SUMMARY
High-density lipoproteins (HDL) prevent atherosclerosis by removing cholesterol from macrophages and by exerting anti-oxidant and anti-inflammatory effects. Oxidation is thought to impair HDL functions, yet certain oxidative modifications may be advantageous; thus, mild oxidation reportedly enhances cell cholesterol uptake by HDL whereas extensive oxidation impairs it. To elucidate the underlying energetic and structural basis, we analyzed the effects of copper and hypochlorite (that preferentially oxidize lipids and proteins, respectively) on thermal stability of plasma spherical HDL. Circular dichroism, light scattering, calorimetry, gel electrophoresis and electron microscopy showed that mild oxidation destabilizes HDL and accelerates protein dissociation and lipoprotein fusion, while extensive oxidation inhibits these reactions; this inhibition correlates with massive protein cross-linking and lipolysis. We propose that mild oxidation lowers kinetic barriers for HDL remodeling due to diminished apolipoprotein affinity for lipids resulting from oxidation of methionine and aromatic residues in apolipoproteins A-I and A-II followed by protein cross-linking into dimers and/or trimers. In contrast, advanced oxidation inhibits protein dissociation and HDL fusion due to lipid re-distribution from core to surface upon lipolysis and to massive protein cross-linking. Our results help reconcile the apparent controversy in the studies of oxidized HDL and suggest that mild oxidation may benefit HDL functions.
Keywords: Kinetic stability, protein dissociation, lipoprotein fusion, cholesterol transport, atherosclerosis
INTRODUCTION
Mature high-density lipoproteins (HDL) are heterogeneous spheroidal particles (d=7–12 nm) containing apolar lipids (mainly cholesterol esters and triglycerides) in the core and polar lipids (mainly phospholipids and cholesterol) and proteins (termed apolipoproteins) in their surface. HDL protect against atherosclerosis via several mechanisms. One is reverse cholesterol transport in which HDL remove excess cholesterol from the foam cells of arterial macrophages to the liver for excretion.1–3 In addition, HDL provide antioxidants for low-density lipoproteins (LDL).4–7 According to the oxidative modification hypothesis, LDL oxidation and retention in the arterial wall triggers formation of atherosclerotic lesions (4,6–8 and references therein). The ability of HDL to inhibit formation of toxic lipid hydroperoxides and to safely remove them from LDL, together with anti-inflammatory action of HDL, is thought to contribute to their atheroprotective effects.4–10 At the same time, failure of antioxidants such as vitamin E to protect against atherosclerosis in clinical trials, together with growing evidence that the anti-atherogenic action of the lipid-lowering drug probucol is not related to its antioxidant effects11 and the notion that such cardioprotective activities as exercise or red wine consumption are pro-oxidant, suggest that not all forms of oxidized lipoproteins are atherogenic. 9,12
Although the central role of LDL oxidation in atherogenesis is widely accepted9, the role of HDL oxidation remains contested. Oxidation is generally thought to impair atheroprotective functions of HDL (13–18 and references therein). Thus, cell cholesterol efflux to HDL, a key early step in reverse cholesterol transport mediated by ATP-binding cassette transporter A1, is reportedly impaired upon HDL oxidation.14–19 This impairment has been ascribed to specific protein modifications such as oxidation of two-to-three Met in the major HDL protein, apolipoprotein A-I (apoA-I), that occurs at initial stages of HDL oxidation,20–22 along with tyrosylation and protein cross-linking via the modified Tyr or Lys.13,16,17,23 However, several studies report that oxidation enhances rather than impairs HDL functions in stimulating lipid efflux from cells22,24–26 and protecting against atherosclerosis.27 Moreover, a recent report suggests that mild oxidation by Cu2+ improves HDL function as cholesterol acceptor from macrophages, while extensive oxidation impairs it.28 Consequently, the discrepancy among functional studies of oxidized HDL may result, at least in part, from the differences in the degree of oxidation and/or in specific modifications produced by various oxidants. To test this hypothesis and to provide the energetic and structural basis for understanding functional consequences of HDL oxidation, we analyzed HDL stability at various stages of oxidation by two widely used oxidants, Cu2+ and OCl−.
Oxidation of apoA-I and apoA-II (that comprise 70% and 20% of HDL protein content, respectively) destabilizes their structures both in lipid-free state and in reconstituted discoidal complexes with phospholipids that provide models for nascent HDL.29,30 This destabilization was attributed to Met sulfoxide (MetO) formation in the apolar faces of amphipathic α-helices that diminishes hydrophobic protein-protein and protein-lipid interactions formed by these helices.29,30 In contrast, studies of plasma spherical HDL, which revealed oxidation-induced structural changes in both proteins (such as cross-linking) and lipids (reduced fluidity),23, 31–34 suggested more stable protein-lipid association upon oxidation.32 To resolve this apparent controversy, we carried out the first comprehensive study of the effects of oxidation on the structural stability of plasma spherical HDL.
Our earlier work has established a kinetic mechanism of lipoprotein stabilization35 and showed that thermal or chemical denaturation of spherical HDL involves protein unfolding, dissociation and particle fusion followed by rupture and release of apolar core lipids.36,37 Importantly, similar protein dissociation and lipoprotein fusion occur during metabolic HDL remodeling by plasma factors (36–39 and references therein); hence our in vitro studies of HDL denaturation provide useful models for understanding metabolic HDL remodeling in vivo. Here, we apply kinetic approach to analyze thermal stability of human HDL that were oxidized by copper (a two-electron oxidant that preferentially reacts with lipids) or hypochlorite (a one-electron oxidant that preferentially reacts with proteins and is a product of myeloperoxidase, an enzyme that oxidizes lipoproteins in vivo.14–17,40) The results reveal that Cu2+ and OCl− cause similar changes in HDL stability: protein dissociation and HDL remodeling and fusion are accelerated upon mild oxidation but inhibited upon extensive oxidation. This result helps reconcile the existing controversy in functional studies of oxidized HDL and provides the basis for understanding functional consequences of HDL oxidation.
RESULTS
Effects of Cu2+ oxidation on HDL stability
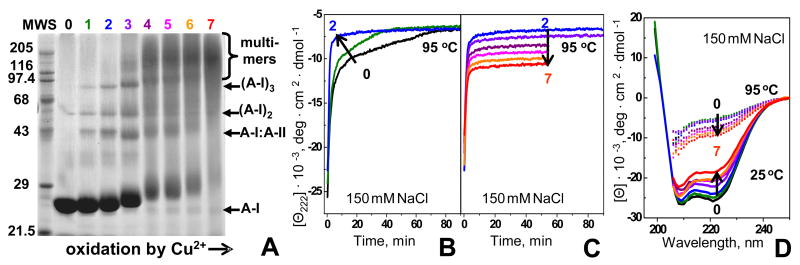
First, we analyzed structure and stability of HDL that were oxidized by Cu2+ to various stages (1–7 in Fig. 1) as described in Materials and Methods. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) of non-oxidized HDL (nHDL) showed major apolipoproteins apoA-I (28 kD) and apoA-II (S-S-linked dimer of 17.4 kD) migrating as single bands. Mild oxidation (stages 1–3) led to formation of three additional bands at 40–90 kD (Fig. 2A); these narrow bands correspond to apoA-I dimer, trimer, and apoA-I:apoA-II heterodimer that are formed upon oxidation-induced cross-linking.23,40,41 Further oxidation led to a gradual replacement of these bands by one broad band (100–200 kD) that encompasses molecular weight of HDL and reflects intra-particle cross-linking of HDL proteins. This massive cross-linking first appeared at stage 3 and became predominant at stages 6, 7 of oxidation. Thus, mild oxidation by Cu2+ leads to limited protein cross-linking into apoA-I-containing dimers and trimers, while more advanced oxidation results in massive cross-linking of the proteins on HDL surface.
Figure 1.
