Abstract
Intercellular tight junctions (TJs) regulate epithelial barrier properties. Claudins are major structural constituents of TJs and belong to a large family of tetra-spanning membrane proteins that have two predicted extracellular loops (ELs). Given that claudin-1 is widely expressed in epithelia, we further defined the role of its EL domains in determining TJ function. The effects of several claudin-1 EL mimetic peptides on epithelial barrier structure and function were examined. Incubation of model human intestinal epithelial cells with a 27-amino acid peptide corresponding to a portion of the first EL domain (Cldn-153–80) reversibly interfered with epithelial barrier function by inducing the rearrangement of key TJ proteins: occludin, claudin-1, junctional adhesion molecule-A, and zonula occludens-1. Cldn-153–80 associated with both claudin-1 and occludin, suggesting both the direct interference with the ability of these proteins to assemble into functional TJs and their close interaction under physiological conditions. These effects were specific for Cldn-153–80, because peptides corresponding to other claudin-1 EL domains failed to influence TJ function. Furthermore, the oral administration of Cldn-153–80 to rats increased paracellular gastric permeability. Thus, the identification of a critical claudin-1 EL motif, Cldn-153–80, capable of regulating TJ structure and function, offers a useful adjunct to treatments that require drug delivery across an epithelial barrier.
Epithelial barrier function is controlled by cell-cell contact sites known as tight junctions (TJs).1,2 TJs are composed of several classes of integral membrane proteins including claudins, occludin,3,4 and junctional adhesion molecule-A (JAM-A)5,6 that are interlinked to the actin cytoskeleton through cytoplasmic scaffold proteins. Among the transmembrane TJ proteins, claudins are required to restrict the paracellular diffusion of small aqueous molecules.7,8 Over two dozen claudin family members have been identified, and most epithelia simultaneously express several different claudin isoforms.
Claudins have molecular mass in the range of 23 kDa and, based on hydropathy analysis, span the bilayer with four transmembrane domains, where the N and C termini are oriented toward the cytoplasm. Interactions between the C terminus and the scaffold proteins zonula occludens-1 (ZO-1) and -2 (ZO-2) are required for claudin assembly into TJs.9 Claudins are also regulated by post-translational modifications, including palmitoylation10 and phosphorylation.11 In addition to restricting the paracellular flux of small aqueous molecules, claudins apparently form paracellular ion channels, because the specific paracellular diffusion of different classes of ions is determined by claudin composition.12,13,14
Because claudins have two extracellular loop (EL) domains, this was a likely motif to regulate paracellular ion permeability. In fact, mutagenesis studies have helped to identify motifs in the first EL domain that determine the charge selectivity of different claudins.15,16 Roles for claudin EL domains in stabilizing TJs are suggested by previous studies that demonstrated that dust mite serine peptidases disrupt TJs by cleaving both EL domains.17 Moreover, Clostridium perfringens enterotxin (CPE) specifically disrupts assembly of claudin-3 and claudin-4 into TJs by binding to the second EL domain.18,19,20,21 This further underscores a role for claudin EL domains in the formation and maintenance of TJ barriers and suggests a pharmacological approach to transiently increase paracellular permeability.22,23,24 Although the specificity of CPE can be an advantage for this approach,20,25 whether CPE interacts with other claudins is still being explored. The recent structural analysis of the CPE claudin binding site is an important step in determining the range of claudins other than claudin-3 and claudin-4 with the ability to interact with CPE.26
Here we examined the ability of a series of claudin-1 EL-mimetic peptides to interfere with TJs formed by T84 intestinal epithelial cells, which abundantly express claudin-1.27 We found that a peptide emulating the second half of the first EL domain of claudin-1 (Cldn-1 53–80) reversibly interferes with TJ structure and function in T84 cells, without affecting adherens junctions (AJs). Using a Cldn-153–80 variant containing biotin and a photoaffinity label,28 we found that this peptide associated strongly with membranes and interacted predominantly with claudin-1 and occludin. Additionally, the importance of the Cldn-1 53–80 peptide was verified by oral administration of this peptide in rats and measurement of gastrointestinal epithelial paracellular permeability. In this model system, Cldn-153–80 enhanced paracellular permeability across the gastric epithelium, but had little effect on the small intestinal epithelium. This lack of permeability effect is likely because of peptide stability, because peptide digestion by duodenal enzymes would render it ineffective. Taken together, these results suggest that the first EL domain of claudin-1 plays a key role in the regulation of TJ function.
Materials and Methods
Peptide Synthesis
Peptides (Table 1) were synthesized as previously described28 using an automated Pioneer Peptide Synthesizer (PE/ABI) with Fmoc-protected amino acids on Fmoc-PEG-PS-resin. The light-sensitive Fmoc-p-benzoylphenylalanine (Advanced ChemTech, Louisville, KY) amino acid was introduced into peptides using HBTU-HOBt/DIPEA. D-Biotin (Sigma, St. Louis, MO) was incorporated into peptides at the N terminus using HBTU-HOBt/DIPEA in Me2SO. Peptide resins were cleaved with a 1-hour exposure of a 95% trifluoroacetic acid/2.5% triisopropylsilane/2.5% H2O solvent mixture. Released peptides were purified to >90% by preparative reversed-phase C18 high performance liquid chromatography, characterized by electrospray ionization mass spectroscopy (Sciex API100), lyophilized to dryness, and stored at −20°C before use. Peptide self-association was performed by incubating 200 μmol/L of peptide at room temperature for 30 minutes in HBSS with Ca2+ (HBSS+) that contained (in g/L) 0.185 CaCl2·2H2O, 0.1 MgSO4, 0.4 KCl, 0.06 KH2PO4, 8.0 NaCl, 0.048 Na2HPO4 (anhydrous), and 1.0 D-glucose. Peptide samples were analyzed by gel electrophoresis of a 1:1 mixture of sample:loading buffer (with or without a reducing agent) on a 16% Tris-Tricine gel, and subsequently silver stained.
Table 1.
Sequences of Peptides Used in This Study
| Peptide | Sequence | mol. wt. |
|---|---|---|
| Cldn-131–53 | Ac-KIYSYAGDNIVT-Ala-QAIYEGLWMS-NH2 | 2635 |
| *Cldn-131–53 | Biotin-KIYSYAGDNIVT-Bpa-QAIYEGLWMS-NH2 | 2818 |
| Cldn-153–80 | Ac-SCVSQSTGQIQCKV-Phe-DSLLNLNSTLQAT-NH2 | 3027 |
| *Cldn-153–80 | Biotin-SCVSQSTGQIQCKV-Bpa-DSLLNLNSTLQAT-NH2 | 3211 |
| Cldn-153–80-CS | Biotin-SSVSQSTGQIQSKV-Phe-DSLLNLSSTLQAT-NH2 | 3152 |
| Cldn-1146–160 | Ac-QEFYDPL-Thr-PINARYE-NH2 | 1897 |
| *Cldn-1146–160 | Biotin-QEFYDPL-Bpa-PINARYE-NH2 | 2080 |
Standard one- or three-letter amino acid codes are used except where Bpa indicates benzoylphenylalanine. Amino acids exchanged for Bpa or cysteine are noted in the alignment. All peptides were amidated (NH2) at the C terminus and either acetylated (Ac) or biotinylated at the N terminus. Photoactivatable (*) peptides were kept in the dark prior to UV-irradiation.
