Abstract
The infection of plants by obligate parasitic nematodes constitutes an interesting model for investigating plant cytoskeleton functions. Root knot nematodes have evolved the ability to manipulate host functions to their own advantage by redifferentiating root cells into multinucleate and hypertrophied feeding cells. These giant cells result from repeated rounds of karyokinesis without cell division. Detailed functional analyses demonstrated that Arabidopsis thaliana Microtubule-Associated Protein65-3 (MAP65-3) was essential for giant cell ontogenesis and that cytokinesis was initiated but not completed in giant cells. In developing giant cells, MAP65-3 was associated with a novel kind of cell plate—the giant cell mini cell plate—that separates daughter nuclei. In the absence of functional MAP65-3, giant cells developed but failed to fully differentiate and were eventually destroyed. These defects in giant cells impaired the maturation of nematode larvae. Thus, MAP65-3 is essential for giant cell development during root knot nematode infection. Subcellular localization of MAP65-3 and analysis of microtubule organization in the dyc283 T-DNA map65-3 mutant demonstrated that MAP65-3 played a critical role in organizing the mitotic microtubule array during both early and late mitosis in all plant organs. Here, we propose a model for the role of MAP65-3 in giant cell ontogenesis.
INTRODUCTION
The spatial organization of microtubules (MTs) is radically modified during cell cycle progression, particularly at the G2/M-phase transition and throughout mitosis. At the onset of mitosis, cortical arrays are progressively replaced by a densely packed ring of MTs encircling the nucleus. This structure is called the preprophase band (PPB) and is later used to guide the outgrowing cell plate along the required division plane (Van Damme et al., 2007). As the cells progress into mitosis, MTs reorganize into an anastral bipolar spindle, ensuring the accurate segregation of chromosomes during anaphase. The phragmoplast forms toward the end of mitosis and directs Golgi-derived vesicles toward the expanding cell plate during cytokinesis (Wasteneys, 2002). Mitotic MT organization is controlled in a dynamic manner by Microtubule-Associated Proteins (MAPs) acting at the growing ends of the MTs (Kline-Smith and Walczak, 2004). The organization of MT arrays is thought to be controlled by motor and nonmotor MAPs, which integrate individual MTs into complex arrays. In plants, several classes of nonmotor MAPs have been characterized by biochemical and genetic approaches: MAP65 (Jiang and Sonobe, 1993), MAP190 (Igarashi et al., 2000), and MAP215/Microtubule Organization1 (MOR1) (Whittington et al., 2001).
Plant MAP65s were first purified from tobacco (Nicotiana tabacum) and carrot (Daucus carota) MT preparations as 65-kD proteins (Jiang and Sonobe, 1993; Chan et al., 1996). Nine genes encoding MAP65s have been identified in Arabidopsis thaliana (Hussey et al., 2002). Biochemical studies have shown that MAP65s bind and bundle MTs in vitro (Jiang and Sonobe, 1993; Chan et al., 1996; Wicker-Planquart et al., 2004). The ability to bind MTs depends on a conserved motif located in the C-terminal half of the protein (Smertenko et al., 2004). Plant MAP65s share a large conserved domain with Saccharomyces cerevisiae Anaphase Spindle Elongation Factor1 (Ase1p) and human Protein Regulation Cytokinesis1 (PRC1) proteins, which are involved in central spindle formation and cytokinesis (Pellman et al., 1995; Mollinari et al., 2002; Schuyler et al., 2003). Recently, Schizosaccharomyces pombe Ase1p was shown to organize antiparallel cytoplasmic MT bundles during both interphase and mitosis (Daga and Chang, 2005; Loiodice et al., 2005).
Plant MAP65s differ with respect to their activities, functions, and target MTs. Transient expression experiments in tobacco cells demonstrated that several members of MAP65s localized to the phragmoplast and to other MT-based structures such as cortical MTs, the PPB, and mitotic spindles (Van Damme et al., 2004). In Arabidopsis, MAP65-6 is associated with mitochondria (Mao et al., 2005), and MAP65-1, which is expressed ubiquitously in all organs and tissues other than the anthers and petals, is associated with interphasic and mitotic MT arrays. MAP65-1 MT binding activity is regulated in a cell cycle–dependent manner (Smertenko et al., 2004; Chang et al., 2005). MAP65-3/PLE was isolated in a genetic screen for root morphogenesis mutants in Arabidopsis (Müller et al., 2002). The map65-3/ple mutants display defects in cytokinesis in the root meristem, presumably because phragmoplast organization is compromised (Müller et al., 2004).
Host–pathogen interactions constitute an interesting model for investigating cytoskeleton function. Nematodes can induce long-term changes in the organization of the plant cytoskeleton (de Almeida Engler et al., 2004). The parasitic nematode Meloidogyne spp establishes and maintains permanent multinucleate giant cells in the root of the host plant. Giant cells provide the developing nematode with nutrients and are essential for the growth and reproduction of this obligate biotrophic pathogen. Nematode larvae penetrate the root and then migrate from cell to cell, inducing the dedifferentiation of five to seven vascular cells in the root. These cells enlarge considerably and become multinucleate through synchronous repeated nuclear divisions without cytokinesis (Jones and Payne, 1978). Hypertrophied mature giant cells contain >100 polyploid nuclei, which have also undergone extensive endoreduplication (Wiggers et al., 1990) and have a dense granular cytoplasm with numerous organelles (Jones, 1981). The parenchyma cells surrounding the giant cells undergo hyperplasia and hypertrophy, which lead to root knot (gall) formation, the most visible symptom of infection. It remains unclear how these nematodes cause such alterations, but it is thought that they secrete proteins that directly affect host cells (Davis et al., 2004). Knowledge of full genome sequences of Meloidogyne incognita (P. Abad; http://meloidogyne.toulouse.inra.fr/) and Meloidogyne hapla (C. Opperman, D. Bird, and V. Williamson; http://www.hapla.org) will provide opportunities for studying plant–nematode interactions.
Characterizing the plant genes required for giant cell development may give insight into how nematodes are able to manipulate host functions to their own advantage. The complexity of giant cell ontogenesis is reflected in the extensive changes in gene expression observed in infected root cells (Gheysen and Fenoll, 2002; Jammes et al., 2005; Caillaud et al., 2008). Genes involved in diverse processes, such as cell cycle activation (de Almeida Engler et al., 1999), cell wall modification (Goellner et al., 2001), and hormone and defense responses (Lohar et al., 2004; Jammes et al., 2005), are differentially expressed during giant cell formation. Large genetic screens and knockouts of genes activated in giant cells have led to the characterization of only a few mutants in which nematode infection was reduced (Gheysen and Fenoll, 2002; Caillaud et al., 2008). Only Ribulose-5-Phosphate Epimerase, which encodes a key enzyme in the pentose phosphate pathway, has been identified in knockout studies as being essential for giant cell formation (Favery et al., 1998). The distribution of MTs and microfilaments in these nematode feeding cells has recently attracted considerable attention. A functional mitotic apparatus containing multiple large spindles is present throughout the multiple mitotic events observed in giant cells. During giant cell expansion, the actin cytoskeleton displays a particular organization, with large numbers of unusual, randomly oriented actin bundles and cables (de Almeida Engler et al., 2004). Recently, candidate genes involved in reorganizing the actin cytoskeleton of giant cells have been characterized (Favery et al., 2004; Jammes et al., 2005).
Here, we investigated the molecular and cellular mechanisms underlying giant cell ontogenesis by carrying out detailed functional analyses of Arabidopsis MAP65-3 in planta. We found that the MAP65-3 gene was expressed during the early stages of nematode-induced giant cell formation. During plant development, MAP65-3 was expressed in all dividing cells and was regulated by both transcriptional and posttranscriptional mechanisms. Phenotypic analysis of map65-3 mutants showed that MAP65-3 played a key role in both karyokinesis and cytokinesis in all plant organs. In the absence of MAP65-3, giant cells were induced but failed to complete their differentiation. Functional analyses in planta and nematode response analysis revealed unpredicted roles for the MAP65-3 protein in dividing plant cells.
