Abstract
Chitin, a polymer of N-acetyl-d-glucosamine, is found in fungal cell walls but not in plants. Plant cells can perceive chitin fragments (chitooligosaccharides) leading to gene induction and defense responses. We identified a LysM receptor-like protein (LysM RLK1) required for chitin signaling in Arabidopsis thaliana. The mutation in this gene blocked the induction of almost all chitooligosaccharide-responsive genes and led to more susceptibility to fungal pathogens but had no effect on infection by a bacterial pathogen. Additionally, exogenously applied chitooligosaccharides enhanced resistance against both fungal and bacterial pathogens in the wild-type plants but not in the mutant. Together, our data indicate that LysM RLK1 is essential for chitin signaling in plants (likely as part of the receptor complex) and is involved in chitin-mediated plant innate immunity. The LysM RLK1-mediated chitin signaling pathway is unique, but it may share a conserved downstream pathway with the FLS2/flagellin- and EFR/EF-Tu–mediated signaling pathways. Additionally, our work suggests a possible evolutionary relationship between the chitin and Nod factor perception mechanisms due to the similarities between their potential receptors and between the signal molecules perceived by them.
INTRODUCTION
Chitin is a polymer of N-acetyl-d-glucosamine found in fungal cell walls, insect exoskeletons, and crustacean shells but not in plants. Plants do not have chitin, but they do have chitin-degrading enzymes. It was hypothesized that plant chitinases can degrade chitin in the fungal cell walls to directly affect the viability of the invading fungal pathogen and to release short chitin fragments (chitooligosaccharides) that can act as a general elicitor of plant innate immunity (Boller, 1995; Stacey and Shibuya, 1997; Shibuya and Minami, 2001; Passarinho and de Vries, 2002). This hypothesis is indirectly supported by the following observations: a number of chitinase genes are induced by fungal infection (Majeau et al., 1990; Roby et al., 1990), and chitinases accumulate at infection sites in planta (Wubben et al., 1992). Chitinases were also shown to degrade fungal cell walls and inhibit fungal growth in vitro, especially in combination with β-1,3-glucanases (Schlumbaum et al., 1986; Mauch et al., 1988; Arlorio et al., 1992; van den Burg et al., 2006). Transgenic plants overexpressing chitinase genes were more resistant to fungal pathogens (Brogue et al., 1991; Jach et al., 1995). In addition, a fungal chitin binding protein (Avr4) was found to specifically bind chitin in fungal cell walls to prevent chitin degradation by plant chitinases (van den Burg et al., 2006).
In agreement with the above hypothesis, exogenously applied chitooligosaccharides enhanced plant resistance against fungal pathogens (Tanabe et al., 2006). Purified chitooligosaccharides were able to induce various defense responses in plants or cultured cells, such as induction of defense-related genes, synthesis of phytoalexin, production of reactive oxygen species, and protein phosphorylation (Felix et al., 1993; Baier et al., 1999; Peck et al., 2001; Shibuya and Minami, 2001; Ramonell et al., 2005). Gene expression profiling studies also demonstrated that chitooligosaccharides can induce a large number of genes (including many defense-related genes) in different plant species (Day et al., 2002; Ramonell et al., 2002, 2005). Mutations in selected chitooligosaccharide-responsive genes (CRGs) led to more susceptibility to a fungal pathogen (Ramonell et al., 2005). Various studies suggested that plants possess a specific system to perceive chitooligosaccharides leading to gene induction and defense (Day et al., 2001; Zhang et al., 2002; Wan et al., 2004; Libault et al., 2007).
Chitin binding sites or proteins were previously identified in membrane preparations from a variety of plants (Baureithel et al., 1994; Ito et al., 1997; Day et al., 2001; Okada et al., 2002). Recently, a LysM domain-containing protein (Chitin Elicitor Binding Protein [CEBiP]) was shown to be involved in the binding and perception of chitooligosaccharides in rice (Oryza sativa; Kaku et al., 2006). The LysM motif was originally identified in enzymes that degrade the bacterial cell wall component peptidoglycan, which is structurally similar to chitin (Joris et al., 1992). Since CEBiP lacks a significant intracellular domain, it likely is only a part of the chitin receptor complex in rice (Kaku et al., 2006). An obvious partner for CEBiP would be a membrane-associated receptor-like kinase.
LysM domain-containing receptor-like kinases (LysM RLKs; e.g., Nod Factor Receptor1 [NFR1] and NFR5) in legumes were shown to be critical for the perception of modified chitooligosaccharides (Nod factors) in the legume–rhizobial symbiotic interaction (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Similar LysM RLK genes are also present in nonleguminous plants (Zhang et al., 2007). For example, there are five such LysM RLK genes in Arabidopsis thaliana. LysM domain-containing proteins are also found in animals, but LysM RLKs appear to be unique to plants (Zhang et al., 2007). Due to the similarity between the Arabidopsis LysM RLKs and legume NFR1 and NFR5 and the similarity between chitooligosaccharides and Nod factors, it is logical to hypothesize that nonleguminous LysM RLKs may be involved in the perception of Nod factor-like molecules (i.e., chitooligosaccharides).
We tested this hypothesis in this work and revealed that one Arabidopsis LysM RLK indeed plays a critical role in chitin signaling.
RESULTS
Identification of a Mutant Blocking the Induction of the Selected CRGs by Chitooligosaccharides
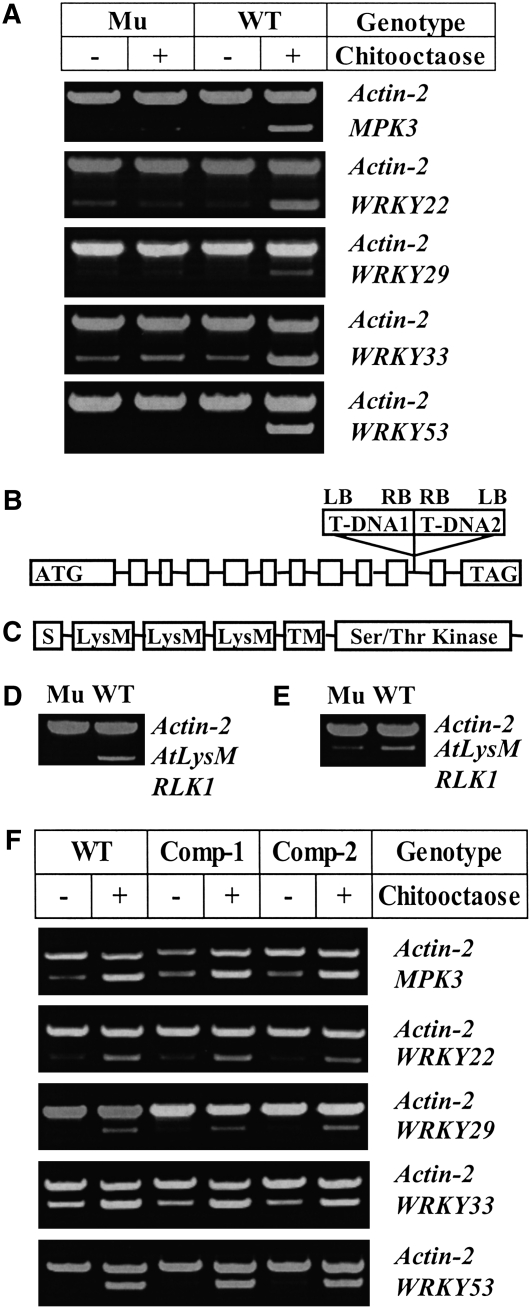
To study the potential role of the Arabidopsis LysM RLK genes in chitin signaling, insertion mutants were obtained for all five LysM RLK genes (i.e., At1g51940, At2g23770, At2g33580, At3g01840, and At3g21630). Homozygous mutants were treated with a purified chitooligosaccharide (chitooctaose), and the expression of known CRGs, MPK3, WRKY22, WRKY33, and WRKY53 (Wan et al., 2004), was examined. Only one mutant, corresponding to At3g21630 (LysM RLK1), appeared to totally block the induction of the selected CRGs (Figure 1A), while mutations in the other four LysM RLK genes had no apparent effect on CRG expression (see Supplemental Figure 1 online). Therefore, the data suggest a critical role for LysM RLK1 in chitin signaling.
