Abstract
In Arabidopsis thaliana, the blue light photoreceptor cryptochromes (CRY) act to promote photomorphogenic development and the transition from vegetative to floral development in long days (LDs). We previously proposed that one of the mechanisms by which CRY regulates light responses is via its physical interaction with COP1, a RING motif–containing E3 ligase. Under LDs, the transcription of FLOWERING LOCUS T (FT) is activated by CONSTANS (CO) in leaf, and the FT protein moves to the shoot apex to induce flowering. CO protein is degraded in darkness, whereas it is stabilized by the CRY-mediated signal. However, the mechanism underlying this process is unknown. We show in this report that CO acts genetically downstream of COP1 and CRY to regulate flowering time. In addition, COP1 physically interacts with CO and functions as an E3 ligase, ubiquitinating CO in vitro and reducing CO levels in vivo. These results suggest that COP1 acts as a repressor of flowering by promoting the ubiquitin-mediated proteolysis of CO in darkness and that CRY-mediated signal may negatively regulate COP1, thereby stabilizing CO, activating FT transcription, and inducing flowering.
INTRODUCTION
In many plants, the transition from vegetative to reproductive development is controlled by the daily duration of light and/or darkness (i.e., the photoperiod). In Arabidopsis thaliana, long days (LDs) accelerate flowering, whereas short days (SDs) delay flowering, and these processes are regulated by multiple photoreceptors. Specifically, flowering time in Arabidopsis is promoted by the blue light photoreceptor cryptochromes (CRY1 and CRY2) but is suppressed by the red light photoreceptor phytochrome B (phyB) (Reed et al., 1993; Guo et al., 1998; Mockler et al., 1999, 2003; Yang et al., 2000; Cerdan and Chory, 2003). In addition to regulating flowering, Arabidopsis cryptochromes act together with phytochromes to regulate photomorphogenic development (Ahmad and Cashmore, 1993; Chory et al., 1996; Ahmad et al., 1998; Guo et al., 1998; Neff and Chory, 1998; Quail, 2002), entrain the circadian clock (Somers et al., 1998), and together with the blue light photoreceptor phototropins, mediate blue light regulation of stomatal opening (Kinoshita et al., 2001; Mao et al., 2005).
Significant progress has been made recently in elucidating the signaling mechanism of Arabidopsis CRY. Cryptochomes typically have an N-terminal PHR (photolyase related) domain that shares sequence similarity with photolyase, a family of flavoproteins that catalyzes the repair of UV light–damaged DNA, and a distinguishing C-terminal domain that is absent in photolyase and has no strong sequence similarity with any known protein domain (Sancar, 1994; Cashmore et al., 1999; Lin and Shalitin, 2003). The C-terminal domain of Arabidopsis CRY1 or CRY2 (CCT1 or CCT2) is shown to mediate the signaling of CRY1 or CRY2 in response to light activation through physical interaction with COP1 (Wang et al., 2001; Yang et al., 2001), a RING finger E3 ubiquitin ligase that also controls various light responses, including photomorphogenesis (Deng et al., 1992), flowering time (McNellis et al., 1994), and stomatal opening (Mao et al., 2005). It has recently been demonstrated that the N-terminal PHR domain of CRY mediates homodimerization of the CRY protein, which is required for light activation of the photoreceptor activity of CRY (Sang et al., 2005; Yu et al., 2007).
In Arabidopsis, CONSTANS (CO) acts as a critical positive regulator of flowering in LDs and encodes a B box–type zinc finger transcriptional activator (Putterill et al., 1995). The co mutants flower late only under LDs, whereas transgenic Arabidopsis plants overexpressing CO flower early in both LDs and SDs (Putterill et al., 1995; Suarez-Lopez et al., 2001). CO mRNA abundance is controlled by multiple regulators, including FLAVIN BINDING, KELCH REPEAT, AND F-BOX1, GIGANTEA (GI), and CYCLING DOF FACTOR1 (Fowler et al., 1999; Imaizumi et al., 2003, 2005; Sawa et al., 2007), and the circadian clock regulates the expression of all these genes (Imaizumi et al., 2005; Sawa et al., 2007). Under LDs, CO mRNA peaks in the afternoon, and CO protein activates transcription of the FLOWERING LOCUS T (FT) gene in leaves (An et al., 2004), which encodes a RAF kinase inhibitor-like protein that promotes flowering (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000; Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002). Recently, it has been demonstrated that the FT protein acts as a florigen that moves from leaves to the shoot apex to induce flowering in Arabidopsis (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Blue light acts to stabilize CO in a CRY-dependent manner, whereas red light acts to reduce CO abundance in a phyB-dependent manner, and under LDs, CRY acts to antagonize the degradation of CO mediated by phyB. In darkness, the CO protein is degraded through the 26S proteasome pathway (Valverde et al., 2004). Recent studies demonstrated that regulated proteolysis plays a pivotal role in light signaling. Specifically, COP1 is shown to interact with HY5 (Osterlund et al., 2000), LAF1 (Seo et al., 2003), and HFR1 (Jang et al., 2005; Yang et al., 2005), all of which are transcriptional activators that act to positively regulate photomorphogenesis and act as an E3 ligase to ubiquitinate and degrade all these proteins in a 26S proteasome-dependent manner (Saijo et al., 2003; Seo et al., 2003; Jang et al., 2005; Yang et al., 2005).
To date, the mechanism of how CO abundance is regulated remains largely unknown. Here, we show by genetic studies that CO acts downstream of COP1 and CRY to regulate flowering time. We also show, by biochemical and transgenic studies, that COP1 physically interacts with CO and functions as an E3 ligase that ubiquitinates CO in vitro and reduces CO levels in vivo. We propose that COP1 acts as a critical negative regulator of flowering that reduces CO abundance in darkness and that the CRY-mediated signal may negatively regulate COP1, thus stabilizing CO and promoting flowering.
RESULTS
COP1 Genetically Acts Downstream of CRY to Regulate Flowering Time
Previous demonstrations that the cop1 mutant plants flower early in SDs (McNellis et al., 1994) and that the cry1 cry2 mutant plants flower late in LDs (Guo et al., 1998; Mockler et al., 1999) prompted us to examine whether CRY and COP1 genetically interact to regulate flowering time. To do this, we first analyzed the flowering time phenotype of the cop1 mutant in LDs or SDs illuminated by white light (LDs or SDs) or by blue light (BL-LD or BL-SD. We found that the cop1 mutant flowered very early in SDs and BL-SDs but slightly early in LDs and BL-LDs (Figures 1A and 1B, Table 1; see Supplemental Figures 1A and 1B and Supplemental Table 1 online). We then analyzed the flowering phenotype of the cry1 cry2 cop1 triple mutant generated previously (Mao et al., 2005) and found that it flowered as early as the cop1 mutant in both LDs and BL-LDs (Figure 1C, Table 1; see Supplemental Figure 1C and Supplemental Table 1 online), indicating that COP1 acts downstream of CRY in regulating flowering time.
Figure 1.
Phenotypes of Plants Grown in SDs or LDs.
(A) and (B) The cop1 mutant flowers significantly and slightly earlier than the wild type in SDs and LDs, respectively.
(C) The cry1 cry2 cop1 triple mutant flowers as early as the cop1 single mutant in LDs.
(D) and (E) The flowering phenotype of the cop1 co double mutant is similar to the co single mutant in both SDs and LDs.
(F) The cry1 cry2 co triple mutant flowers as late as the cry1 cry2 double mutant in terms of days to flowering in LDs. Bars = 1 cm.
Table 1.