Cell Culture
T84 cells (American Type Culture Collection, Manassas, VA; CCL-248) were grown in 5% CO2 in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium with 6% newborn calf serum. To establish polarized monolayers, cells were plated onto 0.33-cm2 or 5-cm2 collagen-coated, transparent permeable polycarbonate supports (0.4 μm pore size) (Transwell Clears, Corning-Costar, Cambridge, MA). Six days after seeding on 0.33-cm2 supports or 12 days after seeding on 5-cm2 supports, confluent T84 monolayers were obtained that had transepithelial resistance (TER) values in the range of 2000 Ω-cm2 and restricted paracellular apical-to-basolateral flux of 3 kDa fluorescein isothiocyanate (FITC)-Dextran (FD-3) to a value of <100 ng/cm2/hr. TER was measured using a volt-ohmmeter (World Precision Instruments, Sarasota, FL).
Protein Labeling with Bait Peptides
T84 cells were grown to confluency on 5-cm2 collagen-coated permeable supports and incubated for 24 hours in media containing 200 μmol/L biotinylated bait peptide (either *Cldn-131–53, *Cldn-153–80, or *Cldn-1146–160) or control medium. For photoactivated bait peptides, cells were then extensively washed with cold HBSS+ and crosslinking was induced by a 15-minute exposure to high-intensity UV light before harvesting. For chemical cross-linking, a nonphotoactivatable, biotinylated Cldn-153–80 peptide was used. Following 24 hours of incubation, cells were washed in sterile HBSS, and BS3 crosslinker (Pierce, Rockford, IL), at 5 mmol/L in PBS (pH 8.0), was added to the cells, which were then incubated for 1 hour at room temperature. Excess crosslinker was quenched by adding 50 μl 1 M Tris buffer (pH 7.5) and further incubating for 15 minutes at room temperature. In both cases, cells were scraped into buffer R (100 mmol/L KCl, 3 mmol/L NaCl, 1 mmol/L Na2ATP, 3.5 mmol/L MgCl2, and 10 mmol/L HEPES, [pH 7.4]) containing freshly prepared 5 mmol/L diisopropyl fluorophosphate, 1.25 mmol/L phenylmethylsulfonyl fluoride, and 10 μg/ml chymostatin (Sigma).
Cells were disrupted at 4°C using a nitrogen cavitation bomb (Parr Instruments, Moline, IL) for 15 minutes at a N2 pressure of 200 p.s.i. Unbroken nuclei and cellular debris were removed by 10 minutes of centrifugation (1000 × g at 4°C). For purification of biotin-peptide bound proteins, the supernatant was passed through a monomeric avidin-Sepharose column (Pierce) and eluted according to manufacturer’s instructions. Biotinylated proteins were resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride, and detected using peroxidase-conjugated streptavidin. To analyze more stringently proteins directly crosslinked to the bait peptides, samples were boiled in 1% SDS for 5 minutes and then diluted 10-fold in SDS-free RIPA buffer (50 mmol/L Tris-HCl [pH 7.4], 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP-40, 0.25% Na-deoxycholate) before avidin-Sepharose affinity isolation. Subsequent elution and analysis was performed as above. For enzyme-linked immunosorbent assay (ELISA), samples were diluted 1:20 into PBS-T (PBS [pH 7.3] + 0.5% Tween 20). Control samples consisted of samples that were not treated with biotinylated peptides. A dilution series was generated and 100 μl/well was added to streptavidin plates (Nunc Immobilizer Streptavidin C8; Nunc, Roskilde, Denmark). The plates were incubated for 1 hour at room temperature on a shaker, washed three times in PBS-T, then further incubated for 1 hour at room temperature with PBS-T containing polyclonal anti-Claudin-1 or anti-Claudin-3 (Invitrogen) diluted to 1:1000. The plates were again washed in PBS-T and incubated with horseradish peroxidase (HRP)-Goat anti-Rabbit IgG (Jackson Immuno Research, Westgrove, PA) at 1:1000 dilution for 1 hour at room temperature. The plates were washed, 50 μl DY999 color substrate solution (R&D Systems, Minneapolis, MN) was added, and the plates were incubated in darkness for 15 minutes. Color development was stopped by adding 50 μl 4 M H2SO4 and A450 values were read using an ELISA plate reader.
Immunofluorescence Microscopy
T84 monolayers were incubated with bait peptides for up to 24 hours, washed, and then fixed/permeabilized for at −20°C for 20 minutes in absolute ethanol. The cells were then washed, incubated for 1 hour at room temperature in HBSS+ containing 5% normal goat serum, and labeled for 1 hour at room temperature with primary antibodies to either human occludin, claudin-1, claudin-3, ZO-1, (Invitrogen/Zymed, Carlsbad, CA), or JAM-A (kindly provided by C. A. Parkos, Emory University). T84 monolayers were washed and incubated for 1 hour at room temperature with Alexa 488-conjugated goat anti-mouse or anti-rabbit IgG (Invitrogen/Molecular Probes, Carlsbad, CA) antibodies, then mounted on glass slides in PBS/glycerol/p-phenylenediamine, 1:1:0.01 (v/v/v). For localization of filamentous actin (F-actin), monolayers were washed, fixed for 10 minutes at room temperature in 3.7% paraformaldehyde, permeabilized for 30 minutes at room temperature in 0.5% Triton X-100, and then labeled with Alexa 568-conjugated phalloidin (Invitrogen/Molecular Probes). To localize bound biotinylated bait peptides, the monolayers were washed, fixed in 3.7% paraformaldehyde, and labeled with FITC-conjugated streptavidin (Jackson Immunoresearch Labs, West Grove, PA). Images were obtained using an LSM510 confocal fluorescence microscope (Zeiss Microimaging, Thornwood, NY). Images shown are representative of at least six experiments, with multiple images taken per slide.
Immunogold Electron Microscopy
Cells were first incubated for 24 hours in either control medium or medium containing *Cldn-153–80. The cells were then washed and fixed for 20 minutes with 4% paraformaldehyde/0.2% glutaraldehyde in 0.1 M PBS (pH 7.2), followed by overnight fixation in 2% paraformaldehyde and treatment with 0.1% sodium borohydride. For better immunoreagent penetration, 0.05% saponin was added to blocking and antibody solutions. Cells were incubated for 1 hour in PBS blocking solution containing 5% normal goat serum, 5% BSA, and 0.1% gelatin, then incubated at 4°C overnight with 0.8 nm gold particle-conjugated goat F(ab)2 anti-biotin (Aurion, Wageningen, The Netherlands) antibody, at 1:100 dilution in PBS containing 0.2% BSA-c (Aurion). The cells were washed and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer. Aurion R-gen SE-electron microscopy silver enhancement was done and the samples were fixed for 15 minutes with 0.5% osmium tetroxide, dehydrated, and embedded in Eponate 12 resin. Ultrathin sections were stained with uranyl acetate and lead citrate, and examined with a Hitachi H-7500 transmission electron microscope.
