Abstract
Gibberellin 3-oxidase (GA3ox) catalyzes the final step in the synthesis of bioactive gibberellins (GAs). We examined the expression patterns of all four GA3ox genes in Arabidopsis thaliana by promoter–β-glucuronidase gene fusions and by quantitative RT-PCR and defined their physiological roles by characterizing single, double, and triple mutants. In developing flowers, GA3ox genes are only expressed in stamen filaments, anthers, and flower receptacles. Mutant plants that lack both GA3ox1 and GA3ox3 functions displayed stamen and petal defects, indicating that these two genes are important for GA production in the flower. Our data suggest that de novo synthesis of active GAs is necessary for stamen development in early flowers and that bioactive GAs made in the stamens and/or flower receptacles are transported to petals to promote their growth. In developing siliques, GA3ox1 is mainly expressed in the replums, funiculi, and the silique receptacles, whereas the other GA3ox genes are only expressed in developing seeds. Active GAs appear to be transported from the seed endosperm to the surrounding maternal tissues where they promote growth. The immediate upregulation of GA3ox1 and GA3ox4 after anthesis suggests that pollination and/or fertilization is a prerequisite for de novo GA biosynthesis in fruit, which in turn promotes initial elongation of the silique.
INTRODUCTION
Bioactive gibberellins (GAs) are a major class of phytohormones that regulates plant growth and development, from seed germination and vegetative growth to fruit and seed set (Davies, 2004; Swain and Singh, 2005). Plants with impaired GA biosynthesis show typical GA-deficient phenotypes, such as dark green leaves, dwarfism, and low fertility, while GA overdose can cause excessive plant growth and increased sterility (Fleet and Sun, 2005). Therefore, it is important for plants to produce and maintain optimal levels of bioactive GAs to ensure normal growth and development. Bioactive GAs in plants are derived from geranylgeranyl diphosphate. Most of the intermediates in this pathway and genes encoding enzymes catalyzing each step have been identified (for reviews, see Hedden and Phillips, 2000; Olszewski et al., 2002). In the last steps of the pathway, two enzymes, GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), catalyze consecutive reactions that convert GA intermediates to the bioactive forms (Figure 1). In Arabidopsis thaliana, GA4 is the major bioactive form, while another bioactive GA (GA1) is present at lower levels (Talón et al., 1990). In addition, GA4 has a higher affinity to the GA receptor GIBBERELLIN-INSENSITIVE DWARF1 (GID1) than GA1 (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). This is consistent with previous observations showing that GA4 is more active than GA1 in promoting seed germination (Yang et al., 1995), flowering (Eriksson et al., 2006), and fertility in Arabidopsis (Fei et al., 2004). Another enzyme, GA 2-oxidase (GA2ox), catalyzes the conversion of bioactive GAs to inactive catabolites. Recently, two novel deactivation mechanisms were identified: epoxidation of non-13-hydroxylated GAs in rice (Oryza sativa) internodes (Zhu et al., 2006) and methylation of GAs in Arabidopsis (Varbanova et al., 2007). It is not clear, however, whether and how these two types of modification are used in different species to modulate active GA content.
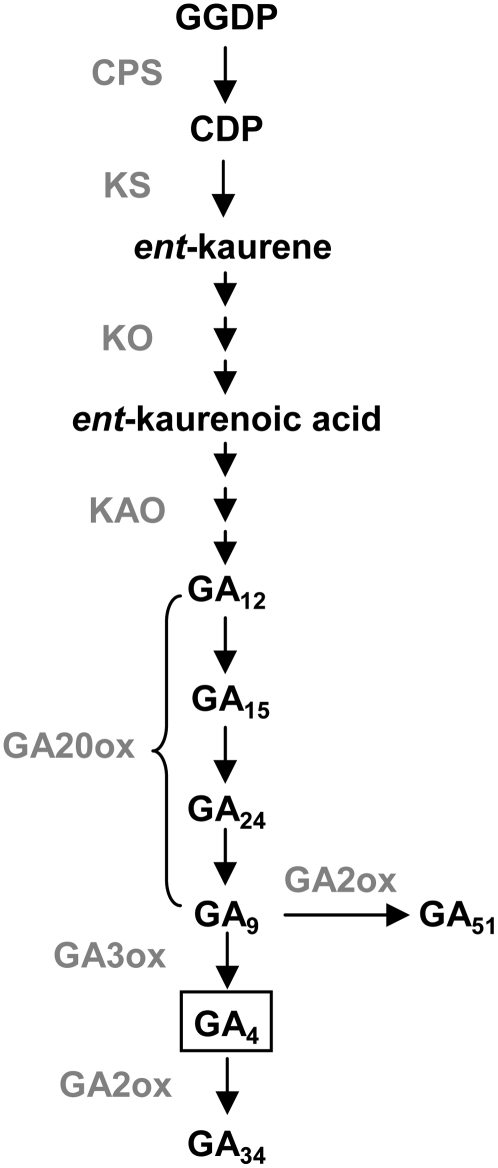
Figure 1.
GA Biosynthetic and Catabolic Pathways in Plants.
This is a simplified diagram only showing the pathways for the synthesis of non-13-hydroxylated GAs. Gray text indicates GA synthetic and catabolic enzymes catalyzing corresponding steps. GGDP, geranylgeranyl diphosphate; CDP, ent-copalyl diphosphate; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase.
The GA metabolic pathway is a dynamic process that is regulated by complex mechanisms. A common mechanism used by various plant species to maintain GA homeostasis is through feedback regulation of the GA20ox (Phillips et al., 1995; Xu et al., 1995) and GA3ox (Chiang et al., 1995; Yamaguchi et al., 1998) genes and feedforward regulation of the GA2ox genes (Thomas et al., 1999). In addition, regulation of GA biosynthesis may be achieved by separating early (ent-copalyl diphosphate synthase [CPS]) and late (ent-kaurene oxidase [KO] and GA3ox) enzymes into different cell types, as was observed in germinating Arabidopsis seeds (Yamaguchi et al., 2001) and elongating roots (Mitchum et al., 2006). GA20ox, GA3ox, and GA2ox are each encoded by a gene family whose members show differential expression patterns (Hedden and Phillips, 2000), reflecting a strategy used by the plant to fine-tune its control of bioactive GA levels during later steps of GA metabolism. Moreover, expression of GA biosynthetic genes is also subjected to regulation by other hormones (Bouquin et al., 2001; Ross et al., 2002; Seo et al., 2006; Achard et al., 2007) and environmental conditions such as low temperature (Yamauchi et al., 2004) and light (Yamaguchi et al., 1998). In most cases, GA3ox and GA20ox genes are targeted, implying that the late steps of GA biosynthesis are the main site for active regulation.
GAs have been found to be more abundant in actively growing tissues, suggesting that GA biosynthesis is tightly linked to the site of action of bioactive GAs (Hedden and Phillips, 2000; Olszewski et al., 2002). However, due to the low abundance of GA, it has been difficult to determine precisely where bioactive GAs are made in plants. Moreover, little is known regarding how plants regulate the levels of bioactive GAs in particular tissues and cells in response to developmental and environmental cues. Recent studies on the expression of GA biosynthetic genes and characterization of GA-deficient mutants are beginning to reveal valuable information on how bioactive GAs are made in plants (Hedden and Phillips, 2000; Olszewski et al., 2002). For example, expression of the Arabidopsis CPS occurs mainly in rapidly growing tissues (Silverstone et al., 1997), suggesting that its expression correlates with cells actively responding to GA. The same study also revealed expression of CPS in the vasculature of mature nonexpanding leaves, suggesting that GA intermediates or active GAs synthesized in this tissue may be transported to support growth of other tissues (Silverstone et al., 1997).
As the enzyme catalyzing the last step in making bioactive GAs, GA3ox is particularly interesting because the location of GA3ox gene expression indicates where bioactive GAs are potentially made in plants. In tobacco (Nicotiana tabacum) and rice, GA3ox genes are expressed in actively dividing and elongating cells, including the rib meristem, shoot apices, tapetum of developing anthers, and root tips (Itoh et al., 1999, 2001; Kaneko et al., 2003). Arabidopsis contains four GA3ox genes (GA3ox1-GA3ox4) that display distinct developmental expression patterns as indicated by quantitative RT-PCR (qRT-PCR) (Mitchum et al., 2006). GA3ox1 is expressed throughout development and has the highest level of mRNA accumulation among GA3oxs in most tissues (except in germinating seeds). GA3ox2 is mainly expressed during seed germination and vegetative growth, while both GA3ox3 and GA3ox4 are mainly expressed in flowers and siliques. The expression patterns suggest that GA3ox1 is the major supplier of the GA3ox enzyme in Arabidopsis, and the other GA3ox genes likely function during specific developmental stages. The tissue- and cell type–specific expression patterns of GA3ox1 and GA3ox2 were also studied using in situ hybridization and promoter-reporter gene fusions (Yamaguchi et al., 2001; Gomez-Mena et al., 2005; Mitchum et al., 2006). Both genes are mainly expressed in actively growing organs or tissues, similar to the tobacco and rice GA3ox genes. Mutant analysis of GA3ox1 and GA3ox2 further verified their physiological functions in GA biosynthesis and plant development (Mitchum et al., 2006). The ga3ox1 single mutant displays a semidwarf phenotype with normal seed germination and fertility. The ga3ox1 ga3ox2 double mutant is much smaller and shorter than ga3ox1 and is nongerminating, confirming the role of GA3ox1 and GA3ox2 in seed germination and vegetative growth. Both ga3ox1 and ga3ox1 ga3ox2 have normal fertility, probably due to expression of GA3ox3 and GA3ox4 in reproductive tissues.
In this study, expression patterns of GA3ox genes during flower and fruit (i.e., silique) development were examined to determine the sites of bioactive GA synthesis during reproductive growth in Arabidopsis. The physiological function of each GA3ox gene was also analyzed by generating multiple ga3ox mutant combinations. In developing flowers, our results suggested that bioactive GAs are made in both stamen filaments (by GA3ox1) and anthers (mainly by GA3ox3). Mutant analysis revealed that GA3ox1 and GA3ox3 play major roles in maintaining sufficient GA synthesis in flowers for anther and petal development, whereas GA3ox2 and GA3ox4 play minor roles. In developing siliques, bioactive GAs are likely synthesized in replums and funiculi (by GA3ox1) and in developing seeds (by all four GA3ox). Mutant and pollination studies further indicated that both maternal tissue-expressed GA3ox1 and seed endosperm-expressed GA3ox4 contribute to silique elongation. Our results also suggest that local transport of bioactive GAs is required to support petal and silique development.