Figure 1.
The Knockout of the LysM RLK1 Gene Blocked the Induction of the Selected CRGs by Chitooctaose.
(A) Analysis of the expression of the selected CRGs in both the mutant and wild-type plants using RT-PCR. Both the mutant (Mu) and wild-type plants were treated with purified chitooctaose or water (as a control) for 30 min, and gene expression of the selected CRGs was then detected using RT-PCR. Actin-2 was used as an internal control, and the amplification of both actin-2 and a CRG was conducted in the same PCR reaction tube with 25 cycles. The experiment was repeated at least three times with similar results.
(B) The gene structure of LysM RLK1 (At3g21630) (not drawn to scale). Square boxes are exons. Solid lines between them are introns. The start codon (ATG) and stop codon (TAG) were included in the first and last exon, respectively. The two T-DNA insertions (T-DNA1 and 2) inserted in the 10th intron in the LysM RLK1 mutant were indicated above the gene. LB, left border; RB, right border.
(C) The predicted domain structure of LysM RLK1. S, signal peptide; LysM, LysM domain; TM, transmembrane domain; WT, wild-type Col-0; Mu, LysM RLK1 mutant.
(D) Analysis of the LysM RLK1 transcript in both the mutant and wild-type plants using RT-PCR. The gene-specific primers were derived from the exons on each side of the insertions.
(E) Analysis of the LysM RLK1 transcript in both the mutant and wild-type plants using RT-PCR. The gene-specific primer pairs were derived from the exons before the insertions. WT, wild-type Col-0; Mu, LysM RLK1 mutant. Actin-2 was used as an internal control.
(F) Restoration of CRG induction in the LysM RLK1 mutant by the ectopic expression of the LysM RLK1 cDNA. The LysM RLK1 cDNA driven by a cauliflower mosaic virus 35S promoter was introduced into the homozygous LysM RLK1 mutant using a modified pCAMBIA1200 binary vector. The complemented plants (homozygous T3) were treated with chitooctaose at a final concentration of 1 μM or water (as a control) for 30 min. RT-PCR analysis showed that the selected CRGs were induced to a similar level in the selected complemented plants (Com-1 and Com-2) to that in the wild type. Com-1 and Com-2, two independent complemented lines; WT, wild-type Col-0. Actin-2 is used as an internal control.
The LysM RLK1 gene is 2988 nucleotides long, with 11 introns (Figure 1B) and a coding sequence of 1854 nucleotides. It encodes a LysM RLK of 617 amino acids, with an extracellular domain containing three predicted LysM motifs, a transmembrane domain, and an intracellular Ser/Thr kinase domain (Figure 1C). Interestingly, LysM RLK1 was recently shown to be phylogenetically related to the Nod factor receptor NFR1 (Zhang et al., 2007).
Two T-DNA insertions arranged tail-to-tail in the same location were identified in the LysM RLK1 mutant in the 10th intron (Figure 1B). RT-PCR analysis using the primers corresponding to the exon regions on the side of the 10th intron failed to detect mRNA expression in the LysM RLK1 mutant (Figure 1D). However, a truncated transcript derived from the gene sequence before the intron was detected by RT-PCR using the gene-specific primer pairs derived from the exons before the insertions (Figure 1E), suggesting that the T-DNA insertions in the intron blocked full-length transcription of the gene.
To confirm that the observed change in CRG expression was caused by the mutation in the LysM RLK1 gene, the mutant was complemented with the full-length LysM RLK1 cDNA driven by the constitutive cauliflower mosaic virus 35S promoter. As expected, complementation restored induction of CRGs (Figure 1F), confirming that it was the insertions in the LysM RLK1 gene that caused the observed change in gene expression. The complementation data also ruled out the possibility that a truncated protein translated from the observed truncated transcript may have affected the expression of the selected CRGs.
To investigate the expression pattern of this gene, RNA from different tissues was analyzed using RT-PCR. Our data showed that the LysM RLK1 gene was expressed ubiquitously in the whole plant, with the weakest expression in pollen (see Supplemental Figure 2 online). This expression pattern is similar to that revealed by DNA microarray analysis (http://www.weigelworld.org/resources/microarray/AtGenExpress and https://www.genevestigator.ethz.ch/at/index.php?page=home).
The Mutation in the LysM RLK1 Gene Blocked the Induction of Virtually All CRGs by Chitooligosaccharides
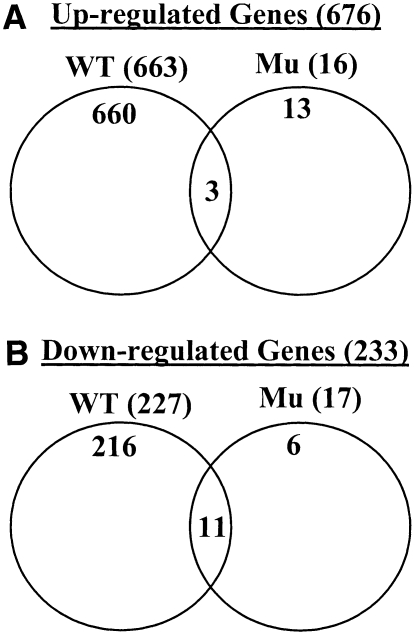
To investigate the effect of the mutation on the genome-wide gene expression in response to chitin, the Affymetrix Arabidopsis whole-genome array ATH1 (with ∼22,000 genes) was used to compare the response of the LysM RLK1 mutant and wild-type plants to purified chitooctaose. Microarray data analysis showed that a total of 890 genes responded >1.5-fold (P < 0.05) to chitooctaose elicitation in the wild-type plants 30 min after the treatment, with 663 upregulated and 227 downregulated (Figure 2; see Supplemental Table 1 online). Out of these 890 genes, 876 (660 upregulated + 216 downregulated) were responsive to chitooctaose in the wild type but not in the mutant. By contrast, only 33 genes were responsive to chitooctaose in the mutant, with 16 upregulated and 17 downregulated (Figure 2; see Supplemental Table 2 online). Among these 33 genes, 14 (three upregulated + 11 downregulated) were also similarly regulated in the wild-type plants, leaving only 19 genes that were specifically regulated by chitooctaose in the mutant. The regulation of this small number of genes by chitooctaose in the mutant may be due to some redundant function provided by one of the other four LysM RLKs and/or due to possible experimental variation, since these genes were only weakly to moderately regulated (−1.7- to 2.7-fold). Therefore, the mutation in the LysM RLK1 gene appeared to block the induction of virtually all CRGs by chitooligosaccharides, suggesting a critical role for LysM RLK1 in chitin signaling.
