Abstract
Although maize (Zea mays) retrotransposons are recombinationally inert, the highly polymorphic structure of maize haplotypes raises questions regarding the local effect of intergenic retrotransposons on recombination. To examine this effect, we compared recombination in the same genetic interval with and without a large retrotransposon cluster. We used three different bz1 locus haplotypes, McC, B73, and W22, in the same genetic background. We analyzed recombination between the bz1 and stc1 markers in heterozygotes that differ by the presence and absence of a 26-kb intergenic retrotransposon cluster. To facilitate the genetic screen, we used Ds and Ac markers that allowed us to identify recombinants by their seed pigmentation. We sequenced 239 recombination junctions and assigned them to a single nucleotide polymorphism–delimited interval in the region. The genetic distance between the markers was twofold smaller in the presence of the retrotransposon cluster. The reduction was seen in bz1 and stc1, but no recombination occurred in the highly polymorphic intergenic region of either heterozygote. Recombination within genes shuffled flanking retrotransposon clusters, creating new chimeric haplotypes and either contracting or expanding the physical distance between markers. Our findings imply that haplotype structure will profoundly affect the correlation between genetic and physical distance for the same interval in maize.
INTRODUCTION
Maize (Zea mays) has a highly polymorphic genome structure (Fu and Dooner, 2002; Song and Messing, 2003; Wang and Dooner, 2006). Retrotransposons, which constitute the bulk of the genome (SanMiguel and Bennetzen, 1998), differ among lines in their makeup and location relative to genes. Consequently, the pattern of interspersion of genes and retrotransposons varies from line to line, defining sharply distinct haplotypes. The extent of sequence variation in the bz1 genomic region is remarkable. In a recent vertical comparison of eight bz1 locus haplotypes, any two haplotypes shared between 25 and 84% of their sequences (Wang and Dooner, 2006). Haplotypic variation is common in the genome (Song and Messing, 2003; Brunner et al., 2005; Yao and Schnable, 2005) and could lead to huge differences in estimates of genetic distance in different backgrounds. This does not happen because the variable retrotransposon component of the genome is recombinationally inert (Fu et al., 2002; Yao et al., 2002). Yet, twofold to threefold variations in estimates of map distances for single genetic intervals have been reported in several maize mapping populations (Beavis and Grant, 1991; Fatmi et al., 1993; Williams et al., 1995). Much of this variation is probably due to trans-acting modifiers, such as the recently demonstrated quantitative trait loci that affect global recombination frequencies in recombinant inbred lines (Esch et al., 2007), but some of it may be due to cis-acting factors.
Cis-acting factors were demonstrated in a study that examined recombination rates across the a1-sh2 genetic interval in three heterozygotes containing the same maize haplotype and different teosinte-derived haplotypes in a common maize background (Yao and Schnable, 2005). This region measures 130 kb in maize inbred UE85, and although most of the intervening teosinte DNA between a1 and sh2 was not sequenced, several large insertion/deletion (indel) polymorphisms relative to maize, including two LTR retrotransposons, one MITE transposon, and one hAT transposon, were uncovered in the sequenced region. The analysis identified up to threefold differences in recombination rates and statistical differences in the distribution of recombination junctions across subintervals among haplotypes. Although levels of sequence polymorphism correlated negatively with rates of recombination in the sequenced region, they did not fully account for the observed results. The authors proposed that other types of cis factors, such as region-specific chromatin structure, may affect the rate and distribution of recombination across the a1-sh2 interval.
The polymorphic chromosomal organization of maize, due mainly to intergenic retrotransposon variation, prompted us to ask the specific question: to what extent does heterozygosity for large retrotransposon insertions, which occurs in every mapping population, affect recombination in the adjacent genes? The highly methylated retrotransposon clusters are probably heterochromatic, as are similar blocks in the knobs of maize (Ananiev et al., 1998) and Arabidopsis thaliana (Lippman et al., 2004), and most likely affect recombination. Genes next to retrotransposon clusters may be less recombinogenic, because the more condensed chromatin state of the retrotransposon cluster may interfere with the access of the recombination machinery to the adjacent euchromatic regions. In order to investigate this, we compared recombination in the same genetic interval in the presence and absence of a large retrotransposon cluster. Fu and Dooner (2002) found that the 1.5-kb bz1-stc1 intergenic segment in the McC bz1 locus haplotype (Fu et al., 2001) was replaced by a 26-kb retrotransposon block in the B73 haplotype (Figures 1A and 1B). We have identified a bz1 haplotype, W22, that resembles McC in its retrotransposon–gene junctions but differs from McC in many single nucleotide polymorphisms (SNPs) and indel polymorphisms. The availability of these three distinct haplotypes has enabled us to examine the effect of retrotransposon heterozygosity on recombination in the adjacent bz1 and stc1 genes. Potential trans effects in such an experiment were eliminated by first introgressing all haplotypes into a common inbred background.
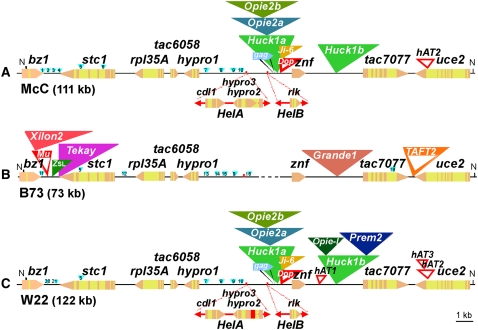
Figure 1.
Organization of McC, B73, and W22 bz1 Haplotypes.