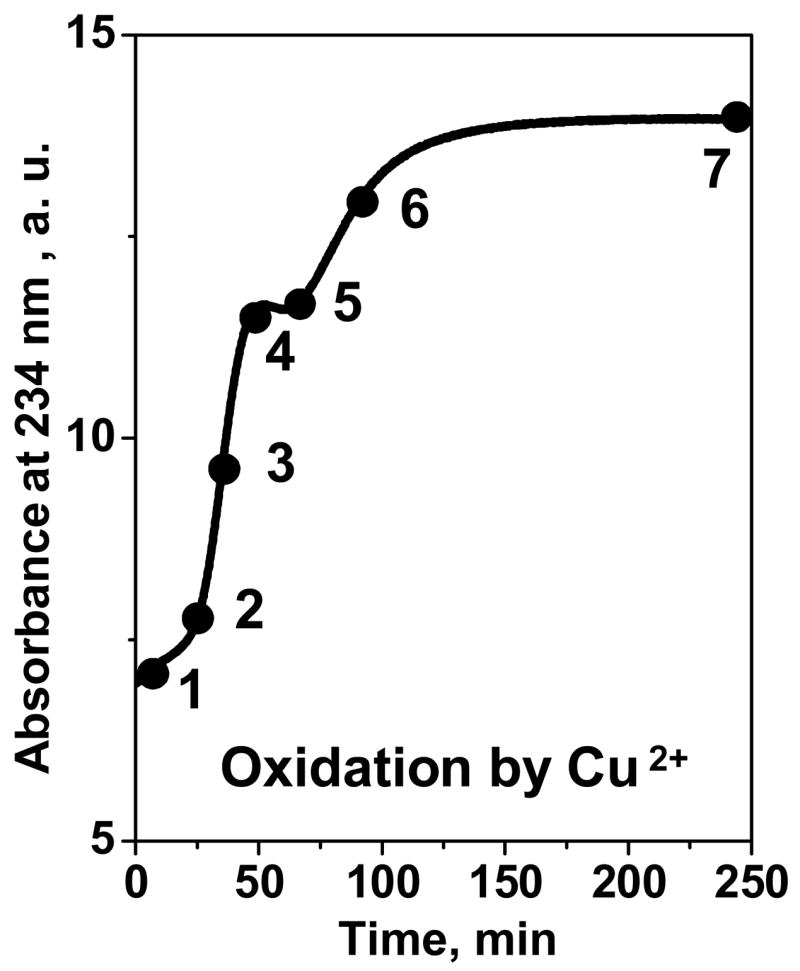
Time course of HDL oxidation by Cu2+. HDL solution (0.1 mg/mL protein in buffer B (10 mM Na phosphate, pH 7.5)) was incubated at 37 °C with 5 μM CuSO4 for up to 240 min; oxidation was monitored by absorbance at 234 nm for conjugated diene formation. Numbers refer to oxidation stages corresponding to different incubation times; HDL oxidation at these stages was stopped by adding EDTA (1:50 Cu2+:EDTA molar ratio, see Materials and Methods).
Figure 2.
HDL protein conformation and kinetic stability at various stages of oxidation by Cu2+. The data were recorded from solutions of HDL in buffer B (2.5 mg/mL protein (A) or 33 μg/mL protein, 150 mM NaCl (B–D)) that were oxidized by incubation with CuSO4 as described in Methods. Numbers correspond to oxidation stages in Fig. 1; 0 stands for nHDL.
(A) Protein cross-linking at various stages of HDL oxidation monitored by SDS PAGE. The gel was stained with Coomassie blue. A-II stands for S-S linked homodimer.
(B, C) Time course of HDL protein unfolding at various oxidation stages. HDL that were oxidized to stages 1–7 were subjected to a T-jump from 25–95 °C; protein unfolding was monitored by CD at 222 nm. Arrows indicate changes in the unfolding rates (B) or amplitudes (C) upon increase in the oxidation degree.
(D) Effect of HDL oxidation on the protein secondary structure before and after heat denaturation. HDL samples were oxidized to stages 1–7 and far-UV CD spectra were recorded at 25 °C. Next, the samples were incubated at 95 °C until CD changes were complete and the spectra were recorded again at 95 °C.
To test the effects of oxidation on the secondary structure and thermal stability, HDL were oxidized by Cu2+ to stages 1–7 and were subjected to temperature jumps (T-jumps). HDL denaturation was triggered by a rapid temperature increase from 25 to 95 °C and the time course of α-helical unfolding was monitored by circular dichroism (CD) spectroscopy at 222 nm (Fig. 2B, C). Earlier analysis of nHDL revealed two-phase unfolding kinetics (suggested by two-exponential fitting of the Θ222(t) data), with the fast phase involving partial unfolding of HDL-bound proteins and slow phase involving further unfolding and partial dissociation of HDL proteins leading to particle fusion followed by rupture.37 Mild oxidation led to acceleration of the slow phase (stages 0–2, Fig. 2B) suggesting faster protein unfolding, dissociation and lipoprotein fusion. Further oxidation (stages 3–7) did not produce additional changes in the unfolding rate but led to a gradual reduction in the unfolding amplitude (increased negative CD, Θ222, at 95 °C, Fig. 2C), suggesting that oxidized HDL retained a larger fraction of their helical structure at high temperatures. This was confirmed by far-UV CD spectra that were recorded from HDL at 25 °C before or after incubation at 95 °C (Fig. 2D). Prior to heating, oxidation to stages 1–7 led to a gradual reduction in far-UV CD amplitude across the spectrum, indicating reduced α-helical protein content upon oxidation. Heating to 95 °C led to irreversible spectral changes indicating partial protein unfolding. CD amplitude after heating (and hence the helical structure retained after heat denaturation) gradually increased with increase in the oxidation degree (Fig. 2C, D). This probably resulted from oxidation-induced protein cross-linking (Fig. 2A) that induced partial unfolding of HDL proteins prior to heating but reduced the extent of their unfolding upon heating. In summary, SDS PAGE and far-UV CD data in Fig. 2 showed that mild oxidation up to stage 2 leads to limited cross-linking of apoA-I and apoA-II into dimers and trimers and accelerates heat-induced protein unfolding. In contrast, further oxidation to stages 3–7 leads to extensive intra-particle cross-linking of HDL proteins and to a reduced extent (rather than increased rate) of their thermal unfolding.
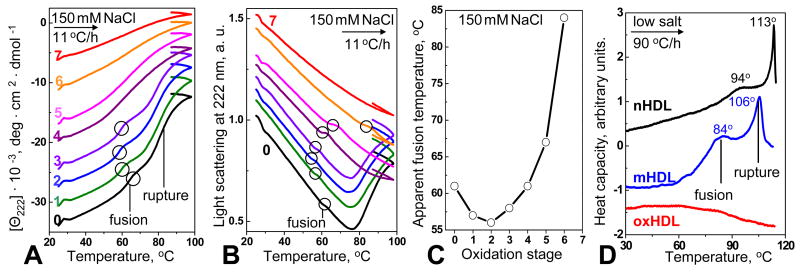
To further test the effects of copper oxidation on lipoprotein stability, CD, light scattering and DSC data were recorded during HDL heating at a constant rate (Fig. 3). CD and 90° light scattering at 222 nm (which monitor protein unfolding and changes in the particle size, respectively) were recorded simultaneously at a scan rate of 11 °C/h (Fig. 3A, B). The results showed two consecutive transitions involving HDL fusion followed by rupture.37 Mild oxidation (stages 1, 2) shifts these transitions to lower temperatures; for example, the apparent temperature of HDL fusion measured by CD or light scattering decreases from 62 °C in nHDL to 56 °C in HDL oxidized to stage 2 (Fig. 3A–C). This is consistent with the kinetic data showing that oxidation to stage 2 accelerates protein unfolding (Fig. 2A). Interestingly, the effect of oxidation on the melting data changes sign upon oxidation to stage 3 and beyond, as evident from the gradual increase in the apparent temperatures of HDL fusion and rupture at these stages (Fig. 3B, C). For example, light scattering heating data of HDL oxidized to stage 6 show an increase in the apparent fusion temperature to about 84 °C (Fig. 3C). Consequently, mild oxidation destabilizes HDL and promotes protein unfolding and lipoprotein fusion, while advanced oxidation inhibits these reactions.