Flowering Time of Mutant Plants
| LDs
|
SDs
|
|||||
|---|---|---|---|---|---|---|
| Genotype | No. of Rosette Leaves | No. of Cauline Leaves | Days to Flowering | No. of Rosette Leaves | No. of Cauline Leaves | Days to Flowering |
| Wild type (Col) | 13.9 ± 0.9 | 2.8 ± 0.4 | 21.8 ± 0.4 | 49.5 ± 1.2 | 7.5 ± 0.7 | 72.9 ± 0.9 |
| Wild type (Ler)a | 9.2 ± 0.6 | 2.5 ± 0.5 | 18.6 ± 0.4 | 23.6 ± 1.2 | 8.1 ± 0.7 | 53.3 ± 0.9 |
| cry1 | 12.6 ± 0.7 | 2.3 ± 0.7 | 22.6 ± 0.5 | 51.1 ± 2.0 | 6.3 ± 0.9 | 71.1 ± 1.5 |
| cry2 | 24.1 ± 1.3 | 3.6 ± 0.7 | 31.4 ± 0.5 | 51.4 ± 1.9 | 7.8 ± 0.6 | 73.9 ± 1.1 |
| cry1 cry2 | 32.6 ± 1.9 | 6.7 ± 0.6 | 41.5 ± 0.9 | 49.9 ± 2.0 | 7.6 ± 0.6 | 74.2 ± 1.2 |
| cop1 | 9.9 ± 0.7 | 1.8 ± 0.5 | 18.2 ± 0.4 | 8.3 ± 0.4 | 1.7 ± 0.4 | 22.3 ± 0.5 |
| co (Col)b | 27.3 ± 1.4 | 4.2 ± 0.7 | 34.7 ± 0.9 | 33.1 ± 2.6 | 8.1 ± 0.8 | 66.8 ± 1.2 |
| co (Ler) | 21.1 ± 1.9 | 4.8 ± 0.5 | 32.1 ± 0.7 | 22.2 ± 1.1 | 7.9 ± 1.9 | 53.4 ± 0.7 |
| cry1 cry2 cop1 | 9.3 ± 0.6 | 1.5 ± 0.5 | 18.7 ± 0.5 | 8.5 ± 0.7 | 1.2 ± 0.7 | 23.0 ± 0.8 |
| cry1 cry2 cob | 22.5 ± 1.9 | 6.1 ± 0.7 | 40.1 ± 0.6 | 33.5 ± 1.9 | 8.5 ± 0.7 | 72.0 ± 1.5 |
| cop1 cob | 21.7 ± 1.7 | 3.2 ± 0.9 | 30.9 ± 0.8 | 23.2 ± 1.8 | 2.2 ± 0.7 | 54.4 ± 0.6 |
| P35S-CO-3xHA#10/cry1 cry2 | 6.6 ± 0.6 | 1.6 ± 0.5 | 16.9 ± 0.8 | 5.0 ± 0.7 | 1.6 ± 0.5 | 21.8 ± 0.4 |
Plants in each experiment were grown under LDs or SDs as indicated. Flowering time was measured as the total number of leaves produced or days to bolting. Data are from at least 20 individuals for each genotype ± sd.
Landsberg erecta (Ler) wild-type plants had fewer rosette leaves than Columbia (Col) wild type at bolting.
The co (Col), cop1 co, and cry1 cry2 co mutants may have substantial Ler background.
CO Genetically Acts Downstream of COP1 and CRY to Regulate Flowering Time
Because CO acts to promote flowering in LDs (Putterill et al., 1995), we asked whether CO might genetically interact with COP1 in regulating floral initiation. To do this, we constructed the cop1 co double mutant and found that its flowering phenotype resembled that of the co single mutant in SDs, LDs, BL-SDs, and BL-LDs (Figures 1D and 1E, Table 1; see Supplemental Figures 1D and 1E and Supplemental Table 1 online), suggesting that CO genetically acts downstream of COP1 to regulate flowering time. Consistent with the flowering phenotype of cop1 and cop1 co mutants, RT-PCR analysis indicated that FT expression in cop1 mutant was enhanced in both LDs and SDs compared with the wild type but reduced in the cop1 co double mutant in both LDs and SDs compared with cop1 mutant (see Supplemental Figure 2 online). The demonstrations that COP1 genetically acts downstream of CRY and that CO acts downstream of COP1 suggest that CO genetically acts downstream of CRY. To confirm this, we generated the cry1 cry2 co triple mutant and found that the flowering phenotype of this triple mutant was similar to that of the cry1 cry2 double mutant in LDs and BL-LDs (Figure 1F, Table 1; see Supplemental Figure 1F and Supplemental Table 1 online). Next, we prepared a construct constitutively expressing CO fused to the 3xHemagglutinin (HA) epitope and transformed it into the cry1 cry2 double mutant plants. More than 20 independent transgenic plants that flowered earlier than cry1 cry2 mutant plants in LDs were obtained, and expression of the CO-HA transgene was determined by RT-PCR (see Supplemental Figure 3A online). We analyzed the flowering phenotype of one line (P35S–CO–3xHA#10/cry1 cry2) in detail and found that the transgenic plants flowered earlier than the cry1 cry2 mutants in LD, SD, BL-LD, and BL-SD conditions (Table 1; see Supplemental Figure 3B and Supplemental Table 1 online). Taken together, these data demonstrate that CO acts downstream of COP1 and CRY in regulating photoperiodic flowering.
CO Interacts with COP1 in Yeast Cells
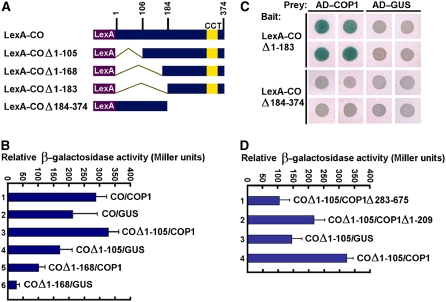
The genetic interaction between COP1 and CO prompted us to investigate whether COP1 physically interacts with CO through the yeast two-hybrid system. We prepared a bait construct expressing the LexA DNA binding domain fused to the full-length CO protein (Figure 2A). This bait construct showed significant background: the level of β-galactosidase activity in yeast cells was similar when coexpressing B42 AD–COP1 or coexpressing the control prey B42 AD–GUS polypeptide (Figure 2B, samples 1 and 2). In an attempt to reduce the strong autonomous transcriptional activity of CO, we made a series of bait constructs expressing CO fragments either containing or lacking the CCT (CO, COL, and TOC1) motif (Strayer et al., 2000) fused to LexA (Figure 2A). The bait constructs expressing CO fragments lacking either amino acids 1 to 105 (COΔ1-105) or 1 to 168 (COΔ1-168) still showed varied degrees of background (Figure 2B, samples 4 and 6). However, β-galactosidase activity increased dramatically when the COΔ1-105 or COΔ1-168 bait was coexpressed with the COP1 prey (Figure 2B, samples 3 and 5), indicating interaction between these domains of CO with COP1. The bait construct expressing CO lacking amino acids 1 to 183 (COΔ1-183) showed little background in the plate assay, and clear interaction was observed between COΔ1-183 and COP1 (Figure 2C). The N-terminal domain of CO lacking the CCT motif (COΔ184-374) did not interact with COP1. These data indicated that CO–COP1 interaction might require the CCT domain of CO.
Figure 2.
Yeast Two-Hybrid Assay of Interaction between CO and COP1.
(A) Yeast two-hybrid bait constructs comprising CO fragments fused to the LexA DNA binding domain (LexA).
(B) Quantitative yeast two-hybrid assays define domains of CO essential for the interaction with COP1. All vector combinations are given as bait/prey. Standard deviations are indicated by error bars (n = 10). The bait constructs showed significant background (samples 2, 4, and 6). However, the β-galactosidase activity increased dramatically when the COΔ1-105 or COΔ1-168 bait was coexpressed with the COP1 prey (samples 3 and 5).
(C) Plate assays showing interaction between the CCT-containing domain of CO (COΔ1-183) and COP1. Blue precipitate represents cumulative β-galactosidase activity resulting from the activation of the lacZ reporter gene by protein–protein interaction. Quadruplicate yeast patches expressing the indicated LexA hybrid (rows) and the indicated AD hybrid (columns) were derived from four independent transformants.
(D) Quantitative yeast two-hybrid assays showing interaction of the C-terminal domain of COP1 with CO. COP1 Δ283-675 and COP1Δ1-209 indicate COP1 fragments containing the N-terminal RING finger and the C-terminal WD40 repeat domain, respectively.
COP1 contains the distinguishing N-terminal RING finger and coiled-coil region (COP1Δ283-675) and the C-terminal WD40 repeat domain (COP1Δ1-209) (Deng et al., 1992), the latter of which is required for the interaction with HY5 (Ang et al., 1998) and CRY1 (Yang et al., 2001). To define which domain is required for the interaction with CO, we conducted a yeast two-hybrid assay using COΔ1-105 as bait and COP1Δ283-675 and COP1Δ1-209 as preys. We found that COΔ1-105 interacted with COP1Δ1-209 (Figure 2D, sample 2) but did not interact with COP1Δ283-675 (Figure 2D, sample 1). This result demonstrated that COP1–CO interaction might require the C-terminal WD40 repeat domain of COP1.
