Abstract
We present high-resolution maps of DNA methylation and H3K4 di- and trimethylation of two entire chromosomes and two fully sequenced centromeres in rice (Oryza sativa) shoots and cultured cells. This analysis reveals combinatorial interactions between these epigenetic modifications and chromatin structure and gene expression. Cytologically densely stained heterochromatin had less H3K4me2 and H3K4me3 and more methylated DNA than the less densely stained euchromatin, whereas centromeres had a unique epigenetic composition. Most transposable elements had highly methylated DNA but no H3K4 methylation, whereas more than half of protein-coding genes had both methylated DNA and di- and/or trimethylated H3K4. Methylation of DNA but not H3K4 was correlated with suppressed transcription. By contrast, when both DNA and H3K4 were methylated, transcription was only slightly reduced. Transcriptional activity was positively correlated with the ratio of H3K4me3/H3K4me2: genes with predominantly H3K4me3 were actively transcribed, whereas genes with predominantly H3K4me2 were transcribed at moderate levels. More protein-coding genes contained all three modifications, and more transposons contained DNA methylation in shoots than cultured cells. Differential epigenetic modifications correlated to tissue-specific expression between shoots and cultured cells. Collectively, this study provides insights into the rice epigenomes and their effect on gene expression and plant development.
INTRODUCTION
In eukaryotic nuclei, DNA associates with proteins to form chromatin. It first wraps around core histones to form nucleosomes that, in turn, are often organized into higher-ordered structures. Chromatin structure plays an essential role in genome organization, transcriptional activity, and memory of developmental state (Bernstein et al., 2002). While all cells in an individual have the same nuclear genome, each cell type may harbor a distinct epigenome, which relies on heritable, often reversible, DNA methylation at cytosines and histone modifications (Richards, 1997).
Cytosine methylation occurs primarily at CG dinucleotides, although significant levels of methylation at CNG and CNN have also been found in plants (Cao and Jacobsen, 2002; Hsieh and Fischer, 2005). As a conserved epigenetic mark, DNA methylation is involved in many important biological processes, including heterochromatin formation, defense against transposon proliferation, genomic imprinting, regulation of endogenous gene expression, and silencing of transgenes (Paszkowski and Whitham, 2001; Bender, 2004; Zhang et al., 2006).
Combinations of covalent modifications of core histones, including acetylation, methylation, phosphorylation, ribosylation, sumoylation, and ubiquitylation, have been proposed to generate a histone code that can be deciphered by specific code-reader proteins (Mellor, 2006). This decoding process profoundly affects gene expression. Acetylation of specific Lys residues in the N termini of H3 or H4 is generally associated with transcriptional activation, while methylation of these residues has been associated with either transcriptional activation or repression, depending on which Lys is methylated and how many methyl groups are added. For example, methylation of H3K9, H3K27, and H4K20 has been reported to affect heterochromatin formation, X chromosome inactivation, and euchromatic gene silencing, whereas methylation of H3K4, H3K36, and H3K79 has been associated with transcriptional activation (Hsieh and Fischer, 2005; Martin and Zhang, 2005). Moreover, monomethylation of H3K9, H3K27, and H3K79 has been linked with transcriptional activation, whereas trimethylation of these three residues has been associated with suppressed gene expression in human T cells (Barski et al., 2007).
Methylation of histone H3 Lys 4 is among the most intensively studied of histone modifications. One to three methyl groups may be added, and each form might have unique yet poorly understood functions (Sims and Reinberg, 2006). For example, dimethylated H3K4 has been correlated with a permissive chromatin state facilitating the initiation of transcription, whereas trimethylated H3K4 is proposed to mark an active state allowing stronger transcriptional activity (Santos-Rosa et al., 2002; Li et al., 2007a). It was recently suggested that di- and trimethylated H3K4 can be generated during transcriptional elongation by RNA polymerase II and do not directly impact elongation per se (Pavri et al., 2006). This finding raised the question whether H3K4 methylation signals that pol II is traveling through the gene is a consequence of this passage or is functionally involved in chromatin remodeling (Li et al., 2007a).
Interplay between DNA methylation and histone modifications has also been well documented. In Neurospora crassa, the H3K9 histone methyltransferase (HMTase) DIM5 is required for the maintenance of DNA methylation (Tamaru and Selker, 2001; Tamaru et al., 2003). The Arabidopsis thaliana H3K9 HMTase KRYPTONITE also directs some CNG and CNN DNA methylation (Jackson et al., 2002). Histone deacetylase HDA6 is needed to enhance RNA-directed DNA methylation (Aufsatz et al., 2002). By contrast, two recent studies have demonstrated that intragenic DNA methylation induces a closed chromatin structure that excludes H3K4me2, H3K4me3, H3K9ac, and H3K14ac (Lorincz et al., 2004; Okitsu and Hsieh, 2007).
Rice (Oryza sativa) is an important model species for cereals and other monocotyledonous plants. Two prominent features of most rice chromosomes are their clear organization into heterochromatic and euchromatic regions and the large amount of pericentromeric heterochromatin. For example, cytological studies using 4′,6-diamidino-2-phenylindole staining indicate that approximately half of chromosomes 4 and 10 is the more densely stained heterochromatin, including their entire short arms and the proximal portions of their long arms (Cheng et al., 2001; Yan and Jiang, 2007). Global repression of transcription in rice heterochromatin has been observed, but the molecular basis is unknown (Jiao et al., 2005; Li et al., 2006). Completion of the rice genome sequence (International Rice Genome Sequencing Project, 2005) provides an unprecedented opportunity to examine epigenetic modifications comprehensively and correlate them with gene expression.
Here, we describe high-resolution mapping of DNA methylation and H3K4me2 and H3K4me3 patterns of rice (spp japonica cv Nipponbare) chromosomes 4 and 10 using tiling-path microarrays. We compare two developmental states: undifferentiated suspension-cultured cells and young light-grown shoots. The large heterochromatic regions on these chromosomes allow a genome-scale investigation of DNA methylation and histone modifications in heterochromatin. The completely sequenced rice centromeres of chromosomes 4 and 8 were also included in this analysis (Nagaki et al., 2004; Zhang et al., 2004). This in-depth, genome-scale analysis provides unprecedented insights into the epigenetic signatures of the rice genome.
RESULTS
Identification of Genomic Regions Associated with DNA Methylation and Two Specific Histone Modifications
To map DNA methylation and histone modifications on rice chromosomes 4 and 10 accurately, a custom oligonucleotide tiling array containing 380,766 36-mer probes with a median resolution of 118 bp was designed (see Methods). Highly repetitive probes except those in centromeres 4 and 8 were excluded such that the tiling probe set covered ∼77.5% of the physical length of rice chromosomes 4 (∼35 Mb) and 10 (∼23 Mb).
To gain insight into DNA methylation and histone modification patterns in different developmental states, two distinct rice tissues, suspension-cultured cells and 7-d-old light-grown shoots, were chosen as starting materials. Methylated DNA was isolated by gel electrophoresis after digestion of total genomic DNA with the restriction enzyme McrBC (see Supplemental Figure 1 online) (Lippman et al., 2004). Genomic regions associated with di- or trimethylated H3K4 were recovered by chromatin immunoprecipitation (ChIP) with antibodies that specifically recognize H3K4me2 or H3K4me3, respectively. DNA fragments recovered by either technique were labeled and hybridized to the tiling arrays using fragmented total genomic DNA as control, which allowed DNA and histone H3K4 methylation to be quantified for each probe.
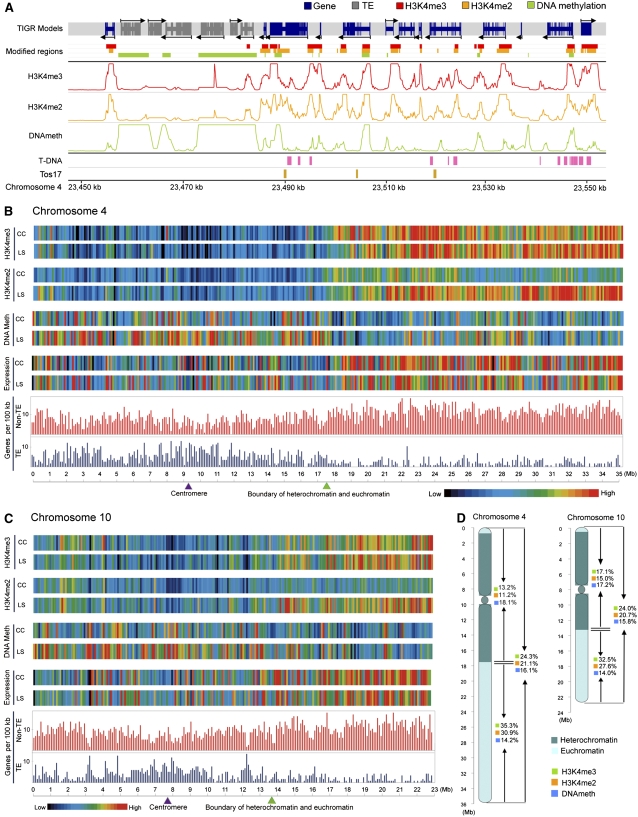
To identify genomic regions significantly enriched in methylated DNA, H3K4me2, or H3K4me3, a Wilcoxon signed rank test (Hollander and Wolfe, 1999) was applied in a sliding window of ±500 bp across the chromosomal tile paths (see Methods). In brief, a methylated DNA or methylated H3K4 region was defined by combining adjacent probes with a significance threshold of P < 0.05, allowing a maximal gap of 150 bp, and requiring a minimal run of two consecutive probes. Figure 1A shows the analysis of a representative region on chromosome 4. Transposable elements (TEs) in general had highly methylated DNA but little H3K4me2 or H3K4me3, whereas non-TE genes had much less methylated DNA and were enriched for H3K4me2 and H3K4me3. A fully dynamic browser for viewing the DNA and H3K4 methylation patterns observed in this study is publicly available at http://plantgenomics.biology.yale.edu.
