Abstract
Primary myelofibrosis (PMF) is a myeloproliferative neoplasia characterized by progressive deposition of extracellular matrix components in the bone marrow. The involvement of members of the bone morphogenetic protein (BMP) family in aberrant bone marrow matrix homeostasis in PMF has not yet been investigated. Therefore, we analyzed expression of BMP1, an activator of latent transforming growth factor β-1 (TGFβ-1) and processor of collagen precursors, and other BMPs in bone marrow from PMF patients and controls (n = 95). Expression of BMP1, BMP6, BMP7, and BMP-receptor 2 was significantly increased in advanced stages of myelofibrosis compared with controls (P ≤ 0.01), and enhanced levels of BMP6 expression were already evident in prefibrotic stages of PMF. Immunohistochemistry showed that bone marrow stromal cells and megakaryocytes were the major cellular sources of BMP1 protein. Because TGFβ-1 and basic fibroblast growth factor have been shown to be important in the development of myelofibrosis, we studied the induction of BMPs by these cytokines in cultured fibroblasts. Fibroblasts treated with TGFβ-1 showed a pronounced up-regulation of BMP6, suggesting that stromal cells may be susceptible to BMP activation by cytokines with a proven role in the pathogenesis of PMF. We conclude that BMP family members may play an important role in the pathogenesis of myelofibrosis in PMF and are apparently induced by cytokines such as TGFβ-1.
Primary myelofibrosis (PMF according to the latest proposals for a revised World Health Organization classification of the myeloproliferative disorders1) belongs to the group of Philadelphia chromosome-negative chronic myeloproliferative diseases. PMF may present in a prefibrotic, hypercellular phase2 that progresses to bone marrow fibrosis during the course of the disease. Other prominent features comprise increased bone marrow angiogenesis, enhanced cellular trafficking of CD34+ progenitors, extramedullary hematopoiesis, and a variable risk for transformation into acute leukemia.3,4 In up to 60% of patients, constitutively activated Janus kinase (JAK2V617F) has been demonstrated to be an underlying molecular defect probably responsible for autonomous proliferation.5 A minor fraction (∼5%) of PMF may show a gain-of-function mutation in the thrombopoietin receptor myeloproliferative leukemia virus oncogene (MPLW515L/K).6
The definite mechanisms leading to myelofibrosis in PMF remain unclear, but it is well accepted that proliferation of fibroblasts represents a reactive and therefore nonclonal process.3 Induction and sustainment of bone marrow fibrosis are mediated by a complex network of cytokines with platelet-derived growth factor, basic fibroblast growth factor (bFGF), and transforming growth factor β-1 (TGFβ-1) being the major effector molecules in this process.7 Besides the action of pro-fibrogenic factors with subsequent overproduction of extracellular matrix (ECM) components, the counteracting proteolytic environment, including collagenases/matrix metalloproteinases and their inhibitors, substantially contribute to the development of PMF.4,8
The family of bone morphogenetic proteins (BMP) currently comprises 20 members9. BMP1 to BMP7 are the most frequently investigated subtypes. The factor discovered first, BMP1, was initially extracted from bone lysates and was found to be capable of inducing ectopic bone formation.10,11 BMP1, unlike BMPs 2 to 7, is not a TGFβ-like protein but acts like a metalloproteinase. Hence, BMP1 is known to process a large set of ECM precursor molecules such as pro-collagen types I to III to mature fibrils. Furthermore, BMP1 cleaves the latent binding complex containing inactive TGFβ-1 in the ECM, thereby allowing matrix metalloproteinases such as matrix metalloproteinase-13 to fully activate TGFβ-1.12,13 Interestingly, BMP1 also seems to mediate resistance of fibroblasts to apoptosis in vitro via processing of the proteoglycan Perlecan.14 In endothelial cells, BMP-1 overexpression was shown to be restricted to areas of tumor angiogenesis in vivo.15
BMP family members other than BMP1 were shown to exert different effects on the development of fibrosis in solid organs and also tumorigenesis. For instance, BMP6 was a predominant mediator of fibrosis in experimental skin tumors,16 whereas BMP7 was shown to protect against glomerular fibrosis and collagen accumulation in an animal model of diabetic nephropathy and in experimental cardiac fibrosis.17,18 Interestingly, in a recent mouse model showing deficiency in GATA-1 expression (GATA-1low), members of the BMP family were shown to be essentially involved in early osteosclerosis.19
Site-directed mutagenesis in vitro provided evidence that a point mutation in human BMP1 (E483K) abolished the ability of BMP1 to cleave pro-collagen type I.20 The entire CUB II domain was lastly identified to be essential for proteinase activity of BMP1.
In summary, members of the BMP family seem to be involved in ECM synthesis and formation, homeostasis of fibroblasts, and angiogenesis rendering them attractive target molecules that contribute to myelofibrosis and angiogenesis, in PMF. To test the hypothesis of a relevant role of BMPs in PMF pathogenesis, we 1) analyzed BMP expression in fibroblasts treated with TGFβ-1 and bFGF in vitro, and 2) consecutively investigated PMF bone marrow samples for BMP expression in correlation with the stage of disease, ie, grade of myelofibrosis. An explorative mutation screening was performed to uncover a potential mutation in the CUB II domain of BMP1. To obtain a much closer insight into dynamics, we monitored BMP1 and BMP6 expression in PMF during the course of disease.
Materials and Methods
Bone Marrow Study Group
Formalin-fixed and paraffin-embedded bone marrow trephines with PMF diagnosed according to the World Health Organization criteria were retrieved from the bone marrow registry of the Institute of Pathology, Hannover Medical School. Bone marrow trephines were routinely fixed in phosphate-buffered formalin (pH 7.4) for 24 hours. The decalcification step was performed in an EDTA-based solution (pH 7.5) for up to 48 hours or by application of an ultrasound bath allowing a shortened processing period of less than 24 hours. The study group (n = 95) comprised hypercellular, prefibrotic PMF (n = 27), advanced PMF with manifest myelofibrosis (n = 43), and 25 control cases showing normal hematopoiesis or a mild reactive hyperplasia of the megakaryocytic lineage. According to the World Health Organization classification in close agreement with clinical data, patients’ bone marrow trephines were diagnosed as PMF in the years 2000 to 2005. No prior history of PMF or a related chronic myeloproliferative disease was recorded in the study group. PMF cases were re-evaluated and subdivided into two groups depending on the degree of myelofibrosis (mf) after silver impregnation (Gomori) as described previously.21,22 In brief, hypercellular PMF cases showing no deposition of fibers were graded as mf 0, and a mild increase of reticulin fibers (collagen type III) led to grade mf 1. Advanced PMF showing a manifest myelofibrosis with extended collagen deposits (type I + III) and intrasinusoidal hematopoiesis were graded as mf 2. In addition, demonstrable osteosclerosis and bone apposition were classified as grade mf 3. For a summary of patients’ clinical data, see Table 1.
Table 1.
