Abstract
Inositol polyphosphate 5-phosphatase (5PTase) is a key enzyme in the phosphatidylinositol metabolic pathway, which plays critical roles in a number of cellular processes in plants. Our previous work implicated the role of 5PTase13, which encodes a WD40-containing type II 5PTase, in hormone-mediated cotyledon vein development. Here, we show that 5PTase13 is also involved in blue light responses in Arabidopsis thaliana. Compared with that in darkness, the expression of 5PTase13 was suppressed by blue light irradiation, and disruption of the gene resulted in shortened hypocotyls and expanded cotyledons. Genetic analysis showed that 5PTase13 acted independently from CRYPTOCHROME1 and CONSTITUTIVE PHOTOMORPHOGENIC1 but interacted functionally with PHOTOTROPIN1 (PHOT1). The expression level of 5PTase13 was significantly enhanced in phot1 single or phot1 phot2 double mutants under blue light, and suppression of 5PTase13 expression rescued the elongated hypocotyls in the phot1 or phot1 phot2 mutants. Further analysis showed that the blue light–induced elevation of cytosolic Ca2+ was inhibited in the phot1 mutant but enhanced in the 5pt13 mutant, suggesting that 5PTase13 antagonizes PHOT1-mediated effects on calcium signaling under blue light.
INTRODUCTION
Plants have evolved a number of mechanisms for responding to a broad spectrum of light through activation of specific sets of photoreceptors (Franklin et al., 2005; Wang, 2005). Recent studies have characterized the phytochrome family of red/far-red light receptors (Schepens et al., 2004; Kim et al., 2005; Rockwell et al., 2006), the cryptochrome (CRY) family of blue light receptors (Lin and Todo, 2005; Li and Yang, 2007), and the phototropin (PHOT) family of blue light receptors (Kimura and Kagawa, 2006).
Arabidopsis thaliana mutants cry1 and cry2 exhibit elongated hypocotyls, and the CRY1 and CRY2 gene products have been identified as blue light receptors that mediate light-dependent inhibition of hypocotyl elongation. Studies have also shown that CRY1 is negatively regulated by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1; a photomorphoregenesis repressor) (Ang et al., 1998) through the interaction of the C-terminal region of CRY1 and the WD40 repeats of COP1 (Yang et al., 2001). In addition, PHOT1 and PHOT2, both of which are Ser/Thr photoreceptor kinases that undergo autophosphorylation in response to blue light (Cho et al., 2007; Christie, 2007), represent another class of blue light receptor controlling plant phototropism, chloroplast orientation, stomatal opening, and rapid inhibition of growth of Arabidopsis etiolated seedlings (Briggs and Christie, 2002; Briggs, 2007; Cho et al., 2007).
The phototropin proteins carry two domains: a C terminus–localized Ser/Thr kinase domain and an N terminus–localized LOV domain (Kasahara et al., 2002; Franklin et al., 2005). Four phototropin-interacting proteins have been identified in Arabidopsis, namely, NON-PHOTOTROPIC HYPOCOTYL3 (Motchoulski and Liscum, 1999; Pedmale and Liscum, 2007), ROOT PHOTOTROPISM2 (RPT2; Inada et al., 2004), PHYTOCHROME KINASE SUBSTRATE1 (Lariguet et al., 2006), and a 14-3-3 protein (Kinoshita et al., 2003), and a fifth in Vicia faba, phot1A-interacting protein (Emi et al., 2005). These five proteins participate in the regulation of phototropic responses (Liscum and Briggs, 1995, 1996; Motchoulski and Liscum, 1999; Sakai et al., 2000; Inada et al., 2004), chloroplast movement (Inada et al., 2004), and stomatal aperture control (Lascève et al., 1999; Inada et al., 2004). Other studies have shown that calcium might be part of the blue light signaling pathway (Baum et al., 1999; Babourina et al., 2002; Folta et al., 2003; Stoelzle et al., 2003; Christie, 2007; Harada and Shimazaki, 2007).
Calcium, a universal second messenger, is involved in diverse cellular functions and is crucial in cell responses to a variety of stimuli (Luan et al., 2002; Yang and Poovaiah, 2003), including circadian oscillations (Xu et al., 2007), gravity (Poovaiah et al., 2002), stresses (Quan et al., 2007), and hormones (Du and Poovaiah, 2005). In particular, studies have revealed the importance of cytosolic Ca2+ concentration ([Ca2+]cyt) in photomorphogenesis (Harada and Shimazaki, 2007). Blue light induces opening of anion and potassium channels (Cho and Spalding, 1996; Suh et al., 2000), resulting in the change of [Ca2+]cyt (Baum et al., 1999). When plants respond to initial blue light illumination, a transient [Ca2+]cyt elevation is detected that is dependent on PHOT1 (Folta et al., 2003) or both PHOT1 (at lower fluence rates) and PHOT2 (at higher fluence rates) (Harada et al., 2003). Blue light–induced calcium influx is also critical for the primary inhibition of hypocotyl growth but not for phototropism (Folta et al., 2003). However, 30 min after blue light illumination, further inhibition of hypocotyl elongation mostly depends on the CRY1-COP1–mediated ubiquitination pathway (Parks et al., 1998; Folta and Spalding, 2001). Participation of several calcium-regulated factors in the regulation of hypocotyl elongation under red or blue light has been validated. For example, SHORT UNDER BLUE LIGHT1, a Ca2+ binding protein, acts as a negative regulator of blue light signaling downstream of CRY1 and CRY2 (Guo et al., 2001). The Ser/Thr protein phosphatase PP7, which binds calmodulin in a Ca2+-dependent manner, functions as a positive regulator of cryptochrome signaling in Arabidopsis (Møller et al., 2003). Despite accumulating evidence of calcium involvement in light signaling, there is a missing link between the photoreceptors and the calcium second messenger.
Phosphatidylinositols play crucial roles in multiple developmental processes, including hormone effects (Lin et al., 2004, 2005; Tan et al., 2007; Xue et al., 2007), cytoskeleton architecture (Sechi and Wehland, 2000), ion transport (Liu et al., 2005), stress responses (Williams et al., 2005), and light regulation (Qin et al., 2005). Inositol polyphosphate 5-phosphatases (5PTases) hydrolyze the phosphate group at the 5′ position of the inositol ring and are key enzymes involved in phosphatidylinositol signaling. The distinct substrates of 5PTases are water-soluble inositol polyphosphates, including inositol 1,4,5-trisphosphate [Ins(1,4,5)P3], inositol 1,3,4,5-tetraphosphate [Ins(1,3,4,5)P4], and membrane-bound phosphoinositides, such as phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] and phosphatidylinositol 3,4,5-trisphosphate. There are 15 genes encoding 5PTases in Arabidopsis that are grouped into two subfamilies based on protein structure: type I (11 members) and type II (four members) (Zhong et al., 2004).
Through genetics approaches, several 5PTases, especially type I members, have been functionally characterized in Arabidopsis, revealing their diverse functions in various aspects of plant growth and development. Disruption of 5PTase1 increased sensitivity to abscisic acid treatment (Berdy et al., 2001; Gunesekera et al., 2007). 5PTase1 and 5PTase2 regulate germination and early seedling development through altering the Ins(1,4,5)P3 level (Gunesekera et al., 2007). COTYLEDON VEIN PATTERN2 (CVP2/5PTase6) is essential for closed venation patterns of foliar organs through modulating the Ins(1,4,5)P3 level (Carland and Nelson, 2004). root hair morphogenesis3 (mrh3/5ptase5) mutants have altered root hair initiation (Jones et al., 2006). Of the type II members, FRAGILE FIBER3 (FRA3/5PTase12) is required for secondary wall synthesis and actin organization in fiber cells (Zhong et al., 2004), and our previous research has shown that 5PTase13 is required for cotyledon vein development through regulating auxin homeostasis (Lin et al., 2005). Here, we identify a role for 5PTase13 in blue light responses and demonstrate crosstalk between PHOT1 and 5PTase13 through the regulation of the calcium transients under blue light.
RESULTS
Blue Light Suppresses 5PTase13 Expression and 5pt13 Mutants Display Shortened Hypocotyls and Expanded Cotyledons in Response to Blue Light Irradiation
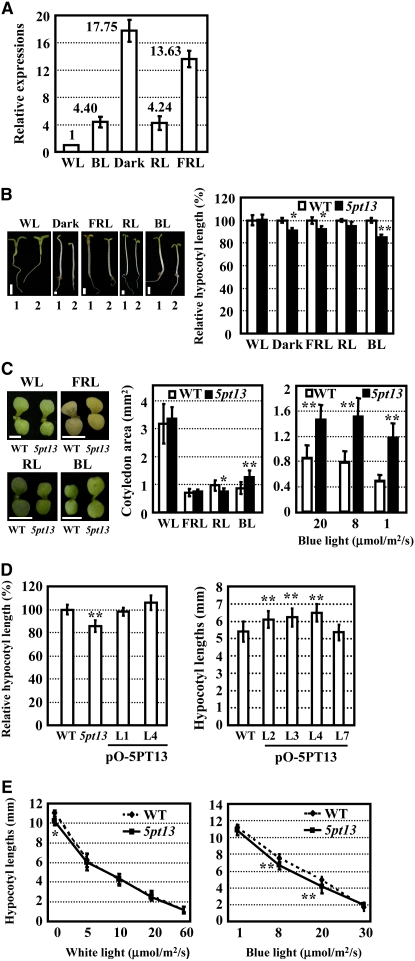
Our previous studies have shown that 5PTase13 expression is differentially regulated under light and dark (Lin et al., 2005). To further investigate the effects of different light spectra on 5PTase13 gene expression, we examined 5PTase13 transcripts under different light spectra through quantitative RT-PCR analysis. As shown in Figure 1A, expression of 5PTase13 was higher in dark-grown seedlings compared with those grown under light. Histochemical analysis of transgenic plants harboring the 5PTase13 promoter-β-glucuronidase (GUS) reporter gene confirmed that expression of 5PTase13 in hypocotyls was inhibited by light at the level of transcription (see Supplemental Figure 1 online).