Figure 1.
Overview of Epigenetic Modifications of Rice Chromosomes 4 and 10.
(A) Distribution of H3K4me3, H3K4me2, DNA methylation, annotated gene models, and Tos17 and T-DNA insertions along a representative 100-kb region of chromosome 4 in light-grown shoots. The three middle tracks show the log10-transformed P values for each probe.
(B) and (C) Epigenetic, gene expression, and gene density landscapes of rice chromosomes 4 and 10 in light-grown shoots (LS) and cultured cells (CC). Color-coded bars represent the percentage of probes within each 100-kb bin that detected the indicated modification (P < 0.05). Average expression level within each bin represents the percentage of signal probes detected in a previous genome-wide transcription analysis using tiling microarrays (Li et al., 2007b).
(D) Summary of the distribution of H3K4me3, H3K4me2, and DNA methylation in heterochromatic and euchromatic regions of chromosomes 4 and 10 in light-grown shoots.
Suitability of the P < 0.05 threshold was assessed by ChIP-PCR, which confirmed 92% (23/25) of H3K4me2- and 100% (24/24) of H3K4me3-enriched regions tested (see Supplemental Figures 2B and 2C online). Methylated DNA regions identified by the P < 0.05 threshold were validated using an McrBC digestion-coupled PCR method (Rabinowicz et al., 2003), and 93% (36/39) of the tested candidates were scored as methylated by this assay (see Supplemental Figure 2A online). We also tested six randomly selected regions with methylated DNA and one without by genomic bisulfite sequencing. All six methylated regions were confirmed to contain methylcytosines in CG, CNG, or CNN contexts (see Supplemental Figures 3A to 3F, 3H, and 3I online), and the hypomethylated region was shown to be devoid of DNA methylation (see Supplemental Figure 3G online). Additional validation comes from the failure to detect any DNA or H3K4 methylation on the two large chloroplast DNA insertions on chromosomes 4 (9.07 ∼ 9.18 Mb) and 10 (10.46 ∼ 10.59 Mb) (International Rice Genome Sequencing Project, 2005), as probes from these regions primarily detect plastid DNA, which lacks these modifications (Ngernprasirtsiri et al., 1988), because it greatly outnumbers nuclear DNA.
Distinct Patterns of DNA and Histone Methylation in Heterochromatic and Euchromatic Regions of Rice Chromosomes Correlate with Differences in Proportions of Transcribed DNA
Rice chromosomes 4 and 10 have similar overall patterns of DNA and H3K4 methylation, with some variation between the two tissues studied (Figures 1B to 1D). In shoots, 5494 methylated DNA, 5137 H3K4me2, and 5050 H3K4me3 regions were identified on chromosome 4, covering 16.1, 21.1, and 24.3% of the chromosome, respectively (Figure 1D; see Supplemental Table 1 online). Similarly, 3482 methylated DNA, 3352 H3K4me2, and 3452 H3K4me3 regions were found on chromosome 10 in shoots, covering 15.8, 20.7, and 24.0% of the chromosome, respectively. The median sizes of methylated DNA, H3K4me2, and H3K4me3 regions were 532, 1064, and 913 bp, respectively, while there were 152, 239, and 152 continuous regions longer than 5.0 kb for DNA methylation, H3K4me2, and H3K4me3, respectively, spanning multiple gene loci (see Supplemental Data Set 1 online).
Further inspection revealed an inverse relationship between the amounts of methylated DNA and methylated H3K4 in heterochromatin and euchromatin on both chromosomes (Figures 1B to 1D; see Supplemental Figure 4 online; Jiao et al., 2005; Li et al., 2006). DNA methylation was more abundant in the heterochromatic half than in the euchromatic half (18.1% versus 14.2% on chromosome 4 in shoots). However, due to the low coverage of highly repetitive sequences in the heterochromain on our tiling array, the real difference in DNA methylation was expected to be bigger than the numbers obtained here. By contrast, the euchromatic half had significantly higher levels of H3K4me2 and H3K4me3 (30.9% versus 11.2% and 35.3% versus 13.2% on chromosome 4 in shoots). Based on the previous genome-wide transcription analysis using tiling microarrays (Li et al., 2007b), transcription of a greater proportion of the DNA was detected in the euchromatic halves of both chromosomes (Figure 1). Elevated DNA methylation and reduced transcription in heterochromatin correlates with the higher density of TEs and related tandem repeats in heterochromatin, which can influence the expression of nearby genes and their DNA methylation (Martienssen and Colot, 2001; Schotta et al., 2003; Lippman et al., 2004). Conversely, the enrichment of H3K4me2 and H3K4me3 and higher transcriptional activity in euchromatin correlates with the greater abundance of non-TE genes.
However, non-TE genes from the euchromatic regions also had significantly higher levels of H3K4me2 and H3K4me3 than those from heterochromatic regions (Figures 2A and 2B). In addition, a higher proportion of non-TE genes from euchromatic regions contained H3K4me2 and H3K4me3 than those from heterochromatic regions (see Supplemental Figures 5A to 5H online). By contrast, no significant difference in DNA methylation of non-TE genes from heterochromatin and euchromatin was detected (Figures 2C; see Supplemental Figure 5 online). Evidently, the higher levels of H3K4me2 and H3K4me3 in euchromatin are related to differences in the chromatin environment as well as the relative proportions of TEs.
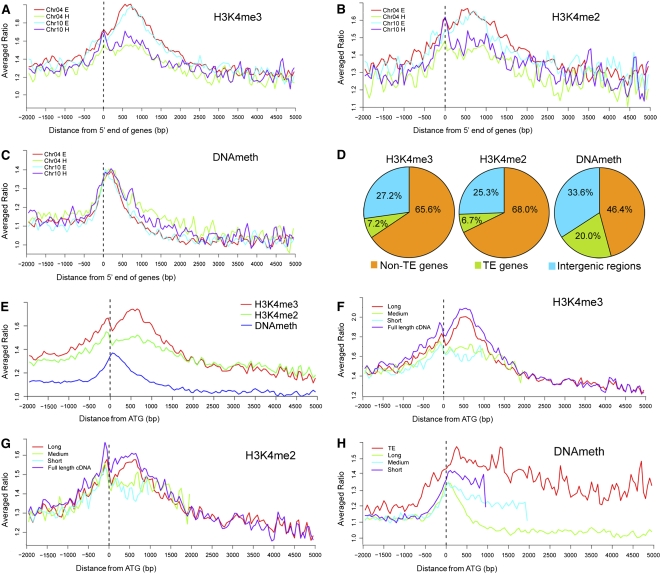
Figure 2.
Distribution of DNA Methylation and Two Histone Modifications within Rice Genes.
(A) to (C) Distribution of H3K4me3, H3K4me2, and DNA methylation levels within non-TE genes located in heterochromatin (H) and euchromatin (E). All genes were aligned at the 5′ end. The y axis shows the averaged ratios of the signals from the samples enriched for each modification to the signal from total genomic DNA.
(D) Frequencies of H3K4me3, H3K4me2, and DNA methylation regions in non-TE genes, TE-related genes, and intergenic regions.
(E) Comparison of the distribution of three epigenetic modifications within genes aligned at their translation start sites (ATG). The y axis shows the averaged ratios of the signals from the samples enriched for each modification to the signal from total genomic DNA.
(F) to (H) Distribution of three epigenetic modifications within different size groups of genes. Genes were aligned at the translation start sites (ATG). The y axis shows the averaged ratios of the signals from the samples enriched for each modification to the signal from total genomic DNA.
DNA Methylation, H3K4me2, and H3K4me3 Preferentially Mark Different Groups of Rice Genes
To characterize the epigenetic modifications at the gene level, we defined the territory of a gene as the body (annotated transcribed region) plus its putative promoter (the 1-kb region upstream of the annotated transcription start site), while DNA between gene territories was designated as the intergenic region. Based on rice genome annotation 4.0 released by The Institute for Genomic Research, our tiling array covered 2248 (of 2432) TE-related loci and 6329 (of 6347) non-TE protein-coding genes from chromosomes 4 and 10. Of the 6329 non-TE genes, 4800 (75.8%), 5008 (79.1%), and 3735 (59.0%) contained H3K4me2, H3K4me3, and DNA methylation, respectively, in shoots (see Supplemental Data Set 2 online). Of the 2248 TEs, 549 (24.4%), 634 (28.2%), and 1611 (71.7%) contained H3K4me2, H3K4me3, and DNA methylation, respectively, in shoots (see Supplemental Data Set 2 online). As shown in Figure 2D, 68.0% of the H3K4me2 and 65.6% of the H3K4me3 regions were found in non-TE gene territories, and 25.3% and 27.2%, respectively, were found in intergenic regions, while only 6.7% and 7.2%, respectively, were found in TE territories. By contrast, 20% of the methylated DNA regions were found in TEs, 46.4% in non-TE genes, and 33.6% in intergenic regions. It is noteworthy that the distribution of DNA methylation in TEs should be higher than revealed by our tiling array, since we excluded certain repeated sequences in TEs. Similar distributions were observed in cultured cells (see Supplemental Figure 7A online).