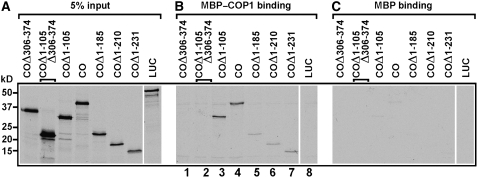
CO Interacts with COP1 in Vitro
To confirm the interaction observed in the yeast two-hybrid assay, we performed protein interaction studies in vitro. A vector was prepared expressing the maltose binding domain (MBP) fused to full-length COP1, and the MBP–COP1 fusion protein was expressed in and purified from Escherichia coli. The full-length, N-, and C-terminal domains of CO, as well as the firefly luciferase (LUC) proteins, were synthesized as radioactively labeled polypeptides by in vitro transcription/translation (Figure 3A). As expected, COP1 bound to full-length CO and various lengths of CO containing the CCT motif (Figure 3B, lanes 3 to 7) but not to the fragments of CO lacking CCT or the control polypeptide LUC (Figure 3B, lanes 1, 2, and 8). None of these fragments bound to the control MBP protein (Figure 3C).
Figure 3.
CO Interacts with COP1 in Vitro.
Radiolabeled CO, six deletion fragments of CO, and LUC polypeptides were incubated with MBP-COP1 and MBP. Bound proteins were identified by SDS-PAGE and autoradiographed on the same x-ray film as 5% of the corresponding input polypeptides (A). The full-length CO and the CCT-containing fragments of CO bound to MBP–COP1 ([B]; lanes 3 to 7) but did not bind to MBP (C).
CO Interacts with COP1 in Vivo
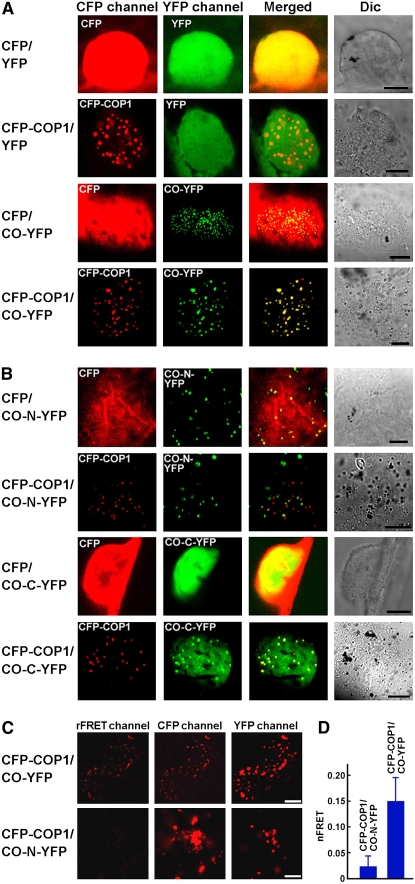
With the demonstration that CO interacts with COP1 in yeast cells and in vitro, we asked whether these two proteins interact in plant cells. Because CO is a nuclear protein and COP1 is found in nuclear bodies (NBs) (Stacey and von Arnim, 1999), we postulated that if these two proteins indeed interact in vivo, they would be localized together. To test this possibility, we transiently expressed CO tagged with yellow fluorescent protein (YFP) and COP1 tagged with cyan fluorescent protein (CFP), either individually or together, in onion epidermal cells. CFP and YFP served as negative controls. As anticipated, COP1 and CO were localized in the same NBs (Figure 4A). Since CO exhibited nuclear speckles (Figure 4A), we examined whether the N- (CO–N) or the C-terminal (CO–C) domain of CO was responsible. The results demonstrated that CO–N was able to form nuclear speckles in the absence of COP1, whereas CO–C did not. However, CO–C was able to form nuclear speckles when COP1 was coexpressed, and the speckles of CO–C colocalized with those of COP1, whereas the speckles formed by CO-N did not colocalize with those of COP1 (Figure 4B). These data indicated that CO might interact with COP1 in vivo and that CO–COP1 interaction requires the CCT domain of CO.
Figure 4.
CO Interacts with COP1 in Vivo.
(A) Colocalization of CO with COP1 in vivo. CO and COP1 localize together to NBs in onion epidermal cells. DIC, differential interference contrast. Bars = 10 μm.
(B) Recruitment of the C-terminal domain of CO into the NBs of COP1. The C-terminal domain of CO (CO–C) does not show nuclear speckles but the N-terminal domain of CO (CO–N) does in onion epidermal cells. However, CO–N and COP1 are not localized in the same NBs. Bars = 10 μm.
(C) FRET analysis between COP1 and CO. Representative nuclear images of the rFRET, CFP, and YFP signals of protoplasts coexpressing CFP–COP1 and CO–YFP or CFP–COP1 and CO–N–YFP. rFRET, real FRET channel image; CFP, CFP channel image; YFP, YFP channel image. Bars = 5 μm.
(D) Intensities of the nFRET signal. Data are mean ± sd of 14 protoplasts.
To further confirm CO–COP1 interaction in vivo, we transiently expressed CFP–COP1 and CO–YFP, either individually or together, in Arabidopsis protoplasts and conducted fluorescence resonance energy transfer (FRET) analysis. Clear real FRET (rFRET) signal was observed for protoplasts coexpressing CFP–COP1 and CO–YFP but not for those coexpressing CFP–COP1 and CO–N–YFP (Figure 4C). Consistent with this result, the intensity of the normalized FRET (nFRET) signal was significantly higher in the protoplasts coexpressing CFP–COP1 and CO–YFP than in those coexpressing CFP–COP1 and CO–N–YFP (Figure 4D). Based on these results, we concluded that CO physically interacts with COP1.
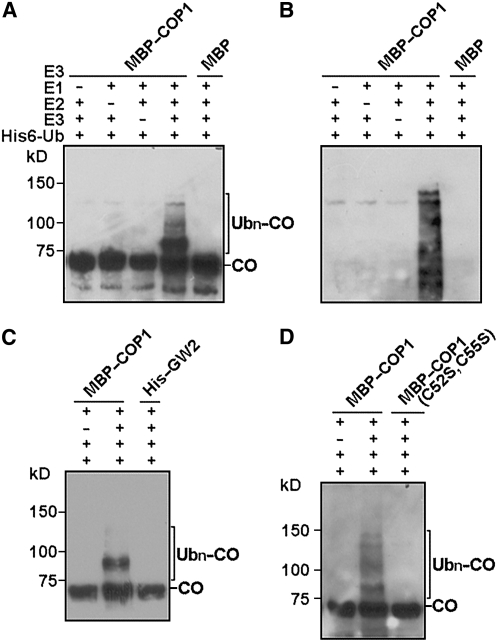
COP1 Ubiquitinates CO in Vitro
Because COP1 is an E3 ligase and CO is degraded in a 26S proteasome–dependent pathway (Valverde et al., 2004), we entertained the possibility that CO might also be a substrate for COP1. To examine this possibility, we prepared a vector containing glutathione S-transferase (GST) fused to CO–3xHA, which we expressed in E. coli. We purified the GST–CO–3xHA fusion protein and conducted in vitro ubiquitination analysis with MBP–COP1 protein. Indeed, CO was found to be ubiquitinated by COP1 in a reaction that was also dependent on E1 and E2 activities (Figures 5A and 5B). To determine whether this reaction was specific, we conducted in vitro ubiquitination analysis of CO using a RING-type E3 ubiquitin ligase, GW2, which is demonstrated to control grain size in rice (Oryza sativa; Song et al., 2007). As shown in Figure 5C, no CO ubiquitination was detected with GW2. It has been reported previously that Cys→Ser substitution in residues 52 and 55 of COP1 abolishes its E3 activity to ubiquitinate LAF1 (Seo et al., 2003). To examine whether the E3 activity of COP1 on CO also requires the RING motif, we produced the MBP–COP1 mutant protein [COP1 (C52S, C55S)] in E. coli and assayed its E3 activity in vitro. Figure 5D shows that COP1 (C52S, C55S) was not able to ubiquitinate CO.
Figure 5.
In Vitro Ubiquitination of CO by COP1.
(A) COP1 E3 activity was assayed in the presence or absence of rabbit E1, human E2 UbcH5b, His6–ubiquitin, and GST–CO–HA. Ubn–CO was detected by protein gel blots using an anti-HA antibody.
(B) Protein gel blot analysis of the same membrane of (A) using an anti-ubiquitin antibody.
(C) Ubiquitination of CO by COP1 but not a rice RING-type E3 ubiquitin ligase GW2.
(D) The MBP–COP1 (C52S, C55S) mutant protein has no E3 ligase activity for CO.