RESULTS
MAP65-3 Is Expressed in Developing Giant Cells Induced by Root Knot Nematodes
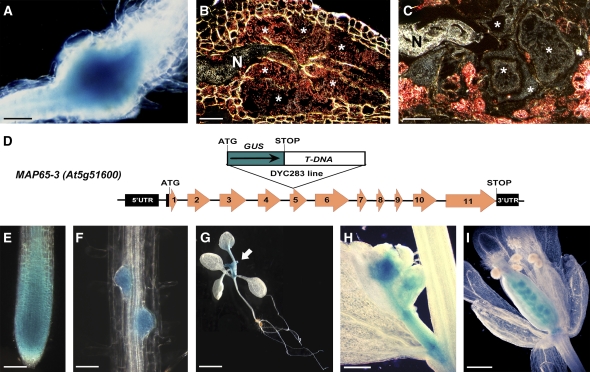
We previously used a promoter trap strategy to isolate genes involved in giant cell formation in response to M. incognita infection. We screened 20,000 T-DNA–tagged Arabidopsis lines by β-glucuronidase (GUS) assay after root knot nematode infection and identified lines showing GUS induction in root galls (Favery et al., 2004). One of these lines, DYC283, displayed early GUS activity in galls (Figure 1A) that was first detected <48 h after giant cell initiation (n > 20 observations; 3 d after infection). During the giant cell maturation phase (5 to 14 d after infection), sections through galls clearly showed GUS staining in developing giant cells and in the surrounding dividing cells (n > 30; 7 d after infection) (Figure 1B). At 21 d after infection, when mature galls contained fully expanded and differentiated giant cells, GUS expression was detected in the surrounding cells but not in the mature giant cells (n > 20; Figure 1C). Thus, the DYC283 line displayed GUS expression restricted to the giant cell initiation and maturation phases.
Figure 1.
The DYC283 T-DNA–Tagged Arabidopsis Line Displays GUS Activity in Giant Cells Induced by M. incognita and during Plant Development.
(A) to (C) GUS expression in galls induced by M. incognita. Localized GUS activity is seen in a root gall at 7 d after infection (A). The sectioned gall shown in (A) is seen by dark-field microscopy (GUS activity is seen as a pink precipitate) in (B). (C) shows a sectioned gall at 21 d after infection observed by dark-field microscopy. Asterisks, giant cells; N, nematode.
(D) Organization of the MAP65-3 gene and molecular analysis of the T-DNA insertion. Boxes indicate exons. GUS corresponds to the coding region of the β-glucuronidase gene present on the T-DNA. The ATG initiation and stop codons are indicated. UTR, untranslated region.
(E) to (H) GUS expression during plant development. Reporter gene activity was observed in root meristem (E), lateral root meristem (F) developing leaves (arrow) of a 14-d-old seedling (G), buds (H), and ovules (I).
Bars = 100 μm in (A), 25 μm in (B) and (C), 50 μm in (E) and (F), 1 cm in (G), and 150 μm in (H) and (I).
The DYC283 line presented a 3:1 segregation of the kanamycin marker carried by the T-DNA. We characterized the insertion site by sequencing the genomic regions flanking the inserted T-DNA (FST project; Samson et al., 2002). Sequence analysis, using The Arabidopsis Information Resource, showed that the T-DNA had integrated into MAP65-3. This gene has 11 exons and encodes a 707–amino acid protein, MAP65-3, from the MAP65 family (Müller et al., 2004). In the DYC283 line, the T-DNA was inserted into the fifth exon, placing the GUS gene in-frame with the MAP65-3 gene and resulting in a functional gene fusion (Figure 1D).
During regular plant development, the GUS gene was expressed in the root meristem, the root elongation zone (Figure 1E), and in lateral root primordia (Figure 1F). In aerial parts of the plant, GUS expression was observed in young leaves, buds, and ovules (Figures 1G to 1I). We confirmed the MAP65-3 expression pattern by transformation with ProMAP65-3:GFP:GUS (see Supplemental Figure 1 online). Green fluorescent protein (GFP) expression analysis showed that MAP65-3 was expressed in all tissues enriched in dividing cells, such as the root and shoot apical meristem, foliar primordia, and young leaves, and during all stages of embryonic development.
MAP65-3 Disruption Leads to Defects in Karyokinesis and Cytokinesis
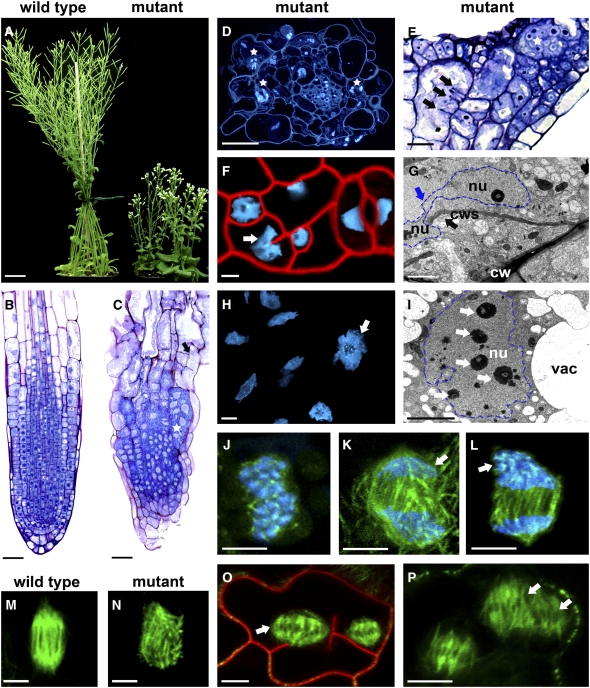
The aerial parts of the plant were very small in plants homozygous for the T-DNA mutation (dyc283 mutant) (Figure 2A). Despite this dwarf phenotype, no organ fusions or abnormal numbers of organs were observed (n > 30). The dyc283 mutant was fertile but had ∼25% fewer seeds per silique than normal. Sections through the dyc283 mutant root revealed polynucleate, hypertrophied cortical and epidermal cells (n = 41; Figures 2B to 2D). Vascular cylinder cells were less affected by the mutation but often showed cell misalignments (n = 17; Figure 2D), as observed in the ethyl methanesulfonate (EMS)–induced ple mutants (Müller et al., 2002). Hypocotyl and leaf primordia had polynucleate cells and aberrant cell wall stubs (Figure 2E), which were also observed during embryogenesis in the dyc283 mutant (see Supplemental Figure 2 online). Strikingly, in some dyc283 mutant cells, the nucleus did not divide and curved around the branched end of the cell wall stub (2.1 ± 0.3 [mean ± sd] per root; n = 7 plants) (Figures 2F and 2G). In young leaves, some nuclei were irregularly lobed and amoeboid in form (4.6 ± 0.6 abnormal nuclei per leaf observed; n = 17 plants) (Figure 2H). Electron microscopy of the dyc283 mutant confirmed the presence of abnormally large nuclei with additional nucleoli (4.7 ± 0.8 nucleoli per enlarged nucleus; n = 29) (Figure 2I).
Figure 2.
MAP65-3 Disruption Leads to Defects in Karyokinesis and Cytokinesis in All Plant Organs.
(A) The aerial parts of the dyc283 mutant were much smaller than those of a wild-type plant grown on soil for a period of 7 weeks.
(B) Longitudinal section through a wild-type root.
(C) Longitudinal section through a dyc283 mutant revealed multinucleate, hypertrophied cells (star) and aberrant cell wall stubs (arrow).
(D) Transverse section through a dyc283 mutant root stained with 4′,6-diamidino-2-phenylindole showed polynucleate (stars) and hypertrophied cells.
(E) Longitudinal section through dyc283 mutant hypocotyl and leaf primordia revealed polynucleate cells (star) and aberrant cell wall stubs (arrows).
(F) A dyc283 mutant expressing H2B:YFP and stained with the membrane dye FM4-64 exhibited multinucleate root cells and cell wall stubs. The arrow shows an aberrant nucleus, which did not divide and curved around the branched end of the cell wall stub.
(G) dyc283 mutant root electron microscopy confirmed the presence of nuclei that did not divide (blue arrow) and curved around the branched end of the cell wall stub (black arrow).
(H) In leaf primordia, nuclei were often irregularly lobed and amoeboid in form (arrow).
(I) Electron microscopy of a dyc283 mutant root showing abnormal enlarged nuclei with greater than normal numbers of nucleoli (arrows).
(J) to (P) Mitotic spindle in dividing cells from leaf primordia of 7-d-old seedlings in dyc283 T-DNA mutant ([J] to [L] and [N] to [P]) and wild-type (M) plants. In some dyc283 mutant cells, MTs appeared mixed with condensed chromosomes in prometaphase (J). At the end of anaphase, aberrant chromosome number was observed at the spindle poles (arrows) ([K] and [L]).