Clinical Parameters, Including Frequency and Mutant T Allele Burden of JAK2V617F Mutations
| PMF mf 0/1 (n = 27)
|
PMF mf 2/3 (n = 43)
|
Control (n = 25)
|
|||
|---|---|---|---|---|---|
| JAK2wt (n = 12) | JAK2V617F (n = 15) | JAK2wt (n = 21) | JAK2V617F (n = 22) | JAK2wt (n = 25) | |
| Mutant T allele burden (%) | — | 34.0 (14.0 to 86.0) | — | 50.0 (6.0 to 94.0) | — |
| Erythrocytes (106/μl) | 3.6 (2.2 to 5.3) | 4.5 (4.2 to 6.6) | 3.4 (2.7 to 4.0) | 4.0 (2.7 to 8.1) | 4.9 (3.7 to 5.7) |
| Hemoglobin (g/dl) | 11.3 (9.6 to 14.2) | 14.1 (10.3 to 16.6) | 8.6 (6.3 to 10.8) | 10.8 (3.0 to 16.8) | 14.8 (9.4 to 16.6) |
| Hematocrit (%) | 34 (30 to 49) | 41 (32 to 52) | 30 (26 to 40) | 33 (26 to 56) | 45 (31 to 50) |
| Leukocytes (103/μl) | 8.2 (5.0 to 31.8) | 11.0 (2.9 to 22.9) | 9.5 (3.1 to 87.5) | 10.8 (3.8 to 48.1) | 6.2 (2.8 to 15.5) |
| Thrombocytes (103/μl) | 696 (530 to 1919) | 711 (430 to 1340) | 343 (16 to 1081) | 257 (84 to 1415) | 212 (22 to 639) |
| Age (years) | 67 (37 to 79) | 64 (34 to 81) | 62 (49 to 83) | 70 (50 to 83) | 59 (11 to 82) |
| Sex (m/f) | 8/4 | 4/11 | 15/6 | 12/10 | 16/9 |
No MPLW515L mutation was detected in the PMF group. The median values are displayed (range). wt, wild type.
Sequential bone marrow trephines in another nine patients with PMF were investigated for individual dynamics of BMP1 and BMP6 mRNA expression during the course of the disease. Note that these nine individual patients are not part of the PMF group summarized in Table 1.
Quantitative Genotyping for a Potential Underlying JAK2V617F and MPLW515L Mutation by Pyrosequencing
We applied pyrosequencing assays that allow the quantification of mutant JAK2V617F or MPLW515L alleles in bone marrow cells.8 Amplicons covering the hotspot point mutations (JAK2G1849T and MPLG1544T, respectively) were amplified by PCR from 25 ng of genomic DNA extracted by means of the Qiagen DNeasy kit (Qiagen, Hilden, Germany) using primers JAK2 forward, 5′-TATGATGAGCAAGCTTTCTCACAAG-3′; JAK2 reverse, 5′-AGAAAGGCATTAGAAAGCCTGTAGTT-3′ (GenBank accession no. AL161450) generating a 102-bp product; MPL forward, 5′-ATCTCCTTGGTGACCGCTCTG-3′; and MPL reverse, 5′-TGGTCCACCGCCAGTCTG-3′ (GenBank accession no. U68161) generating a 130-bp product with an additional 5′-biotin tag on respective reverse primers in a final volume of 50 μl. Pyrosequencing was performed as previously described.23 In brief, single-stranded PCR products were prepared by streptavidin Sepharose beat sorting and JAK2 and MPL specific sequencing (S) primers (JAK2-S 5′-GGTTTTAAATTATGGAGTATGT-3′, nucleotides 55039 to 55060 in GenBank accession no. AL161450, MPL-S 5′-GCCTGCTGCTGCTGAGGT-3′, nucleotides 1085 to 1102 in GenBank accession no. U68161) were applied to perform sequencing analysis and subsequent allele quantification in a PSQ 96MA instrument by using the SNP software (Biotage, Uppsala, Sweden). As described previously,8 samples were scored as heterozygous for JAK2V617F if the percentage of mutant T alleles exceeded 5%. Homozygosity for JAK2V617F was considered when allele burden exceeded 50.0%.
Samples under investigation were accompanied by positive controls (JAK2V617F cell lines HEL and SET-2; PMF patient with MPLW515L as evidenced by direct sequencing) and a negative control (JAK2wild-type/MPLwild-type cell line HL-60).
Frequencies of potential JAK2V617F and MPLW515L mutations are summarized in Table 1.
In Vitro Expression of BMPs by Human Fibroblasts
Primary human fibroblasts (cell line F-18, kindly provided by Dr. Miriam Wittmann, Department of Dermatology and Allergology, Hannover Medical School, and cell line M15D24) were grown as monolayer cultures in RPMI containing 10% fetal calf serum and antibiotics until subconfluence was reached. Cells were then cultured for 24 hours in RPMI 1640 containing 0.5% fetal calf serum and antibiotics in the presence of recombinant bFGF (2 ng/ml, 2 μg/ml stock prepared in 4 mmol/L HCl; 234-FSE; R&D Systems, Minneapolis, MN) and recombinant TGFβ-1 (2 ng/ml, 2 μg/ml stock prepared in 4 mmol/L HCl; 240-B; R&D Systems). Controls were cultured in parallel by using solely the vehicle HCl in RPMI 1640 containing 0.5% fetal calf serum and antibiotics. In vitro experiments were performed in duplicate.
Quantitative Real-Time PCR
Total RNA was extracted from cell cultures by using the TRIzol reagent (Invitrogen, Karlsruhe, Germany). As we previously described,25 total RNA was extracted from total formalin-fixed and paraffin-embedded bone marrow cells after guanidinium isothiocyanate/Proteinase K-based digestion and conventional organic extraction using phenol/chloroform. Total RNA (1 μg), pretreated with RNase free (Rnase-) DNase (1 U/μg RNA; RQ1; Promega, Madison, WI), was transcribed into the complementary DNA using 500 ng of random hexamers (Amersham Pharmacia, Piscataway, NJ) and 200 U of SuperScript II Rnase-Reverse Transcriptase (Invitrogen) in a volume of 20 μl following the manufacturer’s protocol. Negative controls were performed by pipetting water instead of reverse transcriptase. Real-time RT-PCR was performed on an ABI PRISM 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA).
After determination of PCR efficiency and demonstration of linearity of PCR amplification for target and housekeeping genes, a relative quantification approach was performed using the ΔΔCT− method as described previously.8,26 All samples were measured in duplicate. Sequences of PCR primers and TaqMan probes used in this study are listed in Table 2.
Table 2.