Figure 2.
The Mutation in the LysM RLK1 Gene Blocked the Induction of Virtually All CRGs.
(A) Upregulated genes in the wild-type and mutant (Mu) plants in response to chitooctaose, revealed by the Affymetrix Arabidopsis Whole Genome Array ATH1. Upregulated genes: 1.5-fold and P < 0.05.
(B) Downregulated genes in the wild-type and mutant (Mu) plants in response to chitooctaose. Downregulated genes: 1.5-fold and P < 0.05.
The CRGs revealed in the above microarray analysis included many defense-related genes and signal transduction–related genes (see Supplemental Table 1 online), suggesting a potential relationship between gene induction and plant defense mediated by chitooligosaccharides.
To examine whether the mutation in the LysM RLK1 gene causes any changes in gene expression in the absence of chitooligosaccharides, the mutant sample treated with water was compared with the similarly treated wild-type sample. Interestingly, the comparison revealed a significant expression change in 316 genes, with 100 upregulated and 216 downregulated in the mutant (see Supplemental Table 3 online). Out of these 316 genes, 94 overlapped with the above CRGs listed in Supplemental Table 1 online. This finding suggests that LysM RLK1 may also be involved in other events in addition to chitin signaling.
The Mutation in the LysM RLK1 Gene Led to More Susceptibility to Fungal Pathogens but Had No Effect on Bacterial Infection
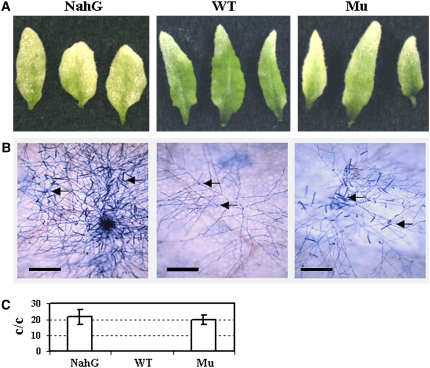
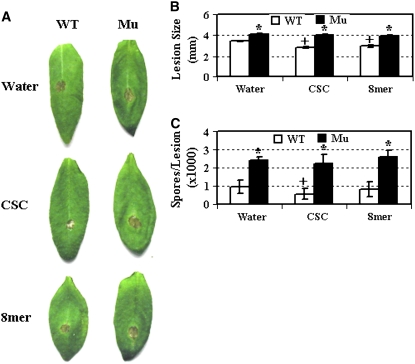
If chitooligosaccharide recognition is an integral part of the response pathway by which plants defend against fungal pathogens, then mutations in the LysM RLK1 gene should affect plant resistance to fungal pathogens. To test this, we inoculated 3-week-old mutant and wild-type plants with the biotrophic powdery mildew fungal pathogen Erysiphe cichoracearum. Ten days later, the mutant plants appeared to support more fungal growth than the wild-type plants. The susceptibility appeared to be less than that observed in NahG plants, which express a bacterial salicylate hydrolase, preventing the accumulation of salicylic acid (SA), making them very susceptible to the fungal pathogen (Figure 3A). Trypan blue staining of the infected leaves also showed that the LysM RLK1 mutant supported more hyphal growth and production of conidiophores earlier than the wild-type plants (Figure 3B). To measure the difference quantitatively, the number of conidiophores (stalks bearing asexual spores) per colony were counted 6 d after inoculation. Many more conidiophores per colony were produced on the LysM RLK1 mutant (and also on the NahG plants) than on the wild-type plants (Figure 3C). Therefore, the LysM RLK1 mutant was more susceptible to the pathogen than the wild-type plants. Additionally, we inoculated 4-week-old plants with the necrotrophic fungus Alternaria brassicicola. Three days after inoculation, the mutant developed slightly bigger lesions than the wild-type plants (Figures 4A and 4B). In agreement with this, the mutant plants also produced more spores per lesion than the wild-type plants (Figure 4C).
Figure 3.
The LysM RLK1 Mutant Was More Susceptible to a Biotrophic Fungal Pathogen Than Wild-Type Plants.
(A) Three-week-old plants were inoculated with E. cichoracearum. Pictures were taken 10 d after inoculation.
(B) Trypan blue staining showing fungal hyphae and conidiophores of E. cichoracearum on Arabidopsis leaves. Arrows indicate sites where conidiophores were forming. Pictures were taken 6 d after inoculation. Only one colony was shown in the pictures. Bars = 0.1 mm.
(C) The quantification of the number of conidiophores (stalks bearing asexual spores) per colony (c/c). Conidiophores were counted 6 d after inoculation. The average value and se were based on at least 15 replications. WT, wild-type Col-0; Mu, LysM RLK1 mutant; NahG, transgenic plants expressing a bacterial salicylate hydrolase.
Figure 4.
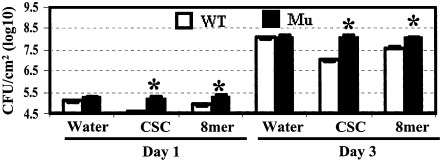
The LysM RLK1 Mutant Was More Susceptible to a Necrotrophic Fungal Pathogen Than Wild-Type Plants, and Exogenously Applied Chitooligosaccharides Enhanced Resistance in the Wild-Type Plants.
(A) Four-week-old plants were inoculated with A. brassicicola. Plants were pretreated twice (24 and 4 h before pathogen inoculation) with CSC (200 μg/mL), purified chitooctaose (8mer, 5 μM), or water. Pictures were taken 3 d after inoculation.
(B) The average diameter of the lesions caused by A. brassicicola on both the mutant and wild-type Arabidopsis leaves. The diameters of lesions from 30 leaves were measured 3 d after inoculation.
(C) The average number of spores of A. brassicicola produced per lesion by both the mutant and wild-type plants 6 d after inoculation. The number of spores per lesion was measured from 30 leaves. WT, wild-type Col-0; Mu, LysM RLK1 mutant; CSC, crab shell chitin; 8mer, chitooctaose; water, distilled water. The asterisk indicates a significant difference between the mutant and wild-type plants under the same treatments based on a Student's t test (P < 0.05). The plus symbol indicates a significance difference between the chitin-treated plants and control (water-treated) plants of the same genotype based on a Student's t test (P < 0.05). Error bars indicate se. Each experiment was repeated at least twice with similar results.