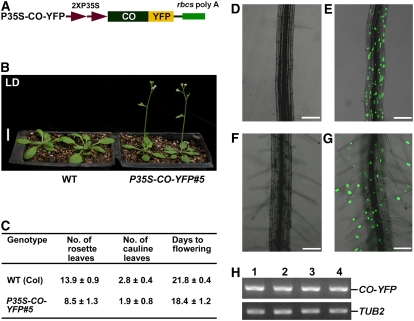
Blue Light Stabilizes CO Protein in Root Cells of Transgenic Plants Expressing CO–YFP
CO–COP1 interaction and the ubiquitination of CO by COP1 predict that CO might be targeted by COP1 for degradation in vivo and that CO abundance might be regulated by the CRY–COP1 signaling system. Our attempts to analyze CO levels using the nuclear extracts prepared from transgenic plants expressing CO tagged with 3xHA by protein gel blot analysis were unsuccessful. In an attempt to circumvent this problem, we prepared a construct constitutively expressing CO fused to YFP (Figure 6A) and transformed it into wild-type Arabidopsis. More than 20 independent transgenic plants exhibiting the early flowering phenotype were obtained. One representative line, P35S–CO–YFP#5, was used for further studies (Figures 6B and 6C). We analyzed CO–YFP accumulation by confocal microscopy. No fluorescence was observed in root cells of either the dark-grown or the dark-adapted P35S–CO–YFP#5 seedlings (Figures 6D and 6F), whereas clear fluorescence was present in seedlings exposed to blue light (Figures 6E and 6G). RT-PCR analysis demonstrated that CO–YFP expressed at similar levels in both dark and blue light conditions (Figure 6H). These data therefore suggest that the CO protein was stabilized by the blue light–mediated signal.
Figure 6.
Blue Light Stabilizes CO in Vivo.
(A) A construct constitutively expressing CO–YFP.
(B) A transgenic P35S–CO–YFP#5 line flowers early in LDs. Bar = 1 cm.
(C) Flowering time of P35S–CO–YFP#5 line in LDs. Flowering time was measured as the total number of leaves produced or days to bolting. Data are from 20 individuals for each genotype ± sd.
(D) Root cells of 8-d-old dark-grown P35S–CO–YFP#5 seedlings do not accumulate CO–YFP fusion protein.
(E) Root cells of 6-d-old dark-grown P35S–CO–YFP#5 seedlings incubated under blue light for 2 d accumulate CO–YFP.
(F) Root cells of 6-d-old white-light-grown P35S–CO–YFP#5 seedlings adapted in darkness for 2 d do not produce CO–YFP.
(G) Root cells of 6-d-old white-light-grown P35S–CO–YFP#5 seedlings incubated under blue light for 2 d accumulate CO–YFP. Bars = 100 μm in (D) to (G).
(H) RT-PCR analysis showing constitutive expression of CO–YFP. Lanes 1 to 4 indicate P35S–CO–YFP#5 samples prepared from (D) to (G), respectively. TUB2 was used as an internal loading control.
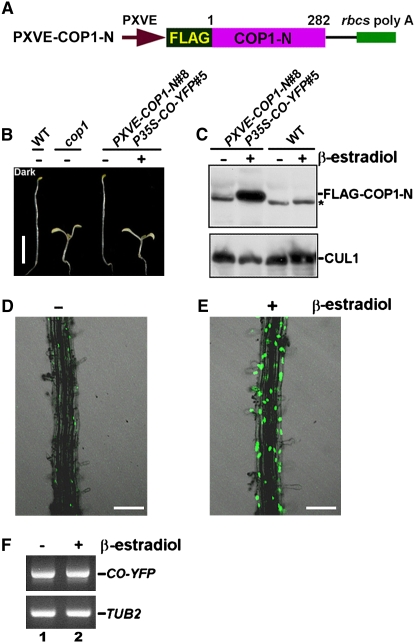
Expression of the Dominant-Negative N-Terminal Fragment of COP1 in Transgenic P35S–CO–YFP#5 Seedlings Results in an Increase in CO Abundance
To determine whether COP1 regulates CO levels in vivo, we prepared a construct expressing the dominant-negative N-terminal fragment of COP1 (COP1–N) (McNellis et al., 1996) fused to a 6xFLAG epitope, which is driven by a chemical-inducible promoter XVE (Zuo et al., 2000) (Figure 7A), and transformed it into wild-type Arabidopsis. A transgenic line, PXVE–COP1–N#8, was obtained and genetically crossed into the P35S–CO–YFP#5 background to generate the double transgenic line PXVE–COP1–N#8 P35S–CO–YFP#5. As shown in Figure 7B, without β-estradiol induction, the PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings were fully etiolated in darkness, similar to the wild type, but displayed a constitutive photomorphogenic phenotype (COP phenotype) with β-estradiol induction, similar to the cop1 mutant. Consistent with the COP phenotype, protein gel blot analysis using an antibody against FLAG indicated that PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings accumulated FLAG–COP1–N fusion protein only in the presence of β-estradiol induction (Figure 7C). Confocal microscopy analysis indicated that almost no CO–YFP accumulated in root cells of the dark-grown PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings in the absence of β-estradiol induction (Figure 7D). However, induction of COP1–N expression in darkness resulted in a dramatic increase in CO accumulation (Figure 7E). RT-PCR analysis demonstrated that CO–YFP expressed at similar levels either with or without β-estradiol induction (Figure 7F). These results suggest that inhibition of the endogenous COP1 activity by expression of the dominant-negative COP1–N fragment might reduce CO degradation.
Figure 7.
Chemical-Induced Expression of COP1 N-terminal Domain Stabilizes CO in Vivo.
(A) A chemical-inducible construct containing FLAG fused to the N-terminal domain of COP1 (COP1–N).
(B) Eight-day-old dark-grown PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings induced with β-estradiol display a constitutive photomorphogenic phenotype. Bar = 5 mm.
(C) Protein gel blot analysis of protein extracts prepared from 8-d-old dark-grown PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings with (+) or without (−) β-estradiol induction using an anti-FLAG antibody. Asterisk indicates a band nonspecifically recognized by the antibody.
(D) and (E) Root cells of 8-d-old dark-grown PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings accumulate much more CO–YFP with induction than without induction. Bars = 100 μm.
(F) RT-PCR analysis showing constitutive expression of CO–YFP. Lanes 1 and 2 denote samples prepared from (D) and (E), respectively.
CO Degradation in Vivo Is Regulated by COP1 and CRY
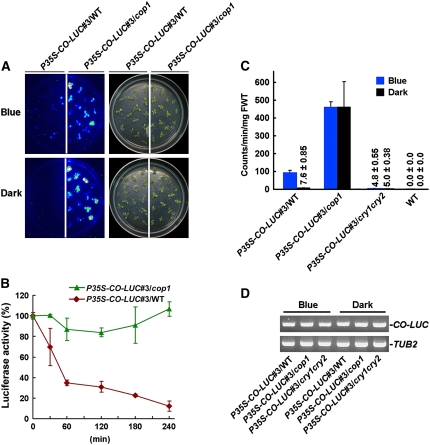
To further confirm whether COP1 and CRY regulate CO degradation in vivo, we prepared a construct constitutively expressing CO fused to LUC (see Supplemental Figure 4A online) and transformed it into wild-type Arabidopsis. More than 20 independent transgenic plants exhibiting early flowering phenotype were obtained. One representative line, P35S–CO–LUC#3 in a wild-type background (P35S–CO–LUC#3/WT; see Supplemental Figure 4B online), was introduced into both cop1 and cry1 cry2 mutant backgrounds by genetic crossing. We found that P35S–CO–LUC#3/cop1 plants flowered earlier than either the cop1 mutant or P35S–CO–LUC#3/WT plants, P35S–CO–LUC#3/WT plants flowered earlier than P35S–CO–LUC#3/cry1 cry2 plants, and P35S–CO–LUC#3/cry1 cry2 plants flowered earlier than cry1 cry2 mutant plants (see Supplemental Figures 4C to 4E online), indicating that CO–LUC was biologically functional but had varied activities in these genetic backgrounds.
Next, using luciferase fluorescence in vivo imaging, we found that the CO levels were dramatically lower in the wild-type background than in the cop1 mutant background under both dark and blue light conditions (Figure 8A), suggesting that CO might be degraded in the wild-type background but hardly degraded in cop1 mutant. Consistent with this result, CO degradation rate was greater in the wild-type background than in the cop1 mutant background during the dark adaptation process (Figure 8B). Further quantification of luciferase activity indicated that more CO accumulated in blue light than in darkness in the wild-type background, whereas high levels of CO accumulated in both blue light and darkness in the cop1 mutant (Figure 8C). Noticeably, the CO levels in cop1 were consistently higher than those in the wild type and cry1 cry2 mutant. Very low levels of CO were detected in the cry1 cry2 double mutant under both dark and blue light conditions (Figure 8C) but sufficient to promote flowering in LDs (see Supplemental Figures 4D and 4E online). Importantly, these measurements of luciferase activity are consistent with the flowering time phenotype of P35S–CO–LUC#3/cop1, P35S–CO–LUC#3/WT, and P35S–CO–LUC#3/cry1 cry2 plants (see Supplemental Figures 4C to 4E online). RT-PCR analysis demonstrated that CO–LUC was expressed at similar levels in the wild type, cop1, and cry1 cry2 mutant backgrounds under both dark and blue light conditions (Figure 8D), indicating that transcription of the P35S–CO–LUC transgene is not affected by either COP1 or CRY. From these data, we concluded that COP1 and CRY are at least partially responsible for negatively and positively regulating CO protein abundance in vivo, respectively.