Primer and Probe Sequences Used throughout This Study
| Primer and probes | Sequence or published reference | GenBank accession number, amplicon size (bp) |
|---|---|---|
| BMP1 forward | 5′-CAGTCCTTTGAGATTGAGCGC-3′ | |
| BMP1 reverse | 5′-TGCTGCTCTCACTGTGCCC-3′ | |
| BMP1 probe | 5′-ACGACAGCTGTGCCTACGACTATCTGGAGGT-3′ | NM 001199, 79 |
| BMP2 forward | 5′-CAACACTGTCGCAGCTTC-3′ | |
| BMP2 reverse | 5′-GAAGAATCTCCGGGTTGTTTTC-3′ | |
| BMP2 probe | 5′-CCATGAAGAATCTTTGGAAGAACTACCAGAAACGAGT-3′ | NM001200.2, 82 |
| BMP4 forward | 5′-ACTGGTCTTGAGTATCCTGAGCGC-3′ | |
| BMP4 reverse | 5′-AAGCAGAGTTTTCACTGGTCCCT-3′ | |
| BMP4 probe | 5′-TTCCACCACGAAGAACATCTGGAGAACATCC-3′ | NM001202.1, 109 |
| BMP6 forward | 5′-GGAAGCATGAGCTGTATGTGAGTTT-3′ | |
| BMP6 reverse | 5′-AGTAATTGGCAGCATAGCATAGCCCTTG-3′ | |
| BMP6 probe | 5′-AAGACCTGGGATGGCAGGACTGGATCA-3′ | NM001218.2, 84 |
| BMP7 forward | 5′-CAAGAACCAGGAAGCCCTGC-3′ | |
| BMP7 reverse | 5′-TTCTTACAGGCCTGCCTCTGG-3′ | |
| BMP7 probe | 5′-ATGGCCAACGTGGCAGAGAACAGCA-3′ | NM001719, 75 |
| BMP-R2 forward | 5′-CTTTACTGAGAATTTTCCACCTCCTG-3′ | |
| BMP-R2 reverse | 5′-GCCAAAGCAATGATTATTGTCTCATC-3′ | |
| BMP-R2 probe | 5′-CACAACACCACTCAGTCCACCTCATTCATTTAAC-3′ | NM001204.5, 90 |
| PLOD2 forward | 5′-ATGGACACAGGATAATGGCTGC-3′ | |
| PLOD2 reverse | 5′-CACCTATTGATACGTTTGGATGGAC-3′ | |
| PLOD2 probe | 5′-CTCTTTGTGAATTCGATACAGTCGACTTGTCTGC-3′ | NM 182943, 89 |
| BMP1-CUBII forward | 5′-ACTGATGAAGCCTCGACCCC-3′ | |
| BMP1-CUBII reverse | 5′-AGAGTAACAGAGGAGGCACCTTTG-3′ | NT023666, 237 |
| COL1 | Ref.8 | NM 000088, 90 |
| β-GUS | Ref.8 | NM 000181, 81 |
| HSP-70.1 | Ref.8 | NM 005345, 62 |
Probes were labeled with 5′-FAM and 3′-TAMRA.
Explorative Screening for a Potential Point Mutation (E483K) in the CUB II Domain of BMP1 in PMF
A 237-bp DNA fragment was amplified covering the potential hotspot mutation G1476A (GenBank accession no. NM001199) in the CUB II domain of BMP1. Amplification of 25 ng DNA derived from total bone marrow cells of advanced PMF (n = 22) and controls (n = 3) was performed in a GeneAmp PCR System 2700 (Applied Biosystems) followed by direct sequencing in an ABI 310 Genetic Analyzer (Applied Biosystems). Primer sequences are listed in Table 2.
Immunohistochemistry
Immunohistochemistry for BMP1 in PMF (mf 0/1, n = 10; mf 2/3, n = 15) and control bone marrows (n = 10) was performed on bone marrow slides with a goat anti-human BMP1/PCP antibody (AF1927; R&D Systems). This antibody recognizes the amino acid sequence 121 to 730 of the mature BMP1 protein. After pretreatment in a pressure cooker at 125°C in citrate buffer (10 mmol/L, pH 6.0) for 3 minutes, the primary antibody was applied in a dilution of 1:50 before incubation for 1 hour at room temperature. Visualization was performed with the DAB Zytomed kit (DAB057; Zytomed Systems, Berlin, Germany) according to the manufacturer’s instructions and by counterstaining with hematoxylin. The protocol was strictly accompanied by a positive control (kidney tissue) and negative controls (samples were processed by omission of primary antibodies).
Statistics and Graphics
To analyze differences of mRNA expression in prefibrotic PMF, advanced PMF, and non-neoplastic hematopoiesis, nonparametric Kruskal-Wallis tests were performed followed by Dunns post test for pairwise group differences. For potential correlation of JAK2 status (allele burden) and BMP expression, unpaired t-tests and one-way analysis of variance tests were performed. P values ≤0.05 were considered as statistically significant. Graphics were designed using GraphPadPrism, Version 5.0 (GraphPad Software, San Diego, CA) and SigmaPlot, Version 8.0 (SPSS, Inc., Chicago, IL).
Results
TGFβ-1 and bFGF Induce Overexpression of BMPs in Cultured Fibroblasts
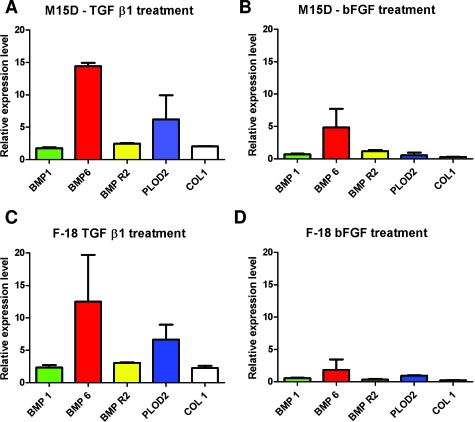
The in vitro treatment of fibroblasts (cell lines M15D and F-18) with TGFβ-1 showed a prominent induction of target gene expression for all BMPs, including BMP-R2, except for BMP7. In contrast to untreated fibroblasts, BMP7 was undetectable by qualitative and quantitative PCR approaches. Fibroblasts cultured in the presence of bFGF showed a different response with restricted induction of BMP6 in M15D and F-18 but not other BMPs. Increased expression of collagen-type 1 (COL1) and procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 (PLOD2) served as markers for sufficient induction of collagen synthesis by TGFβ-1. No induction of COL1 and PLOD2 was observed after stimulation by bFGF (Figure 1; Table 3).
Figure 1.
Bar chart plot of BMP expression in human fibroblasts cultured in the presence of TGFβ-1 and bFGF. Exaggerated expression of BMP6 by up to 14-fold and induction of BMP1 and BMP-R2 could be demonstrated. PLOD2 and COL1 induction was illustrated as a marker for TGFβ-1-induced collagen synthesis. BMP7 was not expressed in the presence of TGFβ-1 and bFGF but in controls. Note that induction was calculated relative to the control cell culture (M15D and F-18, respectively) whose target-gene specific expression was set to 1 (see also Table 3).
Table 3.