Figure 1.
Expression of 5PTase13 Is Regulated by Light, and 5pt13 Mutants Are Less Sensitive to Blue Light.
(A) Expression of 5PTase13 under different light spectra. 5PTase13 mRNA levels were measured under dark, far-red light (FRL; 30 μmol/m2/s), blue light (BL; 20 μmol/m2/s), red light (RL; 30 μmol/m2/s), and white light (WL; 60 μmol/m2/s), respectively.
(B) Hypocotyl length (left panels; bars = 1 mm) and statistical analysis (right panel) of wild-type and 5pt13 mutant seedlings under dark, far-red light (30 μmol/m2/s), or blue light (20 μmol/m2/s). Five-day-old seedlings were measured and statistically analyzed using a Student's t test (*, P < 0.05; **, P < 0.01). Data represent mean ± se (n > 30). Same-age seedlings were used for all hypocotyl measurements throughout this study. The same procedure, data analysis, and presentation were used for production of all panels.
(C) Images of cotyledons (left panels; bars = 1 mm) and statistical analysis (middle panel) indicated the enlarged cotyledons of 5pt13 under red light (30 μmol/m2/s) or blue light (20 μmol/m2/s). The 5pt13 mutant plants showed enlarged cotyledons under different fluence rates of blue light (1, 8, or 20 μmol/m2/s; right panels). Cotyledons of 5-day-old seedlings were measured and statistically analyzed using a Student's t test (*, P < 0.05; **, P < 0.01). Data represent mean ± se (n > 20).
(D) Overexpression of 5PTase13 in 5pt13 (left panel) or wild-type (right panel) plants resulted in an elongated hypocotyl. Hypocotyls of 5-d-old seedlings grown under blue light (20 μmol/m2/s) were measured and statistically analyzed using a one-tailed Student's t test (*, P < 0.05; **, P < 0.01). Data represent mean ± se (n > 30).
(E) The 5pt13 seedlings had shortened hypocotyls under blue light with different fluence rates (1, 8, 20, or 30 μmol/m2/s; right panel) compared with those grown under white light (left panel). Hypocotyl lengths of 5-d-old seedlings were measured and statistically analyzed using a Student's t test (**, P < 0.01). Data represent mean ± se (n > 30).
We further analyzed seedling growth of a 5PTase13 knockout mutant, 5pt13, under various light conditions and observed a small but significant change in blue light–induced hypocotyl shortening in mutant compared with wild-type seedlings. Although the 5pt13 seedlings showed shorter hypocotyls under both dark and light conditions, the hypocotyl shortening phenotype of the mutant was most significant under blue light (Figure 1B). In addition, 5pt13 mutant cotyledons were smaller than the wild type under red light but larger under blue light (Figure 1C, left and middle panels). Under white light or far-red light, there were no significant differences between the mutant and wild type. These results suggest that the 5pt13 mutant may have altered light responses, especially with regard to blue light. Overexpression of 5PTase13 in 5pt13 plants complemented the mutant phenotype under blue light (Figure 1D, left panel; see Supplemental Figure 2A online), indicating that the changes in blue light responses in the mutant were a result of disruption in the 5PTase13 gene. In addition, transgenic wild-type plants that overexpressed 5PTase13 displayed longer hypocotyls under blue light (see Supplemental Figure 2B online; Figure 1D, right panel), further confirming the effects of 5PTase13 on hypocotyl elongation under blue light.
More detailed analysis of hypocotyl lengths revealed no significant difference between the wild type and the 5pt13 mutant under white light at various fluence rates (5, 10, 20, and 60 μmol/m2/s; Figure 1E, left panel). The 5pt13 mutant seedlings had significantly shorter hypocotyls under blue light at the fluence rates of 8 or 20 μmol/m2/s but not at lower or higher fluence rates (Figure 1E, right panel). In addition, enlarged cotyledons were observed under blue light at different fluence rates (1, 8, or 20 μmol/m2/s; Figure 1C, right panel). Taken together, these results suggest that 5PTase13 participates in blue light–mediated regulation of seedling growth under a specific range of blue light fluence rates.
Consistent with the above observations, analysis of transcription profiling revealed altered expression of many light-regulated genes in 5pt13 mutant seedlings compared with those in wild-type seedlings under white light (chip hybridization was described in Lin et al., 2005). These genes, including those encoding EARLY LIGHT-INDUCED PROTEIN (Casazza et al., 2005), SUPPRESSOR OF PHYTOCHROME A (SPA; Hoecker and Quail, 2001; Laubinger et al., 2004), and CHLOROPHYLL A/B BINDING protein (Meehan et al., 1996), were stimulated under 5PTase13 deficiency (Table 1), suggesting that 5PTase13 functions as a negative regulator of light responses. In addition, expression of cell wall synthesis and cytoskeleton-related genes, especially those encoding UDP-glucosyl transferase family proteins (Reiter, 2002) and xyloglucan:xyloglucosyl transferase (Yokoyama et al., 2004; Cosgrove, 2005), were suppressed in 5pt13 seedlings (Table 2), consistent with the finding that 5PTase13 participates in the regulation of hypocotyl growth.
Table 1.
Hybridization with Arabidopsis ATH1 Chips Revealed That Deficiency of 5PTase13 Altered the Expression of Genes Involved in Light and Calcium Signal Transduction and Cation Transport
| Probe Set | Locus No. | Regulation | Descriptions |
|---|---|---|---|
| 259866_at | At1g76640 | I (3.3)* | Calmodulin-related protein, putative |
| 260881_at | At1g21550 | I (3)** | Calcium binding protein, putative |
| 258181_at | At3g21670 | I (3)** | Nitrate transporter (NTP3) |
| 266672_at | At2g29650 | I (2.1)** | Inorganic phosphate transporter, putative |
| 248910_at | At5g45820 | I (2.1)** | CBL-interacting protein kinase 20 (CIPK20) |
| 267516_at | At2g30520 | I (2.2)** | Signal transducer of phototropic response (RPT2) |
| 253172_at | At4g35060 | I (2.2)* | Heavy metal–associated domain-containing protein/copper chaperone (CCH) related |
| 263779_at | At2g46340 | I (1.7)** | Suppressor of phytochrome A-105 (SPA1) |
| 258321_at | At3g22840 | I (1.7)** | Chlorophyll A/B binding family protein/early light-induced protein (ELIP) |
| 258239_at | At3g27690 | I (1.6)** | Chlorophyll A/B binding protein (LHCB2:4) |
| 262940_at | At1g79520 | I (1.6)* | Cation efflux family protein |
| 258350_at | At3g17510 | I (1.5)** | CBL-interacting protein kinase 1 (CIPK1) |
| 259970_at | At1g76570 | I (1.1)** | Chlorophyll A/B binding family protein |
| 260308_at | At1g70610 | I (1)* | ABC transporter (TAP1) |
Data were obtained from the aerial organs of wild-type and 5pt13 seedlings as described in Methods. Results are presented as increased (I) expression in 5pt13 seedlings relative to the wild type, and numbers in parentheses indicate the fold change (log2, with false discovery rate [FDR] values <0.01 [**] or <0.05 [*]).
Table 2.
Hybridization with Arabidopsis ATH1 Chips Revealed That Deficiency of 5PTase13 Suppressed the Expression of Cell Wall Synthesis and Cytoskeleton-Related Genes
| Probe Set | Locus No. | Regulation | Descriptions |
|---|---|---|---|
| 259507_at | At1g43910 | D (−5.7)** | AAA-type ATPase family protein |
| 266877_at | At2g44570 | D (−3.9)* | Glycosyl hydrolase family 9 protein |
| 251791_at | At3g55500 | D (−2.3)** | Expansin, putative (EXP16) |
| 260592_at | At1g55850 | D (−2.3)** | Cellulose synthase family protein |
| 262643_at | At1g62770 | D (−2.1)** | Invertase/pectin methylesterase inhibitor family protein |
| 262978_at | At1g75780 | D (−1.9)** | Tubulin β-1 chain (TUB1) |
| 247189_at | At5g65390 | D (−1.8)** | Arabinogalactan protein (AGP7) |
| 252625_at | At3g44750 | D (−1.7)** | Histone deacetylase, putative (HD2A) |
| 263207_at | At1g10550 | D (−1.7)** | Xyloglucan:xyloglucosyl transferase |
| 257206_at | At3g16530 | D (−1.7)** | Legume lectin family protein |
| 255433_at | At4g03210 | D (−1.7)** | Xyloglucan:xyloglucosyl transferase |
| 258369_at | At3g14310 | D (−1.7)* | Pectinesterase family protein |
| 265499_at | At2g15480 | D (−1.7)* | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
| 253879_s_at | At4g27570 | D (−1.6)** | Glycosyltransferase family protein |
| 261046_at | At1g01390 | D (−1.6)** | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
| 263184_at | At1g05560 | D (−1.5)** | UDP-glucose transferase (UGT75B2) |
| 245052_at | At2g26440 | D (−1.5)* | Pectinesterase family protein |
| 257667_at | At3g20440 | D (−1.5)* | Glycoside hydrolase family 13 protein |
| 255070_at | At4g09020 | D (−1.4)** | Putative/starch debranching enzyme |
| 265340_at | At2g18330 | D (−1.4)** | AAA-type ATPase family protein |
| 257203_at | At3g23730 | D (−1.3)** | Xyloglucan:xyloglucosyl transferase |
| 252997_at | At4g38400 | D (−1.3)* | Expansin family protein (EXPL2) |
| 261825_at | At1g11545 | D (−1.3)* | Xyloglucan:xyloglucosyl transferase |
| 261934_at | At1g22400 | D (−1.2)** | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
| 247246_at | At5g64620 | D (−1.2)* | Invertase/pectin methylesterase inhibitor family protein |
| 261813_at | At1g08280 | D (−1.2)* | Glycosyl transferase family 29 protein |
| 254785_at | At4g12730 | D (−1)** | Fasciclin-like arabinogalactan protein (FLA2) |
Results are presented as decreased (D) expression in 5pt13 seedlings relative to the wild type, and numbers in parentheses indicate the fold change (log2, with FDR values <0.01 [**] or <0.05 [*]). The hybridization performance and data analysis are as described in Methods.