To examine how CRGs responded to A. brassicicola in both the wild-type and mutant plants, MPK3 and WRKY53 were monitored over time. Interestingly, both genes were still induced by the pathogen in the mutant, although to a lower level than that in the wild-type plants (see Supplemental Figure 3 online). As would be expected, these results show that blocking recognition of the chitin response pathway does not totally eliminate the ability of the plant to respond to pathogen invasion. This may explain why the mutant was only moderately susceptible to the fungal pathogens.
To test the specificity of LysM RLK1 in fungal disease resistance, the response of the mutant to the bacterial pathogen Pseudomonas syringae pv tomato DC3000 was also examined. Both the mutant and wild-type plants supported a similar bacterial growth in planta 1 and 3 d after infiltration with the pathogen (Figure 5), indicating that bacterial multiplication was not affected.
Figure 5.
The Mutation in the LysM RLK1 Gene Did Not Affect Growth of a Bacterial Pathogen.
Four-week-old plants were pretreated twice (24 and 4 h before pathogen inoculation) with CSC (200 μg/mL), purified chitooctaose (5 μM), or water before being infiltrated with P. syringae pv tomato DC3000. Leaf discs were collected 1 and 3 d after inoculation to determine bacterial growth. WT, wild-type Col-0; Mu, LysM RLK1 mutant; 8mer, chitooctaose; water, distilled water. Asterisks indicate that the mutant was statistically significantly different from the wild type based on a Student's t test (P < 0.05). Error bars indicate se. Each experiment was repeated at least twice with similar results.
Together, the above data suggest a direct connection between LysM RLK1-mediated chitin signaling and fungal resistance.
Exogenously Applied Chitooligosaccharides Enhanced Plant Resistance to Both Fungal and Bacterial Pathogens
To further test the role of chitin signaling and LysM RLK1 in plant defense, both the wild-type and mutant plants were pretreated with chitooligosaccharides and then tested for their response to different pathogens. Wild-type plants pretreated with a crab shell chitin (CSC) mixture had reduced disease symptoms upon inoculation with the fungal pathogen A. brassicicola, as reflected by the reduced lesion size and spore production (Figures 4A to 4C). However, the protective effect from the purified chitooctaose was not as effective as from CSC. Interestingly, pretreatment also inhibited the growth of the bacterial pathogen P. syringae pv tomato DC3000 inside plants (Figure 5), reflecting a general induction of plant innate immunity by chitooligosaccharides. By contrast, similar pretreatment of the LysM RLK1 mutant plants did not enhance resistance. These data further support the critical role of LysM RLK1 in chitin signaling and plant innate immunity. Similar enhanced fungal resistance upon chitooligosaccharide treatment was also recently observed in rice (Tanabe et al., 2006).
The Mutation in the LysM RLK1 Gene Did Not Affect Other Defense-Related Pathways
To test whether the mutation in the LysM RLK1 gene affected the SA- and jasmonic acid/ethylene-responsive pathways, the LysM RLK1 mutant and wild-type plants were treated with SA, methyl jasmonic acid (MeJA), and 1-aminocyclopropane-1-carboxylic acid (ACC), and expression of the pathway hallmark genes, PR-1 (in the SA pathway) (Ryals et al., 1996) and PDF1.2 (in the jasmonic acid/ethylene pathway) (Penninckx et al., 1996) was examined. Quantitative RT-PCR data demonstrated that both the mutant and wild-type plants showed similar induction of PR-1 by SA and of PDF1.2 by MeJA or ACC (see Supplemental Figure 4 online), suggesting that the mutation did not affect these defense pathways.
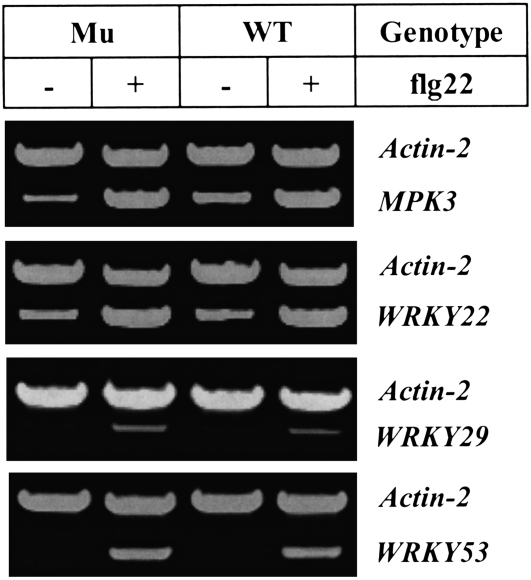
To examine whether the mutation in the LysM RLK1 gene affected the flagellin/FLS2-mediated signaling pathway, both the mutant and wild-type plants were challenged with the flagellin-derived peptide flg22 (Gomez-Gomez and Boller, 2000). The RT-PCR analysis showed that the selected flagellin-responsive genes (MPK3, WRKY22, WRKY29, and WRKY53) were similarly induced by flg22 in the mutant as in the wild-type plants (Figure 6), indicating that the flagellin signaling pathway was not affected in the mutant. Therefore, chitin signaling and flagellin signaling pathways appear to be independent, at least at the initial signal perception stages. However, increasing data suggest that different pathogen-associated molecular patterns (PAMPs) may activate a common downstream pathway to induce pathogen resistance (Asai et al., 2002; Wan et al., 2004; Zipfel et al., 2006).
Figure 6.
The LysM RLK1 Mutation Did Not Block the Induction of Flagellin-Responsive Genes.
Fourteen-day-old, hydroponically grown seedlings were treated with the flagellin-derived flg22 peptide (dissolved in DMSO) at a final concentration of 10 μM or with an equivalent amount of DMSO (as a control). The selected flagellin-responsive genes were detected using RT-PCR. Actin-2 was used as an internal control, and the amplification of both actin-2 and a flagellin-responsive gene was conducted in the same PCR reaction tube with 25 cycles. The experiment was repeated twice with similar results.
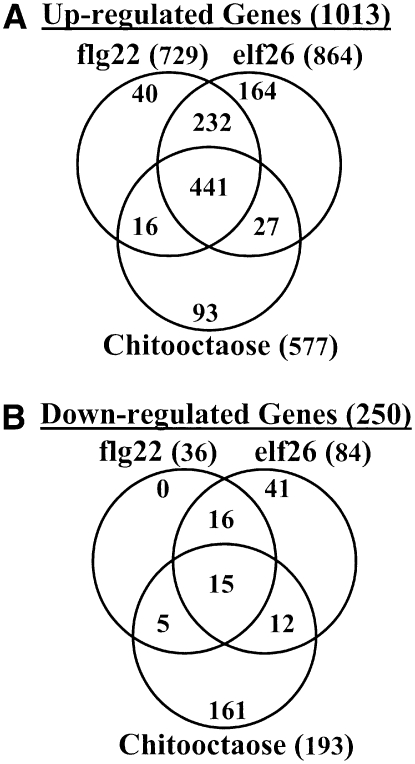
To get a better view of this possible downstream convergence, we compared the genes differentially regulated by chitin (chitooctaose, 8mer), flagellin (flg22), and EF-Tu (elf26) in Arabidopsis (Zipfel et al., 2004, 2006; this work). As expected, a large number of genes (441 genes) were commonly upregulated by all three stimuli (Figure 7; see Supplemental Table 4 online). This overlap supports that different pathways mediated by different PAMPs converge at a downstream step leading to induction of common downstream genes. However, only a small overlap (15 genes) was observed among the downregulated genes (Figure 7). Interestingly, chitooctaose appeared to downregulate many more genes than flg22 and elf26 (Figure 7; see Supplemental Table 4 online), suggesting that the plant chitin response may have unique features, compared with the response to protein-derived PAMPs.
