Abstract
Voltage-dependent L-type Ca2+ (CaV1.2) channels are the principal Ca2+ entry pathway in arterial myocytes. CaV1.2 channels regulate multiple vascular functions and are implicated in the pathogenesis of human disease, including hypertension. However, the molecular identity of CaV1.2 channels expressed in myocytes of myogenic arteries that regulate vascular pressure and blood flow is unknown. Here, we cloned CaV1.2 subunits from resistance size cerebral arteries and demonstrate that myocytes contain a novel, cysteine rich N terminus that is derived from exon 1 (termed “exon 1c”), which is located within CACNA1C, the CaV1.2 gene. Quantitative PCR revealed that exon 1c was predominant in arterial myocytes, but rare in cardiac myocytes, where exon 1a prevailed. When co-expressed with α2δ subunits, CaV1.2 channels containing the novel exon 1c-derived N terminus exhibited: 1) smaller whole cell current density, 2) more negative voltages of half activation (V1/2,act) and half-inactivation (V1/2,inact), and 3) reduced plasma membrane insertion, when compared with channels containing exon 1b. β1b and β2a subunits caused negative shifts in the V1/2,act and V1/2,inact of exon 1b-containing CaV1.2α1/α2δ currents that were larger than those in exon 1c-containing CaV1.2α1/α2δ currents. In contrast, β3 similarly shifted V1/2,act and V1/2,inact of currents generated by exon 1b- and exon 1c-containing channels. β subunits isoform-dependent differences in current inactivation rates were also detected between N-terminal variants. Data indicate that through novel alternative splicing at exon 1, the CaV1.2 N terminus modifies regulation by auxiliary subunits. The novel exon 1c should generate distinct voltage-dependent Ca2+ entry in arterial myocytes, resulting in tissue-specific Ca2+ signaling.
Voltage-dependent L-type Ca2+ (CaV1.2)3 channels are the primary Ca2+ entry pathway in arterial myocytes (1). Ca2+entry through CaV1.2 channels regulates multiple physiological functions, including cell contractility and gene expression (2). In small resistance arteries and arterioles, CaV1.2 channels play a dominant role in myogenic tone development and blood pressure regulation (3, 4). Smooth muscle-specific inactivation of the CaV1.2 gene (CACNA1C) dramatically reduces mean arterial blood pressure in mice, and arterial myocytes from hypertensive subjects exhibit increased L-type Ca2+ channel α1 subunit expression (3, 5). Moreover, a variety of agents that selectively block L-type Ca2+ channels are widely used to treat hypertension because of their ability to reduce arterial myocyte contractility (6). However, despite the central role of CaV1.2 channels in cardiovascular physiology and pathology, the molecular identity of these channels in myocytes of small myogenic arteries that regulate blood pressure and flow is unknown.
In most tissues, native voltage-dependent Ca2+ channels consist of a pore-forming α1 (CaV1.2) subunit and auxiliary β and α2δ subunits (7). CaV1.2 subunits contain the voltage sensor and several regulatory sites that determine the basic biophysical and pharmacological properties of the Ca2+ channel complex, whereas β and α2δ subunits regulate channel trafficking and gating kinetics (8). The human CaV1.2 gene can undergo alternative splicing at 19 out of 55 exons, leading to structural and functional CaV1.2 diversity (9). Vascular CaV1.2 subunits identified so far were cloned either from a whole organ, the rat aorta, or from a smooth muscle layer isolated by laser-capture microdissection from carotid or femoral arteries (10, 11). In each of these studies, clones were derived from conduit vessels that do not develop myogenic tone or critically influence systemic or organ blood pressure (GenBank™ accession numbers: M59786, AY830711-AY830713, Z34811, and Z34812; Refs. 10 and 11). To better understand Ca2+-dependent regulation of arterial smooth muscle physiology and to design more effective interventions for vascular pathologies associated with dysfunctional Ca2+ signaling, it is imperative to determine the molecular identity of CaV1.2 subunits expressed in myocytes of resistance size arteries. Conceivably, myocytes of myogenic arteries may express distinct subunit isoforms that confer tissue-specific physiology.
Here, we cloned full-length CaV1.2 subunits from small, myogenic cerebral arteries. Through the use of 5′-RACE, we found that CaV1.2 subunits expressed in myocytes of these arteries predominantly contain a novel, cysteine-rich N terminus encoded by alternative splicing of the CaV1.2 gene at exon 1. Electrophysiological studies indicate that the novel exon 1-derived N terminus modifies both CaV1.2 subunit current density and regulation by auxiliary subunits. Our study suggests that CaV1.2 exon 1, through alternative splicing, contributes to auxiliary subunit-mediated regulation of arterial myocyte voltage-dependent Ca2+ channels. Thus, splicing of the arterial myocyte CaV1.2 N terminus may contribute to tissue-specific Ca2+ entry and intracellular Ca2+ signaling.
EXPERIMENTAL PROCEDURES
Tissue Preparation
Procedures involving animals were approved by the Animal Care and Use Committee at the University of Tennessee. Sprague-Dawley rats (~250 g) were euthanized by peritoneal injection of sodium pentobarbital solution (150 mg/kg). For total RNA extraction, small (<200 μm) cerebral arteries, isolated cerebral artery myocytes, cardiac myocytes, aorta, heart, and brain, were prepared and placed immediately in TRIzol reagent (Invitrogen). Myocytes were enzymatically dissociated from rat cerebral arteries (~150 μm diameter) using a procedure similar to that previously described (12). Cardiac myocytes were prepared as previously described (13) and kindly provided by Dr. P. A. Hofmann in the Department of Physiology at UTHSC.
Reverse Transcription
First-strand cDNA was synthesized from ~3 μg of total RNA using oligo d(T) and reverse transcriptase (SuperScript™ III, Invitrogen).
Rapid Amplification of CaV1.2 5′-End (5′-RACE) and Cloning of Full-length CaV1.2 Subunits
Primer sequences are provided in Table S1 of supplemental information. 5′-Ends of CaV1.2 subunits were generated using the BD SMART RACE cDNA amplification kit using gene-specific primer 1 (GSP1). Nested PCR was performed using GSP2 and UPM primers. Nested PCR products were purified and ligated into the pGEM-T easy vector (Promega) and transformed in JM109 cells. Plasmids containing inserts were sequenced using a T7 promoter primer (Promega), which revealed two different CaV1.2 5′-end sequences. Full-length CaV1.2 subunits were amplified from cerebral artery cDNA with primers designed to recognize the two different 5′-ends (sense 6 and antisense 4 for the exon 1b-containing subunit, termed CaV1.2e1b, and sense 5 and antisense 4 for the exon 1c-containing subunit, termed CaV1.2e1c) using the Expand Long Template PCR System (Roche Applied Science). Antisense 4 was designed according to the highly conserved 3′-untranslated region of known CaV1.2 sequences (GenBank™ accession number: rat brain, M67515, M67516; rat aorta, M59786). PCR products were ligated into pGEM-T easy vector (Promega) for selection. Plasmids containing DNA inserts were sequenced at the University of Tennessee Molecular Resource Center.
Polymerase Chain Reaction
To identify the presence of exon 1a, exon 1b, exon 1c, β1b, β2a, and β3, two consecutive rounds of PCR were performed. The first PCR round was 20 cycles, and the second PCR round was 40 cycles. PCR reactions were started with a 2-min initial denaturation at 94 °C, then thermocycled at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s in the first round of PCR, or 60 s in the second round of PCR, followed by 72 °C for 3 or 5 min. PCR products were separated in 2% agarose gels. Primer pairs of the first round PCR were: exon 1c-176S1 and exon2-A1 for CaV1.2e1c; exon 1b-97S1 and exon 2-A1 for CaV1.2e1b; actin-2794S1 and actin-3337A1 for β actin. Primer pairs of the second round PCR were: exon 1c-201S2 and exon 1c-382A2 for CaV1.2e1c; exon 1b-137S2 and exon 1b-287A2 for CaV1.2e1b; actin-2901S2 and actin-3037A2 for β actin. A similar procedure was used to identify CaVβ subunits in arterial myocytes. Primers used for amplification of β1b (X61394, Ref. 14), β2a (M80545, Ref. 15), and β3 (M88751, Ref. 16) are listed in supplemental Table S1. pGW-β1b, pcDNA3-β2a, and pcDNA3-β3 were used as positive controls.
Identification of Exons 1a, 1b, 1c, and CaVβ Subunits in Rat Cerebral Artery Myocytes and Cardiac Myocytes
Individual arterial myocytes or cardiac myocytes were visualized using an inverted microscope (Nikon, TS100) and aspirated individually into a small glass pipette, as described previously (17). Reverse transcription was performed with 2–10 myocytes using SuperScript III reverse transcriptase (Invitrogen). Two consecutive PCR rounds were performed to identify the presence of exon 1a, 1b, 1c, β1b, β2a, and β3. PCR products were verified by sequencing.