Figure 3.
Effect of oxidation by Cu2+ on the apparent transition temperatures in HDL. Numbers correspond to oxidation stages in Fig. 1. HDL (2.5 mg/mL protein in buffer B), which were oxidized to various stages by incubation with CuSO4, were used for DSC experiments (D) or were diluted to 33 μg/ml by buffer B containing 150 mM NaCl for CD experiments (A–C); the samples were heated at a constant rate of 11 °C/h (A–C) or 90 °C/h (D). The CD (A) and 90° light scattering data (B) were recorded simultaneously at 222 nm during heating and cooling from 25–98 °C; the data are shifted along Y-axis to avoid overlap. The heating curves in panels B, C show two consecutive transitions that involve protein unfolding/dissociation and HDL fusion followed by HDL rupture.37 Negative slopes in the light scattering melting curves in C may result from an optical artifact and from the temperature dependence of the refractive index.
(C) Temperature of HDL fusion as a function of oxidation degree determined from the melting data in panels A, B.
(D) Heat capacity Cp(T) of HDL that were non-oxidized (nHDL), mildly oxidized (stage 2, mHDL), or extensively oxidized (stage 7, oxHDL) were recorded by DSC during heating at 90 °C/h. The data are shifted along the Y-axis to avoid overlap; HDL fusion and rupture transitions are indicated.
Calorimetric studies further confirmed this notion. Differential scanning calorimetry (DSC) data in Fig. 3D were recorded at a fast heating rate (90 °C/h) using low-salt buffer; as a result, HDL transitions in DSC experiments were observed at higher temperatures than in CD experiments (which were carried out using 150 mM NaCl and slow scan rate to shift the transition temperatures below 100 °C to the range observable by CD). Importantly, the oxidation-induced changes detected in CD and DSC experiments were similar: mild oxidation to stage 2 (mHDL) led to large low-temperature shifts in calorimetric transitions indicating destabilization, while extensive oxidation to stage 7 (oxHDL) eliminated these transitions from the temperature range accessible by DSC (Fig. 3D). Taken together, our kinetic and melting data recorded by CD, light scattering or DSC (Fig. 2B, C; Fig. 3) clearly showed that mild oxidation destabilizes HDL and promotes protein unfolding and lipoprotein fusion and rupture, while advanced oxidation inhibits these reactions.
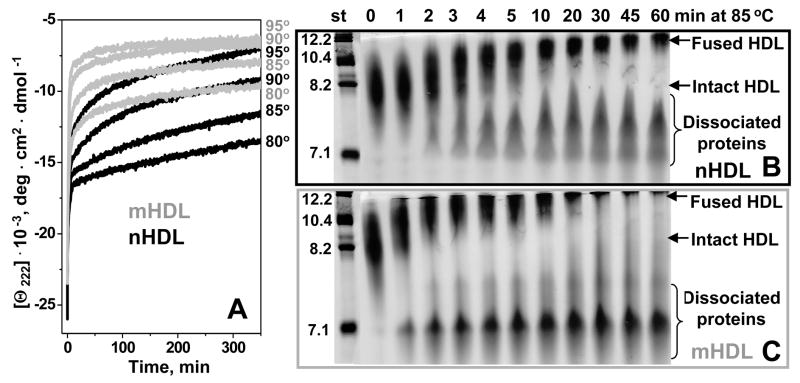
To further test the destabilizing effects of mild oxidation on HDL assembly, we compared T-jump CD data recorded from nHDL and mHDL (oxidized by Cu2+ to stage 2). Compared to nHDL, mHDL showed faster unfolding at 80–95 °C (Fig. 2B; black and grey lines in Fig. 4A). Although T-jump data of nHDL were well-approximated by double exponentials,37 similar data of mHDL could not be fitted with exponential functions, thereby precluding Arrhenius analysis. Nevertheless, acceleration of the slow unfolding phase in mHDL (stage 2, Fig. 2B) suggested faster protein dissociation and particle fusion. To test this notion, nHDL and mHDL were incubated at 85 °C and sample aliquots taken during the first 60 min of incubation were analyzed by non-denaturing gel electrophoresis (Fig. 4B, C). The results showed that protein dissociation and particle fusion in nHDL first occured after 2 min of incubation and reached completion after 30 min (Fig. 4B). In contrast, in mHDL these reactions were well underway after 1 min incubation and reached completion after 5–10 min (Fig. 4C). Faster protein dissociation and particle fusion observed in mHDL further confirm their reduced stability as compared to nHDL.
Figure 4.
Effect of mild oxidation by Cu2+ on the time course of HDL protein unfolding, protein dissociation and particle fusion. (A) HDL samples (2.5 mg/mL protein) that were non-oxidized (nHDL, black) or oxidized to stage 2 (mHDL, grey) were diluted to 33 μg/ml protein in buffer B containing 150 mM NaCl, and were subjected to T-jumps from 25 °C to 80, 85, 90, or 95 °C (shown on the lines). The time course of protein unfolding was monitored by CD at 222 nm. Similar samples of nHDL (B) or mHDL (C) containing 1 mg/mL protein were incubated at 85 °C, the aliquots were taken after 1–60 min of incubation and were subjected to non-denaturing gel electrophoresis to monitor the time course of protein dissociation and particle fusion. Lane numbers show incubation times (in min) at 85 °C; molecular size standards are shown in nm. Normal apoA-I and A-II are highly self-associated at these concentrations, and the proteins that gradually dissociate from HDL at high temperature run as oligomers 7–8 nm in size (B); self-association of mildly oxidized proteins (C) may be affected by their reduced hydrophobicity.
DSC data in Fig. 3D suggest that, in contrast to nHDL and mHDL, oxHDL (oxidized to stage 7 by Cu2+) do not undergo heat-induced protein dissociation and particle fusion and rupture in the experimentally accessible temperature range. To test this notion, nHDL and oxHDL were incubated for 1 h at 95 °C and were analyzed by non-denaturing gel electrophoresis and electron microscopy (Fig. 5). In contrast to nHDL that disintegrated into fused and ruptured particles and dissociated protein after heating to 95 °C, oxHDL showed no protein dissociation or particle fusion even after 60 min incubation at 95 °C (Fig. 5A, right lane). Negative staining electron microscopy confirmed this result: in contrast to nHDL that ruptured and coalesced into large lipid droplets after 1 h incubation at 95 °C, oxHDL showed no large changes in their size and morphology upon heating from 25 to 95 °C (Fig. 5B–E). These results, together with CD, light scattering and DSC data in Fig. 3, clearly show that, in contrast to mild oxidation, extensive oxidation by Cu2+ inhibits heat-induced protein unfolding, dissociation and HDL fusion.
Figure 5.