5PTase13 Acts Independently from CRY1-COP1 but Interacts Functionally with PHOT1
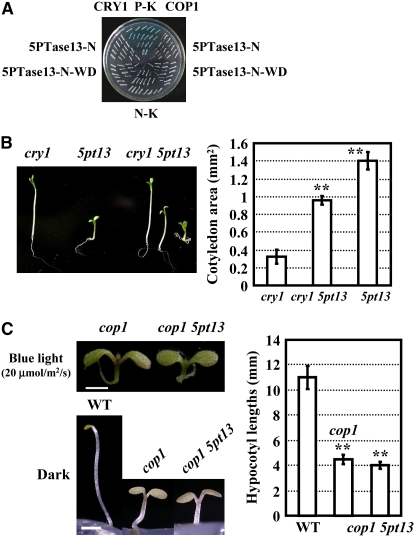
Disruption of 5PTase13 led to changes in blue light responses, including a change in hypocotyl growth. As a blue light receptor, CRY1 plays a key role in the regulation of hypocotyl elongation. Interestingly, COP1, an E3 ligase, regulates blue light response by interacting directly with CRY1. In addition, both COP1 and 5PTase13 contain WD40 repeat domains, which are involved in protein–protein interaction. These observations prompted us to test if there might be functional interactions between 5PTase13 and the other two proteins. We tested for protein–protein interactions using a yeast two-hybrid system and found that 5PTase13 did not interact with either CRY1 or COP1 (Figure 2A). Further genetic analysis of a 5pt13 cry1 double mutant showed that, except for a small proportion (3 to 5%) of seedlings that exhibited long hypocotyls (cry1 mutants) or shortened hypocotyls (5pt13 mutants), the majority (>90%) of seedlings had hypocotyls of an intermediate length (Figure 2B), indicating functional independence of CRY1 and 5PTase13. Additionally, the 5pt13 cop1 double mutant had a similar phenotype to cop1 (Figure 2C), suggesting that COP1 either acts downstream of 5PTase13 or they have no functional relationship.
Figure 2.
5PTase13 Functions Independently of COP1 and CRY1.
(A) Yeast two-hybrid analysis showed no interaction between 5PTase13 and COP1 or CRY1. Interaction between the N-terminal region (until the position before WD40 region) or N-WD40 fragment (until the position at end of the WD40 region) of 5PTase13 with COP1 or CRY1 proteins was determined, respectively. The known interaction between pJG4-5-COP1 and pEG202-CCT1 (CRY1 C-terminal) was used as a positive control (P-K). The interaction between empty vectors pEG202 and pJG4-5 was used as the negative control (N-K).
(B) cry1 5pt13 double mutants had an intermediate phenotype, indicating that there was no direct link between CRY1 and 5PTase13. Hypocotyl lengths were measured and data presented as described in Figure 1B.
(C) cop1 5pt13 double mutants exhibited a cop1 phenotype, indicating that COP1 either acts downstream of 5PTase13 or they have no functional relationship. Hypocotyl lengths were measured and data presented as described in Figure 1B.
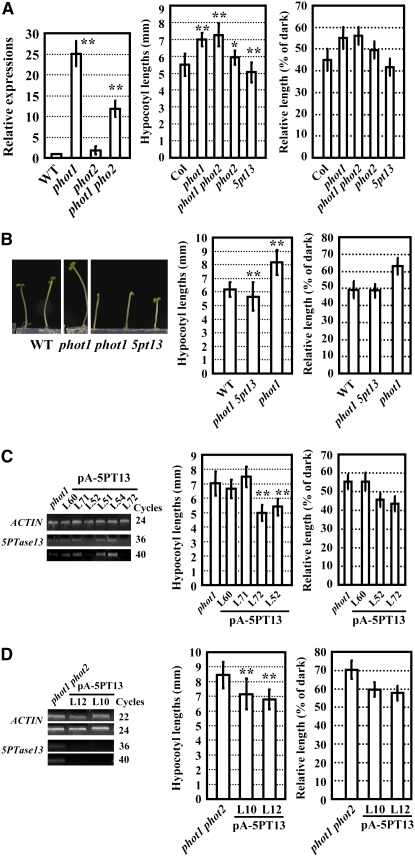
In addition to CRY1 and COP1, other photoreceptors, such as PHOT1 and PHOT2, have been identified as functioning in blue light responses (Briggs and Christie, 2002; Briggs, 2007; Cho et al., 2007). PHOT1 is involved in blue light–mediated hypocotyl growth inhibition through stimulating an increase in [Ca2+]cyt (Folta et al., 2003). Because the expression level of 5PTase13 is regulated by blue light, we decided to measure 5PTase13 expression in the phot1 and phot2 single mutants and the phot1 phot2 double mutant to determine if these blue light receptors are involved in the regulation of 5PTase13 expression. As shown in Figure 3A, under blue light, the transcript level of 5PTase13 in the phot1 or phot1 phot2 double mutants was much higher than that in the wild-type plants, increasing 25- and 12-fold, respectively, whereas no significant change was detected in the phot2 mutant (Figure 3A, left). This result indicates that PHOT1 may be the major blue light receptor mediating blue light–induced repression of 5PTase13 gene expression.
Figure 3.
Deficiency of 5PTase13 Rescues the Shortened Hypocotyls of phot1.
(A) Quantitative real-time RT-PCR analysis showed that 5PTase13 transcriptions were enhanced in phot1 and phot1 phot2 but not in phot2 seedlings under blue light (left panel; error bars represent se of three independent replicates; **, P < 0.01). Compared with elongated hypocotyls of phot1, phot2, and phot1 phot2 under blue light (20 μmol/m2/s), 5pt13 had shortened hypocotyls (middle panel). Relative hypocotyl lengths, compared with that of dark-grown seedlings, are shown in the right panel. See Figure 1B for procedure and data presentation.
(B) Deficiency of 5PTase13 rescued the elongated hypocotyls of phot1 under blue light (20 μmol/m2/s; left panel). The lengths of hypocotyls were measured (middle panel), and the relative length was calculated (right panel).
(C) Suppressed expression of 5PTase13 in phot1 as revealed by RT-PCR analysis (left panel) and correlated with shortened hypocotyls under blue light (20 μmol/m2/s; middle panel). The relative length is shown in the right panel.
(D) RT-PCR analysis showing suppressed expression of 5PTase13 in phot1 phot2 (left panel) correlated with shortened hypocotyls under blue light (20 μmol/m2/s; middle panel). The relative length is shown in the right panel.
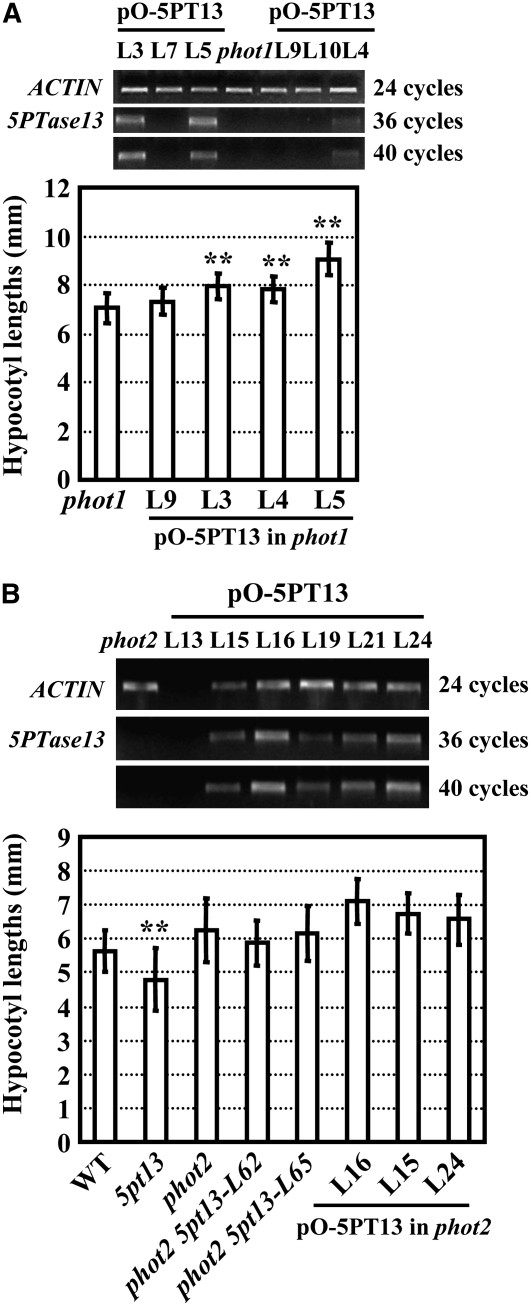
Further studies employing genetic epistatic analysis showed that under blue light, phot1 5pt13 double mutant plants exhibited shortened hypocotyls similar to 5pt13 (Figure 3B), suggesting that 5PTase13 may act downstream of PHOT1 and serve as a negative regulator in blue light signaling. In addition, phot1 single mutant or phot1 phot2 double mutant seedlings with reduced levels of 5PTase13 (due to transgenic expression of antisense RNA) exhibited shortened hypocotyls under blue light (Figures 3C and 3D, right panels), similar to the phot1 5pt13 double mutant seedlings. By contrast, overexpressing 5PTase13 in phot1 seedlings enhanced the long hypocotyl phenotype of the phot1 mutant under blue light (Figure 4A), further supporting the hypothesis that 5PTase13 functions downstream of PHOT1 as a negative regulator of the blue light response. Similar transgenic analysis with phot2 showed no affect on the hypocotyl length of phot2 (Figure 4B), indicating there may not be a functional link between PHOT2 and 5PTase13. Additionally, the yeast two-hybrid assay revealed no physical interaction between PHOT1 and 5PTase13 (see Supplemental Figure 3 online).