Quantitative Real Time PCR
CaV1.2 cDNA was synthesized from 2–10 rat arterial myocytes that were collected as described above. One initial 20-cycle round of PCR was performed, and the PCR products were used as templates for real-time PCR using the QuantiTect SYBR Green PCR kit (Qiagen). The primer pairs used in identification experiments were also used for real time PCR of exon 1b and 1c, except that Exon 1c-201S2 was replaced with Exon 1c-230S2. For exon 1a, Exon 1a-134S1 and Exon 2-A1 were used in initial PCR, and Exon 1a-188S2 and Exon 1a-340A2 were used in real time PCR. Amplification efficiency was evaluated from the slope of the regression curve obtained with several dilutions of cardiac cDNA, and the specificity of primers was tested by melting curve analysis. A mean amplification efficiency of 1.92 was used for calculating the relative percentage of exon 1a, 1b, and 1c mRNA in cerebral artery myocytes and cardiac myocytes.
Construction of Expression Vectors for L-type Ca2+ Channel α1 Subunits
For electrophysiological characterization, full length CaV1.2e1b and CaV1.2e1c were subcloned from the pGEM-T easy vector into the pIRES-hrGFP II vector (Stratagene) using NotI and T4 ligase for expression. EcoRV was used to examine the ligation direction. Constructs with correct nucleotide sequences were transformed into DH5α cells, purified with the HiSpeed plasmid maxi kit (Qiagen), and stored at −20 °C. For generation of EGFP-tagged CaV1.2, CaV1.2e1b and CaV1.2e1c coding sequences were amplified from pIRES-CaV1.2e1b-hrGFP II and pIRES-CaV1.2e1c-hrGFP II, respectively, using PfuUltra II fusion HS DNA polymerase (Stratagene) with the following primers: forward, CACCGTCGACCTGCAGATATCCATCACACTGG; reverse, CACCCCGCGGCCAGGTTGCTGACATAGGACC. cDNAs were purified and ligated into pEGFP-N3 vector using SalI and SacII. The reading frames of pEGFP-CaV1.2e1c-N3 and pEGFP-CaV1.2e1b-N3 vectors were confirmed by sequence analysis.
Cell Culture and Transient Transfection
COS-1 and HEK 293 cells (ATCC) were maintained in DMEM-F12 (Cellgro) supplemented with 10% FBS under standard tissue culture conditions (5% CO2, 37 °C). Endogenous CaV1.2 subunits are not present in significant amounts in COS-1 and HEK 293 cells (18, 19). Cells for transfection were grown in 35-mm Petri dishes and transiently transfected using FuGENE6 (Roche Applied Science). COS-1 cells were used for patch clamp electrophysiology experiments following transfection with different combinations of pIRES-CaV1.2e1c-hrGFP II, pIRES-CaV1.2e1b-hrGFP II, pGW-β1b, pcDNA3-β2a, pcDNA3-β3, and pcDNA3-α2δ-1 (1 μg of each). HEK 293 cells were used for channel localization measurements because plasma membrane fluorescence originating from EGFP-tagged CaV1.2 channels could be more effectively differentiated from that in intracellular compartments than with COS-1 cells. HEK 293 cells were transfected with different combinations of pEGFP-CaV1.2e1b-N3 or pEGFP-CaV1.2e1c-N3 (0.5 μg) and pcDNA3-α2δ-1 and pGW-β1b (1 μg of each). Cells were maintained in a 95% O2/5% CO2 atmosphere at 37 °C and used for electrophysiological and fluorescence experiments between 36 and 72 h after transfection. pGW-β1b was kindly provided by Dr. Henry Colecraft (Johns Hopkins University School of Medicine), pcDNA3-β2a by Dr. Timothy J. Kamp (University of Wisconsin Medical School), and pcDNA3-β3 and pcDNA3-α2δ-1 by Dr. Diane Lipscombe (Brown University).
Patch Clamp Electrophysiology
Whole cell patch clamp recordings were acquired at room temperature using an Axo-patch 200B amplifier (Axon Instruments, Foster City, CA) and pCLAMP 8.2 or 9.2. Borosilicate glass electrodes of resistance 1–3 MΩ were filled with pipette solution containing (in mmol/liter): for COS-1 cells, Cs-MeSO3 135, CsCl 5, EGTA 5, MgATP 4, Na2GTP 0.25, HEPES 10, and glucose 10 (pH 7.2, adjusted with CsOH); for arterial myocytes, CsCl 140, EGTA 5, MgATP 4, HEPES 10, and glucose 10 (pH 7.2, with CsOH). The extra-cellular bath solution contained (in mmol/liter): for COS-1 cells, NaCl 80, TEACl 60, MgCl2 1, HEPES 10, BaCl2 5, and glucose 10 (pH 7.4, adjusted with NaOH), for myocytes, TEACl 140, MgCl2 1, HEPES 10, BaCl2 20, and glucose 10 (pH 7.4, adjusted with TEAOH). Myocyte voltage-dependent Ba2+ currents were isolated by subtracting Cd2+ (250 μM)-insensitive currents from whole cell currents. Solutions were ~300 mOsm, as measured using a Vapor Pressure Osmometer. Isolated cells that were not visibly attached to neighboring cells were used for patch clamp. Cell capacitance was measured by applying a 5-mV test pulse and correcting the capacitance transients with series resistance compensation. For measurement of current-voltage (I-V) relationships, cells were clamped at −80 mV and whole cell currents were evoked every 5 s by 300-ms step depolarizations to between −60 and +60 mV in 10-mV increments. Steady-state inactivation was measured every 15 s by providing 1-s conditioning pulses to between −80 and +60 mV (+30 mV for α1/β3/α2δ) in 10-mV increments before a 200-ms test pulse to 0 mV. A 15-ms return to −80 mV was applied after the conditioning pulse and prior to the test pulse. Tail currents were generated by repolarization to −80 mV after a series of 20-ms test pulses to between −60 mV and +60 mV in 10-mV increments. Whole cell currents were filtered at 1–2 kHz and digitized at 4 – 10 kHz. P/4 protocols were used to subtract leak and capacitive transients. Steady-state inactivation curves and tail currents were fit with the Boltzmann function in Equation 1,
| (Eq. 1) |
where I/Imax is the normalized peak current, V is the conditioning pre-pulse voltage, V1/2 is the voltage for half-inactivation for steady-state inactivation or half-activation for tail currents, k is the slope factor, Rin is the proportion of non-inactivating current, Rmax is the maximal current. Inactivation time constants (τ) were obtained using Equation 2,
| (Eq. 2) |
where It is the inward current at time t, and Iθ is the residual current.
Confocal Microscopy
HEK 293 cells transfected with EGFP-tagged CaV1.2 α1 subunits and either α2δ or α2δ + β1b were visualized using a Zeiss LSM 5 Pascal laser-scanning confocal microscope. EGFP was excited using 488 nm light, and >510 nm light was collected. Analysis methodology similar to that previously described was used to calculate plasma membrane localized fluorescence originating from EGFP-tagged CaV1.2 α1 subunits containing either the exon 1b or exon 1c-derived N terminus (20). Briefly, membrane boundaries were established by visualizing DIC images using a membrane thickness of 0.55 μm. Using these boundaries, membrane localized fluorescence was calculated from background subtracted fluorescence images. For each cell, total membrane-localized fluorescence was calculated by subtracting cytoplasmic fluorescence from total cellular fluorescence. Total membrane fluorescence was then divided by membrane area (in μm2) to establish average membrane fluorescence. Image analysis was performed using Image J software (NIH).
Data Analysis
Electrophysiological data were analyzed using Clampfit 8.2 and 9.2 and Origin 7.5. Values are expressed as means ± S.E. Student’s t-tests were used for comparing paired or unpaired data, and one-way analysis of variance and Student-Newman-Keuls or Dunn posthoc tests were used for comparing multiple data sets. p < 0.05 was considered significant.