Figure 8.
CO Degradation Is Regulated by COP1 and CRY.
(A) Noninvasive in vivo luciferase imaging of P35S–CO–LUC#3/WT and P35S–CO–LUC#3/cop1 seedlings. “Blue” indicates 7-d-old white-light-grown seedlings exposed to blue light for 2 d prior to imaging, and “Dark” indicates seedlings prepared as for “Blue” but then adapted to darkness for 4 h. With blue light exposure, much more CO proteins accumulate in cop1 mutant than in the wild-type background, and after 4 h of dark adaptation, CO is degraded in the wild type but not in the cop1 mutant.
(B) Degradation rate analysis of CO in the wild type and cop1 mutant. Seven-day-old white-light-grown P35S–CO–LUC#3/WT and P35S–CO–LUC#3/cop1 seedlings were exposed to blue light for 2 d and then transferred to darkness for 0, 30, 60, 120, 180, and 240 min, respectively. Luciferase activity was expressed as the percentage of initial activity. CO is degraded rapidly in the wild type but is hardly degraded in the cop1 mutant during dark adaptation. Error bars indicate se.
(C) Analysis of CO–LUC levels in P35S–CO–LUC#3/WT, P35S–CO–LUC#3/cop1, P35S–CO–LUC#3/cry1 cry2, and wild-type seedling. Seedlings were grown in white light for 7 d and then transferred to either blue light or darkness for 2 d. Error bars indicate se. FWT, fresh weight.
(D) RT-PCR analysis showing constitutive expression of CO–LUC. Samples were prepared from seedlings treated in (C).
DISCUSSION
Previous studies demonstrated that Arabidopsis cryptochromes COP1 and CO are involved in regulating flowering time (McNellis et al., 1994; Putterill et al., 1995; Guo et al., 1998; Mockler et al., 1999, 2003) and that cryptochromes act to stabilize the CO protein (Valverde et al., 2004). The photomorphogenic development controlled by CRY is mediated through direct interaction with COP1 (Wang et al., 2001; Yang et al., 2001). In this report, we further the understanding of the mechanism of flowering time regulation involving Arabidopsis CRY, COP1, and CO by demonstrating that COP1 acts genetically downstream of CRY, CO acts downstream of COP1 and CRY, and that COP1 physically interacts with CO in both yeast and plant cells. We demonstrate that COP1 serves as an E3 ligase for CO in vitro and induces CO degradation in Arabidopsis in darkness. We propose that cryptochrome regulation of flowering time might involve negative regulation of COP1, through either direct CRY–COP1 interaction or via other unknown mechanisms, which relieves the E3 ligase activity of COP1 on CO. Once CO accumulates, it activates FT transcription to induce flowering. This proposal is consistent with earlier studies demonstrating that CO abundance is positively regulated by cryptochromes and that CO degradation is mediated by the 26S proteasome pathway (Valverde et al., 2004).
COP1 Acts Downstream of CRY and CO Acts Downstream of COP1 in Regulating Flowering Time
In this study, we confirmed the involvement of COP1, CRY1, and CRY2 in the regulation of flowering time. We demonstrated that the cop1 mutant flowers early in both SDs and LDs (Figures 1A and 1B, Table 1). In the cry1 cry2 double mutant background, the COP1 mutation results in dramatic acceleration of flowering (Figure 1C, Table 1). Arabidopsis CRY1 is the major blue light photoreceptor regulating photomorphogenesis, but it plays a role in regulating flowering time in SDs (Bagnall et al., 1996), and CRY2 is the primary blue light receptor regulating flowering time of Arabidopsis in LDs (Koornneef et al., 1991; Guo et al., 1998). The role for CRY1 in promoting flowering time is evident in this study because the cry1 cry2 double mutant flowers significantly later than the cry2 single mutant in LDs (Table 1). It was shown previously that this double mutant flowers slightly later than the cry2 single mutant (Mockler et al., 1999). This discrepancy might result from different growth conditions. When grown in monochromatic blue light LDs (BL-LDs), the cry1 and cry2 single mutants flower at about the same time as the wild type in blue light, whereas the cry1 cry2 double mutant plants flower significantly later than the cry1 and cry2 single mutants under blue light (Mockler et al., 1999), indicating a redundant role for CRY1 and CRY2 in promoting floral initiation. Furthermore, the involvement of CRY1 in regulating flowering time is supported by our previous demonstration that transgenic Arabidopsis plants expressing the C-terminal domain of either CRY1 (CCT1) fused to β-glucuronidase (GUS) and displaying a constitutive photomorphogenic phenotype exhibit a dramatic early flowering phenotype in SDs (Yang et al., 2000).
To further discern the genetic interaction of CRY and COP1, we analyzed the flowering time phenotypes of cop1, cry1 cry2, and cry1 cry2 cop1 mutants in LD illuminated by monochromatic blue light (BL-LD) in this study. Consistent with the flowering phenotype observed in LDs illuminated by white light, the results demonstrated that the cry1 cry2 cop1 triple mutant plants flower as early as the cop1 single mutant plants in BL-LDs (see Supplemental Figure 1 and Supplemental Table 1 online). Therefore, we conclude that COP1 acts genetically downstream of CRY in regulating flowering time. This conclusion is consistent with previous demonstrations that COP1 acts genetically downstream of CRY in regulating photomorphogenesis (Ang and Deng, 1994) and stomatal opening (Mao et al., 2005).
We further demonstrated that CO acts downstream of COP1 and CRY in regulating flowering time. Our results agree with previous demonstrations that HY5 and HFR1, both of which encode transcriptional activators, act downstream of COP1 in regulating photomorphogenic development (Ang and Deng, 1994; Yang et al., 2005). Based on these studies, we propose a genetic pathway regulating flowering time, in which COP1 lies downstream of CRY and CO lies downstream of COP1. Consistent with this proposal, biochemical studies demonstrated that CRY interacts with COP1 (Wang et al., 2001; Yang et al., 2001), and CO interacts with COP1 (this report). Furthermore, since the cop1 co double mutant plants exhibited a slightly intermediate phenotype to that of their parental mutants in LDs, being more similar to the co mutant (Table 1), CO is likely the dominant but not the sole activator of flowering that lies downstream of the CRY-COP1 signaling system. Other activators might also act together with CO downstream of this system.
CO Physically Interacts with COP1
It has been shown that HY5, LAF1, and HFR1, transcriptional activators that positively regulate photomorphogenic development, all interact with COP1. Specifically, the bZIP protein HY5 and the basic helix-loop-helix protein HFR1 interact with the COP1 WD40 repeat domain (Holm et al., 2001; Jang et al., 2005), while the MYB transcription factor LAF1 binds to the RING motif (Seo et al., 2003). Here, based on our genetic studies between CO and COP1, we show that CO, a B box–type zinc finger transcriptional activator, also localizes to the NBs of COP1 (Figure 4A). Yeast two-hybrid studies, in vitro binding assay, in vivo colocalization studies, and FRET analysis demonstrate that CO–COP1 interaction requires the C-terminal CCT domain of CO (Figures 2C, 3, 4B, and 4C). Although not a bZIP protein, CO also interacts with the COP1 WD40 repeat domain in yeast cells (Figure 2D). COP1 E3 ligase has been previously shown to ubiquitinate LAF1 (Seo et al., 2003), HY5 (Saijo et al., 2003), phyA (Seo et al., 2004), and HFR1 (Jang et al., 2005; Yang et al., 2005). The demonstration that CO can be ubiquitinated in vitro in this study brings the total number of identified COP1 substrates to five.