Each haplotype is identified by the name of the line from which it was extracted, followed by the size of the cloned NotI (N) fragment in parentheses. The eight genes (bz1, stc1, rpl35A, tac6058, hypro1, znf, tac7077, and uce2) are shown as pentagons pointing in the direction of transcription, with exons in peach and introns in yellow. The same symbols are used for gene fragments carried by helitrons HelA and HelB, which are represented as bidirectional arrows below the line in McC and W22. The vacant site for HelA in B73 is marked with a short vertical stroke. Dashed lines represent deletions. Retrotransposons are indicated by closed triangles of different colors. DNA transposons are indicated by open triangles of red and orange. Small insertions are indicated in light blue and numbered as by Wang and Dooner (2006). Only the genes have been drawn to scale.
The confinement of recombination to the genic space in maize protects the genome from the massive disruptive rearrangements that would otherwise occur if the dispersed repetitive retrotransposons (SanMiguel and Bennetzen, 1998) recombined ectopically. However, the orderly exchange of different intergenic retrotransposon clusters by recombination between alleles should occur regularly in populations, leading to nondisruptive genomic changes that amplify the variability created by the explosive increase in retrotransposons in the recent maize ancestry (SanMiguel and Bennetzen, 1998; SanMiguel et al., 1998). We confirm here that recombination in genes shuffles heterozygous retrotransposons flanking them, creating new chimeric haplotypes and expanding or contracting chromosomal segments.
RESULTS
Structure of the W22 bz1 Haplotype
Because of extensive polymorphisms in the content of retrotransposons, helitrons, and other transposons, the size of the bz1 region can vary by more than threefold among maize lines (Wang and Dooner, 2006). NotI fragments containing the bz1 region of W22 are not very different in size from those of McC (Fu and Dooner, 2002), yet the Bz1-McC and Bz1-W22 alleles are known to differ in >1% of their sequences (Ralston et al., 1988), so W22 is an excellent candidate for a contrasting bz1 locus haplotype lacking the retrotransposon cluster in the bz1-stc1 intergenic region. In order to fully characterize the W22 bz1 haplotype, the entire 238-kb Bz1-W22 genomic region was cloned as two adjacent NotI fragments in the pNOBAC1 vector (Fu and Dooner, 2000). Sequencing confirmed that the bz1-stc1 intergenic segment of Bz1-W22 lacked retrotransposons. The structure of the distal 122-kb NotI BAC, which contains the genetic interval analyzed in this study, is presented in Figure 1C.
The overall structure of the W22 bz1 haplotype is remarkably similar to that of McC (Figure 1). The two haplotypes share all of the boundaries between Helitron or retrotransposon insertions and intergenic regions, as well as several hAT, CACTA, and small DNA insertions in either intergenic regions or introns. Surprisingly, the lowest sequence variation between the two haplotypes occurs in the large intergenic region between hypro1 and znf. The percentage divergence in that 67-kb stretch is 0.06%: the two haplotypes share the same four MITEs, the same two Helitrons, although HelA in W22 has experienced a 700-bp deletion of the hypro3 gene fragment, the same Doppia DNA element, and the same 53-kb three-level retrotransposon nest distal to HelB. The 5′ and 3′ LTRs of each of the three retrotransposons differ in both haplotypes, allowing dating of the insertions (SanMiguel et al., 1998; Ma and Bennetzen, 2004) to a time period 0.4 to 1.2 million years ago, from youngest (top) to oldest (bottom) (see Supplemental Table 1 online). Yet, five of the six LTRs in the nest are identical in sequence between W22 and McC, suggesting that this chromosomal segment in the two haplotypes derives from a very recent common ancestor.
The Huck1b retroelement between the znf and tac7077 genes has diverged in W22 relative to McC by the gain of a fractured 1.9-kb Opie-like element, consisting only of the 5′ LTR and the primer binding site, and a 9.5-kb Prem2 element, estimated from its LTR sequence identity to have inserted only 60,000 years ago. These two insertions account for the 11-kb difference in size of the NotI fragment in the two lines. Using LTR sequence information from both haplotypes, Huck1b is estimated to have inserted between 1 and 1.2 million years ago, so it is about as old as Huck1a in the triple-level nest. A comparison of the Huck1b 5′ LTRs and 3′ LTRs in W22 versus McC indicates that they diverged from each other between 0.4 and 0.5 million years ago, pointing to an older common ancestry for this chromosomal segment of the two haplotypes. The subsequent acquisition of retroelement sequences by Huck1b in only one of the haplotypes supports this inference. As discussed below, retrotransposon clusters are shuffled by recombination between the genes that flank them: possibly, two nonconcurrent recombination events in znf and hypro1 in the history of these haplotypes led to the replacement of the hypro1-znf intergenic region of one haplotype by that of the other. Lastly, the 1-kb hAT element in the last intron of uce2, which is present in both haplotypes, contains a second, unrelated 0.5-kb hAT element only in W22.
Remarkably, however, the sequences of most genes and nonrepetitive intergenic segments are just as polymorphic between W22 and McC as they are among other lines (Wang and Dooner, 2006). For example, the transcribed bz1 segments differ by 1.6% in their coding sequences and by 3.2% plus one MITE indel in their noncoding sequences (intron and 3′ untranslated region). The bz1-stc1 intergenic regions differ in six MITEs or other small insertions; excluding those insertions, they differ in 4.6% of their sequences. The transcribed stc1 segments differ by 1.5% in their coding sequences and by 2.2% plus one MITE indel in their noncoding sequences (introns and 3′ untranslated region).
Recombination between bz1 and stc1
In order to examine the effect of retrotransposon heterozygosity on recombination in the adjacent genes, we compared recombination between bz1 and stc1 markers in McC/B73 and McC/W22 heterozygotes. The bz1-stc1 interval in these heterozygotes differs by the presence and absence, respectively, of a 26-kb retrotransposon cluster in the intergenic region. Each haplotype was first introduced into the common genetic background of the inbred W22 to minimize background differences (see Methods).