RESULTS
In Vitro Enzyme Assays for GA3ox3 and GA3ox4
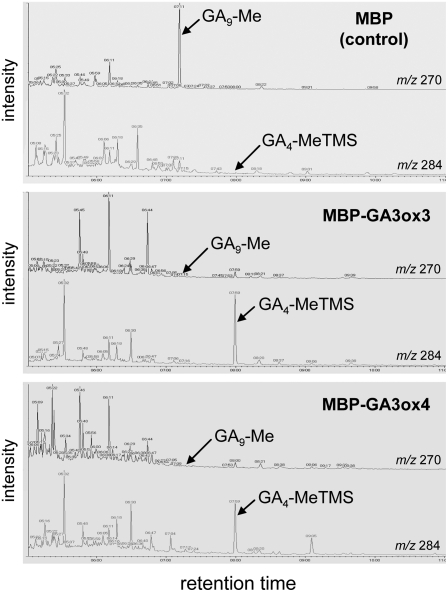
Previously, in vitro enzyme assays have shown that both Arabidopsis GA3ox1 (Williams et al., 1998) and GA3ox2 (Yamaguchi et al., 1998) can convert GA9 to GA4, mimicking the last step of converting inactive precursor to bioactive GA in Arabidopsis. To determine whether GA3ox3 and GA3ox4 have the same activity, we performed in vitro GA3ox enzyme assays. GA3ox3 or GA3ox4 was fused to a maltose binding protein (MBP) and expressed in Escherichia coli. Cell lysates containing the fusion proteins were incubated with GA9. Subsequent full-scan gas chromatography–mass spectrometry analysis showed that GA9 was converted to GA4 by both GA3ox3 and GA3ox4 fusion proteins but not by the MBP protein alone (Figure 2). Likewise, GA20 was converted to GA1 by both MBP-GA3ox3 and MBP-GA3ox4 (see Supplemental Figure 1 online). Therefore, GA3ox3 and GA3ox4 proteins in Arabidopsis have true 3β-hydroxylase activity that converts inactive precursor to bioactive GA.
Figure 2.
In Vitro Enzyme Assays of Wild-Type GA3ox3 and GA3ox4 Proteins.
Gas chromatography–selected ion monitoring showing the conversion of GA9 to GA4 by MBP-GA3ox3 fusion protein (middle panel) and MBP-GA3ox4 fusion protein (bottom panel). Cell lysates containing MBP were used as a negative control (top panel). GA9 and GA4 were identified as GA9-methyl ester (GA9-Me) and GA4-methyl ester-trimethylsilyl ether (GA4-MeTMS), respectively, after derivatization. m/z, mass-to-charge ratio.
Expression Patterns of GA3ox3 and GA3ox4 Genes in Flowers
Because GA3ox catalyzes the last step of the synthesis of bioactive GA, the temporal and spatial expression patterns of GA3ox genes are likely to reflect when and where bioactive GAs are being made in plants. Previous qRT-PCR experiments showed that GA3ox3 and GA3ox4 are mainly expressed during reproductive growth (Mitchum et al., 2006). To further investigate the tissue- and cell-specific expression pattern of these two genes, we generated promoter–β-glucuronidase (GUS) transcriptional constructs (TC-GUS) using the entire intergenic sequence upstream of GA3ox3 (1.8 kb) or GA3ox4 (1.2 kb) (Figure 3). In addition, translational fusions (TL-GUS) were made using the same promoter sequence but included an intron-containing 5′-segment of the coding region fused to GUS. Translational fusions were included because intron sequences have been shown to be necessary for the proper expression of some GA biosynthetic genes (Silverstone et al., 1997; Sakamoto et al., 2001). At least 10 transgenic Arabidopsis lines that contained a single T-DNA insertion site were generated for each construct.
Figure 3.
Structures of GA3ox3 and GA3ox4 Promoter-GUS Reporter Gene Fusion Constructs and a GA3ox3 Genomic DNA Construct.
Black boxes correspond to exons, and the gray boxes correspond to introns of GA3ox genes. p3ox3 contains a genomic GA3ox3 sequence that includes 1.8-kb 5′ promoter sequence and the entire coding region. Asterisks indicate the GUS constructs used for final expression studies. TC, transcriptional fusion; TL, translational fusion.
For GA3ox3, the TC-GUS lines failed to show any expression, while both TL1-GUS and TL2-GUS lines (Figure 3) displayed the same expression patterns in flowers and siliques that are consistent with the previous qRT-PCR data for GA3ox3 (Mitchum et al., 2006). We chose the TL2-GUS line for further studies because it had consistently stronger expression than the TL1-GUS line. These results suggest that the 5′ coding region, including the first two introns of the GA3ox3 gene sequence, is important for its proper expression.
By contrast, none of the GA3ox4-TL-GUS lines showed any GUS activity, but the GA3ox4-TC-GUS lines had a high level of GUS activity in developing seeds and low GUS activities in flowers and germinating seeds that correlated with previous qRT-PCR results (Mitchum et al., 2006). Expression patterns of two independent homozygous lines for each GA3ox3-TL2-GUS and GA3ox4-TC-GUS were examined further. These lines will be referred to as GA3ox3-GUS and GA3ox4-GUS throughout the rest of the article.
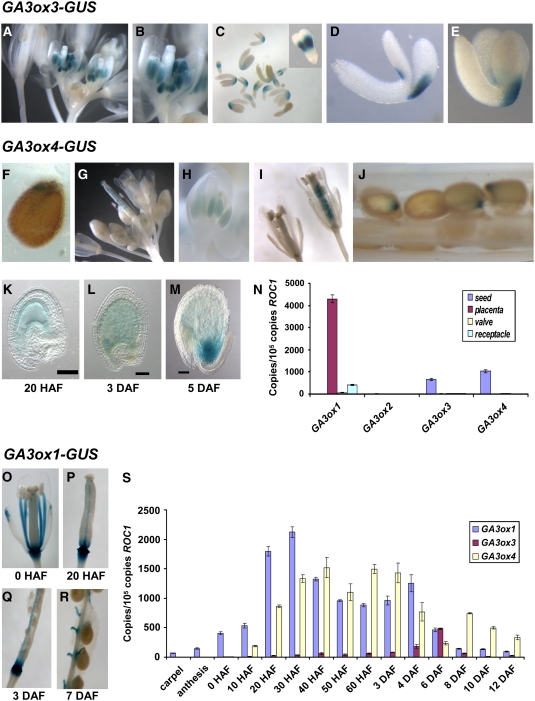
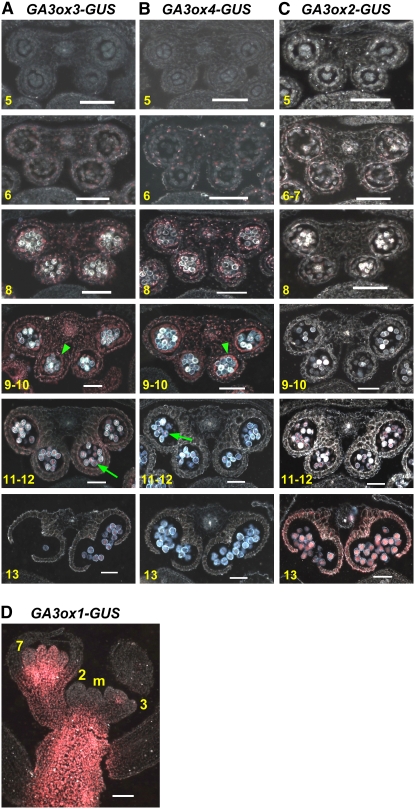
In flowers, both GA3ox3-GUS and GA3ox4-GUS were only expressed in anthers (Figures 4A, 4B, 4G, and 4H). Overall, the expression of GA3ox3-GUS was much stronger than that of GA3ox4-GUS in anthers. This is consistent with our previous qRT-PCR data (Mitchum et al., 2006) and implies that GA3ox3 may contribute more significantly to bioactive GA production in flowers than GA3ox4. To determine the exact timing as well as the cell type specificity of GA3ox3 and GA3ox4 expression during anther development, we made thin sections of X-gluc–stained whole flower clusters of GA3ox3-GUS and GA3ox4-GUS lines (Figures 5A and 5B). Their temporal appearance in flowers was very similar in that expression of both genes reached a peak just prior to anthesis and then slowly disappeared. Anther sections were sorted into developmental stages according to Sanders et al. (1999). As shown in Figures 5A and 5B, both genes began to express weakly in the entire anther at anther stage 6 when pollen mother cells underwent meiosis. This expression in all cell layers surrounding microspores gradually increased and reached its peak around anther stages 9 to 10, right before microspore mitosis occurred. The expression of both GA3ox3 and GA3ox4 was more prominent in the tapetum layer, which is the innermost layer surrounding pollen grains. During this period, expression in microspores was weaker. At anther stages 11 and 12, with the degradation of the tapetum, the expression of both genes in anther walls decreased dramatically while their expression in pollen grains remained at a similar level. By the end of this period, both genes were only expressed in pollen grains. Expression in pollen grains remained after dehiscence in stage 13.
Figure 4.
Temporal and Spatial Expression of GA3ox Genes.
(A) to (E) GA3ox3-GUS line.
(A) Flower cluster.
(B) Close-up of single flower.
(C) Developing embryos between torpedo and upturned-U stages. Inset: close-up of torpedo stage embryo.
(D) Close-up of walking stick stage embryo.
(E) Close-up of upturned-U stage embryo.
(F) to (M) GA3ox4-GUS line.
(F) Seed, 6 h after imbibition.
(G) Flower cluster.
(H) Close-up of a young flower.
(I) Close-up of flowers with young siliques. Silique age: 0 HAF (left) and 20 HAF (right).
(J) Developing seeds in a silique at 7 DAF.
(K) to (M) Clearing of developing GA3ox4-GUS seeds by Hoyer's medium. Bars = 50 μm.
(N) Expression of GA3ox genes in 6- to 8-DAF wild-type siliques as gauged by qRT-PCR. Placenta contains septum, replums, and funiculi.
(O) to (R) GA3ox1-GUS expression in carpel and developing siliques.