Figure 4.
5PTase13 Overexpression Enhances Hypocotyl Elongation of phot1 but Not phot2.
(A) Enhanced expression of 5PTase13 in phot1 (top panel) resulted in elongated hypocotyls compared with phot1 under blue light (bottom panel).
(B) Seedlings with deficiency of 5PTase13 in the phot2 mutant background had similar hypocotyl length with phot2 (bottom panel), and overexpression of 5PTase13 in phot2 (top panel) did not result in shortened hypocotyl elongation under blue light (20 μmol/m2/s; bottom panel).
Deficiency of 5PTase13 Results in Increased [Ca2+]cyt
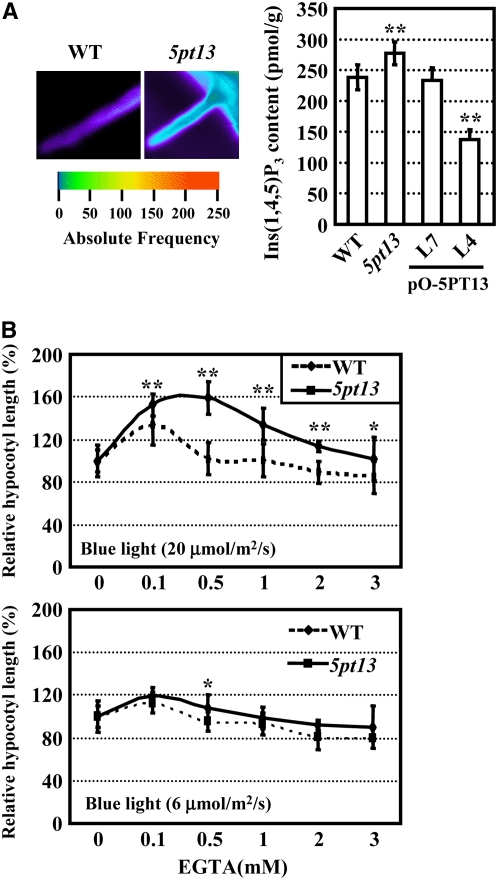
We hypothesize that inositol phosphate metabolism may provide a common link between 5PTase function and calcium fluctuations. It is generally believed that 5PTase13 functions in calcium signaling processes by regulating the level of Ins(1,4,5)P3 and/or Ins(1,3,4,5)P4, the two potent calcium-mobilizing molecules (Zhong et al., 2004; see Supplemental Figure 4 online). Indeed, our experiments indicated that Ins(1,4,5)P3 and calcium levels appeared to be increased in the 5pt13 seedlings (Figure 5A; see Supplemental Figure 5 online).
Figure 5.
5PTase13 Deficiency Results in Increased [Ca2+]cyt and Renders Responses to Exogenous EGTA under Blue Light.
(A) [Ca2+]cyt was increased in 5pt13 (left panel), consistent with the increased content of Ins(1,4,5)P3 in 5pt13 (right panel). The Ins(1,4,5)P3 content was reduced under overexpressed 5PTase13. Image analysis was performed by UV confocal imaging. Root hairs were submerged with 20 μM Indo-1 and pseudocolor according to the cytoplasmic calcium levels. The data were representative of root hairs from >10 individual roots. Five-day-old seedlings grown on Murashige and Skoog (MS) medium under white light were used to measure the Ins(1,4,5)P3 contents. Line 7 without overexpressed 5PTase13 after transformation (see Supplemental Figure 2A online) was used as a negative control. Assays were performed in triplicate, and the experiment was repeated two times and statistically analyzed using a one-tailed Student's t test (P < 0.01).
(B) 5pt13 had altered responses to exogenous EGTA under both high (top panel) and low (low panel) fluence rates of blue light. Hypocotyl lengths of 5-d-old seedlings were measured and statistically analyzed using a one-tailed Student's t test (*, P < 0.05; **, P < 0.01). Error bars represent se (n > 30).
To determine a possible connection between calcium levels and blue light responses in the 5pt13 mutant, we tested the effects of exogenous Ca2+ and its specific chelator EGTA on hypocotyl elongation. Whereas exogenous Ca2+ (10 to 20 mM) did not affect the hypocotyl lengths of the wild type or 5pt13 under blue light, this treatment resulted in shortened hypocotyls of phot1, phot2, and phot1 phot2 mutants (see Supplemental Figure 6 online). The increased [Ca2+]cyt in the 5pt13 mutant resulted in an altered responses to EGTA. In the hypocotyl assay, exogenous EGTA at low concentrations (100 μM) induced hypocotyl elongation in wild-type plants under blue light, especially under a higher fluence rate of blue light (35% longer, 20 μmol/m2/s), and suppressed the elongation when higher concentrations (3 mM EGTA) were used (Figure 5B, top panel). However, 5pt13 mutant hypocotyls displayed reduced sensitivity to EGTA, and hypocotyl length was minimally affected by high concentrations of EGTA (Figure 5B, bottom panel).
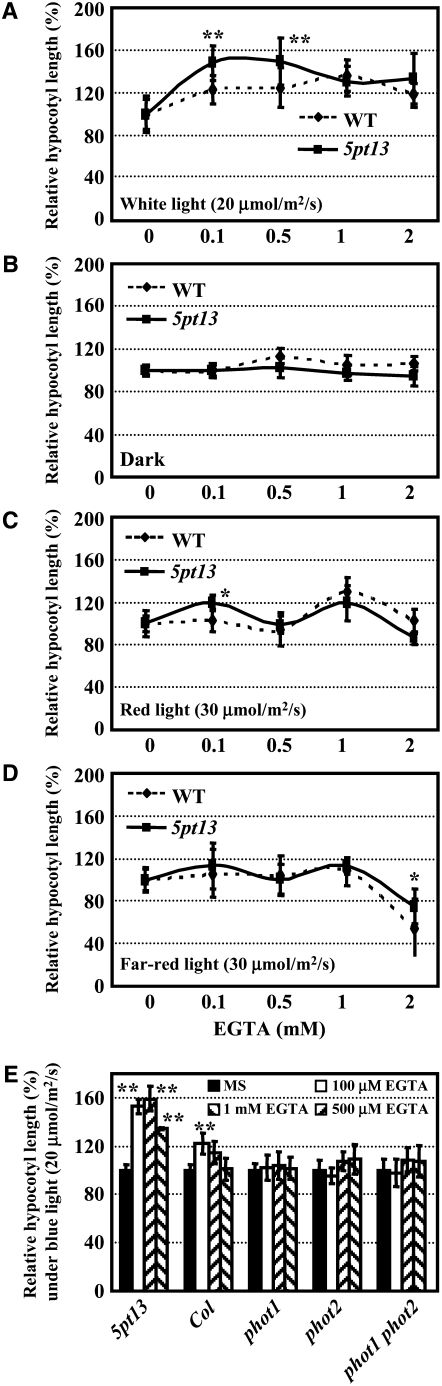
Further analysis showed that in the presence of a high concentration of EGTA (500 μM), the hypocotyl length of 5pt13 was also significantly increased under white light (a 1.5-fold increase; Figure 6A) and blue light (a 1.6-fold increase; Figure 6E) but not under red light, far-red light, or dark (Figures 6B to 6D). This result indicates that blue light and white light normally inhibit hypocotyl growth through increased [Ca2+]cyt; the addition of EGTA under these light conditions reduced calcium elevation, thereby promoting hypocotyl elongation. The observation that the 5pt13 mutant was less sensitive to a high concentration of EGTA (>2 mM) is consistent with the finding that 5PTase13 deficiency leads to an increased calcium elevation in the mutant.
Figure 6.
Responses of Hypocotyl Growth to EGTA under White Light and Blue Light.
Hypocotyl lengths of 5-d-old seedlings grown on the medium supplemented with EGTA under white light (60 μmol/m2/s; [A]), dark (B), red light (30 μmol/m2/s; [C]) far-red light (30 μmol/m2/s; [D]), and blue light (20 μmol/m2/s; [E]) were measured and statistically analyzed using a Student's t test (*, P < 0.05; **, P < 0.01). Error bars represent se (n > 30). Plant materials included wild-type, 5pt13, phot1, phot2, and phot1 phot2 seedlings.
To further test this idea, we also included the phot1 and phot2 mutants in the EGTA hypocotyl assay. Elongation of hypocotyls in the phot1 and phot2 single mutants and phot1 phot2 double mutants, compared with that of 5pt13, was not significantly altered by addition of exogenous EGTA (Figure 6E), further supporting the notion that increased [Ca2+]cyt of 5pt13 may be a critical factor influencing the response of hypocotyls to blue light.
Previous studies have shown that RPT2 transduces signals downstream of PHOT1 and is involved in the phototropic response and stomatal opening (Sakai et al., 2000; Inada et al., 2004). Indeed, analysis of gene expression profiling showed that RPT2 transcript levels were enhanced significantly in 5pt13 seedlings (Table 1), further confirming that 5PTase13 is involved in phototropin signaling.