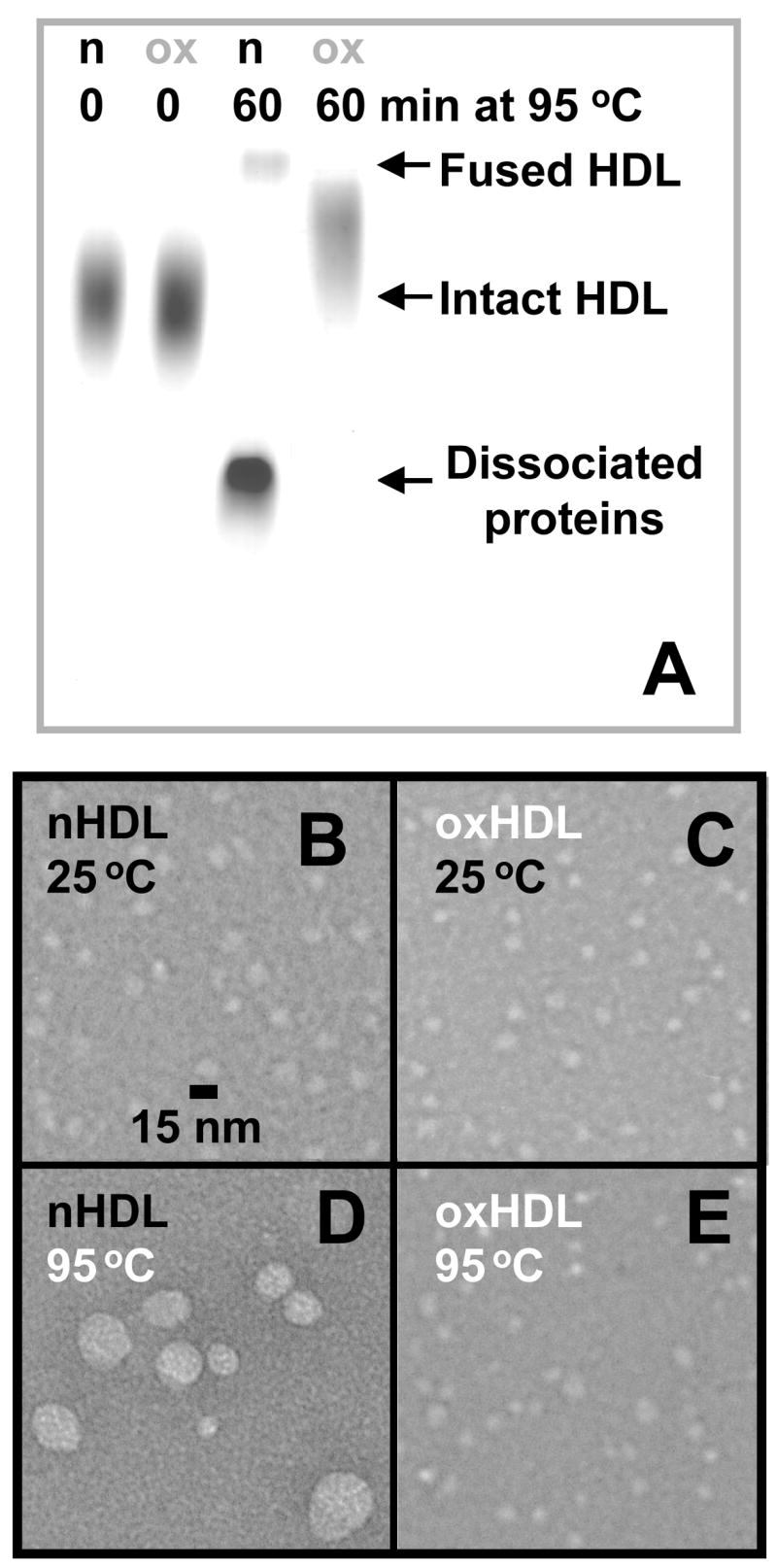
Effect of extensive oxidation by Cu2+ on the heat-induced protein dissociation and HDL fusion. HDL samples (2.5 mg/mL protein in buffer B) that were not oxidized (n) or oxidized to stage 7 (ox) were incubated at 95 °C for 60 min (incubation times are shown on the lines) and were analyzed by non-denaturing gel electrophoresis (A). Electron micrographs of these HDL were recorded at 25 °C before (B, C) and after 1 h incubation at 95 °C (D, E).
Comparison of the effects of Cu2+ and OCl− oxidation
To test whether the effects of copper oxidation on HDL stability extend to other oxidants, we analyzed structure and thermal denaturation of human HDL that were oxidized to various stages by hypochlorite. SDS PAGE of these particles, similar to that of Cu-oxidized HDL, showed formation of apoA-I dimer, trimer and apoA-I:apoA-II heterodimer at early stages of oxidation (Fig. 6, lanes 1, 2), followed by massive intra-particle protein cross-linking into multimers at later stages (lanes 3–6). Kinetic CD data recorded in T-jumps from 25 to 95 °C of OCl-oxidized HDL (Fig. 7A, B) were comparable to similar data of Cu-oxidized HDL (Fig. 2B, C) and showed that mild oxidation (up to stage 2) accelerates protein unfolding, while more advanced oxidation (stage 3 and above) reduces the extent of the unfolding, i. e. increases the amount of residual α-helical structure retained upon heat denaturation. Moreover, CD and light scattering melting data recorded of OCl- and Cu-treated HDL showed similar trends: HDL fusion and rupture shifted to lower temperatures upon mild oxidation (up to stage 2), while more advanced oxidation (stages 3 and above) lead to high-temperature shifts in these transitions (Figs. 3C, 7E). Thus, different oxidants produce similar effects on HDL stability that depend critically on the degree of oxidation and correlate with protein cross-linking.
Figure 6.
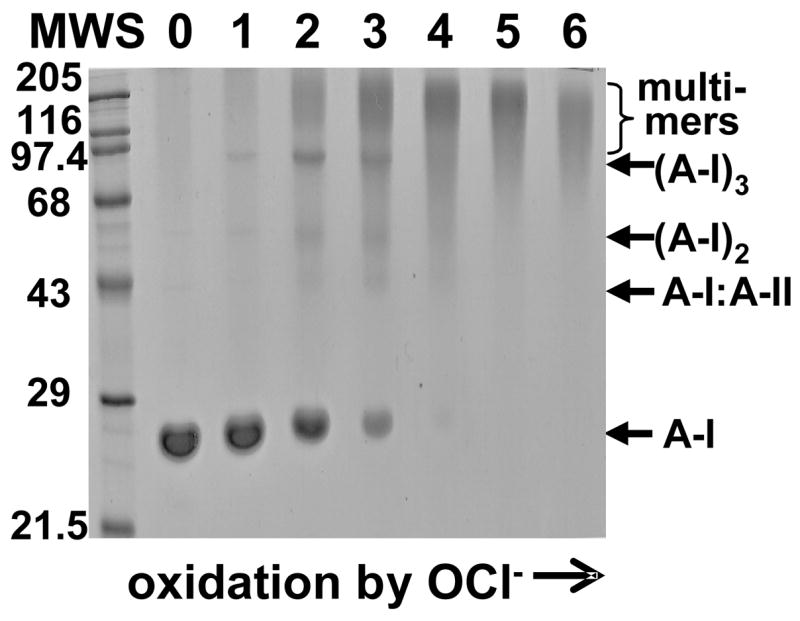
Effects of HDL oxidation to various stages by OCl− on protein cross-linking analyzed by SDS PAGE. HDL (1 mg/mL protein in buffer B) were incubated with various concentrations of NaClO (see Methods); lane numbers correspond to approximate oxidant:HDL molar ratios of (1) 7.5:1, (2) 25:1, (3) 50:1, (4) 100:1, (5) 200:1, (6) 400:1; 0 stands for nHDL. The gel was stained with Coomassie blue.
Figure 7.
Effects of oxidation by OCl− on thermal transitions in HDL. HDL samples were oxidized to stages 1–6 by incubation with various concentrations of NaOCl as described in Fig. 6 and in Methods. The samples were diluted to 33 μg/mL protein in buffer B containing 150 mM NaCl. Protein unfolding was triggered by a T-jump from 25–95 °C and was monitored by CD at 222 nm (A, B); arrows indicate changes in the unfolding rates (A) or amplitudes (B) upon increase in the oxidation degree. In other experiments, similar HDL samples were heated from 25–98 °C at a rate of 10 °C/h, and CD (not shown) and 90° light scattering melting data were recorded at 222 nm (B, C); the data are sifted along Y-axis to avoid overlap. Apparent temperature of HDL fusion as a function of oxidation degree determined from these data is plotted in (E).