Relative Levels of BMP Expression in Fibroblasts (M15D, F-18) Cultured in the Presence of TGFβ-1 and bFGF
| M15D | TGF-β1 [median (range)] | bFGF [median (range)] | F-18 | TGF-β1 [median (range)] | bFGF [median (range)] |
|---|---|---|---|---|---|
| BMP1 | 1.8 (1.6 to 1.9) | 0.7 (0.6 to 0.8) | BMP 1 | 2.3 (1.9 to 2.7) | 0.6 (0.5 to 0.6) |
| BMP6 | 14.4 (13.9 to 14.9) | 4.8 (1.9 to 7.7) | BMP 6 | 12.5 (5.3 to 19.7) | 1.8 (0.2 to 3.5) |
| BMP-R2 | 2.5 (2.4 to 2.6) | 1.2 (1.0 to 1.4) | BMP to R2 | 3.0 (2.9 to 3.1) | 0.3 (0.2 to 0.5) |
| PLOD2 | 6.2 (2.5 to 9.9) | 0.5 (0.1 to 1.0) | PLOD 2 | 6.7 (4.4 to 8.9) | 1.0 (0.9 to 1.0) |
| COL1 | 2.0 (2.0 to 2.1) | 0.3 (0.2 to 0.3) | COL 1 | 2.3 (1.9 to 2.6) | 0.2 (0.2 to 0.3) |
PLOD2 Is Overexpressed in the Advanced Stages of PMF (mf 2/3)
We investigated a series of PMF (mf 0/1, n = 10; mf 2/3, n = 11) and control cases (n = 10) for PLOD2 mRNA expression to reproduce the data of PLOD2 mRNA induction by TGFβ-1 during in vitro culture of fibroblasts. We found a significantly higher PLOD2 mRNA expression level in PMF mf 2/3 (median, 3.0; range, 0.3 to 6.9) compared with controls (median, 1.0; range, 0.5 to 2.6, P = 0.01), and a trend toward a significant difference when compared with PMF mf 0/1 (median, 1.1; range, 0.4 to 5.0; P = 0.05). PMF mf 0/1 did not significantly differ from controls (P = 0.44).
BMP1, -6, -7, and BMP-R2 mRNA Expression in Different Stages of PMF
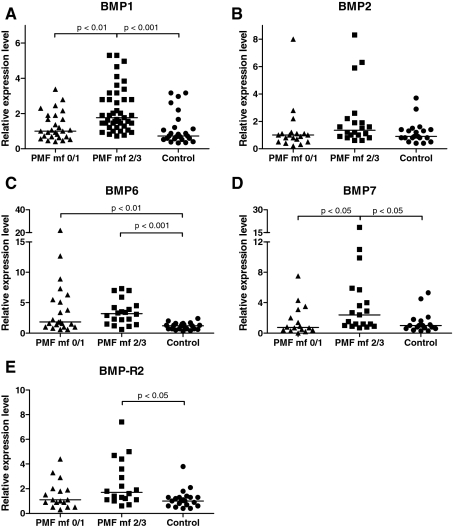
Advanced PMF stages showed a significantly increased BMP1 mRNA expression of up to fivefold compared with the hypercellular, prefibrotic stages (P < 0.01) and non-neoplastic hematopoiesis (P < 0.001). The prefibrotic stage and advanced stages of PMF significantly overexpressed BMP6 compared with control bone marrows (P < 0.01 and 0.001, respectively). BMP7 was increased in advanced stages compared with the prefibrotic stage and controls (P < 0.05). BMP-R2 was overexpressed in advanced PMF compared with controls (P < 0.05). BMP2 expression showed no difference between the groups under investigation (Figure 2).
Figure 2.
Point plot illustration of BMP mRNA expression in bone marrow cells of hypercellular, prefibrotic PMF (PMF mf 0/1), advanced PMF (PMF mf 2/3), and controls. Horizontal bars represent median values of expression. P values denote statistical relevance. Note that BMP mRNA expression in control hematopoiesis was arbitrarily set to 1. A comprehensive illustration of expression levels and potential correlation to grade of myelofibrosis and underlying molecular defects is displayed in Tables 4 and 5.
Of note, only 20% of the cases showed an amplifiable BMP4 mRNA in the qualitative RT-PCR approach. Therefore, no further analysis of BMP4 by quantitative approaches was undertaken. A comprehensive compilation of BMP expression levels is shown in Table 4.
Table 4.
Relative Levels of BMP Expression by Bone Marrow Cells in PMF and Control Hematopoiesis
| PMF mf 0/1 | PMF mf 2/3 | Control | |
|---|---|---|---|
| BMP1 | 1.0 (0.4 to 3.4) | 1.8 (0.7 to 5.3) | 0.7 (0.3 to 3.2) |
| n | 27 | 43 | 25 |
| BMP2 | 1.0 (0.2 to 8.0) | 1.4 (0.6 to 8.3) | 0.9 (0.4 to 3.7) |
| n | 19 | 20 | 20 |
| BMP6 | 1.9 (0.6 to 22.0) | 3.2 (0.6 to 7.3) | 1.2 (0.3 to 2.4) |
| n | 22 | 20 | 20 |
| BMP7 | 0.8 (0.1 to 7.5) | 2.4 (0.7 to 18.5) | 1.0 (0.3 to 5.3) |
| n | 16 | 19 | 17 |
| BMP-R2 | 1.1 (0.3 to 4.4) | 1.7 (0.6 to 7.4) | 1.0 (0.4 to 3.8) |
| n | 17 | 18 | 21 |
Median values (range) and number of cases (n) under investigation are displayed.
Overexpression of BMPs in PMF Bone Marrow Cells Is Not Correlated to the JAK2V617F Status or Mutant Allele Burden
To test a potential relationship between BMP expression and the JAK2 status, we analyzed PMF subgroups according to the grade of myelofibrosis (mf 0/1 and mf 2/3) and the JAK2 status. The allele burden of JAK2V617F cases (percentage of mutant T alleles) was further correlated to mf grade in PMF and BMP expression level, but no correlation was demonstrable. Table 5 exemplarily shows the analysis for BMP1 in PMF.
Table 5.
BMP1 mRNA Expression and Correlation with JAK2V617F Status and Allele Burden
| PMF mf 0/1 (n = 27)
|
|
PMF mf 2/3 (n = 43)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JAK2wt | JAK2V617F | JAK2V617F | JAK2wt | JAK2V617F | JAK2V617F | |||||
| +/− | +/+ | +/− | +/+ | |||||||
| 30.2% (13.7 to 49.2) | 68.1% (56.3 to 86.2) | 30.0% (6.1 to 47.9) | 78.3% (50.4 to 93.9) | |||||||
| n = 12 | n = 11 | n = 4 | n = 21 | n = 10 | n = 12 | |||||
| 0.8 (0.5 to 2.5) | 1.0 (0.4 to 2.8) | 1.8 (0.5 to 3.4) | 1.9 (0.7 to 5.3) | 1.9 (1.0 to 5.3) | 1.6 (0.9 to 4.1) | |||||
| P = 0.44 | P = 0.88 | |||||||||
| P = 0.34 | P = 0.23 | |||||||||
| P = 0.24 | P = 0.46 | |||||||||
As exemplified for BMP1, no correlation of allele burden and BMP expression was demonstrable in PMF cases showing the wild type (wt) of JAK2, heterozygous (+/−) or homozygous (+/+) JAK2V617F mutations. Mutant T allele burden is displayed as the median percentage (range). n, case numbers. X-fold expression of BMP1 is shown as the median value (range).