5PTase13 Regulates Blue Light Response by Increasing Ins(1,4,5)P3 and [Ca2+]cyt
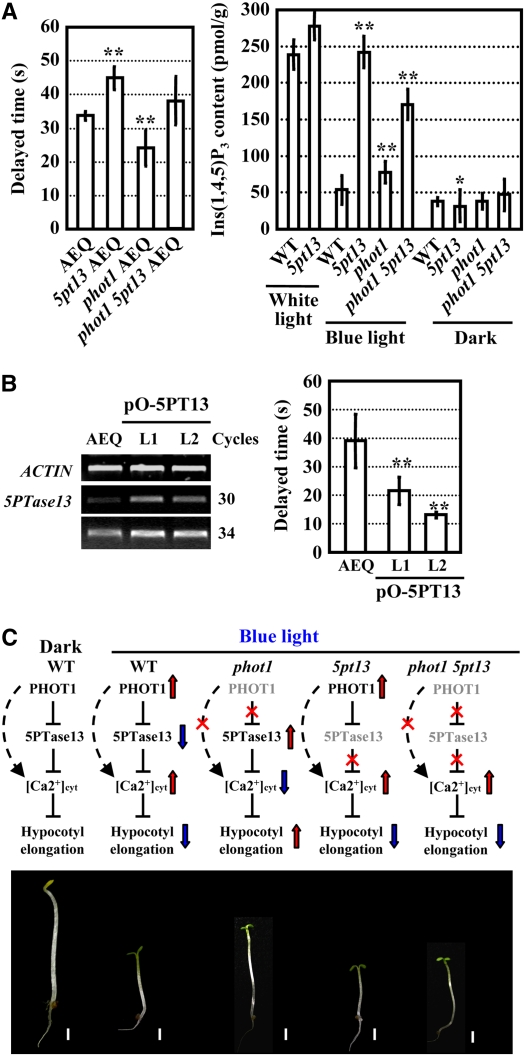
Our observation of increased calcium levels in the 5pt13 mutant and an earlier report on reduced calcium levels in the phot1 mutant under blue light irradiation (Harada et al., 2003) suggest a possible link between 5PTase13 function and the PHOT1 signaling pathways through the regulation of calcium signaling. We tested this idea by monitoring [Ca2+]cyt changes under blue light irradiation using aequorin as a [Ca2+]cyt indicator. Transgenic plants harboring cauliflower mosaic virus (CaMV) 35S promoter:aequorin were crossed with 5pt13, phot1, or 5pt13 phot1 double mutants, and the offspring were analyzed by fluorescence imaging. Under blue light irradiation, the decay of aequorin luminescence took more time in 5pt13 seedlings (∼45 s) compared with wild-type seedlings (∼34 s) and phot1 seedlings (only ∼24 s), indicating a higher level of [Ca2+]cyt in 5pt13 seedlings (the delayed time was calculated through comparison to seedlings without CaMV35S:aequorin under the same irradiation situation). Furthermore, the decreased [Ca2+]cyt in phot1 seedlings was recovered in 5pt13 phot1 double mutants (Figure 7A, left panel). This is also consistent with the Ins(1,4,5)P3 levels under blue light irradiation. As shown in Figure 7A (right panel), compared with the response under white light, Ins(1,4,5)P3 levels decreased (∼20%) under blue light in the wild type but were maintained at a higher level in the 5pt13 mutant. Accordingly, the Ins(1,4,5)P3 levels in the 5pt13 phot1 double mutant were much higher than those in phot1.
Figure 7.
5PTase13 Functions in the Blue Light–Stimulated [Ca2+]cyt Increase.
(A) 5pt13 had enhanced [Ca2+]cyt (left panel) and increased Ins(1,4,5)P3 levels (right panel) under blue light irradiation compared with the wild-type control. The data were statistically analyzed using a Student's t test (*, P < 0.05; **, P < 0.01). Error bars represent se (n > 20).
(B) Enhanced expression of 5PTase13 resulted in reduced [Ca2+]cyt under blue light irradiation. The data were statistically analyzed using a one-tailed Student's t test (**, P < 0.01). Error bars represent se (n > 20).
(C) Hypothetical model depicting 5PTase13 function in the blue light–mediated Ca2+[cyt] increase and hypocotyl growth inhibition.
In addition, seedlings overexpressing 5PTase13 appear to have reduced [Ca2+]cyt under blue light (Figure 7B). These results indicate that 5PTase13 deficiency results in higher levels and longer durations of [Ca2+]cyt elevation upon blue light irradiation, whereas phot1 seedlings have lower levels and a shorter duration of [Ca2+]cyt elevation relative to the wild type. We thus propose that 5PTase13 is involved in the blue light–stimulated increase in [Ca2+]cyt and functions downstream of PHOT1 in blue light signaling (Figure 7C).
DISCUSSION
Phosphatidylinositols regulate calcium fluctuations and other cellular functions. The 5PTases dephosphorylate Ins(1,4,5)P3, Ins(1,3,4,5)P4, or PtdIns(4,5)P2, and often serve as terminators for calcium signaling. In this study, we show that Arabidopsis 5PTase13 plays a role in the regulation of blue light responses. In particular, the expression of 5PTase13 was suppressed by blue light, whereas a deficiency of 5PTase13 resulted in a hypersensitive response to blue light in the hypocotyl assay. Disruption of 5PTase13 rescued the hypocotyl phenotype in the phot1 mutant but not in the cry1 mutant, indicating that 5PTase13 functions downstream of PHOT1 in blue light signaling. The 5pt13 mutant displayed enhanced [Ca2+]cyt, whereas phot1 exhibited reduced [Ca2+]cyt in response to blue light irradiation, leading to a model that PHOT1 suppresses 5PTase13 activity, thereby controlling calcium levels, which in turn regulate hypocotyl elongation.
Our study indicates that PHOT1 mediates blue light suppression of 5PTase13, as this suppression was not evident in the phot2 mutant. A previous study using microarray analysis (Ohgishi et al., 2004) showed that CRY1 and CRY2 appear to be the major receptors that mediate blue light–responsive gene expression, whereas PHOT1 and PHOT2 have very limited effects on the downstream light response genes. We speculate that the expression of some genes may have been altered in the phot1 and phot2 mutants in the microarray study, although such change may be relatively small compared with changes in the cry1 and cry2 mutants. We noted that Ohgishi et al. (2004) used 11-d-old seedlings, whereas 4-d-old seedlings were used in our studies. Along the line of differential function by CRY and PHOT family receptors, it is well documented that CRY proteins plays a major role in hypocotyl growth and PHOT proteins functions in phototropism, chloroplast orientation, and stomatal opening. We speculate that the contribution of PHOT1 to the regulation of hypocotyl growth is less significant compared with the other photoreceptors, such as CRY1, and may explain why 5pt13 mutants displayed a rather subtle hypocotyl growth phenotype. The changes in hypocotyl length in both 5pt13 and phot1 mutants were statistically significant, although small. Furthermore, a deficiency of 5PTase13 in phot1 mutant seedlings rescues the hypocotyl phenotype and causes changes in calcium levels, suggesting that crosstalk between 5PTase13 and PHOT1 plays a role in blue light responses.
Much effort has been focused on the identification of signaling components downstream of blue light receptors and upstream of phenotypic effects, such as inhibition of hypocotyl growth. Several studies have shown that blue light induces elevation of [Ca2+]cyt, and mutations in genes encoding blue light receptors, such as PHOT1 or PHOT2, have compromised calcium elevation, leading to changes in photomorphogenesis (Babourina et al., 2002; Harada et al., 2003; Stoelzle et al., 2003). It is interesting to note that defects in PHOT1 and [Ca2+]cyt appear to affect only the short term initial phase of the hypocotyl growth response, and further inhibition of hypocotyl elongation requires the CRY1-COP1 pathway (Parks et al., 1998; Folta and Spalding, 2001). Our study addresses a missing link between the photoreceptors and the calcium second messenger. More specifically, we identified 5PTase13 as a component that regulates calcium signals in the PHOT1 pathway.
How does PHOT1 regulate the activity of 5PTase13? Our study provides evidence that 5PTase13 is regulated by blue light through PHOT1, at least in part, at the transcriptional level. Our quantitative RT-PCR analysis indicated that 5PTase13 mRNA levels were downregulated by blue light. The finding that the 5PTase13 promoter-GUS reporter negatively responds to blue light confirmed the notion that regulation occurred at the level of transcription. However, we cannot exclude the possibility that 5PTase13 also might be regulated by PHOT1 through other mechanisms, such as direct modification of enzyme activity. The protein–protein interaction by yeast two-hybrid assay did not detect a physical interaction between PHOT1 and 5PTase13 (see Supplemental Figure 3B online); however, the kinase–substrate interaction was often weak and transient, and further studies are needed to determine if 5PTase13 is in fact a substrate of PHOT1.
Blue light suppression of 5PTase13 reveals a double-negative step in blue light signaling, and such negative-negative regulation steps appear to be a common feature of plant signal transduction. For example, similar double-negative steps are found in ethylene signal transduction where ethylene receptors negatively regulate the downstream CONSTITUTIVE TRIPLE RESPONSE1 kinase, which in turn inhibits ethylene responses (Stepanova and Alonso, 2005). We propose that under darkness, the protein level (and activity) of 5PTase13 is high, keeping [Ca2+]cyt low, and hypocotyls elongate rapidly. Upon light irradiation, PHOT1 is activated, leading to suppression of 5PTase13, and an elevation of cytosolic calcium takes place resulting in inhibition of hypocotyl growth.
Several major questions remain to be addressed, including how 5PTase13 regulates calcium, and what the function of calcium is in cell elongation. While little information is available regarding calcium action in cell growth upon blue light irradiation, previous studies have built a connection between 5PTase13 and calcium fluctuations in the cell. At least several inositol phosphates molecules, especially Ins(1,4,5)P3 and Ins(1,3,4,5)P4, have been shown to mediate calcium fluxes across cell membranes. Such transport processes can include release of calcium from intracellular stores or influxes from the apoplast through inward calcium channels. Intracellular calcium is stored in the vacuole, which harbors ligand-gated calcium channels or Ins(1,4,5)P3 receptors. High levels of Ins(1,4,5)P3 activate these calcium channels and then release calcium to the cytosol. The phosphate group at the 5′ position of the inositol ring is thought to be important for ligand specificity of calcium channels, and dephosphorylation at the 5′ position by 5PTases could reduce the level of active ligands, leading to the closure of calcium channels and termination of the signaling process. Therefore, the 5PTases function as terminators for calcium signaling, which might be initiated by various signals, including blue light, as demonstrated here. In this context, earlier studies have identified both plasma membrane channels and intracellular calcium stores that are involved in blue light signaling (Harada et al., 2003; Stoelzle et al., 2003). However, the molecular nature of these channels remains elusive. Toward a better understanding of calcium channels involved in blue light signaling, further identification of 5PTase13 substrates would provide clues about the ligands that activate the calcium channels. Our results constitute substantial progress toward this goal: Ins(1,4,5)P3 levels appear to fluctuate consistently with calcium levels, suggesting the possibility that the Ins(1,4,5)P3 receptor could be the calcium release channel involved in blue light responses. How might [Ca2+]cyt mediate cell elongation? A simple interpretation is that blue light induces Ca2+ and H+ fluxes (Babourina et al., 2002), both of which are critical for cell growth.