Based on the presence of methylated DNA, H3K4me2, or H3K4me3 in the promoter or body of a gene, we further divided all modified genes into three groups: body-modified only, promoter-modified only, and body- and promoter-modified. DNA methylation, H3K4me2, and H3K4me3 tended to occur in the body of a gene; just a small number of genes were modified only in the promoter region (Figure 3A; see Supplemental Figure 8A online). However, annotation of the gene body is not robust for many genes since they are computationally predicted and lack experimental support. Therefore, to define better the differences between promoter and gene body modifications, we further divided the non-TE genes into two groups in three different ways: (1) 2934 genes supported by full-length cDNAs or ESTs (SG) and 3395 unsupported genes (computationally predicted genes [UG]); (2) 4136 known genes (known or putative function predictable) and 2193 unknown genes; and (3) 1684 genes (HH) with high homology and 4645 genes (LH) with low homology to Arabidopsis proteins (Ma et al., 2005). More than 90% of SG, known, and HH genes contained H3K4me2 and H3K4me3 compared with <75% for UG, unknown, and LH genes (see Supplemental Table 2 online), suggesting that these modifications are preferentially associated with canonical, actively transcribed genes in rice.
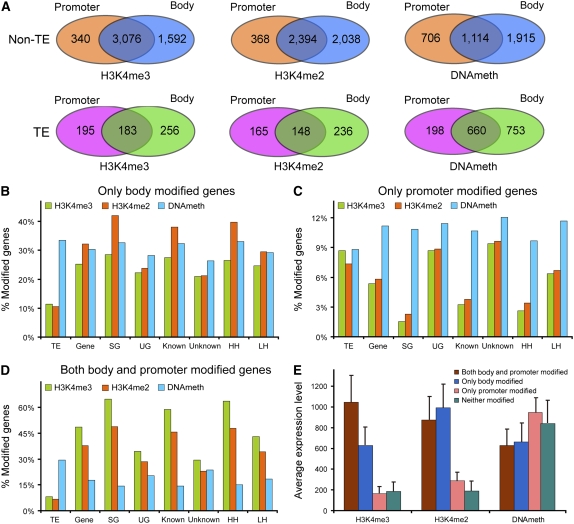
Figure 3.
DNA Methylation and Histone Modifications in Gene Promoter and Body Regions.
(A) Numbers of “only promoter,” “only body,” and “both promoter and body” modified TEs and non-TE genes in light-grown shoots.
(B) to (D) Proportions of “only promoter,” “only body,” and “both promoter and body” modified genes in three pairs of categories. The y axis shows the percentage of genes in the indicated category containing the specified modification.
(E) Expression levels of “only promoter,” “only body,” “both promoter and body” modified, and “neither modified” genes containing H3K4me3, H3K4me2, or methylated DNA. The y axis shows the average expression level calculated from an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). Error bars represent the se from five biological replicate microarray data sets.
We then examined the distribution of methylated DNA, H3K4me2, and H3K4me3 within the territories of these various groups (Figures 3B to 3D; see Supplemental Figures 8B to 8D online). More than 60% of SG and HH genes contained H3K4me3 regions in both their bodies and their promoters (Figure 3D; see Supplemental Table 2 online), and an additional 25% contained H3K4me3 regions only in their bodies (Figure 3B), demonstrating that H3K4me3 is associated with transcribed regions. Less than 60% of UG and unknown genes were modified in this way, perhaps reflecting errors in annotation or low transcription of UG genes. Similar distributions were observed for H3K4me2, except that its relative abundance was greater than H3K4me3 in “only body-modified” genes and less than H3K4me3 in “body- and promoter-modified” genes. Again, these differences were most apparent in SG, known, and HH genes. By contrast, <3.5% of SG and HH genes only contained H3K4me2 or H3K4me3 in their promoters, whereas a much larger fraction of UG, unknown, and LH genes fell in this category (Figure 3C), perhaps because regions annotated as promoter were in fact transcribed.
Much less variation was observed in the distribution of DNA methylation. Between 26 and 33% of all the various groups of non-TE genes fell in the “only body,” 9 to 12% in the “only promoter,” and 15 to 20% in the “body and promoter” categories. SG and HH had the lowest percentages of “only promoter” and “body and promoter” genes and the highest percentages of “only body” genes. More TEs (nearly 30%) were methylated in both body and promoter, whereas the percentages found in the “only body” and “only promoter” categories were similar to non-TE genes (Figure 3B and 3C; see Supplemental Table 2 online). Based on the low level and lack of variation in the percentages of “only promoter” DNA-methylated genes, we speculate that promoter region methylation may not affect transcription.
DNA Methylation, H3K4me2, and H3K4me3 Were Biased toward the 5′ End of Rice Genes but Peaked at Distinct Positions
We further examined the distribution of DNA methylation, H3K4me2, and H3K4me3 within genes. We aligned all the annotated non-TE genes at their translation start sites (ATG) because these are more accurately identified than transcription start sites in unsupported genes and then plotted the relative strength of each modification (the ratio of the signals from the samples enriched for each modification to the signal from total genomic DNA) against the distance from the ATG. This analysis revealed two distinct peaks of H3K4me3 and H3K4me2 within genes: a minor peak located ∼150 bp upstream of the ATG and a major peak centered ∼500 bp downstream (Figure 2E). The major H3K4me3 peak was markedly higher than the H3K4me2 peak. DNA methylation within genes exhibited a single peak near the ATG and between the major and minor peaks of H3K4 methylation (Figure 2E).
To examine these peaks in more detail and potential associations between gene length and epigenetic modifications, we divided the non-TE genes by size into long (> 2.0 kb), medium (1.0 to 2.0 kb), and short groups (<1.0 kb). All the TEs were included as a single group regardless of their size. We then plotted the intensity of H3K4me3, H3K4me2, and DNA methylation against distance from ATG for each group. We also plotted data from genes of all sizes supported by full-length cDNAs separately to serve as a reference group due to their accurate annotation, allowing the identification of the transcription start site. This analysis revealed that the peaks of H3K4me3 and H3K4me2 located ∼500 bp downstream of the ATG were found only in long genes (Figures 2F and 2G; see Supplemental Figures 6 and 7 online). Medium and short genes exhibited only the small peak upstream of the ATG and then H3K4me2 and H3K4me3 gradually decreased. Comparison to the full-length cDNAs showed that the minor peak centers at the putative transcription start sites, which may imply a role in transcriptional initiation. The major peak downstream of the ATG may reflect H3K4 methylation associated with transcriptional elongation (Figures 2E to 2G; see Supplemental Figures 6 and 7 online).
In contrast with H3K4me2 and H3K4me3, DNA methylation was slightly higher in short and medium than in long genes, but in all three groups, it peaked at the ATG and then declined (Figure 2H). Distribution of DNA methylation was quite different in TEs. It peaked ∼200 bp downstream of the ATG and declined much more slowly than in non-TE genes, remaining elevated throughout their territories (Figure 2H).
H3K4me3 Levels Are Positively Correlated with Transcript Abundance, whereas H3K4me2 and DNA Methylation Are Not
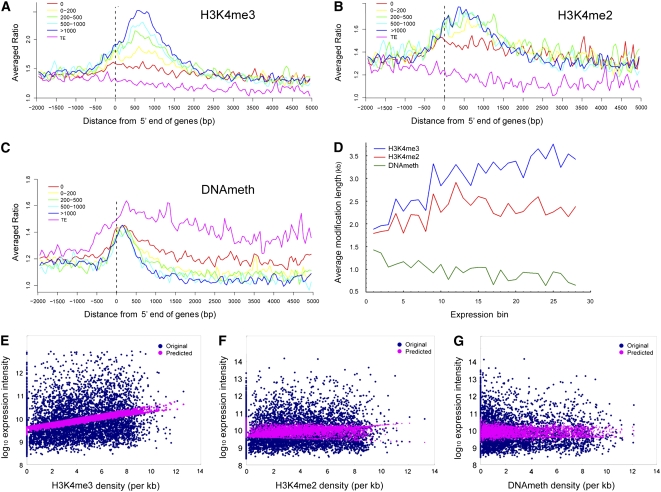
To correlate the three modifications with transcript abundance, we compared our data on rice shoot epigenetic modifications with an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). First, we divided the non-TE genes on chromosomes 4 and 10 into five groups based on their normalized expression intensities: 0, 0 to 200, 200 to 500, 500 to 1000, and >1000 using the TEs as reference group. Then, the average levels of H3K4me3, H3K4me2, and methylated DNA within each group were plotted against the distance from the 5′ end of the gene (Figures 4A to 4C). The TEs exhibited low levels of H3K4me2 and H3K4me3, but elevated DNA methylation over their entire territories. The non-TE genes that were not expressed had the lowest levels of both H3K4me2 and H3K4me3 but the highest levels of DNA methylation. We observed a positive correlation between transcript abundance and H3K4me3 (Figure 4A). By contrast, all four groups of expressed genes had more H3K4me2 than the nonexpressed genes, but there were no significant differences between them (Figure 4B). There were also no significant differences in DNA methylation between the four groups of expressed genes (Figure 4C). We obtained the same results for all three modifications using EST abundance instead of microarray data to estimate transcriptional activity (see Supplemental Figures 9A to 9C online).