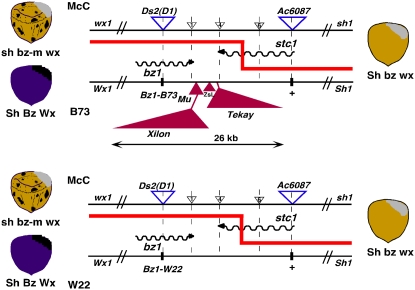
The experimental setup is diagrammed in Figure 2. The 26-kb retrotransposon cluster in B73 is represented in a much smaller scale than the adjacent bz1 and stc1 genes, which are drawn approximately to scale. The cluster is made up of a 9.4-kb Xilon retrotransposon inserted into a 1.7-kb Mu1 element, a 0.7-kb Zeon solo LTR, and a 12.8-kb Tekay retrotransposon. The McC parental haplotype carries bz1-m2(D1), a bz1 allele containing a Ds element in the second exon, and stc1-m1(Ac6087), an stc1 allele with an Ac insertion in the first exon (Shen et al., 2000). The bz1-m2(D1) allele produces a spotted phenotype in the presence of Ac and a stable bronze phenotype in its absence. Flanking bz-m2(D1) and stc1-m1(Ac6087) are the endosperm mutations sh1 and wx1, which serve as recombination markers in the experiment. (Note that the marker order in Figure 2, with the 9S centromere to the left, is the same as in Figure 1 and previous publications [Fu and Dooner, 2002; Wang and Dooner, 2006] but opposite to the more common way of presenting 9S markers with the centromere to the right.) The other parental haplotype, either B73 or W22, carries normal Bz1 and Stc1 alleles and produces a purple phenotype. Flanking Bz1 and Stc1 are the contrasting markers Sh1 and Wx1.
Figure 2.
Genetic Scheme for Identifying Recombinants in McC/B73 (Top) and McC/W22 (Bottom) Heterozygous Haplotypes.
The cartoon depicts the spotted (bz-m) and solid purple (Bz) parental phenotypes at left and the solid bronze (bz) recombinant phenotype at right. The sh1 flanking alleles condition either shrunken or plump seeds; the wx1 flanking alleles condition either waxy or nonwaxy seeds (shown here as staining light or dark with iodine for diagrammatic purposes only). Recombination anywhere between the Ds2(D1) element in bz1 and the Ac6087 element in stc1 gives rise to Sh bz recombinants, most of which will also carry wx. Recombination between Ac6087 and sh1 gives rise to Sh bz-m recombinants. The reciprocal recombinants of both classes are sh Bz. The stc1 and bz transcripts are indicated by the wavy arrows. A 26-kb retrotransposon cluster containing Tekay, a Zeon solo LTR, and a Mu1-Xilon nest separates stc1 from bz in the Bz-B73, but not the Bz-W22, haplotype. The small triangles represent indel polymorphisms (Ins3, Ins5, and Ins6 [Wang and Dooner, 2006]) used to assign Sh bz recombinants to one of four intervals within the larger Ds2(D1)-Ac6087 interval.
The sh1 stc1-m1(Ac6087) bz-m2(D1) wx1/Sh1 Stc1-B73 Bz1-B73 Wx1 and sh1 stc1-m1(Ac6087) bz-m2(D1) wx1/Sh1 Stc1-W22 Bz1-W22 Wx1 heterozygotes were pollinated with a sh1-bz1-X2 wx1 stock, which carries a deletion of the sh1-bz1 region, including the entire bz1-uce2 interval depicted in Figure 1 (Mottinger, 1973; Shen et al., 2000). Use of this deletion allows the recovery of selections in a hemizygous condition, greatly simplifying their molecular analysis. The above test crosses will produce crossover haplotypes conditioning a plump bronze (Sh bz) seed phenotype if recombination occurs between Ds and Ac, as illustrated by the heavy lines in Figure 2. Most of these exceptions will carry a Sh1 wx1 crossover arrangement of flanking markers.
Other exchanges between sh1 and bz1 are also identifiable. Crossovers between Ac6087 and sh1 will produce plump, spotted (Sh bz-m) seed, and the reciprocal crossover class of both Sh bz and Sh bz-m classes will be shrunken and purple (sh Bz). Thus, the sum of Sh bz and Sh bz-m crossovers should equal the number of sh Bz crossovers. Furthermore, the ratio of Sh bz to Sh bz-m kernels should not vary from family to family within a heterozygous haplotype genotype. This expectation provides an internal check that Ac did not transpose from stc1 in any of the McC parent plants. A χ2 analysis (data not shown) revealed that the data were homogeneous for different families of the same genotype and could be combined. The pooled data are shown in Table 1. As can be seen, the reciprocal crossover classes Sh (bz + bz-m) and sh Bz occur in approximately equal numbers in both heterozygotes. The length of the sh1-bz1 interval, estimated from the sum of the last two columns, is significantly greater in the McC/W22 heterozygote than in the McC/B73 heterozygote (2.52 versus 1.89 centimorgan [cM]). In particular, the frequency of the Sh bz class, which provides a rough estimate of the distance between the Ds element in bz1 and the Ac element in stc1, is about twice as high in McC/W22 (0.33%) than in McC/B73 (0.17%). This class includes crossovers of the type shown in Figure 2 as well as rare excisions of either Ds or Ac accompanied by coincidental exchanges in the sh1-bz1 interval. All but 3 of the 283 Sh bz selections from both heterozygotes in Table 1 were also wx, which is not surprising, as interference in the sh1-bz1-wx1 region is very high and double crossovers are rare (Dooner, 1986). The frequency of the Sh bz-m class, which provides an estimate of the distance between Ac and sh1, is not significantly different in the two heterozygotes, suggesting that potential local differences in recombination may average out over the longer stc1-sh1 interval.