5PTase13 expression was similarly suppressed by both blue and red light compared with darkness. However, 5pt13 mutants exhibited shortened hypocotyls and larger cotyledons under blue light but smaller cotyledons under red light (Figure 1C). The phytochrome A–specific signaling intermediate SPA1 (Hoecker and Quail, 2001; Laubinger et al., 2004) was significantly upregulated under 5PTase13 deficiency, suggesting that 5PTase13 might participate in other light-mediated growth processes or signaling pathways in addition to playing a major role in blue light signaling.
METHODS
Plant Growth and Light Conditions
Arabidopsis thaliana (Columbia ecotype) seeds were surface sterilized with 20% bleach for 15 min and washed four times with sterile water and planted aseptically on agar medium containing Murashige and Skoog (1962) salts and 2% sucrose (w/v). In some cases, the medium was supplemented with 25 μg/mL of hygromycin, 15 μg/mL of basta, or 25 μg/mL of kanamycin to screen the resistant transgenic plants or homozygous mutants. After 48 h of vernalization, seedlings were grown at 22°C with a 16-h-white light/8-h-dark cycle, followed by continued growth under the same conditions or transfer to blue light, red light- or far-red light (continuous light) with different fluence rates for 5 d. Exogenous EGTA or calcium treatment was performed by growing the seedlings on MS medium supplemented with different concentrations of EGTA or calcium chloride for 5 d.
All experiments involving blue light, red light, or far-red light illumination were performed in an E-30 LED chamber (Percival) using the blue diodes (λmax = 469 nm), red diodes (λmax = 680 nm), or far-red light (λmax = 730 nm) at 22°C in continuous light. Light fluence rates were measured using an Li250 quantum photometer (Li-Cor).
Hypocotyl length and cotyledon area were measured using the E-ruler software and statistically analyzed. Photos were taken by an SMZ 800 stereoscope (Nikon) equipped with a Nikon digital Coolpix 995 camera. Statistical analyses were performed using Excel tools as described in the figure legends. All experiments were performed in triplicate (n > 30).
Mutant Confirmation and Plant Crosses
The 5pt13 mutant obtained from Syngenta (Garlic 350-F1; http://www.syngenta.com) carried a T-DNA insertion in the fourth exon (Lin et al., 2005). Double mutants were produced by the standard method previously described (Weigel and Glazebrook, 2002). Offspring of the double mutant were analyzed by PCR amplifications with primers 5pt13-1 (5′-TCCGAACGGAATTGTCTCAG-3′) and 5pt13-2 (5′-GCAACATCAAGATCTCCAACA-3′), phot1-s (5′-TCCACTTGCAACCTATGCGTG-3′) and phot1-a (5′-ACCGCTTCATCGATATTCACA-3′), phot2-s (5′-ATGGAGAGGCCAAGAGCCC-3′) and phot2-a (5′-TGTCGGTGTCTGGCCCTTGC-3′) (Mao et al., 2005), cry1-s (5′-GGGAGAGATGTCTTAGTATGCCTTATG-3′) and cry1-a (5′-CCCCTCGAGCCCGGTTTGTGAAAGCCGTCT-3′) (Bruggemann et al., 1996), and aequorin-s (5′-ATGACAAGCAAACAATACTCAGT-3′) and aequorin-a (5′-TTAGGGGACAGCTCCACCG-3′). PCR analysis of the heterozygous phot1/+, phot2/+, cry1/+ cop1/+, and homozygous phot1 phot2 and cry1 cop1 plants was performed as described earlier (Mao et al., 2005; Sang et al., 2005).
GUS Histochemical Staining
Transgenic seedlings harboring a 5PTase13 promoter-GUS reporter gene construct were stained at 37°C overnight in a solution of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (final concentration 1 mg/mL), potassium ferricyanide (final concentration 0.5 mM), potassium ferrocyanide (final concentration 0.5 mM), 0.1% Triton X-100 (final concentration 0.1% [v/v]), and sodium phosphate buffer (final concentration 0.1 M), pH 7.0 (Rook et al., 1998).
Yeast Two-Hybrid Analysis
The cDNA fragment for the complete WD40 repeats of 5PTase13 was amplified by primers 5pt13-11 (5′-CATGCCATGGTATGAGGCTGGGTGTGGGATTG-3′; NcoI site underlined) and 5pt13-12 (5′-CCGCTCGAGGCTTGAAATTAGCTTTCCGTC-3′; XhoI site underlined), and the fragment containing N-terminal and WD40 repeats, designated as N-WD, was amplified by primers 5pt13-13 (5′-CATGCCATGGCGATGGATTCGCTAATTATCG-3′; NcoI site underlined) and 5pt13-12. Resulting fragments were subcloned into the pEG202 vector and confirmed by restriction enzyme digestion and DNA sequencing. Other constructs harboring DNA fragments encoding GUS, Arabidopsis CRY1 C-terminal domain (CCT1), CRY1, or COP1 in pJG4-5 were kindly provided by Hong-Quan Yang (Yang et al., 2001). All combinations of prey and bait constructs were cotransformed into the yeast strain EGY48 through the standard ONE-STEP method (Chen et al., 1992).
The DNA fragment encoding N-terminal and WD40 repeats of 5PTase13 was amplified by primers 5pt13-14 (5′-CGCGGATCCCGATGGATTCGCTAATTATC-3′; BamHI site underlined) and 5pt13-15 (5′-ACGCGTCGACGGGTCCTGGAGATGTCAC-3′; SalI site underlined) and that encoding the 5PTase13 catalytic domain was amplified by primers 5pt13-16 (5′-GGACAACATAATCCGAACGGA-3′) and 5pt13-17 (5′-ACGCGTCGACTCTCGAGTGTCTTCGCACC-3′; SalI site underlined). Resulting fragments were cloned into the pGBKT7 vector and confirmed by restriction enzyme digestion and DNA sequencing. The DNA fragment encoding the LOV domain of PHOT1 was amplified by primers PHOT1-l1 (5′-CGGAATTCATGGAACCAACAGAAAAACCA-3′; EcoRI site underlined) and PHOT1-l2 (5′-CGGGATCCTGGTGTCATGTTGGCATCAG-3′; BamHI site underlined) and that encoding the PHOT1 kinase domain was amplified by primers PHOT1-k1 (5′-CGGAATTCGAGGATTTATGGGCAAACCA-3′; EcoRI site underlined) and PHOT1-k2 (5′-CGGGATCCAACATTTGTTTGCAGATCTTCT-3′; BamHI site underlined). Resulting fragments were cloned into the pGADT7 vector and confirmed by restriction enzyme digestion and DNA sequencing. All combinations of prey and bait constructs were cotransformed into the yeast strain AH109 through the small-scale LiAc yeast transformation method (Clontech).
Interaction analysis, including growth marker-based selection and measurement of the β-galactosidase activity, were essentially performed as described previously (McNellis et al., 1996) or according to the manufacturer's manual (Matchmaker LexA two-hybrid system; Clontech).
Coimmunoprecipitation Analysis
Coimmunoprecipitation was performed using extracts prepared from yeast cells, as described by Nam and Li (2002). The protease inhibitor cocktail (Sigma-Aldrich), anti-MYC monoclonal antibody (NeoMarkers), anti-HA polyclonal antibody (NeoMarkers), nitrocellulose membrane (Schleicher and Schuell), and a nitroblue tetrazolium/bromochloroindolyl phosphate system were used during the process as previously described (Lou et al., 2007).
Constructs and Plant Transformation
Primers 5pt13-3 (5′-ACGCGTCGACCATGGATTCGCTAATTATCGAAG-3′; SalI site underlined) and 5pt13-4 (5′-GGACTAGTTCTCGAGTGTCTTCGCACC-3′; SpeI site underlined) were used to amplify the full-length cDNA of 5PTase13. The resulting 3300-bp cDNA was cloned into the pCAMBIA1302 vector in the sense orientation to generate the overexpression construct. The antisense vector was generated with a 600-bp coding region at the 5′-end of 5PTase13 cDNA, which was amplified by primers 5pt13-5 (5′-ATGGATTCGCTAATTATCG-3′) and 5pt13-6 (5′-GAAGTAGTAACAGACTCATG-3′). This cDNA region was first cloned into the PMD18-T vector and then subcloned into the PHB vector in the antisense orientation. The 5PTase13 promoter was amplified by primers 5pt13-P-1 (5′-ACGCGTCGACACCATCTTGGGTCGTTAATTAGTT-3′; SalI site underlined) and 5pt13-P-2 (5′-CGCGGATCCCGACAATCAGAGGAATTCAAG-3′; BamHI site underlined). The resulting 1.0-kb fragment was cloned into the p1300 vector and confirmed by restriction enzyme digestion. The constructs were transferred into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants (Columbia ecotype) by the floral dip method (Weigel and Glazebrook, 2002).