(S) Temporal expression of GA3ox1, GA3ox3, and GA3ox4 during silique growth. The levels of gene expression in (N) and (S) are represented by the copies of cDNA per 105 copies of ROC1. Means ± se of three technical replicates are shown in both (N) and (S). Each experiment was repeated once using independent samples with similar results. Silique stages were defined according to Bowman (1994) (see Supplemental Table 1 online).
Figure 5.
Cellular Location of GA3ox-GUS Expression during Flower Development.
(A) to (C) Transverse sections of anthers expressing GA3ox3-GUS, GA3ox4-GUS, and GA3ox2-GUS, respectively. Numbers at the bottom left corner of each picture indicate the corresponding anther developmental stages according to Sanders et al. (1999). Arrowhead, tapetum; arrow, pollen grains.
(D) Longitudinal section of inflorescence apex of GA3ox1-GUS. m, inflorescence meristem. 2, stage 2 flower primordia; 3, stage 3 flower; 7, stage 7 flower. Flower stages were determined according to Bowman (1994).
All sections were viewed under dark-field microscopy, and the X-Gluc staining is pink. Bars = 50 μm.
Our previous whole-mount GUS expression study showed that GA3ox2 is also expressed, although very weakly, in anthers during flower development, while GA3ox1 has a strong expression in anther filaments and flower receptacles (Mitchum et al., 2006). To analyze the expression pattern of these two genes in stamens, flower cluster sectioning was performed on their corresponding GUS expression lines. Similar to GA3ox3 and GA3ox4, weak expression of GA3ox2-GUS was observed in all anther layers at anther developmental stages 6 and 7 (Figure 5C). However, unlike GA3ox3 and GA3ox4, GA3ox2 expression disappeared at stage 8 as microspores were released from tetrads and then reappeared at stages 11 and 12 in pollen grains and reached its peak expression at stage 13 when anther dehiscence occurred. The anther expression patterns of the three GA3ox genes suggest that they have distinct functions at different stages of anther development. By contrast, GA3ox1-GUS activity was observed from a very early stage during flower development. As shown in Figure 5D, GA3ox1-GUS expression initiated under the floral apex in stage 3 flowers that underwent sepal initiation (Bowman, 1994). Such expression grew stronger as flowers developed, and it became clear that this expression was located at the flower receptacle when most floral organs were developed (Figure 5D). Once stamen filaments were differentiated, GA3ox1-GUS expression was detected in this tissue and continued throughout flower development (Mitchum et al., 2006). Furthermore, GA3ox1 expression was excluded from the inflorescence meristem but showed strong expression in the young stem below the meristem (Figure 5D), supporting its role in inflorescence stem elongation.
Expression Patterns of GA3ox3 and GA3ox4 Genes in Siliques
In siliques, GA3ox3-GUS and GA3ox4-GUS had distinct expression patterns (Figure 4). GA3ox3-GUS only expressed in developing embryos (Figure 4C). In the heart- and torpedo-stage embryos, GA3ox3-GUS activity was present on both sides of the embryo near the junction between the embryo axis and cotyledons (Figure 4C, inset). When the embryo started to bend, GA3ox3-GUS expression became asymmetric, with the outer side being much stronger (Figure 4D). In mature embryos, only the outer side showed GA3ox3 expression (Figure 4E). By contrast, GA3ox4-GUS activity was detected in the endosperm of early developing seeds, starting ∼20 h after flowering (HAF; Figures 4I, 4K, and 4L). In this study, 0 HAF (in stage 14 flower) is when pollination and fertilization occur and long anthers extend above the stigma, as defined by Bowman (1994) (see Supplemental Table 1 online). By the heart stage (5 d after flowering [DAF]), GA3ox4-GUS expression became localized at the chalazal endosperm (Figure 4M). The occasional weak staining of the inner seed coat and the embryo is likely due to slight diffusion from the endosperm (Figures 4K and 4L). This is supported by the following observations. When wild-type carpels were pollinated by GA3ox4-GUS pollen, similar weak staining of the seed coat was observed in the resulting developing seeds (see Supplemental Figure 2 online). Also, GA3ox4-GUS activity was not detected in the dissected embryo past heart stage. In imbibed seeds, GA3ox4-GUS activity, although very weak, was detected in the chalazal region, similar to the activity in developing seeds (Figures 4F and 4M). Overall, among all vegetative and reproductive tissues examined, GA3ox3-GUS was only expressed in anthers and developing embryos, and GA3ox4-GUS was mainly expressed in anthers and the endosperm of developing seeds. These data correlate closely with our previous real-time PCR results for both genes (Mitchum et al., 2006), supporting that both GA3ox-GUS fusions reflect an accurate expression pattern of the endogenous genes. Furthermore, expression of the GA3ox3 coding sequence under the same regulatory sequence in GA3ox3-GUS (p3ox3, Figure 3) completely rescued the reproductive defects caused by the ga3ox3 mutation (in the ga3ox1 background, described below), suggesting that the regulatory sequence in the GA3ox3-GUS construct is sufficient for proper GA3ox3 expression (see Supplemental Figure 3 online). We did not perform a similar complementation test to verify the regulatory sequence used in GA3ox4-GUS because of the very subtle mutant phenotypes caused by the ga3ox4 mutation (see below).
Comparison of Expression of GA3ox Genes during Silique Development
In developing siliques, all four GA3ox genes appear to be expressed in developing seeds as indicated by expression studies using GA3ox promoter-GUS lines (Mitchum et al., 2006; Figures 4C and 4J). However, none of the GA3ox-GUS fusions are expressed in the seedpods, except for GA3ox1, which is expressed in the silique receptacles (Mitchum et al., 2006). This is surprising because bioactive GAs are needed for silique elongation. To verify expression patterns of GA3ox genes in siliques, we quantified transcript levels of GA3ox genes in seeds, valves, placenta (including septums, replums, and funiculi), and receptacles by real-time RT-PCR. The ROC1 gene, which shows similar expression levels in these tissues (see Supplemental Figure 4A online) was used as the standard to calculate the absolute levels of expression of GA3ox genes (Figure 4N). GA3ox3, GA3ox4, and GA3ox2 (although at a very low level; see Supplemental Figure 4B online) are only expressed in developing seeds, consistent with our GUS data (Figures 4C and 4J). GA3ox1 mRNA was present in the receptacles (Figure 4N) and also in seeds (very low levels; see Supplemental Figure 4B online), similar to our previously reported GA3ox1-GUS staining patterns (Mitchum et al., 2006). However, the highest levels of GA3ox1 mRNA were found to be in the placenta (Figure 4N), where we did not detect any GA3ox1-GUS expression previously. To clarify this discrepancy, we examined GA3ox1-GUS expression again and found that its expression is localized to the replums and funiculi (Figures 4O to 4R), in addition to the previously described receptacles and developing embryos. Detecting GA3ox1-GUS expression in the replum and funiculus requires a lower concentration of potassium ferricyanide (0 to 0.2 mM) in the X-gluc solution, which is the reason we did not see this staining before when 1 mM potassium ferricyanide was used.
We then examined the temporal expression patterns of GA3ox1, GA3ox3, and GA3ox4 in mature carpels and developing siliques (Figure 4S). Before pollination, only GA3ox1 showed weak expression in carpels. After anthesis, GA3ox1 mRNA levels greatly increased and quickly peaked at 30 HAF. GA3ox4 expression was a little delayed, first being detected at 10 HAF and peaking at 40 HAF. By contrast, the GA3ox3 transcript was only present at a very low level between 20 HAF and 3 DAF and peaked around 6 DAF (torpedo stage). The temporal expression of GA3ox genes is highly consistent with their GUS expression pattern. GA3ox2 was not tested due to its low level in siliques.
Identification and Characterization of ga3ox3 and ga3ox4 Mutants
The expression patterns of GA3ox3 and GA3ox4 suggest that they are involved in providing bioactive GAs during flower and silique development. To further investigate their physiological roles in these processes, we isolated mutants that are defective in either gene. A T-DNA insertion mutant of GA3ox3 was identified by screening multidimensional DNA pools of T-DNA insertion mutant populations using a PCR-based method (Alonso et al., 2003). This mutant (ga3ox3-1) has a T-DNA insertion in the first intron of the GA3ox3 gene sequence (Figure 6A). No wild-type GA3ox3 transcript was detectable in this mutant by RT-PCR, indicating that it is a null allele (see Supplemental Figure 5 online). For GA3ox4, we initially identified two alleles (ga3ox4-1 and ga3ox4-4; Figure 6A) from the Arabidopsis TILLING Center at the University of Washington (Henikoff et al., 2004). Both mutants had a point mutation in the second exon, resulting in an amino acid substitution within highly conserved functional domains of 2-oxoglutarate dependent dioxygenases (see Supplemental Figure 6A online; Xu et al., 1995; Hedden and Phillips, 2000). In vitro assays showed that biochemically, ga3ox4-1 is a near-null mutant, while ga3ox4-4 did not dramatically affect the GA3ox enzyme activity (see Supplemental Figure 6B online).
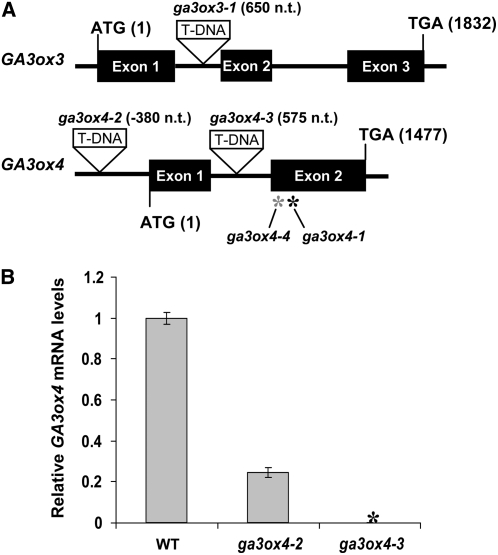
Figure 6.
The ga3ox3 and ga3ox4 Mutant Lines.
(A) Locations of mutations in the ga3ox3 and ga3ox4 mutants. T-DNA insertion sites are depicted as triangles. Numbers next to T-DNA insertions indicate the insertion sites relative to ATG. The black and gray asterisks indicate the positions of ga3ox4-1 and ga3ox4-4 missense mutations at 1094 and 1046 nucleotides after ATG, respectively.
(B) Relative GA3ox4 transcript levels in siliques of ga3ox4-2 and ga3ox4-3 compared with the wild type. The means of three technical replicates of qRT-PCR ± se are shown. The expression level in the wild type was set to 1.0. Black asterisk indicates no wild-type GA3ox4 transcript was detected in ga3ox4-3. Similar results were obtained when qRT-PCR was performed using a second set of samples.
