396. doi: 10.1007/BF00290120. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Leustek T. Plant Mol Biol. 1996;36:1117–1124. doi: 10.1007/BF00041395. [DOI] [PubMed] [Google Scholar]
- 23.Wallsgrove R M, Lea P J, Miflin B J. Plant Physiol. 1983;71:780–784. doi: 10.1104/pp.71.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravanel S, Droux M, Douce R. Arch Biochem Biophys. 1995;316:572–584. doi: 10.1006/abbi.1995.1077. [DOI] [PubMed] [Google Scholar]
- 25.Ravanel S, Gakière B, Job D, Douce R. Biochem J. 1998;331:639–648. doi: 10.1042/bj3310639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Old I A, Phillips S E V, Stockley P G, Saint-Girons I. Prog Biophys Mol Biol. 1991;56:145–185. doi: 10.1016/0079-6107(91)90012-h. [DOI] [PubMed] [Google Scholar]
- 27.Datko A H, Giovanelli J, Mudd S H. J Biol Chem. 1974;249:1139–1155. [PubMed] [Google Scholar]
- 28.Giovanelli J, Veluthambi K, Thompson G A, Mudd S H, Datko A H. Plant Physiol. 1984;76:285–292. doi: 10.1104/pp.76.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curien G, Dumas R, Ravanel S, Douce R. FEBS Lett. 1996;390:85–90. doi: 10.1016/0014-5793(96)00633-3. [DOI] [PubMed] [Google Scholar]
- 30.Curien, G., Job, D., Douce, R. & Dumas, R. (1998) Biochemistry, in press. [DOI] [PubMed]
- 31.Kreft B D, Townsend A, Pohlenz H-D, Laber B. Plant Physiol. 1994;104:1215–1220. doi: 10.1104/pp.104.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datko A H, Mudd S H. Plant Physiol. 1984;75:474–479. doi: 10.1104/pp.75.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giovanelli J, Mudd S H, Datko A H, Thompson G A. Plant Physiol. 1986;81:577–583. doi: 10.1104/pp.81.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holbrook E L, Greene R C, Krueger J H. Biochemistry. 1990;29:435–442. doi: 10.1021/bi00454a019. [DOI] [PubMed] [Google Scholar]
- 35.Brzovic P, Holbrook E L, Greene R C, Dunn M F. Biochemistry. 1990;29:442–451. doi: 10.1021/bi00454a020. [DOI] [PubMed] [Google Scholar]
- 36.Thompson G A, Datko A H, Mudd S H, Giovanelli J. Plant Physiol. 1982;69:1077–1083. doi: 10.1104/pp.69.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giovanelli J, Mudd S H, Datko A H. J Biol Chem. 1978;253:5665–5677. [PubMed] [Google Scholar]
- 38.MacNicol P K, Datko A H, Giovanelli J, Mudd S H. Plant Physiol. 1981;68:619–625. doi: 10.1104/pp.68.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negrutiu I, De Brouwer D, Dirks R, Jacobs M. Mol Gen Genet. 1985;199:330–337. [Google Scholar]
- 40.Thompson G A, Datko A H, Mudd S H. Plant Physiol. 1982;70:1347–1352. doi: 10.1104/pp.70.5.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Droux M, Ravanel S, Douce R. Arch Biochem Biophys. 1995;316:585–595. doi: 10.1006/abbi.1995.1078. [DOI] [PubMed] [Google Scholar]
- 42.Turner W L, Pallett K E, Lea P J. Phytochemistry. 1998;47:189–196. [Google Scholar]
- 43.Ravanel S, Ruffet M-L, Douce R. Plant Mol Biol. 1995;29:875–882. doi: 10.1007/BF00041177. [DOI] [PubMed] [Google Scholar]
- 44.Ravanel S, Job D, Douce R. Biochem J. 1996;320:383–392. doi: 10.1042/bj3200383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander F W, Sandmeier E, Mehta P K, Christen P. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 46.Belfaiza J, Parsot C, Martel A, Bouthier de la Tour C, Margarita D, Cohen G N, Saint-Girons I. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clausen T, Huber R, Messerschmidt A, Pohlenz H-D, Laber B. Biochemistry. 1997;36:12633–12643. doi: 10.1021/bi970630m. [DOI] [PubMed] [Google Scholar]
- 48.Finkelstein J D. J Nutr Biochem. 1990;1:228–236. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 49.Isegawa Y, Watanabe F, Kitaoka S, Nakano Y. Phytochemistry. 1994;35:59–61. [Google Scholar]
- 50.Petersen M, Van Der Straeten D, Bauw G. Plant Physiol. 1995;109:338–338. doi: 10.1104/pp.107.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurvari V, Qian F, Snell W J. Plant Mol Biol. 1995;29:1235–1252. doi: 10.1007/BF00020465. [DOI] [PubMed] [Google Scholar]
- 52.Cossins E A. In: The Biochemistry of Plants. Davies D D, editor. Vol. 11. San Diego: Academic; 1987. pp. 317–353. [Google Scholar]
- 53.Bourguignon J, Neuburger M, Douce R. Biochem J. 1988;255:169–178. doi: 10.1042/bj2550169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Besson V, Neuburger M, Rébeillé F, Douce R. Plant Physiol Biochem. 1995;33:665–673. [Google Scholar]