RT-PCR and Quantitative Real-Time PCR Analysis
Total RNA was extracted from open flowers or seedlings (grown in a 16-h-white light/8-h-dark cycle) using TRIzol reagent (Invitrogen) and used for RT-PCR analysis. The cDNA was produced by reverse transcription with oligo(dT) (ReverTra Ace-α-first-strand synthesis kit; Toyobo), and PCR amplification was performed by specific primers 5pt13-7 (5′-TCCGAACGGAATTGTCTCAG-3′) and 5pt13-8 (5′-GCAACATCAAGATCTCCAACA-3′). The Arabidopsis ACTIN8 gene, amplified by primers actin-1 (5′-GATGCTGATGACATTCAACCT-3′) and actin-2 (5′-GAAGTGAGAAACCCTCATAG-3′), was used as a positive internal control.
Quantitative real-time RT-PCR analysis was performed with the RotorGene 3000 (Corbett Research) using a SYBR green detection protocol (SYBR Premix Ex Taq System; TaKaRa). The product amounts were determined by the method of comparative Δ-Δ Ct. (Fleige et al., 2006). The primers used for 5PTase13 were 5pt13-9 (5′-CCGAGGACAAAATCTAAGTAACG-3′) and 5pt13-10 (5′-CAGCGCCGGTGCTTGGAATAG-3′). Arabidopsis ACTIN7 was amplified by primers actin-3 (5′-CCGGTATTGTGCTCGATTCTG-3′) and actin-4 (5′-TTCCCGTTCTGCGGTAGTGG-3′) and used as internal positive control. The experiments were repeated more than three times.
Measurement of [Ca2+]cyt
[Ca2+]cyt was measured in vivo by luminometry using intact seedlings expressing aequorin as described previously (Harada et al., 2003; Love et al., 2004; Plieth, 2006). Seedlings were grown under a 16-h-white light/8-h-dark cycle for 10 days and clusters of three plants floated in the 200 μL solution containing coelenterazine (final concentration at 1 μM) free base (Sigma-Aldrich) in the dark overnight. The measurements were performed in the luminometer cuvette (2 mL) containing coelenterazine (1 μM)-free base. The reading began after giving a 20-s blue light (20 μmol/m2/s) pulse in the luminometer (FG-200) (Wang and Gu, 1986; Demidchik et al., 2004; Hu et al., 2004; Love et al., 2004). Fluorescence was generated by the sum of autofluorescence of chlorophyll and aequorin luminescence. To detect aequorin fluorescence, the time difference was calculated by comparison to seedlings without CaMV35S:aequorin under the same irradiation situation, which was termed as “delayed time.”
Fluorescence imaging of [Ca2+]cyt was performed according to Legue et al. (1997) and Wymer et al. (1997). Briefly, seedlings were floated in Indo-1 solution (20 μM; in 10 mM dimethylglutaric acid, pH 4.5) and incubated in the dark for 1 h. After washing with liquid MS medium, seedlings were placed on a Zeiss inverted microscope (LSM510 META) for fluorescence imaging (364-nm excision and 400- to 435-nm and 480 ± 20-nm emission). The distribution of Ca2+ was imaged from the root hair or root tip. Calcium levels were pseudocolored according to the scale.
Measurement of Ins(1,4,5)P3 Content
Five-day-old seedlings grown on MS medium under dark, white light, or blue light (20 μmol/m2/s) were used for the measurement of Ins(1,4,5)P3 contents. Three hundred milligrams of freshly ground tissue was used for extraction of Ins(1,4,5)P3 according to the procedure described by Burnette et al. (2003). Ins(1,4,5)P3 content was measured using the d-myo-inositol-1,4,5-trisphosphate [3H] assay kit (Amersham-Pharmacia Biotech). The Ins(1,4,5)P3 content in the samples was interpolated from a standard curve generated with known amounts of Ins(1,4,5)P3. Assays were performed in triplicate, and the experiment was repeated two times.
Expression, Purification, and Biochemical Characterization of Recombinant 5PTase13
Expression of recombinant 5PTase13 in Escherichia coli cells was performed according to Sanchez and Chua (2001). Briefly, the full-length 5PTase13-encoding sequence was N-terminally fused to a polyhistidine-tag and a T7-tag and expressed in E. coli cells according to the supplier's instructions (Gibco BRL). To generate the plasmid pET28b-5PTase13, a DNA fragment encompassing the entire 5PTase13 cDNA was PCR amplified using primers 5pt13-18 (5′-ACGCGTCGACATGGATTCGCTAATTATCGAAG-3′; added SalI site underlined and the start ATG site in italics) and 5pt13-19 (5′-GGGACGATGCGGCCGCTATATCTCGAGTGTC-3′; added NotI site underlined) and subcloned into the pET28b vector (Novagen). The in-frame reading of the construct was confirmed by sequencing to avoid nucleotide substitutions during PCR amplification. Transformed E. coli cells harboring pET28b-5PTase13 were cultured at 37°C, and protein expression was induced by 1 mM isopropylthio-β-galactoside (final concentration) for 8 h. Cells were then harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazol), and sonicated. The resulting supernatants (lysates) were purified on His-tag beads (Sigma-Aldrich), and the purified proteins were analyzed by SDS-PAGE and protein gel blotting with a T7 monoclonal antibody (Novagen). Recombinant 5PTase13 catalytic domain protein was expressed and purified using a similar procedure, employing vector pET28a and primers 5pt13-20 (5′-CGGGGATCCAAGGAAACTTTATATGCCAG-3′; added BamHI site underlined) and 5pt13-21 (5′-CGGCTCGAGTCACAGATTTATCAACATGAG-3′; added XhoI site underlined).
Enzymatic analyses were performed by incubation of purified protein (1.5 μg) at 37°C overnight in solutions containing 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 2 mM MgCl2, supplemented with either 20 mM Ins(1,4,5)P3 and 0.02 μCi [3H]Ins(1,4,5)P3 (10 mM), or 20 mM Ins(1,3,4,5)P4 and 0.016 μCi [3H]Ins(1,3,4,5)P4 (10 mM). The reactions were terminated by heating at 95°C for 5 min (Sanchez and Chua, 2001), and HPLC analysis was performed as described (Xue et al., 1999).
Microarray Hybridization and Analysis
The microarray hybridization using the Arabidopsis ATH1 chip was described by Lin et al. (2005). Briefly, the aerial organs of 4-d-old wild-type and 5pt13 seedlings grown in continuous white light were used for RNA extraction. Furthermore, 15 μg of purified biotin-labeled copy RNA, which was fragmented into 35 to 300 nucleotides, was used for microarray analysis. Two hybridizations (independent biological replicates) were performed for each genotype (wild type or 5pt13) for a total of four hybridizations. Arrays were scanned with the Agilent GeneArray scanner (Affymetrix), and the obtained data were normalized with the Affymetrix Microarray Suite program (version 5.0) and set the algorithm absolute call flag according to P (present), M (marginal), and A (absent). The data were then imported into R software (http://www.R-project.org) and analyzed using the robust test statistics described in the Limma package (Smyth, 2005). The data exhibited intensity-dependent biases; therefore, log2(X) (X = scaling signal) transformed data were further corrected with a global Lowess transformation. FDRs for various P value thresholds were determined by the method of Benjamini and Yekutieli (2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: 5PTase13 (At1g05630, AJ297426), CRY1 (At4g08920, NM_116961), COP1 (At2g32950, NM_128855), Actin7 (At5g09810, NM_121018), Actin8 (At1g49240, NM_103814), PHOT1 (At3g45780, NM_114447), and PHOT2 (At5g58140, NM_125199).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Histochemical Staining of 5PTase13 Promoter:GUS-Containing Seedlings.
Supplemental Figure 2. Overexpression of 5PTase13 in 5pt13 or Wild-Type Plants, as Revealed by RT-PCR Analysis.
Supplemental Figure 3. 5PTase13 Does Not Interact with PHOT1 Protein.
Supplemental Figure 4. 5PTase13 Deficiency Results in Increased [Ca2+]cyt under White Light.
Supplemental Figure 5. The 5PTase13 Protein Exhibits Phosphatase Activities toward Ins(1,3,4,5)P4.
Supplemental Figure 6. Responses of Hypocotyl Growth to Calcium under Blue Light.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 30425029 and 30421001), by the Shanghai municipal government (Grant 055407072), and by a grant from the National Science Foundation (to S.L.). We thank Hong-Quan Yang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing seeds of phot1, phot2, phot1 phot2, cry1, and cop1. We also thank Alex A.R. Webb and Mark Tester (University of Cambridge, Cambridge, UK) for the gifts of Wassilewskija ecotype and Columbia ecotype harboring CaMV35S:aequorin and Jian-Ben Gu (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for the luminometer measurement of [Ca2+]cyt.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hong-Wei Xue (hwxue@sibs.ac.cn).
Online version contains Web-only data.