Table 1.
Recombinant Kernel Selections from Heterozygotes of McC stc1-m1(Ac6087) bz-m2(D1) with either B73 or W22 Stc1 Bz1
| Heterozygous Haplotype | Kernel Population | Recombinant Kernel Class Phenotypes
|
|||
|---|---|---|---|---|---|
| Sh bz | Sh bz-m | Sh (bz + bz-m) | sh Bz | ||
| McC/B73 | 56670 | 96 | 473 | 569 | 507 |
| Percent | 0.17 | 0.83 | 1.00 | 0.89 | |
| 95% C.L. | 0.14 to 0.20 | 0.78 to 0.89 | 0.94 to 1.06 | 0.84 to 0.95 | |
| McC/W22 | 56460 | 187 | 524 | 711 | 710 |
| Percent | 0.33 | 0.93 | 1.26 | 1.26 | |
| 95% C.L. | 0.30 to 0.36 | 0.87 to 0.99 | 1.20 to 1.32 | 1.20 to 1.32 | |
C.L., confidence limit.
The Sh bz selections were first genotyped by PCR for a series of five key indel polymorphisms in the region (shown as open triangles in Figure 2), which allowed us to assign them to the bz gene, the intergenic region, or different parts of the stc1 gene. Selections carrying markers exclusively from the McC haplotype were characterized for the presence of the Ds-bz1 junction, in order to eliminate Ds excisions, and of Ac target site duplication footprints, in order to eliminate Ac excisions. Of the successfully tested Sh bz selections, ∼10% arose from coincidental excisions of Ac or Ds and exchanges in the sh1-bz1 interval (9 of 90 from McC/B73 and 17 of 175 from McC/W22). The vast majority arose from recombination between the Ds element in bz1 and the Ac element in stc1.
Based on the indel marker analysis, the fragments bearing the recombination junctions were PCR-amplified and sequenced in order to precisely locate the recombination junction relative to the nearest flanking polymorphisms, most of which are SNPs. The results from sequencing 239 junctions are shown graphically in Figure 3. In this figure, gene exons are colored peach, introns are colored yellow, and SNPs are shown as vertical lines. The 26-kb retrotransposon cluster in the B73 haplotype is drawn in maroon and in a much smaller scale than the adjacent 5.8-kb genic region. MITEs in noncoding gene sequences or the bz1-stc1 intergenic segment are represented as small blue triangles. As can be seen from the respective SNP densities, the bz1 and stc1 alleles of McC and B73 are somewhat more closely related than those of McC and W22. In both heterozygotes, most intervals are delimited by two SNPs. The smallest interval is a few base pairs in length; the largest one is a 1.8-kb stretch of uninterrupted homology at the 3′ end of stc1 in the McC/B73 haplotype. The number of junctions falling within specific intervals is shown beneath the common McC haplotype in each heterozygote.
Figure 3.
Distribution of Recombination Junctions within the Ds2(D1)-Ac6087 Genetic Interval among Sh bz Recombinants from McC/B73 (Top) and McC/W22 (Bottom) Heterozygotes.
As in Figure 1, exons are in peach and introns are in yellow; the stop codon for each gene is indicated by a red octagon. The retrotransposon cluster in B73 is in maroon and drawn in a smaller scale than the rest of the interval. Polymorphisms are represented as vertical lines (SNPs) or blue triangles (indels), numbered as indicated by Wang and Dooner (2006). The number of recombination junctions in each subinterval defined by these polymorphisms is shown beneath the common McC haplotype in each heterozygote, single-digit numbers in black and double-digit numbers in red. The Ds2(D1)-Ac6087 genetic interval was subdivided into five segments for analysis: bz1, intergenic region, and three roughly equally sized stc1 segments. Significantly different estimates of genetic distances for a segment in the two heterozygotes are indicated in red. See text for additional details.
In order to assess the effect of the heterozygous retrotransposon cluster located between bz1 and stc1 on recombination in the region, we subdivided the Ds2(D1) Ac6087 genetic interval into five segments and, for each segment, compared recombination between McC/B73, which had the retrotransposon cluster, and McC/W22, which did not. The segments, numbered 1 to 5 in the figure, are as follows: the 0.81-kb bz1 interval (1), which corresponds to about half the length of the bz1 gene because of the central location of the Ds insertion in the gene; the 0.82-kb common intergenic sequences between the termination codons of bz1 and stc1 (2); and three similarly sized segments of the stc1 gene (3 to 5), which span practically the entire length of stc1 and are defined by common polymorphisms in the two heterozygotes. The 1.22-kb distal segment 3 extends from the distal-most SNP in the fifth intron of stc1 to a SNP just a few base pairs upstream of the stc1 stop codon and is completely conserved between McC and B73; the 1.37-kb central segment 4 extends from the MITE indel Ins6 in the third intron of stc1-McC to the last SNP in the fifth intron of stc1; and the 1.54-kb proximal segment 5 extends from the Ac6087 marker in the first exon of stc1-McC to Ins6 in the third intron.
Under each of the five segments are given the stretch of homology, in kilobases, the number of identified crossovers, and the genetic length, in centimorgan. The sum for all intervals is shown at right. The Ds2(D1)-Ac6087 genetic distance for each heterozygote is slightly less than would have been calculated from the Sh bz recombinant class of Table 1, which is uncorrected for Ac or Ds excisions and simultaneous exchanges in the sh1-bz1 region (0.30 versus 0.34 cM for McC/B73 and 0.59 versus 0.66 cM for McC/W22). The total length of the Ds2(D1)-Ac6087 interval is significantly smaller in the B73 heterozygote than in the W22 heterozygote (χ2 = 25, 1 df, P < 0.001).