RESULTS
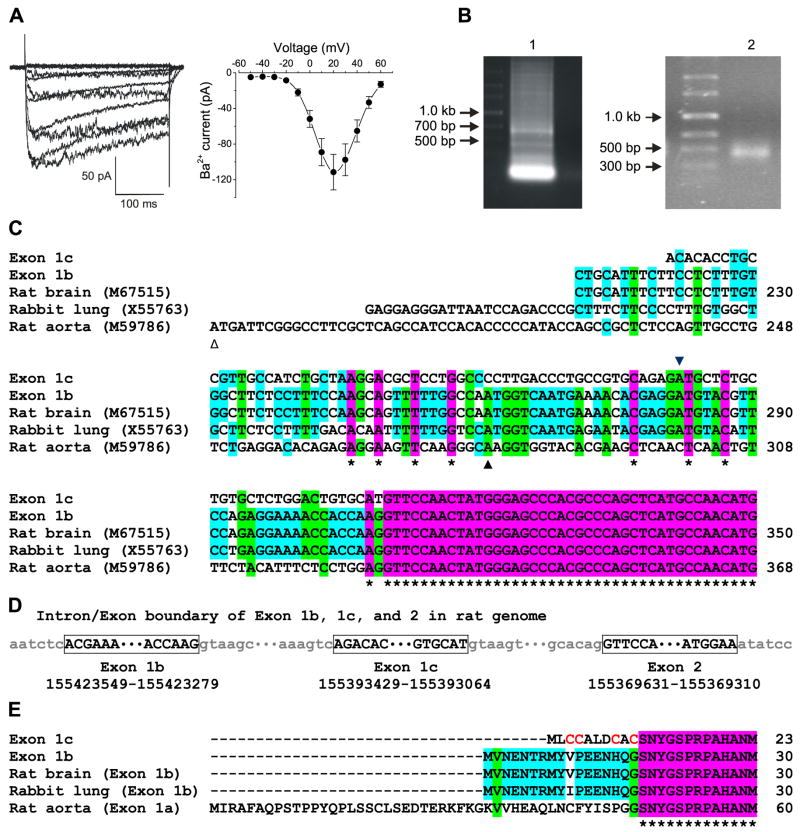
To identify voltage-dependent Ca2+ channels present in myocytes of small cerebral arteries, we first measured currents in these cells using patch clamp electrophysiology. Fig. 1A illustrates Cd2+ (250 μM)-sensitive, voltage-dependent Ba2+ currents characteristic of CaV1.2 that were recorded in cerebral artery myocytes (Fig. 1A). To proceed to clone full-length CaV1.2 subunits from small cerebral artery myocytes using PCR, we first sought to identify the CaV1.2 5′-end sequence(s) expressed in these cells. Two different CaV1.2 N termini have previously been described (21). Exon 1a encodes the CaV1.2 subunit N terminus found in whole heart (22) and whole aorta(10), and exon 1b (also referred to as exon 1) encodes the N terminus detected in whole brain (23), lung (24), and jejunum CaV1.2 subunits (25). CaV1.2 5′-ends expressed in resistance size (<200 μm diameter) cerebral arteries were amplified using 5′-RACE and ligated into pGEM-T easy vector (Fig. 1B). Sequencing analysis of 11 plasmids revealed two different CaV1.2 subunit 5′-ends (Fig. 1C). One did not match any sequence previously described, but was mapped within CACNA1C of the rat genome. The second sequence was identical to the previously described exon 1b. Exon 1a was not detected by 5′-RACE. According to their relative location within the rat genome, the sequence previously described in brain, lung and jejunum (exon 1) was termed “exon 1b”, and the novel 5′-end was termed “exon 1c” (Fig. 1, C and D). Exon 1c is responsible for encoding a 9 amino acid peptide, and substituting glycine at the exon 1/2 junction for cysteine (Fig. 1E). This substitution generates the cysteine-rich N-terminal sequence: MLCCALDCAC.
FIGURE 1. Identification of the CaV1.2 exon 1c in rat cerebral arteries, relative location of exon 1c in the rat genome, and sequence comparison with exons 1a and 1b.
A, voltage-dependent Ba2+ currents recorded in an isolated myocyte by depolarizing voltage steps (left panel) and a mean I-V relationship obtained from 10 cells (right panel). B, 5′-RACE products from cerebral arteries run in 0.8% agarose gel (left panel) with nested PCR products illustrating a band at ~500 bp (right panel). C, homology of exons 1a, 1b, and 1c compared with CaV1.2 nucleotide sequences from rat brain (M67515), rabbit lung (X55763), and rat aorta (M59786). The initiation sites of exon 1a, 1b, and 1c are indicated by △, ▲, and ▼, respectively. Part of exon 2 is also illustrated in purple. D, schematic diagram depicting the relative locations of exon 1b, 1c, and 2 within the rat genome. E, alignment of amino acid sequences encoded by CaV1.2 exon 1a, 1b, and 1c with part of the exon 2-encoded sequence shown in purple. * indicates identical nucleotides or amino acids.
To examine whether exon 1b and 1c were expressed in arterial myocytes, RT-PCR was performed using small groups (~10) of manually selected cerebral artery myocytes (see “Experimental Procedures”). Critically, this methodology avoids contamination from other vascular wall cell types, including fibroblasts and perivascular neurons that express voltage-dependent Ca2+ channel subunits, including CaV1.2 (26, 27). Both exons 1b and 1c were detected in cerebral artery myocytes (Fig. 2A). Next, to examine whether exon 1c exhibits arterial myocyte-specific expression, real-time PCR was performed on dissociated arterial and cardiac myocytes. Data indicated that exon 1c was predominant (~96%) in cerebral artery myocytes (Fig. 2B) Exons 1a, 1b, and 1c were all detected in cardiac myocytes, with exon 1a prevalent (~82%) and exons 1b and 1c each less than 15% of total message (Fig. 2B). Together, these data indicate that in the cardiovascular system, exon 1c message is predominant in arterial myocytes and scarce in cardiac myocytes.
FIGURE 2.
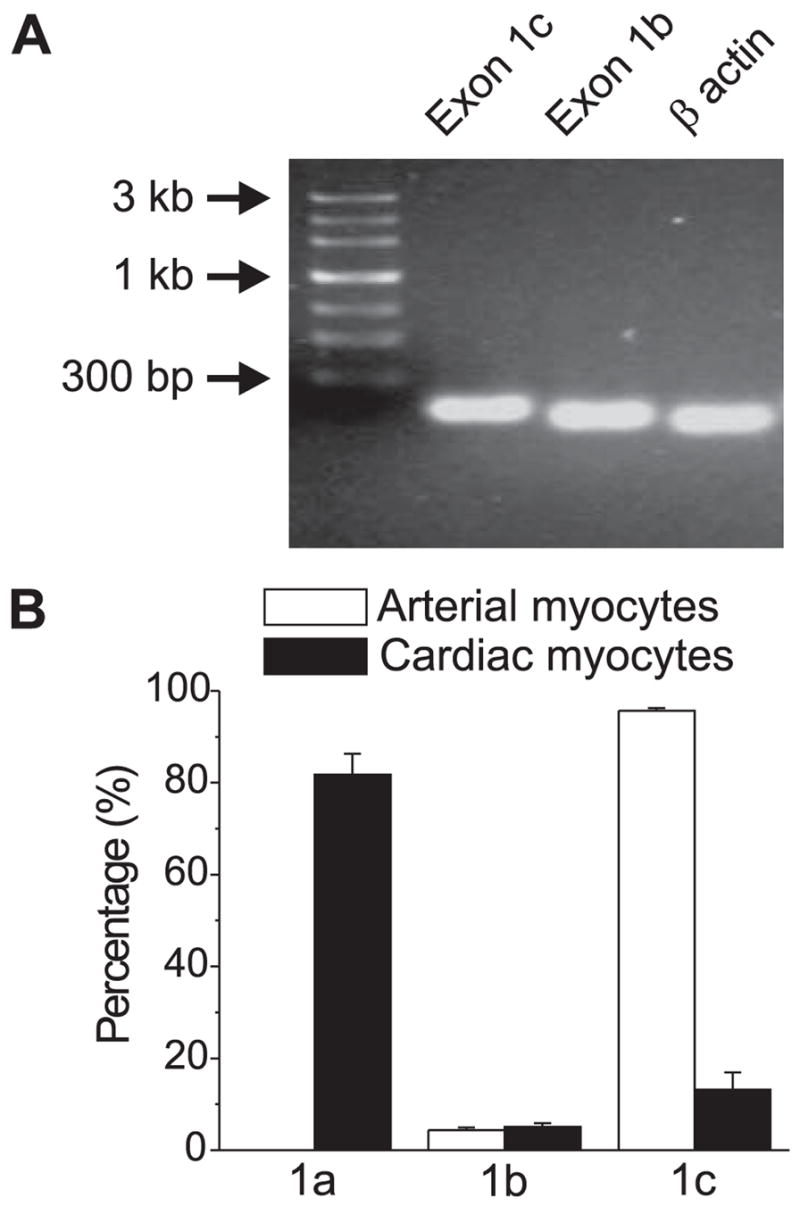
A, RT-PCR revealed the presence of exons 1b and 1c in dissociated cerebral artery myocytes. β Actin is shown as a positive control. B, real-time PCR indicated the relative percentage of exons 1a, 1b, and 1c message in isolated rat cerebral artery myocytes and cardiac myocytes. Exon 1a was not detected in arterial myocytes.