Figure 7.
Comparison of the Genes Regulated by flg22, elf26, and Chitooctaose.
(A) Venn diagram showing the overlap of the upregulated genes (≥2-fold) by flg22, elf26, and chitooctaose.
(B) Venn diagram showing the overlap of the downregulated genes (≥2-fold) by flg22, elf26, and chitooctaose. The genes regulated by flg22 and elf26 were obtained from publications (Zipfel et al., 2004, 2006). The elf26 data (60 min after treatment) were chosen, instead of elf18 data, for comparison due to the following reasons: at 60 min, elf26 induced a comparable number of genes to that by flg22 at 30 min; at 30 min, elf26 only induced approximately half of the genes by flg22; no microarray data were available from the wild-type plants after treated with elf18; elf18 data were obtained using the fls2 mutant instead of the wild type. To be consistent with the twofold cutoff used for flg22 and efl26-regulated genes, only CRGs with a change ≥2-fold were included in the comparison.
Mutations in the Potential Nod Factor Receptors Did Not Affect Chitin Signaling in the Legume Lotus japonicus
To test whether the recently identified legume Nod factor receptors NFR1 and NFR5 are also involved in chitin signaling, the L. japonicus mutants in the these two genes (nfr1 and nfr5) (Limpens et al., 2003; Radutoiu et al., 2003) were challenged with chitin (chitooctaose) and the expression of the selected corresponding CRGs, corresponding to the Arabidopsis CRGs (Wan et al., 2004), was examined using RT-PCR. As expected, the selected CRGs (Lj MPK3, Lj WRKY22, Lj WRKY33, and Lj WRKY53) were similarly induced in the mutants as in the wild-type plants (see Supplemental Figure 5 online), indicating that chitin signaling was not affected by these mutations. Therefore, our data suggest that the Nod factor receptors have evolved to such a level that they can only recognize and perceive the modified lipochitooligosaccharides, Nod factors (Zhang et al., 2007).
DISCUSSION
LysM RLK1 Is Critical for Chitin Signaling
Since the mutation in the LysM RLK1 gene blocked the induction of virtually all CRGs, LysM RLK1 appears to be a critical component in chitin signaling. Considering its structural features (an extracellular LysM motif–containing domain + a transmembrane domain + an intracellular kinase domain), LysM RLK1 likely plays a critical role in chitin perception. The LysM (or lysin motif) domain is a protein module that binds peptidoglycan (Joris et al., 1992; Bateman and Bycroft, 2000), which is structurally similar to chitin. Legume LysM RLKs NFR1 and NFR5 were shown to be the putative receptors of Nod factors (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). However, LysM RLK1 may not function alone in chitin perception. Like many other receptor kinases (Goring and Walker, 2004), LysM RLK1 may also require a partner protein. Indeed, recently, a rice LysM domain–containing protein CEBiP (without an obvious intracellular domain) was shown to bind chitin (Kaku et al., 2006). Therefore, LysM RLK1 may act in concert, either with another LysM RLK or a protein similar to the rice CEBiP. Since mutations in the other four LysM RLK genes had no obvious effect on the expression of selected CRGs, it seems unlikely that the products of these genes are essential for chitin perception. However, there are three CEBiP-like proteins in Arabidopsis encoded by genes At1g21880, At1g77630, and At2g17120. These CEBiP-like proteins lack an obvious intracellular domain to transduce the received chitin signal. Therefore, they may function in chitin perception by interaction with LysM RLK1. We are currently examining this possibility in Arabidopsis.
During the review of this manuscript, a report by Miya et al. (2007) appeared that described similar results also showing a critical role for LysM RLK1 (termed Chitin-Elicitor Receptor Kinase [CERK1]) in chitin signaling. For example, Miya et al. (2007) also found that mutations in LysM RLK1 (CERK1) totally blocked chitin elicitation of CRG expression and conferred greater sensitivity to fungal infection. These authors were also unable to detect specific chitooligosaccharide binding proteins in various membrane preparations from wild-type Arabidopsis plants, although the same technique was able to detect a specific binding protein in the membrane preparation from suspension-cultured rice cells (Kaku et al., 2006). Hence, the question of whether LysM RLK1 (CERK1) directly binds chitin or acts via cooperation with another protein remains to be resolved. We prefer the LysM RLK1 nomenclature since it conforms to that previously used for the legume Nod factor receptors (Limpens et al., 2003) and also conforms to the recent nomenclature suggestions for naming all members of the LysM RLK family (Zhang et al., 2007).
LysM RLK1 Is Involved in Basal Resistance or Plant Innate Immunity
Since the LysM RLK1 mutation led to only moderate susceptibility to fungal pathogens and LysM RLK1 was required for the enhanced resistance to both fungal and bacterial pathogens caused by exogenously applied chitin, LysM RLK1 appears to play a role in basal resistance to pathogens through mediating chitin signaling. This result is not surprising since it is well documented that fungal pathogens produce multiple elicitors that can be recognized by other receptors in plant cells to trigger plant innate immunity (Nürnberger et al., 2004; Zipfel and Felix, 2005). Thus, blocking recognition of the chitin response pathway would not be expected to completely block the plant defense responses. This is supported by our observation that CRGs (e.g., MPK3 and WRKY53) were still induced by a fungal pathogen in the mutant, although to a lower level than that in the wild-type plants (see Supplemental Figure 3 online). This may explain why blocking chitin signaling in the LysM RLK1 mutant only led to moderate susceptibility to fungal pathogens.
The Chitin Signaling Pathway Mediated by LysM RLK1 Is Unique but May Share a Common Downstream Pathway with the Flagellin/FLS2- and EF-Tu/EFR-Mediated Pathways
This work and a previous study (Zhang et al., 2002) demonstrated that the chitin signaling pathway is independent of the SA, jasmonic acid, and ethylene pathways. Chitin perception and flagellin perception also appear to be two independent processes, since the LysM RLK1 mutant was fully responsive to the flagellin-derived peptide flg22. However, similar downstream signaling components are found in these two pathways, such as the MAPK cascade and WRKY transcription factors (Asai et al., 2002; Wan et al., 2004), suggesting that a common downstream pathway may be activated by these two different PAMPs to induce pathogen resistance. A similar overlap was also revealed between the flagellin and EF-Tu (another bacterial PAMP) signaling pathways (Zipfel et al., 2006).
The comparison of the genes differentially regulated by chitooctaose (8mer), flagellin (flg22), and EF-Tu (elf26) in Arabidopsis (Zipfel et al., 2004, 2006; this work) revealed a large number of genes (441 genes) that were commonly upregulated by all three stimuli (Figure 7A; see Supplemental Table 4 online). These data suggest that different pathways mediated by different PAMPs converge at a downstream step leading to induction of common downstream genes.