An examination of the distribution of junctions in Figure 3 reveals that exchanges occur only in the bz1 and stc1 genes. No recombination at all occurs in the intergenic region in either heterozygote. This is not surprising in the McC/B73 heterozygote, given that its intergenic region contains the large retrotransposon insertion, but the absence of crossovers in the McC/W22 intergenic region suggests that recombination is inhibited by the high density of SNP and indel heterologies in the interval, an effect previously documented for recombination within bz1 (Dooner and Martinez-Ferez, 1997; Dooner, 2002) and a1 (Yao et al., 2002; Yandeau-Nelson et al., 2006). Recombination was significantly higher in the bz1 interval and two of the three stc1 intervals of the McC/W22 heterozygote. The size of the bz1 genetic interval 1 is about four times larger in W22 than in B73 (χ2 = 8.15, 1 df, P < 0.01). The size of the distal stc1 segment is twice as large (χ2 = 6.05, 1 df, P < 0.01) and the size of the proximal stc1 segment is three times larger (χ2 = 18.6, 1 df, P < 0.001). Only in the central stc1 segment did recombination apparently not differ. Although both heterozygotes differ in multiple SNPs, which are known to have an inhibitory effect on recombination, SNP density in the bz1 and stc1 genes is actually lower in the McC/B73 heterozygote than in the McC/W22 heterozygote. Thus, the overall lower recombination observed in the McC/B73 heterozygote is most likely due to the presence of the large retrotransposon cluster in the B73 bz1-stc1 intergenic region.
Recombination in the common gene space of heterozygous haplotypes has been proposed to shuffle retrotransposon blocks, creating new chimeric haplotypes (Wang and Dooner, 2006). In the McC/B73 heterozygote, recombination events within the stc1 gene should produce a recombinant arrangement of retrotransposon blocks, as shown in Figure 4A. Like McC, they should lack the 26-kb retrotransposon cluster located between bz1 and stc1 in B73, and like B73, they should lack the 53-kb retrotransposon nest and the Helitrons of McC. To verify this, we characterized the size of the stc1-hybridizing NotI band in several stc1 crossovers by CHEF gel DNA gel blot analysis. Representative data from two crossovers in segments 3 and 4 of stc1 (Figure 3) are shown in Figure 4B. These crossovers were known from the initial analysis of the recombination junctions to lack the 26-kb retrocluster from B73. As expected, the parental McC haplotype gives a 111-kb NotI fragment and the parental B73 haplotype gives a 73-kb NotI fragment, both in the W22-introgressed line used in the recombination experiment and in the original B73 inbred. The two different stc1 crossovers give a smaller, ∼50-kb NotI fragment, the size expected from the recombinational loss of the large retroclusters of B73 and McC and the retention of the sole distal Grande1 retrotransposon of B73.
Figure 4.
Generation of New Chimeric Haplotypes in McC/B73 Heterozygotes by Recombination within the stc1 Gene.
(A) Diagram illustrating how an exchange event in stc1 recombines the flanking retrotransposon clusters, producing a novel haplotype that lacks both clusters.
(B) CHEF gel DNA gel blot analysis of parental and recombinant haplotypes (NotI digest, stc1 probe). M, size markers in kilobases; McC, bz-m2(D1) stc1-M1(Ac6087); B73 (left), B73 Bz1-B73 stc1-B73 introgressed into W22; B73 (right), B73 Bz1-B73 stc1-B73 from the original B73 inbred; CO (left) and CO (right), Sh bz wx crossovers resulting from exchanges in stc1 gene intervals 3 and 4 (see Figure 3), respectively.
DISCUSSION
In this study, we investigated whether retrotransposon polymorphisms, which are widespread in maize (Wang and Dooner, 2006), affect recombination in neighboring genes. We compared recombination between markers in the adjacent bz1 and stc1 genes in heterozygotes between haplotypes that differed by the presence or absence of a heterozygous retrotransposon cluster in the intergenic region and found that the genetic distance between the markers was twice as large in the absence of the cluster. To monitor recombination, we made use of Ac and Ds markers that affected seed pigmentation, allowing high-resolution analysis of the interval. In McC and W22, haplotypes that lack the retrotransposon cluster, the physical distance between the Ds2(D1) marker in bz1 and the Ac6087 marker in stc1 is ∼6 kb, and the measured size of the genetic interval in an McC/W22 heterozygote is 0.59 cM. In the B73 haplotype, the physical distance is ∼32 kb because of the insertion in the bz1-stc1 intergenic region of a retrotransposon cluster, consisting of Tekay, a Zeon solo LTR, and a Xilon-Mu1 nest, and the measured size of the genetic interval in an McC/B73 heterozygote is 0.28 cM.
The heterozygous structure within the genetic interval is more than five times the size of the interval itself, so it could reduce recombination by interfering with the normal pairing of the adjacent genic sequences. Furthermore, the retrotransposon cluster is heavily methylated and probably more condensed than the adjacent euchromatin. Recombination is reduced by fivefold in the 0.81-kb segment of bz1 located between Ds2(D1) and the cluster and by twofold overall in the ∼4-kb segment of stc1 located between Ac6087 and the cluster. However, the pattern of reduction within stc1 is somewhat unexpected. Recombination is reduced in the proximal (3′) one-third and distal (5′) one-third, but not in the middle. This is not the pattern one would expect if the retrotransposon effect diminished with distance from the cluster. It is unlikely that the observed differences result from a small experimental sample, as 77 and 141 stc1 recombinants were characterized in McC/B73 and McC/W22 heterozygotes, respectively. Differences in the density of heterologies, which shows a negative correlation with intragenic recombination in maize (Dooner and Martinez-Ferez, 1997; Dooner, 2002; Yao et al., 2002; Yandeau-Nelson et al., 2006), could, in principle, account for some of the observed differences, but the bz1 and stc1 alleles of B73 are less polymorphic with McCs in intervals 3 and 5 than in those of W22. One possible explanation for the higher recombination in the 5′ stc1 segment of McC/W22 heterozygotes would be that that segment is normally more recombinogenic in W22 than in B73, perhaps because of sequence differences in the two Stc1 promoters, although the enhancement of recombination by promoter-adjacent sequences, as in yeast and mammals (Petes, 2001), has not been demonstrated in plants.