Subsequently, two more GA3ox4 mutants, ga3ox4-2 and ga3ox4-3, were obtained from the SALK T-DNA collection (Figure 6A). One of them, ga3ox4-2, has a T-DNA insertion in the 5′-untranslated region. Although this mutant was previously reported to be a null allele (Kim et al., 2005), our qRT-PCR analysis using primers downstream from the insertion site indicated that the mutant siliques still accumulated ∼25% of GA3ox4 transcript that contains the entire coding region (Figure 6B). Another T-DNA mutant, ga3ox4-3, had a T-DNA insertion in the intron. No wild-type GA3ox4 transcript was detected in this mutant, indicating that it is a null allele (Figure 6B). Both ga3ox4-1 and ga3ox4-3 exhibited similar phenotypes in further mutant characterization (see Supplemental Table 2 online). In the rest of this article, we only present data collected from the ga3ox4-3 mutant, referred to as ga3ox4.
Phenotypic Analysis of GA3ox Mutants
The ga3ox3 (i.e., ga3ox3-1) and ga3ox4 single mutants have no obvious phenotype under normal growth conditions, probably due to functional redundancy with each other and with the other two GA3ox genes. To reveal the physiological function of GA3ox3 and GA3ox4, we generated double and triple mutant combinations by making crosses among ga3ox3 and ga3ox4 and the previously characterized ga3ox1-3 and ga3ox2-1 null mutants (Mitchum et al., 2006). However, because GA3ox2 and GA3ox4 are only 1.2 kb apart on chromosome 1, mutants containing both ga3ox2 and ga3ox4 were not obtained by genetic crosses in this study.
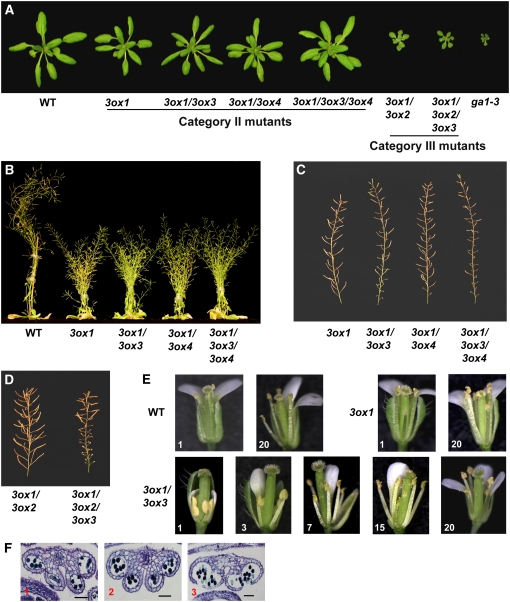
Based on rosette size and final height, we divided all available GA3ox single, double, and triple mutant combinations into three categories. Category I mutants include ga3ox2, ga3ox3, ga3ox4, ga3ox2 ga3ox3, and ga3ox3 ga3ox4. All of these mutants contain the wild-type GA3ox1 allele and displayed a wild-type phenotype (see Supplemental Table 3 online). This is not surprising because GA3ox1 has the highest expression in most tissues, except in germinating seeds (Mitchum et al., 2006), and its presence appears to provide sufficient synthesis of bioactive GAs for normal plant growth. Category II includes all mutants that have the homozygous ga3ox1 mutation but still have the wild-type GA3ox2 allele (i.e., ga3ox1, ga3ox1 ga3ox3, ga3ox1 ga3ox4, and ga3ox1 ga3ox3 ga3ox4). Mutants in this category have partial GA-deficient phenotypes, including semidwarfism (∼50% of the wild-type height), slightly smaller rosettes, and delayed flowering, compared with wild-type plants (Figures 7A and 7B, Table 1). Within this category, all mutants have similar phenotypes as the ga3ox1 mutant during vegetative growth. This is consistent with previous qRT-PCR data showing that GA3ox3 and GA3ox4 have no expression or very weak expression during vegetative growth (Mitchum et al., 2006). Thus, vegetative growth is not affected when either or both of these genes are nonfunctional. Category III includes ga3ox1 ga3ox2 and ga3ox1 ga3ox2 ga3ox3, and these plants exhibited more severe dwarf phenotypes (26% of the wild-type height) than the mutants in Category II (Figures 7A and 7B, Table 1). As before, removing GA3ox3 function did not further reduce vegetative growth. In all three categories, both ga3ox3 and ga3ox4 mutations have no effect on rosette size, plant height, or flowering time (Table 1). This can be explained by the fact that both genes are mainly expressed in flowers and siliques.
Figure 7.
Phenotypic Characterization of the ga3ox Mutants.
(A) The 24-d-old rosettes of wild-type and homozygous mutants as labeled.
(B) The 44-d-old plants as labeled.
(C) Primary inflorescence stems of Category II ga3ox mutants.
(D) Primary inflorescence stems of Category III ga3ox mutants.
(E) Gradual improvement in the development of ga3ox1 ga3ox3 mutant flowers on primary inflorescence. Top row: wild-type and ga3ox1 flowers. Bottom row: ga3ox1 ga3ox3 flowers. Some of the floral organs were removed to show the anther phenotype.
(F) Toluidine blue–stained transverse sections of anthers of the first three flowers on the primary inflorescence stem of ga3ox1 ga3ox3. Numbers at the bottom left corner of each picture in (E) and (F) indicate the positions of the flowers on the primary inflorescence stem. Bar = 50 μm.
Table 1.
Phenotypes of the ga3ox Mutants Compared with the Wild Type and ga1-3
| Mutant Category | Flowering Time (d) | Flowering Time (Rosette Leaf No.) | Rosette Diameter (cm) | Final Height (cm) | Percentage of Fertile Siliques | Seed per Fertile Silique | |
|---|---|---|---|---|---|---|---|
| Wild type | 21.3 ± 0.1a | 8.7 ± 0.1a | 5.60 ± 0.11a | 42.0 ± 0.7a | 98.0 ± 0.4b/c | 51.5 ± 0.4a | |
| 3ox1 | II | 22.6 ± 0.2b | 9.2 ± 0.1b | 4.83 ± 0.09b | 20.8 ± 0.6d | 99.2 ± 0.3b | 49.0 ± 1.0a |
| 3ox1 3ox3 | II | 23.6 ± 0.2c | 9.2 ± 0.1b | 4.88 ± 0.13b | 23.9 ± 0.6c | 59.9 ± 1.9d | 33.4 ± 1.0c |
| 3ox1 3ox4 | II | 20.8 ± 0.2c | 8.5 ± 0.1b | 4.92 ± 0.09b | 23.0 ± 0.3c | 97.0 ± 0.4c | 49.3 ± 0.6a |
| 3ox1 3ox3 3ox4 | II | 23.1 ± 0.2c | 8.8 ± 0.1b | 5.07 ± 0.18b | 25.8 ± 0.3b | 51.2 ± 1.5e | 29.2 ± 0.7d |
| 3ox1 3ox2 | III | 27.4 ± 0.5d | 10.3 ± 0.2c | 2.03 ± 0.08c | 11.1 ± 0.3e | 100 ± 0.0a | 46.1 ± 0.6b |
| 3ox1 3ox2 3ox3 | III | 30.3 ± 0.3e | 11.2 ± 0.2d | 1.95 ± 0.06c | 10.9 ± 0.3e | 45.8 ± 1.5f | 32.8 ± 1.2c |
| ga1-3 | 46.3 ± 0.3f | 18.7 ± 0.3e | 1.08 ± 0.04d | NA | None | NA |
For the percentage of fertile siliques data, fertile siliques contain at least one seed. All measurements are the means ± se from 15 to 20 plants per genotype. Rosette diameter was measured 24 d after sowing. Measurements of seeds in individual siliques were taken on primary inflorescence stems only. When two values are marked with different letters (a to f), the difference between them is significant based on Student's t test (P < 0.01). NA, not applicable.
In Arabidopsis, severely GA-deficient mutants (such as ga1-3) are male-sterile (with short stamen filaments and no viable pollen) and have poorly developed petals, indicating that bioactive GAs are essential for stamen and petal development (Koornneef and van der Veen, 1980; Cheng et al., 2004). Previously, we showed that the ga3ox1 and ga3ox1 ga3ox2 mutants have no fertility defect, suggesting that expression of GA3ox3 and/or GA3ox4 genes is sufficient to support flower development (Mitchum et al., 2006). Flowers and fertility (average numbers of seeds per silique) of all mutants in Category I that contain functional GA3ox1 appeared similar to the wild type (see Supplemental Table 3 online). Among the semidwarf mutants in Category II, dramatic phenotypic differences were observed during reproductive development (Figure 7C, Table 1). All flowers on the primary inflorescence in ga3ox1 had near wild-type fertility, whereas removing GA3ox3 function in the ga3ox1 background caused a significant fertility defect and reduced petal growth (Figures 7C and 7E, Table 1). The first 10 siliques on the primary stem of the ga3ox1 ga3ox3 double mutant were mostly sterile with only one to two fertile siliques formed randomly near the 10th silique (Figure 7C). These early fertile siliques usually contained very few seeds. Surprisingly, later siliques of this double mutant gradually increased in fertility, and the majority became fertile after the 20th to 25th silique (see Supplemental Table 4 online). In addition, siliques on secondary inflorescences displayed the same fertility pattern, but the transition from sterile to fertile siliques occurred earlier (data not shown). By contrast, most of the siliques on the primary stem of the ga3ox1 ga3ox4 double mutant were fertile, similar to those of ga3ox1, although the first two siliques were often sterile. The ga3ox1 ga3ox3 ga3ox4 triple mutant has a slightly more severe fertility defect than ga3ox1 ga3ox3, as judged by the percentage of fertile siliques and seed number per fertile silique (Figure 7C, Table 1). In the third mutant category, the ga3ox1 ga3ox2 ga3ox3 mutant also had a similar fertility phenotype as that of ga3ox1 ga3ox3, yet more severe (Figure 7D, Table 1). By contrast, ga3ox1 ga3ox2 had 100% fertile siliques. These results indicate that GA3ox1 and GA3ox3 have major and overlapping functions in providing bioactive GAs during flower development, while GA3ox2 and GA3ox4 play minor roles.