LysM RLK Family Members Are Critical for Both Nod Factor and Chitin Signaling, Suggesting a Possible Evolutionary Relationship between Two Pathways
Recently, specific legume LysM RLKs were proposed to act as the receptors for Nod factors, which are critical for rhizobial bacteria to invade and induce nodulation on their legume hosts (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Nod factors are lipochitooligosaccharides, usually composed of 3-6 N-acetyl-d-glucosamine residues (Stacey et al., 2006). This study and the work from Miya et al. (2007) have now demonstrated that another LysM RLK (LysM RLK1/ CERK1) is essential for chitin signaling (likely as a part of the receptor complex) and the induction of plant innate immunity, further supporting that LysM receptors can recognize both friend and foe (Knogge and Scheel, 2006). Collectively, these results suggest that Nod factor and chitin signaling pathways are evolutionarily related, especially at the initial perception stages. However, Nod factor recognition has become highly specific, since mutations in either of the L. japonicus NFR1 and NFR5 genes did not block the induction of the selected CRGs by chitin in this plant (see Supplemental Figure 5 online). Since chitin elicitation is widespread in plants, while Nod factor recognition is largely limited to legumes, it appears that the latter was derived from the former within the legumes. This hypothesis is also supported by phylogenetic analyses, which shows a possible evolutionary relationship between the LysM RLK1 and the Nod factor receptors (Zhang et al., 2007).
METHODS
Insertion Mutant
The LysM RLK1 insertion mutant (096F09) used in this work was generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (Max Planck Institute for Plant Breeding Research, Cologne, Germany) (Rosso et al., 2003). The homozygous plants were identified by genotyping using the gene-specific primers 5′-AGAATATATCCACGAGCACACGGTTCCAG-3′ (forward) and 5′-GACGAAAAGAGAGTGGATAAAGCAACCAC-3′ (reverse), together with the T-DNA left border primer, 5′-CCCATTTGGACGTGAATGTAGACAC-3′.
These two primers were also used to detect the expression of the LysM RLK1 gene via RT-PCR. The other primers used to detect the transcript 5′ of the insertion site were as follows: 5′-ATGAAGCTAAAGATTTCTCTAATCGCTC-3′ and 5′-GAAATGCACCATTTGGATCTCTTCCAG-3′.
Complementation
To complement the LysM LRK1 mutant, the full-length cDNA was obtained from total RNA isolated from seedlings via RT-PCR (see below for RNA isolation and cDNA synthesis). After confirmation by sequencing, the cDNA was cloned into the modified binary vector pCAMBIA1200 that contains a 35S promoter-multiple cloning site poly(A) signal downstream of the 35S promoter. The final construct was electroporated into Agrobacterium tumafaciens EHA105. The resultant A. tumafaciens was then used to transform the homozygous LysM RLK1 mutant via floral dipping (Clough and Bent, 1998). Multiple transgenic lines were obtained and raised to homozygous T3 lines.
Growth of Seedlings and Treatment with Chitooligosaccharides or Other Chemicals
Arabidopsis thaliana seedlings were grown in liquid medium as described (Zhang et al., 2002). Fourteen-day-old seedlings were treated with chitooctaose (Sigma-Aldrich) at a concentration of 1 μM or with distilled water (as a control) for 30 min. To test flagellin-responsive genes, 14-d-old seedlings were also treated with the flagellin-derived flg22 peptide (dissolved in DMSO) at a final concentration of 10 μM or with an equivalent amount of DMSO (as a control) for 30 min. To test other defense pathways, Arabidopsis seedlings were also treated for 24 h with 5 mM SA, 100 μM MeJA, and 0.5 mM ACC (all obtained from Sigma-Aldrich) and dissolved in 0.1% ethanol. The control plants were similarly treated with an equivalent amount of ethanol. After treatment, the seedlings were collected and frozen in liquid nitrogen for RNA isolation.
RNA Isolation and cDNA Synthesis
Total RNA was isolated using the Trizol reagent according to the manufacturer's instructions (Invitrogen). The isolated RNA was further purified using Qiagen RNeasy mini columns according to the manufacturer's instructions and treated with Turbo DNase (Ambion). cDNA was synthesized using M-MLV reverse transcriptase according to the manufacturer's instructions (Promega).
RT-PCR
The gene-specific primer pairs (forward and reverse) for detecting the following CRGs are as follows: 5′-CTCACGGAGGACAGTTCATAAG-3′ and 5′-GAGATCAGATTCTGTCGGTGTG-3′ for MPK3 (At3g45640); 5′-GTAAGCTCATCAGCTACTACCAC-3′ and 5′-ACCGCTAGATGATCCTCAACAG-3′ for WRKY22 (At4g01250); 5′-ATGGACGAAGGAGACCTAGAAG-3′ and 5′-CCGCTTGGTGCGTACTCGTTTC-3′ for WRKY29 (At4g23550); 5′-CTCCGACCACAACTACAACTAC-3′ and 5′-GGCTCTCTCACTGTCTTGCTTC-3′ for WRKY33 (At2g38470); 5′-CCTACGAGAGATCTCTTCTTCTG-3′ and 5′-AGATCGGAGAACTCTCCACGTG-3′ for WRKY53 (At4g23810). As an internal control, the following forward and reverse primers of actin-2 (At3g18780) were designed: 5′-GACTAAGAGAGAAAGTAAGAGATAATCCAG-3′ and 5′-CAGCCTTTGATTTCAATTTGCATGTAAGAG-3′.
To analyze gene expression, equivalent amounts of total RNA from both control and treated samples were input in cDNA synthesis (see above), and then equivalent amounts of cDNA were included in PCR reactions that contained both a gene-specific primer pair and the actin-2 primer pair under the following PCR conditions: 94°C, 3 min; 94°C, 30 s; 55°C, 30 s; 72°C, 1.5 min; 25 cycles; 72°C, 3 min. The resultant PCR products were resolved on a 1.2% agarose gel for comparison. The actin-2 band served as an internal control.
Quantitative PCR
To quantify gene expression using quantitative PCR, the forward and reverse primers of each gene were as follows: 5′- AACACGTGCAATGGAGTTTGTGGTCACT-3′ and 5′- ACCATTGTTACACCTCACTTTGGCACAT-3′ for PR-1 (At2g14610); 5′-AGTGCATTAACCTTGAAGGAGCCAAACAT-3′ and 5′-AACAGATACACTTGTGTGCTGGGAAGACA-3′ for PDF1.2 (At5g44420); 5′-TGGCCATTGATCTTGTTGACAGAATGTTGA-3′ and 5′-TCGTGCAATTTAGCAAGGTACTGGTGATT-3′ for MPK3 (At3g45640); 5′-TTTAGGCGCCAAATTCCCAAGGAGTTATT-3′ and 5′-TCTGGACTTGTTTCGTTGCCCAACAGTTT-3′ for WRKY53 (At4g23810); 5′-GGTATTCTTACCTTGAAGTATCCTATTG-3′ and 5′-CTCATTGTAGAAAGTGTGATGCCAGATC-3′ for actin-2 (At3g18780). Actin-2 was used as an internal control to normalize gene expression across different samples. The reactions were conducted on a 7500 Real-Time PCR system (Applied Biosystems) using SYBR Green Master Mix (Applied Biosystems) with the following conditions: 95°C, 10 min; 95°C, 15 s; 60°C, 1 min; 40 cycles, followed by the dissociation curve analysis to verify the single amplicon. The fold change in the target gene, normalized to actin-2 and relative to the gene expression in the control sample, was calculated as described (Libault et al., 2007).