(O) and (P) MT spindle morphogenesis defect in multinucleate cells. Arrows show metaphase spindles enlarged and/or aberrant in shape. The plasma membrane was stained by FM4-64 (O).
cw, cell wall; cws, cell wall stub; nu, nucleus; vac, vacuole. Bars = 2 cm in (A), 50 μm in (B) and (C), 20 μm in (D) and (E), 1 μm in (G) and (I), and 5 μm in (F), (H), and (J) to (P).
We used the MAP4 microtubule binding domain (MBD):GFP reporter protein to visualize dynamic changes in the organization of the MT cytoskeleton in living cells of the dyc283 mutant and wild-type plants. In a subset of dyc283 mutant premitotic cells (n = 5 of 50 cells), MTs were mixed with condensed chromosomes during prometaphase (Figure 2J). In addition, some dyc283 dividing cells had MT spindle defects (n = 10 of 100 spindles examined in 10 plants; Figures 2K to 2L and 2N). No spindle defects were observed in MBD-GFP control cells (n = 100; Figure 2M). In dyc283 multinucleate cells, metaphase spindles were frequently enlarged (n > 15 of 100 examined in 10 plants; Figure 2O) and/or aberrant in shape (n > 15 of 100 examined in 10 plants; Figure 2P). Thus, abnormal, enlarged, irregularly lobed, amoeboid nuclei in the dyc283 mutant seem to result from a defect in MT organization in early mitosis.
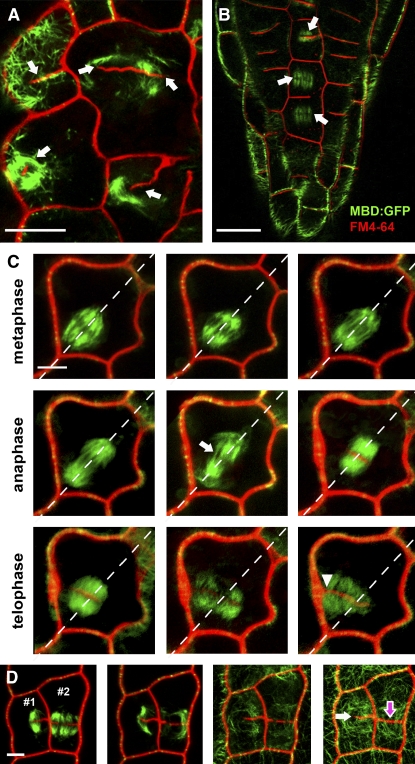
We performed a real-time analysis of cell plate deposition and MT organization to investigate the formation of cell wall stubs in affected cells (Figure 3). In the dyc283 mutant, phragmoplasts were often misplaced or wavy (n > 30 of 75 examined; Figures 3A and 3B). Mitotic MT dynamics were analyzed from metaphase to late cytokinesis (Figure 3C). In dyc283 mutant cells with a metaphase spindle of normal appearance (n = 4; Figure 3C), the late anaphase spindle became disorganized among the phragmoplast initials. However, the cell plate seemed to be correctly located in the midline of the early phragmoplast. During telophase, phragmoplast MTs expanded until one side of the phragmoplast contacted the cell's perimeter. On the other side, phragmoplast MTs collapsed, preventing complete fusion of the cell plate with the mother cell wall (Figures 3C and 3D). New MTs then extended from the collapsed phragmoplast toward the cell cortex and new MT bundles formed (Figure 3D). Thus, in the dyc283 mutant, the cell wall stubs originate from a failure of phragmoplast MTs to complete cytokinesis.
Figure 3.
In the dyc283 Mutant, the Cell Wall Stubs Originate from a Failure of Phragmoplast MTs to Complete Cytokinesis.
MT organization during cell wall stub formation stained by FM4-64 (red channel) and in the dyc283 mutant expressing the GFP:MAP4 MBD (green channel).
(A) and (B) In the dyc283 mutant, phragmoplasts were often misplaced or wavy (arrows).
(C) Time-lapse analysis from metaphase to late cytokinesis of a dyc283 mutant cell expressing GFP-MBD, showing a disorganized late anaphase spindle (arrow). During telophase, the cell plate fused with the mother cell wall on one side of the cell only (arrowhead).
(D) Time-lapse analysis from late cytokinesis to interphase in the dyc283 mutant cell. In cell 1, phragmoplast expansion led to the fusion of the cell plate with the mother cell wall on one side of the cell only. MT bundles formed around the aberrant cell wall stub (white arrow). In cell 2, phragmoplast expansion led to the formation of a complete cell wall (pink arrow) separating the two daughter cells.
Bars = 10 μm in (A), (B), and (D) and 5 μm in (C).
In the Absence of MAP65-3, Giant Cells Are Induced but Do Not Differentiate Fully
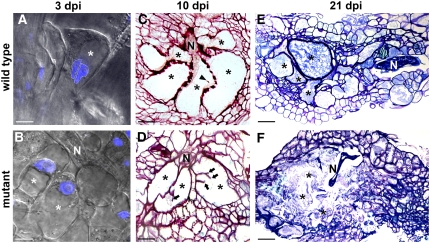
We examined the response of dyc283 to the nematode M. incognita. The infective second-stage juvenile (J2) was able to invade the dyc283 mutant root tissue by penetrating the zone of elongation, migrating along the vascular cylinder, and inducing a gall, similar to that observed in wild-type plants. In wild-type plants, the first sign of giant cell induction is the formation of vascular binucleate cells (n = 20; Figure 4A). At this early stage, a single enlarged nucleus was observed in dyc283 mutant giant cells (Figure 4B). Observations of gall sections at 10 d after infection showed that the nematode had initiated feeding sites composed of three to five giant cells in both the wild type and the dyc283 mutant (n > 20; Figures 4C and 4D). The giant cells of the dyc283 mutant were slightly smaller than those of the wild type (Figure 4D). At early stages of wild-type giant cell ontogenesis, structures resembling cell wall fragments were frequently observed between two daughter nuclei (n > 40 of 35 giant cell sections examined; Figure 4C; see Supplemental Figure 3 online). However, these small cell wall structures were never observed in dyc283 mutant giant cells. Instead, unusual cell wall stubs were observed in the dyc283 mutant giant cells (n > 20; Figure 4D). Once the nematode feeding site had been initiated, J2 nematodes became sedentary and developed into third-stage juveniles. Later in the interaction, nematodes developed into the fourth developmental stage in the wild type, whereas nematode development remained arrested at the third juvenile stage in the dyc283 mutant (n > 30; see Supplemental Figure 4 online). Neither fourth-stage juvenile nematodes nor mature giant cells resembling those of the wild type (Figure 4E) were ever observed in the dyc283 mutant. Instead, dead nematodes and degenerated giant cells were observed at 21 d after infection (n = 10; Figure 4F). Consequently, no females were observed on the surface of dyc283 mutant roots (see Supplemental Figure 4 online). Incomplete nematode development indicates a defect in giant cell formation in the dyc283 mutant.
Figure 4.
Defects in Giant Cell Ontogenesis in the Absence of MAP65-3.
(A) and (B) Nuclei in young giant cells (3 d after infection [dpi]) induced by M. incognita in plants expressing H2B:YFP (blue channel). In the control wild-type plant, the giant cell was binucleate (A). In the dyc283 mutant plant, a single enlarged nucleus was observed (B).
(C) and (D) Cross sections at 10 d after infection through the galls of a wild-type plant (C) and a dyc283 plant (D). The wild-type giant cell presented structures resembling cell wall fragments (arrowhead) (C). Unusual cell wall stubs (arrows) were observed in the dyc283 mutant giant cells (D).
(E) and (F) Cross sections at 21 d after infection through the galls of a wild-type plant (E) and a dyc283 mutant plant (F). In the wild-type plant, giant cells were mature and nematode developed into the fourth juvenile stage (E). In the dyc283 mutant plant, giant cells decayed and nematode development remained arrested at the third juvenile stage (F).
Asterisks, giant cells; N, nematode. Bars = 20 μm in (A) to (D) and 40 μm in (E) and (F).