Calcium Switch
Confluent, polarized T84 monolayers were transiently exposed for 20 minutes at 37°C to 2 mmol/L ethylene glycol-bis(aminoethyl ether) N,N,N′,N′-tetraacetic acid (EGTA) in Ca2+/Mg2+-free HBSS containing 10 mmol/L HEPES (HBSS−) chelate extracellular calcium that facilitated junction disassembly.6,29 After washing, monolayers were allowed to recover in complete cell culture media (containing Ca2+) in either control medium or medium containing 200 μmol/L of Cldn-131–53, Cldn-153–80, or Cldn-1146–160 peptide and barrier function was measured using either TER or the FD-3 paracellular permeability assay.
Paracellular Permeability Assays
Confluent, polarized T84 monolayers on permeable supports, and monolayers subjected to the Ca2+ switch assay, were exposed to bait peptides for 0, 6, or 24 hours. Paracellular permeability to 3 kDa FD-3 was assessed at these time points according to previously published methods.30 Briefly, after measuring TER, monolayers were washed in HBSS+ and then equilibrated for 10 minutes at 37°C on an orbital shaker. Monolayers were loaded from the apical side with 1 mg/ml FD-3, samples removed from the basolateral side of the well were taken at t = 0 and 120 minutes, and fluorescence intensity was analyzed using a CytoFluor 2350 Fluorescence Measurement System (Millipore, Cambridge, MA). FD-3 concentrations were calculated using a standard curve as ng FD-3 transported/cm2/hour. Numerical values from individual experiments were pooled and expressed as mean ± SE. Control and test values at each time point were compared by two-tailed unpaired Student’s t-test, with statistical significance at P < 0.05.
Claudin-1 Depletion
SK-CO15 cells were used for claudin-1 depletion experiments. The cells were plated at 30% confluence on Transwell permeable supports and incubated for 24 hours overnight in DMEM + 10% newborn calf serum. Claudin-1 siRNA (Qiagen –Hs_CLDN1_3_HP, target sequence ATG GTACTT CAT AAT AAA CTA) or control siRNA (laminin A/C) was mixed with HiPerFect transfection reagent (Qiagen, Valencia, CA) and diluted into serum-free OptiMEM at a final concentration of 50 nmol/L, and incubated at room temperature for 15 minutes, and then 0.1 ml of siRNA solution was mixed with 0.9 ml of serum-free OptiMEM. The cells were incubated for 24 hours at 37°C with 0.2 ml of siRNA/OptiMeM on the apical side and 0.8 ml on the basolateral site of the chamber. The cells were then washed and the medium replaced with DMEM + 10% serum and monitored over the next 3 days for changes in TER. Cldn-153–80 peptide was diluted in serum-containing medium to 0.1 mmol/L and added to both the apical and basolateral site of the filter. Changes in TER were measured over a 28-hour time span and then the cells were fixed, permeabilized, immunostained, and analyzed for changes in localization of Claudin-1 or Claudin-3 by confocal fluorescence microscopy as described above.
In Vivo Gastrointestinal Permeability Studies
Rats were assayed in vivo for gastrointestinal paracellular permeability as previously described.31 In brief, rats (nine per group) were fasted for 2 hours before oral gavage of disaccharide permeability probes (500 mg/ml sucrose, 60 mg/ml lactulose, 40 mg/ml mannitol in 0.2 ml water), and placed into individual metabolic cages (Nalge Nunc International, Rochester, NY). For the permeability measurements, urine was collected gravimetrically. Concentration of sucrose, lactulose, and mannitol in specimens was determined by pulsed amperometric detection, using a Dionex high-pressure liquid chromatography (HPLC) with MA-1 columns (Dionex, Bannockburn, IL) and NaOH-based elution, and expressed as urinary fractional excretion.31 Peptides were administered at the same time as the oral probes. Baseline values for intestinal permeability were determined after the rats acclimatized to the laboratory and stabilized to a constant baseline value. Permeability changes were determined over an 18-hour time period. Animals were subsequently sacrificed; intestines were harvested, fixed in 2% paraformaldehyde, embedded in paraffin, sectioned, and stained with H&E.
Results
Modulation of TJs by a Specific Claudin-1 EL Peptide Mimetic
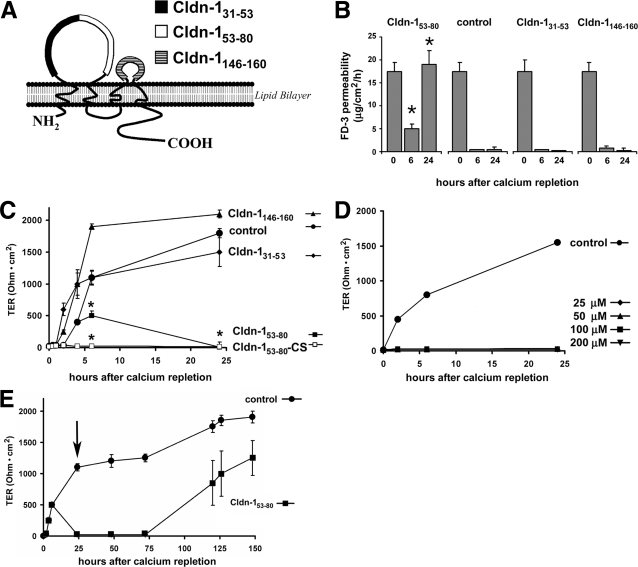
A series of peptides were synthesized to emulate three discrete EL domains of claudin-1 (Table 1 and Figure 1A): the proximal half of the first extracellular loop from Arg-31 to Ser-53 (Cldn-131–53), the distal half of the first extracellular loop from Ser-53 to Thr-80 (Cldn-153–80), and the entire second loop from Gln-146 to Glu-160 (Cldn-1146–160). To determine whether these claudin-1 EL mimetic peptides had the capacity to disrupt TJs, we tested the ability of three different EL peptides to interfere with TJ assembly using a calcium switch protocol.32,33,34 As shown in Figure 1B, the Cldn-153–80 peptide was effective at inhibiting the recovery of T84 cell barrier function, because FD-3 permeability remained high. In contrast, FD-3 permeability of either control cells or cells incubated with either Cldn-131–53 or Cldn-1146–160 was markedly reduced, suggesting that TJs were completely reformed under these conditions.
Figure 1.
Specific claudin extracellular loop (EL) peptides interfere with TJ formation. A: Shown on the backbone of claudin-1 is the location of motifs corresponding to the Cldn-131–53, Cldn-153–80 and Cldn-1146–160 synthetic peptides. Peptide sequences and variants are shown in Table 1. Confluent T84 monolayers were subjected to a calcium depletion/repletion protocol in absence or presence of claudin-1 EL mimetic peptides. B: Paracellular flux of FD-3 was measured at 0, 6 and 24 hours following calcium repletion. As indicated by high levels of FD-3 flux across the monolayers, Cldn-153–80 significantly inhibited the re-establishment of TJs (*, P < 0.05) as compared to cells that were not treated with peptides (control) or were treated with Cldn-131–53 or Cldn-1146–160. C: TER was measured at varying times following calcium repletion. As was the case in a, Cldn-153–80 significantly inhibited the re-establishment of high resistance TJs as compared to controls (*, P < 0.05). A peptide variant of Cldn-153–80 where the cysteines were mutated to serines (Cldn-153–80-CS) was also effective. D: Dose response of Cldn-153–80. Peptide concentrations as low as 25 μmol/L were effective at inhibiting TJ reformation. E: The effect of Cldn-153–80 on TJ permeability was reversible, because changing to peptide-free medium (arrow) allowed the monolayers to re-establish high-resistance barriers.