Microarray Experiment and Data Analysis
Double-stranded cDNA was synthesized from 8 μg of purified total RNA and purified and turned into cRNA according to the Affymetrix GeneChip Expression Analysis Technical Manual. The purified biotin-labeled cRNA was fragmented and hybridized with microarrays according to the manufacturer's instructions (Affymetrix GeneChip Expression Analysis Technical Manual). Three biological replicates were included in the microarray experiment.
The scanned data (CEL or DAT files) were analyzed using the software DNA-Chip Analyzer (dChip) (version release September 23, 2005) (Li and Wong, 2001). The default setting were employed for normalization using the default array with median brightness (i.e., Mu-8mer-I) as the baseline. Model-based expression values were computed using the default settings. To identify significantly regulated genes between the two different samples, the following criteria were selected: 1.5-fold, with a t test P value < 0.05, the absolute signal intensity difference between a baseline and its corresponding treatment >100, and the P (presence) call% in the samples involved ≥20%. Additionally, the false discovery rate (FDR) was also computed by applying the above comparison criteria to the sample-wise permuted data sets and recording the number of obtained genes at each permutation. After 200 permutations, the median of the 200 values was reported as the median FDR, and the 90th percentile (90th largest value) of the 200 values was reported as 90% FDR (see the dChip website for details; www.dchip.org).
The microarray data were deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo/; accession number GSE8319).
Disease Assays
Disease assay with Erysiphe cichoracearum isolate UCSC1 was conducted as previously described (Wilson et al., 2001). Trypan blue staining of fungal structures was conducted as described (Ramonell et al., 2005). The conidiophores (stalks bearing asexual spores) per colony were counted 6 d after inoculation. Disease assay with Alternaria brassicicola was conducted as described (Van Wees et al., 2003) with a spore suspension of 5 × 105 spores/mL by dot-inoculating 5 μL of the spore solution. The disease assays with Pseudomonas syringae pv tomato DC3000 were conducted as previously described (Zhang and Gassmann, 2003) with a bacterial concentration of 5 × 104 colony-forming units/mL.
To test whether exogenously applied chitooligosaccharides can enhance defense, plants were pretreated twice (24 and 4 h before pathogen inoculation) by spraying with either CSC mixture at a final concentration of 200 μg/mL supplemented with 0.01% of Silwet L-77 or purified chitooctaose supplemented with 0.01% of Silwet L-77 at a final concentration of 5 μM until runoff. Control plants were similarly treated with water supplemented with 0.01% of Silwet L-77.
Identification and Detection of the Selected CRGs in the Legume Plant Lotus japonicus
The corresponding CRG genes in L. japonicus were identified by searching the cDNA sequences of the selected Arabidopsis CRGs (and also actin-2) used in this work against The Institute for Genomic Research Lotus japonicus Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=l_japonicus). The closest hits were chosen and arbitrarily named after their Arabidopsis counterparts with the prefix Lj (for L. japonicus). The following primer pairs were designed to detect the corresponding genes: 5′-CACCCTTGCGTAGAGAGTTTACTGATGTC-3′ and 5′-GTTGACGAGGATATTGAGGAAGTTGTCTG-3′ for Lj MPK3 (TC8079); 5′-TCACCTTGCTGGTTCTGGTTCTGGTTCTG-3′ and 5′-TCTGATAGGGGTGCAACCCCATCTTCTTC-3′ for Lj WRKY22 (AV423663); 5′-AGTTGTGGTTCAGACCACCAGTGACATTG-3′ and 5′-ACCCCATTGAGTTTCCAAACCCTGATGAG-3′ for Lj WRKY33 (TC14849); 5′-CCCATCAAAAGAACCAACCACAACAAGAG-3′ and 5′-ATCCGCACGCACTTGAACCATGTATTGTG-3′ for Lj WRKY53 (TC9074); and 5′-AAGGTTCGTAAACGATGGCTGATGCTGAG-3′ and 5′-ACCTTGATCTTCATGCTGCTAGGAGCAAG-3′ for Lj Actin-2 (TC14247). Lj Actin-2 was used as an internal control in RT-PCR reactions.
Accession Numbers
The Arabidopsis Genome Initiative accession numbers for the genes used in this study are as follows: LysM RLK1, At3g21630; LysM RLK2, At1g51940; LysM RLK3, At2g33580; LysM RLK4, At2g23770; LysM RLK5, At3g01840; MPK3, At3g45640; WRKY22, At4g01250; WRKY29, At4g23550; WRKY33, At2g38470; WRKY53, At4g23810; PR-1, At2g14610; PDF1.2, At5g44420; and actin-2, At3g18780. The accession numbers of some corresponding genes in L. japonicus were as follows: Lj MPK3, TC8079; Lj WRKY22, AV423663; Lj WRKY33, TC14849; Lj WRKY53, TC9074; and Lj Actin-2, TC14247.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Initial Screen of the Five LysM RLK Mutants in Response to Chitin Treatment.
Supplemental Figure 2. Expression Analysis of the LysM RLK1 Gene in Different Tissues or Organs.
Supplemental Figure 3. The Selected CRGs Were Still Induced in the LysM RLK1 Mutant by a Fungal Pathogen but to a Reduced Level.
Supplemental Figure 4. The Mutation in the LysM RLK1 Gene Did Not Affect Other Defense-Related Pathways.
Supplemental Figure 5. The Mutations in the Nod Factor Receptor Genes NFR1 and NFR5 in the Legume Lotus japonicus Did Not Affect the Induction of the Selected CRGs in the Plant.
Supplemental Table 1. The 890 Genes Regulated in the Wild Type by Chitooctaose.
Supplemental Table 2. The 33 Genes Regulated in the Mutant by Chitooctaose.
Supplemental Table 3. The 316 Genes Affected by the Mutation in the LysM RLK1 Gene in Absence of Chitooctaose.
Supplemental Table 4. The 456 Genes Commonly Regulated by flg22, elf26, and Chitooctaose.