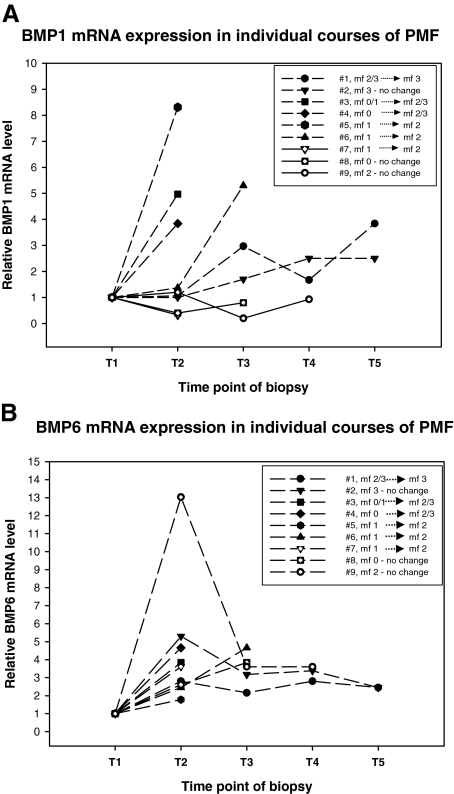
Increased mRNA Levels of BMP1 and BMP6 Are Detectable During Progressive Myelofibrosis in Individual Courses of PMF
We investigated nine individual courses of PMF with follow-ups ranging from 12 to 48 months (median, 24 months) and two to five sequential trephine biopsies (median, three biopsies) for BMP1 and BMP6 mRNA expression. Three patients showed a full-blown myelofibrosis (mf 2/3) at diagnosis and remained stable, and three cases started with a mild reticulin fiber deposition (mf 0/1) and developed a more severe myelofibrosis (mf 2). Two patients showed progressive myelofibrosis (mf 2/3) after initial diagnosis of hypercellular, prefibrotic PMF (mf 0), and another case revealed persistence of the hypercellular phase during the follow-up (mf 0). Six of nine individual courses of PMF revealed a modest to strong increase of BMP1 mRNA expression by bone marrow cells (median, 4.4; range, 2.5 to 8.3). In most cases, increasing BMP1 levels paralleled increasing grades of myelofibrosis. In only one of nine cases was a mild decrease of BMP1 expression during mf 1 to mf 2 transition (no. 7) noticed. The patient (no. 8) with a persisting hypercellular stage (mf 0) showed a slight decrease of BMP1 mRNA level (Figure 3A; Table 6).
Figure 3.
Line and scatter plot of BMP1 (A) and BMP6 (B) mRNA expression in individual courses of PMF. T1 to T5 represent the time points where sequential biopsies were taken. Expression level for each biopsy was expressed as the mean of two independent measurements. Increased levels are displayed as long dashed lines, and decreased levels are displayed as solid lines. Figure should be interpreted by concomitant review of Table 6, which shows relevant histological parameters such as the grade of fibrosis. Starting level of BMP1 and BMP6 in individual courses was set to 1.
Table 6.
Time Points of Sequential Biopsies as Displayed in Figure 2
| Case | Month/year of biopsy | BMP1 mRNA level | BMP6 mRNA level | Grade of fibrosis | Allele burden (%) JAK2V617F/MPLW515L |
|---|---|---|---|---|---|
| 1 | 05/2002 | 1.0 | mf 2/3 | 75 JAK2V617F | |
| 08/2002 | 1.1 | 2.8 | mf 2/3 | 56 JAK2V617F | |
| 11/2002 | 3.0 | 2.2 | mf 2/3 | 56 JAK2V617F | |
| 05/2003 | 1.7 | 2.8 | mf 2/3 | 66 JAK2V617F | |
| 07/2003 | 3.8 | 2.5 | mf 3 | 61 JAK2V617F | |
| 2 | 06/2002 | 1.0 | mf 3 | 77 JAK2V617F | |
| 11/2002 | 1.0 | 5.3 | mf 3 | 35 JAK2V617F | |
| 05/2003 | 1.7 | 3.2 | mf 3 | 65 JAK2V617F | |
| 12/2003 | 2.5 | 3.4 | mf 3 | 76 JAK2V617F | |
| 06/2004 | 2.5 | 2.5 | mf 3 | 69 JAK2V617F | |
| 3 | 12/2002 | 1.0 | mf 0/1 | 40 JAK2V617F | |
| 12/2003 | 5.0 | 3.8 | mf 2/3 | 38 JAK2V617F | |
| 4 | 02/2001 | 1.0 | mf 0 | 24 JAK2V617F | |
| 07/2004 | 3.8 | 4.7 | mf 2/3 | 7 JAK2V617F* | |
| 5 | 11/2000 | 1.0 | mf 1 | 37 JAK2V617F | |
| 01/2002 | 8.3 | 1.8 | mf 2 | 7 JAK2V617F* | |
| 6 | 11/2000 | 1.0 | mf 1 | 85 MPLW515L | |
| 08/2002 | 1.4 | 2.5 | mf 1 | 87 MPLW515L | |
| 07/2004 | 5.3 | 4.7 | mf 2 | 100 MPLW515L | |
| 7 | 05/2000 | 1.0 | mf 1 | 59 MPLW515L | |
| 12/2003 | 0.3 | 3.6 | mf 2 | 95 MPLW515L | |
| 8 | 05/2001 | 1.0 | mf 0 | JAK2wt, MPLwt | |
| 06/2002 | 0.4 | 2.6 | mf 0 | JAK2wt, MPLwt | |
| 08/2002 | 0.8 | 3.8 | mf 0 | JAK2wt, MPLwt | |
| 9 | 09/2002 | 1.0 | mf 2 | JAK2wt, MPLwt | |
| 01/2003 | 1.2 | 13.0 | mf 2 | JAK2wt, MPLwt | |
| 06/2003 | 0.2 | 3.6 | mf 2 | JAK2wt, MPLwt | |
| 10/2003 | 0.9 | 3.6 | mf 2 | JAK2wt, MPLwt |
If present, allele frequencies for JAK2V617F and MPLW515L are illustrated. wt, wild type.
A decline of JAK2V617F before rapid transformation into acute leukemia.