Using 5′-end primers specific to either exon 1b or exon 1c, full length CaV1.2 cDNAs were amplified (forward primers: sense6 for exon 1b, sense5 for exon 1c; reverse primer: anti-sense4), and termed “CaV1.2e1b” (GenBank™ accession number: DQ538522) and “CaV1.2e1c” (AY974797), according to their exon 1 sequences (Fig. 3, A and B). The coding region of the CaV1.2e1c subunit contains 6,477 nucleotides (including the stop codon) and encodes a 2,158-amino acid protein, whereas CaV1.2e1b contains 6,498 nucleotides and encodes a 2,165-amino acid protein. CaV1.2e1b and CaV1.2e1c are identical except for their exon 1-derived N termini. Each variant contains exon 8, 9 + 9a, 21, and 32 + 33, consistent with CaV1.2 subunits identified in large, conduit arteries (11, 28).
FIGURE 3. Full-length CaV1.2 cDNAs cloned from rat small cerebral arteries.
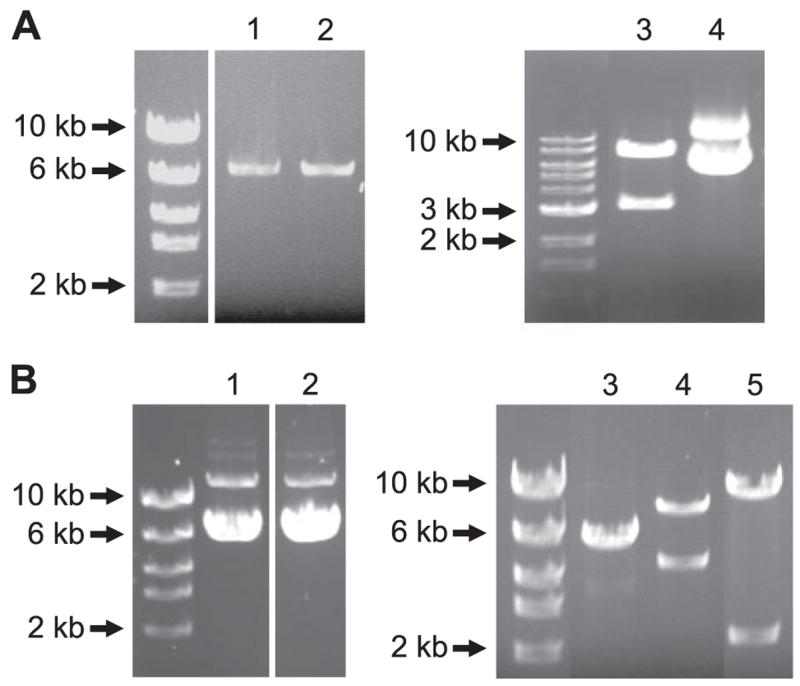
A, full-length CaV1.2e1b and CaV1.2e1c were cloned by RT-PCR and sub-cloned into pGEM-T Easy vector. Lane 1, CaV1.2e1b; lane 2, CaV1.2e1c; lane 3, CaV1.2e1c was released from pGEM-T easy vector by NotI digestion; lane 4, pGEM-T easy-CaV1.2e1c. B, CaV1.2e1b and CaV1.2e1c were subcloned into pIRES-hrGFP II vector, and the orientation direction of CaV1.2 in the expression vector was revealed by EcoRV digestion. Lane 1, pIRES-CaV1.2e1c-hrGFP II; lanes 2 and 3, pIRES-CaV1.2e1b-hrGFP II; lane 4, EcoRV digestion product of pIRES-CaV1.2e1b(+)-hrGFP II; and lane 5, EcoRV digestion product of pIRES-CaV1.2e1b(−)-hrGFP II.
To investigate whether exon 1 splicing imprints a distinct functionality to the voltage-dependent Ca2+ channel in arterial myocytes, CaV1.2e1b or CaV1.2e1c were subcloned into pIRES-hrGFP II vector and transfected in COS-1 cells for electrophysiological characterization. Voltage-dependent Ba2+ currents (IBa) were absent in non-transfected cells (data not shown). When co-expressed with α2δ, maximal IBa for both CaV1.2e1b and CaV1.2e1c occurred at +10 mV (Fig. 4, A and B, Table 1). However, the mean peak current density of CaV1.2e1b/α2δ currents was ~2-fold larger than for CaV1.2e1c/α2δ currents (Fig. 4, B and C, Table 1). Voltages of steady-state half-inactivation (V1/2,inact) and half-activation (V1/2,act) of CaV1.2e1c/α2δ currents were also ~7 and ~5 mV more negative than those of CaV1.2e1b/α2δ currents, respectively (Fig. 4, B and C, Table 1). In contrast, inactivation rate constants of CaV1.2e1c/α2δ and CaV1.2e1b/α2δ currents were similar (Fig. 4D). These data indicate that when co-expressed with α2δ, the novel exon 1c-derived CaV1.2 N terminus attenuates membrane current density and induces a negative shift in both V1/2,act and V1/2,inact, when compared with currents mediated by channels containing the CaV1.2 exon 1b-derived N terminus.
FIGURE 4. Current-voltage (I-V) relationships of CaV1.2e1b and CaV1.2e1c channels when expressed with auxiliary subunits.
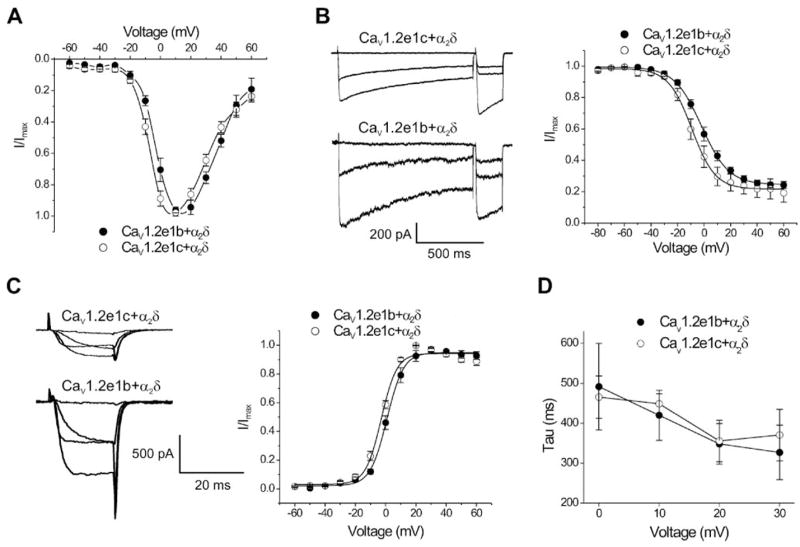
Ba2+ currents were normalized to facilitate comparison. A, I-V relationships of CaV1.2e1b and CaV1.2e1c when co-expressed with α2δ. B, steady-state inactivation of CaV1.2e1c and CaV1.2e1b currents when co-expressed with α2δ. Exemplar current traces of 1-s conditioning depolarizing pulses evoked at −80, +10, and +30 mV followed by 200-ms test pulses to 0 mV (left panel). Cell capacitances for original recordings were: CaV1.2e1c + α2δ, 90 pF; CaV1.2e1b + α2δ, 66 pF. Mean steady-state inactivation fit with a Boltzmann function (right panel). C, voltage-dependent activation of CaV1.2e1c and CaV1.2e1b currents when co-expressed with α2δ. Exemplar tail currents evoked by repolarization to −80 mV after depolarizing test pulses to −20, 0, 20, and 40 mV (left panel). Cell capacitances for original recordings were: CaV1.2e1c + α2δ, 98 pF; CaV1.2e1b + α2δ, 66 pF. Mean voltage-dependent current activation (right panel). D, inactivation of CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents were best fit with a single exponential function. Tau was similar for CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents.
TABLE 1.