As is evident from a comparison of the three haplotypes in Figure 1, B73 and W22 differ in other large insertion polymorphisms besides the 26-kb retrotransposon cluster in the bz1-stc1 intergenic region, so the reduction in recombination observed between bz1 and stc1 could be partly the result of heterozygosity for other large insertions in the introgressed segment. However, recombination in the bz1-stc1 interval, which contains the retrotransposon cluster and constitutes about one-fourth of the bz1-sh1 distance, is reduced by twofold, whereas recombination in the stc1-sh1 interval, which includes the polymorphic sequences to the right of stc1 in Figure 1, is not reduced significantly in McC/B73 relative to McC/W22 heterozygotes. These observations argue that the reduction in recombination reported here arises principally from the heterozygosity of the large retrotransposon cluster in the bz1-stc1 interval.
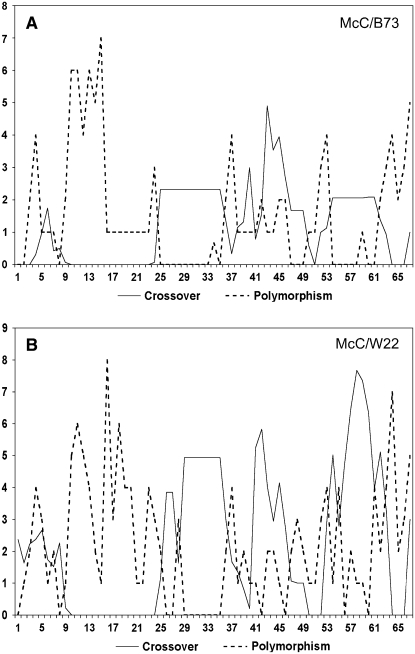
The distribution of recombination junctions in the Ds2(D1)-Ac6087 interval correlates negatively with the density of polymorphisms in both heterozygotes. In Figure 5, frequencies of crossovers and polymorphisms in the 6.7-kb interval are plotted as a moving average every 100 bp from the proximal to the distal end. The highly polymorphic intergenic region lies between intervals 9 and 24 and contains no junctions in either heterozygote. Almost all of the polymorphisms outside of this region are SNPs. As can be seen, there is a general inverse relationship between the frequencies of crossovers and heterologies across the entire interval. These data confirm and extend the earlier observations at bz1 and a1 cited above.
Figure 5.
Frequencies of Recombinants (Solid Lines) and Polymorphisms (Dashed Lines) in the 6.7-kb Interval between Ds2(D1) and Ac6087.
bz1 is to the left and stc1 is to the right. The 6.7-kb interval has been divided into 67 100-bp segments. Segments that make up fractions of an interval have been assigned a number of crossovers in proportion to the fraction of the interval constituted by that segment. The intergenic region lies between intervals 9 and 24.
All of the recombination junctions fell within introns or exons of bz1 or stc1 (Figure 3). The bz1-stc1 intergenic region does not include the 5′ end of either gene, the end often associated with high conversion and the initiation of recombination in yeast (Petes, 2001). However, analysis of recombination junctions in a 100-kb stretch extending upstream (distal) of stc1 that included several intergenic regions confirms that most crossovers fall within genes rather than in intergenic regions (L. He and H.K. Dooner, unpublished data). Similar observations have been made in the maize a1-sh2 region (Yao et al., 2002). In Arabidopsis, a much less polymorphic species than maize, most crossover junctions fall in intergenic regions (Mezard, 2006), so the distribution of recombination appears to differ in the two plants.
The methylated retrotransposons that are interspersed with genes in the maize genome are probably heterochromatic, as they are in the knobs of maize and Arabidopsis (Ananiev et al., 1998; Lippman et al., 2004), and can affect recombination in neighboring genes. The general recombinational inertness of heterochromatin in many organisms (Harper and Cande, 2000) and the reduction of recombination in euchromatic regions adjacent to heterochromatin in Drosophila (Baker, 1958; Westphal and Reuter, 2002) are well-known phenomena. The distribution of LTR retrotransposons in the euchromatic portions of Drosophila chromosomes varies among wild-type strains (Franchini et al., 2004), but the effect of this variability on recombination has not been studied. More relevant to this study is a recent finding in rye (Secale cereale), showing that a polymorphic interstitial heterochromatic sequence in chromosome 2R significantly suppressed recombination in the arm (Kagawa et al., 2002). What sets maize apart from other species studied to date is the extensive variation in the distribution of retrotransposons, hence interspersed heterochromatin, from line to line. Our results suggest that this variability will affect the local distribution of recombination events across the genome.