MAP65-3:Yellow Fluorescent Protein Fusions Complement the Mutant Phenotype
Analysis of heterozygous DYC283 plants revealed no aberrant cellular phenotype, confirming the recessive nature of the mutation. We screened Arabidopsis mutant collections for loss-of-function insertion mutants and identified an additional independent map65-3 allele from the Institut National de la Recherche Agronomique Versailles collection (Samson et al., 2002). The EBJ96 line carries a single T-DNA insertion in the 11th exon of MAP65-3. As expected, plants homozygous for the ebj96 mutation had a phenotype similar to that of the dyc283 mutant. We complemented the mutant phenotype by constructing two MAP65-3:YFP (for Yellow Fluorescent Protein) fusions, YFP:MAP65-3 and MAP65-3:YFP, under ProMAP65-3 control. When introduced into dyc283 or ebj96 mutants, both of these constructs restored the wild-type root and shoot phenotype (see Supplemental Figure 5 online). In these dyc283 and ebj96 transformants (n = 10), giant cells developed normally and nematodes had a complete life cycle (see Supplemental Figure 4 online). These results demonstrate that the map65-3 recessive mutation was responsible for the observed phenotype.
MAP65-3 Is Restricted to Mitotic Cells in Planta
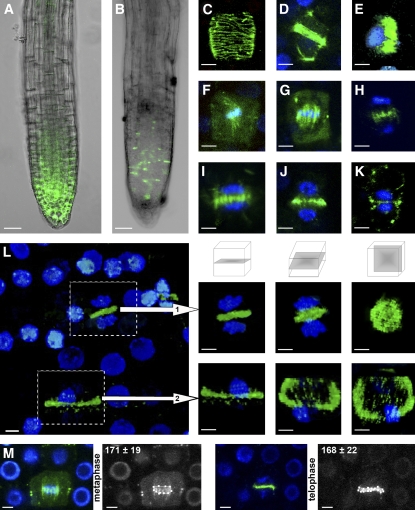
We characterized the specific cellular distribution of MAP65-3 in planta using Arabidopsis plants transformed with the functional ProMAP65-3:GFP:MAP65-3 fusion. Unlike the ProMAP65-3:GFP:GUS fusion, which was strongly expressed in all meristematic root cells (Figure 5A), the GFP:MAP65-3 signal was detected specifically in cells undergoing cell division (Figure 5B). No signal was detected in interphase cells. Just before mitosis, MAP65-3 was associated with cortical MT arrays (n > 20; Figure 5C). In early prophase, MAP65-3 was present in the PPB (n > 7; Figures 5D and 5E). During metaphase, cell division was mostly oriented transversely or longitudinally with respect to the cell axis, as predicted by the transverse or longitudinal position of the PPB. At this stage, MAP65-3 was associated with MT spindles (n > 25; Figures 5F and 5G). MAP65-3 was also distributed diffusely through the cytoplasm during metaphase only (Figures 5F and 5G). After chromosome separation, MAP65-3 was found in the anaphase midline (n > 5; Figure 5H). During the formation of daughter nuclei in early telophase, an intense signal for MAP65-3 was associated with the early phragmoplast array (n > 30; Figure 5I). During cytokinesis, MAP65-3 began to accumulate at the cell center, with a fluorescence intensity peak near the phragmoplast midline (n > 30; Figure 5J). Punctate labeling of MAP65-3 was observed, extending from the center to the periphery, separating the two daughter cells (n > 30; Figure 5K). Rotated projections of the MAP65-3 signal revealed that, in telophase, MAP65-3 was associated with the entire phragmoplast midline (n > 20; Figure 5L). At the end of cytokinesis, the MAP65-3 signal was observed only at the cell periphery, forming a ring around the newly formed cell plate (n > 20; Figure 5L). Total MAP65-3:GFP fluorescence quantification during metaphase (171 ± 19) and telophase (168 ± 22) within a given cell (n = 5) of ProMAP65-3:GFP:MAP65-3 plants revealed no significant difference (t test, P = 0.47; Figure 5M). This suggests that MAP65-3 is relocalized from the metaphase spindle MTs and the cytoplasm of the cell to the phragmoplast midline.
Figure 5.
MAP65-3 in Planta Localization throughout the Cell Cycle.
(A) Expression pattern of ProMAP65-3:GFP:GUS fusion in an Arabidopsis root (green channel).
(B) Localization of the MAP65-3:GFP fusion under the control of the MAP65-3 promoter in an Arabidopsis root (green channel).
(C) to (K) Changes in the distribution of MAP65-3 in Arabidopsis root meristematic cells expressing H2B:YFP (blue channel) and GFP:MAP65-3 (green channel). MAP65-3 labeled cortical MTs in G2 (C), PPB in preprophase ([D] and [E]), metaphase spindle ([F] and [G]), anaphase spindle (H), and phragmoplast ([I] to [K]).
(L) Rotated projections of the MAP65-3 signal. In telophase (1), MAP65-3 was associated with the entire phragmoplast midline. At the end of cytokinesis (2), MAP65-3 was observed only in the cell periphery, forming a ring around the newly formed cell plate.
(M) MAP65-3:GFP fluorescence quantification during both metaphase and telophase within a given cell. The region corresponding to the MAP65-3 signal was defined using LSM Image Browser software. The total MAP65-3:GFP fluorescence intensity was similar during metaphase and telophase within a given cell (t test).
Bars = 20 μm in (A) and (B) and 5 μm in (C) to (M).
MAP65-3 Colocalizes with All Mitotic MT Arrays and Cell Plate Deposition in Somatic Cells
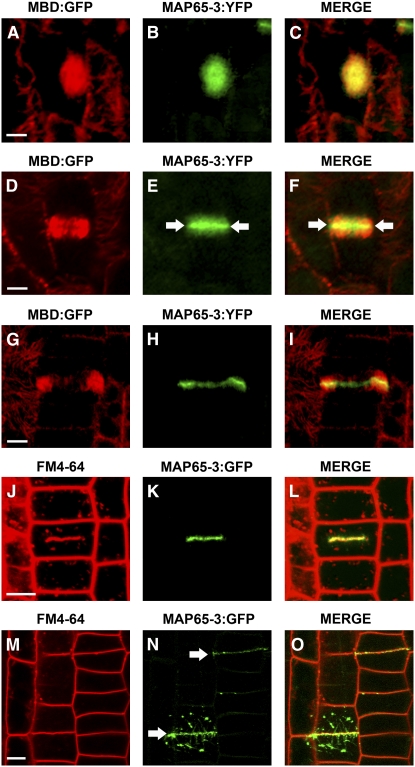
Imaging of cells simultaneously expressing both MBD:GFP and MAP65-3:YFP demonstrated the presence of MAP65-3 in all MT arrays during mitosis and early cytokinesis in planta (Figure 6). At metaphase, MAP65-3 colocalized with the MT arrays forming the bipolar spindle (n > 10; Figures 6A to 6C). After anaphase, MTs were organized into a phragmoplast array with a dark midline between the two mirror halves (n = 15; Figure 6D). At this stage, MAP65-3 was concentrated at the phragmoplast midline and appeared to diffuse away from this region (n > 10; Figures 6E and 6F). When the phragmoplast was fully expanded, MTs depolymerized from the central region of the phragmoplast, as indicated by weak MT labeling (Figure 6G). MAP65-3 remained associated with phragmoplast MTs, and its signal was most pronounced toward the midline (Figures 6H and 6I). The MAP65-3 signal was most intense toward the periphery of the phragmoplast, where most of the MTs were present (n > 10; Figure 6I).
Figure 6.
MAP65-3 Colocalizes with Mitotic MT Arrays and Newly Formed Cell Plates in Planta.
(A) to (I) Localization of MAP65-3 in Arabidopsis root meristematic cells coexpressing MBD:GFP (red channel) and MAP65-3:YFP (green channel).
(A) to (C) During metaphase, MBD (A) and MAP65-3 (B) signals colocalized in the metaphase spindle (C).
(D) to (F) During cytokinesis, MBD labeled the two mirror halves of the phragmoplast (D). MAP65-3 labeled the phragmoplast (E) but also the center of the forming cell plate (arrows) (F).
(G) to (I) MTs depolymerized from the central region of the phragmoplast, as indicated by the weak MBD signal (G). MAP65-3 remained associated with phragmoplast MTs, and its signal was most pronounced toward the midline (H). The MAP65-3 signal was most intense toward the periphery of the phragmoplast, where most of the MTs were present (I).
(C), (F), and (I) are merged images in which the yellow coloration corresponds to MBD:GFP and MAP65-3:YFP colocalization.
(J) to (O) Cell plate formation in an Arabidopsis root meristematic cell expressing GFP:MAP65-3.
(J) to (L) The membrane dye FM4-64 (red channel) (J) and GFP:MAP65-3 signal (green channel) (K) colocalized at the newly formed cell plate (L).