To monitor oxidative modifications in HDL core, UV/visible absorption spectra were recorded from HDL that were oxidized to different stages by Cu2+ or OCl− (Fig. 8). In nHDL, absorption peaks with triple maxima extending from 430 to 485 nm are characteristic of carotenoids that are present in small amount in the lipoprotein core.42,43 These peaks rapidly declined in amplitude upon copper oxidation to stages 1, 2 and disappeared at stage 3 and above; a more gradual decline was observed in hypochlorite-oxidized HDL in which the peaks disappeared at stage 4. This is consistent with the notion that carotenoids (which are potent antioxidants preventing or repairing oxidative damage in HDL lipids and proteins) are consumed at early stages of oxidation,43 and their consumption is faster in Cu- as compared to OCl-oxidized HDL. Comparison with thin-layer chromatography data shows that carotenoid consumption is closely followed by lipolysis of phosphatidylcholine (PC) and production of lysoPC that becomes significant at oxidation stages where the carotenoids disappear (stage 3 in Cu- or stage 4 in OCl-oxidized HDL, Fig. 8).
Figure 8.
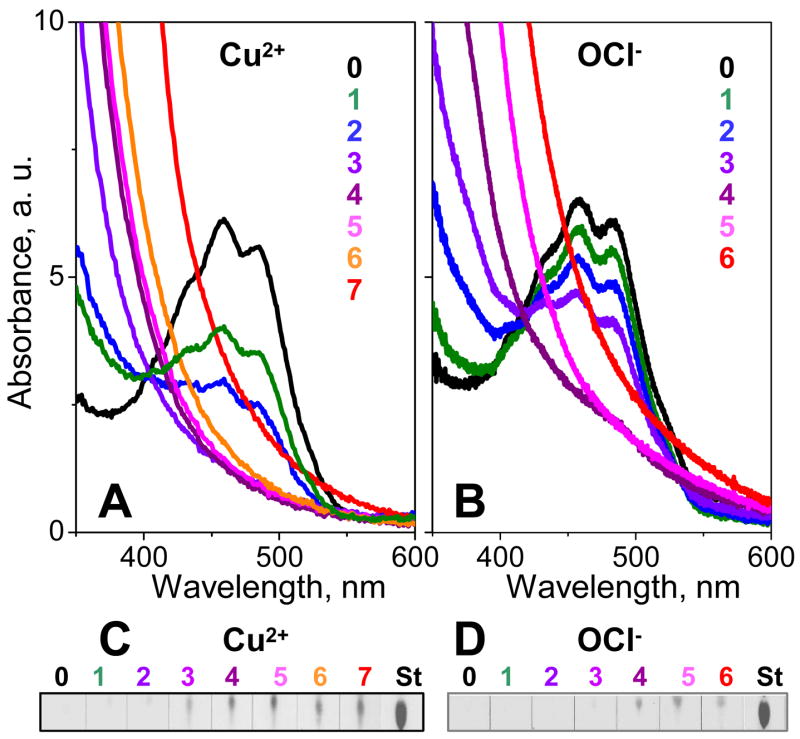
Lipid modifications at various stages of oxidation. HDL were oxidized by Cu2+ (A, C) or OCl− (B, D) to various stages (shown by numbers and color coded as in Figs. 1, 2, 7). Absorption spectra at 25 °C were recorded from 300–600 nm to monitor carotenoid consumption (A, B); at shorter wavelengths, the overall oxidation-induced increase in absorption is largely due to formation of conjugated diens (Fig. 1). LysoPC formation upon oxidation-induced PC lypolysis was monitored by thin-layer chromatography (C, D). 0 stands for nHDL, St for lysoPC standard.
In summary, copper and hypochlorite produce similar effects on HDL stability that change sign upon progression of oxidation beyond stage 2 (Figs. 1–9).
DISCUSSION
Changes in HDL stability correlate with protein modifications
Our results show, for the first time, that mild oxidation destabilizes HDL and accelerates protein dissociation and lipoprotein fusion while extensive oxidation inhibits these reactions. These results are not limited to a particular oxidant since they are valid for two different reagents: Cu2+, a one-electron oxidant that directly reacts with HDL lipids but not proteins, and OCl−, a two-electron oxidant that preferentially reacts with proteins, especially at low oxidant:HDL ratios corresponding to oxidation stages 1, 2 in Fig. 6–8.44 Thus, oxidation to stages 1, 2 in OCl-treated HDL involves almost exclusively protein modifications, while similar stages in Cu-treated HDL also involve significant modifications to the lipids. This suggests that maximal destabilization of HDL observed at oxidation stage 2 by either reagent is mainly due to protein modifications.
Oxidation of Met and aromatic groups and limited apoA-I cross-linking destabilize mHDL
What HDL modifications induced by Cu2+ or OCl− can promote or inhibit protein dissociation and particle fusion? A common protein modification by various reagents is conversion of two-to-three Met in apoA-I and the sole Met26 in apoA-II to MetO;20–22 Met86 and Met112 in apoA-I on HDL are modified by most oxidants at early stages (below stage 2), while Met 148 is modified by OCl− at later stages 21,45). Our observation of reduced thermal stability in mHDL (stage 2, Figs. 2B, 3, 4) is consistent with the earlier studies showing that MetO formation in apoA-I and apoA-II destabilizes these proteins in solution and in discoidal lipoproteins.29,30 This probably results from the introduction of polar sulfoxide (S=O) group in the lipid-binding faces of the amphipathic apolipoprotein α-helices, which weakens the hydrophobic interactions formed by these helices.29 Therefore, reduced hydrophobicity of MetO-containing apoA-I and apoA-II probably contributes to their enhanced dissociation from spherical mHDL observed upon HDL remodeling by lipid transfer protein24 or by thermal denaturation (Figs. 2B, 4).
Since MetO are formed at initial stages of oxidation yet maximal HDL destabilization is observed at stage 2, additional modifications must be involved. Such destabilizing oxidative modifications may involve aromatic residues located in the lipid-binding helical faces, such as Phe37 and Phe73 in apoA-I (whose oxidation on HDL was detected by mass spectrometry45); Trp oxidation in apoA-I (evident from the gradual reduction in the intrinsic Trp fluorescence intensity16 at oxidation stages 1–7, data not shown); and Tyr modifications by OCl−.16,19 In addition, HDL stability may be affected by apolipoprotein cross-linking into dimers and trimers observed at stage 2 (Figs 2A, 6). Such cross-linking is expected to increase the enthalpy of protein dissociation from the particle surface and hence may stabilize HDL. On the other hand, limited cross-linking of apoA-I and apoA-II on the surface of spherical HDL causes conformational changes that increase solvent exposure of particular αhelices,23,34 which may reduce protein affinity for lipid and hence destabilize HDL. In fact, oxidation of apoA-I:dimyristoyl PC complexes by OCl−, which affects solely apoA-I leading to dimer and trimer formation, progressively destabilizes these discoidal HDL, suggesting that spherical HDL are also destabilized by apoA-I cross-linking into dimers and trimers (S. J., submitted). Oxidative protein cross-linking in lipoproteins may occur via Lys or Tyr, yet Tyr are cross-linked by oxidants other than Cu2+ or OCl−,46 suggesting that HDL proteins in our studies may be cross-linked via Lys. In summary, HDL destabilization upon mild oxidation results, at least in part, from oxidation of Met and aromatic groups in apolar helical faces followed by apoA-I cross-linking into dimers and trimers.