The implication of the finding reported here is that, for very closely linked markers, local variation in haplotype structure will have a strong influence on estimates of genetic distance. Intergenic retrotransposons add physical length to the interval and reduce recombination in adjacent genes, thus doubly affecting the ratio of genetic to physical length in some mapping populations relative to others. Heterozygosity for intergenic retrotransposons is probably extensive in maize mapping populations. For example, the B73 and Mo17 bz1 locus haplotypes differ from each other by the presence of the 26-kb bz1-stc1 intergenic retrocluster (Fu and Dooner, 2002; Brunner et al., 2005), so the bz1-stc1 distance in a B73×Mo17 map, such as the widely used IBM map (Lee et al., 2002; Sharopova et al., 2002), will be different from that in a W22/McC or W22/Mo17 map. When sequenced, the B73 genome will become the standard for map-based cloning efforts in maize. Using it as the reference for the physical map, one would compute a centimorgan:kilobase ratio of 0.0087 for the Ds2(D1)-Ac6087 interval in an McC/B73 heterozygote (0.28 cM:32 kb). By contrast, the centimorgan:kilobase ratio for the same interval in an McC/W22 heterozygote would be 10-fold higher (0.59 cM:6 kb, or 0.098). Over longer genetic distances, variations in centimorgan:kilobase ratios between different mapping populations may average out.
Recombination within genes can lead to either the loss or gain of intergenic retrotransposons in the recombinants. We showed here that recombination within stc1 in an McC/B73 heterozygote leads to the joint loss of flanking retrotransposons and to a calculated reduction of the distance between the bz1 and znf genes from 82 and 44 kb in McC and B73, respectively, to 18 kb in the recombinant. The recombinant now carries a bz1-znf gene island uninterrupted by retrotransposons. In the reciprocal product, which should be present in the unanalyzed sh Bz recombinant class, both retroclusters would be retained and the distance between bz1 and znf would expand to 126 kb. Again, taking B73 as the reference maize genome, the reciprocal products would have intraspecific expansion factors of 0.4 and 2.9, similar to those reported in an interspecific comparison of longer syntenic blocks from two maize (B73) homeologous regions with rice (Oryza sativa) as the standard (Bruggmann et al., 2006). Whether similar contraction:expansion ratios will occur when comparing megabase-sized distances within maize will have to await a fuller characterization of the genome from other maize lines.
METHODS
Plant Materials
All of the maize (Zea mays) stocks used in this study shared the common genetic background of the inbred W22. The bronze1 alleles and the aleurone phenotypes of the various stocks are described below. Except for the W22 stock carrying a Bz1-B73 allele, the derivation of the other stocks has been described previously.
Bz1-McC (purple): the normal allele of the McC haplotype (Ralston et al., 1988). bz1-m2(DI) (bronze in the absence of Ac, spotted in its presence) harbors a 3.3-kb Ds element at positions 755 to 762 in the second exon of Bz1-McC (McClintock, 1962; Dooner et al., 1986). sh1-bz1-X2 (shrunken, bronze): an x-ray–induced deletion of a large chromosomal fragment that includes the sh1 and bz1 loci (Mottinger, 1973). stc1-m1(Ac6087): an insertion mutation in the first exon of stc1-McC produced by the transposition of Ac from the nearby bz1 locus in McC (Shen et al., 2000). Bz1-W22 (purple): the normal allele of the color-converted version of the inbred W22 (Ralston et al., 1988). This stock traces back to the W22 R1-r:standard stock derived by Brink (1956) for his now classical studies of R1 locus paramutation. Bz1-B73 (purple): the normal allele of the inbred B73. It was introgressed as part of the Sh1 Bz1 segment of that inbred by repeated backcrosses to a W22 stock carrying the sh1-bz1-X2 deletion, thus precluding any recombination within the introgressed segment. The B73 parent was genotypically c1 Sh1 Bz1, and the recurrent W22 parent was C1 sh1-bz1-X2. Plump, purple kernels were selected in each generation, and after three backcrosses, a line carrying a C1 Sh1 Bz1 recombinant chromosome was identified and selfed twice to establish homozygotes. The line was genotyped for bnl1401 and wx1 markers, located 13 and 25 cM proximal to bz1, respectively, and determined to carry W22 alleles at both of those loci. Thus, the Sh1 Bz1 introgressed fragment from B73 is small. Its distal crossover junction lies in the 4-cM c1-sh1 interval and its proximal junction lies in the 13-cM bz1-bnl1401 interval, so the size of the fragment ranges from a minimum of 2 cM to a maximum of 17 cM.
Selection and Analysis of Crossovers
The mutations sh1 (shrunken endosperm) and wx1 (waxy endosperm) were used as markers flanking bz1. They map, respectively, ∼2 cM distal and 25 cM proximal to bz1 in 9S. The sh1-wx1 region exhibits high chiasma interference (Dooner, 1986), so double crossovers in the region are rare. The sh1 stc1-m1(Ac6087) bz-m2(D1) wx1/Sh1 Stc1-B73 Bz1-B73 Wx1 and sh1 stc1-m1(Ac6087) bz-m2(D1) wx1/Sh1 Stc1-W22 Bz1-W22 Wx1 heterozygotes were hand-pollinated with a sh1-bz1-X2 wx1 stock. Sh bz recombinants were selected as single seeds with a plump, solid bronze phenotype in ears segregating spotted and purple seed and grown in the greenhouse for analysis. Leaf DNA was made from all selections by a urea extraction procedure (Greene et al., 1994) and used for subsequent PCR amplification. The selections were genotyped with a series of molecular markers, as described in Results.
PCR and Sequencing
PCR was performed using Qiagen Taq polymerase (Qiagen). The PCR products were run on 1% agarose gels or 8% polyacrylamide gels based on their size and the polymorphisms to be discriminated. For sequencing, PCR products were purified by isopropanol precipitation and 70% ethanol washing. The same PCR primers were used as sequencing primers to directly sequence purified PCR products using ABI BigDye Terminator V3.1 reagent (Applied Biosystems). DNA sequencing was performed in an ABI 3700 DNA analyzer.