Supplementary Material
Acknowledgments
We thank GABI-Kat for the T-DNA insertion mutant 096F09 (Max Plank Institute for Plant Breeding Research, Cologne, Germany), Christopher Lawrence for A. brassicicola, Walter Gassmann for P. syringae pv tomato DC3000, Scott Peck for the flg22 peptide, and Jens Stougaard for L. japonicus mutants nfr1-1 and nfr5-1. The microarray hybridization and scanning was conducted by the Keck Center of the University of Illinois at Urbana-Champaign. The research was funded by a grant from the U.S. Department of Energy (DE-FG02-02ER15309) to G.S. and a grant from the National Institutes of Health (National Institute of General Medical Sciences Grant 1 R15 GM073630-01) to K.M.R. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gary Stacey (staceyg@missouri.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arlorio, M., Ludwig, A., Boller, T., and Bonafonte, P. (1992). Inhibition of fungal growth by plant chitinases and b-1,3-glucanases: A morphological study. Protoplasma 171 34–43. [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Baier, R., Schiene, K., Kohring, B., Flaschel, E., and Niehaus, K. (1999). Alfalfa and tobacco cells react differently to chitin oligosaccharides and Sinorhizobium meliloti nodulation factors. Planta 210 157–164. [DOI] [PubMed] [Google Scholar]
- Bateman, A., and Bycroft, M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299 1113–1119. [DOI] [PubMed] [Google Scholar]
- Baureithel, K., Felix, G., and Boller, T. (1994). Specific, high affinity binding of chitin fragments to tomato cells and membranes: Competitive inhibition of binding by derivatives of chitooligosaccharides and a Nod factor of Rhizobium. J. Biol. Chem. 269 17931–17938. [PubMed] [Google Scholar]
- Boller, T. (1995). Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 189–214. [Google Scholar]
- Brogue, K., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., Mauvais, C.J., and Broglie, R. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254 1194–1197. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Day, R.B., et al. (2002). Large-scale identification of elicitor-responsive genes in suspension-cultured rice cells by DNA microarray. Plant Biotechnol. 19 153–155. [Google Scholar]
- Day, R.B., Okada, M., Ito, Y., Tsukada, N.K., Zaghouani, H., Shibuya, N., and Stacey, G. (2001). Binding site for chitin oligosaccharides in the soybean plasma membrane. Plant Physiol. 126 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., Regenass, M., and Boller, T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation and establishment of a refractory state. Plant J. 4 307–316. [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goring, D.R., and Walker, J.C. (2004). Self-rejection – A new kinase connection. Science 303 1474–1475. [DOI] [PubMed] [Google Scholar]
- Ito, Y., Kaku, H., and Shibuya, N. (1997). Identification of a high-affinity binding protein for N-acetylchitooligosaccharide elicitor in the plasma membrane of suspension-cultured rice cells by affinity labeling. Plant J. 12 347–356. [DOI] [PubMed] [Google Scholar]
- Jach, G., Gornhardt, B., Mundy, J., Logemann, J., Pinsdorf, E., Leah, R., Schell, J., and Maas, C. (1995). Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8 97–109. [DOI] [PubMed] [Google Scholar]
- Joris, B., Englebert, S., Chu, C.P., Kariyama, R., Daneo-Moore, L., Shockman, G.D., and Ghuysen, J.M. (1992). Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70 257–264. [DOI] [PubMed] [Google Scholar]
- Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., Minami, E., and Shibuya, N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knogge, W., and Scheel, D. (2006). LysM receptors recognize friend and foe. Proc. Natl. Acad. Sci. USA 18 10829–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., and Wong, W.H. (2001). Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault, M., Wan, J., Czechowski, T., Xu, D., Udvardi, M., and Stacey, G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 20 900–911. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Majeau, N., Trudel, J., and Asselin, A. (1990). Diversity of cucumber chitinase isoforms and characterization of one seed basic chitinase with lysozyme activity. Plant Sci. 68 9–16. [Google Scholar]
- Mauch, F., Mauch-Mani, B., and Boller, T. (1988). Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and b-1,3-glucanase. Plant Physiol. 88 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Okada, M., Matsumura, M., Ito, Y., and Shibuya, N. (2002). High-affinity binding proteins for N-acetylchitooligosaccharide elicitor in the plasma membranes from wheat, barley and carrot cells: Conserved presence and correlation with the responsiveness to the elicitor. Plant Cell Physiol. 43 505–512. [DOI] [PubMed] [Google Scholar]
- Passarinho, P., and de Vries, S.C. (2002). Arabidopsis chitinase: A genomic survey. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.002, http://www.aspb.org/publications/arabidopsis/.
- Peck, S.C., Nuhse, T.S., Hess, D., Iglesias, A., Meins, F., and Boller, T. (2001). Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Ramonell, K., Berrocal-Lobo, M., Koh, S., Wan, J., Edwards, H., Stacey, G., and Somerville, S. (2005). Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 138 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell, K.M., Zhang, B., Ewing, R.M., Chen, Y., Xu, D., Stacey, G., and Somerville, S. (2002). Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol. Plant Pathol. 3 301–311. [DOI] [PubMed] [Google Scholar]
- Roby, D., Broglie, K., Cressman, R., Biddle, P., Chet, I., and Broglie, R. (1990). Activation of a bean chitinase promoter in transgenic tobacco plants by phytopathogenic fungi. Plant Cell 2 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumbaum, A., Mauch, F., Vögeli, U., and Boller, T. (1986). Plant chitinases are potent fungal growth inhibitors. Nature 324 365–367. [Google Scholar]
- Shibuya, N., and Minami, E. (2001). Oligosaccharide signalling for defence responses in plant. Physiol. Mol. Plant Pathol. 59 223–233. [Google Scholar]
- Stacey, G., Libault, M., Brechenmacher, L., Wan, J., and May, G.D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9 110–121. [DOI] [PubMed] [Google Scholar]
- Stacey, G., and Shibuya, N. (1997). Chitin recognition in rice and legumes. Plant Soil 194 161–169. [Google Scholar]
- Tanabe, S., Okada, M., Jikumaru, Y., Yamane, H., Kaku, H., Shibuya, N., and Minami, E. (2006). Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosaccharide. Biosci. Biotechnol. Biochem. 70 1599–1605. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A., Harrison, S.J., Joosten, M.H., Vervoort, J., and de Wit, P.J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19 1420–1430. [DOI] [PubMed] [Google Scholar]
- Van Wees, S.C.M., Chang, H.-S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J., Zhang, S., and Stacey, G. (2004). Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 5 125–135. [DOI] [PubMed] [Google Scholar]
- Wilson, I.W., Schiff, C.L., Hughes, D.E., and Somerville, S.C. (2001). Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics 158 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubben, J.P., Joosten, M.H.A.J., Van Kan, J.A.L., and De Wit, P.J.G.M. (1992). Subcellular localization of plant chitinases and 1,3-beta-glucanases in Cladosporium fulvum (syn. Fulvia fulva) infected tomato leaves. Physiol. Mol. Plant Pathol. 41 23–32. [Google Scholar]
- Zhang, B., Ramonell, K., Somerville, S., and Stacey, G. (2002). Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 15 963–970. [DOI] [PubMed] [Google Scholar]
- Zhang, X.-C., and Gassmann, W. (2003). RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 15 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.-C., Wu, X., Findley, S., Wan, J., Libault, M., Nguyen, H.T., Cannon, S.B., and Stacey, G. (2007). Molecular evolution of LysM type receptor-like kinases in plants. Plant Physiol. 144 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., and Felix, G. (2005). Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 8 353–360. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J.D., Boller, T., and Felix, G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.