Massive protein cross-linking and lipid re-distribution inhibit oxHDL remodeling in vitro
What protein or lipid modifications may prevent protein dissociation and HDL remodeling and fusion upon advanced oxidation? One possibility is that extensive intra-particle cross-linking of HDL proteins at oxidation stages 3–7 (Fig. 2A, 6) may prevent their dissociation from the particle surface and thereby inhibit HDL remodeling. Another cause for fusion inhibition is increase in lipid charge and polarity upon lipolysis of unsaturated acyl chains; this lipolysis becomes significant upon completion of antioxidant consumption at oxidation stages 3–4, leading to accumulation of such products as sterols, lysoPC, and free fatty acids at these and later stages (Fig. 8). For example, extensive oxidation converts apolar core lipids such as cholesterol esters into sterols and free fatty acids; these newly formed polar and charged lipids are expected to move from lipoprotein core to its surface, thereby creating excess surface material at the expense of the core, which is expected to inhibit fusion47. In summary, increased resistance of oxHDL to protein dissociation and lipoprotein fusion may result from lipid re-distribution from core to surface upon oxidative lipolysis and from extensive protein cross-linking on HDL surface.
Comparison with other lipoproteins
Lipoprotein destabilization upon mild oxidation followed by stabilization upon extensive oxidation is apparently unique to spherical HDL. In contrast, model discoidal HDL are progressively destabilized upon protein oxidation (S.J., submitted). We propose that this difference between HDL disks and spheres results mainly from oxidative modifications to core lipids that are present only in spherical particles.
Our oxidation studies of spherical plasma LDL are consistent with this notion. They show that, in contrast to HDL, LDL oxidation in vitro leads to progressive stabilization against lipoprotein fusion. We propose that this difference results, at least in part, from the differences in size and hydrophobicity of the exchangeable HDL proteins, apoA-I (243 a. a.) and apoA-II (70 a. a.), as compared to the non-exchangeable LDL protein, apoB (4536 a. a.). In contrast to apoA-I and apoA-II whose oxidative modifications, such as MetO formation or limited cross-linking, promote protein dissociation and lipoprotein fusion at early stages of oxidation, oxidative modifications in apoB are apparently insufficient to cause dissociation of this large hydrophobic non-exchangeable apolipoprotein from LDL surface. As a result, protein oxidative modifications in LDL are effectively counteracted by the antifusogenic lipid modifications, such as increased lipid charge and polarity upon lipolysis.47
Functional implications
Lipoprotein fusion and apolipoprotein dissociation are essential steps in metabolic HDL remodeling in plasma leading to formation of HDL subclasses with distinct functional properties (5,13,36,38,39 and references therein). Hence, the effects of oxidation on these reactions may have important functional implications. Our results show that these effects depend critically on the degree of HDL oxidation and change sign as oxidation progresses beyond mild stages (Fig. 3C, 7E). Since lipoprotein oxidation in vivo44 probably falls within the range of mild oxidation (stages 0–2 in our studies), functional consequences of such oxidation may also depend on its extent. In fact, Pirillo and colleagues recently reported that mild oxidation of HDL improves its function as cholesterol acceptor by increasing transporter-mediated lipid efflux from macrophages, apparently via the transient formation of small pre-β-migrating particles that include lipid-poor apoA-I, while extensive oxidation inhibits this function.28 Our results help explain this observation; we show that mild oxidation accelerates protein dissociation and HDL fusion. Thus, mild oxidation is expected to accelerate HDL metabolism and shift the population distribution among HDL subclasses towards larger particles and dissociated lipid-poor apoA-I. This lipid-poor apoA-I, which is the primary acceptor of cell phospholipids and cholesterol (28,48 and references therein), may contribute to the enhanced cholesterol uptake upon mild oxidation. In addition, protein dissociation that is promoted by mild oxidation may create packing defects in HDL surface that facilitate cholesterol incorporation into mHDL. As a result, mild oxidation in vivo may enhance HDL functions in reverse cholesterol transport. In contrast, extensive oxidation (beyond stage 3) inhibits protein dissociation and lipoprotein remodeling, which would impair reverse cholesterol transport.
In summary, oxidation by various agents may alter HDL functions not only by chemically modifying proteins and lipids but also by altering kinetic stability of the lipoprotein assembly, and thereby altering the rate of HDL remodeling and/or shifting the population distribution among HDL subclasses. A unique property of spherical HDL is that the effect of oxidation on the particle stability and remodeling changes sign, which helps explain the apparent controversy in the existing reports on HDL oxidation.
MATERIALS AND METHODS
HDL isolation
Human HDL from five healthy volunteers were used. Single-donor HDL were isolated from fresh EDTA-treated plasma by density gradient ultracentrifugation in the density range 1.063–1.21 g/mL.48 HDL migrated as a single band on agarose gel. HDL stock solution of 7.5–10 mg/mL protein concentration (measured by modified Lowry assay) was dialyzed against buffer A (10 mM Na phosphate, pH 7.5, 0.25 mM EDTA, 0.05% NaN3). The stock solution was stored in the dark at 4 °C and was used in 8 weeks during which no protein degradation was detected by SDS PAGE and no changes in the protein secondary structure or lipoprotein stability was observed by CD spectroscopy and DSC.
HDL oxidation and biochemical characterization
HDL oxidation by Cu2+ or ClO− was performed following established protocols.44,46 Briefly, HDL stock solution was dialyzed extensively at 4 °C against buffer B (10 mM Na phosphate, pH 7.5), which is the standard buffer used throughout this study. Copper oxidation of HDL (0.1–2.5 mg/mL protein in buffer B) was initiated by addition of CuSO4 (5–125 μM) to the final ratio of 50 μM oxidant to 1 mg/mL protein. Conjugated diene formation during lipid peroxidation at 37 °C was monitored by absorbance at 234 nm using Varian Cary-300 UV/visible spectrometer with thermoelectric temperature control. Oxidation was stopped by adding EDTA (using 1:50 Cu2+ to EDTA molar ratio) at different stages (marked 1–7 in Fig. 1).
Hypochlorite-induced oxidation to various stages was achieved by incubating HDL (1 mg/mL protein in buffer B) for 12 h at 37 °C with NaOCl solutions of 0.09, 0.35, 0.7, 1.3, 2.7, or 5.3 mM concentrations (marked as oxidation stages 1–6, respectively) that correspond to approximate NaOCl:HDL molar ratios of 7.5:1, 25:1, 50:1, 100:1, 200:1 or 400:1.44 NaOCl concentration was determined spectrophotometrically using molar absorption coefficient of 350 cm−1 at 292 nm.
Thin-layer chromatography was used as described47 to monitor lysoPC formation. Varian Cary-300 absorption spectrometer was used to monitor carotenoid consumption at various stages of HDL oxidation. Non-denaturing gel electrophoresis of HDL at various stages of oxidation and/or thermal denaturation was carried out by using 10% polyacrylamide homogeneous gel.37 SDS PAGE was performed using 12% or 15% homogeneous system. The gels were run at 80 V for 20 min and at 120 V for 2 h and were stained with Coomassie blue.
Circular dichroism spectroscopy and light scattering
CD data were recorded using an AVIV 215 spectropolarimeter with thermoelectric temperature control as described.37 Briefly, far-UV CD spectra (185–250 nm) and the kinetic and melting data were recorded from HDL solutions of 33 μg/mL protein concentration in buffer B containing 150 mM NaCl. Using this salt concentration destabilizes HDL and facilitates spectroscopic observation of thermal transitions below 100 °C.37 In melting experiments, the CD data were recorded at 222 nm during sample heating and cooling with 1 °C increment and 300 s accumulation time per data point, which corresponds to the scan rate of 11 °C/h; 90° light scattering at 222 nm was monitored simultaneously with the CD signal by using fluorescence accessory.50 In T-jump experiments, the sample temperature was rapidly increased at time t=0 from 25 °C to higher values, and the time course of the μ-helical unfolding was monitored at 222 nm. The CD data were normalized to protein concentration and expressed as molar residue ellipticity [Θ].
Electron microscopy
HDL were visualized at room temperature by negative staining electron microscopy using a CM12 transmission electron microscope (Philips Electron Optics) as described.36 Particle size analysis was carried out in PHOTOSHOP computer graphics using 200–300 particles per image.