Figure 4.
H3K4me3, H3K4me2, and DNA Methylation Mark Different Levels of Gene Expression.
(A) to (C) Distribution of H3K4me3, H3K4me2, and DNA methylation within five groups of genes with different levels of expression intensity from an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). TEs were used as reference. The y axis shows the averaged ratios of the signals from the samples enriched for each modification to the signal from total genomic DNA.
(D) Effect of length of modified region on gene expression. The x axis shows the expression bin number. Genes were sorted into 30 equal bins based on increasing expression intensity from an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). The y axis shows the average length of modified region of the genes in each bin.
(E) to (G) Multivariate linear regression analysis of the effects of three kinds of epigenetic modifications on gene expression. The x axis shows modification density calculated as described in the Supplemental Methods online. The y axis shows the log10-transformed expression intensity calculated from an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org).
We next performed a multivariate regression analysis to correlate the density of H3K4me3, H3K4me2, and methylated DNA coverage with transcript abundance assessed using the same microarray data set as above. This demonstrated a linear positive relationship between H3K4me3 levels and transcript abundance, whereas no correlation between H3K4me2 or DNA methylation and transcript abundance was detected (Figures 4E to 4G). However, an analysis of the correlation between the length of the modified regions within genes and transcript abundance uncovered more complex interactions. We divided expressed genes into 30 equal bins based on expression levels and plotted transcript abundance against the average length of the methylated DNA, H3K4me2, and H3K4me3 regions in the genes within each bin (Figure 4D). We observed a positive correlation between H3K4me3 and transcript abundance, indicating that more actively transcribed genes bear longer modified regions. We also observed a positive correlation between H3K4me2 length and transcript abundance for genes with low transcriptional activity. However, this leveled by bin 12 (moderate transcript abundance) so it was masked when all genes were considered at once. By contrast, we observed a negative correlation between length of DNA methylation and transcript abundance (Figure 4D).
H3K4me3 Levels Are Positively Correlated with the Number of Tissues in Which a Gene Is Expressed, whereas H3K4me2 and DNA Methylation Levels Are Not
To examine the effects of these three modifications on tissue specificity of gene expression, we chose the four distinct tissues with the largest pools of ESTs: panicle (150,845), leaf (204,353), root (79,340), and callus (184,189) (Rice Gene Expression Anatomy Viewer; http://www.tigr.org/). We then divided the genes on chromosomes 4 and 10 into four groups based on whether their ESTs were found in none, one, two, or three to four of these tissues and plotted the average intensities of H3K4me3, H3K4me2, and DNA methylation for each group. H3K4me3 levels were positively correlated with the number of tissues a in which a gene was expressed, whereas we did not observe any correlations between DNA methylation and H3K4me2 levels and the number of tissues in which a gene was expressed (see Supplemental Figure 10 online).
DNA Methylation and Histone Modifications of the Gene Body Correlate Better with Transcript Abundance Than DNA Methylation and Histone Modifications of the Promoter
To study the correlation between the positions of modified regions and transcript abundance, we first divided the genes into four groups for each modification, (1) only promoter modified, (2) only body modified, (3) both body and promoter modified, and (4) unmodified, and then compared the average transcript abundance of genes in each group derived from an analysis of rice seedling expression using an Affymetrix microarray (http://www.ricearray.org). Genes with H3K4me3 on “both promoter and body” had the most abundant transcripts, transcript abundance of those with H3K4me3 “only on body” was 40% lower, and transcript abundance of genes without H3K4me3 or with it only on the promoter was very low (Figure 3E). For genes carrying H3K4me2, those with H3K4me2 only on their bodies had the most transcripts, transcript abundance of “both body and promoter” modified genes was slightly lower, while transcript abundance of genes without H3K4me2 or with it only on their promoters was very low. By contrast, genes bearing DNA methylation only on their promoters had the most transcripts, while transcript abundance of genes with DNA methylation on both body and promoter or only on body was ∼30% lower (Figure 3E). These data imply that DNA methylation of the body is associated with a suppressive effect on transcription.
Methylation of DNA Correlates with Suppressed Gene Expression
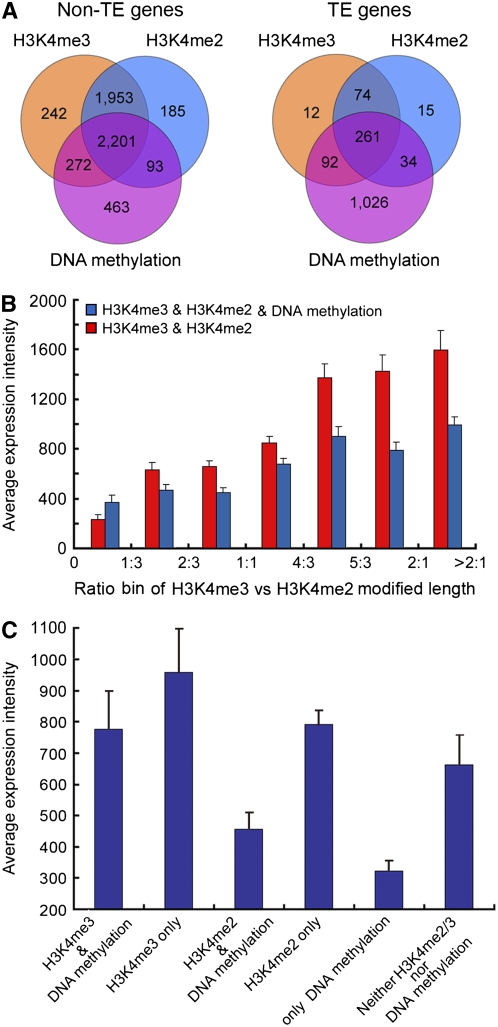
Most genes (TE and non-TE) with H3K4me2 on their bodies also had H3K4me3, and most non-TE genes with DNA methylation also had H3K4me2 or H3K4me3, but only 27% of TEs with DNA methylation also had methylated H3K4 (Figures 5A; see Supplemental Figure 11 online). To correlate effects of combinations of modifications with gene expression, we calculated the average expression levels of the non-TE genes with each combination of epigenetic modifications based on an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). In genes containing both H3K4me2 and H3K4me3, we observed that gene expression increased with the ratio of H3K4me3 to H3K4me2 and that gene expression was slightly reduced if the DNA was also methylated (Figure 5B). Genes that only contained H3K4me3 were expressed at higher levels than those that only contained H3K4me2. Genes that combined either modification with DNA methylation were expressed at slightly lower levels, whereas genes with DNA methylation but no H3K4 modification were expressed at very low levels (Figure 5C). By contrast, genes that did not contain any methylated DNA, H3K4me2, or H3K4me3 were expressed at levels comparable to those that only contained H3K4me2, showing that DNA methylation, by itself, is linked strongly to suppressed gene expression.
Figure 5.
Combinatorial Effects of Epigenetic Modifications on Transcription in Light-Grown Rice Shoots.
(A) Numbers of TEs and non-TE genes containing H3K4me3, H3K4me2, and DNA methylation regions and various combinations of these modifications.
(B) Transcription of non-TE genes containing both H3K4me2 and H3K4me3 regions with and without DNA methylation. The x axis shows the ratio of H3K4me3 to H3K4me2. Genes were classified into seven bins according to the ratio of the lengths of the H3K4me3 and H3K4me2 regions. The y axis shows the average transcript abundance within each group.
(C) Expression of non-TE genes with only one or two types of epigenetic modification. The y axis shows the average transcript abundance within each group. Average transcript abundances in (B) and (C) were calculated from an analysis of rice seedling expression using Affymetrix microarrays (http://www.ricearray.org). Error bars represent the se from five biological replicate microarray data sets.
Elevated H3K4me3 and Reduced DNA Methylation Are Correlated with Differential Gene Expression in Rice Shoots and Cultured Cells
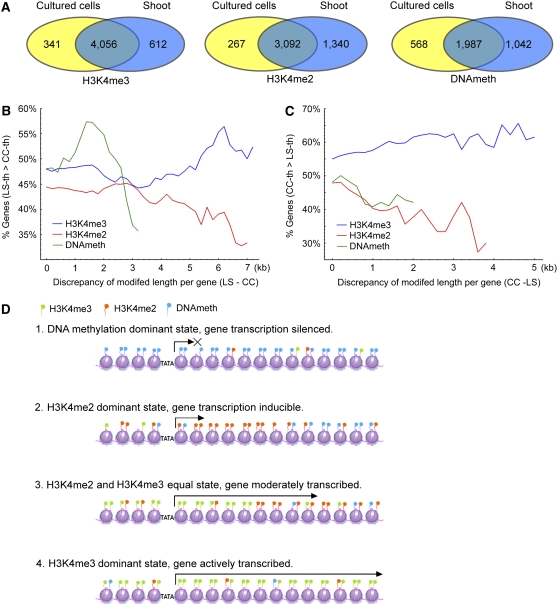
We examined tissue-specific epigenetic modifications and their possible effects on gene expression on a global scale by comparing undifferentiated cultured cells with 7-d-old light-grown shoots. Cultured cells had less DNA methylation in heterochromatin and H3K4me2 in euchromatin than shoots (Figures 1B and 1C). Most genes were modified the same way in both tissues; however, for all three modifications, substantially more genes were modified in shoots than in cultured cells (Figure 6A; see Supplemental Figures 5 and 12 online). For instance, 3092 genes contained H3K4me2 in both tissues, 1340 genes contained H3K4me2 in shoots and not cultured cells, while only 267 showed the opposite pattern. Examples of differential modifications of specific genes are presented in Supplemental Figure 13 online. These results are consistent with the hypothesis that epigenetic modifications help mark developmental states, since shoots are more differentiated than cultured cells.