Electrophysiological properties of CaV1.2 channels containing e1b or e1c when expressed alone or co-expressed with auxiliary subunits
| Combinations | I-V
|
Steady-state inactivation
|
Activation
|
|||||
|---|---|---|---|---|---|---|---|---|
| Ipeak density | n | V1/2,inact | ΔV1/2,inacta | n | V1/2,act | ΔV1/2,actb | n | |
| pA/pF | mV | mV | mV | mV | ||||
| CaV1.2e1cα1/α2δ | −5.1 + 0.9 | 6 | −9.3 + 1.8 | 5 | −3.0 + 0.8 | 5 | ||
| CaV1.2e1cα1/β1b/α2δ | −18.8 + 4.2c | 9 | −14.6 + 0.9c | −5.3 | 8 | −7.3 + 1.6 | −4.3 | 7 |
| CaV1.2e1cα1/β2a/α2δ | −19.2 + 3.9c | 8 | −12.9 + 1.0 | −3.6 | 7 | −14.9 + 0.7c | −12.0 | 6 |
| CaV1.2e1cα1/β3/α2δ | −28.6 + 6.8c | 11 | −19.4 + 1.3c | −10.1 | 8 | −11.3 + 1.0c | −8.4 | 6 |
| CaV1.2e1bα1/α2δ | −10.1 + 1.7d | 5 | −1.9 + 2.0d | 5 | 1.5 + 1.2d | 5 | ||
| CaV1.2e1bα1/β1b/α2δ | −20.9 + 4.7e | 8 | −17.7 + 0.7e | −15.8 | 8 | −8.1 + 1.3e | −9.6 | 8 |
| CaV1.2e1bα1/β2a/α2δ | −17.1 + 3.6e | 10 | −14.1 + 1.4e | −12.2 | 9 | −15.7 + 1.2e | −17.2 | 9 |
| CaV1.2e1bα1/β3/α2δ | −27.1 + 5.9e | 10 | −16.2 + 1.7e | −14.3 | 7 | −7.5 + 1.1d,e | −9.0 | 5 |
ΔV1/2,inact, β subunit-induced shift in V1/2,inact of corresponding CaV1.2α1/α2δ channels.
ΔV1/2,act: β subunit-induced shift in V1/2,act of corresponding CaV1.2α1/α2δ channels.
p < 0.05 CaV1.2e1cα1/β/α2δ versus CaV1.2e1cα1/α2δ.
p < 0.05 CaV1.2e1b versus corresponding CaV1.2e1c.
p < 0.05 CaV1.2e1bα1/β/α2δ versus CaV1.2e1bα1/α2δ.
Ca2+ channel β subunits regulate CaV1.2 channel voltage-dependence in an isoform-specific manner. For example, β2a causes slow inactivation when compared with other β subunits (29, 30). Expression of β subunits has been investigated in whole aorta (31), but not in myocytes of small, resistance size arteries. Here, we investigated the transcription of three different β subunits. Using RT-PCR, we studied β1b and β2a, which are membrane associated, and β3, which is cytosolic (8). β1b and β3 were detected in dissociated cerebral artery myocytes, whole aorta, and brain (Fig. 5, A and C). Although β2a was present in whole cerebral artery, aorta, and brain, message was not detected in isolated cerebral artery myocytes (Fig. 5B). Data suggest that β2a expression in arterial myocytes is either very low, or that β2a is expressed predom inantly in cerebral artery wall cell types other than myocytes (e.g. neurons).
FIGURE 5. RT-PCR of CaVβ subunits expressed in dissociated rat cerebral arterial myocytes.
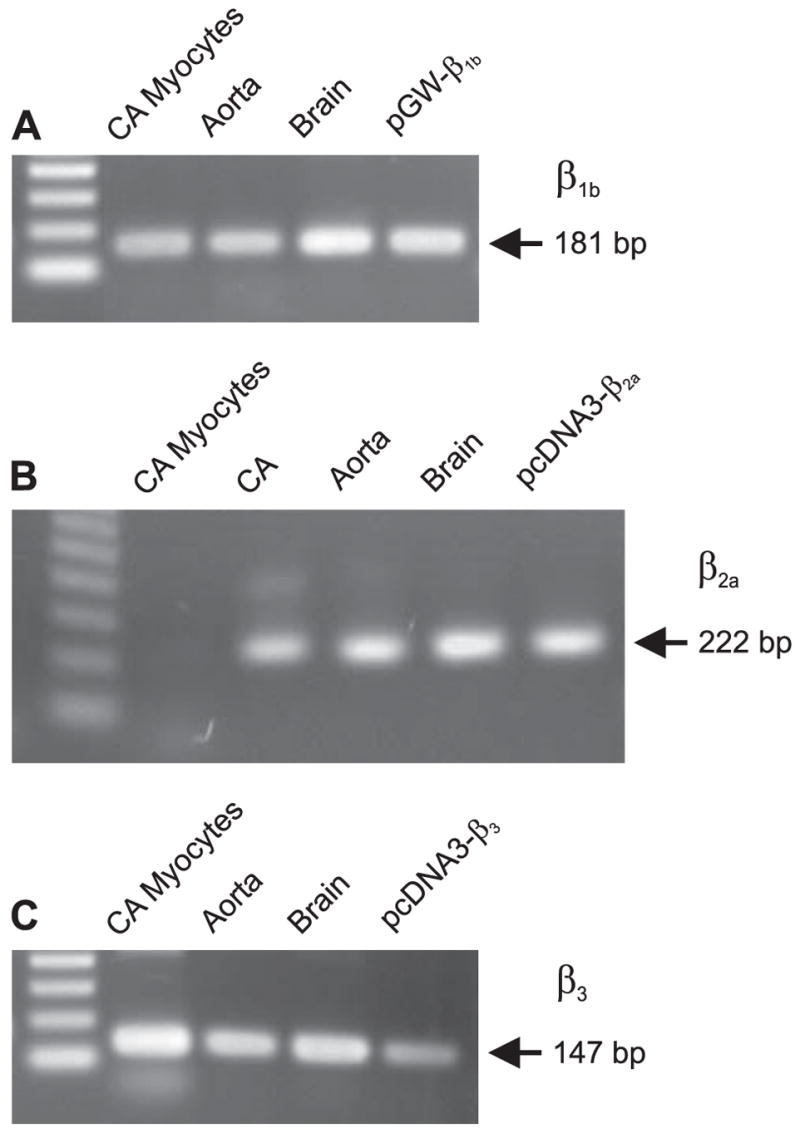
A, β1b subunit. B, β2a subunit. C, β3 subunit. CA indicates cerebral artery.
We sought to explore the functional effects of N-terminal splicing on β subunit regulation of CaV1.2. Although β2a was not detected in cerebral artery myocytes, β2a may be expressed in other cell types that contain the exon 1c CaV1.2 variant. Thus, β1b, β2a, or β3 regulation of CaV1.2e1b and CaV1.2e1c currents was investigated. Fig. 6 illustrates raw current traces for CaV1.2e1c and mean data obtained with CaV1.2e1b and CaV1.2e1c when co-expressed with α2δ and each β subunit isoform. Co-expression of β1b, β2a, or β3 with α2δ increased CaV1.2e1b and CaV1.2e1c current density and eliminated the current density difference that occurred between CaV1.2e1b/α2δ and CaV1.2e1c/α2δ (Table 1). β subunits also shifted I-V relationships further to the left, with maximal IBa occurring at ~0 mV. However, β1b shifted the V1/2,inact of CaV1.2e1b/α2δ currents by ~−16 mV, compared with only an ~−5 mV shift in the V1/2,inact of CaV1.2e1c/α2δ currents. At 0 and +10 mV, the slow component of inactivation (τslow) decayed more rapidly for CaV1.2e1b/α2δ/β1b currents than for CaV1.2e1c/α2δ/β1b currents, whereas the fast component (τfast) was similar (Fig. 6G). β2a caused an ~−12 mV shift in the V1/2,inact of CaV1.2e1b/α2δ currents, but had no effect on the V1/2,inact of CaV1.2e1c/α2δ currents. Current inactivation rate was similar for CaV1.2e1b/α2δ/β2a and CaV1.2e1c/α2δ/β2a (Fig. 6H). β3 similarly shifted the V1/2,inact of CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents; by −14 and −10 mV, respectively (Table 1). In contrast to effects with β1b, between −10 and +10 mV τslow decayed more slowly for CaV1.2e1b/α2δ/β3 currents than for CaV1.2e1c/α2δ/β3 currents, although τfast was similar (Fig. 6I).
FIGURE 6. Inactivation of CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents when expressed with β1b, β2a, or β3 subunits.