(M) to (O) When the two daughter cells were completely separated, as indicated by the FM4-64 stain (M), MAP65-3 colocalized with the edge of the fully expanded newly formed cell plate (arrows) (N). Organelle-like structures were seen in the cytoplasm of the two daughter cells at the end of cytokinesis (N).
(L) and (O) are merged images in which the yellow coloration corresponds to FM4-64 and MAP65-3:YFP colocalization.
Bars = 5 μm.
As MAP65-3 localized to the phragmoplast midline in late telophase, we examined the possible colocalization of MAP65-3 with the newly formed cell plate. During the centrifugal growth of the cell plate from the center of the cell to the periphery, MAP65-3 colocalized with the fluorescent membrane dye FM4-64 (n > 30; Figures 6J to 6L). When the two daughter cells were completely separated, MAP65-3 colocalized with the edge of the fully expanded newly formed cell plate (n > 30; Figures 6M to 6O). A punctate, organelle-like MAP65-3 signal was also observed in the cytoplasm of the two daughter cells at the end of cytokinesis (n > 20; Figures 6N and 6O).
MAP65-3 Colocalizes with Mini Cell Plates in Developing Giant Cells
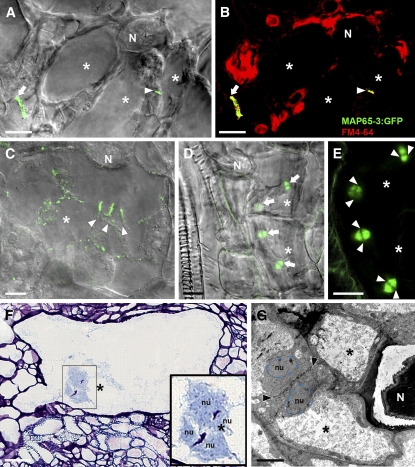
We investigated the subcellular distribution of MAP65-3 during giant cell formation by inoculating ProMAP65-3:MAP65-3:GFP plants with M. incognita. In vivo confocal microscopy of gall sections at 10 d after infection revealed that MAP65-3 was associated with mitotic MT arrays in surrounding cells, as described above for root meristematic cells. After these cells had completed cytokinesis, MAP65-3 colocalized with the FM4-64–stained newly formed cell plate separating the two daughter cells (n > 20; Figures 7A and 7B). In developing giant cells, we detected a MAP65-3 signal in mini cell plates stained with FM4-64 (n > 15; Figures 7A and 7B). In vivo confocal analysis showed that this MAP65-3 signal did not develop further (Figure 7C) and finally disappeared. To determine the origin of the giant cell mini cell plates labeled by MAP65-3:GFP, we performed a detailed analysis of the relationship between mini cell plates, the phragmoplast, and nuclei. Microscopy confirmed the presence of early phragmoplasts in mitotic giant cells (Figure 7D), whereas late phragmoplasts were never observed in giant cells. The dark, poorly labeled phragmoplast midlines (Figure 7E) corresponded to the mini cell plate deposits. Optical and electron microscopy demonstrated that giant cell mini cell plates frequently occurred between two nuclei in developing wild-type giant cells (n > 40; Figures 7F and 7G; see Supplemental Figure 3 online).
Figure 7.
Subcellular Localization of MAP65-3 in Giant Cells.
(A) to (E) In vivo confocal microscopy of giant cells induced by M. incognita.
(A) to (C) MAP65-3:GFP (green channel) localization in giant cells at 10 d after infection.
(A) MAP65-3 localization to the mini cell plate in a giant cell.
(B) Merged image in which the yellow coloration corresponds to the colocalization of FM4-64 (red channel) and MAP65-3:GFP (green channel). The arrow represents a cell plate in surrounding cells, and the arrowhead represents a mini cell plate in giant cells.
(C) Localization of MAP65-3:GFP into giant cell mini cell plates (arrowheads) at 14 d after infection.
(D) and (E) Phragmoplast MT organization in giant cells of a plant expressing MBD:GFP (green channel).
(D) Early synchronous MT phragmoplast arrays were detected in mitotic giant cells (arrows).
(E) The dark, poorly labeled phragmoplast midlines (arrowheads) corresponded to mini cell plates.
(F) Optical microscopic analysis of a developing giant cell at 10 d after infection showed the mini cell plate separating daughter nuclei.
(G) Electron microscopic analysis of a developing giant cell at 7 d after infection. Arrowheads show the giant cell mini cell plate that separates the daughter nuclei (blue dotted lines).
Asterisks, giant cells; N, nematode; nu, nucleus. Bars = 20 μm in (A) to (C) and (F), 10 μm in (D) and (E), and 5 μm in (G).
DISCUSSION
Role of MAP65-3 in Organizing Mitotic MT Arrays in Dividing Plant Cells
We show here that MAP65-3 is expressed specifically in tissues enriched in dividing cells, such as root and shoot apical meristems, embryos, and organ primordia. Interestingly, MAP65-3 presented a distinct expression peak at the G2/M boundary in Arabidopsis cell cultures, indicating possible involvement in early mitosis (Menges et al., 2005). MAP65-3 is coregulated with mitosis-specific genes, Arabidopsis homologs of the genes encoding the spindle-assembly checkpoint proteins MAD2, BUB3, and BUBR1, and cytokinesis-related genes, encoding for example the syntaxin KNOLLE and the MT-associated proteins EB1C and MAP65-4 (Menges et al., 2005). Studies of the pattern of expression in planta and the subcellular distribution of MAP65-3 showed that MAP65-3 was regulated by both transcriptional and posttranscriptional mechanisms. A similar pattern of regulation has been reported for the S. cerevisiae homolog Ase1p (Juang et al., 1997). We demonstrated that the distribution of MAP65-3 changed in vivo with changes in the arrangement of MTs during cell cycle progression. During the G2/M transition, MAP65-3 was associated with cortical MTs, the PPB, and perinuclear MTs. MAP65-3 was also detected on the metaphase MT spindle array, suggesting a possible role in spindle morphogenesis. MAP65-3 was subsequently relocalized to the early phragmoplast MT array. A similar subcellular distribution has been described for the tobacco kinesin TKRP125 (Asada et al., 1997) and its Arabidopsis homolog KRP125c (Bannigan et al., 2007).
Only a few mutant phenotypes resulting from MAP mutations have been characterized in plants (Asada et al., 1997; Walker and Smith, 2002; Shoji et al., 2004; Müller et al., 2006). A MAP65 mutant phenotype has been reported only for MAP65-3. The EMS-induced map65-3/ple mutants display cell wall stubs and multiple nuclei in the root meristem, a characteristic feature of cytokinesis-defective mutants (Müller et al., 2002). In our study, phenotypic analyses of two independent map65-3 knockout T-DNA mutants demonstrated that MAP65-3 played a key role in root, embryo, and shoot cell development. The map65-3 mutant phenotype observed in all plant cells closely resembles that described for some mor1 alleles. The mor1 mutant exhibits cell wall stubs, multinucleate cells, and aberrant chromosomal arrangements, such as disorganized spindles (Kawamura et al., 2006). The analysis of MT array organization in map65-3 knockout T-DNA mutants revealed that some dividing cells underwent abnormal karyokinesis. One possibility is that MAP65-3 is required to correct cases of defective spindle formation. A surveillance mechanism called the spindle-assembly checkpoint allows cells to prevent defects during karyokinesis. This ubiquitous safety device ensures the fidelity of chromosome segregation in mitosis (reviewed in Musacchio and Salmon, 2007). Since MAP65-3 expression is coregulated with that of spindle-assembly checkpoint genes (Menges et al., 2005) and observed in the metaphase/anaphase spindle, we suggest that MAP65-3 may be part of the spindle-assembly checkpoint complex.
MAP65-3 Is Essential for Cytokinesis in Somatic Cells
The EMS-induced map65-3/ple mutants display cytokinesis defects, presumably due to compromised phragmoplast organization (Müller et al., 2004). We demonstrate here that the cell wall stubs, formed in the absence of MAP65-3, originate from a failure of phragmoplast MTs to complete cytokinesis. Based on in vivo confocal imaging of MT organization in the dyc283 mutant, we propose a model for cell wall stub formation resulting from defects during cytokinesis (see Supplemental Figure 6 online). Briefly, cell wall stubs form when phragmoplast expansion is disrupted on one side of the cell, as is the case in mutant cells that lack MAP65-3.