The entirety of courses under investigation showed an up-regulation of BMP6 mRNA expression (nos. 3 to 8) or the persistence of a steady state after an initial increase (nos. 1, 2, and 9). Interestingly, also in course no. 8 showing the prefibrotic stage of PMF in three biopsies taken between 2001 and 2002, a considerable increase of BMP6 mRNA was demonstrable (Figure 3B; Table 6).
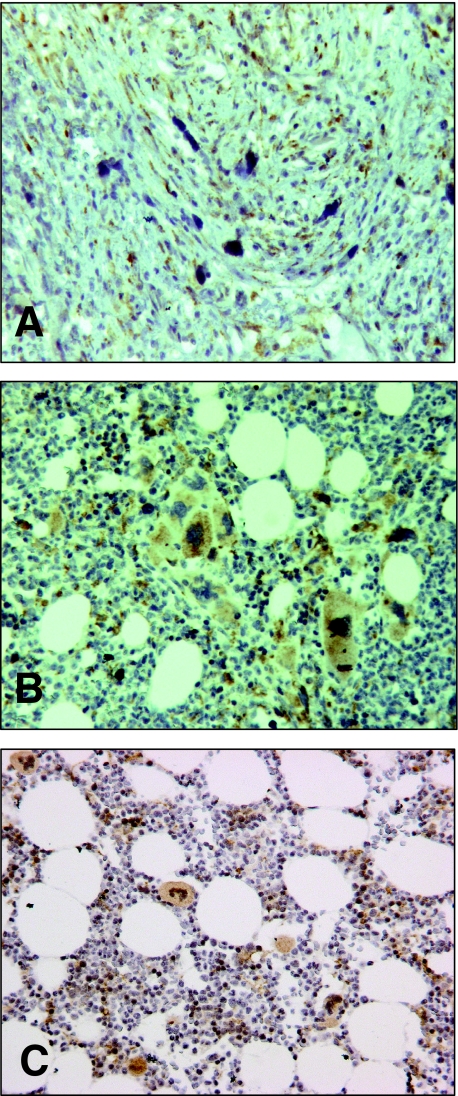
Bone Marrow Stromal Cells in Advanced Stages of PMF and Megakaryocytes Provide the Cellular Source of BMP1 in Advanced Stages
In advanced stages of PMF, bone marrow stromal cells, ie, fibroblasts, showed a strong positivity for BMP1 in their cytoplasm and represented the predominant cellular source. Besides fibroblasts, megakaryocytes were labeled in advanced stages (Figure 4, A and B).
Figure 4.
Immunohistochemical localization of BMP1 protein expression by bone marrow cells in PMF. BMP1 was visualized by DAB brown staining and showed strong labeling of bone marrow fibroblasts in advanced PMF (A) and megakaryocytes (B). Bone marrows showing normal hematopoiesis or a mild reactive hyperplasia of the megakaryocytic lineage showed BMP1 labeling of megakaryocytes and occasionally stroma cells stained for BMP1 (C).
In bone marrow samples of PMF and control hematopoiesis, megakaryocytes were also labeled for BMP1 protein in a similar cytoplasmic manner but with different staining intensity, ie, because of their large numbers in PMF bone marrow samples, BMP1-positive megakaryocytes dominated over megakaryocytes in normal hematopoiesis. In control bone marrows, interstitial cells were infrequently stained for BMP1.
No Evidence for a BMP1E483K Mutation or Other Mutation Sites in the CUBII Domain of BMP1 in Bone Marrow Cells of PMF
We sequenced a region of the CUBII domain of BMP1 that is known to be important for full proteinase activity and cleavage of collagen precursor molecules. In a series of advanced PMF (n = 22), we could not detect any mutation site, neither the G1476A (GenBank accession no. NM 001199) leading to E483K nor other mutations.
Discussion
PMF bone marrows are enriched by fibrogenic cytokines such as TGFβ-1, bFGF, platelet-derived growth factor, and other growth factors.3,7,27 We and others showed that exaggerated expression of these cytokines contribute not only to the process of fibrogenesis but also to autocrine stimulation of cellular lineages.7,28,29 In fact, megakaryocytes contain large amounts of nuclear bFGF, suggesting an autocrine mechanism.30 Megakaryocytes in PMF per se were shown to be involved in fibrogenesis because of either active secretion or decay-associated passive efflux of cellular cytokines into the bone marrow stroma.31,32
We first aimed to simulate the bone marrow environment in PMF by treating primary fibroblasts with TGFβ-1 and bFGF to determine a potential induction of those BMP members for which a role in fibrogenesis has been suggested.9,19
Fibroblasts treated with TGFβ-1 showed an exaggerated induction of BMP6 but also of BMP1 and BMP-R2 (Figure 1, A and C). This induction was also notable in fibroblasts cultured in the presence of bFGF but was restricted to BMP6 (Figure 1, B and D). As shown previously, TGFβ-1 is potent in inducing collagen genes and synthesis.33 Therefore, we also analyzed the induction of PLOD2 and COL1 expression by TGFβ-1 that was not demonstrable in bFGF-treated cultures. Interestingly, BMP7 was no longer detectable on the RT-PCR level under both TGFβ-1 and bFGF culture conditions. BMPs are thought to be differentially involved in the process of fibrogenesis. In contrast to BMP1, -2, and -6, BMP7 was shown to act in the prevention of fibrosis in kidneys and models of cardiac fibrosis.17,18 In our study, BMP7 was increased in advanced PMF (Figure 2D). If this increase reflects a counteracting mechanism against fibrogenesis also in the bone marrow, this needs to be clarified.
In our study, BMP1 was rather moderate but significantly increased in vitro and in vivo. We decided to further investigate this particular subtype because of its exclusive role in collagen processing. BMP1 cleaves precursor forms of type I and III collagens to produce mature fibrils. BMP1 also enables the activation of latent forms of TGFβ-1 that then in turn induce expression of collagens and other ECM molecules by fibroblasts.9,12 At best, our in vitro data suggest that BMP1 itself is inducible by TGFβ-1. Therefore, a positive feedback between these two factors seems to be likely in the process of myelofibrosis.
At the protein level, fibroblasts in advanced stages were uncovered as a major cellular source for BMP1. Megakaryocytes were also strongly positive for BMP1, but this was not restricted to advanced PMF and was also demonstrable in hypercellular PMF and even in normal bone marrows (Figure 4C). BMP1 expression is therefore not fibroblast-specific, and the role of megakaryocyte-derived BMP1 needs to be further clarified. It is likely that megakaryocytes actively secrete BMP1 and, because of their high number in PMF bone marrows, essentially contribute to the development of myelofibrosis, including osteosclerosis. BMP1 is also possibly involved in the accumulation of PMF megakaryocytes through anti-apoptotic mechanisms, as shown for BMP1-associated processing of Perlecan in fibroblasts in vitro.14 Of note, in normal bone marrows of our control group, some interstitial cells, including fibroblasts, expressed BMP1, underlining the physiological impact of BMP1 in matrix homeostasis.
An explorative screening for a potential mutation in the CUBII domain of BMP1 as an underlying molecular defect could be excluded by direct sequencing in a series of PMF bone marrows.