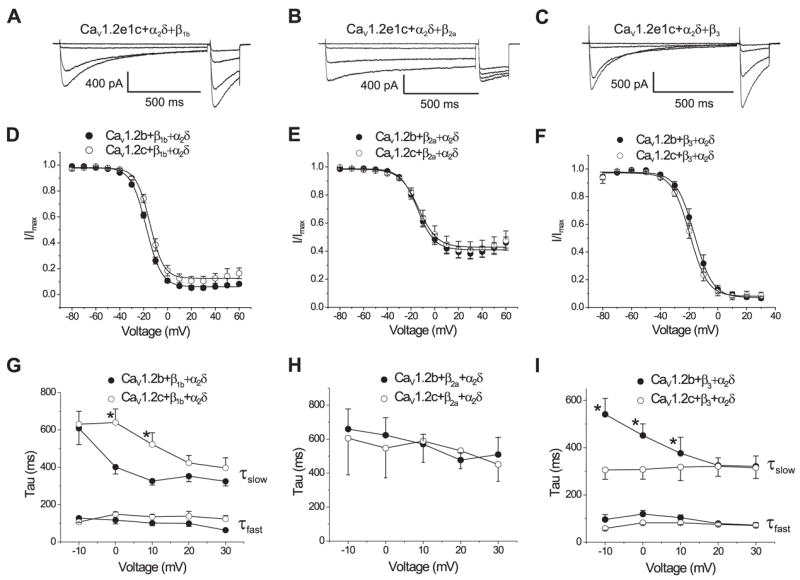
A–C, original recordings of 1-s depolarizing pulses to −80, −20, −10, and 0 mV followed by 200-ms test pulses to 0 mV for each subunit combination indicated. D–F, voltage dependence of steady-state inactivation for each subunit combination. G–I, voltage dependence of inactivation constants for each combination specified. Inactivation of CaV1.2/α2δ/β1b and CaV1.2/α2δ/β3 currents were best fit with a bi-exponential function representing τfast and τslow, whereas CaV1.2/α2δ/β2a current inactivation was best fit with a single exponential. * illustrates p < 0.05.
A similar pattern was observed with β subunit-mediated regulation of voltage-dependent activation. β1b and β2a caused a larger negative shift in the V1/2,act of CaV1.2e1b/α2δ currents than in CaV1.2e1c/α2δ currents (Table 1). In contrast, β3 similarly shifted the V1/2,act of CaV1.2/α2δ currents, regardless of the exon 1 splice variant. Collectively, these data indicate that alternative splicing of the exon 1-derived CaV1.2 N terminus modifies regulation by β subunits and suggest that modifications in current phenotype are specific to particular β subunit isoforms.
To further study membrane insertion of CaV1.2 containing exon 1b or 1c, vectors that express EGFP fused to the C terminus of CaV1.2e1b or CaV1.2e1c channels were constructed. Membrane localization of expressed channels was studied using confocal microscopy. In agreement with current density measurements, when co-expressed with α2δ membrane fluorescence intensity of CaV1.2e1b-EGFP channels was higher than for CaV1.2e1c-EGFP channels (Fig. 7, A and B). Co-expression of β1b further increased CaV1.2e1b-EGFP and CaV1.2e1c-EGFP fluorescence intensity and normalized the difference between the N-terminal isoforms that occurred when each was co-expressed with only α2δ (Fig. 7, A and B). These data indicate that alternative splicing between exon 1b and 1c modifies CaV1.2 channel membrane insertion in the presence of α2δ, and this difference can be normalized by co-expression of a β subunit.
FIGURE 7. Membrane localization of EGFP-tagged CaV1.2e1b and CaV1.2e1c channels when expressed with α2δ or with α2δ + β1b subunits in HEK 293 cells.
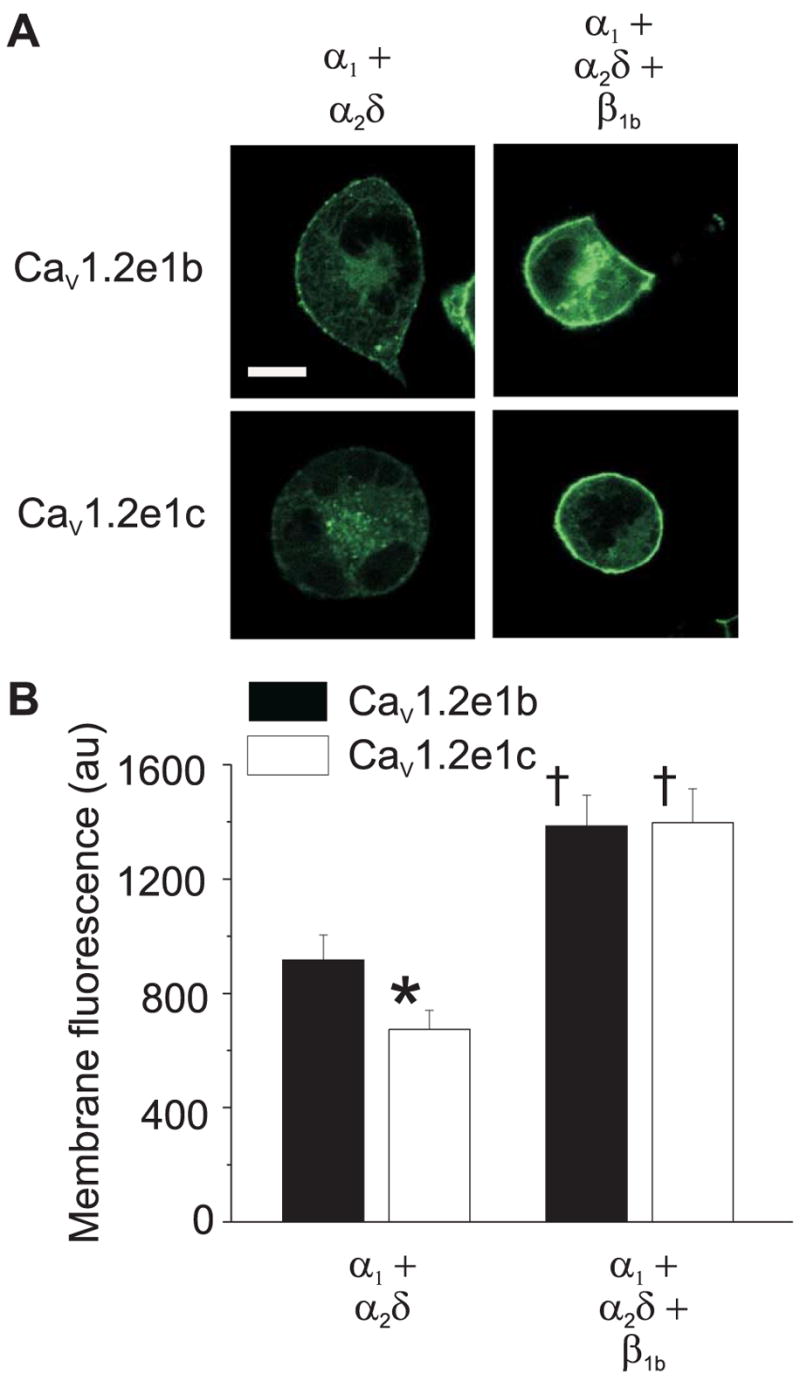
A, representative images illustrating cellular fluorescence from EGFP-tagged CaV1.2e1b and CaV1.2e1c channels when expressed with subunit combinations indicated. Scale bar, 10 μm. B, mean data illustrating that co-expression of α2δ subunits elevated localized membrane fluorescence intensity of CaV1.2e1b more than for CaV1.2e1c, and that co-expression with α2δ + β1b subunits further increased membrane fluorescence and normalized the difference between CaV1.2e1b/α2δ and CaV1.2e1c/α2δ. au indicates arbitrary units. Number of cells: CaV1.2e1b/α2δ, 18; CaV1.2e1c/α2δ, 16; CaV1.2e1b/α2δ/β1b, 17; CaV1.2e1c/α2δ/β1b, 20. * indicates p < 0.05 when compared with CaV1.2e1b + α2δ, and †, p < 0.05 when compared with the same α1 subunit + α2δ.
DISCUSSION
In the present study, we have for the first time cloned full-length CaV1.2 subunits expressed in small, resistance size, arteries and discovered a novel 5′-end sequence generated by alternative splicing at exon 1. Termed “exon 1c”, this sequence encodes a unique N terminus in which 4 out of 9 amino acids are cysteine residues. Quantitative PCR indicates that the vast majority (~96%) of CaV1.2 mRNA in isolated cerebral artery myocytes contains exon 1c, with the residual proportion containing exon 1b, the previously reported “brain” CaV1.2 subunit exon 1 (23). In contrast, exon 1a was entirely absent in cerebral artery myocytes, as determined by both 5′-RACE and quantitative PCR. While exons 1a, 1b, and 1c were all detected in isolated cardiac myocytes, exon 1a was prevalent and the relative expression of exon 1c (~13%) was much lower than in arterial myocytes. Therefore, in the cardiovascular system, exon 1c is likely to be predominant in arterial myocyte CaV1.2 subunits, whereas exon 1a would be prevalent in cardiac myocyte CaV1.2. Two different promoters drive the expression of CaV1.2 channels containing exon 1a- and 1b-derived N termini in heart and smooth muscle, which may explain their different expression profiles (32, 33). However, promoters which drive exon 1c expression are unclear.