The effect of the EL peptides on reformation of high-resistance TJs was also assessed by TER measurements (Figure 1, C–E). Consistent with the FD-3 permeability assay, Cldn-153–80 was significantly more effective at inhibiting the recovery of high-resistance monolayers than either Cldn-131–53 or Cldn-1146–160. We further confirmed specificity of the Cldn-153–80 peptide in a dose-response assay, where concentrations of Cldn-153–80 as low as 25 μmol/L effectively inhibited the recovery of TJs for at least 24 hours following calcium repletion. Moreover, because the Cldn-153–80 contains two cysteine residues, one concern was that Cldn-153–80 might act as a reducing agent. To rule out this possibility, we produced a variant of Cldn-153–80 where both cysteines were replaced with serines (Cldn-153–80-CS). As shown in Figure 1C, Cldn-153–80-CS was as effective as Cldn-153–80 at inhibiting the return of high-resistance barriers, suggesting that this EL peptide does not function as a reducing agent and implies that a disulfide bond between these cysteines is not required for it to inhibit TJ recovery. Additionally, studies using smaller overlapping peptide fragments of Cldn-153–80 (SCVSQSTGQ, STGQIQCKV, CKVFDSL, VFDSLLNLN, NLNSTLQAT) failed to affect TER or FD-3 permeability, suggesting that the entire Cldn-153–80 peptide was required for the inhibitory effect (data not shown).
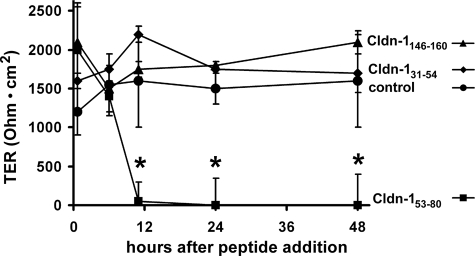
Importantly, when monolayers treated with Cldn-153–80 peptide were subsequently washed and incubated further in control medium, monolayer barrier function was re-established, suggesting that the Cldn-153–80 peptide did not irreversibly damage the cells. Although the effect of Cldn-153–80 was reversible, complete recovery required several days following peptide withdrawal. Because there was no decrease in cell number or viability in cells treated with Cldn-153–80 (eg, Figure 2Ae vs. Figure 2Aj), this requirement for an extended recovery period suggested that the peptide had a strong interaction with T84 cells. Given these observations, we examined whether the claudin EL peptides had the ability to disrupt established TJs. As demonstrated in Figure 3, Cldn-153–80 significantly decreased TER 10 hours after addition to established T84 monolayers, an effect that persisted for 2 days. In contrast, neither Cldn-131–53 nor Cldn-1146–160 caused a significant decrease in TER.
Figure 2.
Cldn-153–80 binds to T84 cells and induces TJ disassembly. A: Confluent T84 monolayers were incubated in either control medium (a–e) or in the presence of 200 μmol/L Cldn-153–80 (f–j) for 24 hours and the effect on localization of claudin-1 (a,f), occludin (b,g), ZO-1 (c,h), JAM-A (d,i), and E-cadherin (e,j) was assessed by immunofluorescence confocal microscopy. Cldn-153–80 treatment altered the localization of the TJ proteins (f–i), but not E-cadherin. Bar = 10 μm. B: Cells were incubated in either control medium (a,e,i) or in the presence of 200 μmol/L *Cldn-131–53 (b,f,j), *Cldn-153–80 (c,g,k), or *Cldn-1146–160 (d,h,l) for 24 hours. The cells were then washed, UV-crosslinked, fixed, and treated with FITC-streptavidin to label bound peptide (a–d) and Alexa588-phalloidin to label F-actin (e–h). Merged images are in (i–l). Only *Cldn-153–80 showed high levels of binding to the cells, while the other peptides only show occasional cell association (arrows). Scale bar = 10 μm. C: Cells were either incubated in control medium (a) or in the presence of *Cldn-153–80 (b) for 24 hours and then processed for avidin gold electron microscopy. Black arrowheads indicate abundant association of *Cldn-153–80 with the cells, arrows show internalized *Cldn-153–80 and white arrowheads indicate TJs. Altered TJ morphology was observed in cells treated with *Cldn-153–80 as compared to untreated cells. Scale bar = 667 nm.
Figure 3.
Cldn-153–80 decreases barrier function of intact TJs. Confluent T84 monolayers were incubated in either control medium or in the presence of 200 μmol/L Cldn-131–53, Cldn-153–80, or Cldn-1146–160 for varying amounts of time. After overnight incubation, only Cldn-153–80 significantly decreased TER, indicating that this peptide specifically increased paracellular permeability. *significantly different from control values (P < 0.05).
Cldn-153–80 Peptide Disrupts TJ Morphology
Given the effect of Cldn-153–80 on barrier function, the effect of claudin-1 EL peptides on TJ morphology was assessed. In contrast to untreated control cells, cells incubated for 24 hours with Cldn-153–80 showed mislocalization of claudin-1, occludin, JAM-A, and ZO-1 (Figure 2A). This correlated well with the effect of Cldn-153–80 on TER shown in Figure 1C. Although Cldn-153–80 influenced several TJ proteins, it had little, if any, effect on the AJ protein E-cadherin, indicating that the effect of the peptide was TJ-specific. Note that cells incubated for 24 hours with either Cldn-131–53 or Cldn-1146–160 were comparable to untreated controls (data not shown), consistent with their inability to disrupt TER.
To visualize sites where claudin-1 EL peptides bind to T84 cells, we used biotinylated, photoactivatable peptide analogues (Table 1), which were detected using FITC-conjugated streptavidin (Figure 2B). After 24-hour incubation, UV crosslinking, and extensive washing, *Cldn-153–80 was stably associated with the cells, whereas *Cldn-131–53 and *Cldn-1146–160 were minimally associated with cells. *Cldn-153–80 binding also caused F-actin rearrangements, most likely induced by TJ disassembly. Conversely, cells treated with the other two EL peptides showed little effect on F-actin, and were comparable to controls. Binding of *Cldn-153–80 to lateral membranes was further confirmed by immunogold electron microscopy (Figure 2C) staining. Although TJs were disrupted, other classes of junctions, including desmosomes, were preserved in the presence of *Cldn-153–80. Interestingly, *Cldn-153–80 localized to vesicle-like structures just under the plasma membrane, suggesting that the peptide was most likely internalized after binding to the cell surface.
Cldn-153–80 Peptide Interacts with Claudin-1 and Occludin
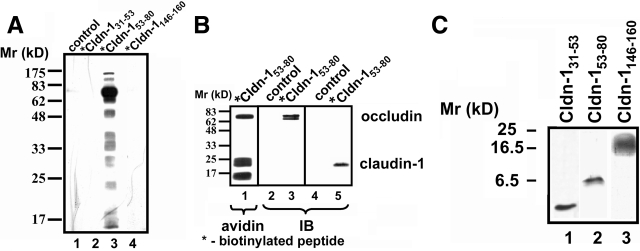
Given that Cldn-153–80 influenced TJ function and structure, and to further examine the mechanisms underlying this effect on TJs, we biochemically analyzed the ability of the different claudin-1 EL peptides to stably interact with cell proteins. Cells were incubated for 24 hours in media containing either *Cldn-131–53 ,*Cldn-153–80, or *Cldn-1146–160, extensively washed, and then subjected to UV crosslinking before harvesting. Proteins associated with biotinylated claudin-1 EL bait peptides were isolated using avidin-Sepharose, resolved by SDS-PAGE, transferred to nitrocellulose and blotted with peroxidase-conjugated avidin. Under these labeling conditions, we found that several proteins were associated with biotinylated *Cldn-153–80 (Figure 4A). By contrast, cells incubated with either *Cldn-131–53 or *Cldn-1146–160 did not show significant protein labeling. These results were consistent with the functional and morphological studies above and further suggest a specific interaction between Cldn-153–80 and cell proteins.