Differential scanning calorimetry
Excess heat capacity, Cp(T), was measured from degassed HDL solutions of 2.5–4 mg/mL protein concentration in buffer B using an upgraded Microcal MC-2 differential scanning microcalorimeter. The data, which were recorded during sample heating from 5–115 °C at a rate of 90 °C/h, were corrected for buffer baseline and normalized to protein concentration. ORIGIN software was used for the analysis and display of CD and DSC data.
Fluorescence spectroscopy
Intrinsic Trp fluorescence was measured using a Fluoromax-2 spectrofluorometer. The spectra from HDL that were oxidized to various stages were recorded at 25 °C from 300–500 nm using 280 nm excitation wavelength and 5 nm excitation and emission slit widths.
All experiments in this study were repeated 3–5 times to ensure reproducibility.
Acknowledgments
We thank Cheryl England and Michael Gigliotti for help with lipoprotein isolation and biochemical characterization, Donald L. Gantz for expert help with electron microscopy, and Dr. Donald M. Small for reading the manuscript prior to publication. This work was supported by the National Institutes of Health grants GM067260 and HL026355.
ABBREVIATIONS
- HDL
high-density lipoprotein
- nHDL
native (non-oxidized) HDL
- mHDL
mildly oxidized HDL
- oxHDL
extensively oxidized HDL
- LDL
low-density lipoprotein
- apo
apolipoprotein
- PC
phosphatidylcholine
- MetO
methionine sulfoxide (S=O double bond)
- CD
circular dichroism
- DSC
differential scanning calorimetry
- T-jump
temperature jump
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fielding CJ, Fielding PE. Molecular physiology of reverse cholesterol transport. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 2.von Eckardstein A, Nofer JR, Assman G. High density lipoproteins and arteriosclerosis. Role of cholesterol efflux and reverse cholesterol transport. Arterioscler Thromb Vasc Biol. 2001;21:13–27. doi: 10.1161/01.atv.21.1.13. [DOI] [PubMed] [Google Scholar]
- 3.Rader DJ. Molecular regulation of HDL metabolism and function: implications for novel therapies. J Clin Invest. 2006;116(12):3090–3100. doi: 10.1172/JCI30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parthasarathy S, Barnett J, Fong LG. High-density lipoprotein inhibits the oxidative modification of low-density lipoprotein. Biochim Biophys Acta. 1990;1044:275–283. doi: 10.1016/0005-2760(90)90314-n. [DOI] [PubMed] [Google Scholar]
- 5.Kontush A, Chantepie S, Chapman MJ. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler Thromb Vasc Biol. 2003;23(10):1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 6.Stocker R, Keaney JF., Jr Role of oxidative modifications in atherosclerosis. Physiol Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- 7.Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fonarow GC, Vahabzadeh K, Hama S, Hough G, Kamranpour N, Berliner JA, Lusis AJ, Fogelman AM. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res. 2004;45(6):993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Witztum JL, Steinberg D. The oxidative modification hypothesis of atherosclerosis: does it hold for humans? Trends Cardiovasc Med. 2001;11(3–4):93–102. doi: 10.1016/s1050-1738(01)00111-6. [DOI] [PubMed] [Google Scholar]
- 9.Rohrer L, Hersberger M, von Eckardstein A. High density lipoproteins in the intersection of diabetes mellitus, inflammation and cardiovascular disease. Curr Opin Lipidol. 2004;15(3):269–278. doi: 10.1097/00041433-200406000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res. 2004;95(8):764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 11.Wu BJ, Kathir K, Witting PK, Beck K, Choy K, Li C, Croft KD, Mori TA, Tanous D, Adams MR, Lau AK, Stocker R. Antioxidants protect from atherosclerosis by a heme oxygenase-1 pathway that is independent of free radical scavenging. J Exp Med. 2006;203(4):813–816. doi: 10.1084/jem.20052321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergt C, Oram JF, Heinecke JW. Oxidized HDL: the paradox-idation of lipoproteins. Arterioscler Thromb Vasc Biol. 2003;23(9):1488–1490. doi: 10.1161/01.ATV.0000090570.99836.9C. [DOI] [PubMed] [Google Scholar]
- 13.Pussinen PJ, Metso J, Keva R, Hirschmugl B, Sattler W, Jauhiainen M, Malle E. Plasma phospholipid transfer protein-mediated reactions are impaired by hypochlorite-modification of high density lipoprotein. Int J Biochem Cell Biol. 2003;35(2):192–202. doi: 10.1016/s1357-2725(02)00130-9. [DOI] [PubMed] [Google Scholar]
- 14.Bergt C, Pennathur S, Fu X, Byun J, O’Brien K, McDonald TO, Singh P, Anantharamaiah GM, Chait A, Brunzell J, Geary RL, Oram JF, Heinecke JW. The myeloperoxidase product hypochlorous acid oxidizes HDL in the human artery wall and impairs ABCA1-dependent cholesterol transport. Proc Natl Acad Sci USA. 2004;101(35):13032–13037. doi: 10.1073/pnas.0405292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls SJ, Zheng L, Hazen SL. Formation of dysfunctional high-density lipoprotein by myeloperoxidase. Trends Cardiovasc Med. 2005;15(6):212–219. doi: 10.1016/j.tcm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Peng DQ, Wu Z, Brubaker G, Zheng L, Settle M, Gross E, Kinter M, Hazen SL, Smith JD. Tyrosine modification is not required for myeloperoxidase-induced loss of apolipoprotein A-I functional activities. J Biol Chem. 2005;280(40):33775–33784. doi: 10.1074/jbc.M504092200. [DOI] [PubMed] [Google Scholar]
- 17.Shao B, Oda MN, Oram JF, Heinecke JW. Myeloperoxidase: an inflammatory enzyme for generating dysfunctional high density lipoprotein. Curr Opin Cardiol. 2006;21(4):322–328. doi: 10.1097/01.hco.0000231402.87232.aa. [DOI] [PubMed] [Google Scholar]
- 18.Kontush A, Chapman MJ. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol Rev. 2006;58(3):342–374. doi: 10.1124/pr.58.3.1. [DOI] [PubMed] [Google Scholar]
- 19.Shao B, Oda MN, Bergt C, Fu X, Green PS, Brot N, Oram JF, Heinecke JW. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J Biol Chem. 2006;281(14):9001–9004. doi: 10.1074/jbc.C600011200. [DOI] [PubMed] [Google Scholar]
- 20.Garner B, Witting PK, Waldeck AR, Christison JK, Raftery M, Stocker R. Oxidation of high density lipoproteins. I Formation of methionine sulfoxide in apolipoproteins AI and AII is an early event that accompanies lipid peroxidation and can be enhanced by alpha-tocopherol. J Biol Chem. 1998;273(11):6080–6087. doi: 10.1074/jbc.273.11.6080. [DOI] [PubMed] [Google Scholar]
- 21.Pankhurst G, Wang XL, Wilcken DE, Baernthaler G, Panzenbock U, Raftery M, Stocker R. Characterization of specifically oxidized apolipoproteins in mildly oxidized high density lipoprotein. J Lipid Res. 2003;44(2):349–355. doi: 10.1194/jlr.M200256-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Panzenböck U, Stocker R. Formation of methionine sulfoxide-containing specific forms of oxidized high-density lipoproteins. Biochim Biophys Acta. 2005;1703(2):171–181. doi: 10.1016/j.bbapap.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Artola RL, Conde CB, Bagatolli L, Pécora RP, Fidelio GD, Kivatinitz SC. High-density lipoprotein from hypercholesterolemic animals has peroxidized lipids and oligomeric apolipoprotein A-I: its putative role in atherogenesis. Biochem Biophys Res Commun. 1997;239(2):570–574. doi: 10.1006/bbrc.1997.7496. [DOI] [PubMed] [Google Scholar]