BAC Isolation and Sequencing
NotI BAC clones of the bz genomic region of W22 were isolated as described previously (Fu and Dooner, 2000). The W22 BAC clones were sequenced by the shotgun sequencing strategy, assembled, analyzed, and annotated as described for other bz BAC clones from different maize inbreds and land races (Wang and Dooner, 2006).
DNA Gel Blot Analysis
DNA was prepared from isolated nuclei of 4-week-old shoots and leaves. Agarose plugs containing ∼10 μg of high molecular weight DNA from different haplotypes were digested to completion with NotI. The digested genomic DNAs were resolved on 1% agarose gels by pulsed-field gel electrophoresis (CHEF-DR II system; Bio-Rad). The gels were blotted to a Hybond+ nylon membrane (Amersham Pharmacia Biotech), and the membranes were hybridized with random primer–labeled 32P probes from stc1-McC. Conditions for hybridization, high-stringency washing, and exposure to x-ray film were standard.
Accession Number
The GenBank accession number for the 238-kb W22 bz region sequence is EU338354.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Estimated Times of Insertion of LTR Retrotransposons in the McC and W22 Haplotypes.
Supplementary Material
Acknowledgments
We thank members of the Dooner laboratory for comments on the manuscript, Zsuzana Swigonova for dating the retrotransposon insertions, and Charles Du for Figure 5. This research was supported by National Science Foundation Grants MCB 02-12785 and MCB 05-23103 to H.K.D.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hugo K. Dooner (dooner@waksman.rutgers.edu).
Online version contains Web-only data.
References
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin. Genetics 149 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, W.S. (1958). Crossing over in heterochromatin. Am. Nat. 92 59–60. [Google Scholar]
- Beavis, W.D., and Grant, D. (1991). A linkage map based on information from four F2 populations of maize (Zea mays L.). Theor. Appl. Genet. 82 636–644. [DOI] [PubMed] [Google Scholar]
- Brink, R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann, R., et al. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., Fengler, K., Morgante, M., Tingey, S., and Rafalski, A. (2005). Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. (1986). Genetic fine structure of the bronze locus in maize. Genetics 113 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. (2002). Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., English, J., Ralston, E., and Weck, E. (1986). A single genetic unit specifies two transposition functions in the maize element Activator. Science 234 210–211. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., and Martinez-Ferez, I.M. (1997). Recombination occurs uniformly within the bronze locus, a meiotic recombination hotspot in the maize genome. Plant Cell 9 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch, E., Szymaniak, J.M., Yates, H., Pawlowski, W.P., and Buckler, E.S. (2007). Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics 177 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatmi, A., Poneleit, C.G., and Pfeiffer, T.W. (1993). Variability of recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L.). Theor. Appl. Genet. 86 859–866. [DOI] [PubMed] [Google Scholar]
- Franchini, L.F., Ganko, E.W., and McDonald, J.F. (2004). Retrotransposon-gene associations are widespread among D. melanogaster populations. Mol. Biol. Evol. 21 1323–1331. [DOI] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2000). A gene enriched BAC library for cloning large allele-specific fragments from maize: Isolation of a 240-kb contig of the bronze region. Genome Res. 10 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Park, W., Yan, X., Zheng, Z., Shen, B., and Dooner, H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions differ by two orders of magnitude. Proc. Natl. Acad. Sci. USA 99 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, B., Walko, R., and Hake, S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, L.C., and Cande, W.Z. (2000). Mapping a new frontier: Development of integrated cytogenetic maps in plants. Funct. Integr. Genomics 1 89–98. [DOI] [PubMed] [Google Scholar]
- Kagawa, N., Nagaki, K., and Tsujimoto, H. (2002). Tetrad-FISH analysis reveals recombination suppression by interstitial heterochromatin sequences in rye (Secale cereale). Mol. Genet. Genomics 267 10–15. [DOI] [PubMed] [Google Scholar]
- Lee, M., Sharopova, N., Beavis, W.D., Grant, D., Katt, M., Blair, D., and Hallauer, A. (2002). Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol. 48 453–461. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1962). Topographical relations between elements of control systems in maize. Carnegie Inst. Washington Year Book 61 448–461. [Google Scholar]
- Mezard, C. (2006). Meiotic recombination hotspots in plants. Biochem. Soc. Trans. 34 531–534. [DOI] [PubMed] [Google Scholar]
- Mottinger, J. (1973). Unstable mutants of bronze induced by premeiotic X-ray treatment in maize. Theor. Appl. Genet. 43 190–195. [DOI] [PubMed] [Google Scholar]
- Petes, T.D. (2001). Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2 360–369. [DOI] [PubMed] [Google Scholar]
- Ralston, E.J., English, J., and Dooner, H.K. (1988). Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., and Bennetzen, J.L. (1998). Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. (Lond.) 82 37–44. [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20 43–45. [DOI] [PubMed] [Google Scholar]
- Sharopova, N., et al. (2002). Development and mapping of SSR markers for maize. Plant Mol. Biol. 48 463–481. [DOI] [PubMed] [Google Scholar]
- Shen, B., Zheng, Z., and Dooner, H.K. (2000). A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: Characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 97 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., and Dooner, H.K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal, T., and Reuter, G. (2002). Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 160 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C.G., Goodman, M.M., and Stuber, C.W. (1995). Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics 141 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson, M.D., Xia, Y., Li, J., Neuffer, M.G., and Schnable, P.S. (2006). Unequal sister chromatid and homolog recombination at a tandem duplication of the A1 locus in maize. Genetics 173 2211–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., and Schnable, P.S. (2005). Cis-effects on meiotic recombination across distinct a1-sh2 intervals in a common Zea genetic background. Genetics 170 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B.J., and Schnable, P.S. (2002). Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.