MAP65-3 localizes to the phragmoplast midline (Müller et al., 2004; Smertenko et al., 2004; Van Damme et al., 2004; this study). We showed that MAP65-3 colocalized in planta with the nascent cell plate in early telophase. At the end of cytokinesis, the MAP65-3 signal was found only at the periphery of the cell, forming a ring around the newly formed cell plate, corresponding to the midline of the late MT–phragmoplast array. It is generally assumed that antiparallel phragmoplast MTs interdigitate at the cell plate midline (Müller et al., 2004; Smertenko et al., 2004; Van Damme et al., 2004). This has led to the logical suggestion that MAP65-3 binds overlapping regions of phragmoplast MTs, thereby stabilizing the MTs (Müller et al., 2004; Van Damme et al., 2004). As Austin et al. (2005) recently demonstrated that phragmoplast MTs do not interdigitate at the midline during cytokinesis, an alternative explanation is now required for the localization of MAP65-3 along the phragmoplast midline. We suggest that, during cytokinesis, MAP65-3 is associated with the MT plus-ends and/or cell plate matrix material. High-resolution electron microscopy studies have shown that the MT plus-ends are embedded in, and stabilized by, the cell plate assembly-matrix material in the phragmoplast midline (Segui-Simarro et al., 2004; Austin et al., 2005). Therefore, MT plus-end binding proteins probably form part of the MT plus-end capture complex during phragmoplast expansion (Austin et al., 2005). The best candidates for involvement in the MT plus-end capture complex identified to date are the end binding proteins EB1 (Chan et al., 2003; Dhonukshe et al., 2005) and Microtubule Organization1 (Bisgrove et al., 2004; Kawamura et al., 2006). The similar localization of these nonmotor MAPs along the phragmoplast midline is consistent with our hypothesis that MAP65-3 is associated with the MT plus-end–capture complex during cytokinesis.
MAP65-3 Is Required for Giant Cell Ontogenesis
We describe here a defect in nematode feeding cell formation. In the absence of functional MAP65-3, giant cells started to develop but did not complete their differentiation and were destroyed. These giant cell defects impaired the maturation of the infecting nematodes, which are dependent on the nutrients supplied by fully developed giant cells. Thus, MAP65-3 plays a critical role in susceptible plant–nematode interaction, as shown by the requirement of this protein for giant cell development.
In the early stages of giant cell formation, the activation of MAP65-3 transcription reflects rapid cell cycle reactivation. As reported previously for cell cycle marker genes (de Almeida Engler et al., 1999), we showed that MAP65-3 was expressed in the initial phases of giant cell formation and that the expression of this gene rapidly declined before the development of fully mature giant cells. The patterns of mitotic gene expression observed are consistent with cytological observations of repeated synchronous nuclear division in developing giant cells (Jones and Payne, 1978). As MAP65-3 is present in dividing cells, we suggest that this protein is involved in giant cell mitotic MT array organization.
It is generally assumed that giant cells result from repeated nuclear divisions without cytokinesis (Huang and Maggenti, 1969). We report here that cytokinesis is initiated in giant cells and is essential for giant cell ontogenesis. In vivo confocal and electron microscopy analyses of the first giant cell nuclear division showed that the newly formed cell plate initially lined up between the two daughter nuclei but did not develop further. Our data are consistent with the observations of Jones and Payne (1978), describing the normal alignment of cell plate vesicles in giant cells followed by the dispersal of these vesicles and cytokinesis arrest. We demonstrated that MAP65-3 was associated with a novel kind of cell plate—the giant cell mini cell plate—formed between daughter nuclei during cytokinesis initiation. Microscopy confirmed the presence of early phragmoplast MTs in giant cells that do not develop further. Therefore, we hypothesize that the giant cell mini cell plate is deposited by phragmoplast MTs.
Giant cell mini cell plates were never observed in the absence of MAP65-3. Instead, we observed aberrant cell wall stubs in the map65-3 mutant giant cells. Cell wall stubs were often observed in giant cells after treatment with cell cycle inhibitors, such as hydroxyurea (de Almeida Engler et al., 1999). Interestingly, immunolocalization of the MT cytoskeleton in giant cells demonstrated that phragmoplasts were never observed close to the cell wall (de Almeida Engler et al., 2004).
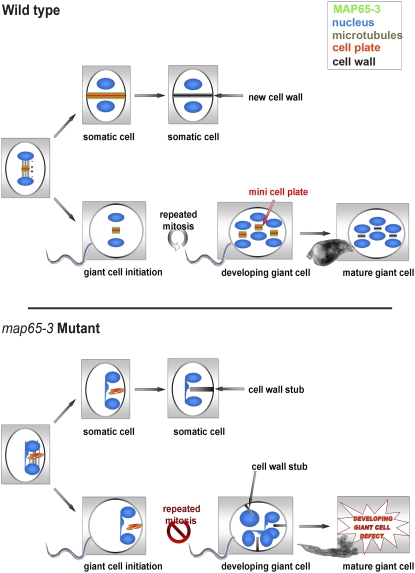
We propose a model of the role of MAP65-3 during giant cell ontogenesis (Figure 8). As in somatic cells, MAP65-3 decorates the nascent cell plate created from the phragmoplast MT array in giant cells. However, the nascent cell plate does not expand further to complete cytokinesis in giant cells. The giant cell mini cell plate may form a physical barrier separating the two daughter nuclei and be required for the multiple rounds of mitosis that occur in developing giant cells, resulting in the formation of a functional feeding site. In the absence of MAP65-3, giant cell formation is initiated, but a defect in giant cell mini cell plate formation leads to the formation of aberrant cell wall stubs. The accumulation of mitosis defects (i.e., cell wall stubs and connected nuclei) during repeated mitoses prevents the development of functional feeding cells, resulting in the death of the nematode. Thus, MAP65-3 is essential for giant cell ontogenesis.
Figure 8.
Role of MAP65-3 during Giant Cell Ontogenesis.
In a wild-type dividing somatic cell, MAP65-3 (green) is associated with the expanding cell plate (red) during cytokinesis. In map65-3 mutant somatic cells, the disorganized phragmoplast leads to the formation of aberrant cell wall stubs and connected daughter nuclei (blue). In wild-type giant cells, MAP65-3 is associated with mini cell plates required for the formation of a functional nematode feeding cell. In giant cell map65-3 mutants, a defect in mini cell plate formation prevents the development of functional feeding cells, resulting in the death of the nematode.
METHODS
Plant Materials, Growth Conditions, and Nematode Infection
The T-DNA mutagenized Arabidopsis thaliana line collection (ecotype Wassilewskija) was generated at the Institut National de la Recherche Agronomique Versailles for promoter trap and gene tagging (Bechtold et al., 1993). The lines were screened individually for GUS expression after Meloidogyne incognita infection, as described previously (Favery et al., 1998). For in vitro analyses, seeds were surface-sterilized and grown on Murashige and Skoog medium containing 1% sucrose, 0.7% plant cell culture–tested agar (Sigma-Aldrich), and 50 μg/mL kanamycin. Plates were inclined at an angle of 60° to allow the roots to grow along the surface. Kanamycin resistance was scored in 2-week-old seedlings. For nematode infection in vitro, 100 surface-sterilized freshly hatched M. incognita J2 larvae were added to each 2-week-old seedling. The plates were kept at 20°C with a 16-h photoperiod. All observations were obtained from three independent experiments.
Histochemical Localization of GUS Activity and Microscopic Analyses
GUS activity was assayed histochemically with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as described by Favery et al. (1998). Galls, root apex, and shoot apical meristems were dissected from GUS-stained plants, fixed in 1% glutaraldehyde and 4% formaldehyde in 50 mM sodium phosphate buffer, pH 7.2, dehydrated, and embedded in Technovit 7100 (Heraeus Kulzer) as described by the manufacturer. Sections (4 μm) were stained with 0.05% ruthenium red or toluidine blue and mounted in dibutyl phthalate xylen (BDH Laboratory Supplies, VWR International). Sections were observed with a Zeiss Axioplan 2 microscope.