- 24.Panzenböck U, Kritharides L, Raftery M, Rye KA, Stocker R. Oxidation of methionine residues to methionine sulfoxides does not decrease potential antiatherogenic properties of apolipoprotein A-I. J Biol Chem. 2000;275(26):19536–19544. doi: 10.1074/jbc.M000458200. [DOI] [PubMed] [Google Scholar]
- 25.Francis GA, Mendez AJ, Bierman EL, Heinecke JW. Oxidative tyrosylation of high density lipoprotein by peroxidase enhances cholesterol removal from cultured fibroblasts and macrophage foam cells. Proc Natl Acad Sci USA. 1993;90(14):6631–6635. doi: 10.1073/pnas.90.14.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang WQ, Merriam DL, Moses AS, Francis GA. Enhanced cholesterol efflux by tyrosyl radical-oxidized high density lipoprotein is mediated by apolipoprotein AI-AII heterodimers. J Biol Chem. 1998;273(28):17391–17398. doi: 10.1074/jbc.273.28.17391. [DOI] [PubMed] [Google Scholar]
- 27.Macdonald DL, Terry TL, Agellon LB, Nation PN, Francis GA. Administration of tyrosyl radical-oxidized HDL inhibits the development of atherosclerosis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2003;23:1583–1588. doi: 10.1161/01.ATV.0000085840.67498.00. [DOI] [PubMed] [Google Scholar]
- 28.Pirillo A, Uboldi P, Pappalardo G, Kuhn H, Catapano AL. Modification of HDL3 by mild oxidative stress increases ATP-binding cassette transporter 1-mediated cholesterol efflux. Cardiovasc Res. 2007;75(3):566–574. doi: 10.1016/j.cardiores.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Anantharamaiah GM, Hughes TA, Iqbal M, Gawish A, Neame PJ, Medley MF, Segrest JP. Effect of oxidation on the properties of apolipoproteins A-I and A-II. J Lipid Res. 1988;29(3):309–318. [PubMed] [Google Scholar]
- 30.Sigalov A, Stern LJ. Oxidation of methionine residues affects the structure and stability of apolipoprotein A-I in reconstituted high density lipoprotein particles. Chem Phys Lipids. 2001;113(1–2):133–146. doi: 10.1016/s0009-3084(01)00186-4. [DOI] [PubMed] [Google Scholar]
- 31.Ferretti G, Taus M, Dousset N, Solera ML, Valdiguié P, Curatola G. Physicochemical properties of copper-oxidized high density lipoprotein: a fluorescence study. Biochem Mol Biol Int. 1993;30(4):713–719. [PubMed] [Google Scholar]
- 32.Shoukry MI, Gong EL, Nichols AV. Apolipoprotein-lipid association in oxidatively modified HDL and LDL. Biochim Biophys Acta. 1994;1210(3):355–360. doi: 10.1016/0005-2760(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 33.Kontush A, Kohlschütter A, Beisiegel U. Physical properties of oxidized lipoproteins. Biochem Mol Biol Int. 1995;37(4):707–716. [PubMed] [Google Scholar]
- 34.Greilberger J, Jurgens G. Oxidation of high-density lipoprotein HDL3 leads to exposure of apo-AI and apo-AII epitopes and to formation of aldehyde protein adducts, and influences binding of oxidized low-density lipoprotein to type I and type III collagen in vitro. Biochem J. 1998;331(1):185–191. doi: 10.1042/bj3310185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gursky O, Ranjana &, Gantz DL. Complex of human apolipoprotein C-1 with phospholipid: Thermodynamic or kinetic stability? Biochemistry. 2002;41:7373–7384. doi: 10.1021/bi025588w. [DOI] [PubMed] [Google Scholar]
- 36.Mehta R, Gantz DL, Gursky O. Human plasma high-density lipoproteins are stabilized by kinetic factors. J Mol Biol. 2003;328(1):183–192. doi: 10.1016/s0022-2836(03)00155-4. [DOI] [PubMed] [Google Scholar]
- 37.Jayaraman S, Gantz DL, Gursky O. Effects of salt on thermal stability of human plasma high-density lipoproteins. Biochemistry. 2006;45:4620–4628. doi: 10.1021/bi0524565. [DOI] [PubMed] [Google Scholar]
- 38.Rye KA, Clay MA, Barter PJ. Remodelling of high density lipoproteins by plasma factors. Atherosclerosis. 1999;145(2):227–238. doi: 10.1016/s0021-9150(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 39.Barter PJ. Hugh Sinclair Lecture: The regulation and remodeling of HDL by plasma factors. Atheroscler Suppl. 2002;3(4):39–47. doi: 10.1016/s1567-5688(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 40.Malle E, Marsche G, Panzenboeck U, Sattler W. Myeloperoxidase-mediated oxidation of high-density lipoproteins: fingerprints of newly recognized potential proatherogenic lipoproteins. Arch Biochem Biophys. 2006;445(2):245–255. doi: 10.1016/j.abb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Huuskonen J, Olkkonen VM, Jauhiainen M, Sareneva T, Somerharju P, Ehnholm C. Oxidative modification of HDL3 in vitro and its effect on PLTP-mediated phospholipid transfer. Biochim Biophys Acta. 1998;1391(2):181–192. doi: 10.1016/s0005-2760(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 42.Gómez SL, Turchiello RF, Jurado MC, Boschcov P, Gidlund M, Neto AM. Characterization of native and oxidized human low-density lipoproteins by the Z-scan technique. Chem Phys Lipids. 2004;132(2):185–195. doi: 10.1016/j.chemphyslip.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Boullier A, Mazière JC, Filipe P, Patterson LK, Bartels DM, Hug GL, Freitas JP, Santus R, Morlière P. Interplay of oxygen, vitamin E, and carotenoids in radical reactions following oxidation of Trp and Tyr residues in native HDL3 apolipoproteins. Biochemistry. 2007;46(17):5226–5237. doi: 10.1021/bi602530g. [DOI] [PubMed] [Google Scholar]
- 44.Panzenböck U, Raitmayer S, Reicher H, Lindner H, Glatter O, Malle E, Sattler W. Effects of reagent and enzymatically generated hypochlorite on physicochemical and metabolic properties of high density lipoproteins. J Biol Chem. 1997;272(47):29711–29720. doi: 10.1074/jbc.272.47.29711. [DOI] [PubMed] [Google Scholar]
- 45.Bergt C, Oettl K, Keller W, Andreae F, Leis HJ, Malle E, Sattler W. Reagent or myeloperoxidase-generated hypochlorite affects discrete regions in lipid-free and lipid-associated human apolipoprotein A-I. Biochem J. 2000;346(2):345–354. [PMC free article] [PubMed] [Google Scholar]
- 46.Leeuwenburgh C, Rasmussen JE, Hsu FF, Mueller DM, Pennathur S, Heinecke JW. Mass spectrometric quantification of markers for protein oxidation by tyrosyl radical, copper, and hydroxyl radical in low density lipoprotein isolated from human atherosclerotic plaques. J Biol Chem. 1997;272(6):3520–3526. doi: 10.1074/jbc.272.6.3520. [DOI] [PubMed] [Google Scholar]
- 47.Jayaraman S, Gantz DL, Gursky O. Effects of oxidation on the structure and stability of human low-density lipoprotein. Biochemistry. 2007;46(19):5790–5797. doi: 10.1021/bi700225a. [DOI] [PubMed] [Google Scholar]
- 48.Rye KA, Barter PJ. Formation and metabolism of prebeta-migrating, lipid-poor apolipoprotein A-I. Arterioscler Thromb Vasc Biol. 2004;24(3):421–428. doi: 10.1161/01.ATV.0000104029.74961.f5. [DOI] [PubMed] [Google Scholar]
- 49.Schumaker VN, Puppione DL. Sequential flotation ultracentrifugation. Methods Enzymol. 1986;128:155–170. doi: 10.1016/0076-6879(86)28066-0. [DOI] [PubMed] [Google Scholar]
- 50.Benjwal S, Verma S, Röhm KH, Gursky O. Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 2006;15:635–639. doi: 10.1110/ps.051917406. [DOI] [PMC free article] [PubMed] [Google Scholar]