The gradual recovery of fertility in mutants that lacked both GA3ox1 and GA3ox3 was interesting. To investigate the mechanism involved, individual flowers on the primary stems of ga3ox1 ga3ox3 were compared with those of ga3ox1. After anthesis, length of anther filaments and size of petals of ga3ox1 flowers are similar to those of the wild type (Figure 7E). All flowers of the ga3ox1 mutant exhibited this wild-type phenotype and were fertile. By contrast, the first two flowers of ga3ox1 ga3ox3 had very short anther filaments, and anther surfaces were smooth until stamens senesced, implying the lack of anther dehiscence (Figure 7E). Further examination of the transverse sections indicated that the epidermal layer of the anther remained intact, although the tapetum layer had disappeared (Figure 7F), suggesting that anther development was arrested around stages 11 and 12. Furthermore, petals in these flowers were much smaller than those of wild-type flowers after anthesis. All defects (anther filament, petal elongation, and pollen release) gradually decreased in the later flowers (Figures 7E and 7F). Starting with the 5th flower, pollen grains were released normally. By the 20th flowers, the stamens consistently reached the stigma. However, anther filaments never grew above the stigma, as was observed in ga3ox1. Similarly, petals in later flowers were still shorter than the wild type. Early flowers of ga3ox1 ga3ox3 ga3ox4 and ga3ox1 ga3ox2 ga3ox3 exhibited similar phenotypes as those of ga3ox1 ga3ox3, but these two triple mutants needed longer transition periods to reach consistently fertile flowers (see Supplemental Table 4 online). In particular, ga3ox1 ga3ox2 ga3ox3 showed the slowest improvement in flower morphology among these three mutants, which was reflected by their low percentage of fertile siliques (Table 1).
Endogenous GA Contents in Rosette Leaves and Flowers of the ga3ox Mutants
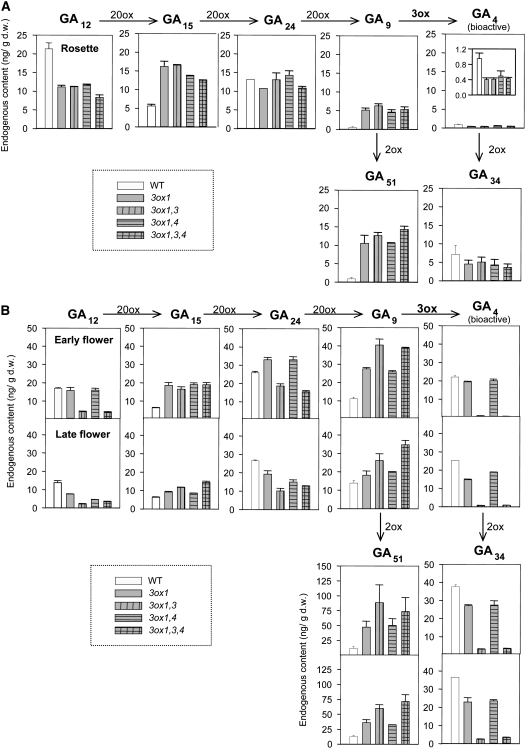
To evaluate the effect of ga3ox mutation(s) on the production of GAs, we determined concentrations of precursor, bioactive, and deactivated GAs in rosette leaves and flowers of wild-type and Category II mutants by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. As discussed above, all ga3ox mutants in Category II exhibited similar vegetative phenotypes, and they possessed slightly smaller rosette leaves than wild-type plants (Table 1, Figure 7A). In agreement with the phenotype data, the levels of bioactive GA4 in the rosette leaves of these mutants (harvested at anthesis of the first flower) were ∼50% of those in wild-type plants (Figure 8A). In contrast with the levels of GA4, those of GA9 and GA51 (2β-hydroxylated GA9) were highly elevated in Category II ga3ox mutants relative to those in wild-type plants. These data are consistent with the premise that GA3ox activity is partly impaired in these mutants. Decreased and increased levels of GA12 and GA15, respectively, in the ga3ox mutants suggest that the conversion of GA12 to GA15 by GA20ox might be activated through feedback regulation. Although the levels of GA24 were not elevated, both upregulation of GA20ox expression and reduced GA3ox activity are likely to have contributed to the increased levels of GA9 and GA51.
Figure 8.
Endogenous GA levels in Rosette Leaves and Flowers of Wild-Type and Category II ga3ox Mutant Plants.
(A) GA levels (nanograms per gram dry weight [ng/g d.w.]) in rosette leaves at anthesis of the first flower. The data for GA4 are also shown in the inset with a different y axis scale for clarity. Data are means ± se of biological duplicates.
(B) GA levels in early (top graph) and late (bottom graphs) flowers. Note that the y axis scale for GA51 is different from that for other GAs. Means ± se of biological triplicates are shown. Results of 13-OH GAs are shown in Supplemental Figure 7 online.
To investigate the correlation between the reproductive phenotype and the status of the GA biosynthetic pathway in ga3ox mutants, we determined endogenous GA levels in flowers (Figure 8B). Because early and late flowers displayed different fertility phenotypes in some ga3ox mutants (Figures 7C and 7E, Table 1), we harvested early (collected immediately before the anthesis of the first flowers) and late (harvested when 20 to 25 siliques were present on primary stems) flower clusters for GA measurements. Both flower clusters were harvested from the primary inflorescence stems. In comparison with the wild type, ga3ox1 and ga3ox1 ga3ox4 early flowers contained only slightly lower levels of GA4 (Figure 8B). By contrast, the amounts of GA4 in early flowers of ga3ox1 ga3ox3 and ga3ox1 ga3ox3 ga3ox4 mutants were substantially reduced. Similar GA profiles were observed for 13-OH GAs (see Supplemental Figure 7 online). These results are consistent with the sterility of early flowers of these mutants and indicate a major role for GA3ox1 and GA3ox3 in the synthesis of bioactive GAs during flower development.
Although, in contrast with the early flowers, the late flowers of ga3ox1 ga3ox3 and ga3ox1 ga3ox3 ga3ox4 mutants were fertile (Figures 7C and 7E, Table 1), no discernible increase in the levels of bioactive GAs was detected with our LC-MS/MS system (Figure 8B; see Supplemental Figure 7 online). It is still possible that a subtle increase in GA levels, which is below the detection limit of our system, or an increase in GA signaling rescued the flower defect. However, no increased expression of GA3ox2 or GA3ox4 was observed in the late flowers in mutants lacking both GA3ox1 and GA3ox3 (see Supplemental Figure 8 online). This is consistent with a recent study showing that GA3ox1 is the only GA3ox gene under feedback regulation in Arabidopsis flowers (Matsushita et al., 2007). We also examined the expression of GA receptor genes (GID1s) and GA signaling repressor genes (DELLAs) in ga3ox1 ga3ox3 ga3ox4 mutant flowers by qRT-PCR. Again, no significant change in expression was detected for these GA signaling genes in late flowers compared with early ones (see Supplemental Figure 9 online). Therefore, the improved fertility of the late flowers is unlikely caused by elevated early GA signaling.
Production of Bioactive GAs in Seeds and Maternal Tissues for Silique Elongation
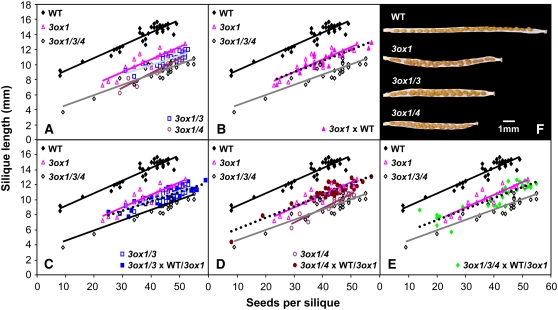
GA promotes fruit set and subsequent growth in many species (Gillaspy et al., 1993; Ozga and Reinecke, 2003). However, it is unclear whether bioactive GAs produced in seeds are needed for fruit growth. As different GA3ox genes are expressed in distinct tissues in the developing fruit, we used the ga3ox mutants to determine whether maternal tissues and/or seeds synthesize bioactive GAs for silique elongation. Although some of the ga3ox mutant combinations described above exhibited defective anther development and sterile siliques, under slightly different growth conditions (see Methods) the transition period from sterile to consistently fertile flowers was much shorter (data not shown). This allowed silique elongation to be compared between the various mutants to investigate the role of the different GA3oxs in this process. Flowers were self-pollinated (by hand) with varying amounts of pollen to generate a range of seed numbers. When examined in this way, ga3ox1, ga3ox1 ga3ox3, ga3ox1 ga3ox4, and ga3ox1 ga3ox3 ga3ox4 flowers all produced shorter siliques than self-pollinated wild-type flowers (comparing siliques containing the same number of seeds; Figure 9A). In other words, the extent of seed-promoted silique growth was reduced in the ga3ox mutants, suggesting that GA is involved in this process. Consistent with this conclusion, applied GA largely restored silique growth (data not shown). GA3ox1 and GA3ox4 play major roles in silique elongation because mutants lacking these two gene functions displayed the most severe phenotype.
Figure 9.
GA3ox1 and GA3ox4 Are Required for Normal Silique Growth.
(A) to (E) Relationship between seed number per silique and final silique length for wild-type and ga3ox mutants either self-pollinated or cross-pollinated with pollen from wild-type or ga3ox1 plants. All data are from hand-pollinated flowers with at least 19 siliques for each combination. Some flowers were pollinated with a reduced amount of pollen to vary the number of seeds set. Data from self-pollinated wild-type, ga3ox1, and ga3ox1 ga3ox3 ga3ox4 flowers are shown in each panel to make comparisons easier. In (C) to (E), data from the pollination with wild-type and ga3ox1 pollen have been combined as both pollen genotypes gave very similar silique lengths. Lines of best fit are shown by solid lines of different colors (self-pollinated flowers) and by dotted black lines for flowers pollinated with wild-type and/or ga3ox1 pollen.
(F) Dry siliques of self-pollinated wild-type and ga3ox mutants with the valve removed to reveal the seeds. Each carpel shown contains 23 to 24 seeds, which are more crowded in the mutants due to reduced silique growth. Siliques of the ga3ox1 ga3ox3 ga3ox4 triple mutant resemble those of the ga3ox1 ga3ox4 double mutant.