Figure 4.
Crosslinking of Cldn-153–80 to claudin-1 and occludin. A: Cells were incubated in either control medium (lane 1) or for 24 hours in the presence of biotinylated bait peptides at 200 μmol/L (*Cldn-131–53 [lane 2], *Cldn-153–80 [lane 3], or *Cldn-1146–160 [lane 4]). The cells were then washed, UV-crosslinked, harvested, and homogenized by nitrogen cavitation. Biotinylated bait peptide-protein conjugates were isolated by avidin affinity chromatography, resolved by SDS-PAGE, transferred to nitrocellulose, and detected with HRP-streptavidin. Only cells incubated with *Cldn-153–80 showed significant proteins crosslinked to the biotinylated peptide (lane 3). B: Cells treated with *Cldn-153–80 as described above were further subjected to boiling in 1% SDS before avidin affinity chromatography and streptavidin or immunoblot. Samples probed with HRP-streptavidin showed three prominent bands at ∼68 kDa, 23 kDa, and 17 kDa (lane 1). By immunoblot, the 68-kDa band was found to correspond to occludin (lane 3) and the 23-kDa band corresponded to claudin-1 (lane 5). Neither protein was isolated from control incubated cells (lanes 2 and 4). C: Cldn-131–53 (lane 1), Cldn-153–80 (lane 2), or Cldn-1146–160 (lane 3) at 200 μmol/L in HBSS+ were incubated for 30 minutes at room temperature and then resolved using Tris-tricine gel electrophoresis. Peptides were visualized directly in the gel by silver staining. Note that Cldn-153–80 primarily formed dimers (lane 2), whereas Cldn-131–53 was monomeric (lane 1) and Cldn-1146–160 formed a higher order complex (lane 3).
To identify proteins covalently attached to Cldn-153–80, protein extracts labeled with *Cldn-153–80 and isolated with avidin-Sepharose were boiled in 1% SDS before SDS-PAGE and strepavidin immunoblot analysis. There were three major bands detected with streptavidin-HRP with Mr of ∼68,000, 23,000, and 17,000 (Figure 4A). Based on the Mr values of the associated proteins, we investigated whether these bands corresponded to occludin and claudin-1. As shown in Figure 4B, the 68-kDa protein band was recognized by anti-occludin and the 23-kDa protein band was recognized by anti-claudin-1, consistent with binding of *Cldn-153–80 to these two TJ proteins. The identity of the 17-kDa band is not known at present.
The ability of *Cldn-153–80 to bind claudin-1 is particularly intriguing and suggests a specific interaction. To examine this possibility further, we examined the ability of different EL peptides to stably self-associate in aqueous solution (Figure 4C). Here we used native, rather than photoactivatable, peptides. Interestingly, Cldn-153–80 migrated on tricine gels at ∼6.5 kDa, suggesting that it was dimerized. Because these were reducing gels, Cldn-153–80 dimer formation was not likely to be due to the formation of disulfide bonds. In contrast to Cldn-153–80, *Cldn-131–53 remained monomeric. Also, Cldn-1146–160 formed considerably larger complexes, in sizes ranging from hexamers to octamers. Additional studies using mixtures of Cldn-131–53, Cldn-153–80, and/or Cldn-1146–160 did not reveal any specific heterologous interactions (data not shown). These data indicate that the EL peptides differ in the ability to self-associate. In particular, the stable formation of dimers by Cldn-153–80 suggests that the peptide can directly interact with the homologous EL domain of claudin-1.
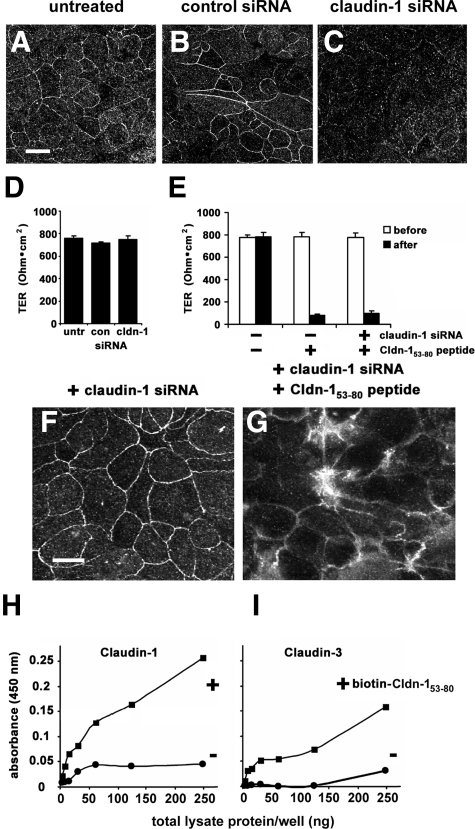
Cldn-153–80 Inhibits TJ Formation and Function in Claudin-1-Depleted Cells
To determine whether the effect of Cldn-153–80 required a direct interaction with claudin-1, we used siRNA to interfere with claudin-1 expression. Because T84 cells are difficult to transfect with siRNA, we used a model intestinal epithelial cell line, SK-CO15, which form well developed TJs. By immunoblot, claudin-1 siRNA depleted claudin-1 protein expression to 63.8 ± 0.1% of control values (n = 3), as opposed to claudin-3 protein, which was largely unchanged (101.1 ± 0.1% of control [n = 3]). When assessed by confocal immunofluorescence microscopy, cells treated with claudin-1 siRNA showed a specific decrease in claudin-1 expression and TJ localization, as compared with untreated or control siRNA-treated cells (Figure 5, A–C). However, there was little effect of claudin-1 siRNA on TER (Figure 5D), suggesting that other claudins expressed by SK-CO15 cells contribute to TJ barrier function. Consistent with this, claudin-3 localization to TJs was largely unaffected by claudin-1 siRNA (Figure 5F). Moreover, claudin-1-depleted cells remained sensitive to further treatment with Cldn-153–80, as indicated by the effect of the peptide on claudin-3 localization (Figure 5G) and TER (Figure 5E). To determine whether Cldn-153–80 directly interacted with claudin-3, we examined peptide binding to claudin-1 and claudin-3 in T84 cells, using ELISA (Figure 5I). Biotinylated Cldn-153–80 was found to bind to both claudin-1 and claudin-3 using this approach. These data are consistent with the notion that the Cldn-153–80 EL peptide has the capacity to interact with other claudins besides claudin-1. In particular, the ability of Cldn-153–80 to bind to claudin-3 is consistent with previous reports that claudin-1 and claudin-3 heterotypically interact.35,36,37
Figure 5.