COP1 Is an E3 Ligase Responsible for Targeted Degradation of CO
In this study, we demonstrate that CO physically interacts with COP1 and that COP1 exhibits E3 ubiquitin ligase activity for CO in vitro. CO accumulation is higher in both transgenic plants expressing the dominant-negative N-terminal domain of COP1 (Figure 7E) and in the cop1 mutant (Figures 8A to 8C), indicating that COP1 is required for targeted degradation of CO in vivo. COP1 activity can be regulated by multiple components of the COP/DET/FUS proteins, which are negative regulators of plant development (Chory et al., 1989; Deng et al., 1991; Misera et al., 1994; Wei et al., 1994; Wei and Deng, 2003). Among them, DET1 was initially shown to be involved in regulating chromatin remodeling and gene expression (Benvenuto et al., 2002; Schroeder et al., 2002). The COP9 signalosome (CSN) is a constitutively nuclear-enriched protein complex consisting of eight distinct subunits that is required for the nuclear accumulation of COP1 (Chamovitz et al., 1996). COP10 is a ubiquitin conjugating enzyme variant (Suzuki et al., 2002), which forms a complex (CDD) with DDB1 and DET1 and interacts with both CSN and COP1 (Yanagawa et al., 2004). The E3 ligase activity of COP1 also requires SPA1 (Seo et al., 2003), initially identified as a nuclear-localized repressor of far-red light signaling (Hoecker et al., 1998, 1999). Indeed, SPA1 interacts with both COP1 and CO and acts to destabilize CO (Saijo et al., 2003; Laubinger et al., 2006).
COP1 activity can also be regulated by photoreceptors. In the dark, COP1 localizes to the nucleus, whereas in the light, it localizes to the cytoplasm (von Arnim and Deng, 1994), and this nucleo-cytoplasmic translocation is regulated by CRY1, phyA, and phyB (Osterlund and Deng, 1998). Based on the observations that CRY physically interacts with COP1 (Wang et al., 2001; Yang et al., 2001), that blue light acts to stabilize CO in vivo, and that CO abundance is dramatically reduced in the cry1 cry2 mutant (Valverde et al., 2004; this report), we propose that the E3 ligase activity of COP1 on CO and/or the removal of COP1 from the nucleus during the day time in LDs might be regulated by cryptochromes. In agreement with this, another substrate of COP1 E3 ligase, HY5, is shown to accumulate in dark-grown transgenic plants overexpressing either GUS–CCT1 or GUS–CCT2. These transgenic plants display a cop1 mutant-like phenotype because GUS–CCT1 or GUS–CCT2 suppresses COP1 activity through direct interaction with COP1 (Wang et al., 2001; Yang et al., 2001). In addition, COP1 has been shown to physically interact with the red/far-red photoreceptor phyA and to act as an E3 ligase for phyA, resulting in elimination of phyA and subsequent termination of phyA signaling (Seo et al., 2004). Thus, it is conceivable that phyA might also regulate COP1 activity. These possibilities need to be investigated in future studies.
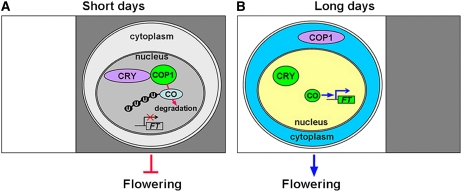
A Mode of COP1 and CRY Action
The circadian clock sets peak expression of CO in the afternoon in LDs, and light stabilizes and activates CO to induce FT expression. In SDs, the peak expression of CO occurs at midnight, and the CO protein is degraded in darkness (Suarez-Lopez et al., 2001; Valverde et al., 2004). Based on previous studies and our results, we propose that in SDs when CO mRNA peaks at midnight, COP1 is predominantly localized to the nucleus, where it leads to CO ubiquitination and degradation (Figure 9A). By contrast, when expression of CO peaks in the afternoon during LDs, light activation of CRY might mediate the translocation of a large proportion of COP1 from the nucleus to the cytoplasm, presumably through direct CRY–COP1 interaction or via other unknown mechanisms, and CO would then be able to accumulate in the nucleus and activate FT expression to promote flowering (Figure 9B). Although we showed in this report that the CO protein's stability is regulated by a CRY- and COP1-dependent pathway, our results do not exclude additional regulatory mechanisms associated with photoperiodic regulation of flowering. For example, it has recently been shown that microRNA172 acts to promote photoperiodic flowering in a CO-independent manner (Jung et al., 2007). Furthermore, CRY and phyB act antagonistically in the regulation of floral induction (Mockler et al., 1999) and CO stability (Valverde et al., 2004), and CRY2 and phyB are shown to physically interact (Mas et al., 2000). The mechanism of flowering time regulation involving phyB is not discussed in our model, and further investigation will be needed to elucidate its mode of action. Taken together, the results described here provide an important mechanism of photoperiodic control of flowering time involving regulation of CO protein stability by the CRY–COP1 signaling system and are consistent with the previous proposal that CO abundance is regulated by a posttranslational mechanism (Valverde et al., 2004; Imaizumi and Kay, 2006).
Figure 9.
A Working Model Illustrating Photoperiodic Regulation of CO by CRY and COP1.
When CO mRNA peaks at midnight in SDs, COP1 is predominantly localized in the nucleus. CRY interacts with COP1 but is not able to repress its activity. COP1–CO interaction results in ubiquitination and degradation of CO (A), whereas when CO mRNA peaks in the afternoon in LDs, light activation of CRY during the daytime might mediate translocation of COP1 from nucleus to cytoplasm. Consequently, CO is able to accumulate and activate the transcription of FT to promote flowering (B). U, ubiquitin.
METHODS
Construction of Expression Cassettes, Transformation, and Growth Conditions of Plants
Col or Ler lines of Arabidopsis thaliana were used as the wild type. Seeds were sown on Murashige and Skoog (MS) medium, cold-treated for 3 d at 4°C, and then transferred to controlled environment cabinets under either LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions with a fluence rate of 120 μmol·s−1·m−2 of white light (produced by cool-white fluorescent lamps) at 22°C. Plants were all grown in LDs or SDs of white light unless stated otherwise. Experiments involving blue light illumination were performed in an LED growth chamber (Percival) using the blue diodes (maximum wavelength of 469 nm) at 22°C, and the light intensity was ∼35 μmol·s−1·m−2. Light spectra and intensity were measured with a HandHeld spectroradiometer (ASD) and a Li250 quantum photometer (Li-Cor).
PCR-amplified fragments encoding YFP, 3xHA, and firefly LUC were cloned into SpeI and SacI sites of pBluescript SK+ (abbreviated as pBS). The PCR-amplified full-length CO fragment was cloned into the EcoRI and SpeI sites of pBS–YFP, pBS–3xHA, and pBS–LUC, resulting in pBS–CO–YFP, pBS–CO–3xHA, and pBS–CO–LUC, respectively. The PCR-amplified 3xFLAG fragment was cloned into HindIII and EcoRI sites of pBS. The PCR-amplified cDNA fragment encoding Arabidopsis COP1 N-terminal domain (COP1–N) (lacking amino acids 283 to 675) was cloned into EcoRI and SpeI of pBS-3xFLAG, resulting in pBS–3xFLAG–COP1–N. All of the constructs used were confirmed by DNA sequencing. Fragments encoding CO–YFP, CO–3xHA, and CO–LUC were excised by HindIII and SacI and cloned into the plant expression vector pKYL71 (Schardl et al., 1987) or pHB (Mao et al., 2005) to generate pKYL71–CO–YFP, pKYL71–CO–3xHA, and pHB–CO–LUC, respectively. The fragment encoding 3xFLAG–COP1–N was excised from pBS–3xFLAG–COP1–N by digestion with XhoI and SpeI and cloned into pER8 (Zuo et al., 2000) to generate pER8-3xFLAG–COP1–N, in which 3xFLAG–COP1–N is under the control of the β-estradiol–inducible promoter XVE. All constructs were introduced into the Agrobacterium tumefaciens strain GV3101. The floral dipping method (Bechtold et al., 1993) was used to create independent transformants in either the wild-type background or the cry1 cry2 double mutant. Transgenic seeds were screened on MS plates containing either 100 μg/mL kanamycin or 50 μg/mL hygromycin. Homozygous T4 seeds were used for phenotypic analysis.
Construction of Double and Triple Mutants
The co-2 mutant in a Ler background was introduced into a Col background by two rounds of crossing. The cop1-4 (Col) (Deng and Quail, 1992), cry1 cry2 (Col) (Mockler et al., 1999; Mao et al., 2005), and co-2 (Ler) (Putterill et al., 1993) mutants and transgenic lines P35S–CO–YFP#5 (Figures 6B and 6C), P35S–CO–LUC#3, (see Supplemental Figure 4B online), and PXVE–COP1–N#8 in Col background were used as parental materials to construct the cop1 co double and cry1 cry2 co triple mutants, and transgenic PXVE–COP1–N#8 P35S–CO–YFP#5, P35S–CO–LUC#3/cop1, and P35S–CO–LUC#3/cry1 cry2 lines by genetic crossing. The initial cop1 co double mutant was backcrossed with cop1-4 mutant twice, resulting in the cop1 co double mutant with more Col genetic background. The CO mutation was verified by having the late flowering phenotype and by DNA sequencing. The COP1 mutation was analyzed by phenotypic analysis of dark-grown seedlings and adult plants. The mutations of CRY1 and CRY2 were characterized by phenotypic analysis (long hypocotyl and late flowering) and further verified by PCR using primers described previously (Mao et al., 2005). The CO–YFP or CO–LUC transgene was confirmed by early flowering and a kanamycin or basta resistance phenotype. The PXVE–COP1–N transgene was verified by phenotypic analysis of dark-grown seedlings with 10 μM β-estradiol induction and then by protein gel blots using a monoclonal anti-FLAG antibody (Sigma-Aldrich). The cry1 cry2 cop1 triple mutant was generated previously (Mao et al., 2005).