Electrophysiological and imaging studies revealed that when co-expressed with α2δ subunits, current density generated by CaV1.2e1b was significantly greater than for CaV1.2e1c. In addition, when co-expressed with α2δ, membrane fluorescence intensity of EGFP-tagged CaV1.2e1b channels was higher than that of CaV1.2e1c channels. Four genes encode α2δ subunits, and α2δ-1, the isoform used here, increases CaV1.2 membrane insertion and alters channel inactivation (34). α2 is an extracellular domain that associates with the plasma membrane through a disulfide-bond with the membrane-spanning δ domain (8). Glycosylation of the α2 domain is essential for α2δ-mediated membrane insertion of CaV1.2 subunits (35). In contrast, δ mimics the effects of full-length α2δ on channel kinetics (36). Each N terminus might interact differently with α2δ, leading to the observed differences between CaV1.2e1b and CaV1.2e1c channels. Alternatively, differences in CaV1.2e1b and CaV1.2e1c channels might be interpreted as different conformational channel states controlled by the different N termini of CaV1.2 subunits, which only become evident in the presence of α2δ subunits. Regardless of the molecular underpinnings, our study indicates that alternative splicing of exon 1 alters both channel voltage sensitivity and current density.
β subunits regulate CaV1.2 properties by binding to the Alpha Interaction Domain (AID) in the I-II linker (37). However, because a single amino acid mutation in the AID does not eliminate β-subunit-mediated modulation of CaV1.2 subunits, additional interaction sites between α1 and β subunits likely exist (38). Our data revealed that β1b and β3 were present in isolated cerebral artery myocytes, whereas β2a was found only in intact cerebral arteries. Only one rat β2a sequence has been described, but multiple splice variants of human β2a exist (NCBI search, November 6, 2006) (39, 40). The primers used in our study to detect β2a message recognize a rat β2a sequence that corresponds to a highly conserved region in all five human splice variants. Thus, it appears to be unlikely that β2a splice variants not recognized by the primers used here exist in rat arterial smooth muscle. These data underscore the need for caution when extrapolating RT-PCR data obtained using whole arterial cDNA to message levels present in myocytes. Future studies will determine whether exon 1c is also predominant in myocytes of other arteries, or is a specific cerebral artery splice variant.
β3 is a cytosolic-localized protein, whereas β1b and β2a associate with the plasma membrane through either an acidic motif within the C terminus or palmitoylation sites in the N terminus, respectively (41, 42). When compared with currents obtained with α2δ alone, all three β subunits elevated current density, with a greater increase seen in CaV1.2 channels containing exon 1c than in channels containing exon 1b, which abolished the current density difference between CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents. β1b also normalized the difference in membrane fluorescence intensity between CaV1.2e1b/α2δ and CaV1.2e1c/α2δ channels. β3 caused similar shifts in both the V1/2,inact and V1/2,act of CaV1.2e1b/α2δ and CaV1.2e1c/α2δ currents, while β1b and β2a caused larger negative shifts in CaV1.2e1b/α2δ currents than in CaV1.2e1c/α2δ currents. When co-expressed with either β1b or β3, the decay of CaV1.2/α2δ currents containing e1b or e1c was best fit by a bi-exponential function. Substitution of e1b for e1c decelerated the slow decay component of CaV1.2/α2δ/β1b currents. In contrast, e1c accelerated the slow decay of CaV1.2/α2δ/β3 currents, when compared with e1b. The rates of decay of CaV1.2/α2δ or CaV1.2/α2δ/β2a currents were best fit with a single exponential function and were similar in CaV1.2 currents containing either the exon 1b or exon 1c-derived N terminus. Collectively, these data suggest that the CaV1.2 N terminus modifies β subunit-mediated channel regulation. These findings also indicate that in the presence of a β subunit, not only is the rate of slow inactivation modulated by splicing of the N terminus, but also that kinetic alterations depend on the β subunit isoform involved. In contrast, exon 1b or 1c-derived N termini do not appear to significantly alter fast inactivation.
CaV1.2e1c contains 4 cysteine residues, which are putative palmitoylation sites. One function of palmitoylation is to aid protein targeting to lipid rafts in the plasma membrane (43). Conceivably, different local lipid microenvironments may explain distinctions between CaV1.2e1b and CaV1.2e1c currents. It is also possible that the cysteine-rich N terminus regulates CaV1.2e1c channels by interacting with other cellular molecules and regulatory proteins, since cysteine-cysteine interactions that occur through disulfide bridging can alter channel folding and protein-protein interactions (44, 45). Immobilization of the CaV1.2 N terminus also inhibits both voltage- and Ca2+-dependent inactivation (19). Although not defined here, our results pave the way for future studies to determine the full range of mechanisms by which exon 1b and 1c encoded N termini regulate CaV1.2 channel phenotypes.
In native arterial myocytes, the current phenotype produced by channels containing CaV1.2 subunits and thus, voltage-dependent Ca2+ influx, is the result of an orchestration of factors. A majority of CaV1.2 channels containing an N terminus derived from exon 1c rather than exon 1b could modify voltage-dependent Ca2+ influx through several mechanisms. First, CaV1.2e1c/α2δ current density was smaller than for CaV1.2e1b/α2δ. Second, CaV1.2e1c/α2δ currents inactivated at more negative voltages than CaV1.2e1b/α2δ currents. Third, when co-expressed with α2δ, less CaV1.2e1c channels localized to the plasma membrane than did CaV1.2e1b channels. Collectively, these exon 1c-derived effects would reduce voltage-dependent Ca2+ influx in arterial myocytes. Arterial myocytes do not generate action potentials, but undergo steady-state changes in membrane potential. In arteries at physiological pressures, myocyte membrane potential is between ~−60 and −40 mV (46). Thus, arterial myocytes maintain relatively depolarized potentials when compared with many other cell types, particularly those that generate action potentials. A predominance of CaV1.2 channels containing exon 1c may serve to limit Ca2+ influx under the steady depolarized potentials that occur in arterial myocytes. Because arterial myocyte membrane potential changes steadily rather than rapidly (as in the case of action potentials), differences in inactivation rates between CaV1.2e1b/α2δ/β and CaV1.2e1c/α2δ/β channels would presumably have a smaller influence on Ca2+ influx than the exon 1c-induced alterations in current density, steady-state inactivation, and membrane insertion. Because CaV1.2 α1 sub-units undergo splicing at 19 out of 55 exons, and some of these variants have also been described to modify channel electro-physiological properties, it is premature to provide a detailed comparison of properties of the splice variants described here and what is likely to be a heterogeneous CaV1.2 splice variant population in myocytes (9). Furthermore, auxiliary subunit expression in arterial myocytes, which would modify CaV1.2 currents, is also unclear. Even though we detected β1b and β3 message in arterial myocytes, it is uncertain which other β subunit isoforms are expressed, the relative proportions of each isoform, the relative amount of β to α2δ subunits, the relative proportions of each auxiliary subunit to α1 subunits, and whether β and α2δ subunit isoforms exhibit defined cellular localization that could specifically modify the properties of α1 subunits containing exon 1b- or 1c-derived N termini. The electrophysiological properties of arterial myocyte CaV1.2 currents result from the combination of these additional, multiple factors. Nevertheless, our data indicate that electrophysiological properties of CaV1.2 currents are modified by splicing of the CaV1.2 α1 subunit N terminus. Although β subunits normalized the isoform specific effects of α2δ on CaV1.2e1b andCaV1.2e1c channels in an overexpression system, it remains to be determined whether this occurs in arterial myocytes in physiology and disease. Given that alternative splicing of CaV1.2 subunits has been observed in disease (11, 47), and that CaV1.2 subunit expression is up-regulated in arterial myocytes of hypertensive animals (5), modifications in exon 1 splicing could occur during vascular disease and may contribute to increased CaV1.2 expression in hypertension.
In the human genome, sequences highly homologous to exon 1a and 1b are present in the CaV1.2 gene, yet a sequence identical to 1c is not found. A sequence that shares high homology with the 5′-untranslated region upstream of exon 1c is present in the human genome (p13.33 of chromosome 12), raising the possibility that CaV1.2 channels in human arterial myocytes contain an N terminus different to that described in human jejunum (25). Future studies will be required to identify human CaV1.2 α1 subunit exon 1 splice variants in arterial myocytes and other cell types.
In summary, we have established that CaV1.2 subunits which are expressed in myocytes of small, resistance size, cerebral arteries contain a novel exon 1c-derived splice variant that alters current regulation by auxiliary subunits. Considering the critical importance of CaV1.2 channels in the regulation of blood pressure and flow, and their role in vascular pathologies, including hypertension (5), exon 1c likely contributes to vascular specific Ca2+ signaling and Ca2+-dependent physiology.
Supplementary Material
Acknowledgments
We thank Dr. Steven J. Tavalin for helpful discussions on the manuscript.
Footnotes
This study was supported by grants from the National Institutes of Health (to J. H. J., HL67061 and HL77678; and A. M. D., AA11560 and HL77424) and the American Heart Association National Center (to J. H. J.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
The abbreviations used are: CaV1.2, voltage-dependent L-type Ca2+; RACE, rapid amplification of cDNA ends; EGFP, enhanced green fluorescent protein; I-V, current-voltage.