Cldn-153–80 interacts with both claudin-1 and claudin-3. a-c. SK-CO15 cells were either untreated (A) or treated with either control siRNA (B) or claudin-1 siRNA (C), further cultured for 72 hours and claudin-1 localization was analyzed by confocal immunofluorescence microscopy. Scale bar = 20 μm. D: Cells were either untreated (untr), treated with control siRNA (con) or claudin-1 siRNA (cldn-1), further cultured for 72 hours and barrier function was assessed by TER measurement. Neither control nor claudin-1 siRNA had a measurable effect on barrier function. E: Cells either untreated or treated with claudin-1 siRNA were further cultured for 48 hours and then cultured for 24 hours in either the presence or absence of Cldn-153–80 peptide. Claudin-1 depleted cells showed a comparable decrease in TER following Cldn-153–80 treatment as cells that were not treated with siRNA. Cells treated with either claudin-1 siRNA alone (F) or claudin-1 siRNA + Cldn-153–80 peptide (G) were fixed and immunostained for claudin-3. Cldn-153–80 peptide treated cells showed a near uniform effect on TJ localization of claudin-3, even in the absence of claudin-1. Scale bar = 20 μm. H and I: T84 cells were subjected to a calcium switch and then incubated in either the presence (+) or absence (−) of biotinylated Cldn-153–80 peptide in culture medium for 24 hours. The cells were then isolated and treated with a chemical crosslinker, and samples were processed and analyzed by ELISA as described in Methods. Biotinylated Cldn-153–80 peptide specifically bound to both claudin-1 and claudin-3, consistent with the ability of the peptide to interact with multiple classes of claudins.
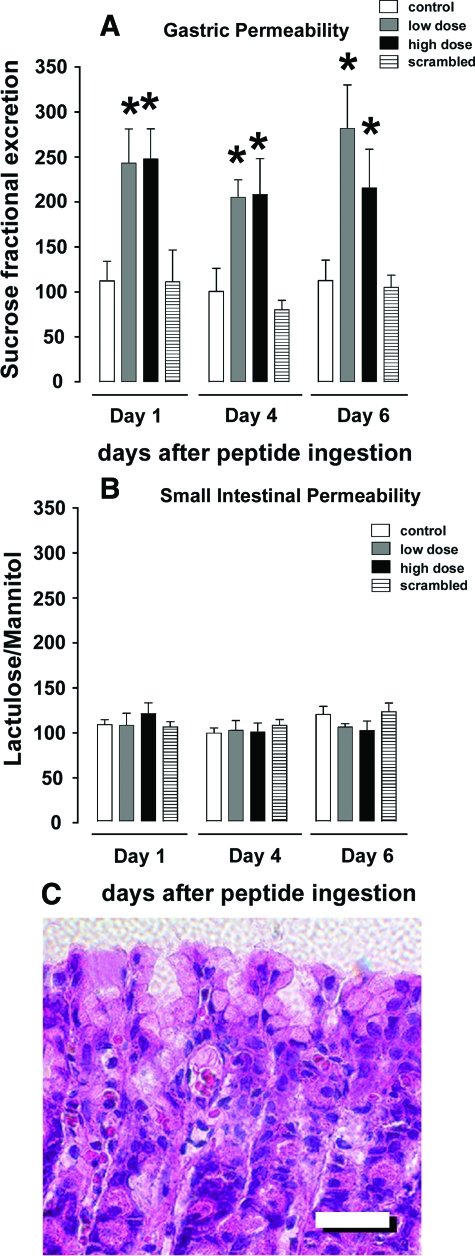
Cldn-153–80 Enhances Gastric Epithelial Permeability in Vivo
To determine whether the ability of Cldn-153–80 to influence TJs in cultured cells translates to in vivo models, we examined the effect of this peptide on gastrointestinal paracellular permeability in rats, using a previously validated approach31 based on use of disaccharide probes for gastric (sucrose) and small intestinal permeability (lactulose and mannitol). A solution containing a mixture of the three disaccharides and either a low (100 μg/kg body weight) or high dose (1 mg/kg body weight) of the Cldn-153–80 peptide was administered to rats by oral gavage, and urinary disaccharide excretion was determined over an 18-hour period by HPLC (Figure 6).
Figure 6.
Oral administration of Cldn-153–80 increased gastric permeability in vivo. A and B: Rats were administered sucrose, lactulose, and mannitol alone (control, white bars) or along with Cldn-153–80 at either 0.1 mg/kg body weight (low dose, gray bars), 1 mg/kg body weight (high dose, black bars), or with 1 mg/kg scrambled peptide (stippled bars) by oral gavage. Permeability of the stomach (A) and small intestine (B) were assessed by measuring urine disaccharide content as described in Methods. Cldn-153–80 induced a significant increase in gastric permeability at both low and high dose (*, P < 0.05). C: H&E-stained section of gastric tissue from rats treated with high dose Cldn-153–80. Peptide administration did not induce any epithelial loss or altered mucosal architecture. Scale bar =100 μm.
Consistent with our cultured cell studies, rats treated with either low-dose or high-dose Cldn-153–80 had significant increases in gastric permeability. The effect of Cldn-153–80 on small intestine was less dramatic, although there was a significant increase in permeability on day 1 following high-dose administration of the peptide. We suspect that the latter effect is likely secondary to peptide cleavage by proteases in the duodenum. Interestingly, administration of Cldn-153–80 and the resultant increases in paracellular permeability in gastric epithelial cells did not induce epithelial damage/denudation, nor did this treatment incite an inflammatory response in the gastric mucosa. Histological analysis confirmed the presence of an intact epithelium and normal mucosal architecture (Figure 6). Overall, these results suggest that Cldn-153–80 has the capacity to increase paracellular permeability in vitro and in vivo.
Discussion
Here we report that a claudin-1 EL peptide mimetic, Cldn-153–80, has the capacity to disrupt TJs and gastrointestinal barrier function both in cultured cells and in vivo. Several lines of evidence indicate that this is a specific interaction related to the region of the first EL domain corresponding to the Cldn-153–80 peptide, particularly because the Cldn-131–53 peptide that emulates another segment of the first EL domain was not effective. For instance, Cldn-153–80 bound to cells and enabled affinity isolation of claudin-1 and occludin. The interaction between Cldn-153–80 and claudin-1 is particularly intriguing and, because the Cldn-153–80 peptide spontaneously dimerizes in solution, it suggests that this interface helps stabilize head-to-head interactions between claudins. This suggests that head-to-head interactions between claudins are also required to stabilize these proteins in TJ strands, in addition to previously defined roles for ZO-1 and ZO-2 in regulating claudin assembly.9 A role for head-to-head interactions between claudin EL domains on adjacent cells in stabilizing TJs would be analogous to a comparable role for connexin EL domains in stabilizing gap junctions.38,39 A structural role for the first claudin EL domain in maintaining TJ architecture also complements previous work demonstrating that the first EL domain regulates TJ ion permeability.15,16 However, we cannot rule out the possibility that these results reflect a role for the first EL domain in mediating lateral interactions between claudins in the same cell, because it is impossible to discriminate the orientation of these interactions within the cell membrane.