T-DNA Insertion Site Analysis and Isolation of Homozygous dyc283 and ebj96 Plants
We characterized the insertion site by sequencing the genomic regions flanking the inserted T-DNA as described by Samson et al. (2002). To isolate homozygous dyc283/dyc283 and ebj96/ebj96 plants, we analyzed the segregation of the kanamycin marker carried by the T-DNA on progeny resulting from each of 20 plants. Progeny of five plants segregated 100% kanamycin-resistant plants, indicating that they were homozygous for the T-DNA–tagged allele. To confirm this result, PCR experiments were done with the MAP65-3 primers that span the T-DNA insertion site and a third primer, GUS (5′-TCCAGACTGAATGCCCACAG-3′), specific for the sequence of the T-DNA. The primers DYCR (5′-GCAGTTCAGAAGCTGATGGAGG-3′) and DYC5RC (5′-CCTGCCTGAGTATGTTATACTCC-3′) were used for DYC283, and DYC6 (5′-GGAGTATAACATACTCAGGCAGG-3′) and DYC9RC (5′-GATGATCAAACCAAACGACATTCAG-3′) were used for EBJ96. When genomic DNA from homozygous plants was used as a template, no amplification was obtained with MAP65-3 primers, which span the T-DNA. A 273-bp PCR product for dyc283 or a 625-bp PCR product for ebj96 was obtained from amplifications with GUS and DYCR or GUS and DYC6, respectively. Phenotypic analysis of dyc283 and ebj96 mutants was always performed compared with wild-type plants of the same genetic background (Wassilewskija). For these two alleles, progeny of multiple homozygous mutants were examined.
Transgenic Plants and Crosses
For ProMAP65-3:GFP:GUS fusion, a fragment of 1240 bp upstream of the start codon was amplified by PCR using the primers Gw5pdyc (5′-AAAAAGCAGGCTTCACACTCTTCCCTACACAAAACCGC-3′) and Gw3pdyc (5′-AGAAAGCTGGGTGTTCGAAATGCTTAAGCCTGTAACAGGG-3′). The PCR fragment was inserted into the pDON207 donor vector and then in the plant expression vector pKGWFS7 (Karimi et al., 2002) using Gateway technology (Invitrogen). For the subcellular localization of MAP65-3, the Pro35S HindIII/SpeI fragment of the pK7WGF2, pK7FWG2, pH7WGY2, and pH7YWG2 vectors (Karimi et al., 2002) was replaced by ProMAP65-3. The coding sequence of MAP65-3 was amplified by PCR, using wild-type plant cDNA as the template. The primers Gw5dycB (5′-AAAAAGCAGGCTTCACCATGGCAAGTGTTCAAAAAGATCCG-3′) and Gw3dycK (5′-AGAAAGCTGGGTGTCAAACCAAACGACATTCAGACTG-3′) were used for GFP and YFP:MAP65-3 fusion. The primers Gw5dycB and Gw3dycL (5′-AGAAAGCTGGGTGAACCAAACGACATTCAGACTG-3′) were used for MAP65-3:GFP and YFP fusion. These sequences were inserted into the pDON207 donor vector and then in the ProMAP65-3 plant expression vector using Gateway technology (Invitrogen). These constructs were sequenced by Genome Express and transformed into Agrobacterium tumefaciens strain GV3101. Wild-type Wassilewskija and homozygous dyc283 or ebj96 Arabidopsis plants were transformed using the dipping method (Clough and Bent, 1998) and selected on Murashige and Skoog medium 0.7% agar plates containing 50 μg/mL kanamycin or hygromycin. Transformed plants were transferred to soil, and seeds were collected. For each construct, 10 independent primary T1 transformants were verified by PCR, and T2 plants were obtained for subsequent analysis. Transgenic plants expressing ProMAP65-3:MAP65-3:GFP or the N-terminal domain of the MT binding domain of MAP4 fused to the GFP (Pro35S:MBD:GFP) were crossed with Pro35S:H2B:YFP Arabidopsis plants. The dyc283 mutants were crossed with Pro35S:MBD:GFP or Pro35S:H2B:YFP Arabidopsis plants. Plants expressing the two constructs were obtained, and homozygous progeny was used for microscopy analysis. We screened for MT array organization labeled by MDB-GFP in 10 wild-type and 10 dyc283 7-d-old seedlings. Division figures were difficult to observe in mutant roots, and they were observed mainly from leaf primordia at the apex.
Confocal Microscopy
Optical sections were obtained on fresh roots using an inverted confocal microscope (model LSM510; Zeiss). YFP and GFP fluorescence were monitored in Lambda mode with a 499- to 550-nm beam path (488-nm excitation line). The fluorescent dye FM4-64 (Molecular Probes) was used at 1 μM final concentration. GFP or YFP and FM4-64 fluorescence were monitored in Lambda mode using the 488-nm line of an argon ion laser, and the emitted light was filtered through a 499- to 620-nm band-pass filter. All observations were obtained from at least three independent experiments. To visualize the MT cytoskeleton and nuclei in the nematode feeding site, galls at 7 and 14 d after infection were excised and embedded in 7% agar. Vibroslices of 100 μm (for galls at 7 d after infection) or 300 μm (for galls at 14 d after infection) were obtained using a HM560V vibratome (Microm). Fresh roots and vibroslices were observed with a 63× water-immersion Apochromat objective (Zeiss). MAP65-3:GFP fluorescence quantification was performed during both metaphase and telophase within a given cell (n = 5) of two independent transformed ProMAP65-3:GFP:MAP65-3 plants using a 499- to 550-nm beam path (488-nm excitation line), set at 5-s exposure time and 20-μm z-step intervals. Z-sections were projected, and nonsaturated images of spindles and phragmoplasts labeled with GFP:MAP65-3 were analyzed using LSM Image Browser software. In the histogram display mode, the region corresponding to the MAP65-3 signal was defined using the mask tool and the average intensities of the entire GFP:MAP65-3 signal were calculated using the measure tool. MAP65-3:GFP fluorescence intensity data were compared with a Student's t test.
Electron Microscopy Method
Root knots of Arabidopsis were isolated and fixed with 2% (v/v) glutaraldehyde, 50 mM PIPES, pH 7.05, 5 mM CaCl2, and 0.1% (w/v) tannic acid at 4°C. After rinsing in buffer containing 50 mM PIPES and 5 mM CaCl2 (30 min), root knots were then postfixed for 1 h at room temperature with 1% OsO4 and 0.8% potassium ferricyanide in the same buffer. After rinsing in water, samples were stained with 2% aqueous uranyl acetate for 2 h and then rinsed with distilled water, dehydrated through an increasing acetone series, and embedded in epoxy resin. Ultrathin sections were collected on Formvar-coated copper grids, stained with uranyl acetate and lead citrate, and examined with a Philips CM12 transmission electron microscope.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At5g51600 (MAP65-3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MAP65-3 Expression Pattern during Plant Development Observed Using the ProMAP65-3:GFP:GUS Fusion.
Supplemental Figure 2. Cytokinetic Defects during Embryogenesis in the dyc283 Mutant (Embryogenesis Is Observed on Whole-Mount Cleared Seeds).
Supplemental Figure 3. Giant Cell Mini Cell Plates.
Supplemental Figure 4. dyc283 and ebj96 Mutant Phenotypes during Plant Nematode Interaction and Complementation by MAP65-3:YFP under ProMAP65-3 Control.
Supplemental Figure 5. Complementation of the dyc283 and ebj96 Root and Shoot Mutant Phenotypes by MAP65-3:YFP under ProMAP65-3 Control.
Supplemental Figure 6. Model for Cell Wall Stub Formation in the Absence of MAP65-3 during Cytokinesis in Somatic Cells.
Supplementary Material
Acknowledgments
We thank Richard Cyr (Pennsylvania State University) for the generous gift of Pro35S:MBD:GFP seeds and Frédéric Berger (Temasek LifeSciences Laboratory, Singapore) for Pro35S:H2B:YFP seeds. We thank Marylin Vantard and Laetitia Perfus-Barbeoch Zurletto for fruitful discussions. We thank Gilbert Engler for helping with confocal microscopy and critical reading of the manuscript. We are grateful to Mansour Karimi (Plant Systems Biology, Vlaams Instituut voor Biotechnologie University of Gent) for the Gateway destination vectors. The T-DNA lines were generated at and obtained from the Institut National de la Recherche Agronomique-Versailles Genomic Resource Center, France (http://www-ijpb.versailles.inra.fr/en/sgap/equipes/variabilite/crg). This work was supported by Institut National de la Recherche Agronomique and GENOPLANTE contracts AF2001032 and ANR05GPLA020 AFINDIS. M.-C.C. was supported by a fellowship from the Ministère de la Recherche et l'Enseignement Supérieure.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bruno Favery (favery@antibes.inra.fr).
Online version contains Web-only data.