- 55.Daubner S C, Matthews R G. J Biol Chem. 1982;257:140–145. [PubMed] [Google Scholar]
- 56.Clark J E, Ljungdahl L G. J Biol Chem. 1984;259:10845–10849. [PubMed] [Google Scholar]
- 57.Sumner J M, Jencks D A, Khani S, Matthews R G. J Biol Chem. 1986;261:7697–7700. [PubMed] [Google Scholar]
- 58.Fedec P, Cossins E A. Phytochemistry. 1976;15:359–362. [Google Scholar]
- 59.Wong K, Cossins E A. Phytochemistry. 1976;15:921–925. [Google Scholar]
- 60.Poulton J E. In: The Biochemistry of Plants. Coon E E, editor. Vol. 7. New York: Academic; 1981. pp. 667–723. [Google Scholar]
- 61.Kumar A, Taylor M A, Mad Arif S A, Davies H V. Plant J. 1996;9:147–158. [Google Scholar]
- 62.Stoner G L, Eisenberg M A. J Biol Chem. 1975;250:4037–4043. [PubMed] [Google Scholar]
- 63.Baldet P, Gerbling H, Axiotis S, Douce R. Eur J Biochem. 1993;217:479–485. doi: 10.1111/j.1432-1033.1993.tb18267.x. [DOI] [PubMed] [Google Scholar]
- 64.Shellhammer J, Meinke D. Plant Physiol. 1990;93:1162–1167. doi: 10.1104/pp.93.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chou T-C, Talalay P. Biochemistry. 1972;11:1065–1073. doi: 10.1021/bi00756a019. [DOI] [PubMed] [Google Scholar]
- 66.Aarnes H. Plant Sci Lett. 1977;10:381–390. [Google Scholar]
- 67.Lee J-H, Chae H S, Lee J-H, Hwang B, Hahn K W, Kang B G, Kim W T. Biochim Biophys Acta. 1997;1354:13–18. doi: 10.1016/s0167-4781(97)00114-0. [DOI] [PubMed] [Google Scholar]
- 68.Schröder G, Eichel J, Breinig S, Schröder J. Plant Mol Biol. 1997;33:211–222. doi: 10.1023/a:1005711720930. [DOI] [PubMed] [Google Scholar]
- 69.Chiang P K, Gordon R K, Tal J, Zeng G C, Doctor B P, Pardhasaradhi K, McCann P P. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 70.Tanaka H, Masuta C, Kataoka J, Kuwata S, Koiwai A, Noma M. Plant Sci. 1996;113:167–174. [Google Scholar]
- 71.Kawalleck P, Plesch G, Hahlbrock K, Somssich I E. Proc Natl Acad Sci USA. 1992;89:4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitsui S, Wakasugi T, Sugiura M. Plant Cell Physiol. 1993;34:1089–1096. [Google Scholar]
- 73.Masuta C, Tanaka H, Uehara K, Kuwata S, Koiwai A, Noma M. Proc Natl Acad Sci USA. 1995;92:6117–6121. doi: 10.1073/pnas.92.13.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aarnes H. Plant Sci Lett. 1980;19:81–89. [Google Scholar]
- 75.Staton A L, Mazelis M. Arch Biochem Biophys. 1991;290:46–50. doi: 10.1016/0003-9861(91)90589-b. [DOI] [PubMed] [Google Scholar]
- 76.Giovanelli J, Mudd S H, Datko A H. Plant Physiol. 1985;77:450–455. doi: 10.1104/pp.77.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Datko A H, Mudd S H. Plant Physiol. 1982;69:1070–1076. doi: 10.1104/pp.69.5.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muhitch M J. J Plant Physiol. 1997;150:16–22. [Google Scholar]
- 79.Boerjan W, Bauw G, van Montagu M, Inzé D. Plant Cell. 1994;6:1401–1414. doi: 10.1105/tpc.6.10.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Good X, Kellogg J A, Wagoner W, Langhoff D, Matsumura W, R. K. Bestwick R K. Plant Mol Biol. 1994;26:781–790. doi: 10.1007/BF00028848. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka H, Masuta C, Uehara K, Kataoka J, Koiwai A, Noma M. Plant Mol Biol. 1997;35:981–986. doi: 10.1023/a:1005896711321. [DOI] [PubMed] [Google Scholar]
- 82.Finnegan E J, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallsgrove R M, Lea P J, Miflin B J. Plant Physiol. 1983;71:780–784. doi: 10.1104/pp.71.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clandinin M T, Cossins E A. Phytochemistry. 1974;13:585–591. [Google Scholar]
- 85.Shah S P J, Cossins E A. FEBS Lett. 1970;7:267–270. doi: 10.1016/0014-5793(70)80177-6. [DOI] [PubMed] [Google Scholar]
- 86.McGuire J J, Bertino J R. Mol Cell Biochem. 1981;38:19–48. doi: 10.1007/BF00235686. [DOI] [PubMed] [Google Scholar]
- 87.McGuire J J, Coward J K. In: Folates and Pterins. Blakley R L, Benkovic S J, editors. Vol. 1. New York: Wiley Interscience; 1984. pp. 135–190. [Google Scholar]
- 88.Lin B F, Huang R F S, Shane B. J Biol Chem. 1993;268:21674–21679. [PubMed] [Google Scholar]
- 89.Neuburger M, Rébeillé F, Jourdain A, Nakamura S, Douce R. J Biol Chem. 1996;271:9466–9472. doi: 10.1074/jbc.271.16.9466. [DOI] [PubMed] [Google Scholar]