Flowering Studies
Seedlings were germinated on MS medium and then transferred to either soil and grown in LDs or SDs illuminated by white light at a fluence rate of 120 μmol·s−1·m−2 or to MS medium in 77 × 77 × 97-mm clear acrylic containers (Sigma-Aldrich; Magenta vessel GA-7 V8505) and grown in an E-30 LED growth chamber in LDs or SDs illuminated by blue light at a fluence rate of 35 μmol·s−1·m−2. Flowering time was measured by scoring the number of rosette leaves and cauline leaves at flowering and/or the number of days from germination to bolting. At least 20 plants were analyzed.
Construction of Vectors for the LexA Yeast Two-Hybrid System
Bait Constructs
PCR-amplified cDNA fragments of Arabidopsis CO, COΔ1-105 (lacking amino acids 1 to 105), COΔ1-168 (lacking amino acids 1 to 168), COΔ1-183 (lacking amino acids 1 to 183), and COΔ184-374 (lacking amino acids 184 to 374) were cloned into EcoRI and XhoI sites of pLexA.
Prey Constructs
The prey constructs expressing GUS, COP1, COP1Δ283-675 (lacking amino acids 283 to 675), and COP1Δ1-209 (lacking amino acids 1 to 209) were made previously (Ang et al., 1998; Yang et al., 2001).
Yeast Two-Hybrid Assay
Yeast transformation and the calculation of relative β-galactosidase activities were performed as described previously (Yang et al., 2001). The plate assay was performed as described previously (Sang et al., 2005) with minor modifications: the induction plate was grown at 30°C for 24 h and then photographed. At least three independent experiments were performed, and the result of one representative is shown.
Expression and Purification of Recombinant Proteins
The pBS–CO–3xHA clone was used as PCR template to amplify the CO–3xHA fragment, which was finally cloned into the EcoRI and XhoI sites of pGEX-4T-1 (Amersham Biosciences) to express the GST–CO–3xHA fusion protein. Full-length PCR-amplified COP1 fragment was cloned into EcoRI and HindIII sites of pMal-c2 (New England Biolabs). The resulting pMal–COP1 clone was used as a template to generate the vector expressing MBP–COP1 (C52S, C55S) mutant protein with the Stratagene in vitro mutagenesis kit.
After confirmed by DNA sequencing, these constructs were transformed into Escherichia coli BL21 cells, and expression of the fusion protein was induced by 0.5 mM isopropyl-β-d-thiogalactoside. The induced cells were lysed by sonication in purification buffer (50 mM Tris-HCl, pH 7.5, and 200 mM NaCl) containing 1% Triton X-100, 5 mM DTT, 2 mM phenylmethylsulphonyl fluoride, and a proteinase inhibitor cocktail (Roche), and MBP, MBP–COP1, and MBP–COP1 (C52S, C55S) proteins were purified on amylose resins (New England Biolabs). For purification of GST–CO–3xHA fusion protein, treated cells were lysed by sonication in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3) containing 1% Triton X-100, 5 mM DTT, 2 mM phenylmethylsulphonyl fluoride, and a proteinase inhibitor cocktail, and then the GST–CO–3xHA fusion protein was purified on glutathione sepharose 4B resins (Amersham Biosciences).
In Vitro Transcription/Translation and in Vitro Binding Assay
Full-length CO, six deletion fragments (COΔ306-374 [lacking amino acids 306 to 374], COΔ1-105Δ306-374 [lacking amino acids 1 to 105 and 306 to 374], COΔ1-105 [lacking amino acids 1 to 105], COΔ1-185 [lacking amino acids 1 to 185], COΔ1-210 [lacking amino acids 1 to 210], and COΔ1-231 [lacking amino acids 1 to 231]), and LUC polypeptides were synthesized and labeled with [35S]-Met in the TNT T7 Quick for PCR DNA transcription/translation system (Promega). Ten microliters of the translation mixture was incubated with 30 μL of amylase resins, to which equal amounts of MBP or MBP–COP1 fusion protein were added, for ∼1 h at 25°C in 500 μL binding buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, and 0.5 mM DTT). After six washes with 1 mL wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.2% Nonidet P-40), the bound proteins were eluted by heating and analyzed by SDS-PAGE and autoradiography.
In Vitro Ubiquitination Assays
In vitro ubiquitination assays were performed in a total volume of 30 μL consisting of 50 mM Tris, pH 7.4, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 400 ng protein substrate (GST–CO–3xHA), 100 ng rabbit E1 (Boston Biochem), 100 ng human E2 UbcH5b (Boston Biochem), 1 μg His6-ubiquitin (Sigma-Aldrich), and 200 ng MBP, MBP–COP1, MBP–COP1 (C52S, C55S), or HIS6–GW2 protein. Reactions were performed at 30°C for 2.5 h with shaking and were terminated by adding SDS sample buffer. The reaction mixtures were separated on 6% SDS-PAGE, and ubiquitinated GST–CO–3xHA protein was detected by protein gel blotting using an antibody against HA (Santa Cruz Biotechnology) or ubiquitin (Boston Biochem). The exposure time for enhanced chemiluminescence of different immunoblots was not controlled precisely, so the signal intensities from different immunoblots are not directly comparable.
Subcellular Colocalization Study
The CFP and YFP coding sequences were fused in frame to the 5′ end of COP1 and to the 3′ end of CO, CO–N (amino acids 1 to 305), and CO–C (amino acids 106 to 374) to generate CFP–COP1, CO–YFP, CO–N–YFP, and CO–C–YFP fusions, respectively. Expression of the fragments encoding CFP, CFP–COP1, YFP, CO–YFP, CO–N–YFP, and CO–C–YFP were driven by the 35S promoter. Onion epidermal cells were bombarded with different combinations of constructs using a particle gun–mediated system (PDS-1000/He; Bio-Rad) and analyzed by confocal microscopy (Carl Zeiss; LSM 510 Meta).
FRET Analysis
Arabidopsis mesophyll protoplasts isolated from 4-week-old wild-type plants were either transfected with a construct expressing CO–YFP or CFP–COP1 or cotransfected with constructs expressing CO–YFP and CFP–COP1, or CO–N–YFP and CFP–COP1 by following procedures as described (http://genetics.mgh.harvard.edu/sheenweb/protocols_reg.html). The protoplasts expressing the transfected constructs were imaged on a Zeiss LSM510 META NLO laser scanning confocal microscope using a Plan-Apochromat ×100 /1.4 oil objective. FRET analysis was performed as described previously (Gordon et al., 1998). Each protoplast image was taken with FRET, CFP, and YFP channels, respectively. The FRET, CFP, and YFP channels were excited at 458, 458, and 514 nm and collected at emission signals of 535 to 590 nm, 480 to 520 nm, and 535 to 590 nm, respectively. For bleed-through correction, the images of cells expressing CFP–COP1 only or CO–YFP only were taken, and the rFRET, YFP, and CFP images were determined by Auto Deblur and AutoVisualize software (Media Cybemetics) using the Gordon and Herman method. The total intensities of rFRET, YFP, and CFP were measured by Image-Pro Plus software (Media Cybemetics). The nFRET signals were calculated using the equation nFRET = rFRET/sqrt (YFP × CFP).
Chemical Induction and YFP Fluorescence Images
Root cells of 8-d-old P35S–CO–YFP#5 seedlings grown on MS medium under different light conditions were analyzed using a Carl Zeiss LSM 510 Meta confocal microscope with YFP filter sets. YFP fluorescence was excited with a 514-nm argon laser, and images were collected in the 530 to 600-nm range. All fluorescence images were taken with identical exposure. Seven-day-old dark-grown PXVE–COP1–N#8 P35S–CO–YFP#5 seedlings were treated with DMSO or inducer (10 μM β-estradiol) for 1 d in liquid MS medium in darkness and then subjected to confocal microscopy. More than 20 seedlings were analyzed for each treatment in each one of the three independent trials.