References
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. [DOI] [PubMed] [Google Scholar]
- Babourina, O., Newman, I., and Shabala, S. (2002). Blue light-induced kinetics of H+ and Ca2+ fluxes in etiolated wild-type and phototropin-mutant Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 99 2433–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, G., Long, J.C., Jenkins, G.I., and Trewavas, A.J. (1999). Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+. Proc. Natl. Acad. Sci. USA 96 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 29 1165–1188. [Google Scholar]
- Berdy, S.E., Kudla, J., Gruissem, W., and Gillaspy, G.E. (2001). Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol. 126 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R. (2007). The LOV domain: A chromophore module servicing multiple photoreceptors. J. Biomed. Sci. 14 499–504. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7 204–210. [DOI] [PubMed] [Google Scholar]
- Bruggemann, E., Handwerger, K., Essex, C., and Storz, G. (1996). Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J. 10 755–760. [DOI] [PubMed] [Google Scholar]
- Burnette, R.N., Gunesekera, B.M., and Gillaspy, G.E. (2003). An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol. 132 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., and Nelson, T. (2004). Cotyledon vascular pattern2-mediated inositol (1,4,5) trisphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casazza, A.P., Rossini, S., Rosso, M.G., and Soave, C. (2005). Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol. Biol. 58 41–51. [DOI] [PubMed] [Google Scholar]
- Chen, D.C., Yang, B.C., and Kuo, T.T. (1992). One-step transformation of yeast in stationary phase. Curr. Genet. 21 83–84. [DOI] [PubMed] [Google Scholar]
- Cho, H.Y., Tseng, T.S., Kaiserli, E., Sullivan, S., Christie, J.M., and Briggs, W.R. (2007). Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 143 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, M.H., and Spalding, E.P. (1996). An anion channel in Arabidopsis hypocotyls activated by blue light. Proc. Natl. Acad. Sci. USA 93 8134–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58 21–45. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- Demidchik, V., Essah, P.A., and Tester, M. (2004). Glutamate activates cation currents in the plasma membrane of Arabidopsis root cells. Planta 219 167–175. [DOI] [PubMed] [Google Scholar]
- Du, L., and Poovaiah, B.W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437 741–745. [DOI] [PubMed] [Google Scholar]
- Emi, T., Kinoshita, T., Sakamoto, K., Mineyuki, Y., and Shimazaki, K. (2005). Isolation of a protein interacting with Vfphot1a in guard cells of Vicia faba. Plant Physiol. 138 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige, S., Walf, V., Huch, S., Prgomet, C., Sehm, J., and Pfaffl, M.W. (2006). Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett. 28 1601–1613. [DOI] [PubMed] [Google Scholar]
- Folta, K.M., Lieg, E.J., Durham, T., and Spalding, E.P. (2003). Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol. 133 1464–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Unexpected roles for cryptochrome2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26 471–478. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Larner, V.S., and Whitelam, G.C. (2005). The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49 653–664. [DOI] [PubMed] [Google Scholar]
- Gunesekera, B., Torabinejad, J., Robinson, J., and Gillaspy, G.E. (2007). Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiol. 143 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Mockler, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291 487–490. [DOI] [PubMed] [Google Scholar]
- Harada, A., Sakai, T., and Okada, K. (2003). Phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc. Natl. Acad. Sci. USA 100 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, A., and Shimazaki, K. (2007). Phototropins and blue light-dependent calcium signaling in higher plants. Photochem. Photobiol. 83 102–111. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., and Quail, P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hu, X.Y., Neill, S.J., Cai, W.M., and Tang, Z.C. (2004). Induction of defence gene expression by oligogalacturonic acid requires increases in both cytosolic calcium and hydrogen peroxide in Arabidopsis thaliana. Cell Res. 14 234–240. [DOI] [PubMed] [Google Scholar]
- Inada, S., Ohgishi, M., Mayama, T., Okada, K., and Sakai, T. (2004). RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.A., Raymond, M.J., and Smirnoff, N. (2006). Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45 83–100. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., et al. (2002). Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.I., Park, J.E., Zarate, X., and Song, P.S. (2005). Phytochrome phosphorylation in plant light signaling. Photochem. Photobiol. Sci. 4 681–687. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and Kagawa, T. (2006). Phototropin and light-signaling in phototropism. Curr. Opin. Plant Biol. 9 503–508. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Emi, T., Tominaga, M., Sakamoto, K., Shigenaga, A., Doi, M., and Shimazaki, K. (2003). Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 133 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U.V., Trevisan, M., Kami, C., de Carbonnel, M., Alonso, J.M., Ecker, J.R., Liscum, E., and Fankhauser, C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103 10134–10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascève, G., Leymarie, J., Olney, M.A., Liscum, E., Christie, J.M., Vavasseur, A., and Briggs, W.R. (1999). Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol. 120 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S., Fittinghoff, K., and Hoecker, U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell 16 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legue, V., Blancaflor, E., Wymer, C., Perbal, G., Fantin, D., and Gilroy, S. (1997). Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.H., and Yang, H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83 94–101. [DOI] [PubMed] [Google Scholar]
- Lin, C., and Todo, T. (2005). The cryptochromes. Genome Biol. 6 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.H., Wang, Y., Mueller-Roeber, B., Brearley, C.A., Xu, Z.H., and Xue, H.W. (2005). At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiol. 139 1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.H., Ye, R., Ma, H., Xu, Z.H., and Xue, H.W. (2004). DNA chip-based expression profile analysis indicates involvement of the phosphatidylinositol signaling pathway in multiple plant responses to hormone and abiotic treatments. Cell Res. 14 34–45. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1996). Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Li, L., and Luan, S. (2005). An essential function of phosphatidylinositol phosphates in activation of plant shaker-type K+ channels. Plant J. 42 433–443. [DOI] [PubMed] [Google Scholar]
- Lou, Y., Gou, J.Y., and Xue, H.W. (2007). PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19 163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, J., Dodd, A.N., and Webb, A.A. (2004). Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 16 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14(Suppl): S389–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, J., Zhang, Y.C., Sang, Y., Li, Q.H., and Yang, H.Q. (2005). From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., Torii, K.U., and Deng, X.W. (1996). Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan, L., Harkins, K., Chory, J., and Rodermel, S. (1996). Lhcb transcription is coordinated with cell size and chlorophyll accumulation (studies on fluorescence-activated, cell-sorter-purified single cells from wild-type and in mutans Arabidopsis thaliana). Plant Physiol. 112 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, S.G., Kim, Y.S., Kunkel, T., and Chua, N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286 961–964. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212. [DOI] [PubMed] [Google Scholar]
- Ohgishi, M., Saji, K., Okada, K., and Sakai, T. (2004). Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, B.M., Cho, M.H., and Spalding, E.P. (1998). Two genetically separable phases of growth inhibition induced by blue light in Arabidopsis seedlings. Plant Physiol. 118 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale, U.V., and Liscum, E. (2007). Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 282 19992–20001. [DOI] [PubMed] [Google Scholar]
- Poovaiah, B.W., Yang, T., and van Loon, J.J. (2002). Calcium/calmodulin-mediated gravitropic response in plants. J. Gravit. Physiol. 9 211–214. [PubMed] [Google Scholar]
- Qin, Z.X., Chen, Q.J., Tong, Z., and Wang, X.C. (2005). The Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase, AtItpk-1, is involved in plant photomorphogenesis under red light conditions, possibly via interaction with COP9 signalosome. Plant Physiol. Biochem. 43 947–954. [DOI] [PubMed] [Google Scholar]
- Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., Shang, M., Chen, S., Pardo, J.M., and Guo, Y. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19 1415–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell, N.C., Su, Y.S., and Lagarias, J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Gerrits, N., Kortstee, A., van Kampen, M., Borrias, M., Weisbeek, P., and Smeekens, S. (1998). Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15 253–263. [DOI] [PubMed] [Google Scholar]
- Plieth, C. (2006). Aequorin as a reporter gene. Methods Mol. Biol. 323 307–327. [DOI] [PubMed] [Google Scholar]
- Reiter, W.D. (2002). Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5 536–542. [DOI] [PubMed] [Google Scholar]
- Sakai, T., Wada, T., Ishiguro, S., and Okada, K. (2000). RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, J.P., and Chua, N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Y., Li, Q.H., Rubio, V., Zhang, Y.C., Mao, J., Deng, X.W., and Yang, H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi, A.S., and Wehland, J. (2000). The actin cytoskeleton and plasma membrane connection: PtdIns(4,5)P(2) influences cytoskeletal protein activity at the plasma membrane. J. Cell Sci. 21 3685–3695. [DOI] [PubMed] [Google Scholar]
- Schepens, I., Duek, P., and Fankhauser, C. (2004). Phytochrome-mediated light signalling in Arabidopsis. Curr. Opin. Plant Biol. 7 564–569. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentlemen, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420.
- Stepanova, A.N., and Alonso, J.M. (2005). Arabidopsis ethylene signaling pathway. Sci. STKE 2005 cm4. [DOI] [PubMed] [Google Scholar]
- Stoelzle, S., Kagawa, T., Wada, M., Hedrich, R., and Dietrich, P. (2003). Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc. Natl. Acad. Sci. USA 100 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, S., Moran, N., and Lee, Y. (2000). Blue light activates potassium-efflux channels in flexor cells from Samanea saman motor organs via two mechanisms. Plant Physiol. 123 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I., Sharon, M., Zheng, C., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Wang, H. (2005). Signaling mechanisms of higher plant photoreceptors: A structure-function perspective. Curr. Top. Dev. Biol. 68 227–261. [DOI] [PubMed] [Google Scholar]
- Wang, W.G., and Gu, J.B. (1986). Comparison of the methods for extracting ATP from plant leaves. Plant Physiol Commun. 5 54–55. [Google Scholar]
- Weigel, D., and Glazebrook, J., eds (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Williams, M.E., Torabinejad, J., Cohick, E., Parker, K., Drake, E.J., Thompson, J.E., Hortter, M., and Dewald, D.B. (2005). Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of stress-response pathway. Plant Physiol. 138 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer, C.L., Bibikova, T.N., and Gilroy, S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12 427–439. [DOI] [PubMed] [Google Scholar]
- Xu, X., Hotta, C.T., Dodd, A.N., Love, J., Sharrock, R., Lee, Y.W., Xie, Q., Johnson, C.H., and Webb, A.A. (2007). Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell 19 3474–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, H.W., Chen, X., and Li, G. (2007). Involvement of phospholipid signaling in plant growth and hormone effects. Curr. Opin. Plant Biol. 10 483–489. [DOI] [PubMed] [Google Scholar]
- Xue, H.W., Pical, C., Brearley, C., Elge, S., and Müller-Röber, B. (1999). A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure. Cloning and functional expression in baculovirus-infected insect cells. J. Biol. Chem. 274 5738–5745. [DOI] [PubMed] [Google Scholar]
- Yang, H.Q., Tang, R.H., and Cashmore, A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T., and Poovaiah, B.W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8 505–512. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., Rose, J.K., and Nishitani, K. (2004). A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 134 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., Morrison III, W.H., and Ye, Z.H. (2004). FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16 3242–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.