To determine the relative contributions of maternal (e.g., carpel, placenta, funiculus, and seed coat) and nonmaternal (embryo and endosperm) tissues to silique growth, the different genotypes were also pollinated with wild-type or ga3ox1 pollen. For the ga3ox1 mutant, seed-promoted silique growth was unaffected by the pollen genotype and therefore by the presence or absence of a wild-type GA3ox1 allele in the embryo/endosperm (Figure 9B). This result suggests that the reduced silique growth of the ga3ox1 mutant is due to reduced activity of this gene in maternal tissues, consistent with the expression of this gene in replums, funiculi, and silique receptacles.
Similar experiments were conducted to investigate the site of action of the GA3ox3 and GA3ox4 genes. Silique growth was slightly reduced in the ga3ox1 ga3ox3 double mutant compared with the ga3ox1 single mutant, and this phenotype was not affected by the pollen genotype (Figure 9C). Thus, GA3ox3 plays at most a minor role in silique growth in maternal tissues (in the presence of wild-type GA3ox2 and GA3ox4). By contrast, seed-promoted silique growth for the ga3ox1 ga3ox4 double mutant was similar to that of the ga3ox1 ga3ox3 ga3ox4 triple mutant when self-pollinated but similar to the ga3ox1 single mutant when pollinated with either wild-type or ga3ox1 pollen (Figure 9D). Thus, the effect of ga3ox4 could be completely overcome by the introduction of a wild-type GA3ox4 allele to the embryo/endosperm. As described earlier, GA3ox4 is expressed in the endosperm (but not in the embryo), suggesting that GA3ox4 acts in this tissue to promote silique growth. Similar results were obtained for the ga3ox1 ga3ox3 ga3ox4 triple mutant using different pollen genotypes (Figure 9E), consistent with a role for GA3ox1 and a very minor (or no) role for GA3ox3 in maternal tissues and a requirement for GA3ox4 in the endosperm.
DISCUSSION
GA deficiency or GA overdose lead to impaired flower and fruit development (Fleet and Sun, 2005), indicating that the presence of optimal concentrations of bioactive GAs is critical to ensure normal reproductive growth in plants. Developmental expression analysis of GA3ox genes in Arabidopsis allowed us to monitor where bioactive GAs are likely to be synthesized during different stages of reproductive growth, provided that GA3ox enzymes are made and their substrates are available. Our previous qRT-PCR study showed that all four GA3ox genes in Arabidopsis are expressed in reproductive organs, although GA3ox2 mRNA is only present at extremely low levels in both flowers and siliques (Mitchum et al., 2006). By promoter-GUS gene fusions and qRT-PCR analyses (Mitchum et al., 2006; this study), we now have a comprehensive view of the sites of bioactive GA synthesis in Arabidopsis reproductive organs. In addition, the physiological roles of all four GA3ox genes during reproductive growth were defined by isolation and characterization of the single, double, and triple mutants. These results, in comparison with known GA action sites, also reveal some role for local and long-distance transport of bioactive GAs during flower and fruit development in Arabidopsis.
Sites of Synthesis of Bioactive GAs and the Role of Local GA Transport during Reproductive Growth
In Arabidopsis flowers, bioactive GAs have long been known to promote stamen and petal development, as severely GA-deficient mutants produce male-sterile flowers with poorly developed petals (Koornneef and van der Veen, 1980). Consistently, all four GA3ox genes are almost exclusively expressed in stamens during flower development, with the exception that GA3ox1 is also expressed in flower receptacles and the vasculature of sepal tips. In stamens, GA3ox1 is expressed in the filament, whereas the other three GA3oxs are expressed in anthers and pollen (with GA3ox3 showing the highest expression and GA3ox2 and GA3ox4 having very low levels of expression). Therefore, bioactive GAs are likely synthesized in stamen filaments and anthers, both of which require GA for normal development. Our mutant studies indicated that GA3ox1 and GA3ox3 are the major contributors of GA3ox function for stamen development, consistent with their relatively high levels of expression in this organ. These results also support the idea that for normal stamen development (at least for the early flowers), local de novo synthesis of bioactive GAs is necessary. On the other hand, none of the GA3ox genes is expressed in petals, although bioactive GAs are also required for petal growth (Koornneef and van der Veen, 1980). It is likely that bioactive GAs, mainly GA4 in Arabidopsis, made in the flower receptacles and/or stamens need to be transported to petals to promote their growth. This notion was confirmed by the severe petal defects in mutants that lack both GA3ox1 and GA3ox3 functions. These results are consistent with previous studies in petunia (Petunia hybrida) showing that emasculation of stamens caused reduced corolla growth, which could be rescued by GA application (Weiss and Halevy, 1989). It is not clear, though, whether plants mobilize bioactive GAs by diffusion or active transport. Similar anther expression patterns of GA3ox genes were also observed in tobacco (Itoh et al., 1999) and rice (Itoh et al., 2001). Taken together, stamens appear to be a common site for bioactive GA synthesis in flowers of many species, and bioactive GAs made in stamens not only function locally but are also translocated to nearby tissues, like petals, to support their growth.
The differential expression patterns of the four GA3ox genes in stamens suggest their distinctive roles in the development of this organ. As the only gene expressed in stamen filaments, GA3ox1 is likely responsible for producing bioactive GAs to promote the fast elongation of stamen filaments in wild-type plants. Unlike GA3ox1, the other three GA3ox genes are expressed in anthers starting when meiosis of microspore mother cells begins (around anther development stage 6). In the severely GA-deficient mutant ga1-3, anther development is arrested after meiosis during microsporogenesis (stage 7; Cheng et al., 2004), implying that bioactive GAs are not essential for anther development prior to this stage. The timing of GA3ox2, GA3ox3, and GA3ox4 expression in anthers is therefore consistent with the requirement for bioactive GAs in anther development after meiosis is complete. Nevertheless, GA3ox3 is the major contributor of GA3ox function in developing anthers because the transcripts of the other two anther-expressed GA3ox genes are only present at extremely low levels in this tissue. In addition, only the ga3ox3 but not the ga3ox2 or ga3ox4 mutation dramatically affected anther development in the ga3ox1 background. Furthermore, the ga3ox3 mutation, but not the ga3ox4 mutation, drastically decreased the levels of bioactive GAs in flowers in the ga3ox1 mutant background (Figure 8B; see Supplemental Figure 7 online). Our ga3ox mutant studies also suggest that bioactive GAs made in the anther and stamen filament can be transported between these two tissues to support their development because stamen filaments in the ga3ox1 mutant are similar to the wild type (Figure 7E), and the ga3ox mutant plants containing functional GA3ox1 produce normal flowers.
In developing siliques, GA3ox1 is mainly expressed in the replums, funiculi, and the silique receptacles. The other three GA3ox genes are only expressed in developing seeds, with GA3ox2 and GA3ox3 expressed in embryos and GA3ox4 expressed in the endosperm (Figure 4). This expression pattern is consistent with the mutant analysis showing that GA3ox1 plays a major role in promoting fruit elongation by acting in maternal tissues. Furthermore, based on its expression pattern and the results from controlled pollinations, bioactive GAs produced by GA3ox4 in the endosperm of developing seeds also contribute to silique elongation (Figure 9). This result suggests that active GAs are transported from the seed endosperm to the surrounding maternal tissues where they promote growth. Although it is possible that GA3ox4 acts on silique growth indirectly via effects on seed development, seed GA levels were not sufficiently reduced in any of the mutant combinations examined here to cause seed abortion (Singh et al., 2002). Thus, in Arabidopsis, silique growth is promoted by a combination of GA produced in maternal tissues as well as in the endosperm of developing seeds (Figure 9E). Furthermore, the production of active GAs by seeds, due to GA3ox4 activity in the endosperm, can partially explain the localized effect of seeds on silique growth (Cox and Swain, 2006). The immediate upregulation of GA3ox1 in the placenta (Figure 4S) and of GA3ox4 in the endosperm after anthesis suggests that pollination or ovule fertilization is a prerequisite to initiate de novo GA biosynthesis in fruit, which in turn promotes initial elongation of the silique. This result is consistent with a previous study showing that unfertilized carpels of emasculated wild-type Arabidopsis flowers fail to elongate, which could be rescued to ∼66% of the pollinated control by GA application (Vivian-Smith and Koltunow, 1999).
Within developing seeds, GA3ox1, GA3ox2, and GA3ox3 are expressed in distinct locations in the embryo, suggesting that they promote growth of specific regions. Expression of GA3ox3 at the junction of the cotyledon and radicle appears to correlate with differential cell elongation rates when the embryo started to bend (Figure 4D). However, none of the ga3ox mutant combinations displayed any defects during embryo development, suggesting a role for GA movement within the embryo in supporting growth of the entire embryo. GA3ox4-GUS was expressed in the endosperm (Figures 4K to 4M), which is unique among the GA3ox genes. Although we were unsuccessful in detecting the location of the GA3ox4 transcript in seeds by in situ hybridization, this endosperm staining pattern is supported by pollination studies (Figure 9; see Supplemental Figure 2 online). Our results are inconsistent with a recent report showing localization of GA3ox4 mRNA to the outer integument of developing seeds and the embryo surface by in situ hybridization (Kim et al., 2005). In addition, they observed an abnormal seed coat phenotype in the ga3ox4-2 allele, which we were not able to detect in any of the three ga3ox4 alleles (see Supplemental Figure 10 online).
It is difficult to determine the actual function of each GA3ox gene in the development of the Arabidopsis seed. This is partly due to the overlapping expression of GA3ox genes in developing seeds. In addition, because each GA3ox gene has dual expression in flowers and seeds, impaired seed development could be a result of defects in flower development.
During the vegetative phase of plant growth, GA3ox1 is mainly expressed in the rib region near the shoot apical meristem (Mitchum et al., 2006). During reproductive growth, the strong expression of GA3ox1 in the stem near the inflorescence meristem (Figure 5D) suggests that bioactive GAs are synthesized locally to promote the fast elongation of the inflorescence stem. Therefore, newly differentiated cells near the meristems are likely to be a site for GA biosynthesis and for GA action. The rib meristem expression of GA3ox1 correlates with the semidwarf phenotype of ga3ox1 (Figure 7B). Further removal of GA3ox3 and/or GA3ox4 functions did not lead to additional stem growth defects, suggesting that bioactive GAs made in anthers by these genes are not transported to support inflorescence stem growth, although they can be transferred within the same flower to promote petal growth. In fact, the inflorescence stems of the double ga3ox1 ga3ox3 and triple ga3ox1 ga3ox3 ga3ox4 mutants are slightly taller than that of ga3ox1, probably due to their reduced fertility (Hensel et al., 1994).