References
- 1.Gollasch M, Nelson MT. Kidney Blood Press Res. 2000;20:355–371. doi: 10.1159/000174250. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Lipp P, Bootman MD. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 3.Moosmang S, Schulla V, Welling A, Feil R, Feil S, Wegener JW, Hofmann F, Klugbauer N. EMBO J. 2003;22:6027–6034. doi: 10.1093/emboj/cdg583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson MT, Patlak JB, Worley JF, Standen NB. Am J Physiol. 1990;259:C3–18. doi: 10.1152/ajpcell.1990.259.1.C3. [DOI] [PubMed] [Google Scholar]
- 5.Sonkusare S, Palade PT, Marsh JD, Telemaque S, Pesic A, Rusch NJ. Vascul Pharmacol. 2006;44:131–142. doi: 10.1016/j.vph.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Triggle DJ. Curr Pharm Des. 2006;12:443–457. doi: 10.2174/138161206775474503. [DOI] [PubMed] [Google Scholar]
- 7.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 8.Arikkath J, Campbell KP. Curr Opin Neurobiol. 2003;13:298–307. doi: 10.1016/s0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 9.Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, Soong TW. J Biol Chem. 2004;279:44335–44343. doi: 10.1074/jbc.M407023200. [DOI] [PubMed] [Google Scholar]
- 10.Koch WJ, Ellinor PT, Schwartz A. J Biol Chem. 1990;265:17786–17791. [PubMed] [Google Scholar]
- 11.Tiwari S, Zhang Y, Heller J, Abernethy DR, Soldatov NM. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0606539103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaggar JH. Am J Physiol. 2001;281:C439–448. doi: 10.1152/ajpcell.2001.281.2.C439. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Hofmann PA. Am J Physiol Heart Circ Physiol. 2003;285:H97–103. doi: 10.1152/ajpheart.00956.2002. [DOI] [PubMed] [Google Scholar]
- 14.Pragnell M, Sakamoto J, Jay SD, Campbell KP. FEBS Lett. 1991;291:253–258. doi: 10.1016/0014-5793(91)81296-k. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Reyes E, Castellano A, Kim HS, Bertrand P, Baggstrom E, Lacerda AE, Wei XY, Birnbaumer L. J Biol Chem. 1992;267:1792–1797. [PubMed] [Google Scholar]
- 16.Castellano A, Wei X, Birnbaumer L, Perez-Reyes E. J Biol Chem. 1993;268:3450–3455. [PubMed] [Google Scholar]
- 17.Jaggar JH, Li A, Parfenova H, Liu J, Umstot ES, Dopico AM, Leffler CW. Circ Res. 2005;97:805–812. doi: 10.1161/01.RES.0000186180.47148.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meir A, Bell DC, Stephens GJ, Page KM, Dolphin AC. Biophys J. 2000;79:731–746. doi: 10.1016/S0006-3495(00)76331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobrinsky E, Tiwari S, Maltsev VA, Harry JB, Lakatta E, Abernethy DR, Soldatov NM. J Biol Chem. 2005;280:12474–12485. doi: 10.1074/jbc.M412140200. [DOI] [PubMed] [Google Scholar]
- 20.Pietrzykowski AZ, Martin GE, Puig SI, Knott TK, Lemos JR, Treistman SN. J Neurosci. 2004;24:8322–8332. doi: 10.1523/JNEUROSCI.1536-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao P, Yong TF, Liang MC, Yue DT, Soong TW. Cardiovasc Res. 2005;68:197–203. doi: 10.1016/j.cardiores.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Mikami A, Imoto K, Tanabe T, Niidome T, Mori Y, Takeshima H, Narumiya S, Numa S. Nature. 1989;340:230–233. doi: 10.1038/340230a0. [DOI] [PubMed] [Google Scholar]
- 23.Snutch TP, Tomlinson WJ, Leonard JP, Gilbert MM. Neuron. 1991;7:45–57. doi: 10.1016/0896-6273(91)90073-9. [DOI] [PubMed] [Google Scholar]
- 24.Biel M, Ruth P, Bosse E, Hullin R, Stuhmer W, Flockerzi V, Hofmann F. FEBS Lett. 1990;269:409–412. doi: 10.1016/0014-5793(90)81205-3. [DOI] [PubMed] [Google Scholar]
- 25.Lyford GL, Strege PR, Shepard A, Ou Y, Ermilov L, Miller SM, Gibbons SJ, Rae JL, Szurszewski JH, Farrugia G. Am J Physiol Cell Physiol. 2002;283:C1001–1008. doi: 10.1152/ajpcell.00140.2002. [DOI] [PubMed] [Google Scholar]
- 26.Soldatov NM. Proc Natl Acad Sci U S A. 1992;89:4628–4632. doi: 10.1073/pnas.89.10.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang ZZ, Hong X, Wang J, Soong TW. Cell Calcium. 2007;41:417–428. doi: 10.1016/j.ceca.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Restituito S, Cens T, Rousset M, Charnet P. Biophys J. 2001;81:89–96. doi: 10.1016/S0006-3495(01)75682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens GJ, Page KM, Bogdanov Y, Dolphin AC. J Physiol. 2000;525:377–390. doi: 10.1111/j.1469-7793.2000.t01-1-00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hullin R, Singer-Lahat D, Freichel M, Biel M, Dascal N, Hofmann F, Flockerzi V. EMBO J. 1992;11:885–890. doi: 10.1002/j.1460-2075.1992.tb05126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saada N, Dai B, Echetebu C, Sarna SK, Palade P. Biochem Biophys Res Commun. 2003;302:23–28. doi: 10.1016/s0006-291x(03)00097-4. [DOI] [PubMed] [Google Scholar]
- 33.Saada NI, Carrillo ED, Dai B, Wang WZ, Dettbarn C, Sanchez J, Palade P. Cell Calcium. 2005;37:301–309. doi: 10.1016/j.ceca.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Klugbauer N, Marais E, Hofmann F. J Bioenerg Biomembr. 2003;35:639–647. doi: 10.1023/b:jobb.0000008028.41056.58. [DOI] [PubMed] [Google Scholar]
- 35.Sandoval A, Oviedo N, Andrade A, Felix R. FEBS Lett. 2004;576:21–26. doi: 10.1016/j.febslet.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 36.Felix R, Gurnett CA, De WM, Campbell KP. J Neurosci. 1997;17:6884–6891. doi: 10.1523/JNEUROSCI.17-18-06884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bichet D, Cornet V, Geib S, Carlier E, Volsen S, Hoshi T, Mori Y, De WM. Neuron. 2000;25:177–190. doi: 10.1016/s0896-6273(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 38.Gerster U, Neuhuber B, Groschner K, Striessnig J, Flucher BE. J Physiol. 1999;517:353–368. doi: 10.1111/j.1469-7793.1999.0353t.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi SX, Mittman S, Colecraft HM. Biophys J. 2003;84:3007–3021. doi: 10.1016/S0006-3495(03)70027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foell JD, Balijepalli RC, Delisle BP, Yunker AM, Robia SL, Walker JW, McEnery MW, January CT, Kamp TJ. Physiol Genomics. 2004;17:183–200. doi: 10.1152/physiolgenomics.00207.2003. [DOI] [PubMed] [Google Scholar]
- 41.Bogdanov Y, Brice NL, Canti C, Page KM, Li M, Volsen SG, Dolphin AC. Eur J Neurosci. 2000;12:894–902. doi: 10.1046/j.1460-9568.2000.00981.x. [DOI] [PubMed] [Google Scholar]
- 42.Chien AJ, Gao T, Perez-Reyes E, Hosey MM. J Biol Chem. 1998;273:23590–23597. doi: 10.1074/jbc.273.36.23590. [DOI] [PubMed] [Google Scholar]
- 43.Resh MD. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 44.Arien H, Wiser O, Arkin IT, Leonov H, Atlas D. J Biol Chem. 2003;278:29231–29239. doi: 10.1074/jbc.M301401200. [DOI] [PubMed] [Google Scholar]
- 45.Cho HC, Tsushima RG, Nguyen TT, Guy HR, Backx PH. Biochemistry. 2000;39:4649–4657. doi: 10.1021/bi992469g. [DOI] [PubMed] [Google Scholar]
- 46.Davis MJ, Hill MA. Physiol Rev. 1999;79:387–423. doi: 10.1152/physrev.1999.79.2.387. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Chen X, Margulies K, Jeevanandam V, Pollack P, Bailey BA, Houser SR. J Mol Cell Cardiol. 2000;32:973–984. doi: 10.1006/jmcc.2000.1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.