Figure 6.
Differential Modifications and the Effects on Gene Differential Expression between Shoots and Suspension-Cultured Cells.
(A) Numbers of non-TE genes containing the indicated epigenetic modifications in shoots and/or cultured cells.
(B) and (C) Correlations of differential epigenetic modification and differential gene expression in light-grown shoots (LS) and cultured cells (CC). The y axis shows the percentage of genes that are more highly expressed in the indicated tissue based on a published rice microarray data set (Jiao and Deng, 2007). The x axis shows the difference in length of the indicated modification between the two tissues.
(D) Model of the association between DNA methylation, H3K4me2, and H3K4me3 and transcriptional activity.
Similarly, more TEs also contained DNA methylation and H3K4me2 in shoots than cultured cells, whereas there was no significant difference between the tissues in the numbers of TEs with H3K4me3 (see Supplemental Figures 5 and 12 online). These differences in epigenetic modification of TEs may contribute to the activation of some transposons in cultured cells (see Supplemental Figures 15B and 18 online), such as the Tos17 retrotransposon, which was activated during tissue culture but inactivated again in regenerated rice plants (Miyao et al., 2003).
An analysis of differential epigenetic modifications and tissue-specific expression was performed using a published rice microarray data set (Jiao and Deng, 2007). We first sorted all genes into two groups for each modification: those with longer modified regions in shoots and those with longer modified regions in cultured cells. We then plotted the percentages of genes with higher expression in one tissue against the difference in length of their modified regions. In both cases (shoots > cultured cells and cultured cells > shoots), we observed a positive correlation between the difference in length of the H3K4me3 regions and tissue-specific expression, whereas we observed the opposite trend for H3K4me2 and DNA methylation (Figures 6B and 6C).
To verify these observations, a number of specific samples were selected for corroboration by ChIP-PCR and RT-PCR. First, we substantiated the positive correlation between H3K4me3 and transcript abundance by detecting more transcripts from 36 non-TE genes, including 13 photosynthesis-related genes, and 22 TEs that varied in H3K4me3 between shoots and cultured cells in the tissue with more elevated H3K4me3 (see Supplemental Figures 14 and 15 online). Second, we corroborated the interactions between H3K4me2 and H3K4me3 using (1) a sample of 24 genes whose transcripts were more abundant in the tissue where they contained more H3K4me3 (see Supplemental Figure 16 online) and (2) a sample of 24 genes containing equal amounts of H3K4me3 in both tissues whose transcripts were less abundant in the tissue where they contained more H3K4me2 as well (see Supplemental Figure 17 online). Third, we validated the correlation between DNA methylation and reduced transcription in shoots by detecting transcripts of eight non-TE genes and 15 TEs in cultured cells, where they are hypomethylated, but not in shoots, where they are hypermethylated (see Supplemental Figure 18 online).
Genomic bisulfite sequencing of a DNA helicase-related gene and a TE from shoots and cultured cells showed that CG methylation was similar in both tissues, but methylation at CNG and CNN sites was significantly lower in cultured cells (see Supplemental Figures 3E, 3F, 3H, and 3I online). This result is consistent with the observation that symmetric methylation of CG is more stable and mainly associated with constitutive silencing of TEs and pseudogenes in Arabidopsis, while methylation at CNG and CNN is more dynamic and correlates with tissue-specific gene expression (Zhang et al., 2006).
Analysis of Epigenetically Modified Noncoding RNA Elements in Rice
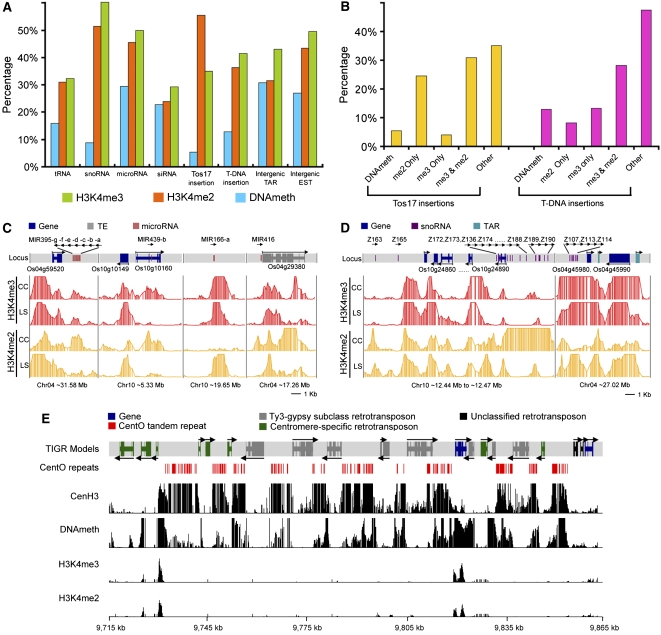
To investigate epigenetic modifications of noncoding RNA (ncRNA) loci, we collected tRNA, microRNA, small nucleolar RNA (snoRNA), and endogenous small interfering RNA (siRNA) sequences from public sources (see Methods) and mapped the epigenetic modifications in the expanded ncRNA locus region (including 0.5 kb on each side). All ncRNA loci had variable proportions of epigenetic modifications (Figure 7A; see Supplemental Figure 19 online). The snoRNA group, which is mostly involved in pre-rRNA processing, had a high percentage of H3K4 methylation but very little DNA methylation. By contrast, the siRNA group, which is involved in RNA-directed DNA methylation (Chan et al., 2004) and regulation of gene expression, had similar amounts of methylated DNA, H3K4me2, and H3K4me3.
Figure 7.
Mapping H3K4me3, H3K4me2, and DNA Methylation Regions in Relation to Other Genomic Elements and in Centromere 4 in Light-Grown Shoots.
(A) Percentages of tRNA, snoRNA, microRNA, siRNA, Tos17 and T-DNA insertions, intergenic TARs, and ESTs that overlap with H3K4me3, H3K4me2, and DNA methylation regions.
(B) Frequencies of Tos17 and T-DNA insertions occurring in H3K4me3, H3K4me2, and DNA methylation regions.
(C) and (D) Representative examples of microRNA and snoRNA loci differentially modified by H3K4me3 and H3K4me2 in light-grown shoots (LS) and cultured cells (CC). Rows 2 to 5 show log10-transformed P values for each probe.
(E) Epigenetic modifications of rice centromere 4. Rows 3 to 6 show log10-transformed P values for each probe. The locus ID numbers for those TE and non-TE gene models are summarized in Supplemental Table 3 online.
Interestingly, we also found some ncRNA loci that are modified tissue-specifically by H3K4 methylation. For example, the chromosomal neighborhood of microRNA MIR166-a, which targets mRNAs coding for HD-Zip transcription factors (PHABULOSA and PHAVOLUTA), is highly modified by H3K4me2 in shoots but not in cultured cells (Figure 7C). Similarly, a ∼6.0-kb region on chromosome 10 containing a cluster of 26 snoRNAs is highly enriched for H3K4me2 in cultured cells but not in shoots (Figure 7D).
Significant Fractions of Intergenic DNA Methylation and Histone Modification Regions Overlap with Signatures for Active Transcription
Hypothesizing that intergenic epigenetic modifications might mark coding units missed by the current genome annotation, we examined the overlap of intergenic H3K4me3, H3K4me2, and DNA methylation regions with two indicators of intergenic transcription: intergenic ESTs and transcriptionally active regions (TARs) (Li et al., 2007b). We found that 20.1, 18.9, and 10.8% of intergenic H3K4me3, H3K4me2, and DNA methylation regions, respectively, overlapped with intergenic ESTs (see Supplemental Figures 20A and 13G online) and that 30.5, 24.9, and 17.2% of intergenic H3K4me3, H3K4me2, and DNA methylation regions, respectively, overlapped with TARs (see Supplemental Figure 20B online). Combining ESTs and TARs with noncoding elements residing between annotated genes, ∼30% of intergenic DNA methylation and ∼50% intergenic H3K4 modification regions could be related to a transcribed sequence in shoots, although these percentages are lower in cultured cells (see Supplemental Figures 20C and 20D online).
Tos17 Retrotransposons Are More Abundant in H3K4me2 Regions, whereas T-DNA Insertions Are More Abundant in H3K4me3 Regions
Rice Tos17 and T-DNA integration sites are reported to be correlated with chromatin structure (Miyao et al., 2003; Jeong et al., 2006). We therefore mapped Tos17 and T-DNA insertions on rice chromosomes 4 and 10 and found 2148 Tos17 and 2280 T-DNA insertions in the euchromatic regions but only 294 Tos17 and 1210 T-DNA sites in the heterochromatic regions. Since DNA methylation, H3K4me2, and H3K4me3 are associated with different chromatin states, we examined relationships between integration sites and epigenetic modifications. Of the 2442 Tos17 insertion sites, 1356 (55.5%) and 854 (35.0%) are in H3K4me2 or H3K4me3 regions, respectively, while only 32 (5.4%) are in DNA methylation regions (Figure 7A; see Supplemental Figure 19A online).