Physical Separation of Early and Late GA Biosynthesis in Reproductive Tissues
Previous studies have demonstrated that early (CPS) and late (GA3ox1 and GA3ox2) GA biosynthetic genes in Arabidopsis are expressed in distinct cell types during seed germination (Yamaguchi et al., 2001) and in elongating roots (Mitchum et al., 2006). Additionally, expression of GA3 (encoding KO) is colocalized with that of GA3ox in germinating seeds, suggesting that intercellular translocation of an intermediate (likely ent-kaurene) is required for the synthesis of active GAs (Yamaguchi et al., 2001). Comparison of expression patterns of CPS (Silverstone et al., 1997) with the four GA3ox genes reveals that translocation of GA intermediate(s) also occurs in developing flowers. CPS expression overlaps with GA3oxs in anthers and flower receptacles but not in the stamen filaments where GA3ox1 is expressed. Such physical separation of the early and late steps in the GA biosynthetic pathway may add another layer of regulation to GA production.
Potential Source-to-Sink Movement of Bioactive GAs during Plant Reproductive Growth
Application of bioactive GAs rescues most GA-deficient phenotypes in plants (Hedden and Phillips, 2000). However, the physiological role of transport of endogenously made GAs in regulating plant growth and development is not well understood. Previous grafting experiments using wild-type and GA-deficient mutants in pea (Pisum sativum; Proebsting et al., 1992) and in maize (Zea mays; Katsumi et al., 1983) showed that some GA precursors and bioactive GAs were transported from rootstocks to scions. But conflicting results were also obtained from these studies concerning whether sufficient bioactive GAs were transported from the rootstocks to grafted shoots to rescue the GA-deficient phenotype in shoots. Recently, by feeding labeled GA4 to leaves and then measuring GA4 levels in the shoot apical meristem, Eriksson et al. (2006) showed that GA4 is transported from rosette leaves to the shoot apex to facilitate flower initiation in Arabidopsis.
As discussed earlier, our current study suggests that local transport of bioactive GAs within the flowers and siliques is necessary for the development of these organs. Our studies also raise the possibility for a role of long-distance transport of bioactive GAs in supporting reproductive growth. We observed that the timing of cessation of rosette expansion correlated with anthesis of the first flower in mutants lacking both GA3ox1 and GA3ox3 (see Supplemental Figure 11 online). At this stage, considerable amounts of GA4 (50% of the wild type) were still produced in the rosette leaves of these mutants (Figure 8). It is possible that increasing amounts of bioactive GAs may be transported from source tissue (mature rosette) to sink tissue (inflorescence), thus causing the continuous improvement of flower development in ga3ox1 ga3ox3 mutants. The more severe fertility defect in ga3ox1 ga3ox2 ga3ox3 (than in ga3ox1 ga3ox3 and ga3ox1 ga3ox3 ga3ox4) may also be attributed to the lower GA levels in rosettes of this mutant. Alternatively, a gradual increase in GA responsiveness of stamens and petals could also rescue the floral defects in the ga3ox1 ga3ox3 mutant background. Future studies on GA signaling in developing flowers will help to elucidate the mechanism involved.
This study suggests that local and perhaps also long-distance transport of bioactive GAs occurs during reproductive growth. Our findings clearly illustrate the plasticity of the plant in mobilizing bioactive GAs to meet its developmental needs. It will be important to investigate the molecular mechanism that controls GA transport in plants.
METHODS
Plant Material and Growth Conditions
The Columbia (Col-0) genetic background was used as the wild type in this study. The ga1-3 (backcrossed six times to Col-0), ga3ox1-2, ga3ox2-1, and ga3ox1 ga3ox2 mutants used in this study were described previously (Mitchum et al., 2006). To induce germination, seeds of the GA-deficient mutants ga1-3, ga3ox1 ga3ox2, and ga3ox1 ga3ox2 ga3ox3 were incubated with 100 μM GA3 for 7 d at 4°C and then washed thoroughly with water before planting. Wild-type seeds were incubated in water at 4°C for 3 d prior to planting. Plants were grown on MetroMix 200 (Sungro Horticulture) at 22°C under long-day conditions (16 h light/8 h darkness). Plants used for analysis of silique growth were grown on growool as described (Swain et al., 2004). For hand-pollinations, the sepals, petals, and anthers were removed 1 d before anthesis, and flowers were pollinated with pollen from selected genotypes. For individual flowers, between ∼10 and 100% of the stigma was covered with pollen grains to vary the number of seeds per set. Between one and three flowers per inflorescence were pollinated, depending on the number of receptive flower buds. Generally only a single inflorescence was used per plant, but in some cases a second inflorescence was also used. Inflorescences were decapitated immediately above pollinated flowers, and seed number and silique length were recorded once the silique was fully desiccated.
Identification of T-DNA Insertion and TILLING Mutant Lines
The ga3ox3-1 mutant was isolated by screening the multidimensional DNA pools of T-DNA insertion mutant populations using a PCR-based method, as described previously (Alonso et al., 2003). The T-DNA left and right border primers (Alonso et al., 2003) and GA3ox3 gene–specific primers (3ox3F and 3ox3R) were used for PCR (see Supplemental Table 5 online). Mutant lines were identified by analyzing PCR products by DNA blot analyses using a gene-specific cDNA probe. The location of the T-DNA insertion in the mutant was determined by DNA sequence analysis of the PCR product. This mutant was later deposited in the ABRC as seed stock Salk_115634. The ga3ox4-2 (Salk_035893) and ga3ox4-3 (Salk_152479) mutants were isolated by searching the SALK Institute Genomic Analysis Laboratory T-DNA express database (Alonso et al., 2003). Homozygous ga3ox3 and ga3ox4 T-DNA insertion mutants were identified by PCR using allele-specific primers (see Supplemental Table 5 online). To detect any remaining transcripts produced in each mutant, real-time RT-PCR was performed using gene-specific primers (see Supplemental Table 5 online) as previously described (Mitchum et al., 2006). The housekeeping gene UBQ11 was used to normalize different samples. Once verified, all single ga3ox3 and ga3ox4 T-DNA mutants were backcrossed once to Col-0.
The ga3ox4-1 and ga3ox4-4 mutants were identified via the Arabidopsis TILLING (Targeting Induced Local Lesions in Genomes) Project (Henikoff et al., 2004). Procedures for mutant identification and genotyping are described in the Supplemental Methods online.
Generation of Double and Triple Mutant Combinations
Homozygous ga3ox3-1 was crossed to ga3ox1-2 and ga3ox2-1 mutants (Mitchum et al., 2006) to generate double and triple mutants containing these three mutations. Homozygous ga3ox4-1 and ga3ox4-3 mutants were then crossed to ga3ox1 ga3ox3 to generate the rest of the double and triple mutants. Allele-specific primers for each locus (see Supplemental Table 5 online) were used to identify homozygous mutant alleles by PCR.
In Vitro Enzyme Assays for GA3ox Activity
Plasmids expressing MBP-GA3ox3, MBP-GA3ox4, MBP-ga3ox4-1, and MBP-ga3ox4-4 fusions were constructed as described in the Supplemental Methods online. For enzyme assays, Escherichia coli cells from a 20 mL of culture were collected by centrifugation, and soluble proteins were resuspended in 1 mL of Tris-HCl, pH 7.5. GA3ox activity assays were conducted as reported previously (Yamaguchi et al., 1998). The levels of MBP fusion proteins in bacterial lysates were estimated by immunoblot analysis using a rabbit anti-MBP polyclonal antibody (New England Biolabs) and anti-rabbit IgG antibody conjugated with horseradish peroxidase (Sigma-Aldrich). Signals were detected using a SuperSignal Pico Chemiluminescence Substrate (Pierce). When quantification of the substrate and the product was needed, deuterium-labeled GAs were added to the samples before GA extraction. Gas chromatography–mass spectrometry analysis was performed using a mass spectrometer (Automass Sun; JEOL) equipped with a gas chromatograph (6890N; Agilent Technologies).
Analysis of Endogenous GAs in Wild-Type and ga3ox Mutants
Three types of tissues were collected for measuring endogenous GA contents in wild-type and ga3ox category II mutants: rosette leaves harvested at anthesis of the first flower, early flower clusters immediately before the anthesis of the first flower, and late flower clusters after ∼20 to 25 siliques formed on primary stems. All flower clusters were harvested from primary inflorescence stems. Endogenous GA levels were determined by LC-MS/MS analysis using a quadrupole/time-of-flight tandem mass spectrometer (Q-tof Premier; Waters) and an Acquity Ultra Performance liquid chromatograph (Waters) as detailed in Varbanova et al. (2007).
Mutant Characterization
The longest rosette leaf was determined as a measure of rosette radius. The flowering time was scored as the number of days after planting when the flower bud was visible by the naked eye or as total number of rosette leaves produced by the primary inflorescence stem after bolting. Final height is the final length of the primary stem. Seeds were counted for a minimum of 10 fertile siliques on the primary stem for each wild-type or Category I plant. For the ga3ox mutants with reduced fertility, seed numbers in all fertile siliques on the primary stem were scored. Images of individual mutant flowers were taken under a Leica MZDLIII dissecting microscope (Leica Microsystems).
Construction of GA3ox3 and GA3ox4-GUS Gene Fusions and Plant Transformation
Transcriptional and translational GA3ox3-GUS and GA3ox4-GUS gene fusions were constructed using the pBI101.1 vector. p3ox3-TC-GUS was a transcriptional fusion of 1789 bp of the GA3ox3 promoter region upstream of the translational start site fused to the GUS reporter gene. p3ox3-TL1-GUS was a translational fusion of 1789 nucleotides of the GA3ox3 promoter region upstream of the translational start site containing the first exon, first intron, and 79 nucleotides of the second exon fused to the GUS reporter gene. p3ox3-TL2-GUS was a translational fusion of 1789 nucleotides of the GA3ox3 promoter region upstream of the translational start site containing exon 1, intron 1, exon 2, intron 2, and 38 nucleotides of exon 3 fused to the GUS reporter gene. p3ox4-TC-GUS was a transcriptional fusion of 1157 nucleotides of the GA3ox4 promoter region upstream of the translational start site fused to the GUS reporter gene. p3ox4-TL-GUS was a translational fusion containing 1157 nucleotides of the GA3ox4 promoter region upstream of the translational start site including exon 1, the first intron, and 201 nucleotides of the second exon fused to the GUS reporter gene. Detailed procedures for making the GA3ox3-GUS and GA3ox4-GUS gene fusion constructs are described in the Supplemental Methods online. Plant transformations using GA3ox-GUS plasmids and isolation of homozygous transgenic lines that contain a single insertion site were conducted as previously described (Mitchum et al., 2006).