References
- Asada, T., Kuriyama, R., and Shibaoka, H. (1997). TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 110 179–189. [DOI] [PubMed] [Google Scholar]
- Austin, J.R., II, Segui-Simarro, J.M., and Staehelin, L.A. (2005). Quantitative analysis of changes in spatial distribution and plus-end geometry of MTs involved in plant-cell cytokinesis. J. Cell Sci. 118 3895–3903. [DOI] [PubMed] [Google Scholar]
- Bannigan, A., Scheible, W.R., Lukowitz, W., Fagerstrom, C., Wadsworth, P., Somerville, C., and Baskin, T.I. (2007). A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 120 2819–2827. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Elis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana. C. R. Acad. Sci. Paris 316 1194–1199. [Google Scholar]
- Bisgrove, S.R., Hable, W.E., and Kropf, D.L. (2004). +TIPs and MT regulation. The beginning of the plus end in plants. Plant Physiol. 136 3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud, M.C., Dubreuil, G., Quentin, M., Perfus-Barbeoch, L., Lecomte, P., de Almeida Engler, J., Abad, P., Rosso, M.-N., and Favery, B. (2008). Root-knot nematodes manipulate plant cell functions during a compatible interaction. J. Plant Physiol. 165 104–113. [DOI] [PubMed] [Google Scholar]
- Chan, J., Calder, G.M., Doonan, J.H., and Lloyd, C.W. (2003). EB1 reveals mobile MT nucleation sites in Arabidopsis. Nat. Cell Biol. 5 967–971. [DOI] [PubMed] [Google Scholar]
- Chan, J., Rutten, T., and Lloyd, C.W. (1996). Isolation of microtubule associated proteins from carrot cytoskeletons: A 120kDa MAP decorates all four microtubule arrays and the nucleus. Plant J. 10 251–259. [DOI] [PubMed] [Google Scholar]
- Chang, H.Y., Smertenko, A.P., Igarashi, H., Dixon, D.P., and Hussey, P.J. (2005). Dynamic interaction of NtMAP65-1a with MTs in vivo. J. Cell Sci. 118 3195–3201. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Daga, R.R., and Chang, F. (2005). Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA 102 8228–8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, E.L., Hussey, R.S., and Baum, T.J. (2004). Getting to the roots of parasitism by nematodes. Trends Parasitol. 20 134–141. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler, J., De Vleesschauwer, V., Burssens, S., Celenza, J.L., Inzé, D., Van Montagu, M., Engler, G., and Gheysen, G. (1999). Molecular markers and cell cycle inhibitors show the importance of the cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler, J., Van Poucke, K., Karimi, M., De Groodt, R., Gheysen, G., Engler, G., and Gheysen, G. (2004). Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant J. 38 12–26. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., Mathur, J., Hulskamp, M., and Gadella, T.W., Jr. (2005). MT plus-ends reveal essential links between intracellular polarization and localized modulation of endocytosis during division-plane establishment in plant cells. BMC Biol. 3 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B., Chelysheva, L.A., Lebris, M., Jammes, F., Marmagne, A., De Almeida Engler, J., Lecomte, P., Vaury, C., Arkowitz, R.A., and Abad, P. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B., Lecomte, P., Gil, N., Bechtold, N., Bouchez, D., Dalmasso, D., and Abad, P. (1998). RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 17 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen, G., and Fenoll, C. (2002). Gene expression in nematode feeding sites. Annu. Rev. Phytopathol. 40 191–219. [DOI] [PubMed] [Google Scholar]
- Goellner, M., Wang, X., and Davis, E.L. (2001). Endo-beta-1,4-glucanase expression in compatible plant-nematode interactions. Plant Cell 13 2241–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.S., and Maggenti, A.R. (1969). Mitotic aberrations and nuclear changes of developing giant cells in Vicia faba caused by root knot nematode, Meloidogyne javanica. Phytopathology 59 447–455. [PubMed] [Google Scholar]
- Hussey, P.J., Hawkins, T.J., Igarashi, H., Kaloriti, D., and Smertenko, A. (2002). The plant cytoskeleton: Recent advances in the study of the plant MT-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50 915–924. [DOI] [PubMed] [Google Scholar]
- Igarashi, H., Orii, H., Mori, H., Shimmen, T., and Sonobe, S. (2000). Isolation of a novel 190 kDa protein from tobacco BY-2 cells: Possible involvement in the interaction between actin filaments and MTs. Plant Cell Physiol. 41 920–931. [DOI] [PubMed] [Google Scholar]
- Jammes, F., Lecomte, P., de Almeida Engler, J., Bitton, F., Martin-Magniette, M.L., Renou, J.P., Abad, P., and Favery, B. (2005). Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 44 447–458. [DOI] [PubMed] [Google Scholar]
- Jiang, C.-J., and Sonobe, S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant MT-associated protein. J. Cell Sci. 105 891–901. [DOI] [PubMed] [Google Scholar]
- Jones, M.G. K. (1981). The development and function of plant cells modified by endoparasitic nematodes. In Plant Parasitic Nematodes, B.M. Zuckerman and R.A. Rhode, eds (New York: Academic Press), pp. 225–279.
- Jones, M.G.K., and Payne, H.L. (1978). Early stages of nematode-induced giant cell formation in roots of Impatiens balsamina. J. Nematol. 10 70–84. [PMC free article] [PubMed] [Google Scholar]
- Juang, Y.L., Huang, J., Peters, J.M., McLaughlin, M.E., Tai, C.Y., and Pellman, D. (1997). APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275 1311–1314. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kawamura, E., Himmelspach, R., Rashbrooke, M.C., Whittington, A.T., Gale, K.R., Collings, D.A., and Wasteneys, G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of MT arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S.L., and Walczak, C.E. (2004). Mitotic spindle assembly and chromosome segregation: Refocusing on MT dynamics. Mol. Cell 15 317–327. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Schaff, J.E., Laskey, J.G., Kieber, J.J., Bilyeu, K.D., and Bird, D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214. [DOI] [PubMed] [Google Scholar]
- Loiodice, I., Staub, J., Setty, T.G., Nguyen, N.P., Paoletti, A., and Tran, P.T. (2005). Ase1p organizes antiparallel MT arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, T., Jin, L., Li, H., Liu, B., and Yuan, M. (2005). Two MT-associated proteins of the Arabidopsis MAP65 family function differently on MTs. Plant Physiol. 138 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.P., Jiang, W., Schoehn, G., Hunter, T., and Margolis, R.L. (2002). PRC1 is a MT binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Fuchs, E., Ovecka, M., Wysocka-Diller, J., Benfey, P.N., and Hauser, M.T. (2002). Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Han, S., and Smith, L.G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16 888–894. [DOI] [PubMed] [Google Scholar]
- Müller, S., Smertenko, A., Wagner, V., Heinrich, M., Hussey, P.J., and Hauser, M.T. (2004). The plant MT-associated protein AtMAP65–3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and Salmon, E.D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8 379–393. [DOI] [PubMed] [Google Scholar]
- Pellman, D., Bagget, M., Tu, Y.H., Fink, G.R., and Tu, H. (1995). Two MT-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., Liu, J.Y., and Pellman, D. (2003). The molecular function of Ase1p: Evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Simarro, J.M., Austin, J.R., II, White, E.A., and Staehelin, L.A. (2004). Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16 836–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, T., Narita, N.N., Hayashi, K., Asada, J., Hamada, T., Sonobe, S., Nakajima, K., and Hashimoto, T. (2004). Plant-specific MT-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol. 136 3933–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A.P., Chang, H.Y., Wagner, V., Kaloriti, D., Fenyk, S., Sonobe, S., Lloyd, C., Hauser, M.T., and Hussey, P.J. (2004). The Arabidopsis MT-associated protein AtMAP65-1: Molecular analysis of its MT bundling activity. Plant Cell 16 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, D., Bouget, F.Y., Van Poucke, K., Inze, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40 386–398. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., Vanstraelen, M., and Geelen, D. (2007). Cortical division zone establishment in plant cells. Trends Plant Sci. 12 458–464. [DOI] [PubMed] [Google Scholar]
- Walker, K.L., and Smith, L.G. (2002). Investigation of the role of cell-cell interactions in division plane determination during maize leaf development through mosaic analysis of the tangled mutation. Development 129 3219–3226. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). MT organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115 1345–1354. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical MTs in plants. Nature 411 610–613. [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart, C., Stoppin-Mellet, V., Blanchoin, L., and Vantard, M. (2004). Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J. 39 126–134. [DOI] [PubMed] [Google Scholar]
- Wiggers, R.J., Starr, J.L., and Price, H.J. (1990). DNA content and variation in chromosome number in plant cells affected by Meloidogyne incognita and M. arenaria. Phytopathology 80 1391–1395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.