Further division of the insertion sites into “H3K4me2 only,” “H3K4me3 only,” and “both H3K4me2 and H3K4me3” revealed striking differences between Tos17 and T-DNA. More than 50% of Tos17 insertions were in regions containing H3K4me2, and very few were found in regions containing only H3K4me3 or DNA methylation (Figure 7B; see Supplemental Figures 13E, 13F, and 19B online). A significant fraction of T-DNA insertions was also found in regions containing H3K4me2 or H3K4me3, but, unlike Tos17, >15% were found in regions containing only H3K4me3 and 13% were found in methylated DNA (Figure 7A; see Supplemental Figure 19A online). These findings suggest that Tos17 and T-DNA may have distinct mechanisms for selecting host sites in the integration process.
Rice Centromeres 4 and 8 Have Unique Epigenetic Modifications
Centromeres are essential for the proper segregation of chromosomes during eukaryotic cell division, and specific histone modifications have been reported to be involved in assembling the mitotic chromosomal apparatus (de la Barre et al., 2000). Inclusion of the complete sequences of rice centromeres 4 and 8 on the tiling array allowed us to examine epigenetic modifications of centromeres at high resolution.
The core centromeres consist of numerous clusters of centromere-specific CentO satellite repeats, interspersed with retrotransposons, and a few non-TE genes (Figure 7E; see Supplemental Figure 21 and Supplemental Table 3 online). We mapped H3K4me2, H3K4me3, and DNA methylation within centromeres 4 and 8, together with the binding sites of CenH3, a centromere-specific histone H3 variant needed for proper assembly of the kinetochore (Nagaki et al., 2004).
H3K4me2 and H3K4me3 were virtually absent from the core centromeric region, except for a short region on a non-TE gene in each core centromere (Figure 7E; see Supplemental Figure 21 online). This result is consistent with two prior low-resolution ChIP-PCR analyses in which H3K4me2 was not detected in most of the core region of rice centromere 8 except on a few active protein coding genes (Nagaki et al., 2004).
By contrast, we detected extensive binding of CenH3 within both core centromeric regions. CenH3 primarily bound CentO repeats and was rarely detected on centromeric retrotransposons (Figures 7E; see Supplemental Figure 21 online), suggesting that kinetochores might be formed through the interaction of CenH3 with CentO repeats.
CentO repeats were highly methylated, whereas retrotransposon DNA was not, in contrast with the hypermethylation of retrotransposon DNA in other parts of the genome. Among 19 annotated TEs in the core centromere 4 region (see Supplemental Table 3 online), only 12 were defined as DNA methylated either in the body or promoter region (see Supplemental Data Set 2 online). Moreover, the methylated region of DNA on centromeric TE is shorter than typical noncentromeric TE (Figures 2H and 4C) and not comparable with the broad DNA methylation domain of CentO repeats (Figure 7E). The relative hypomethylation of centromeric retrotransposons is consistent with their constitutive transcription in rice (Neumann et al., 2007). This may contribute to evolutionary expansion of centromeres through a transcription-mediated mechanism (Zhang et al., 2004).
DISCUSSION
The data presented here display the comprehensive landscapes of three distinct epigenetic modifications on two fully sequenced rice chromosomes and reveal the combinatorial interplay between epigenetic modifications and gene expression during development. The inclusion of two very large blocks (tens of Mb) of heterochromatin and two completely sequenced centromeres in our analysis provided insight into the epigenetic makeup of these unique eukaryotic chromosomal regions. This data set should therefore be useful generally for investigations in any biological process in rice and other crops, particularly cereals.
Combinatorial Interactions of DNA Methylation and Histone Modifications, Particularly within Gene Coding Regions, Are Correlated with Transcription and Chromatin State
DNA methylation is a heritable epigenetic mark implicated in gene silencing, perhaps by altering chromatin structure indirectly to a form recalcitrant to transcription (Lorincz et al., 2004; Okitsu and Hsieh, 2007). Our analysis confirmed that the DNA of most rice TEs was heavily methylated and also demonstrated that the DNA of more than half of the non-TE genes was also methylated in either the promoter or transcribed regions. Furthermore, more than two-thirds of non-TE genes with methylated DNA also contain H3K4me2 and H3K4me3. Comparison of groups of genes with varying amounts of DNA methylation, H3K4me2, and H3K4me3 showed that DNA methylation by itself is correlated with suppressed transcription. However, this effect is attenuated when DNA methylation is combined with H3K4me2 and/or H3K4me3, although genes with DNA methylation are still transcribed at lower levels than those without in the presence of methylated H3K4.
We observed a complex relationship between H3K4me2 and H3K4me3. There is a positive correlation between H3K4me2 levels and transcription of poorly expressed genes, but this effect plateaus at moderate levels of gene expression. By contrast, there is a positive linear correlation between H3K4me3 content and transcription (Figure 4; see Supplemental Figure 9 online). Previous studies of a few specific genes in yeast and chicken demonstrated that H3K4me2 occurs on both active and inactive genes, whereas H3K4me3 is present exclusively on active genes (Santos-Rosa et al., 2002; Schneider et al., 2004). Our results confirm this finding on a genomic scale and extend it to the plant kingdom. Analysis of transcription in the presence of varying proportions of H3K4me2 and H3K4me3 showed that transcript abundance decreased with increasing H3K4me2. The nature of this seemingly competitive interaction between H3K4me2 and H3K4me3 needs further investigation.
A recent genome-wide study of DNA methylation in Arabidopsis indicated that genes with DNA methylation in the body region as a whole tended to be constitutively active (Zhang et al., 2006). Another Arabidopsis study demonstrated that moderately expressed genes were most likely to have methylated DNA, while genes with either high or low levels of expression were relatively hypomethylated (Zilberman et al., 2007). These observations could be explained if, in Arabidopsis, most genes with methylated DNA also contain methylated H3K4 and the highly expressed genes in the latter study contain H3K4me3 regions.
Our data support a model that in rice chromatin genes are marked by different epigenetic modifications whose combinations determine distinct gene expression states. There are four typical chromatin states with respect to the three epigenetic modifications examined in this study (Figure 6D). DNA methylation in the absence of methylated H3K4 (state 1) marks a gene for silencing, resulting in a condensed chromatin structure that impedes transcription. The presence of H3K4me2, even in the presence of DNA methylation (state 2), alters the chromatin structure to a form permissive for initiation of transcription. The presence of moderate amounts of H3K4me3 (state 3) adjusts the chromatin to a state permitting more active transcription. Finally, if H3K4me3 is the dominant modification (state 4), the chromatin adopts a conformation permitting maximal transcription.
Characteristic Patterns of DNA Methylation, H3K4me2, and H3K4me3 in Rice Genes
All three epigenetic modifications examined in this study are located mainly in the gene territories. Previous studies of DNA methylation in a small number of Arabidopsis genes, such as FWA and FIS2, have shown that DNA methylation in the promoter region plays an important role in suppressing gene expression (Soppe et al., 2000; Jullien et al., 2006). However, in our global analysis of DNA methylation in rice, only a small number of genes was methylated only in their promoters. Instead, most were methylated in both body and promoter or only in the body. Closer analysis showed that DNA methylation preferentially occurs closely downstream from the transcription start site. Moreover, DNA methylation of the transcribed region is associated with a larger suppressive effect than DNA methylation of the promoter region, since genes with DNA methylation only in their promoters have higher expression levels than genes methylated only in their bodies or in both bodies and promoters. This may be due to the lack of DNA methylation in their bodies.
This result is consistent with the conclusion from two recent experiments using patched DNA methylation on stable episomes or transgenes in mammalian cells, which demonstrated that DNA methylation impacts transcriptional elongation much more than initiation (Lorincz et al., 2004; Okitsu and Hsieh, 2007). In the filamentous fungus N. crassa, transcriptional initiation was not significantly inhibited by dense DNA methylation in the promoter sequences, while transcriptional elongation was completely blocked by intragenic methylation (Rountree and Selker, 1997). In plant protoplasts, DNA methylation of the coding region alone, particularly near the 5′ end, can also interfere with transcriptional elongation (Hohn et al., 1996). Thus, our observation that DNA methylation in the gene body has a larger effect on transcription than DNA methylation in the promoter is conserved in many organisms.
The two histone H3K4 modifications examined also preferentially occurred at the 5′ ends of genes, but both showed a key difference to the DNA methylation pattern. Maps of H3K4me2 and H3K4me3 within longer genes showed two peaks: a minor peak located at putative transcription start sites, ∼150 to 200 bp upstream of the ATG, and a major peak centered 500 bp downstream of the ATG. The positions of the two peaks of both H3K4me2 and H3K4me3 suggest a role in or relationship with transcriptional initiation and elongation. It is noteworthy that the single peak of DNA methylation is located between the minor and major peaks of the two H3K4 histone modifications, centering near the ATG. At present we do not know the implications of these distribution patterns, although they probably indicate some interplay between DNA methylation and the histone modifications.