Histochemical Staining with X-Gluc
GUS assays on GA3ox3-GUS and GA3ox4-GUS lines were conducted as described previously (Jefferson et al., 1987; Yamaguchi et al., 2001). The concentration of potassium ferricyanide applied in the assay was determined empirically: GA3ox3-GUS flowers (0 mM), siliques (1 mM), and developing embryos (1 mM); GA3ox4-GUS flowers (0 mM), siliques (1 mM), developing seeds (1 mM), and germinating seeds (0.1 mM). The concentrations of potassium ferricyanide used for flowers of GA3ox1-TC-GUS and GA3ox2-TL-GUS were the same as described in Mitchum et al. (2006), except that 0 mM was used for GA3ox1-TC-GUS siliques. Whole-mount photographs were taken under the same dissecting microscope described above. Homozygous lines #11-3-1 and #14-2-11 of GA3ox3-TL-GUS and #7-1-6 and #9-1-6 of GA3ox4-TC-GUS were chosen for detailed analysis due to their representative GUS patterns within each group. Figures 4 and 5 contain images of #14-2-11 of GA3ox3-GUS and #7-1-6 of GA3ox4-GUS. Clearing of GA3ox4-GUS developing seeds in Hoyer's medium was performed according to Stangeland and Salehian (2002). X-Gluc–stained seeds were fixed in ethanol/acetic acid (1:1) for 1 h, washed three times in absolute ethanol and once in 70% ethanol, and then cleared in Hoyer's medium (chloral hydrate:glycerol:water = 100:5:30). Seeds were then mounted in Hoyer's medium containing gum arabicum (gum arabicum:chloral hydrate:glycerol:water = 7.5:100:5:30) and observed using bright-field microscopy under a Zeiss Axioplan2 microscope (Carl Zeiss MicroImaging) with Nomarski optics. Images were taken with a Zeiss Axiocam HRc camera. For sectioning, X-gluc–stained tissues were fixed and embedded in paraffin according to the protocol described in Sessions et al. (1999). Sections (12 μm) were prepared and viewed with dark-field microscopy using the same Zeiss Axioplan2 microscope.
Histology of GA3ox Mutant Flowers
Single flowers from primary inflorescence stems were collected. Tissue fixation, embedding, and sectioning were performed as described above. Tissue sections (10 μm) were incubated in 0.25% Toluidine blue in 0.1 M PBS, pH 7.0, for 15 min and rinsed in water once prior to deparaffinization. Bright-field microscopy was performed as described above.
Quantitative Real-Time PCR
Total RNA extraction, cDNA synthesis, and real-time PCR were performed as described previously (Mitchum et al., 2006). The same primer sets were used for GA3ox genes as before (Mitchum et al., 2006). ROC1 primer sequences are listed in Supplemental Table 5 online. Absolute expression levels for GA3ox genes were determined as described in the Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g15550, At1g80340, At4g21690, and At1g80330 for Arabidopsis GA3ox1 to GA3ox4, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Wild-Type GA3ox3 and GA3ox4 Proteins Convert GA20 to GA1 in in Vitro Enzyme Assays.
Supplemental Figure 2. GA3ox4-GUS Expression in Embryos Resulted from Crosses between GA3ox4-GUS Plants (Pollen Donors) and Wild-Type Plants.
Supplemental Figure 3. The p3ox3/ga3ox1 ga3ox3 Transformants Rescued the Sterile Phenotype of ga3ox1 ga3ox3.
Supplemental Figure 4. Expression of ROC1 and GA3ox Genes as Determined by qRT-PCR.
Supplemental Figure 5. The ga3ox3 Mutant Has No Wild-Type GA3ox3 Transcript.
Supplemental Figure 6. In Vitro Enzyme Assays of GA3ox4 and Mutant Proteins ga3ox4-1 and ga3ox4-4.
Supplemental Figure 7. Levels of Endogenous 13-OH GAs in Flowers of Wild-Type and Category II ga3ox Mutant Plants.
Supplemental Figure 8. GA3ox2 and GA3ox4 Expression in Flowers of ga3ox Mutants.
Supplemental Figure 9. Transcript Levels of DELLA and GID1 Genes in ga3ox1 ga3ox3 ga3ox4 Flowers.
Supplemental Figure 10. Scanning Electron Microscopy of Wild-Type and ga3ox4 Mutant Seeds.
Supplemental Figure 11. Rosette Diameters of ga3ox1 ga3ox3 after Flower Initiation.
Supplemental Table 1. List of Flower and Silique Developmental Stages.
Supplemental Table 2. Phenotype Comparison of ga3ox4-1 and ga3ox4-3 in the ga3ox1 and ga3ox1 ga3ox3 Backgrounds.
Supplemental Table 3. Phenotypic Characterization of Category I ga3ox Mutants Compared with the Wild Type.
Supplemental Table 4. Comparison of Fertility in ga3ox1 ga3ox3, ga3ox1 ga3ox3 ga3ox4, and ga3ox1 ga3ox2 ga3ox3 Mutants.
Supplemental Table 5. List of Primers and Their Uses.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Andy Phillips for providing the GA3ox4 cDNA clone, Charles Gasser for the ROC1 cDNA clone, Leslie Eibest for assistance with scanning electron microscopy, and Jose Dinneny for suggestions on histology experiments. This work was supported by USDA National Initiative Competitive Grant 03-35304-13284 to T.-p.S. and by a CSIRO OCE Fellowship to M.O.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tai-ping Sun (tps@duke.edu).
Online version contains Web-only data.
References
- Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., Moritz, T., and Harberd, N.P. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Meier, C., Foster, R., Nielsen, M.E., and Mundy, J. (2001). Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127 450–458. [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. (1994). Arabidopsis: An Atlas of Morphology and Development. (New York: Springer-Verlag).
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Chiang, H.-H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, C.M., and Swain, S.M. (2006). Goldacre paper. Localised and non-localised promotion of fruit development by seeds in Arabidopsis. Funct. Plant Biol. 33 1–8. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (2004). Plant Hormones: Biosynthesis, Signal Transduction, Action! (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Eriksson, S., Bohlenius, H., Moritz, T., and Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, H., Zhang, R., Pharis, R.P., and Sawhney, V.K. (2004). Pleiotropic effects of the male sterile33 (ms33) mutation in Arabidopsis are associated with modifications in endogenous gibberellins, indole-3-acetic acid and abscisic acid. Planta 219 649–660. [DOI] [PubMed] [Google Scholar]
- Fleet, C.M., and Sun, T.P. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena, C., de Folter, S., Costa, M.M., Angenent, G.C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132 429–438. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Till, B.J., and Comai, L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel, L.L., Nelson, M.A., Richmond, T.A., and Bleecker, A.B. (1994). The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol. 106 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Kawaide, H., Chen, X., Kamiya, Y., and Matsuoka, M. (1999). The gene encoding tobacco gibberellin 3β-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J. 20 15–24. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., and Kobayashi, M. (2001). Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., Itoh, H., Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2003). Where do gibberellin biosynthesis and signaling occur in rice plants? Plant J. 34 1–12. [DOI] [PubMed] [Google Scholar]
- Katsumi, M., Foard, D.E., and Phinney, B.O. (1983). Evidence for the translocation of gibberellin A3 and gibberellin-like substances in grafts between normal, dwarf1 and dwarf5 seedlings of Zea mays L. Plant Cell Physiol. 24 379–388. [Google Scholar]
- Kim, Y.C., Nakajima, M., Nakayama, A., and Yamaguchi, I. (2005). Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol. 46 1317–1325. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Matsushita, A., Furumoto, T., Ishida, S., and Takahashi, Y. (2007). AGF1, an AT-Hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G., Yamaguchi, S., Hanada, A., Kuwahara, A., Yoshioka, Y., Kato, T., Tabata, S., Kamiya, Y., and Sun, T.P. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45 804–818. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling:Biosynthesis, catabolism, and response pathways. Plant Cell 14 S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga, J.A., and Reinecke, D.M. (2003). Hormonal interactions in fruit development. J. Plant Growth Regul. 22 73–81. [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E.J., Lange, T., Huttly, A., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proebsting, W.M., Hedden, P., Lewis, M.J., Croker, S.J., and Proebsting, L.N. (1992). Gibberellin concentration and transport in genetic lines of pea: Effect of grafting. Plant Physiol. 100 1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.J., O'Neill, D.P., Wolbang, C.M., Symons, G.M., and Reid, J.B. (2002). Auxin-gibberellin interactions and their role in plant growth. J. Plant Growth Regul. 20 346–353. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.C., Lee, P.Y., and Truong, M.T. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11 297–322. [Google Scholar]
- Seo, M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20 259–263. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Chang, C.-w., Krol, E., and Sun, T.-p. (1997). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12 9–19. [DOI] [PubMed] [Google Scholar]
- Singh, D.P., Jermakow, A.M., and Swain, S.M. (2002). Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14 3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland, B., and Salehian, Z. (2002). An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol. Biol. Rep. 20 107–114. [Google Scholar]
- Swain, S.M., Muller, A.J., and Singh, D.P. (2004). The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol. 134 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., and Singh, D.P. (2005). Tall tales from sly dwarves: Novel functions of gibberellins in plant development. Trends Plant Sci. 10 123–129. [DOI] [PubMed] [Google Scholar]
- Talón, M., Koornneef, M., and Zeevaart, J.A.D. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Varbanova, M., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith, A., and Koltunow, A.M. (1999). Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 121 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D., and Halevy, A.H. (1989). Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta 179 89–96. [DOI] [PubMed] [Google Scholar]
- Williams, J., Phillips, A.L., Gaskin, P., and Hedden, P. (1998). Function and substrate specificity of the gibberellin 3β-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 117 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.-L., Li, L., Wu, K., Peeters, A.J.M., Gage, D.A., and Zeevaart, J.A.D. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T.-p. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28 443–453. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-p. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.-Y., Nagatani, A., Zhao, Y.-J., Kang, B.-J., Kendrick, R.E., and Kamiya, Y. (1995). Effects of gibberellins on seed germination of phytochrome-deficient mutants of Arabidopsis thaliana. Plant Cell Physiol. 36 1205–1211. [PubMed] [Google Scholar]
- Zhu, Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.