The binding of Cldn-153–80 to occludin suggests a close interaction between claudin-1 and occludin. Whether Cldn-153–80 specifically binds to occludin is less clear. For instance, a Cldn-153–80 bound to claudin-1 that, in turn, is co-clustered with occludin (due to interactions with the cytoplasmic scaffold) might enable the probe to be close enough to occludin for crosslinking. In fact, interactions between Cldn-153–80 and occludin may be required for disrupting TJs, because several studies have demonstrated that occludin EL mimetic peptides also interfere with TJs.27,40,41,42,43,44,45,46,47 Nonetheless, the ability of Cldn-153–80 to simultaneously disrupt several distinct classes of TJ proteins underscores the notion that these proteins are coordinately regulated.
The peptide corresponding to the second EL domain, Cldn-1146–160, did not affect TJ permeability or structure. Because Cldn-1146–160 formed large complexes or aggregates in solution, this may reduce the ability of this peptide to interact with cells and does not necessarily rule out a role for the second EL domain in mediating claudin-claudin interactions. In fact, studies demonstrating that CPE binds to the second EL domain of claudin-3 and claudin-4 and disrupts TJs, argue in favor of a role for the second EL domain in stabilizing TJs.18,19,20 The second EL domain was also implicated in a recent analysis of claudin-5 assembly into TJs.48 Also, heterotypic claudin-claudin specificity is dictated by motifs in both the first and second EL domains.36 Thus, a likely model is that both claudin EL domains act in concert to mediate heterotypic binding of claudins between adjacent cells.
Several motifs within Cldn-153–80 provide clues to the potential mode of action for this peptide. Cldn-153–80 contains a SLLN motif identified in the Vps27 protein that functions as a membrane insertion loop.49,50 Cldn-153–80 also contains a LNLNSTL motif that has been shown to act as a similar membrane insertion motif in phospholipase A2.51 Although these domains may reflect the ability of Cldn-153–80 to strongly associate and possibly integrate into membranes, these domains may also mediate interactions between Cldn-153–80 and hydrophobic elements of claudin-1 itself. However, several peptide fragments, including one consisting of SLLNLNLSTL, were not sufficient to disrupt TJs. Instead, an additional motif, IQCKVF, was also required. One possibility is that the IQCKVF motif may serve to limit the hydrophobicity of the first EL domain, by forming a charged β-sheet domain, comparable to the role of a similar YQCKVY β-sheet motif that restricts the membrane insertion of Semliki Forest Virus fusion protein.52 Whether this is the case and how claudin EL domains interact will require detailed structural information about the conformation of claudin EL domains. However, if fairly nonspecific hydrophobic interactions help mediate interactions between claudin EL domains, this would suggest that peptides such as Cldn-153–80 have the ability to interact with other claudins in addition to claudin-1. Moreover, claudin-1 has the ability to heterotypically interact with at least one other claudin, claudin-3.35,36,37 Consistent with this, our data suggest that Cldn-153–80 interacts with claudin-3 and is likely to interact with other claudins as well. Finally, Cldn-153–80 could indirectly affect claudins associated with claudin-1 via scaffold proteins, such as ZO-1 or ZO-2.9
We found that Cldn-153–80 has the ability to disrupt established TJs in cultured cell models and increase paracellular gastric permeability in vivo. Importantly Cldn-153–80 was not toxic and the effect on TJ permeability was reversible. These properties, and the efficacy of orally administered Cldn-153–80, suggest that claudin EL-mimetic peptides might provide a useful adjunct to treatments that require drug delivery across an epithelial barrier. Future work will evaluate the utility of this approach.
Acknowledgments
We thank Charles A. Parkos for critical evaluation of the manuscript. We also thank Sian Owens, Jeffrey Tom, and Susan Voss for experimental assistance.
Footnotes
Address reprint requests to Asma Nusrat, Department of Pathology & Laboratory Medicine, Emory University School of Medicine, Whitehead Biomedical Research Building (Rm 115), 615 Michael St, Atlanta GA 30322. E-mail: anusrat@emory.edu.
Supported by the National Institutes of Health (grants DK 55679 and DK 59888 to A.N., and HL 083120 to M.K.); Emory University Digestive Diseases Research Development Center (Minicenter grant DK 64399), Medical Research Council of Canada, and the Crohn’s and Colitis Foundation of Canada (J.B.M.); and the Alberta Children’s Hospital Research Foundation (A.G.B.).
References
- Balda MS, Matter K. Transmembrane proteins of tight junctions. Semin Cell Dev Biol. 2000;11:281–289. doi: 10.1006/scdb.2000.0177. [DOI] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- Chen YH, Lu Q, Goodenough DA, Jeansonne B. Nonreceptor tyrosine kinase c-Yes interacts with occludin during tight junction formation in canine kidney epithelial cells. Mol Biol Cell. 2002;13:1227–1237. doi: 10.1091/mbc.01-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki Y, Hirai S, Manabe N, Izumi Y, Hirose T, Nakaya M, Suzuki A, Mizuno K, Akimoto K, Tsukita S, Shuin T, Ohno S. Dynamic changes in protein components of the tight junction during liver regeneration. Cell Tissue Res. 2001;305:399–409. doi: 10.1007/s004410100397. [DOI] [PubMed] [Google Scholar]
- Liang TW, DeMarco RA, Mrsny RJ, Gurney A, Gray A, Hooley J, Aaron HL, Huang A, Klassen T, Tumas DB, Fong S. Characterization of huJAM: evidence for involvement in cell-cell contact and tight junction regulation. Am J Physiol Cell Physiol. 2000;279:C1733–1743. doi: 10.1152/ajpcell.2000.279.6.C1733. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. Human junction adhesion molecule regulates tight junction resealing in epithelia. J Cell Sci. 2000;113 (Pt 13):2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci. 2005;118:1427–1436. doi: 10.1242/jcs.01735. [DOI] [PubMed] [Google Scholar]
- Fujibe M, Chiba H, Kojima T, Soma T, Wada T, Yamashita T, Sawada N. Thr203 of claudin-1, a putative phosphorylation site for MAP kinase, is required to promote the barrier function of tight junctions. Exp Cell Res. 2004;295:36–47. doi: 10.1016/j.yexcr.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Tang VW, Goodenough DA. Paracellular ion channel at the tight junction. Biophys J. 2003;84:1660–1673. doi: 10.1016/S0006-3495(03)74975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Gomes AS, Paul DL, Goodenough DA. Study of claudin function by RNA interference. J Biol Chem. 2006;281:36117–36123. doi: 10.1074/jbc.M608853200. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Fanning AS, Anderson JM. Reversal of charge selectivity in cation or anion-selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol. 2003;285:F1078–1084. doi: 10.1152/ajprenal.00116.2003. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Van Itallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol. 2003;284:C1346–1354. doi: 10.1152/ajpcell.00547.2002. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol. 2002;283:C142–147. doi: 10.1152/ajpcell.00038.2002. [DOI] [PubMed] [Google Scholar]