Differences between Epigenetic Modifications of Heterochromatin and Euchromatin
Our analysis of DNA methylation, H3K4me2, and H3K4me3 in two rice chromosomes and centromeres covered heterochromatin on a remarkably large scale because approximately half of rice chromosomes 4 and 10 are cytologically defined as heterochromatin, including the entire short arms and extensions into the long arms (Cheng et al., 2001; Yan and Jiang, 2007). This analysis is an improvement in both scale and resolution on a prior epigenetic modification analysis of a 1.5-Mb region encompassing the 730-kb heterochromatic knob on chromosome 4 in Arabidopsis (Lippman et al., 2004). Our data revealed that heterochromatin has higher levels of DNA methylation than euchromatin, for example, 18.1% versus 14.2% on rice chromosome 4 in shoots. However, the difference in DNA methylation is not as large as expected, which may be due to the low coverage of highly repetitive sequences in the heterochromain on our tiling array. By contrast, the amounts of H3K4me2 or H3K4me3 in euchromatin and heretrochromatin are significantly different: for example, 30.9% versus 11.2% for H3K4me2 and 35.3% versus 13.2% for H3K4me3 on chromosome 4 in shoots. This is consistent with the report from Arabidopsis (Lippman et al., 2004) and suggests that histone modifications, such as H3K4me2 and H3K4me3, play important roles in discriminating heterochromatin and euchromatin. However, not all genes in heterochromatin are silenced. More than 10% of the heterochromatic region is modified by H3K4me2 or H3K4me3, and transcriptional activity has been detected in the heterochromatin of rice chromosomes 4 and 10 (Jiao et al., 2005; Li et al., 2006).
These data will be useful for studies in many other crops because of the high synteny between rice and other cereal genomes, which also contain large heterochromatic regions (Kurata et al., 2002). Indeed, they may also be useful for dicotyledenous crops that have large blocks of heterochromatin, such as tomato (Solanum lycopersicum; Yan and Jiang, 2007).
Differences in Epigenetic Modifications and Transcription in Rice Shoots and Cultured Cells
We observed that many more protein-coding genes contained H3K4me2, H3K4me3, and DNA methylation in light-grown shoots than in cultured cells, although there were also many genes that contained these modifications in cultured cells and not in light-grown shoots (Figure 6A; see Supplemental Figures 5 and 12 online). Similar differences were observed in the epigenetic modifications of TEs and noncoding RNAs in the two tissues. These results illustrate that each cell type has its own epigenome that can vary quite dramatically between tissues. We also demonstrated that these differences in epigenetic modifications are correlated with differences in transcript abundance (Figure 6B; see Supplemental Figures 14 to 18 online).
This suggests that differential modification of the epigenome may be an important component of developmental regulation of gene expression, which could be tested by studying changes in the epigenome during plant regeneration from cultured cells.
This knowledge could also be used to influence the activity of transposons or other genetic elements, such as has been done in the case of Tos17.
A Comparison of Histone H3K4 Di- and Trimethylation Patterns in Rice and Humans
High-resolution maps of histone H3K4me2 and H3K4me3 patterns in rice enable us to compare the epigenetic patterns in a plant with a map of the same histone modifications in the nonrepetitive portions of human chromosomes 21 and 22 (Bernstein et al., 2005). In rice shoots, ∼21 and ∼24% of chromosomes 4 and 10 are covered by H3K4me2 and H3K4me3, respectively; while in humans, only ∼1.0% of the two chromosomes are covered by H3K4me2 or H3K4me3 (Bernstein et al., 2005). The huge difference between rice and humans in the percentage of chromosomal coverage of H3K4me2 and H3K4me3 may be due to the differences in gene organization and structure, such as intron size of the two genomes. Data from this study indicated that H3K4me3 is a mark for expressed rice genes and covers only one small region in the coding sequence. The seemingly large difference of H4K4me3 modification rate in rice and human can thus be explained by the combination of two factors. First, the gene density is much greater in the rice genome, which has ∼40,000 genes in ∼390 Mb compared with the 20,000 genes in 3000 Mb in humans (International Rice Genome Sequencing Project, 2005). Second, the median size of H3K4me3 regions in rice (913 bp) is larger than that in humans (659 bp). These two factors would bring a combined 20-fold difference in the total genome coverage of the H3K4me3-modified regions, whereas the actual association between H3K4me3 and expressed sequences is similar in the two organisms. This interpretation is strengthened by recent studies (Barski et al., 2007; Heintzman et al., 2007) that show that the positioning of H3K4me2 and H3K4me3 relative to the transcription start site and within the coding region in humans is very similar to what we observed in rice (Figure 2; see Supplemental Figure 6 online).
The Epigenetic Modification Patterns of the First Two Completely Sequenced Centromeres of Multicellular Organisms
It has been reported that centromeres in human (and fruit fly) cells have a chromatin structure distinct from both euchromatin and heterochromatin. Specifically, neither heterochromatin markers, such as H3K9me2 and H3K9me3, nor euchromatin markers, such as H3K9ac and H3K4me3, were detected in the centromere region of extended chromatin fibers from interphase cells, although H3K4me2 was detected (Sullivan and Karpen, 2004). In our analysis, the complete sequences of rice centromeres 4 and 8 were tiled at high resolution. However, H3K4me2 was not detected in either rice centromere 4 or 8 in our study nor in the core region of rice centromere 8 in a previous study (Nagaki et al., 2004). This suggests that centromere chromatin structure may differ among organisms. Interestingly, we found that both CenH3 binding and DNA methylation are not continuous in the core centromere regions. Instead, both are strongly enriched at the CentO satellite repeats but relatively depleted at the interspersed retrotransposons. This finding is consistent with a prior low-resolution analysis (Nagaki et al., 2004; Yan et al., 2006) but provides high-resolution insights not discernable previously, such as the discrete binding patterns of CenH3 on CentO repeat regions. The specific interactions between CenH3 and CentO repeats likely play important roles during kinetochore assembly and thus provide a foundation for further analysis of centromere function.
METHODS
Plant Materials and Growth Conditions
All plants used in this study were rice strain Oryza sativa ssp japonica cv Nipponbare. Dehusked seeds were surface-sterilized and sown on solidified Murashige and Skoog medium with 3.0% sucrose. Plants were grown in chambers at 28°C with continuous white light (220 μmol·m−2·s−1) for 7 d, and the entire shoots were harvested. Suspension-cultured cells were derived from the same rice strain and maintained as described previously (Su et al., 2007).
Isolation of Epigenetically Modified Genomic DNA Fragments
Methylated DNA was isolated from total genomic DNA prepared using the DNeasy plant mini kit (Qiagen) by the McrBC digestion method (Lippman et al., 2004). DNA bearing modified histones was isolated by ChIP with antibodies that specifically recognize H3K4me2 (Upstate), H3K4me3 (Abcam), and CenH3 (Nagaki et al., 2004). See Supplemental Methods online for detailed experiment protocols.
Tiling Microarray Design, Hybridization, Scanning, and Data Analysis
Tiling probes were selected by the NASA Oligonucleotide Selection Algorithm (NOPSA) (Stolc et al., 2005). The algorithm for filtering the repetitive probes is described in the Supplemental Methods online. Microarrays were hybridized with Cy3- or Cy5-labeled DNA for 16 to 20 h at 50°C and then washed as described in the Supplemental Methods online. Hybridization images were generated by a GenePix 4200A scanner (Axon). Raw data were sequentially processed by LOESS normalization and Quantile normalization, and then regions bearing epigenetic modifications were identified using the Wilcoxon signed rank test (see Supplemental Methods online for specific procedure and parameters). For DNA and histone methylation, two or three biological replicates, respectively, of each tissue were performed.
McrBC-PCR, ChIP-PCR, RT-PCR, and Genomic Bisulfite Sequencing
The McrBC-PCR method was adapted from a previous study (Rabinowicz et al., 2003). One aliquot (McrBC+) of 1.0 μg of genomic DNA was digested overnight with 10 units of McrBC (Biolabs) and another aliquot (McrBC−) was incubated overnight with heat-inactivated McrBC. PCR was performed on equal amounts of DNA from both aliquots. ChIP-PCR was performed on DNA samples from input, mock (no antibody), and ChIP. For RT-PCR, total RNA was extracted using the RNeasy plant mini kit (Qiagen), and then first-strand cDNA was synthesized from 1.0 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) primed by oligo(dT)19, and PCR was performed using first-strand cDNA as template. For genomic bisulfite sequencing, 1.0 μg of genomic DNA was treated with sodium bisulfite using the CpGenome DNA modification kit (CHEMICON). PCR products amplified from the bisulfite-treated DNA were cloned into the vector pGEM-T easy (Promega), and 10 individual clones were sequenced. All the PCR primers are listed in Supplemental Table 4 online.
Accession Number
All the microarray data and specifications were deposited in the National Center for Biotechnology Information microarray database under accession number GSE9925.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Isolation of Methylated DNA by the McrBC Digestion Method.
Supplemental Figure 2. Experimental Confirmation of DNA Methylation, H3K4me3, and H3K4me2 Regions.
Supplemental Figure 3. Experimental Validation of DNA Methylation Regions by Genomic Bisulfite Sequencing.
Supplemental Figure 4. Coverage of Rice Chromosomes 4 and 10 by Three Types of Epigenetic Modifications in Suspension-Cultured Cells.
Supplemental Figure 5. Proportions of Epigenetically Modified Protein-Coding and TE Genes in Heterochromatin and Euchromatin.
Supplemental Figure 6. Distribution of H3K4me3, H3K4me2, and DNA Methylation within Aligned Genes in Light-Grown Rice Shoots.
Supplemental Figure 7. Distributions of H3K4me3, H3K4me2, and DNA Methylation within Aligned Genes in Cultured Rice Cells.
Supplemental Figure 8. Occurence of DNA Methylation, H3K4me3, and H3K4me2 in Promoter or Body Regions of Genes in Cultured Rice Cells.