Luciferase Activity Assays
For noninvasive in vivo LUC imaging, 7-d-old LD-grown P35S–CO–LUC#3/WT and P35S–CO–LUC#3/cop1 seedlings were exposed to blue light at a fluence rate of 35 μmol·s−1·m−2 for 2 d and then analyzed directly or 240 min after being transferred to darkness. Seedlings were sprayed with 3 mM d-Luciferin (BT11-500; BioThema) and kept in the dark for 6 min before images were taken. The imaging system (Roper Scientific; 7383-0001) consists of a high-performance CDD camera mounted in a dark chamber, a camera controller, and a computer. Image acquisition and processing were performed with the WinView software provided by the camera manufacturer. Exposure time was set at 6 min.
Quantification of luciferase activity was assayed with a TD-20/20 Luminometer (Turner Designs). Seven-day-old LD-grown P35S–CO–LUC#3/WT, P35S–CO–LUC#3/cop1, and P35S–CO–LUC#3/cry1 cry2 seedlings were either exposed to blue light at a fluence rate of 35 μmol·s−1·m−2 for 2 d or transferred to darkness for 2 d and then analyzed to generate the data shown in Figure 8C. For CO degradation rate analysis, 7-d-old white-light-grown P35S–CO–LUC#3/WT and P35S–CO–LUC#3/cop1 seedlings were exposed to blue light for 2 d and analyzed 0, 30, 60, 120, 180, and 240 min after being transferred to darkness. Luciferase activity was expressed as the percentage of initial activity. Seedlings (150 mg) were collected in liquid nitrogen, and total protein was extracted using 100 μL 1× Luciferase Cell Culture Lysis Reagent (Promega). The luciferase activity was measured in duplicates with Luciferase Assay reagent (E1500; Promega). At least three independent trials were performed, and one representative is shown.
RT-PCR
To analyze FT, CO–YFP, and CO–LUC transcript levels, total RNA was extracted from the seedlings using RNArosereagent (Watson), and the residual genomic DNA was removed using the DNA-free kit (Ambion). The concentration of RNA was determined by spectrophotometric measurements. The cDNA was synthesized using oligo(dT)18 primer and Rever-Tra Ace M-MLV reverse transcriptase RNaseH− (Toyobo) according to the manufacturer's recommendation. The cDNAs were amplified for 30 cycles except for FT, which was amplified for 35 cycles, at 94°C for 35 s, 55°C for 35 s, and 72°C for 30 s. The forward and reverse primers for FT (P1 and P2), CO–YFP (P3 and P4), CO–LUC (P5 and P6), and CO–HA (P7 and P8) were as follows: P1, 5′-CAAGTCCTAGCAACCCTCACC-3′; P2, 5′-ATAGGCATCATCACCGTTCGT-3′; P3, 5′-AAATATGGCTCCTCAGGGACTCA-3′; P4, 5′-ACTTGTGGCCGTTTACGTCG-3′; P5, 5′-CCCAAAGGGACAGTAGAGC-3′; P6, 5′-CCAGGAACCAGGGCGTAT-3′; P7, 5′-GGAGATAGAGTTGTTCCGCTT-3′; P8, 5′-CGTATGGGTAGAATGAAGGAA-3′. To avoid the interference of the endogenous CO expression, the antisense primers were based on the sequence of YFP (P4), LUC (P6), and HA (P8), respectively. A fragment encoding Tubulin 2 (TUB2) was amplified as an internal standard with primers P9 and P10: P9, 5′-GACTGTCTCCAAGGGTTCCAG-3′; P10, 5′-CACCATGAAGAAGTGAAGACG-3′. The cDNAs were diluted 10-fold and amplified for TUB2. The PCR products were analyzed through agarose gel electrophoresis using ethidium bromide staining. The expected lengths of products are 423 bp for TUB2 and FT, 499 bp for CO–YFP, 398 bp for CO–LUC, and 502 bp for CO–HA. All RT-PCR analyses were repeated three times, and one representative is shown.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CRY1 (At4g08920), CRY2 (At1g04400), COP1 (At2g32950), FT (At1g65480), CO (At5g15840), and TUB2 (At5g62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of Plants Grown in LDs or SDs Illuminated by Blue Light (BL-LD or BL-SD).
Supplemental Figure 2. RT-PCR Analysis of FT Expression.
Supplemental Figure 3. Phenotype of P35S-CO–3xHA/cry1 cry2 Plants in LDs.
Supplemental Figure 4. Phenotypes of cop1 and cry1 cry2 Mutant Plants Expressing CO–LUC in LDs.
Supplemental Table 1. Flowering Time of Mutant Plants Grown in LDs or SDs Illuminated by Blue Light (BL-LD or BL-SD).
Supplementary Material
Acknowledgments
We thank X.W. Deng, C.T. Lin, and W.R. Briggs for helpful comments; Q. Hu for FRET analysis; N.H. Chua for pER8 vector; and H.X. Lin for GW2 protein. This work was supported by the National Natural Science Foundation of China (Grants 90208005 and 30325007 to H.-Q.Y. and 30421001) and the Shanghai Leading Academic Discipline Project (B209).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hong-Quan Yang (hqyang@sjtu.edu.cn).
Online version contains Web-only data.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., and Cashmore, A.R. (1998). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., and Deng, X.W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J., King, R.W., and Hangarter, R.P. (1996). Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta 200 278–280. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, Sci. Vie Life Sci. 316 1194–1199. [Google Scholar]
- Benvenuto, G., Formiggini, F., Laflamme, P., Malakhov, M., and Bowler, C. (2002). The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 12 1529–1534. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423 881–885. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chory, J., Chatterjee, M., Cook, R.K., Elich, T., Fankhauser, C., Li, J., Nagpal, P., Neff, M., Pepper, A., Poole, D., Reed, J., and Vitart, V. (1996). From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc. Natl. Acad. Sci. USA 93 12066–12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., and Quail, P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2 83–95. [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, G.W., Berry, G., Liang, X.H., Levine, B., and Herman, B. (1998). Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74 2702–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Hardtke, C.S., Gaudet, R., and Deng, X.W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, T., and Kay, S.A. (2006). Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 11 550–558. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306. [DOI] [PubMed] [Google Scholar]
- Jaeger, K.E., and Wigge, P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J.H., Seo, Y.H., Seo, P.J., Reyes, J.L., Yun, J., Chua, N.H., and Parka, C.M. (2007). The GIGANTEA-regulated MicroRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414 656–660. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Marchal, V., Le Gourrierec, J., Wenkel, S., Adrian, J., Jang, S., Kulajta, C., Braun, H., Coupland, G., and Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. [DOI] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Mao, J., Zhang, Y.C., Sang, Y., Li, Q.H., and Yang, H.Q. (2005). From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P., Devlin, P.F., Panda, S., and Kay, S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408 207–211. [DOI] [PubMed] [Google Scholar]
- Mathieu, J., Warthmann, N., Kuttner, F., and Schmid, M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17 1055–1060. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., Torii, K.U., and Deng, X.W. (1996). Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misera, S., Muller, A.J., Weiland-Heidecker, U., and Jurgens, G. (1994). The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol. Gen. Genet. 244 242–252. [DOI] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., and Deng, X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., and Coupland, G. (1993). Chromosome walking with YAC clones in Arabidopsis: Isolation of 1700 kb of contiguous DNA on chromosome 5, including a 300 kb region containing the flowering-time gene CO. Mol. Gen. Genet. 239 145–157. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sancar, A. (1994). Structure and function of DNA photolyase. Biochemistry 33 2–9. [DOI] [PubMed] [Google Scholar]
- Sang, Y., Li, Q.H., Rubio, V., Zhang, Y.C., Mao, J., Deng, X.W., and Yang, H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, M., Nusinow, D.A., Kay, S.A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl, C.L., Byrd, A.D., Benzion, G., Altschuler, M.A., Hildebrand, D.F., and Hunt, A.G. (1987). Design and construction of a versatilesystem for the expression of foreign genes in plants. Gene 61 1–11. [DOI] [PubMed] [Google Scholar]
- Schroeder, D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12 1462–1472. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z., and Lin, H.X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 623–630. [DOI] [PubMed] [Google Scholar]
- Stacey, M.G., and von Arnim, A.G. (1999). A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J. Biol. Chem. 274 27231–27236. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (2003). The cop9 signalosome. Annu. Rev. Cell Dev. Biol. 19 261–286. [DOI] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.Q., Wu, Y.J., Tang, R.H., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103 815–827. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yu, X., Shalitin, D., Liu, X., Maymon, M., Klejnot, J., Yang, H., Lopez, J., Zhao, X., Bendehakkalu, K.T., and Lin, C. (2007). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA 104 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.