Taking into account the role of BMP1 expression in the development of myelofibrosis, BMP1 expression level might serve as a potentially useful diagnostic adjunct allowing a better discrimination between prefibrotic PMF and essential thrombocythemia (ET). As described previously,1,2 prefibrotic PMF and ET are distinctive entities that may show some overlaps in clinical and histopathological presentation. Even more so, the discovery of disease-specific markers even in early stages of PMF and ET would be of particular importance. In an exploratory analysis, we found significantly lower BMP1 expression levels in ET cases (n = 20) compared to prefibrotic PMF (P = 0.02). However, demonstration of a clear-cut range of BMP1 expression allowing a definite diagnostic support was hampered because of outliers and resulting overlaps in both groups. However, these hitherto preliminary data on BMP1 in ET will be further investigated in a larger series.
In mice deficient for the megakaryocytic transcription factor GATA-1 (GATA-1low), a severe osteosclerosis could be demonstrated after 3 to 5 months of age.34 A recent study showed that osteosclerosis in this animal model is obviously triggered by aberrant expression of BMP members by megakaryocytes, eg, BMP6 showed a considerable overexpression.19 Our data show the significant increase of BMP6 in human PMF bone marrows, thereby supporting the thesis that this BMP family member is also involved in advanced ECM formation. Of note, some of the prefibrotic stages of PMF showed outliers overexpressing BMP6, suggesting that BMP6 might serve as a marker for a more progressive development of fibrosis and osteosclerosis.
Interestingly, another study using the similar GATA-1low mouse model showed development of myelofibrosis later than demonstrable osteosclerosis, ie, at 1 year of age.34 This observation may indicate some artificial characteristics of this particular model because in humans osteosclerosis rarely, if ever, precedes the stage of myelofibrosis. However, some patients rapidly develop a full-blown PMF where some overlaps between the different stages of myelofibrosis might be demonstrable.
Recently, an interesting study revealed that CD34+ progenitor cells from peripheral blood of patients with PMF showed an aberrant expression of BMPs and the BMP inhibitor Noggin.35 This study revealed an aberrant BMP expression profile different from our investigation, ie, a down-regulation of BMP-R2. We showed a moderate but significant increase of BMP-R2 in advanced PMF (Figure 2E), which fits in the model of activated BMP signaling in this disease. These discrepancies might be due to the origin and type of cells analyzed (CD34+ cells isolated from peripheral blood).
In individual courses of PMF, the majority of biopsies revealed an increasing level of BMP1 accompanying the process of ongoing aberrant matrix deposition but obviously without indicating the definite switch from the hypercellular phase (mf 0) to manifest myelofibrosis (mf 2/3). Therefore, BMP1 does not represent a clear-cut progress marker in this particular setting. BMP6 also showed an up-regulation in prefibrotic stages of course no. 8, which fits to increased expression level revealed in some other PMF cases with mf 0/1 (Figure 2C). But also BMP6 appears not to be the one and only indicator useful for estimation of the “fibrotic switch.” Nevertheless, its involvement in the process of myelofibrosis is beyond doubt.
Because BMP1 is highly expressed in areas of tumor angiogenesis15 we were highly interested in proving BMP1 expression by endothelial cells in exaggerated angiogenesis of PMF. We thoroughly reviewed the slides for BMP1 labeling of endothelial cells but could not detect any considerable staining. BMP1 is therefore less likely to be involved in endothelial cell proliferation in PMF.
Another histological hallmark of advanced stages of PMF is aberrant apposition of bone matrix and osteosclerosis. BMP1 was initially demonstrated to be capable of inducing bone formation,10 and the early steps of osteogenesis include the processing of collagen molecules (in particular type I). Therefore, it appears that overexpressed BMP1 in PMF in the network with other BMPs such as BMP6 not only contributes to exaggerated ECM deposition but also to aberrant bone matrix formation.
In conclusion, BMPs are likely to be involved in pathogenesis of PMF through direct activation of latent TGFβ-1 thereby indirectly inducing their own expression followed by exaggerated processing of precursor collagens and contribution to ECM formation.
Acknowledgments
We gratefully acknowledge the skilled technical assistance of Ms. Anna-Lena Becker, Ms. Henriette Bruchhardt, and Ms. Sabine Schroeter.
Footnotes
Address reprint requests to Oliver Bock, M.D., Assistant Professor, Institute of Pathology, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany. E-mail: Bock.Oliver@MH-Hannover.de.
Supported by Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung 10-2191 (to O.B. and H.K.), and Deutsche Forschungsgemeinschaft (DFG/Bo 1954/1-1 to O.B. and H.K.),
References
- Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, Barosi G, Verstovsek S, Birgegard G, Mesa R, Reilly JT, Gisslinger H, Vannucchi AM, Cervantes F, Finazzi G, Hoffman R, Gilliland DG, Bloomfield CD, Vardiman JW. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood. 2007;110:1092–1097. doi: 10.1182/blood-2007-04-083501. [DOI] [PubMed] [Google Scholar]
- Thiele J, Kvasnicka HM. Prefibrotic chronic idiopathic myelofibrosis: a diagnostic enigma? Acta Haematol. 2004;111:155–159. doi: 10.1159/000076524. [DOI] [PubMed] [Google Scholar]
- Tefferi A. Pathogenesis of myelofibrosis with myeloid metaplasia. J Clin Oncol. 2005;23:8520–8530. doi: 10.1200/JCO.2004.00.9316. [DOI] [PubMed] [Google Scholar]
- Xu M, Bruno E, Chao J, Huang S, Finazzi G, Fruchtman SM, Popat U, Prchal JT, Barosi G, Hoffman R, MPD Research Consortium Constitutive mobilization of CD34+ cells into the peripheral blood in idiopathic myelofibrosis may be due to the action of a number of proteases. Blood. 2005;105:4508–4515. doi: 10.1182/blood-2004-08-3238. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355:2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, Steensma DP, Elliott MA, Wolanskyj AP, Hogan WJ, McClure RF, Litzow MR, Gilliland DG, Tefferi A. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- Le Bousse-Kerdilès MC, Martyre MC. Dual implication of fibrogenic cytokines in the pathogenesis of fibrosis and myeloproliferation in myeloid metaplasia with myelofibrosis. Ann Hematol. 1999;78:437–444. doi: 10.1007/s002770050595. [DOI] [PubMed] [Google Scholar]