Supplemental Figure 9. Correlation between H3K4me3, H3K4me2, and DNA Methylation and Gene Expression Estimated from EST Frequencies in Rice Shoots.
Supplemental Figure 10. Correlation of H3K4me3, H3K4me2, and DNA Methylation with Tissue Specificity of Gene Expression.
Supplemental Figure 11. Numbers of Transposable Elements and Non-TE Genes Containing H3K4me3, H3K4me2, and DNA Methylation Regions and Various Combinations thereof in Cultured Rice Cells.
Supplemental Figure 12. Numbers of TEs and Non-TE Genes That Are Differentially Modified by H3K4me3, H3K4me2, or DNA Methylation in Light-Grown Rice Shoots and Cultured Cells.
Supplemental Figure 13. Examples of Differentially Modified Genes, Tos17 Insertion Sites, and Intergenic Modifications in Light-Grown Rice Shoots and Cultured Cells.
Supplemental Figure 14. Higher Levels of H3K4me3 in Light-Grown Shoots Than in Cultured Cells Correlate with Higher Gene Expression Levels in Light-Grown Shoots Than in Cultured Cells.
Supplemental Figure 15. Higher Levels of H3K4me3 in Suspension-Cultured Cells Than in Light-Grown Shoots Correlate with Higher Gene Expression Levels in Cultured Cells Than in Light-Grown Shoots.
Supplemental Figure 16. Higher Levels of H3K4me3 Than H3K4me2 Correlate with Higher Levels of Gene Expression.
Supplemental Figure 17. Higher Levels of H3K4me2 Correlate with Reduced Gene Expression in Light-Grown Shoots and Cultured Cells if Both Tissues Have Equal Levels of H3K4me3.
Supplemental Figure 18. DNA Hypomethylation Correlates with Increased Gene Expression in Cultured Rice Cells Compared with Light-Grown Shoots.
Supplemental Figure 19. Mapping H3K4me3, H3K4me2, and DNA Methylation Regions onto Other Genomic Elements in Cultured Rice Cells.
Supplemental Figure 20. Mapping Intergenic Epigenetically Modified Regions against Intergenic TARs and ESTs.
Supplemental Figure 21. Epigenetic Modifications of Rice Centromere 8.
Supplemental Table 1. Coverage of Three Epigenetic Modifications on Chromosomes 4 and 10.
Supplemental Table 2. Percentages of Different Classes of Genes That Are Modified Only in Body, Only in Promoter, or in Both Body and Promoter.
Supplemental Table 3. Annotated Gene Models in the Core Region of Centromeres 4 and 8.
Supplemental Table 4. Primers Used in McrBC-PCR, ChIP-PCR, RT-PCR, and Genomic Bisulfite Sequencing.
Supplemental Data Set 1. Modifed Regions on Rice Chromosomes 4 and 10.
Supplemental Data Set 2. Gene Modification Status in Body Region and Promoter Region.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Lei Li for comments and discussions of the manuscript. This work was supported by a grant from the 863 program on rice functional genomics of the Ministry of Science and Technology of China and National Institute of Biological Sciences at Beijing and in part by the National Science Foundation Plant Genome Program (DBI-0421675) to X.W.D. X.L. is a long-term postdoctoral fellow of the Human Frontier Science Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
References
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J.M. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D.E., Wang, Z., Wei, G., Chepelev, I., and Zhao, K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Bender, J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55 41–68. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B.E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D.K., Huebert, D.J., McMahon, S., Karlsson, E.K., Kulbokas III, E.J., Gingeras, T.R., Schreiber, S.L., and Lander, E.S. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120 169–181. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W.-L., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1336. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Buell, C.R., Wing, R.A., Gu, M., and Jiang, J. (2001). Toward a cytological characterization of the rice genome. Genome Res. 11 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Barre, A. E., Gerson, V., Gout, S., Creaven, M., Allis, C.D., and Dimitrov, S. (2000). Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 19 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman, N.D., et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39 311–318. [DOI] [PubMed] [Google Scholar]
- Hohn, T., Corsten, S., Rieke, S., Muller, M., and Rothnie, H. (1996). Methylation of coding region alone inhibits gene expression in plant protoplasts. Proc. Natl. Acad. Sci. USA 93 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander, M., and Wolfe, D.A. (1999). Nonparametric Statistical Methods. (New York: John Wiley & Sons).
- Hsieh, T.-F., and Fischer, R.L. (2005). Biology of chromatin dynamics. Annu. Rev. Plant Biol. 56 327–351. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Jeong, D.-H., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45 123–132. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., and Deng, X.W. (2007). A genome-wide transcriptional activity survey of rice transposable element-related genes. Genome Biol. 8 R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., et al. (2005). A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription. Plant Cell 17 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien, P.E., Kinoshita, T., Ohad, N., and Berger, F. (2006). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata, N., Nonomura, K.-I., and Harushima, Y. (2002). Rice genome organization: The centromere and genome interactions. Ann. Bot. (Lond.) 90 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Carey, M., and Workman, J.L. (2007. a). The role of chromatin during transcription. Cell 128 707–719. [DOI] [PubMed] [Google Scholar]
- Li, L., et al. (2007. b). Global identification and characterization of transcriptionally active regions in the rice genome. PLoS ONE 2 e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Wang, X., Stolc, V., Li, X., Zhang, D., Su, N., Tongprasit, W., Li, S., Cheng, Z., Wang, J., and Deng, X.W. (2006). Genome-wide transcription analyses in rice using tiling microarrays. Nat. Genet. 38 124–129. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., Gendrel, A.V., Black, M., Vaughn, M.W., Dedhia, N., McCombie, W. R., Lavine, K., Mittal, V., May, B., Kasschau, K.D., Carrington, J.C., Doerge, R.W., Colot, V., and Martienssen, R. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476. [DOI] [PubMed] [Google Scholar]
- Lorincz, M.C., Dickerson, D.R., Schmitt, M., and Groudine, M. (2004). Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11 1068–1075. [DOI] [PubMed] [Google Scholar]
- Ma, L., et al. (2005). A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 15 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A., and Colot, V. (2001). DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293 1070–1074. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Zhang, Y. (2005). The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6 838–849. [DOI] [PubMed] [Google Scholar]
- Mellor, J. (2006). It takes a PHD to read the histone code. Cell 126 22–24. [DOI] [PubMed] [Google Scholar]
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozuka, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Neumann, P., Yan, H., and Jiang, J. (2007). The centromeric retrotransposons of rice are transcribed and differentially processed by RNAi. Genetics 176 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngernprasirtsiri, J., Kobayashi, H., and Akazawa, T. (1988). DNA methylation as a mechanism of transcriptional regulation in nonphotosynthetic plastids in plant cells. Proc. Natl. Acad. Sci. USA 85 4750–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu, C.Y., and Hsieh, C.-L. (2007). DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 27 2746–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski, J., and Whitham, S.A. (2001). Gene silencing and DNA methylation processes. Curr. Opin. Plant Biol. 4 123–129. [DOI] [PubMed] [Google Scholar]
- Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., and Reinberg, D. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125 703–717. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Palmer, L.E., May, B.P., Hemann, M.T., Lowe, S.W., McCombie, W.R., and Martienssen, R.A. (2003). Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 13 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J. (1997). DNA methylation and plant development. Trends Genet. 13 319–323. [DOI] [PubMed] [Google Scholar]
- Rountree, M.R., and Selker, E.U. (1997). DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C.T., Schreiber, S.L., Mellor, J., and Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419 407–411. [DOI] [PubMed] [Google Scholar]
- Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C., and Kouzarides, T. (2004). Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6 73–77. [DOI] [PubMed] [Google Scholar]
- Schotta, G., Ebert, A., Dorn, R., and Reuter, G. (2003). Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14 67–75. [DOI] [PubMed] [Google Scholar]
- Sims III, R.J., and Reinberg, D. (2006). Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 20 2779–2786. [DOI] [PubMed] [Google Scholar]
- Soppe, W.J.J., Jacobsen, S.E., Alonso-Blanco, C., Jackson, J.P., Kakutani, T., Koornneef, M., and Peeters, A.J.M. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6 791–802. [DOI] [PubMed] [Google Scholar]
- Stolc, V., et al. (2005). A pilot study of transcription unit analysis in rice using oligonucleotide tiling-path microarray. Plant Mol. Biol. 59 137–149. [DOI] [PubMed] [Google Scholar]
- Su, N., He, K., Jiao, Y., Chen, C., Zhou, J., Li, L., Bai, S., Li, X., and Deng, X.W. (2007). Distinct reorganization of the genome transcription associates with organogenesis of somatic embryo, shoots, and roots in rice. Plant Mol. Biol. 63 337–349. [DOI] [PubMed] [Google Scholar]
- Sullivan, B.A., and Karpen, G.H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru, H., and Selker, E.U. (2001). A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., Zhang, X., McMillen, D., Singh, P.B., Nakayama, J.-i., Grewal, S.I., Allis, C.D., Cheng, X., and Selker, E.U. (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34 75–79. [DOI] [PubMed] [Google Scholar]
- Yan, H., et al. (2006). Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18 2123–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H., and Jiang, J. (2007). Rice as a model for centromere and heterochromatin research. Chromosome Res. 15 77–84. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W.-L., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1–13. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Huang, Y., Zhang, L., Li, Y., Lu, T., Lu, Y., Feng, Q., Zhao, Q., Cheng, Z., Xue, Y., Wing, R.A., and Han, B. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.