- Wan H, Winton HL, Soeller C, Taylor GW, Gruenert DC, Thompson PJ, Cannell MB, Stewart GA, Garrod DR, Robinson C. The transmembrane protein occludin of epithelial tight junctions is a functional target for serine peptidases from faecal pellets of Dermatophagoides pteronyssinus. Clin Exp Allergy. 2001;31:279–294. doi: 10.1046/j.1365-2222.2001.00970.x. [DOI] [PubMed] [Google Scholar]
- Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000;476:258–261. doi: 10.1016/s0014-5793(00)01744-0. [DOI] [PubMed] [Google Scholar]
- Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 1999;147:195–204. doi: 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh M, Masuyama A, Takahashi A, Asano N, Mizuguchi H, Koizumi N, Fujii M, Hayakawa T, Horiguchi Y, Watanbe Y. A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol Pharmacol. 2005;67:749–756. doi: 10.1124/mol.104.008375. [DOI] [PubMed] [Google Scholar]
- Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P, Sukumar S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NN, Eddington ND, Fasano A. Tight junction modulation and its relationship to drug delivery. Adv Drug Deliv Rev. 2006;58:15–28. doi: 10.1016/j.addr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty AL, Mrsny RJ. Emerging technologies that overcome biological barriers for therapeutic protein delivery. Expert Opin Biol Ther. 2003;3:1071–1081. doi: 10.1517/14712598.3.7.1071. [DOI] [PubMed] [Google Scholar]
- Kominsky SL, Tyler B, Sosnowski J, Brady K, Doucet M, Nell D, Smedley JG, 3rd, McClane B, Brem H, Sukumar S. Clostridium perfringens enterotoxin as a novel-targeted therapeutic for brain metastasis. Cancer Res. 2007;67:7977–7982. doi: 10.1158/0008-5472.CAN-07-1314. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Betts L, Smedley JG, 3rd, McClane BA, Anderson JM. Structure of the claudin-binding domain of clostridia perfringens enterotoxin, J Biol Chem. 2008 doi: 10.1074/jbc.M708066200. DOI: 10.1074/jbc.M708066200. [DOI] [PubMed] [Google Scholar]
- Nusrat A, Brown GT, Tom J, Drake A, Bui TT, Quan C, Mrsny RJ. Multiple protein interactions involving proposed extracellular loop domains of the tight junction protein occludin. Mol Biol Cell. 2005;16:1725–1734. doi: 10.1091/mbc.E04-06-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrat A, Chen JA, Foley CS, Liang TW, Tom J, Cromwell M, Quan C, Mrsny RJ. The coiled-coil domain of occludin can act to organize structural and functional elements of the epithelial tight junction. J Biol Chem. 2000;275:29816–29822. doi: 10.1074/jbc.M002450200. [DOI] [PubMed] [Google Scholar]
- Parkos CA, Colgan SP, Bacarra AE, Nusrat A, Delp-Archer C, Carlson S, Su DH, Madara JL. Intestinal epithelia (T84) possess basolateral ligands for CD11b/CD18-mediated neutrophil adherence. Am J Physiol. 1995;268:C472–479. doi: 10.1152/ajpcell.1995.268.2.C472. [DOI] [PubMed] [Google Scholar]
- Sanders SE, Madara JL, McGuirk DK, Gelman DS, Colgan SP. Assessment of inflammatory events in epithelial permeability: a rapid screening method using fluorescein dextrans. Epithelial Cell Biol. 1995;4:25–34. [PubMed] [Google Scholar]
- Meddings JB, Gibbons I. Discrimination of site-specific alterations in gastrointestinal permeability in the rat. Gastroenterology. 1998;114:83–92. doi: 10.1016/s0016-5085(98)70636-5. [DOI] [PubMed] [Google Scholar]
- Nigam SK, Rodriguez-Boulan E, Silver RB. Changes in intracellular calcium during the development of epithelial polarity and junctions. Proc Natl Acad Sci USA. 1992;89:6162–6166. doi: 10.1073/pnas.89.13.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AM, Walsh SV, Verkade P, Boquet P, Nusrat A. Constitutive activation of Rho proteins by CNF-1 influences tight junction structure and epithelial barrier function. J Cell Sci. 2003;116:725–742. doi: 10.1242/jcs.00300. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Contreras RG, Bolivar JJ, Ponce A, Chavez De Ramirez B, Cereijido M. Role of calcium in tight junction formation between epithelial cells. Am J Physiol. 1990;259:C978–986. doi: 10.1152/ajpcell.1990.259.6.C978. [DOI] [PubMed] [Google Scholar]
- Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1166–1178. doi: 10.1152/ajplung.00182.2003. [DOI] [PubMed] [Google Scholar]
- Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem. 2007;282:3005–3013. doi: 10.1074/jbc.M703547200. [DOI] [PubMed] [Google Scholar]
- Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TW, Paul DL, Goodenough DA, Bruzzone R. Functional analysis of selective interactions among rodent connexins. Mol Biol Cell. 1995;6:459–470. doi: 10.1091/mbc.6.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote CI, Zhou L, Zhu X, Nicholson BJ. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J Cell Biol. 1998;140:1187–1197. doi: 10.1083/jcb.140.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CH, Mruk DD, Lee WM, Cheng CY. Targeted and reversible disruption of the blood-testis barrier by an FSH mutant-occludin peptide conjugate. FASEB J. 2007;21:438–448. doi: 10.1096/fj.05-4144com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Mandell KJ, Parkos CA, Mrsny RJ, Nusrat A. The second loop of occludin is required for suppression of Raf1-induced tumor growth. Oncogene. 2005;24:4412–4420. doi: 10.1038/sj.onc.1208634. [DOI] [PubMed] [Google Scholar]
- Tavelin S, Hashimoto K, Malkinson J, Lazorova L, Toth I, Artursson P. A new principle for tight junction modulation based on occludin peptides. Mol Pharmacol. 2003;64:1530–1540. doi: 10.1124/mol.64.6.1530. [DOI] [PubMed] [Google Scholar]
- Blaschuk OW, Oshima T, Gour BJ, Symonds JM, Park JH, Kevil CG, Trocha SD, Michaud S, Okayama N, Elrod JW, Alexander JS, Sasaki M. Identification of an occludin cell adhesion recognition sequence. Inflammation. 2002;26:193–198. doi: 10.1023/a:1016571830091. [DOI] [PubMed] [Google Scholar]
- Vietor I, Bader T, Paiha K, Huber LA. Perturbation of the tight junction permeability barrier by occludin loop peptides activates beta-catenin/TCF/LEF-mediated transcription. EMBO Rep. 2001;2:306–312. doi: 10.1093/embo-reports/kve066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NP, Mruk D, Mo MY, Lee WM, Cheng CY. A 22-amino acid synthetic peptide corresponding to the second extracellular loop of rat occludin perturbs the blood-testis barrier and disrupts spermatogenesis reversibly in vivo. Biol Reprod. 2001;65:1340–1351. doi: 10.1095/biolreprod65.5.1340. [DOI] [PubMed] [Google Scholar]
- Wong V, Gumbiner BM. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RS, Vanhook MK, Barozzi N, Toth I, Johnson LG. Specific modulation of airway epithelial tight junctions by apical application of an occludin peptide. Mol Pharmacol. 2006;69:492–500. doi: 10.1124/mol.105.017251. [DOI] [PubMed] [Google Scholar]
- Piontek J, Winkler L, Wolburg H, Muller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Sultzman LA, Martin DM, Kriz RW, Towler PS, Knopf JL, Clark JD. Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca(2+)-dependent lipid-binding domain and a Ca(2+)-independent catalytic domain. J Biol Chem. 1994;269:18239–18249. [PubMed] [Google Scholar]
- Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]