- Bock O, Neuse J, Hussein K, Brakensiek K, Buesche G, Buhr T, Wiese B, Kreipe H. Aberrant collagenase expression in chronic idiopathic myelofibrosis is related to the stage of disease but not to the JAK2 mutation status. Am J Pathol. 2006;169:471–481. doi: 10.2353/ajpath.2006.060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Urist MR, Iwata H, Ceccotti PL, Dorfman RL, Boyd SD, McDowell RM, Chien C. Bone morphogenesis in implants of insoluble bone gelatin. Proc Natl Acad Sci USA. 1973;70:3511–3515. doi: 10.1073/pnas.70.12.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol. 2006;175:111–120. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M, Billings PC, Pacifici M, Leboy PS, Kirsch T. Authentic matrix vesicles contain active metalloproteases (MMP): A role for matrix vesicle-associated MMP-13 in activation of transforming growth factor-beta. J Biol Chem. 2001;276:11347–11353. doi: 10.1074/jbc.M009725200. [DOI] [PubMed] [Google Scholar]
- Laplante P, Raymond MA, Labelle A, Abe J, Iozzo RV, Hébert MJ. Perlecan proteolysis induces an alpha2beta1 integrin- and Src family kinase-dependent anti-apoptotic pathway in fibroblasts in the absence of focal adhesion kinase activation. J Biol Chem. 2006;281:30383–30392. doi: 10.1074/jbc.M606412200. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Gitelman SE, Kobrin MS, Ye JQ, Lopez AR, Lee A, Derynck R. Recombinant Vgr-1/BMP-6-expressing tumors induce fibrosis and endochondral bone formation in vivo. J Cell Biol. 1994;126:1595–1609. doi: 10.1083/jcb.126.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, de Caestecker M, Kopp J, Mitu G, Lapage J, Hirschberg R. Renal bone morphogenetic protein-7 protects against diabetic nephropathy. J Am Soc Nephrol. 2006;17:2504–2512. doi: 10.1681/ASN.2006030278. [DOI] [PubMed] [Google Scholar]
- Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- Garimella R, Kacena MA, Tague SE, Wang J, Horowitz MC, Anderson HC. Expression of bone morphogenetic proteins and their receptors in the bone marrow megakaryocytes of GATA-1(low) mice: a possible role in osteosclerosis. J Histochem Cytochem. 2007;55:745–752. doi: 10.1369/jhc.6A7164.2007. [DOI] [PubMed] [Google Scholar]
- Hartigan N, Garrigue-Antar L, Kadler KE. Bone morphogenetic protein-1 (BMP-1): identification of the minimal domain structure for procollagen C-proteinase activity. J Biol Chem. 2003;278:18045–18049. doi: 10.1074/jbc.M211448200. [DOI] [PubMed] [Google Scholar]
- Thiele J, Kvasnicka HM, Facchetti F, Franco V, van der Walt J, Orazi A. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica. 2005;90:1128–1132. [PubMed] [Google Scholar]
- Buhr T, Buesche G, Choritz H, Länger F, Kreipe H. Evolution of myelofibrosis in chronic idiopathic myelofibrosis as evidenced in sequential bone marrow biopsy specimens. Am J Clin Pathol. 2003;119:152–158. doi: 10.1309/PTVG-B3DX-B8A8-M7KD. [DOI] [PubMed] [Google Scholar]
- Hussein K, Brakensiek K, Buesche G, Buhr T, Wiese B, Kreipe H, Bock O. Different involvement of the megakaryocytic lineage by the JAK2 (V617F) mutation in Polycythemia vera, essential thrombocythemia and chronic idiopathic myelofibrosis. Ann Hematol. 2007;86:245–253. doi: 10.1007/s00277-007-0252-3. [DOI] [PubMed] [Google Scholar]
- Schönermark MP, Bock O, Buechner A, Steinmeier R, Benbow U, Lenarz T. Quantification of tumor cell invasion using confocal laser scan microscopy. Nat Med. 1997;3:1167–1171. doi: 10.1038/nm1097-1167. [DOI] [PubMed] [Google Scholar]
- Bock O, Kreipe H, Lehmann U. One-step extraction of RNA from archival biopsies. Anal Biochem. 2001;295:116–117. doi: 10.1006/abio.2001.5188. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Kuter DJ, Bain B, Mufti G, Bagg A, Hasserjian RP. Bone marrow fibrosis: pathophysiology and clinical significance of increased bone marrow stromal fibres. Br J Haematol. 2007;139:351–362. doi: 10.1111/j.1365-2141.2007.06807.x. [DOI] [PubMed] [Google Scholar]
- Chagraoui H, Komura E, Tulliez M, Giraudier S, Vainchenker W, Wendling F. Prominent role of TGF-beta 1 in thrombopoietin-induced myelofibrosis in mice. Blood. 2002;100:3495–3503. doi: 10.1182/blood-2002-04-1133. [DOI] [PubMed] [Google Scholar]
- Bock O, Loch G, Schade U, von Wasielewski R, Schlue J, Kreipe H. Aberrant expression of transforming growth factor beta-1 (TGF beta-1) per se does not discriminate fibrotic from non-fibrotic chronic myeloproliferative disorders. J Pathol. 2005;205:548–557. doi: 10.1002/path.1744. [DOI] [PubMed] [Google Scholar]
- Bock O, Schlue J, Lehmann U, von Wasielewski R, Langer F, Kreipe H. Megakaryocytes from chronic myeloproliferative disorders show enhanced nuclear bFGF expression. Blood. 2002;100:2274–2275. doi: 10.1182/blood-2002-06-1811. [DOI] [PubMed] [Google Scholar]
- Martyré MC, Romquin N, Le Bousse-Kerdiles MC, Chevillard S, Benyahia B, Dupriez B, Demory JL, Bauters F. Transforming growth factor-beta and megakaryocytes in the pathogenesis of idiopathic myelofibrosis. Br J Haematol. 1994;88:9–16. doi: 10.1111/j.1365-2141.1994.tb04970.x. [DOI] [PubMed] [Google Scholar]
- Castro-Malaspina H, Rabellino EM, Yen A, Nachman RL, Moore MA. Human megakaryocyte stimulation of proliferation of bone marrow fibroblasts. Blood. 1981;57:781–787. [PubMed] [Google Scholar]
- Kimura A, Katoh O, Hyodo H, Kuramoto A. Transforming growth factor-beta regulates growth as well as collagen and fibronectin synthesis of human marrow fibroblasts. Br J Haematol. 1989;72:486–491. doi: 10.1111/j.1365-2141.1989.tb04310.x. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Bianchi L, Cellai C, Paoletti F, Rana RA, Lorenzini R, Migliaccio G, Migliaccio AR. Development of myelofibrosis in mice genetically impaired for GATA-1 expression (GATA-1(low) mice). Blood. 2002;100:1123–1132. doi: 10.1182/blood-2002-06-1913. [DOI] [PubMed] [Google Scholar]
- Andrieux J, Roche-Lestienne C, Geffroy S, Desterke C, Grardel N, Plantier I, Selleslag D, Demory JL, Laï JL, Leleu X, Le Bousse-Kerdiles C, Vandenberghe P. Bone morphogenetic protein antagonist gene NOG is involved in myeloproliferative disease associated with myelofibrosis. Cancer Genet Cytogenet. 2007;178:11–16. doi: 10.1016/j.cancergencyto.2007.06.001. [DOI] [PubMed] [Google Scholar]